Published online Jun 15, 2024. doi: 10.4251/wjgo.v16.i6.2646
Peer-review started: October 27, 2023
First decision: January 6, 2024
Revised: January 24, 2024
Accepted: March 27, 2024
Article in press: March 27, 2024
Published online: June 15, 2024
Processing time: 231 Days and 19.7 Hours
Colon cancer (CC) occurrence and progression are considerably influenced by the tumor microenvironment. However, the exact underlying regulatory mechanisms remain unclear.
To investigate immune infiltration-related differentially expressed genes (DEGs) in CC and specifically explored the role and potential molecular mechanisms of complement factor I (CFI).
Immune infiltration-associated DEGs were screened for CC using bioinformatics. Quantitative reverse transcription polymerase chain reaction was used to examine hub DEGs expression in the CC cell lines. Stable CFI-knockdown HT29 and HCT116 cell lines were constructed, and the diverse roles of CFI in vitro were assessed using CCK-8, 5-ethynyl-2’-deoxyuridine, wound healing, and transwell assays. Hematoxylin and eosin staining and immunohistochemistry staining were employed to evaluate the influence of CFI on the tumorigenesis of CC xenograft models constructed using BALB/c male nude mice. Key proteins associated with glycolysis and the Wnt pathway were measured using western blotting.
Six key immune infiltration-related DEGs were screened, among which the expression of CFI, complement factor B, lymphoid enhancer binding factor 1, and SRY-related high-mobility-group box 4 was upregulated, whereas that of fatty acid-binding protein 1, and bone morphogenic protein-2 was downregulated. Furthermore, CFI could be used as a diagnostic biomarker for CC. Functionally, CFI silencing inhibited CC cell proliferation, migration, invasion, and tumor growth. Mechanistically, CFI knockdown downregulated the expression of key glycolysis-related proteins (glucose transporter type 1, hexokinase 2, lactate dehydrogenase A, and pyruvate kinase M2) and the Wnt pathway-related proteins (β-catenin and c-Myc). Further investigation indicated that CFI knockdown inhibited glycolysis in CC by blocking the Wnt/β-catenin/c-Myc pathway.
The findings of the present study demonstrate that CFI plays a crucial role in CC development by influencing glycolysis and the Wnt/β-catenin/c-Myc pathway, indicating that it could serve as a promising target for therapeutic intervention in CC.
Core Tip: We identified six hub immune infiltration-related differentially expressed genes, of which the expression of complement factor I (CFI), complement factor B, lymphoid enhancer binding factor 1, and SRY-related high-mobility-group box 4 were upregulated, whereas that of fatty acid-binding protein 1 and bone morphogenic protein-2 were downregulated. CFI knockdown inhibited HT29 and HCT116 cell proliferation, migration, invasion, and tumor growth in a colon cancer (CC) mouse model constructed from HT29 cells. CFI knockdown suppressed the expression of glycolysis-related proteins (glucose transporter type 1, hexokinase 2, lactate dehydrogenase A, and pyruvate kinase M2) and Wnt/β-catenin/c-Myc-related protein-expression levels (β-catenin and c-Myc). CFI knockdown suppressed glycolysis by inhibiting the Wnt/β-catenin/c-Myc signaling pathway. These findings delineate a potential therapeutic strategy for the clinical management of CC.
- Citation: Du YJ, Jiang Y, Hou YM, Shi YB. Complement factor I knockdown inhibits colon cancer development by affecting Wnt/β-catenin/c-Myc signaling pathway and glycolysis. World J Gastrointest Oncol 2024; 16(6): 2646-2662
- URL: https://www.wjgnet.com/1948-5204/full/v16/i6/2646.htm
- DOI: https://dx.doi.org/10.4251/wjgo.v16.i6.2646
Colorectal cancer (CRC) is a highly prevalent malignancy that is a substantial public health concern[1]. In 2020, CRC accounted for 10% of all human cancers diagnosed, with colon cancer (CC) constituting approximately 5% of CRC cases
The complement system includes three pathways, the classical, alternative, and lectin pathways, which are crucial for both innate and acquired inflammatory immune responses[7]. Activation of these pathways leads to cleavage of the central complement components C3 and C5, forming a membrane attack complex, and ultimately triggering cell lysis[8-10]. Complement factor I (CFI), a serine protease, is an important negative controller of the complement system. The C3b and C4b components are cleaved by CFI, leading to inhibition of the activation of all three pathways[11,12]. Recently, several investigations have demonstrated that CFI may contribute to the advancement of tumors such as cutaneous squamous carcinoma, glioma, and ovarian cancer[13-15]. However, the roles and potential mechanisms of action of CFI in CC remain unclear.
The dysregulation of cellular energy metabolism is highly relevant to cancer development, and one of the key changes in cancer metabolic pathways is the enhancement of aerobic glycolysis, known as the Warburg effect[16]. Glycolysis meets a substantial portion of the energy demand of cancer cells and converts glucose to lactate, which generates only two ATP molecules, enhances nucleotide and amino acid production, and is insensitive to hypoxia in invasive cancer cells[17,18]. Complement proteins involved in the glycolytic processes. C3aR and C5aR1, are expressed on the mitochondrial outer membrane and control Ca2+ flux and the direction of the electron transport chain, thereby influencing glycolysis and oxidative phosphorylation[19-21]. However, the correlation between the complement system and glycolysis in CC has rarely been reported.
In the present study, we found that CFI knockdown suppressed CC cell proliferation, migration, invasion, and xenograft tumor growth. Further studies demonstrated that silencing CFI inhibited tumor progression by suppressing the Wnt/β-catenin/c-Myc pathway and glycolysis, which was validated in both in vivo and in vitro assays. This implies that CFI is a prospective therapeutic target for CC.
GSE33113 and GSE44861 from the Gene Expression Omnibus (GEO) database (https://www.ncbi.nlm.nih.gov/geo/) were used. The GSE33113 dataset (GPL570 platform) comprises six normal samples and 90 CC tumor samples. The GSE44861 dataset (GPL3921 platform) consists of 55 normal samples and 56 tumor samples. GEO2R (https://www.ncbi.nlm.nih.gov/geo/geo2r) was employed to screen for differentially expressed genes (DEGs) in the two datasets with the criteria of |log2 fold change| ≥ 1 and P value ≤ 0.05. DEGs were visualized using volcano plots and heat maps, which were generated using the ggplot2 and pheatmap packages in R software (version 4.3.0). Immune infiltration-related genes were obtained from the GeneCards database (https://www.genecards.org/). Then, immune infiltration-related DEGs were screened using the Venn tool (https://bioinfogp.cnb.csic.es/tools/venny/).
The Database for Annotation, Visualization, and Integrated Discovery (https://david.ncifcrf.gov/) was used to conduct the functional enrichment studies, including Gene Ontology (GO) and Kyoto Encyclopedia of Genes and Genomes (KEGG) analyses. GO enrichment analysis includes molecular function (MF), biological process (BP), and cellular component (CC).
The DEGs were entered into the STRING database (https://string-db.org/) to construct protein-protein interaction (PPI) networks with interaction scores greater than 0.4. Cytoscape (version 3.6.1) was used to visualize the PPI network. The Molecular Complex Detection (MCODE) plugin was used to identify hub modules. A maximum depth of 100, a k-score of 2, a degree cutoff of 2, and a node score cutoff of 0.2 were the selection criteria for module identification. The CytoHubba plugin was used to determine key genes in the hub modules.
GO enrichment, gene-expression patterns, principal component analysis (PCA), and receiver operating characteristic (ROC) curve analysis of key genes were performed using the circlize, ggridges, factoextra, and pROC packages in R software, respectively, based on the GSE33113 dataset. In the ROC curve analysis, the area under the ROC curve (AUC) value exceeding 0.9 was deemed statistically significant. Subsequently, CFI and complement factor B (CFB) expression in CC tissues was evaluated using the GEPIA 2 database (http://gepia2.cancer-pku.cn/#index). The TIMER database (https://cistrome.shinyapps.io/timer/) and TIMER 2.0 (http://timer.comp-genomics.org) were used to assess the association between CFI expression and infiltrating immune cells in the tumor microenvironment.
HT29 and HCT116 cells (human CC cell lines) and NCM460 cells (normal human colon epithelial cell line) were acquired from cell bioscience Biotechnology Co., Ltd. (Shanghai, China). Cells were cultivated in RPMI 1640 medium (Thermo Fisher Scientific, Waltham, Massachusetts, United States) with the addition of 10% fetal bovine serum (Thermo Fisher Scientific) and 100 U/mL penicillin/streptomycin in a humidified environment with 5% CO2 at 37 °C.
As indicated in a previous study[22], the mRNA expression profile of the target genes was assessed using quantitative reverse transcription polymerase chain reaction (qRT-PCR). Briefly, cDNA was rapidly reverse-transcribed following RNA extraction. Using the 2-ΔΔCt method, the mRNA levels were computed. The GAPDH protocol was used as an internal reference. The primer sequences used herein are listed in Supplementary Table 1.
Two short hairpin RNA (shRNA) sequences, including sh-CFI-1 and sh-CFI-2, targeting CFI, and sh-NC (blank control) were designed and synthesized by Obio Technology (Shanghai) Corp., Ltd. (Shanghai, China). HT29 and HCT116 cells were divided into two groups: sh-NC and sh-CFI. These shRNA sequences were integrated into the pSuper-retro-puro vector using the Lipofectamine 2000 reagent (Thermo Fisher Scientific), according to the manufacturer’s instructions. These cells were administered with puromycin (2-3 g/mL) for two wk to generate stably transfected cell lines. The knockdown efficiency was assessed by qRT-PCR after 48 h of infection.
