Published online Jun 15, 2024. doi: 10.4251/wjgo.v16.i6.2404
Revised: February 27, 2024
Accepted: April 3, 2024
Published online: June 15, 2024
Processing time: 168 Days and 10.7 Hours
Research on gastrointestinal mucosal adenocarcinoma (GMA) is limited and controversial, and there is no reference tool for predicting postoperative survival.
To investigate the prognosis of GMA and develop predictive model.
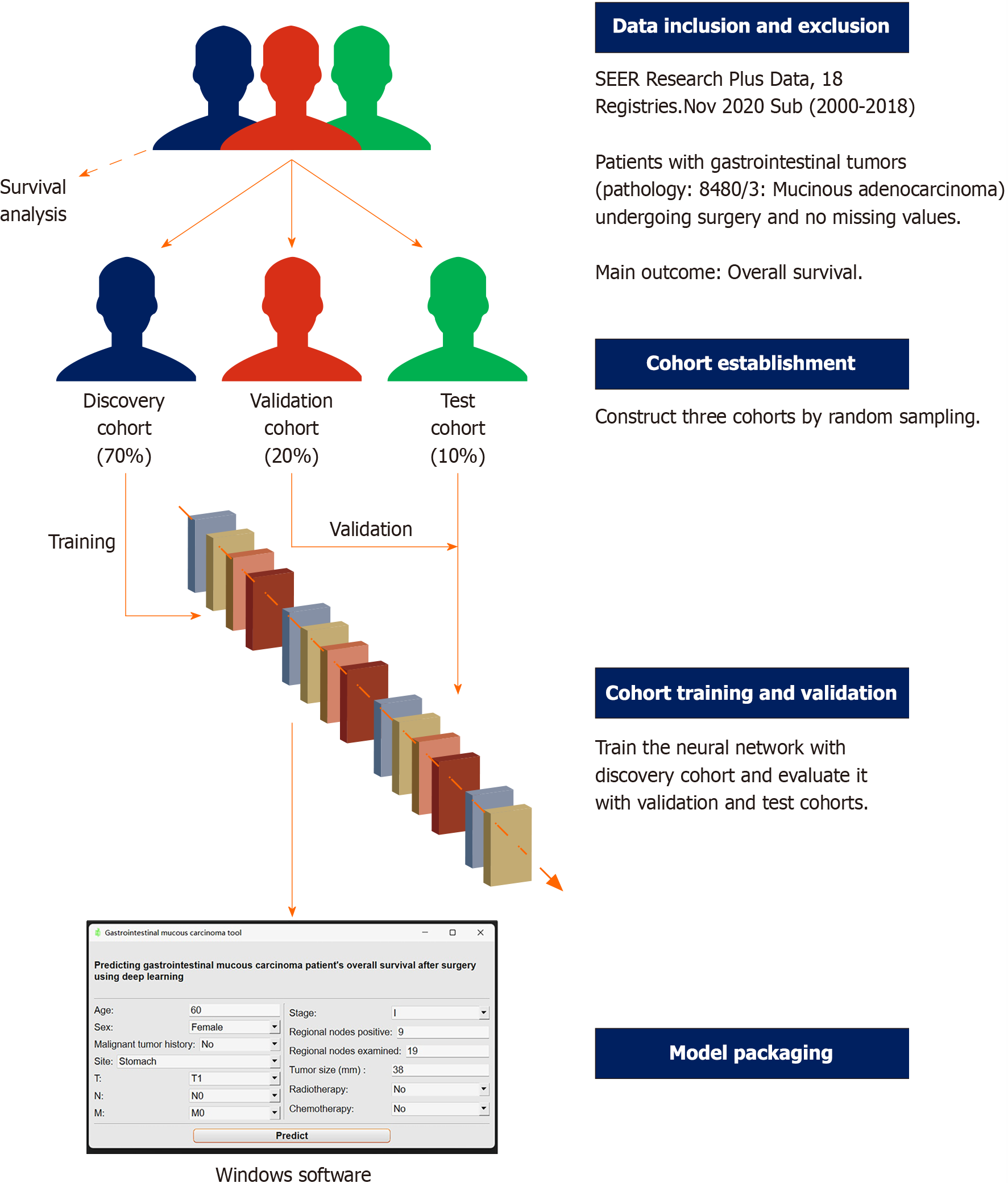
From the Surveillance, Epidemiology, and End Results database, we collected clinical information on patients with GMA. After random sampling, the patients were divided into the discovery (70% of the total, for model training), validation (20%, for model evaluation), and completely blind test cohorts (10%, for further model evaluation). The main assessment metric was the area under the receiver operating characteristic curve (AUC). All collected clinical features were used for Cox proportional hazard regression analysis to determine factors influencing GMA’s prognosis.
This model had an AUC of 0.7433 [95% confidence intervals (95%CI): 0.7424-0.7442] in the discovery cohort, 0.7244 (GMA: 0.7234-0.7254) in the validation cohort, and 0.7388 (95%CI: 0.7378-0.7398) in the test cohort. We packaged it into Windows software for doctors’ use and uploaded it. Mucinous gastric adenocarcinoma had the worst pro
The deep learning-based tool developed can accurately predict the overall survival of patients with GMA postoperatively. Combining surgery, chemotherapy, and adequate lymph node dissection during surgery can improve pat
Core Tip: After surgery, some patients can be diagnosed with gastrointestinal mucous adenocarcinoma (GMA) by pathology, a rare subtype cancer. However, research on GMA is limited and controversial, and there is no reference tool for their postoperative survival prediction. We searched Surveillance, Epidemiology, and End Results database and collected 11390 GMA patients’ clinical information. Then we constructed a deep learning-based tool to predict GMA patients’ overall survival after surgery, and the tool has been uploaded. After our analysis, combining surgery, chemotherapy, and adequate lymph node dissection during surgery can improve patient outcomes.
- Citation: Song J, Yan XX, Zhang FL, Lei YY, Ke ZY, Li F, Zhang K, He YQ, Li W, Li C, Pan YM. Unveiling the secrets of gastrointestinal mucous adenocarcinoma survival after surgery with artificial intelligence: A population-based study. World J Gastrointest Oncol 2024; 16(6): 2404-2418
- URL: https://www.wjgnet.com/1948-5204/full/v16/i6/2404.htm
- DOI: https://dx.doi.org/10.4251/wjgo.v16.i6.2404
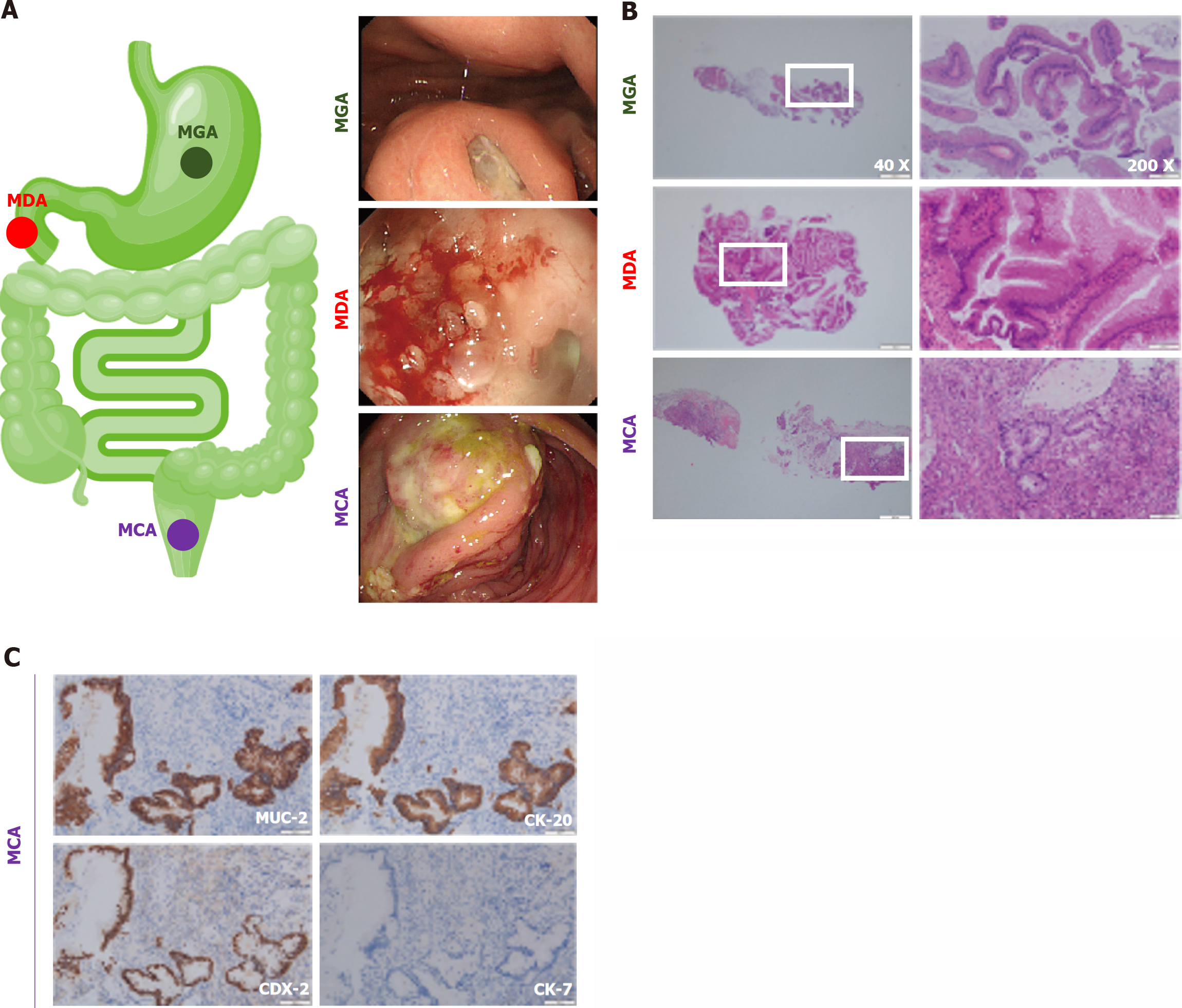
Gastrointestinal cancer is one of the most common fatal tumors in the United States, and colorectal cancer is the third most frequent malignant tumor and the third most deadly tumor[1,2]. Surgery is one of the most popular therapies[3,4]. However, after surgery, some patients can be pathologically diagnosed with gastrointestinal mucous adenocarcinoma (GMA), a rare subtype represented by mucinous gastric adenocarcinoma (MGA), mucinous duodenal adenocarcinoma (MDA), and mucinous colorectal adenocarcinoma (MCA). Figure 1 shows typical endoscopic and pathological images of the GMA, including the MGA, MDA, and MCA. To further identify GMA, immunohistochemistry is used frequently, and common antibody combinations include MUC-2, CK-20, CDX-2, and CK-7[5,6]. Taking the MCA as an example, MUC-2, CK-20, and CDX-2 were positive, whereas CK-7 was negative (Figure 1C).
Research on the GMA remains limited, and some conclusions from related studies are contradictory[7,8]. For example, there is conflicting information in the literature regarding the prognosis and overall survival (OS) of patients with MCA in the literature[7]. Consequently, awareness of GMA among doctors and researchers is limited, including some necessary expertise, such as a dearth of pertinent research to support additional preoperative or postoperative treatment for GMA. Large-scale clinical data analyses are required, particularly in randomized controlled clinical trials with high levels of evidence. The postoperative prognosis is another matter that concerns doctors, patients, and their families. Prognostic information currently available for GMA is scarce, especially because an individualized survival prediction system is lacking.
The Surveillance, Epidemiology, and End Results (SEER) database is the largest tumor database in the United States, with over 50 years of history. It covers approximately 48.0% of the American population. It is especially well-suited for studies on uncommon illnesses and cancer epidemiology surveys because of its wide coverage and authority.
In this study, we searched the SEER database, retrospectively analyzed the clinical data of patients with GMA using a large amount of clinical data, developed an OS prediction model for patients with GMA based on deep learning algorithms, and packaged it for simple usage by clinicians. In addition, we conducted statistical analyses and reviewed studies on the GMA to identify the risk and protective factors related to prognosis.
We searched the SEER database and collected the clinical information of patients with GMA. Data originated from SEER Research Plus Data, 18 Registries, Nov 2020 Sub (2000-2018) sub-database, which covers approximately 27.8% of the American population. Detailed inclusion and exclusion criteria were as follows: (1) ICD-O-3 Hist/behav was 8480/3: Mucinous adenocarcinoma; (2) primary sites were gastrointestinal tract (C16.0-Cardia, NOS, C16.1-Fundus of stomach, C16.2-Body of stomach, C16.3-Gastric antrum, C16.4-Pylorus, C16.5-Lesser curvature of stomach NOS, C16.6-Greater curvature of stomach NOS, C16.8-Overlapping lesion of stomach, C16.9-Stomach, NOS, C17.0-Duodenum, C17.1-Jejunum, C17.2-Ileum, C17.8-Overlapping lesion of small intestine, C17.9-Small intestine, NOS, C18.0-Cecum, C18.1-Appendix, C18.2-Ascending colon, C18.3-Hepatic flexure of colon, C18.4-Transverse colon, C18.5-Splenic flexure of colon, C18.6-Descending colon, C18.7-Sigmoid colon, C18.8-Overlapping lesion of colon, C18.9-Colon, NOS, C19.9-Rectosigmoid junction or C20.9-Rectum, and NOS); (3) patients have gotten surgery; (4) complete American Joint Committee on Cancer TNM stage and other clinical features needed; and (5) no missing values (Table 1).
| Option in SEER | Value |
| Database | SEER Research Plus Data, 18 registries. Nov 2020 Sub (2000-2018) |
| ICD-O-3 Hist/behav | 8480/3: Mucinous adenocarcinoma |
| Primary site-labeled | C16.0-Cardia, NOS |
| C16.1-Fundus of stomach | |
| C16.2-Body of stomach | |
| C16.3-Gastric antrum | |
| C16.4-Pylorus | |
| C16.5-Lesser curvature of stomach NOS | |
| C16.6-Greater curvature of stomach NOS | |
| C16.8-Overlapping lesion of stomach | |
| C16.9-Stomach, NOS | |
| C17.0-Duodenum | |
| C17.1-Jejunum | |
| C17.2-Ileum | |
| C17.8-Overlapping lesion of small intestine | |
| C17.9-Small intestine, NOS | |
| C18.0-Cecum | |
| C18.1-Appendix | |
| C18.2-Ascending colon | |
| C18.3-Hepatic flexure of colon | |
| C18.4-Transverse colon | |
| C18.5-Splenic flexure of colon | |
| C18.6-Descending colon | |
| C18.7-Sigmoid colon | |
| C18.8-Overlapping lesion of colon | |
| C18.9-Colon, NOS | |
| C19.9-Rectosigmoid junction | |
| C20.9-Rectum, NOS | |
| Other | Receive surgery and records without missing value |
This retrospective study was designed for diagnostic testing. After screening according to the inclusion and exclusion criteria, all patients were randomly assigned to the discovery (70%), validation (20%), and test (10%) cohorts. The discovery cohort was used to train the deep learning survival model, which was evaluated in the validation cohort and another completely blind test cohort. The primary outcome was the OS of the patients with GMA (Figure 2).
The data for this research came from the publicly accessible SEER database, and patients’ information was anonymized and untraceable. Consequently, this study was exempt from ethical approval and written permission.
Age, sex, tumor site, history of malignant tumors, and TNM stage are potential risk factors for gastrointestinal cancer[7,9-11]. A larger tumor diameter or more positive lymph nodes generally indicates a more advanced tumor stage, and additional lymph node examinations can help determine this stage. Therefore, they are also considered conceivable predictors. Radiotherapy and chemotherapy are the most commonly used treatment strategies in addition to surgery.
The variables listed above were entered into the least absolute shrinkage and selection operator (LASSO) regression with 10-fold cross-validation to find the lowest lambda value. Clinical features with nonzero coefficients in the regression model were selected as final predictor variables based on this lambda value.