Cell viability was evaluated using the Cell counting kit-8 (CCK-8) kit (Solarbio, Beijing, China). Transfected HT29 and HCT116 cells were seeded in 96-well plates and incubated for the specified time intervals (0, 24, 48, 72 h). A total of 10 μL of CCK-8 reagent was introduced into each well, following which it was placed in an incubator at 37 °C for 1 h. Optical density was measured at 450 nm using a microplate reader (DALB, Shanghai, China).
CC cell proliferation was measured using the 5-ethynyl-2’-deoxyuridine (EdU) assay. In 24-well plates, the cells were seeded at a density of 1 × 105 cells/well and incubated for 24 h. An EdU analysis kit (Beyotime Biotechnology, Shanghai, China) was used to assess cell proliferation according to the manufacturer’s instructions. Images were acquired using a microscope (Olympus, Tokyo, Japan).
CC cells were seeded into six-well plates and allowed to grow for 24 h until reaching 95%-100% confluence. Subsequently, a sterile 200 μL pipette tip was used to vertically scrape the cells. The scraped cells were then incubated for 24 h. Images of the cells were captured under a light microscope (Olympus) at 0 and 24 h, and the migration distance was analyzed using the ImageJ software (National Institutes of Health, Bethesda, MD, United States).
Cell invasion was evaluated using the transwell assay, as previously described, with slight modifications[23]. Briefly, CC cells were implanted in 100 mL of serum-free medium in the Matrigel-coated upper chamber. After 24 h, the cells were fixed in 4% paraformaldehyde for 20 min and stained with crystal violet. The number of invading cells was estimated in five randomly selected microscopic fields and analyzed using ImageJ software (National Institutes of Health).
Radioimmunoprecipitation assay buffer containing a protease inhibitor (Solarbio) was used to harvest complete protein. Sodium dodecyl sulfate-polyacrylamide gel electrophoresis was used to separate the proteins. Based on protein size, the proteins were transferred from the gel to polyvinylidene fluoride membranes (Roche, Basel, Switzerland) using a 200 mA current for 50 min and then incubated with 5% skim milk. The membranes were subsequently exposed to primary antibodies, including, anti-glucose transporter type 1 (GLUT1) (1:2000; Abcam, Cambridge, United Kingdom), anti-hexokinase 2 (HK2) (1:2000; Abcam), anti-lactate dehydrogenase A (LDHA) (1:2000; Abcam), anti-pyruvate kinase M2 (PKM2) (1:2000; Abcam), anti-β-catenin (1:2000; Abcam), anti-c-Myc (1:2000; Abcam), anti-β-actin (1:2000; Abcam), and GAPDH (1:2000; Abcam) and incubated overnight at 4 °C. Then they were treated with HRP-conjugated secondary antibodies for 1 h at room temperature. An improved enhanced chemiluminescence detection kit (Amersham, Little Chalfont, United Kingdom) was utilized to visualize protein bands. Band intensities were quantified using ImageJ software (National Institutes of Health).
Glycolysis in CC cells was measured using Glucose Uptake Fluorometric Assay Kits (Solarbio) and Lactate Colorimetric Assay Kits (Solarbio) according to the manufacturer’s instructions.
All animal experiments were performed in accordance with the protocols specified in the National Institutes of Health Guide for the Care and Use of Laboratory Animals. Animal studies were approved by the Animal Ethics Committee of the Affiliated Hospital of Chengdu University of Traditional Chinese Medicine (2023DL-039). Twelve BALB/c male nude mice (weighing 18-20 g, aged 5 wk) were acquired from specific pathogen-free Biotechnology Co. (Beijing, China). Mice were kept in a controlled setting with a 12 h light/dark cycle, with five to six mice per cage, and they had access to unlimited food and water supplies. Using a completely randomized design approach, the mice were sequentially numbered and group assignments were made using a random number table. The mice were then separated into the Lv-sh-NC (n = 6) and Lv-sh-CFI (n = 6) groups. CC tumor models were established by injecting 100 μL HT29 cells suspended in phosphate buffered saline subcutaneously into the right flank of the mice. The tumor size was assessed by monitoring the length (l) and width (w) of the tumors weekly using Vernier calipers. The formula used to calculate tumor volume was as follows: V (mm3) = (w2 × l)/2. The mice were euthanized with 50 mg/kg sodium pentobarbital after four wk, and the xenografted tumor tissues were removed. Tumor weights were recorded, and tumor tissues were processed for hematoxylin and eosin (HE) staining and immunohistochemical examination.
Pathologic changes in mouse tumor tissues were assessed by HE and immunohistochemistry staining. Mouse tumor tissues were fixed in a solution containing 4% paraformaldehyde and embedded in paraffin. Tissue sections, 4 μM thick, were then prepared using a paraffin slicer. HE staining was performed on tissue sections (Beyotime Biotechnology). Paraffin-embedded tissues were heated to 98 °C for antigen retrieval, blocked with goat serum, and incubated with anti-Ki67 antibody (1:600; Abcam) overnight at 4 °C to investigate the expression of Ki67 in CC tissues. The slices were then incubated with secondary antibodies processed with the 3,3’-diaminobenzidine reagent kit and finally photographed.
HT29 cells transfected with sh-CFI at a concentration of 20 mmol/L were treated with lithium chloride (LiCl), a Wnt/β-catenin signaling pathway activator, and incubated. The HT29 cells were categorized into three groups: sh-NC, sh-CFI, and sh-CFI + LiCl.
Each set of data was collected from a minimum of three separate measurements and displayed as mean ± SD. Student’s t test was used to compare the two groups. A one-way ANOVA with Tukey’s multiple comparison test was used for group comparisons. Statistical significance was set at P value < 0.05.
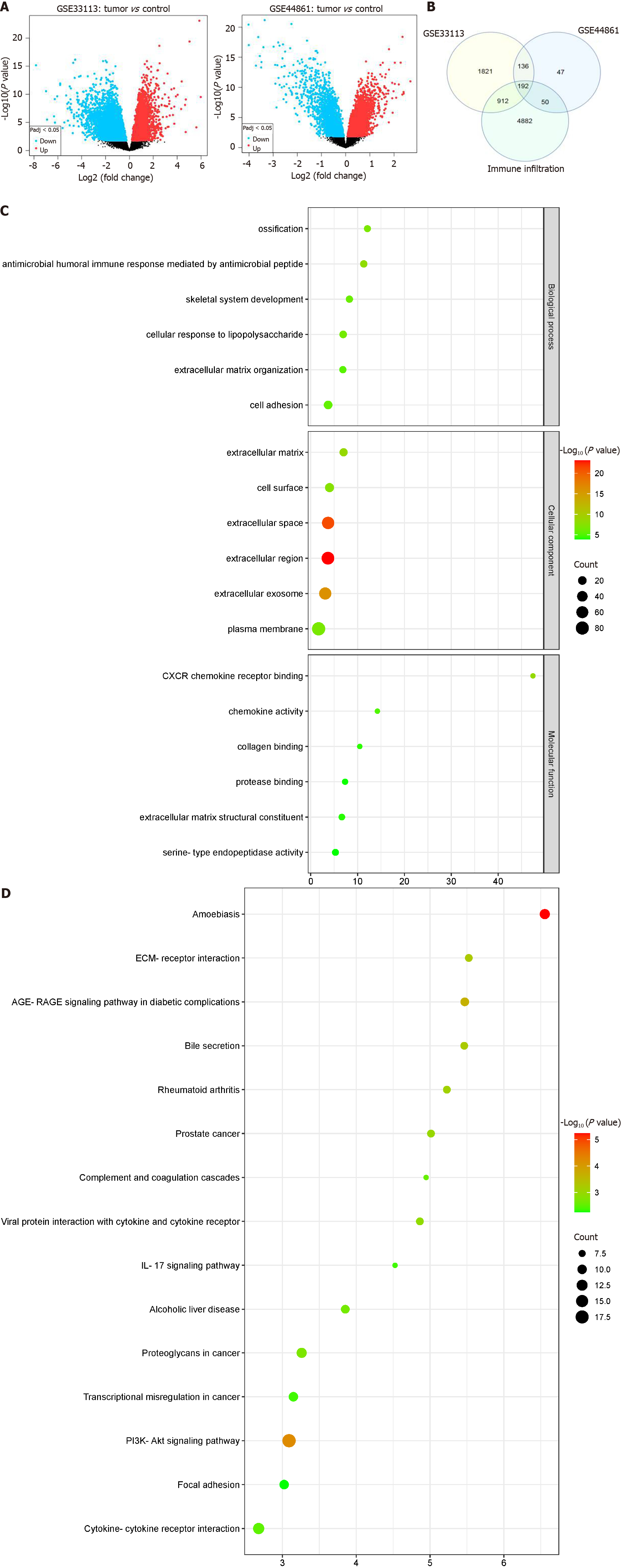
The GSE33113 and GSE44861 datasets were initially standardized (Supplementary Figure 1A). Subsequently, the DEGs were screened using GEO2R according to the criteria of P ≤ 0.05 and |logfold change| ≥ 1. We identified 3061 DEGs in the GSE33113 dataset and 425 DEGs in the GSE44861 dataset, respectively. DEGs were visualized using volcano plots for the two datasets (Figure 1A). The top 25 genes with the most significant up- and down-regulation in each dataset were then selected for cluster analysis and visualized using heatmaps (Supplementary Figure 1B). Next, DEGs from the GSE33113 and GSE44861 datasets were subjected to intersection with immune infiltration-related genes, resulting in the identification of 192 shared DEGs (Figure 1B). Thereafter, GO functional annotation and KEGG pathway analyses were performed to clarify the potential biological roles of DEGs. In the BP terms, the DEGs were substantially involved in humoral immune response-mediated antimicrobial response, ossification, and skeletal system development. In the CC terms, the DEGs were predominantly abundant in the extracellular matrix, cell surface, and extracellular space. In the MF terms, the DEGs were principally involved in CXCR chemokine receptor binding, chemokine activity, and collagen binding (Figure 1C and Supplementary Figure 2A). KEGG analysis revealed that the DEGs were mostly implicated in amoebiasis, the P13K-AKT signaling pathway, and cytokine-cytokine receptor interactions (Figure 1D and Supplementary Figure 2B).
The PPI network of DEGs was established using the STRING tool. The PPI network model was visually represented using Cytoscape software (Supplementary Figure 3A). Seven highly interconnected modules were identified from the PPI network as potential functional molecular complexes for CC using the MCODE plug-in (Supplementary Figure 3B-H). In total, 22 key genes were subsequently screened using the degree algorithm from the seven important modules. To better represent the relationship between proteins and pathways and indicate changes in the functional strength of the pathways, GO-enriched chordal plots of the 22 key genes were generated (Supplementary Figure 4).
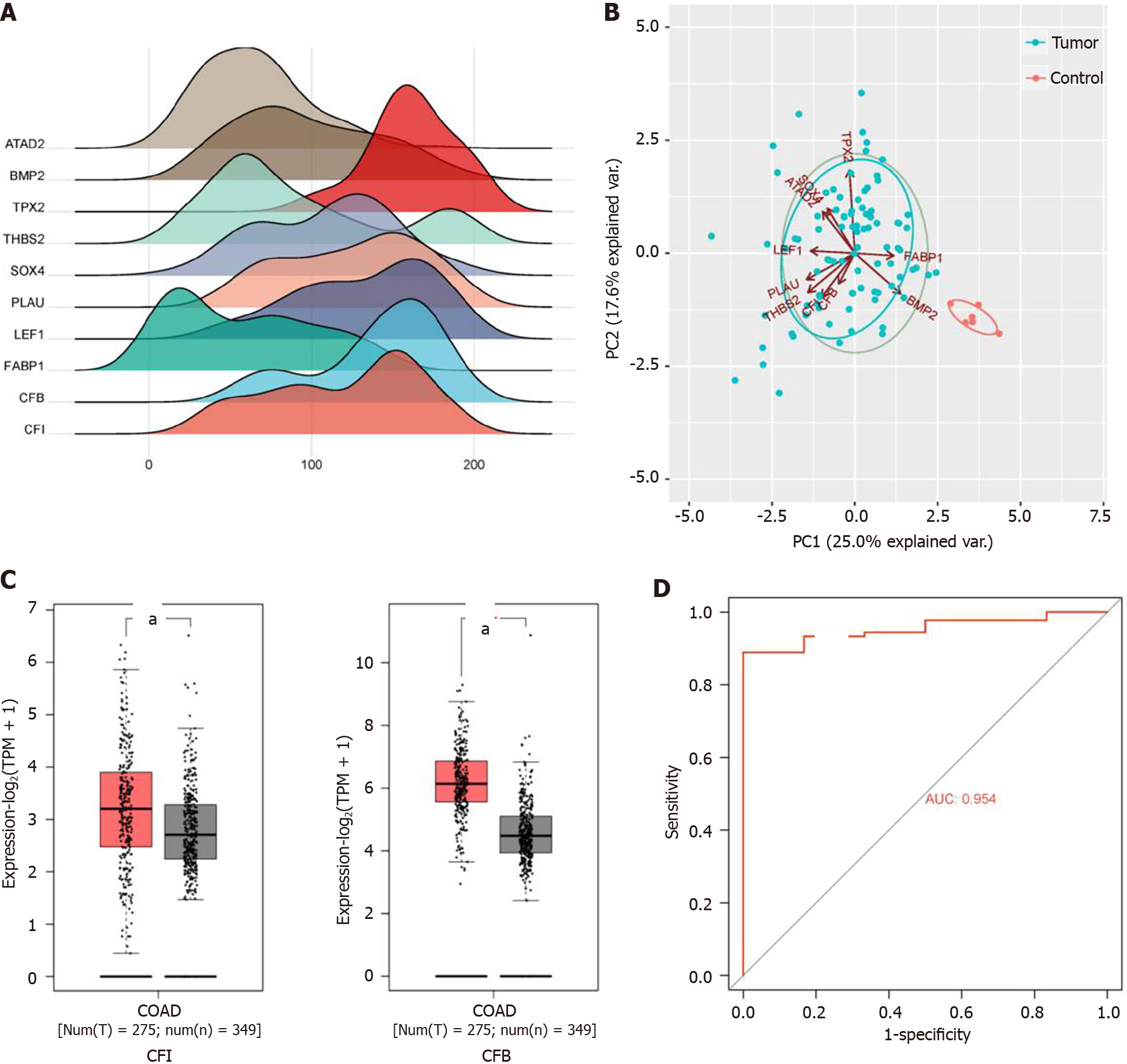
After reviewing the literature, we selected 10 hub genes [ATPase family AAA domain containing 2 (ATAD2), bone morphogenic protein-2 (BMP2), TPX2 microtubule nucleation factor (TPX2), thrombospondin 2 (THBS2), SRY-related high-mobility-group box 4 (SOX4), plasminogen activator, urokinase (PLAU), lymphoid enhancer binding factor 1 (LEF1), fatty acid-binding protein 1 (FABP1), CFB, and CFI] for subsequent analyses based on their innovativeness. Expression ridgeline maps were generated using R software with raw data from the GSE33113 dataset (Figure 2A). Two principal components, PC1 and PC2, were obtained using hub gene expression as variables for PCA. Together, these components explained 42.6% of the variance and could be used to distinguish the control group from the model group (Figure 2B). Studies have shown that complement is crucial for tumor development[24]. Therefore, the expression levels of the complement regulators CFI and CFB were explored using the GEPIA 2 database. These results suggested that CFI and CFB expression levels were markedly elevated in CC tissues compared with the corresponding expression levels in normal tissues (Figure 2C). ROC curve analysis was performed to explore whether CFI and CFB could be employed as diagnostic biomarkers for CC. The findings revealed that the AUC value of the ROC curve for the CFI was 95.4%, demonstrating that the CFI could effectively differentiate CC samples from normal colon samples (Figure 2D).
Subsequently, we explored the relationship between CFI expression and the extent of infiltration of various immune cell types into the CC microenvironment using the TIMER database. The results revealed a strong correlation between CFI expression and immune cells, such as CD8+, CD4+, and Macrophages (Supplementary Figure 5A). Additionally, using the TIMER 2.0 database, we found that high CFI expression actively promoted the infiltration of CD8+ and CD4+ immune cells (Supplementary Figure 5B-E).
The expression levels of the hub genes were assessed using qRT-PCR in human CC cells (HT29 and HCT116) and normal human colon epithelial cells (NCM460). Our results indicated that CFI, CFB, LEF1, and SOX4 were noticeably upregulated; conversely, FABP1 and BMP2 expression levels were substantially decreased in human CC cells compared with the corresponding expression levels in NCM460 cells (Figure 3).
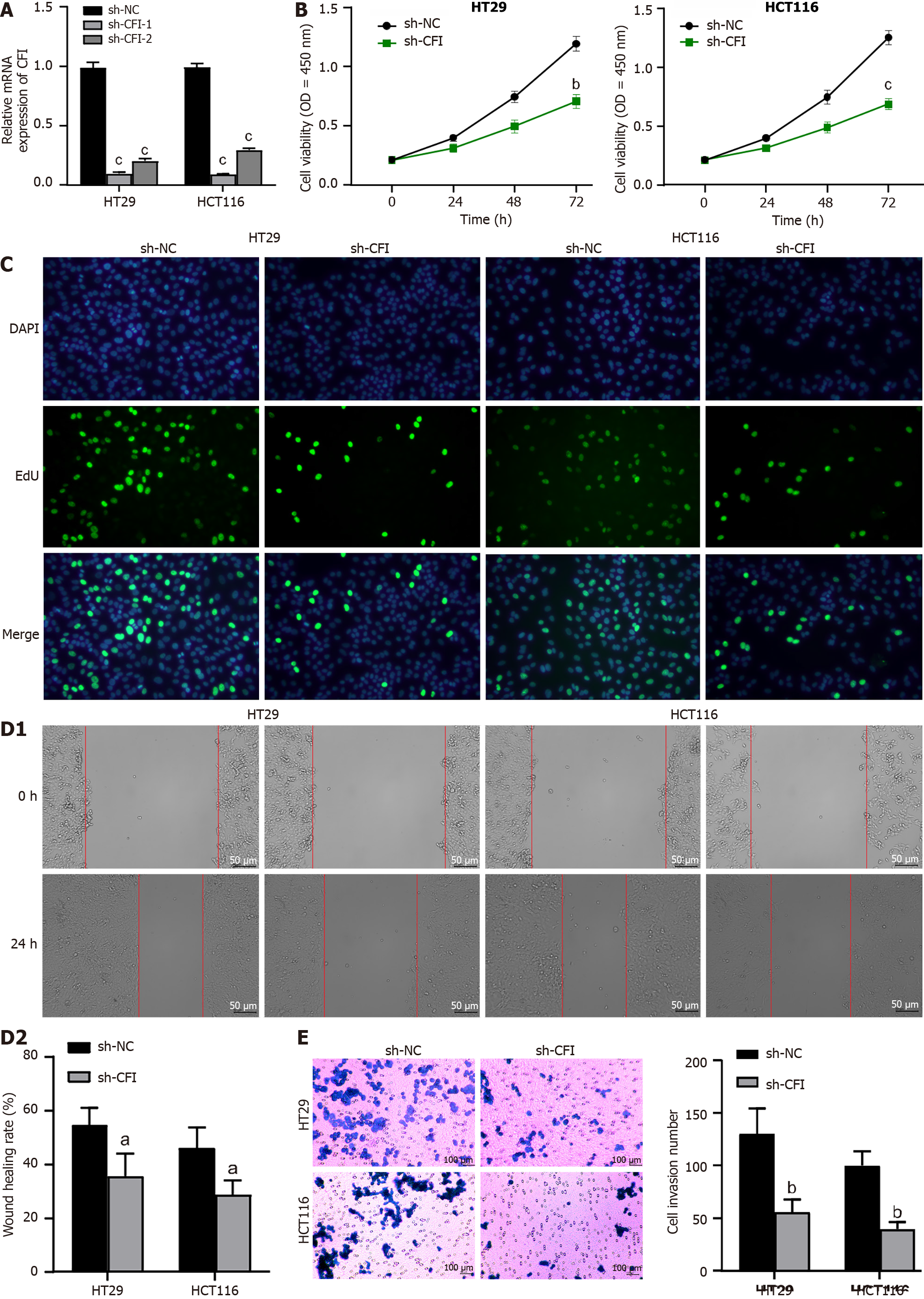
HT29 and HCT116 cells were transfected with sh-CFI vectors, and the transfection efficiency was confirmed using qRT-PCR. The findings revealed that CFI expression was greatly reduced in the sh-CFI-1 and sh-CFI-2 groups compared with that in the sh-NC group (Figure 4A). Owing to its high knockdown efficiency, sh-CFI-1 was selected for subsequent assays. Subsequently, the CCK-8 assay was used to determine the viability of CC cells. Notably, CFI knockdown in both HT29 and HCT116 cell lines resulted in a significant decrease in CC cell viability (Figure 4B). The EdU assay further indicated that the suppression of CFI expression effectively reduced CC cell proliferation (Figure 4C). Furthermore, we assessed the migration and invasion of CC cells using wound-healing and transwell assays. The findings showed that CC cell migration and invasion were significantly decreased in the sh-CFI group compared with the corresponding rates of CC cell migration and invasion in the sh-NC group (Figure 4D and E). These results indicated that CFI knockdown significantly inhibited the proliferation, migration, and invasion of CC cells.