According to SEER rules, tumors with a diameter of 989 mm or larger are still recorded as 989 mm. Patients older than 100 years were listed as such. The tumor sites were merged according to their records. Those who survived for less than one month were still regarded as one month.
The training process was completed in python 3.9 (using Pytorch, Torchtuples, Sklearn, Pandas, Numpy, and Pycox). Unlike the typical classification, survival prediction has two variables: Survival time and status. This model was built based on DeepSuvr theory[12]. To obtain a better training effect, we transformed categorical clinical features (sex, malig
Model performance was evaluated using the area under the receiver operating characteristic curve (AUC). The closer the AUC is to 1.0, the better the model performance. The closer the AUC is to 0.5, the more inclined the model is toward random guessing. The bootstrap method was used to obtain the AUC and 95% confidence interval (CI). The model was truncated at 1, 3, and 5 years to obtain a more comprehensive assessment. Other evaluation metrics included specificity, sensitivity, accuracy, positive predictive value (PPV), and negative predictive value (NPV). We also used a Cox proportional hazards (CPH) model using the same clinical features for comparison.
Finally, the model was packaged into a Windows tool that doctors could use more conveniently. This process was completed in Pycharm, using the pyside6 and pyinstaller package.
We compared the prognosis of GMA at different sites using Kaplan-Meier curves and log-rank tests. All collected clinical features were utilized to conduct multivariate CPH regression to identify the protective and risk factors for GMA. Some clinical features (T, N, M, and stage) were reintegrated before this process.
All statistical analyses were performed using R version 4.2.0. The Chi-square test was used for categorical variables, and the Kruskal-Wallis test was used for continuous variables with a non-normal distribution. A two-sided P value less than 0.05 was considered statistically significant. The following R packages were used for data analysis and visualization: glmnet, pROC, ggsci, ggplot2, survminer, survival, forest model, epiDisplay, circlize, and ggridge.
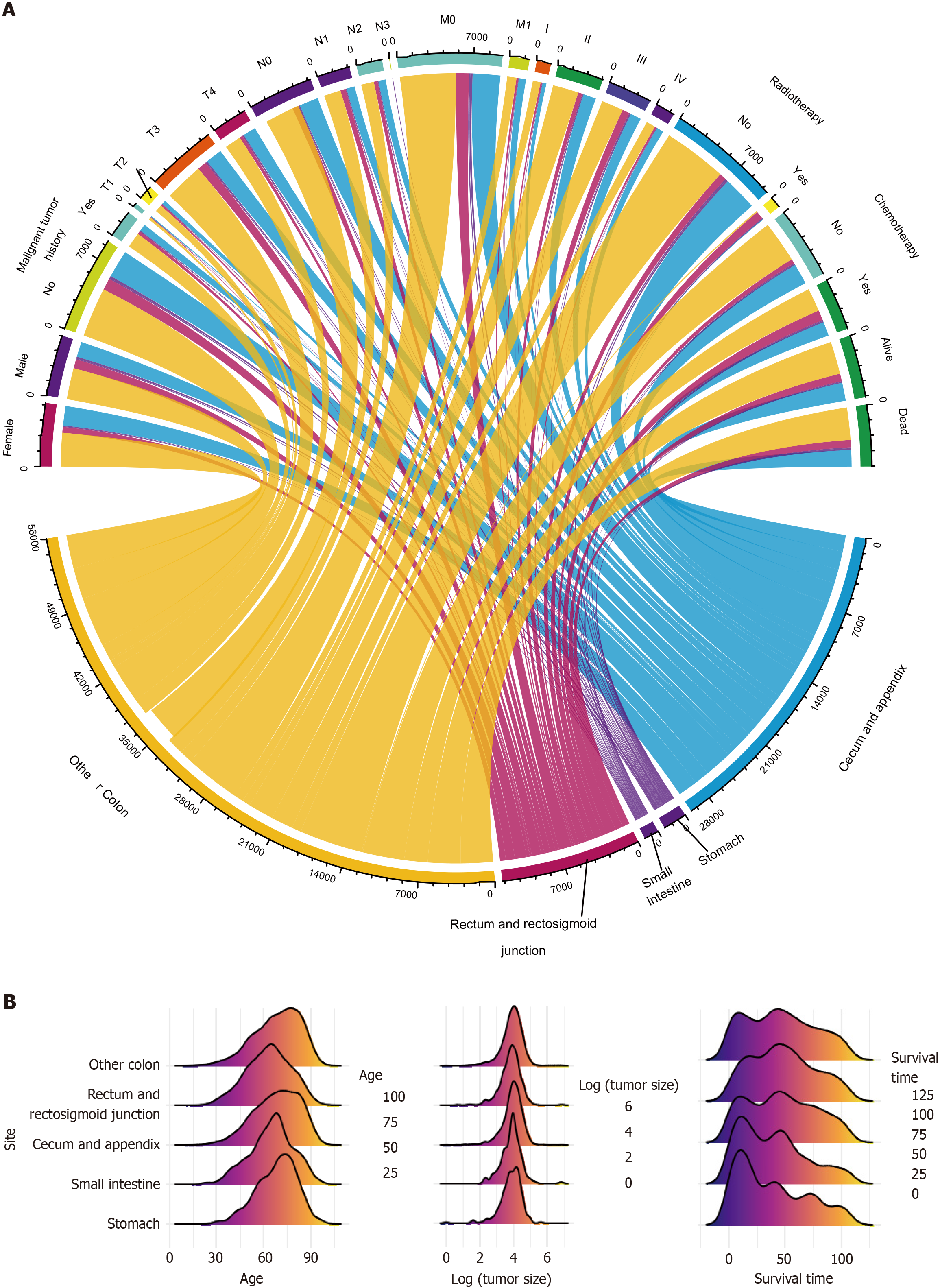
Ultimately, 11390 patients were included in the study. They were then randomly assigned to one of the three cohorts. There were 7972 patients in the discovery cohort, 2392 in the validation cohort, and 1026 in the test cohort. There were no significant differences among the three cohorts. The median ages of the discovery and test cohorts were 69 years, whereas that of the validation cohort was 70 years. Among the three cohorts, most patients with GMA were female among three cohorts and had no history of malignant tumors. In the three cohorts, the most common GMA tumor sites were the other parts of the colon (not the rectum and rectosigmoid junction, or the cecum and appendix). Most patients were evaluated as T3, N0, or M0; therefore, most patients were staged as IIA. The median tumor size was 53.5 mm in the discovery cohort, 53.0 mm in the validation cohort, and 51.0 mm in the test cohort. The median number of positive regional nodes in the three cohorts was 0. The median number of regional nodes examined was 18 in the discovery and validation cohorts and 17 in the test cohort. Most patients did not receive radiotherapy or chemotherapy. The median survival time in the discovery and validation cohorts was 45 months, while test cohort patients had a 48-month median survival time. Almost 50% of the patients in the three groups were alive at the end of the follow-up period (Table 2).
| Discovery cohort (n = 7972) | Validation cohort (n = 2392) | Test cohort (n = 1026) | Statistical test method | P value | |
| Age | Kruskal-Wallis | 0.5211 | |||
| Median (IQR) | 69 (58, 79) | 70 (59, 79) | 69 (58, 80) | ||
| Sex | Chi-square | 0.4781 | |||
| Female | 4029 (50.54) | 1219 (50.96) | 539 (52.53) | ||
| Male | 3943 (49.46) | 1173 (49.04) | 487 (47.47) | ||
| Malignant tumor history | Chi-square | 0.2368 | |||
| No | 6217 (77.99) | 1830 (76.51) | 807 (78.65) | ||
| Yes | 1755 (22.01) | 562 (23.49) | 219 (21.35) | ||
| Site | Chi-square | 0.6989 | |||
| Stomach | 180 (2.26) | 46 (1.92) | 18 (1.75) | ||
| Small intestine | 105 (1.32) | 37 (1.55) | 11 (1.07) | ||
| Cecum and appendix | 2312 (29.00) | 729 (30.48) | 293 (28.56) | ||
| Rectum and rectosigmoid junction | 1003 (12.58) | 295 (12.33) | 129 (12.57) | ||
| Other colon | 4372 (54.84) | 1285 (53.72) | 575 (56.04) | ||
| T | Chi-square | 0.3557 | |||
| T1 | 265 (3.32) | 75 (3.14) | 27 (2.63) | ||
| T1a | 2 (0.03) | 2 (0.08) | 0 (0.00) | ||
| T1b | 16 (0.20) | 4 (0.17) | 1 (0.10) | ||
| T2 | 883 (11.08) | 301 (12.58) | 107 (10.43) | ||
| T3 | 4505 (56.51) | 1331 (55.64) | 591 (57.60) | ||
| T4 | 54 (0.68) | 20 (0.84) | 8 (0.78) | ||
| T4a | 1323 (16.60) | 356 (14.88) | 173 (16.86) | ||
| T4b | 924 (11.59) | 303 (12.67) | 119 (11.60) | ||
| N | Chi-square | 0.6443 | |||
| N0 | 4123 (51.72) | 1284 (53.68) | 543 (52.92) | ||
| N1 | 260 (3.26) | 74 (3.09) | 22 (2.14) | ||
| N1a | 832 (10.44) | 235 (9.82) | 96 (9.36) | ||
| N1b | 920 (11.54) | 267 (11.16) | 126 (12.28) | ||
| N1c | 137 (1.72) | 29 (1.21) | 16 (1.56) | ||
| N2 | 104 (1.30) | 31 (1.30) | 13 (1.27) | ||
| N2a | 654 (8.20) | 208 (8.70) | 99 (9.65) | ||
| N2b | 898 (11.26) | 255 (10.66) | 107 (10.43) | ||
| N3 | 11 (0.14) | 4 (0.17) | 2 (0.19) | ||
| N3a | 20 (0.25) | 3 (0.13) | 1 (0.10) | ||
| N3b | 13 (0.16) | 2 (0.08) | 1 (0.10) | ||
| M | Chi-square | 0.4620 | |||
| M0 | 6689 (83.91) | 2022 (84.53) | 873 (85.09) | ||
| M1 | 96 (1.20) | 22 (0.92) | 10 (0.97) | ||
| M1a | 563 (7.06) | 162 (6.77) | 79 (7.70) | ||
| M1b | 624 (7.83) | 186 (7.78) | 64 (6.24) | ||
| Stage | Chi-square | 0.8700 | |||
| I | 883 (11.08) | 292 (12.21) | 102 (9.94) | ||
| IA | 10 (0.13) | 3 (0.13) | 0 (0.00) | ||
| IB | 21 (0.26) | 3 (0.13) | 2 (0.19) | ||
| II | 3 (0.04) | 1 (0.04) | 0 (0.00) | ||
| IIA | 2236 (28.05) | 675 (28.22) | 305 (29.73) | ||
| IIB | 405 (5.08) | 112 (4.68) | 52 (5.07) | ||
| IIC | 303 (3.80) | 111 (4.64) | 46 (4.48) | ||
| III | 2 (0.03) | 1 (0.04) | 0 (0.00) | ||
| IIIA | 222 (2.78) | 67 (2.80) | 28 (2.73) | ||
| IIIB | 1742 (21.85) | 506 (21.15) | 227 (22.12) | ||
| IIIC | 862 (10.81) | 251 (10.49) | 111 (10.82) | ||
| IV | 96 (1.20) | 22 (0.92) | 10 (0.97) | ||
| IVA | 523 (6.56) | 145 (6.06) | 73 (7.12) | ||
| IVB | 603 (7.56) | 188 (7.86) | 63 (6.14) | ||
| IVC | 61 (0.77) | 15 (0.63) | 7 (0.68) | ||
| Tumor size (mm) | Kruskal-Wallis | 0.4812 | |||
| Median (IQR) | 53.5 (38.0, 72.0) | 53.0 (38.0, 75.0) | 51.0 (40.0, 70.0) | Median (IQR) | |
| Regional nodes positive | Kruskal-Wallis | 0.4721 | |||
| Median (IQR) | 0 (0, 3) | 0 (0, 3) | 0 (0, 3) | ||
| Regional nodes examined | Kruskal-Wallis | 0.4691 | |||
| Median (IQR) | 18 (13, 24) | 18 (13, 24) | 17 (13, 23) | ||
| Radiotherapy | Chi-square | 0.4074 | |||
| No | 7231 (90.70) | 2180 (91.14) | 943 (91.91) | ||
| Yes | 741 (9.30) | 212 (8.86) | 83 (8.09) | ||
| Chemotherapy | Chi-square | 0.6545 | |||
| No | 4615 (57.89) | 1378 (57.61) | 608 (59.26) | ||
| Yes | 3357 (42.11) | 1014 (42.39) | 418 (40.74) | ||
| Survival time | Kruskal-Wallis | 0.2472 | |||
| Median (IQR) | 45 (20.00, 69.00) | 45 (20.75, 69.00) | 48 (21.00, 71.75) | ||
| Status | Chi-square | 0.8302 | |||
| Alive | 4034 (50.60) | 1195 (49.96) | 522 (50.88) | ||
| Dead | 3938 (49.40) | 1197 (50.04) | 504 (49.12) |
The characteristics of all three cohorts of patients were visually displayed in Figure 3, including categorical (Figure 3A) and numerical variables (Figure 3B). They described the sources and general distribution of GMA at different tumor sites.
LASSO Cox regression was used to filter the collected clinical features. After 10-fold cross-validation, the minimum lambda value was 0.0031 (Supplementary Figure 1A). The model’s variable coefficients were examined with this lambda value, and none was equal to zero (Supplementary Table 3). This means that age, sex, malignant tumor history, tumor site, TNM stage, tumor size, regional lymph node positivity, regional lymph nodes examined, radiotherapy, and chemotherapy could predict the OS of patients with GMA. Therefore, they were all used in subsequent modeling.
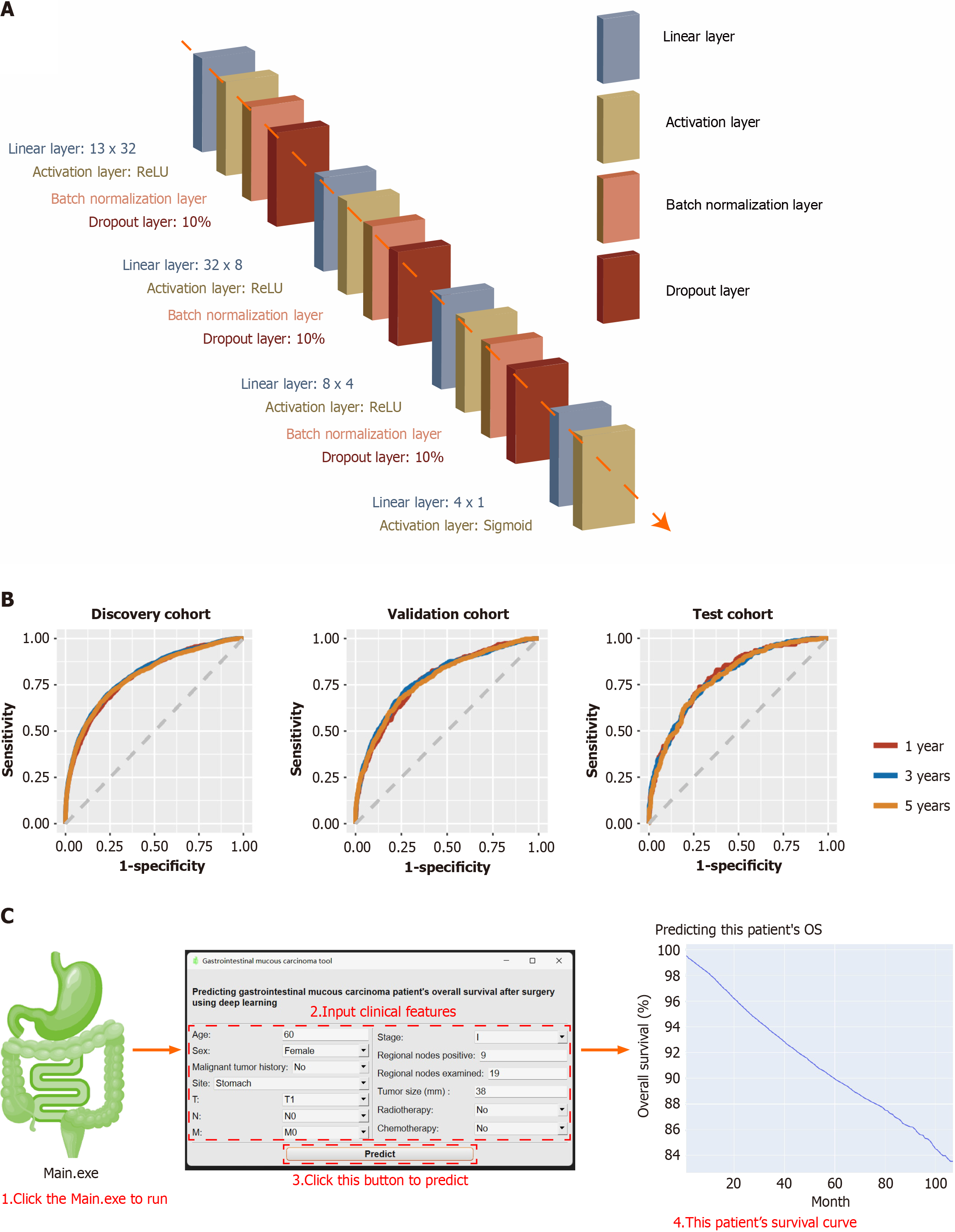
After 100 epochs, the early stopping function abruptly terminated training. The training curves are presented in Supplementary Figure 1B. Finally, the deep learning model had 14 layers. It included a linear layer (13 × 32), an activation layer (ReLU), a batch normalization layer, a dropout layer (10%), a second linear layer (32 × 8), a second activation layer (ReLU), second batch normalization layer, second dropout layer (10%), third linear layer (8 × 4), third activation layer (ReLU), third batch normalization layer, third dropout layer (10%), fourth linear layer (4 × 1), and fourth activation layer (Sigmoid) (Figure 4A). The final output was a GMA patient’s OS probability for the next 1-107 months. The model parameters are shown in Supplementary Figure 2.