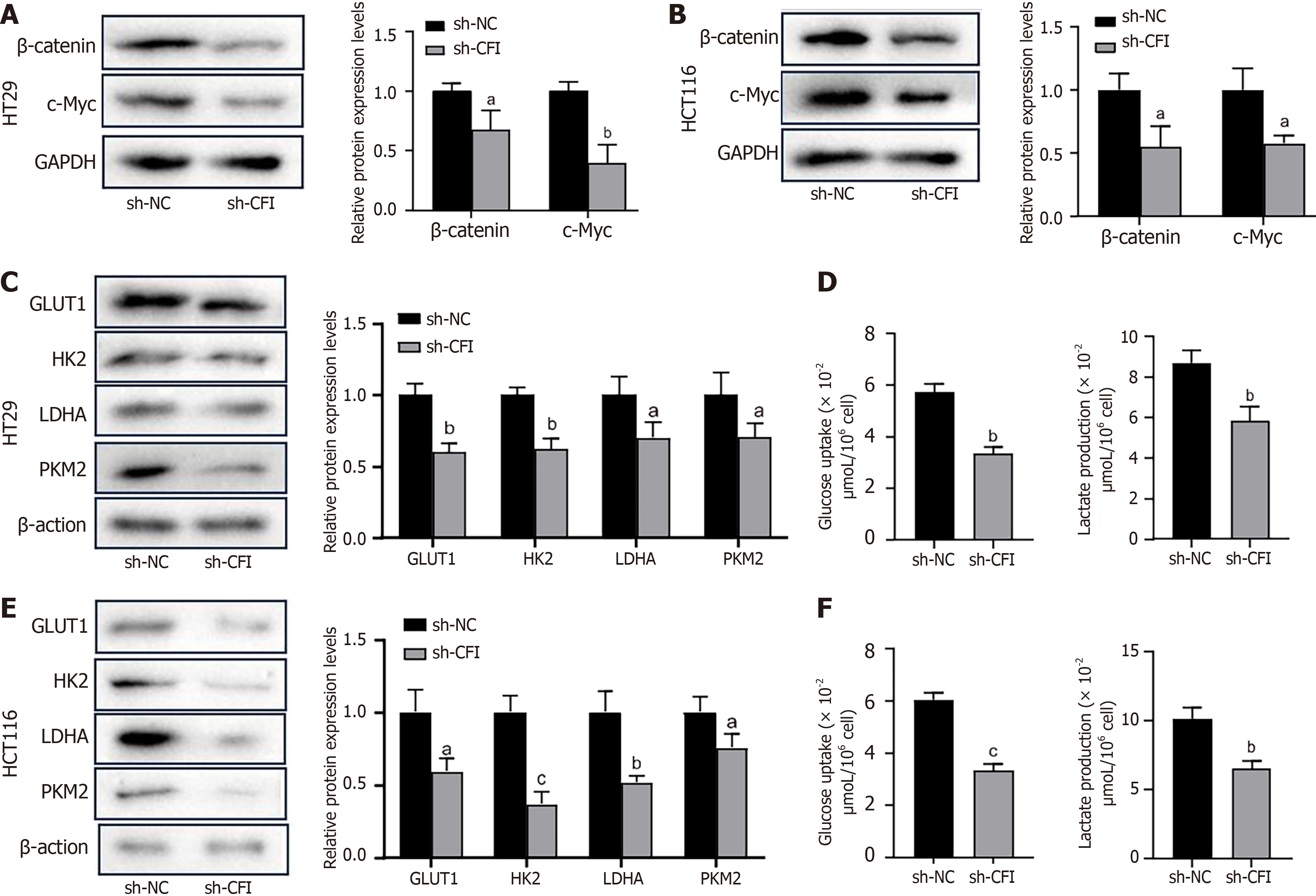
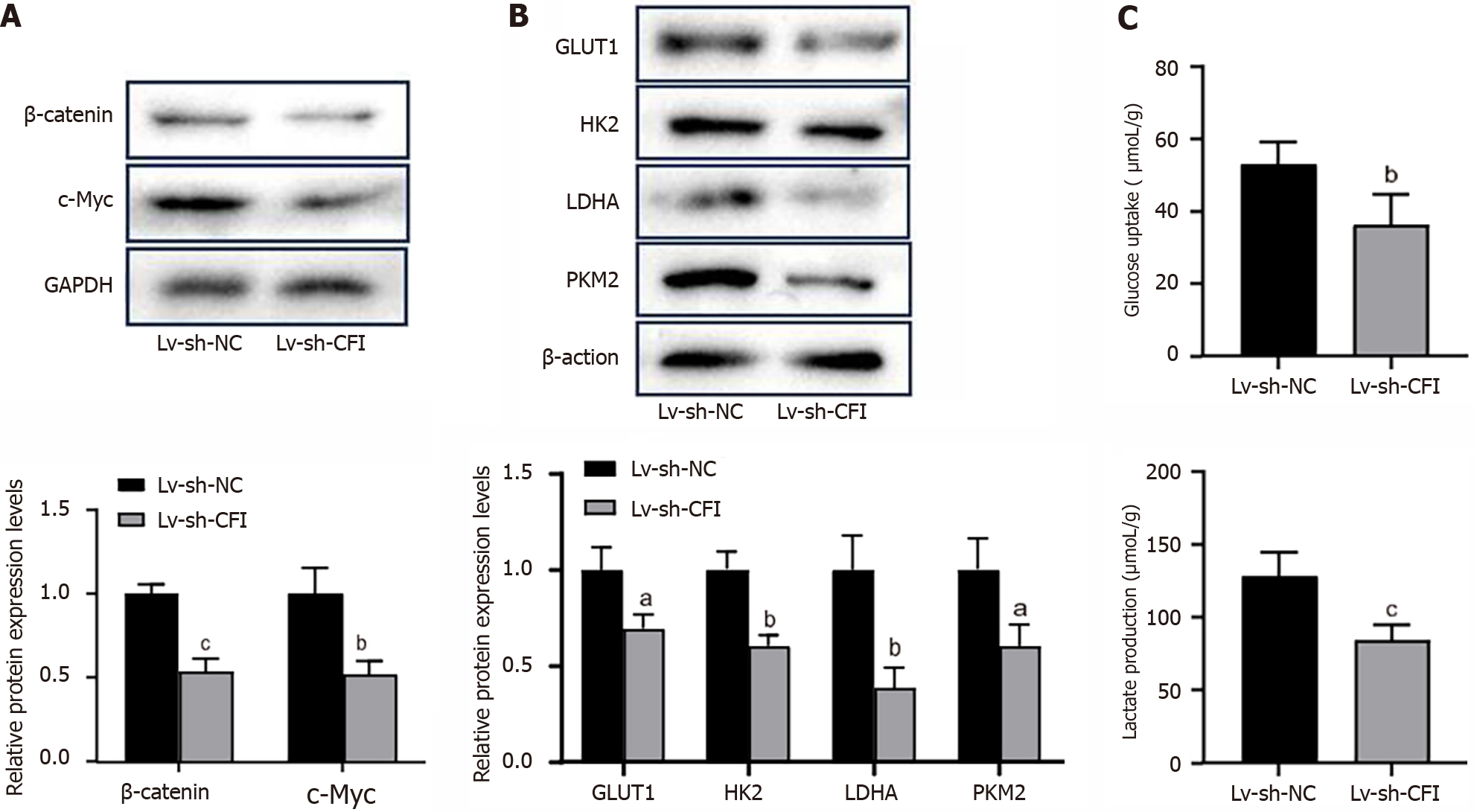
We examined the effect of CFI on the Wnt signaling pathway and glycolysis in CC cells. Western blotting showed that CFI knockdown inhibited the expression of key proteins of the Wnt pathway, including β-catenin and c-Myc, in HT29 and HCT116 cells (Figure 5A and B). Furthermore, the findings indicated that the expression of GLUT1, HK2, LDHA, and PKM2, which are key rate-limiting enzymes in glycolysis, was downregulated in the sh-CFI group compared with that in the sh-NC group (Figure 5C and E). The levels of glucose uptake and lactate production were evaluated, and the results showed that CFI knockdown inhibited glucose uptake and lactate production (Figure 5D and F). These data suggested that CFI might mediate CC development by managing the Wnt/β-catenin/c-Myc pathway and glycolytic metabolism.