This model had a 0.7433 (95%CI: 0.7424-0.7442) AUC in the discovery cohort, 0.7244 (95%CI: 0.7234-0.7254) AUC in the validation cohort, 0.7388 (95%CI: 0.7378-0.7398) AUC in the test cohort (Table 3). The receiver operating characteristic curves are shown in Figure 4B.
| AUC | AUC, 95%CI | ||
| Mean | Low | High | |
| Discovery cohort | 0.7433 | 0.7424 | 0.7442 |
| Validation cohort | 0.7244 | 0.7234 | 0.7254 |
| Test cohort | 0.7388 | 0.7378 | 0.7398 |
In comparison, the same variables and the CPH method were used to fit the data. It only had an AUC of 0.7155 (95%CI: 0.7145-0.7166) in the discovery cohort, 0.6942 (95%CI: 0.6932-0.6953) in the validation cohort, and 0.7178 (95%CI: 0.7168-0.7188) in the test cohort (Supplementary Table 4). Regardless of the mean or 95%CI of the AUC, it is evident that the deep-learning-based model performs better than the CPH.
After 1, 3, and 5 years, we thoroughly assessed the performance of the deep-learning-based model. In discovery cohort, it had 0.7953 AUC (95%CI: 0.7817-0.8090), 0.7688 specificity, 0.6742 sensitivity, 0.7536 accuracy, 0.9250 NPV and 0.3581 PPV in 1-year OS prediction; 0.8034 AUC (95%CI: 0.7933-0.8136), 0.7675 specificity, 0.6963 sensitivity, 0.7421 accuracy, 0.8200 NPV and 0.6243 PPV in the 3-year OS prediction; 0.7971 AUC (95%CI: 0.7873-0.8069), 0.7985 specificity, 0.6595 sensitivity, 0.7365 accuracy, 0.7441 NPV and 0.7253 PPV in 5-year OS prediction. The validation cohort showed 0.7757 AUC (95%CI: 0.7501-0.8012), 0.6493 specificity, 0.7775 sensitivity, 0.6697 accuracy, 0.9388 NPV, and 0.2964 PPV in the 1-year OS prediction; 0.7843 AUC (95%CI: 0.7650-0.8036), 0.7260 specificity, 0.7242 sensitivity, 0.7253 accuracy, 0.8234 NPV and 0.5987 PPV in the 3-year OS prediction; 0.7772 AUC (95%CI: 0.7587-0.7958), 0.7586 specificity, 0.6746 sensitivity, 0.7203 accuracy, 0.7355 NPV and 0.7010 PPV in the 5-year OS prediction. The test cohort showed 0.7938 AUC (95%CI: 0.7566-0.8310), 0.7182 specificity, 0.7386 sensitivity, 0.7212 accuracy, 0.9400 NPV and 0.3148 PPV in 1-year OS prediction; 0.7888 AUC (95%CI: 0.7603-0.8173), 0.6869 specificity, 0.7507 sensitivity, 0.7086 accuracy, 0.8424 NPV and 0.5527 PPV in 3-year OS prediction; 0.7871 AUC (95%CI: 0.7597-0.8146), 0.7296 specificity, 0.7127 sensitivity, 0.7222 accuracy, 0.7655 NPV and 0.6723 PPV in 5-year OS prediction (Table 4).
| Discovery cohort | Validation cohort | Test cohort | |||||||
| 1 yr | 3 yr | 5 yr | 1 yr | 3 yr | 5 yr | 1 yr | 3 yr | 5 yr | |
| AUC | 0.7953 | 0.8034 | 0.7971 | 0.7757 | 0.7843 | 0.7772 | 0.7938 | 0.7888 | 0.7871 |
| AUC, 95%CI | 0.7817-0.8090 | 0.7933-0.8136 | 0.7873-0.8069 | 0.7501-0.8012 | 0.7650-0.8036 | 0.7587-0.7958 | 0.7566-0.8310 | 0.7603-0.8173 | 0.7597-0.8146 |
| Specificity | 0.7688 | 0.7675 | 0.7985 | 0.6493 | 0.7260 | 0.7586 | 0.7182 | 0.6869 | 0.7296 |
| Sensitivity | 0.6742 | 0.6963 | 0.6595 | 0.7775 | 0.7242 | 0.6746 | 0.7386 | 0.7507 | 0.7127 |
| Accuracy | 0.7536 | 0.7421 | 0.7365 | 0.6697 | 0.7253 | 0.7203 | 0.7212 | 0.7086 | 0.7222 |
| NPV | 0.9250 | 0.8200 | 0.7441 | 0.9388 | 0.8234 | 0.7355 | 0.9400 | 0.8424 | 0.7655 |
| PPV | 0.3581 | 0.6243 | 0.7253 | 0.2964 | 0.5987 | 0.7010 | 0.3148 | 0.5527 | 0.6723 |
For convenience, we packaged the model into Windows software. After unzipping, users can double-click Main.exe to start. After inputting the GMA patient’s clinical characteristics, click Predict to run the built-in pre-trained neural network. After the calculation, the prediction results were automatically drawn into a survival curve (Kaplan-Meier curve). The horizontal axis represents a certain month, and the vertical axis represents the OS probability that the predicted patient is still alive in that month. The curve can be zoomed in or out using the mouse, and a specific value is displayed when hovering (Figure 4C).
Overall, the incidence rate of GMA is declining, about 1.7% (1.9% in male and 1.5% in female) (Supplementary Table 5). Moreover, the 1-year survival rate of patients with GMA is about 84% (95%CI: 83%-85%), the 3-year survival rate of them is about 64% (95%CI: 63%-65%) and the 5-year survival rate of them is about 53% (95%CI: 52%-54%) (Supplementary Table 6).
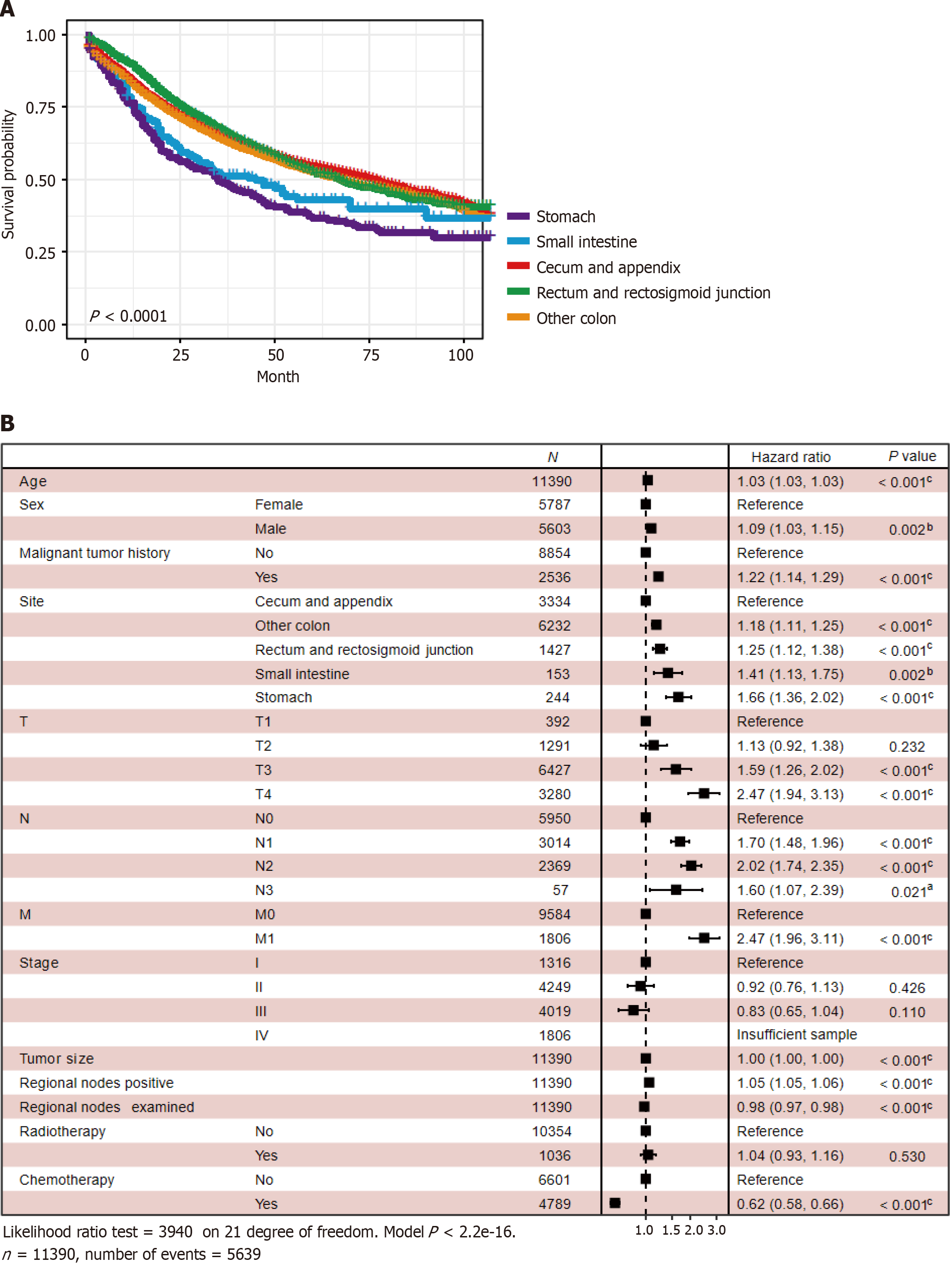
Survival analysis showed that patients with GMA in the stomach had the worst prognosis (P < 0.0001) (Figure 5A). Multivariate CPH regression displayed that these clinical features were risk factors: older age [hazard ratio (HR): 1.03; 95%CI: 1.03-1.03, P < 0.001), male (HR: 1.09, 95%CI: 1.03-1.15, P = 0.002), malignant tumor history (HR: 1.22, 95%CI: 1.14-1.29, P < 0.001), rectum and rectosigmoid junction (HR: 1.25, 95%CI: 1.12-1.38, P < 0.001), small intestine (HR: 1.41, 95%CI: 1.13-1.75, P < 0.002), stomach (HR: 1.66, 95%CI: 1.36-2.02, P < 0.001), other colon sites (HR: 1.18, 95%CI: 1.11-1.25, P < 0.001), T3 (HR: 1.59, 95%CI: 1.26-2.02, P < 0.001), T4 (HR: 2.47, 95%CI: 1.94-3.13, P < 0.001), N1 (HR: 1.70, 95%CI: 1.48-1.96, P < 0.001), N2 (HR: 2.02, 95%CI: 1.74-2.35, P < 0.001), N3 (HR: 1.60, 95%CI: 1.07-2.39, P = 0.021), M1 (HR: 2.47, 95%CI: 1.96-3.11, P < 0.001), larger tumor size (HR: 1.00, 95%CI: 1.00-1.00, P < 0.001), regional nodes positive (HR: 1.05, 95%CI: 1.05-1.06, P < 0.001). These were protective factors: Regional nodes examined (HR: 0.98, 95%CI: 0.97-0.98, P < 0.001) and chemotherapy (HR: 0.62, 95%CI:0.58-0.66, P < 0.001) (Figure 5B).
After surgical resection, some patients may be pathologically diagnosed with GMA, with approximately 1%-20% in the colorectum and 7% in the stomach[13-16]. GMA is distinguished by the presence of many mucinous components that account for approximately 50% of the tumor volume[7,17]. More mucinous components may indicate a poor prognosis[18]. Several factors, including younger age, advanced tumor stage, female sex, microsatellite instability (MSI), and molecular mutations (such as KRAS and BRAF), have been linked to the development of GMA according to earlier investigations[15,16,18-20]. It is still debatable whether GMA and common gastrointestinal tumors have similar OS, as previous studies have produced conflicting reports[7,21]. For example, Warschkow et al[22] observed that MCA had a similar prognosis to other colorectal cancers. Hugen argued that stage III mucinous rectal adenocarcinoma instead of MCA had a worse prognosis. However, more studies, especially a retrospective analysis with a larger sample size (222256 patients), demonstrated that MCA increased mortality risk by 2%-8%[16,22-25]. Similarly, Rokutan et al[15] noticed that MGA was related to poor outcomes, but Hsu stated the opposite conclusion[26]. Most studies have reported that the prognosis of GMA is poor, although additional research and attention are needed.
Since GMA is mainly diagnosed during postoperative pathological examination and there is currently no effective prognostic model, we searched the SEER database and constructed a deep learning algorithm. In the medical field, classic survival prediction is based on the CPH. However, the biggest shortcoming of this theory is that it assumes that the impact of covariates on survival is linear. Although it is simple and easy to implement, this ideal assumption is unsuitable for the intricate changes in the real world. Machine learning, especially deep learning, has been gradually applied in medicine in recent years, including clinical data, medical imaging data, pathological slides, and genomics[27,28]. Due to the presence of a time series and the surviving state, survival prediction is neither a typical classification nor a regression problem. Katzman et al[12] proposed the DeepSurv method to solve this problem, which has been applied in some tumor prognosis studies, such as lung cancer and head and neck cancer[12,29,30]. Our previous study also demonstrated that it is better than traditional algorithms such as CPH[31]. Therefore, we built a deep-learning-based model based on the DeepSurv algorithm.
In this study, we collected the clinical data of 11390 patients. We divided them into three cohorts (7972 patients in the discovery cohort for model training, 2392 in the validation cohort, and 1026 in the test cohort for model evaluation) to predict the OS of patients with GMA after surgery. This model had a 0.7433 (95%CI: 0.7424-0.7442) AUC in the discovery cohort, 0.7244 (95%CI: 0.7234-0.7254) AUC in the validation cohort, 0.7388 (95%CI: 0.7378-0.7398) AUC in the test cohort, which showed predictive value to prognosis and was packaged into a Windows tool. Multivariate survival analysis revealed that chemotherapy and more regional lymph nodes examined were protective factors for GMA, which means that clinicians should consider a combination therapy of surgery and chemotherapy and perform adequate lymph node dissection during surgery.