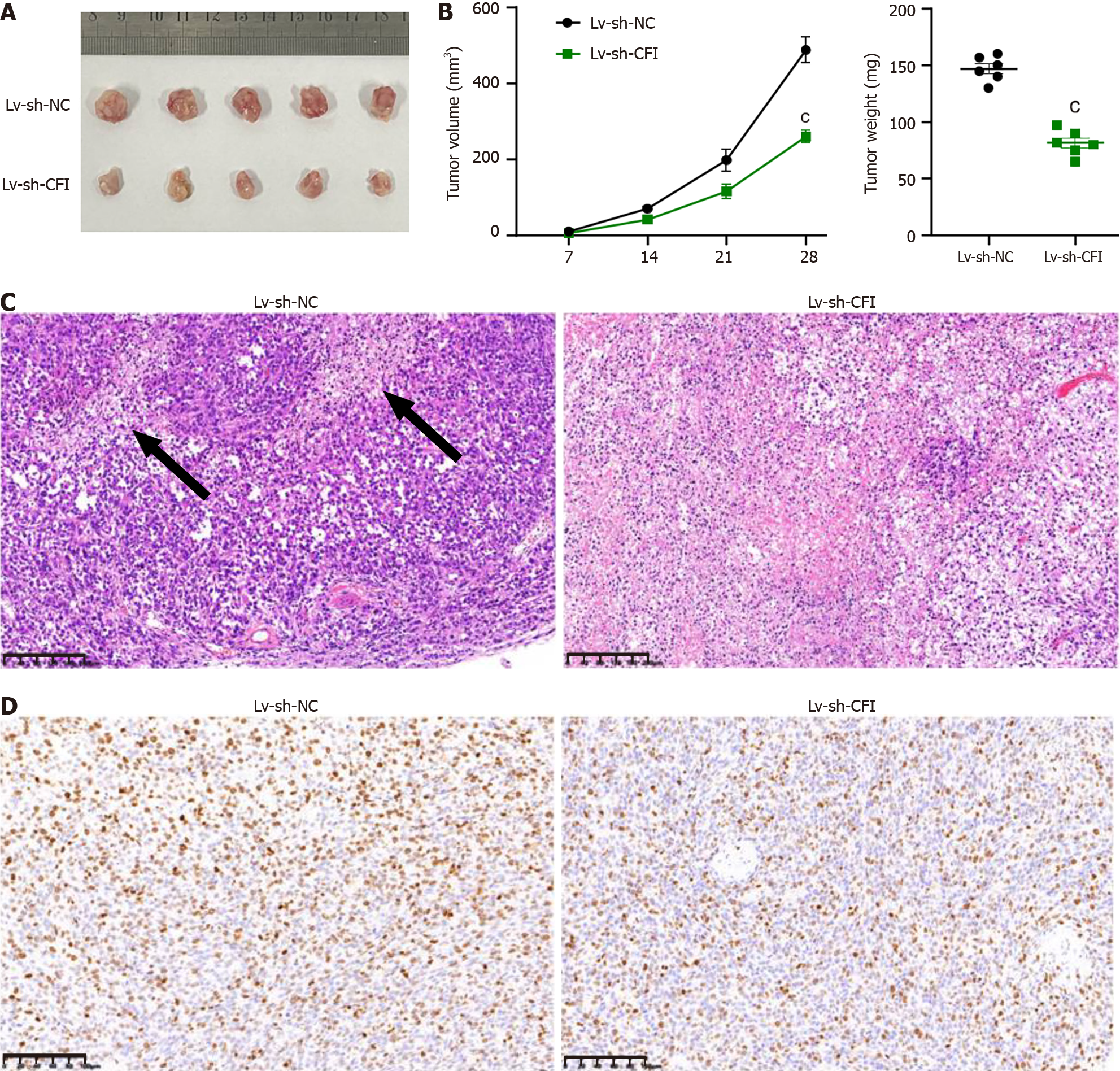
Xenograft experiments using HT29 cells were performed to elucidate the role of CFI in tumor growth in vivo. These findings demonstrated that the volume and weight of mouse tumors were significantly reduced in the Lv-sh-CFI group compared with the corresponding parameters in the Lv-sh-NC group (Figure 6A and B). The internal structure of the tumors was observed by HE staining, and the tumor tissues in the Lv-sh-NC group showed extensive structural damage. Tumor tissues in the Lv-sh-CFI group showed fewer pathological changes and a more normal tissue structure than those in the Lv-sh-NC group (Figure 6C). Immunohistochemistry assay results showed that Ki67 expression was reduced in the Lv-sh-CFI group compared with that in the Lv-sh-NC group (Figure 6D). The above findings indicated that the downregulation of CFI inhibited the growth of tumors in CC model mice.
Subsequently, the effects of knocking down CFI on Wnt/β-catenin/c-Myc signaling and glycolytic metabolism in vivo were explored. The results showed that the protein-expression levels of β-catenin, c-Myc, GLUT1, HK2, LDHA, and PKM2 were significantly reduced in the Lv-sh-CFI group compared with the corresponding expression levels in the Lv-sh-NC group (Figure 7A and B). In addition, reduced CFI expression inhibited glucose uptake and lactate production in tumor tissues (Figure 7C). The in vivo experiments further increased the plausibility that CFI affects CC development through glycolytic metabolism and Wnt/β-catenin/c-Myc signaling pathway regulation.
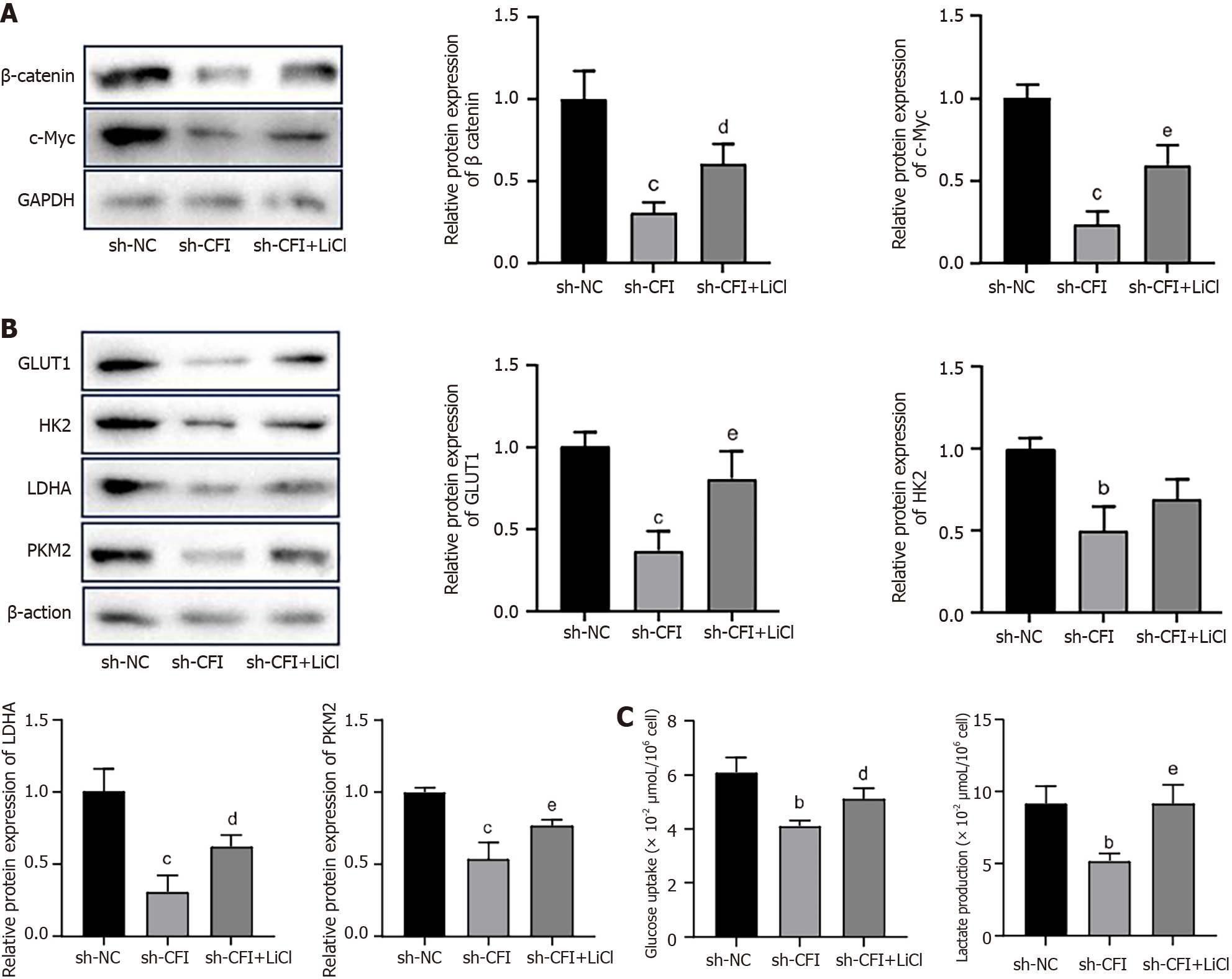
Our prior research showed that knocking down CFI inhibited the Wnt/β-catenin/c-Myc signaling pathway and glycolytic metabolism. Subsequently, the correlation between glycolysis and the Wnt/β-catenin/c-Myc pathway was further explored. HT29 cells were treated with LiCl, which activated the Wnt signaling pathway. The western blot results showed that LiCl intervention reversed the decrease in the expression of β-catenin and c-Myc caused by the CFI knockdown (Figure 8A). The expression of the main glycolysis-related enzymes, GLUT1, HK2, LDHA, and PKM2, was notably higher in the sh-CFI + LiCl group than in the sh-CFI group (Figure 8B). Moreover, the glucose-uptake and lactate-production abilities of cells in the sh-CFI + LiCl group were elevated compared with those in the sh-CFI group (Figure 8C), which indicated that CFI regulated glycolysis in CC via the Wnt/β-catenin/c-Myc pathway.