According to previous studies, the diagnosis of GMA, including MCA and MGA[7,8]. This is consistent with the find
Currently, no GMA patient survival model is available. Existing prognostic models mostly focus on common patho
Some researchers have reported that new targeted drugs for GMA are in progress[7,37,38]. Simultaneously, as GMA usually has a higher MSI, immunotherapy may bring better efficacy to GMA[7]. These factors are expected to enhance GMA the prognosis of patients with GMA.
This study had some limitations. It has been observed that some gene mutations are related to GMA prognosis, which was not considered at this time[15]. Besides, other potential factors like co-morbidity, immunohistochemistry, family history/genetic syndromes, and type of surgery (open/min access) may have potential influence on GMA, but not recorded in SEER database. This retrospective study inevitably has selection bias and information bias. And more Asian data and prospective data can validate our model better. Subsequent researchers may consider further improvements in the above areas.
The deep learning-based tool developed in this study can accurately predict the OS of patients with gastrointestinal mucous carcinoma after surgery. Combining surgery, chemotherapy, and adequate lymph node dissection during surgery can improve patient outcomes.
The authors thank the Aerospace Center Hospital, Peking University Aerospace School of Clinical Medicine, for providing endoscopic and pathological photographs.
Provenance and peer review: Unsolicited article; Externally peer reviewed.
Peer-review model: Single blind
Specialty type: Oncology
Country/Territory of origin: China
Peer-review report’s classification
Scientific Quality: Grade B
Novelty: Grade B
Creativity or Innovation: Grade B
Scientific Significance: Grade B
P-Reviewer: Govindarajan KK, India S-Editor: Chen YL L-Editor: A P-Editor: Zhao YQ
| 1. | Tong Y, Gao H, Qi Q, Liu X, Li J, Gao J, Li P, Wang Y, Du L, Wang C. High fat diet, gut microbiome and gastrointestinal cancer. Theranostics. 2021;11:5889-5910. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 84] [Cited by in RCA: 122] [Article Influence: 30.5] [Reference Citation Analysis (0)] |
| 2. | Siegel RL, Miller KD, Wagle NS, Jemal A. Cancer statistics, 2023. CA Cancer J Clin. 2023;73:17-48. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 116] [Cited by in RCA: 9903] [Article Influence: 4951.5] [Reference Citation Analysis (2)] |
| 3. | Kuipers EJ, Grady WM, Lieberman D, Seufferlein T, Sung JJ, Boelens PG, van de Velde CJ, Watanabe T. Colorectal cancer. Nat Rev Dis Primers. 2015;1:15065. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1014] [Cited by in RCA: 1120] [Article Influence: 112.0] [Reference Citation Analysis (0)] |
| 4. | Machlowska J, Baj J, Sitarz M, Maciejewski R, Sitarz R. Gastric Cancer: Epidemiology, Risk Factors, Classification, Genomic Characteristics and Treatment Strategies. Int J Mol Sci. 2020;21. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 897] [Cited by in RCA: 852] [Article Influence: 170.4] [Reference Citation Analysis (0)] |
| 5. | Chu PG, Chung L, Weiss LM, Lau SK. Determining the site of origin of mucinous adenocarcinoma: an immunohistochemical study of 175 cases. Am J Surg Pathol. 2011;35:1830-1836. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 82] [Cited by in RCA: 93] [Article Influence: 7.2] [Reference Citation Analysis (0)] |
| 6. | Shin JH, Bae JH, Lee A, Jung CK, Yim HW, Park JS, Lee KY. CK7, CK20, CDX2 and MUC2 Immunohistochemical staining used to distinguish metastatic colorectal carcinoma involving ovary from primary ovarian mucinous adenocarcinoma. Jpn J Clin Oncol. 2010;40:208-213. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 42] [Cited by in RCA: 52] [Article Influence: 3.3] [Reference Citation Analysis (0)] |
| 7. | Luo C, Cen S, Ding G, Wu W. Mucinous colorectal adenocarcinoma: clinical pathology and treatment options. Cancer Commun (Lond). 2019;39:13. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 89] [Cited by in RCA: 189] [Article Influence: 31.5] [Reference Citation Analysis (0)] |
| 8. | Meng NL, Wang YK, Wang HL, Zhou JL, Wang SN. Research on the Histological Features and Pathological Types of Gastric Adenocarcinoma With Mucinous Differentiation. Front Med (Lausanne). 2022;9:829702. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 1] [Cited by in RCA: 1] [Article Influence: 0.3] [Reference Citation Analysis (0)] |
| 9. | Zuo D, Li C, Liu T, Yue M, Zhang J, Ning G. Construction and validation of a metabolic risk model predicting prognosis of colon cancer. Sci Rep. 2021;11:6837. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 2] [Cited by in RCA: 12] [Article Influence: 3.0] [Reference Citation Analysis (0)] |
| 10. | Li X, Wen D, Li X, Yao C, Chong W, Chen H. Identification of an Immune Signature Predicting Prognosis Risk and Lymphocyte Infiltration in Colon Cancer. Front Immunol. 2020;11:1678. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 28] [Cited by in RCA: 70] [Article Influence: 14.0] [Reference Citation Analysis (0)] |
| 11. | Xiaobin C, Zhaojun X, Tao L, Tianzeng D, Xuemei H, Fan Z, Chunyin H, Jianqiang H, Chen L. Analysis of Related Risk Factors and Prognostic Factors of Gastric Cancer with Bone Metastasis: A SEER-Based Study. J Immunol Res. 2022;2022:3251051. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 1] [Cited by in RCA: 12] [Article Influence: 4.0] [Reference Citation Analysis (0)] |
| 12. | Katzman JL, Shaham U, Cloninger A, Bates J, Jiang T, Kluger Y. DeepSurv: personalized treatment recommender system using a Cox proportional hazards deep neural network. BMC Med Res Methodol. 2018;18:24. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 398] [Cited by in RCA: 724] [Article Influence: 103.4] [Reference Citation Analysis (0)] |
| 13. | Glasgow SC, Yu J, Carvalho LP, Shannon WD, Fleshman JW, McLeod HL. Unfavourable expression of pharmacologic markers in mucinous colorectal cancer. Br J Cancer. 2005;92:259-264. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 58] [Cited by in RCA: 44] [Article Influence: 2.2] [Reference Citation Analysis (0)] |
| 14. | Leopoldo S, Lorena B, Cinzia A, Gabriella DC, Angela Luciana B, Renato C, Antonio M, Carlo S, Cristina P, Stefano C, Maurizio T, Luigi R, Cesare B. Two subtypes of mucinous adenocarcinoma of the colorectum: clinicopathological and genetic features. Ann Surg Oncol. 2008;15:1429-1439. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 95] [Cited by in RCA: 116] [Article Influence: 6.8] [Reference Citation Analysis (0)] |
| 15. | Rokutan H, Hosoda F, Hama N, Nakamura H, Totoki Y, Furukawa E, Arakawa E, Ohashi S, Urushidate T, Satoh H, Shimizu H, Igarashi K, Yachida S, Katai H, Taniguchi H, Fukayama M, Shibata T. Comprehensive mutation profiling of mucinous gastric carcinoma. J Pathol. 2016;240:137-148. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 24] [Cited by in RCA: 36] [Article Influence: 4.0] [Reference Citation Analysis (0)] |
| 16. | Verhulst J, Ferdinande L, Demetter P, Ceelen W. Mucinous subtype as prognostic factor in colorectal cancer: a systematic review and meta-analysis. J Clin Pathol. 2012;65:381-388. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 203] [Cited by in RCA: 189] [Article Influence: 14.5] [Reference Citation Analysis (0)] |
| 17. | Nagtegaal ID, Odze RD, Klimstra D, Paradis V, Rugge M, Schirmacher P, Washington KM, Carneiro F, Cree IA; WHO Classification of Tumours Editorial Board. The 2019 WHO classification of tumours of the digestive system. Histopathology. 2020;76:182-188. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 2554] [Cited by in RCA: 2423] [Article Influence: 484.6] [Reference Citation Analysis (3)] |
| 18. | Yan C, Yang H, Chen L, Liu R, Shang W, Yuan W, Yang F, Sun Q, Xia L. Clinical significance of mucinous component in colorectal adenocarcinoma: a propensity score-matched study. BMC Cancer. 2021;21:1286. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in RCA: 9] [Reference Citation Analysis (0)] |
| 19. | Chew MH, Yeo SA, Ng ZP, Lim KH, Koh PK, Ng KH, Eu KW. Critical analysis of mucin and signet ring cell as prognostic factors in an Asian population of 2,764 sporadic colorectal cancers. Int J Colorectal Dis. 2010;25:1221-1229. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 64] [Cited by in RCA: 85] [Article Influence: 5.7] [Reference Citation Analysis (0)] |
| 20. | Reynolds IS, Furney SJ, Kay EW, McNamara DA, Prehn JHM, Burke JP. Meta-analysis of the molecular associations of mucinous colorectal cancer. Br J Surg. 2019;106:682-691. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 31] [Cited by in RCA: 51] [Article Influence: 8.5] [Reference Citation Analysis (0)] |
| 21. | Kanda M, Oba K, Aoyama T, Kashiwabara K, Mayanagi S, Maeda H, Honda M, Hamada C, Sadahiro S, Sakamoto J, Saji S, Yoshikawa T; Japanese Foundation for Multidisciplinary Treatment of Cancer. Clinical Signatures of Mucinous and Poorly Differentiated Subtypes of Colorectal Adenocarcinomas by a Propensity Score Analysis of an Independent Patient Database from Three Phase III Trials. Dis Colon Rectum. 2018;61:461-471. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 10] [Cited by in RCA: 17] [Article Influence: 2.4] [Reference Citation Analysis (0)] |
| 22. | Warschkow R, Tarantino I, Huttner FJ, Schmied BM, Guller U, Diener MK, Ulrich A. Predictive value of mucinous histology in colon cancer: a population-based, propensity score matched analysis. Br J Cancer. 2016;114:1027-1032. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 18] [Cited by in RCA: 28] [Article Influence: 3.1] [Reference Citation Analysis (0)] |
| 23. | Hugen N, Verhoeven RH, Radema SA, de Hingh IH, Pruijt JF, Nagtegaal ID, Lemmens VE, de Wilt JH. Prognosis and value of adjuvant chemotherapy in stage III mucinous colorectal carcinoma. Ann Oncol. 2013;24:2819-2824. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 56] [Cited by in RCA: 85] [Article Influence: 7.1] [Reference Citation Analysis (0)] |
| 24. | Mekenkamp LJ, Heesterbeek KJ, Koopman M, Tol J, Teerenstra S, Venderbosch S, Punt CJ, Nagtegaal ID. Mucinous adenocarcinomas: poor prognosis in metastatic colorectal cancer. Eur J Cancer. 2012;48:501-509. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 95] [Cited by in RCA: 124] [Article Influence: 9.5] [Reference Citation Analysis (0)] |
| 25. | Kim SH, Shin SJ, Lee KY, Kim H, Kim TI, Kang DR, Hur H, Min BS, Kim NK, Chung HC, Roh JK, Ahn JB. Prognostic value of mucinous histology depends on microsatellite instability status in patients with stage III colon cancer treated with adjuvant FOLFOX chemotherapy: a retrospective cohort study. Ann Surg Oncol. 2013;20:3407-3413. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 52] [Cited by in RCA: 72] [Article Influence: 6.0] [Reference Citation Analysis (0)] |
| 26. | Hsu JT, Wang CW, Le PH, Wu RC, Chen TH, Chiang KC, Lin CJ, Yeh TS. Clinicopathological characteristics and outcomes in stage I-III mucinous gastric adenocarcinoma: a retrospective study at a single medical center. World J Surg Oncol. 2016;14:123. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 9] [Cited by in RCA: 10] [Article Influence: 1.1] [Reference Citation Analysis (0)] |
| 27. | Kang M, Ko E, Mersha TB. A roadmap for multi-omics data integration using deep learning. Brief Bioinform. 2022;23. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 102] [Cited by in RCA: 155] [Article Influence: 51.7] [Reference Citation Analysis (0)] |
| 28. | Jiang Y, Yang M, Wang S, Li X, Sun Y. Emerging role of deep learning-based artificial intelligence in tumor pathology. Cancer Commun (Lond). 2020;40:154-166. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 173] [Cited by in RCA: 238] [Article Influence: 47.6] [Reference Citation Analysis (0)] |
| 29. | Howard FM, Kochanny S, Koshy M, Spiotto M, Pearson AT. Machine Learning-Guided Adjuvant Treatment of Head and Neck Cancer. JAMA Netw Open. 2020;3:e2025881. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 64] [Cited by in RCA: 96] [Article Influence: 19.2] [Reference Citation Analysis (0)] |
| 30. | She Y, Jin Z, Wu J, Deng J, Zhang L, Su H, Jiang G, Liu H, Xie D, Cao N, Ren Y, Chen C. Development and Validation of a Deep Learning Model for Non-Small Cell Lung Cancer Survival. JAMA Netw Open. 2020;3:e205842. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 53] [Cited by in RCA: 168] [Article Influence: 33.6] [Reference Citation Analysis (0)] |
| 31. | Li W, Lin S, He Y, Wang J, Pan Y. Deep learning survival model for colorectal cancer patients (DeepCRC) with Asian clinical data compared with different theories. Arch Med Sci. 2023;19:264-269. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in RCA: 13] [Reference Citation Analysis (0)] |
| 32. | Wang L, Hirano Y, Heng G, Ishii T, Kondo H, Hara K, Obara N, Asari M, Kato T, Yamaguchi S. Mucinous Adenocarcinoma as a High-risk Factor in Stage II Colorectal Cancer: A Propensity Score-matched Study from Japan. Anticancer Res. 2020;40:1651-1659. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 10] [Cited by in RCA: 16] [Article Influence: 3.2] [Reference Citation Analysis (0)] |
| 33. | Cai C, Yang L, Zhuang X, He Y, Zhou K. A five-lncRNA model predicting overall survival in gastric cancer compared with normal tissues. Aging (Albany NY). 2021;13:24349-24359. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 2] [Cited by in RCA: 5] [Article Influence: 1.3] [Reference Citation Analysis (0)] |
| 34. | Yue T, Chen S, Zhu J, Guo S, Huang Z, Wang P, Zuo S, Liu Y. The aging-related risk signature in colorectal cancer. Aging (Albany NY). 2021;13:7330-7349. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 15] [Cited by in RCA: 77] [Article Influence: 19.3] [Reference Citation Analysis (0)] |
| 35. | Hung YS, Chang SC, Liu KH, Hung CY, Kuo YC, Tsai CY, Hsu JT, Yeh TS, Chen JS, Chou WC. A prognostic model based on lymph node metastatic ratio for predicting survival outcome in gastric cancer patients with N3b subclassification. Asian J Surg. 2019;42:85-92. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 12] [Cited by in RCA: 9] [Article Influence: 1.5] [Reference Citation Analysis (0)] |
| 36. | Miao Y, Xu Z, Feng W, Zheng M, Gao H, Li W, Zhang Y, Zong Y, Lu A, Zhao J. Platelet infiltration predicts survival in postsurgical colorectal cancer patients. Int J Cancer. 2022;150:509-520. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 5] [Cited by in RCA: 19] [Article Influence: 4.8] [Reference Citation Analysis (1)] |
| 37. | Pereira de Sousa I, Cattoz B, Wilcox MD, Griffiths PC, Dalgliesh R, Rogers S, Bernkop-Schnürch A. Nanoparticles decorated with proteolytic enzymes, a promising strategy to overcome the mucus barrier. Eur J Pharm Biopharm. 2015;97:257-264. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 91] [Cited by in RCA: 109] [Article Influence: 10.9] [Reference Citation Analysis (0)] |
| 38. | Marxen E, Mosgaard MD, Pedersen AML, Jacobsen J. Mucin dispersions as a model for the oromucosal mucus layer in in vitro and ex vivo buccal permeability studies of small molecules. Eur J Pharm Biopharm. 2017;121:121-128. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 18] [Cited by in RCA: 23] [Article Influence: 2.9] [Reference Citation Analysis (0)] |