The incidence and mortality rates of CC are increasing leading to serious public health concerns[25]. Recent studies have revealed that immunotherapy, a treatment that employs the body’s innate immune system to treat cancer, shows promise for the treatment of CC[26]. Therefore, it is crucial to further investigate potential therapeutic targets for immunotherapy. In the present study, bioinformatics analysis and experimental validation revealed that six key immune infiltration-associated DEGs were differentially expressed in CC. Additionally, the findings suggested that CFI knockdown inhibited CC cell proliferation, migration, invasion and tumor growth in vivo, which was regulated by inhibition of Wnt/β-catenin/c-Myc pathway activation and glycolysis metabolism.
In present study, six key immune infiltration-associated DEGs were screened, including LEF1, SOX4, FABP1, BMP2, CFB, and CFI. LEF1 is a transcription factor that enhances antitumor immunity by creating memories in invariant natural killer T cells[27]. Upregulation of LEF-1 expression in CC activates the Wnt/β-catenin pathway, further promoting tumor progression[28,29]. SOX4 is a member of the SRY-related HMG-box gene family that promotes CC cell proliferation, migration, and epithelial-mesenchymal transition[30,31]. The study findings showed that CD39 expression in regulatory T-cells is mediated via the transforming growth factor-β/SOX4 axis and ROS-driven autophagy[32]. FABP1 belongs to the fatty acid-binding protein family and is preferentially depleted in CC with microsatellite instability[33]. BMP2 is involved in the regulation of cancer stem cells, epithelial-mesenchymal transition, cancer angiogenesis, and the tumor microenvironment, and is a promising target for tumor therapy[34]. In CC, BMP2 inhibits tumor growth and enhances chemosensitivity[35]. In the present study, we identified elevated LEF1 and SOX4 expression and found that FABP1 and SOX4 expression were reduced in CC, further clarifying the carcinogenic role of LEF1 and SOX4 in tumors and the antitumor effects of FABP1 and BMP2, which may affect tumor progression by regulating the immune response.
CFI and CFB are the regulatory proteins involved in the complement system[36]. As a fundamental component of the innate immune system, the complement system is one of the first lines of defense against foreign infections or stressed cells[37]. It consists of more than 50 soluble, membrane and intracellular proteins, including C1-C9, C3aR, C5aR, CR2, CFI, and CFH[38]. Studies have shown that various complement-targeting drugs are available for tumor treatment. Anti-CD20 mAbs, such as rituximab and ofatumumab, are effective in treating lymphoid malignancies[39]. The human anti-distinctive antibody 105AD7, which mimics CD55, has demonstrated efficacy in treating CRC, resulting in a low relapse rate in responsive patients[40]. Complement receptors C3aR and C5aR function as novel immune checkpoint receptors. Preclinical and clinical data suggest that the C3aR/C5aR/interleukin-10 axis plays a crucial role in regulating T cell-mediated antitumor immunity[41]. These studies suggest that the complement system is a promising target for tumor therapy. In the present investigation, we identified two immune infiltration-associated complement proteins, CFI and CFB, both of which are significantly overexpressed in CC. CFB expression has been detected in normal colorectal mucosa, adenomas, and carcinomas and may be involved in normal immune and inflammatory responses over a large-intestinal mucosal surface area[42]. CFI is a serine protease with molecular weight of 88 kDa. It was initially synthesized as a solitary polypeptide chain and subsequently crosslinked via disulfide bonds to form glycosylated heavy and light chains of 50 and 38 kDa, respectively[11]. CFI participates in the progression of multiple tumors, such as breast and stomach cancers[43,44]. Through ROC curve analysis, we found that the CFI could serve as a diagnostic marker for CC. Therefore, we chose CFI to further explore its role and mechanism of action in CC.
Through both in vivo and in vitro experiments, we found that CFI knockdown hindered CC cell proliferation, migration, invasion, and tumor growth. Glycolysis is significantly correlated with the tumor microenvironment in CC. Glycolysis- and lactate-related gene signatures can be used to predict colon adenocarcinoma prognosis and correlate with the immune microenvironment[45]. The interplay between the glycolytic-cholesterol biosynthesis axis and the tumor microenvironment suggests that gamma-glutamyl hydrolase inhibits glycolysis in CC[46]. In the current study, the results indicated that CFI silencing inhibited glycolysis. One study showed that some oncogenes upregulate the expression of particular glucose transporters and glycolytic enzymes to augment glycolysis and facilitate tumor progression[47]. In the present study, CFI knockdown inhibited the expression of key rate-limiting enzymes in the glycolytic metabolic pathway, including GLUT1, HK2, LDHA, and PKM2. Furthermore, lactate is a glycolytic metabolite generated by most tumor cells and is correlated with metastasis and unfavorable prognoses in patients with cancer[48]. CFI knockdown inhibited glucose uptake and lactate production in HT29 cells. Previous studies have demonstrated that, during the type 1 T helper cell response, complement proteins regulates nutrient influx and metabolic reprogramming, and CD46 activation promotes LAMTOR5 expression, leading to enhance glycolysis and oxidative phosphorylation[49]. Altogether, these results proved that the complement system influences tumor progression by regulating glycolysis.
Abnormal Wnt signaling alters glycolytic metabolism[50]. A previous study showed that the stimulation of the Wnt/β-catenin signaling pathway by FAM83F promoted cervical cancer development and aerobic glycolysis[51]. By blocking the Wnt/β-catenin/c-Myc signaling pathway, butyrate suppresses the migration, invasion, and aerobic glycolysis in gastric cancer[52]. Therefore, we explored whether CFI knockdown inhibits glycolysis by modulating the Wnt signaling pathway. We found that CFI knockdown significantly reduced the expression of key proteins in the Wnt signaling pathway: β-catenin and c-Myc, which further inhibited glycolytic metabolism. Importantly, inhibitors of Wnt signaling can be used to treat various tumors. Vantictumab and ipafricept, which are Wnt pathway inhibitors, combined with nab-paclitaxel and gemcitabine, can be used to treat metastatic pancreatic cancer[53]. Inhibitors of the Wnt signaling pathway enhance the therapeutic activity of low concentrations of glycolytic modulators in tongue cancer cells[54]. Taken together, in our study, we found that the complement protein CFI knockdown inhibits glycolysis by suppressing the Wnt/β-catenin/c-Myc signaling pathway, which may provide a theoretical basis for targeting the complement, glycolysis, and Wnt pathways cascade treatment therapy in CC.
However, there were certain limitations to the present study. First, an in vivo study was performed in BALB/c mice using a cell line-derived xenograft. PDX models developed from patients might be more useful for understanding the crucial role of CFI in CC progression. Second, as CFI was identified as an immune infiltration-related gene herein, it was investigated for its involvement in the Wnt/β-catenin/c-Myc signaling pathway and glycolysis bioprocesses. However, the specific role of CFI in the tumor microenvironment requires further investigation. Additionally, while six key genes related to immune infiltration were identified, our study specifically focused on CFI for in-depth analysis. The roles and underlying molecular mechanisms of the other key genes in CC require further investigation in future studies.
In the present study, we identified six immune infiltration-associated DEGs, of which CFI, CFB, LEF1, and SOX4 expression was upregulated, whereas FABP1 and BMP2 expression was downregulated. More importantly, CFI knockdown inhibited CC cell proliferation, migration and invasion, as well as tumor growth, which was mediated by inhibition of the Wnt/β-catenin/c-Myc signaling pathway and subsequently inhibiting glycolysis. The findings of the present study offer novel perspectives and possible treatment strategies for the clinical management of CC.
Provenance and peer review: Unsolicited article; Externally peer reviewed.
Peer-review model: Single blind
Specialty type: Oncology
Country/Territory of origin: China
Peer-review report’s scientific quality classification
Grade A (Excellent): 0
Grade B (Very good): B
Grade C (Good): 0
Grade D (Fair): 0
Grade E (Poor): 0
P-Reviewer: Vasilescu A, Romania S-Editor: Wang JJ L-Editor: A P-Editor: Yu HG
| 1. | Morgan E, Arnold M, Gini A, Lorenzoni V, Cabasag CJ, Laversanne M, Vignat J, Ferlay J, Murphy N, Bray F. Global burden of colorectal cancer in 2020 and 2040: incidence and mortality estimates from GLOBOCAN. Gut. 2023;72:338-344. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 511] [Cited by in RCA: 903] [Article Influence: 451.5] [Reference Citation Analysis (1)] |
| 2. | Zhou CB, Fang JY. The role of pyroptosis in gastrointestinal cancer and immune responses to intestinal microbial infection. Biochim Biophys Acta Rev Cancer. 2019;1872:1-10. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 63] [Cited by in RCA: 117] [Article Influence: 19.5] [Reference Citation Analysis (0)] |
| 3. | Sung H, Ferlay J, Siegel RL, Laversanne M, Soerjomataram I, Jemal A, Bray F. Global Cancer Statistics 2020: GLOBOCAN Estimates of Incidence and Mortality Worldwide for 36 Cancers in 185 Countries. CA Cancer J Clin. 2021;71:209-249. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 75126] [Cited by in RCA: 64450] [Article Influence: 16112.5] [Reference Citation Analysis (176)] |
| 4. | Shinji S, Yamada T, Matsuda A, Sonoda H, Ohta R, Iwai T, Takeda K, Yonaga K, Masuda Y, Yoshida H. Recent Advances in the Treatment of Colorectal Cancer: A Review. J Nippon Med Sch. 2022;89:246-254. [RCA] [PubMed] [DOI] [Full Text] [Cited by in RCA: 112] [Reference Citation Analysis (1)] |
| 5. | Singh D, Vignat J, Lorenzoni V, Eslahi M, Ginsburg O, Lauby-Secretan B, Arbyn M, Basu P, Bray F, Vaccarella S. Global estimates of incidence and mortality of cervical cancer in 2020: a baseline analysis of the WHO Global Cervical Cancer Elimination Initiative. Lancet Glob Health. 2023;11:e197-e206. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 409] [Cited by in RCA: 678] [Article Influence: 339.0] [Reference Citation Analysis (0)] |
| 6. | Hou W, Yi C, Zhu H. Predictive biomarkers of colon cancer immunotherapy: Present and future. Front Immunol. 2022;13:1032314. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 97] [Cited by in RCA: 89] [Article Influence: 29.7] [Reference Citation Analysis (0)] |
| 7. | Pio R, Ajona D, Ortiz-Espinosa S, Mantovani A, Lambris JD. Complementing the Cancer-Immunity Cycle. Front Immunol. 2019;10:774. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 136] [Cited by in RCA: 150] [Article Influence: 25.0] [Reference Citation Analysis (0)] |
| 8. | Gros P, Milder FJ, Janssen BJ. Complement driven by conformational changes. Nat Rev Immunol. 2008;8:48-58. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 234] [Cited by in RCA: 233] [Article Influence: 13.7] [Reference Citation Analysis (0)] |
| 9. | Ricklin D, Hajishengallis G, Yang K, Lambris JD. Complement: a key system for immune surveillance and homeostasis. Nat Immunol. 2010;11:785-797. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 2932] [Cited by in RCA: 2747] [Article Influence: 183.1] [Reference Citation Analysis (0)] |
| 10. | Serna M, Giles JL, Morgan BP, Bubeck D. Structural basis of complement membrane attack complex formation. Nat Commun. 2016;7:10587. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 178] [Cited by in RCA: 178] [Article Influence: 19.8] [Reference Citation Analysis (0)] |
| 11. | Goldberger G, Arnaout MA, Aden D, Kay R, Rits M, Colten HR. Biosynthesis and postsynthetic processing of human C3b/C4b inactivator (factor I) in three hepatoma cell lines. J Biol Chem. 1984;259:6492-6497. [PubMed] |
| 12. | Zipfel PF, Skerka C. Complement regulators and inhibitory proteins. Nat Rev Immunol. 2009;9:729-740. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 857] [Cited by in RCA: 928] [Article Influence: 58.0] [Reference Citation Analysis (0)] |
| 13. | Trudel D, Avarvarei LM, Orain M, Turcotte S, Plante M, Grégoire J, Kappelhoff R, Labbé DP, Bachvarov D, Têtu B, Overall CM, Bairati I. Proteases and their inhibitors as prognostic factors for high-grade serous ovarian cancer. Pathol Res Pract. 2019;215:152369. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1] [Cited by in RCA: 1] [Article Influence: 0.2] [Reference Citation Analysis (0)] |
| 14. | Cai X, Qiu W, Qian M, Feng S, Peng C, Zhang J, Wang Y. A Candidate Prognostic Biomarker Complement Factor I Promotes Malignant Progression in Glioma. Front Cell Dev Biol. 2020;8:615970. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 3] [Cited by in RCA: 4] [Article Influence: 1.0] [Reference Citation Analysis (0)] |
| 15. | Rahmati Nezhad P, Riihilä P, Piipponen M, Kallajoki M, Meri S, Nissinen L, Kähäri VM. Complement factor I upregulates expression of matrix metalloproteinase-13 and -2 and promotes invasion of cutaneous squamous carcinoma cells. Exp Dermatol. 2021;30:1631-1641. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 2] [Cited by in RCA: 17] [Article Influence: 4.3] [Reference Citation Analysis (0)] |
| 16. | Vaupel P, Multhoff G. Revisiting the Warburg effect: historical dogma versus current understanding. J Physiol. 2021;599:1745-1757. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 117] [Cited by in RCA: 500] [Article Influence: 125.0] [Reference Citation Analysis (0)] |
| 17. | Fukushi A, Kim HD, Chang YC, Kim CH. Revisited Metabolic Control and Reprogramming Cancers by Means of the Warburg Effect in Tumor Cells. Int J Mol Sci. 2022;23. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 106] [Cited by in RCA: 119] [Article Influence: 39.7] [Reference Citation Analysis (0)] |
| 18. | Icard P, Shulman S, Farhat D, Steyaert JM, Alifano M, Lincet H. How the Warburg effect supports aggressiveness and drug resistance of cancer cells? Drug Resist Updat. 2018;38:1-11. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 182] [Cited by in RCA: 404] [Article Influence: 57.7] [Reference Citation Analysis (0)] |
| 19. | Ishii M, Beeson G, Beeson C, Rohrer B. Mitochondrial C3a Receptor Activation in Oxidatively Stressed Epithelial Cells Reduces Mitochondrial Respiration and Metabolism. Front Immunol. 2021;12:628062. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 5] [Cited by in RCA: 37] [Article Influence: 9.3] [Reference Citation Analysis (0)] |
| 20. | Arbore G, West EE, Spolski R, Robertson AAB, Klos A, Rheinheimer C, Dutow P, Woodruff TM, Yu ZX, O'Neill LA, Coll RC, Sher A, Leonard WJ, Köhl J, Monk P, Cooper MA, Arno M, Afzali B, Lachmann HJ, Cope AP, Mayer-Barber KD, Kemper C. T helper 1 immunity requires complement-driven NLRP3 inflammasome activity in CD4⁺ T cells. Science. 2016;352:aad1210. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 290] [Cited by in RCA: 412] [Article Influence: 45.8] [Reference Citation Analysis (0)] |
| 21. | Niyonzima N, Rahman J, Kunz N, West EE, Freiwald T, Desai JV, Merle NS, Gidon A, Sporsheim B, Lionakis MS, Evensen K, Lindberg B, Skagen K, Skjelland M, Singh P, Haug M, Ruseva MM, Kolev M, Bibby J, Marshall O, O'Brien B, Deeks N, Afzali B, Clark RJ, Woodruff TM, Pryor M, Yang ZH, Remaley AT, Mollnes TE, Hewitt SM, Yan B, Kazemian M, Kiss MG, Binder CJ, Halvorsen B, Espevik T, Kemper C. Mitochondrial C5aR1 activity in macrophages controls IL-1β production underlying sterile inflammation. Sci Immunol. 2021;6:eabf2489. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 11] [Cited by in RCA: 100] [Article Influence: 25.0] [Reference Citation Analysis (0)] |
| 22. | Watanabe K, Shiga K, Maeda A, Harata S, Yanagita T, Suzuki T, Ushigome H, Maeda Y, Hirokawa T, Ogawa R, Hara M, Takahashi H, Matsuo Y, Mitsui A, Kimura M, Takiguchi S. Chitinase 3-like 1 secreted from cancer-associated fibroblasts promotes tumor angiogenesis via interleukin-8 secretion in colorectal cancer. Int J Oncol. 2022;60. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 28] [Cited by in RCA: 28] [Article Influence: 9.3] [Reference Citation Analysis (0)] |
| 23. | Wang L, Li S, Luo H, Lu Q, Yu S. PCSK9 promotes the progression and metastasis of colon cancer cells through regulation of EMT and PI3K/AKT signaling in tumor cells and phenotypic polarization of macrophages. J Exp Clin Cancer Res. 2022;41:303. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in RCA: 138] [Reference Citation Analysis (0)] |
| 24. | Lu P, Ma Y, Wei S, Liang X. The dual role of complement in cancers, from destroying tumors to promoting tumor development. Cytokine. 2021;143:155522. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 3] [Cited by in RCA: 13] [Article Influence: 3.3] [Reference Citation Analysis (0)] |
| 25. | Verhaar S, Vissers PA, Maas H, van de Poll-Franse LV, van Erning FN, Mols F. Treatment-related differences in health related quality of life and disease specific symptoms among colon cancer survivors: results from the population-based PROFILES registry. Eur J Cancer. 2015;51:1263-1273. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 16] [Cited by in RCA: 20] [Article Influence: 2.0] [Reference Citation Analysis (0)] |
| 26. | Lichtenstern CR, Ngu RK, Shalapour S, Karin M. Immunotherapy, Inflammation and Colorectal Cancer. Cells. 2020;9. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 88] [Cited by in RCA: 201] [Article Influence: 40.2] [Reference Citation Analysis (0)] |
| 27. | Van Kaer L. LEF1 Creates Memories in iNKT Cells That Potentiate Antitumor Immunity. Cancer Immunol Res. 2023;11:144. [RCA] [PubMed] [DOI] [Full Text] [Reference Citation Analysis (0)] |
| 28. | Gu S, Liu F, Xie X, Ding M, Wang Z, Xing X, Xiao T, Sun X. β-Sitosterol blocks the LEF-1-mediated Wnt/β-catenin pathway to inhibit proliferation of human colon cancer cells. Cell Signal. 2023;104:110585. [RCA] [PubMed] [DOI] [Full Text] [Cited by in RCA: 17] [Reference Citation Analysis (0)] |
| 29. | Chen X, Tu J, Liu C, Wang L, Yuan X. MicroRNA-621 functions as a metastasis suppressor in colorectal cancer by directly targeting LEF1 and suppressing Wnt/β-catenin signaling. Life Sci. 2022;308:120941. [RCA] [PubMed] [DOI] [Full Text] [Cited by in RCA: 8] [Reference Citation Analysis (0)] |
| 30. | Lu C, Xie T, Guo X, Wu D, Li S, Li X, Lu Y, Wang X. LncRNA DSCAM-AS1 Promotes Colon Cancer Cells Proliferation and Migration via Regulating the miR-204/SOX4 Axis. Cancer Manag Res. 2020;12:4347-4356. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 7] [Cited by in RCA: 13] [Article Influence: 2.6] [Reference Citation Analysis (0)] |
| 31. | Li L, Liu J, Xue H, Li C, Liu Q, Zhou Y, Wang T, Wang H, Qian H, Wen T. A TGF-β-MTA1-SOX4-EZH2 signaling axis drives epithelial-mesenchymal transition in tumor metastasis. Oncogene. 2020;39:2125-2139. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 53] [Cited by in RCA: 71] [Article Influence: 14.2] [Reference Citation Analysis (0)] |
| 32. | Gerner MC, Ziegler LS, Schmidt RLJ, Krenn M, Zimprich F, Uyanik-Ünal K, Konstantopoulou V, Derdak S, Del Favero G, Schwarzinger I, Boztug K, Schmetterer KG. The TGF-b/SOX4 axis and ROS-driven autophagy co-mediate CD39 expression in regulatory T-cells. FASEB J. 2020;34:8367-8384. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 31] [Cited by in RCA: 34] [Article Influence: 6.8] [Reference Citation Analysis (0)] |
| 33. | Wood SM, Gill AJ, Brodsky AS, Lu S, Friedman K, Karashchuk G, Lombardo K, Yang D, Resnick MB. Fatty acid-binding protein 1 is preferentially lost in microsatellite instable colorectal carcinomas and is immune modulated via the interferon γ pathway. Mod Pathol. 2017;30:123-133. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 9] [Cited by in RCA: 14] [Article Influence: 1.8] [Reference Citation Analysis (0)] |
| 34. | Li TT, Lai YW, Han X, Niu X, Zhang PX. BMP2 as a promising anticancer approach: functions and molecular mechanisms. Invest New Drugs. 2022;40:1322-1332. [RCA] [PubMed] [DOI] [Full Text] [Cited by in RCA: 15] [Reference Citation Analysis (0)] |
| 35. | Vishnubalaji R, Yue S, Alfayez M, Kassem M, Liu FF, Aldahmash A, Alajez NM. Bone morphogenetic protein 2 (BMP2) induces growth suppression and enhances chemosensitivity of human colon cancer cells. Cancer Cell Int. 2016;16:77. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 26] [Cited by in RCA: 44] [Article Influence: 4.9] [Reference Citation Analysis (0)] |
| 36. | Magrini E, Minute L, Dambra M, Garlanda C. Complement activation in cancer: Effects on tumor-associated myeloid cells and immunosuppression. Semin Immunol. 2022;60:101642. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 2] [Cited by in RCA: 28] [Article Influence: 9.3] [Reference Citation Analysis (0)] |
| 37. | Olcina MM, Kim RK, Melemenidis S, Graves EE, Giaccia AJ. The tumour microenvironment links complement system dysregulation and hypoxic signalling. Br J Radiol. 2019;92:20180069. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 6] [Cited by in RCA: 12] [Article Influence: 1.7] [Reference Citation Analysis (0)] |
| 38. | Kemper C, Pangburn MK, Fishelson Z. Complement nomenclature 2014. Mol Immunol. 2014;61:56-58. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 48] [Cited by in RCA: 48] [Article Influence: 4.4] [Reference Citation Analysis (0)] |
| 39. | Gao Y. Complement system in Anti-CD20 mAb therapy for cancer: A mini-review. Int J Immunopathol Pharmacol. 2023;37:3946320231181464. [RCA] [PubMed] [DOI] [Full Text] [Cited by in RCA: 1] [Reference Citation Analysis (0)] |
| 40. | Ullenhag GJ, Spendlove I, Watson NF, Indar AA, Dube M, Robins RA, Maxwell-Armstrong C, Scholefield JH, Durrant LG. A neoadjuvant/adjuvant randomized trial of colorectal cancer patients vaccinated with an anti-idiotypic antibody, 105AD7, mimicking CD55. Clin Cancer Res. 2006;12:7389-7396. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 24] [Cited by in RCA: 24] [Article Influence: 1.3] [Reference Citation Analysis (0)] |
| 41. | Wang Y, Zhang H, He YW. The Complement Receptors C3aR and C5aR Are a New Class of Immune Checkpoint Receptor in Cancer Immunotherapy. Front Immunol. 2019;10:1574. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 28] [Cited by in RCA: 57] [Article Influence: 9.5] [Reference Citation Analysis (0)] |
| 42. | Andoh A, Fujiyama Y, Sakumoto H, Uchihara H, Kimura T, Koyama S, Bamba T. Detection of complement C3 and factor B gene expression in normal colorectal mucosa, adenomas and carcinomas. Clin Exp Immunol. 1998;111:477-483. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 32] [Cited by in RCA: 37] [Article Influence: 1.4] [Reference Citation Analysis (0)] |
| 43. | Zhao L, Zhang Z, Lin J, Cao L, He B, Han S, Zhang X. Complement receptor 1 genetic variants contribute to the susceptibility to gastric cancer in chinese population. J Cancer. 2015;6:525-530. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 10] [Cited by in RCA: 15] [Article Influence: 1.5] [Reference Citation Analysis (0)] |
| 44. | Okroj M, Holmquist E, Nilsson E, Anagnostaki L, Jirström K, Blom AM. Local expression of complement factor I in breast cancer cells correlates with poor survival and recurrence. Cancer Immunol Immunother. 2015;64:467-478. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 21] [Cited by in RCA: 26] [Article Influence: 2.6] [Reference Citation Analysis (0)] |
| 45. | Liu C, Liu D, Wang F, Xie J, Liu Y, Wang H, Rong J, Wang J, Zeng R, Zhou F, Peng J, Xie Y. Identification of a glycolysis- and lactate-related gene signature for predicting prognosis, immune microenvironment, and drug candidates in colon adenocarcinoma. Front Cell Dev Biol. 2022;10:971992. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in RCA: 12] [Reference Citation Analysis (0)] |
| 46. | Chen YJ, Guo X, Liu ML, Yu YY, Cui YH, Shen XZ, Liu TS, Liang L. Interaction between glycolysis‒cholesterol synthesis axis and tumor microenvironment reveal that gamma-glutamyl hydrolase suppresses glycolysis in colon cancer. Front Immunol. 2022;13:979521. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in RCA: 16] [Reference Citation Analysis (0)] |
| 47. | Kondaveeti Y, Guttilla Reed IK, White BA. Epithelial-mesenchymal transition induces similar metabolic alterations in two independent breast cancer cell lines. Cancer Lett. 2015;364:44-58. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 67] [Cited by in RCA: 76] [Article Influence: 7.6] [Reference Citation Analysis (0)] |
| 48. | Ippolito L, Morandi A, Giannoni E, Chiarugi P. Lactate: A Metabolic Driver in the Tumour Landscape. Trends Biochem Sci. 2019;44:153-166. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 140] [Cited by in RCA: 347] [Article Influence: 49.6] [Reference Citation Analysis (0)] |
| 49. | Kolev M, Dimeloe S, Le Friec G, Navarini A, Arbore G, Povoleri GA, Fischer M, Belle R, Loeliger J, Develioglu L, Bantug GR, Watson J, Couzi L, Afzali B, Lavender P, Hess C, Kemper C. Complement Regulates Nutrient Influx and Metabolic Reprogramming during Th1 Cell Responses. Immunity. 2015;42:1033-1047. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 151] [Cited by in RCA: 204] [Article Influence: 20.4] [Reference Citation Analysis (0)] |
| 50. | El-Sahli S, Xie Y, Wang L, Liu S. Wnt Signaling in Cancer Metabolism and Immunity. Cancers (Basel). 2019;11. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 56] [Cited by in RCA: 92] [Article Influence: 15.3] [Reference Citation Analysis (0)] |
| 51. | Zhang C, Liu L, Li W, Li M, Zhang X, Zhang C, Yang H, Xie J, Pan W, Guo X, She P, Zhong L, Li T. Upregulation of FAM83F by c-Myc promotes cervical cancer growth and aerobic glycolysis via Wnt/β-catenin signaling activation. Cell Death Dis. 2023;14:837. [RCA] [PubMed] [DOI] [Full Text] [Cited by in RCA: 11] [Reference Citation Analysis (0)] |
| 52. | Liang Y, Rao Z, Du D, Wang Y, Fang T. Butyrate prevents the migration and invasion, and aerobic glycolysis in gastric cancer via inhibiting Wnt/β-catenin/c-Myc signaling. Drug Dev Res. 2023;84:532-541. [RCA] [PubMed] [DOI] [Full Text] [Cited by in RCA: 15] [Reference Citation Analysis (0)] |
| 53. | Dotan E, Cardin DB, Lenz HJ, Messersmith W, O'Neil B, Cohen SJ, Denlinger CS, Shahda S, Astsaturov I, Kapoun AM, Brachmann RK, Uttamsingh S, Stagg RJ, Weekes C. Phase Ib Study of Wnt Inhibitor Ipafricept with Gemcitabine and nab-paclitaxel in Patients with Previously Untreated Stage IV Pancreatic Cancer. Clin Cancer Res. 2020;26:5348-5357. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 15] [Cited by in RCA: 50] [Article Influence: 10.0] [Reference Citation Analysis (0)] |
| 54. | Kleszcz R, Paluszczak J. The Wnt Signaling Pathway Inhibitors Improve the Therapeutic Activity of Glycolysis Modulators against Tongue Cancer Cells. Int J Mol Sci. 2022;23. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 1] [Cited by in RCA: 5] [Article Influence: 1.7] [Reference Citation Analysis (0)] |