Published online Jun 15, 2024. doi: 10.4251/wjgo.v16.i6.2350
Revised: April 2, 2024
Accepted: April 24, 2024
Published online: June 15, 2024
Processing time: 113 Days and 24 Hours
The prevention and early diagnosis of liver cancer remains a global medical challenge. During the malignant transformation of hepatocytes, a variety of oncogenic cellular signalling molecules, such as novel high mobility group-Box 3, angiopoietin-2, Golgi protein 73, glypican-3, Wnt3a (a signalling molecule in the Wnt/β-catenin pathway), and secretory clusterin, can be expressed and secreted into the blood. These signalling molecules are derived from different signalling pathways and may not only participate in the malignant transformation of hepatocytes but also become early diagnostic indicators of hepatocarcinogenesis or specific targeted molecules for hepatocellular carcinoma therapy. This article reviews recent progress in the study of several signalling molecules as sensitive biomarkers for monitoring hepatocarcinogenesis.
Core Tip: The early monitoring or diagnosis of hepatocellular carcinoma (HCC) are still medical challenge, and identifying novel biomarkers with high sensitivity and specificity for HCC are urgently needed. Recent progress in several oncogenic cellular signalling molecules that derived from different signalling pathways were reviewed, such as novel high mobility group-Box 3, angiopoietin-2, Golgi protein 73, glypican-3, Wnt3a (a signalling molecule in the Wnt/β-catenin pathway), and secretory clusterin. They might not only participate in the malignant transformation of hepatocytes but also become early diagnostic indicators of hepatocarcinogenesis or specific targeted molecules for HCC therapy.
- Citation: Yao M, Fang RF, Xie Q, Xu M, Sai WL, Yao DF. Early monitoring values of oncogenic signalling molecules for hepatocellular carcinoma. World J Gastrointest Oncol 2024; 16(6): 2350-2361
- URL: https://www.wjgnet.com/1948-5204/full/v16/i6/2350.htm
- DOI: https://dx.doi.org/10.4251/wjgo.v16.i6.2350
The prevention, early monitoring or diagnosis of hepatocellular carcinoma (HCC) are still urgent medical problems[1,2]. Hepatocarcinogenesis is mainly related to chronic persistent hepatitis virus infection [hepatitis B virus (HBV), hepatitis C virus (HCV) or hepatitis D virus][3,4], chemical carcinogen intake, and increased metabolic dysfunction-associated fatty liver disease (MAFLD)[5,6]. Recently, MAFLD has become one of the most chronic liver diseases and a possible etio
Liver cancer is still the sixth most common cancer in the world and the third leading cause of cancer death. HCC is the 6th most common cancer worldwide and is the 3rd leading cause of tumour death. HCC is the 3rd most common cause of cancer-related deaths worldwide[15]. Early diagnosis of HCC or monitoring of hepatocyte malignant transformation is of utmost importance[16]. To date, few markers have early monitoring or specific diagnostic value. It is therefore essential to reduce the incidence and impact of these diseases by understanding risk factors and prevention strategies at early stage[17]. Clinically, therapeutic options for advanced HCC at the time of diagnosis are limited; in many cases, these trea
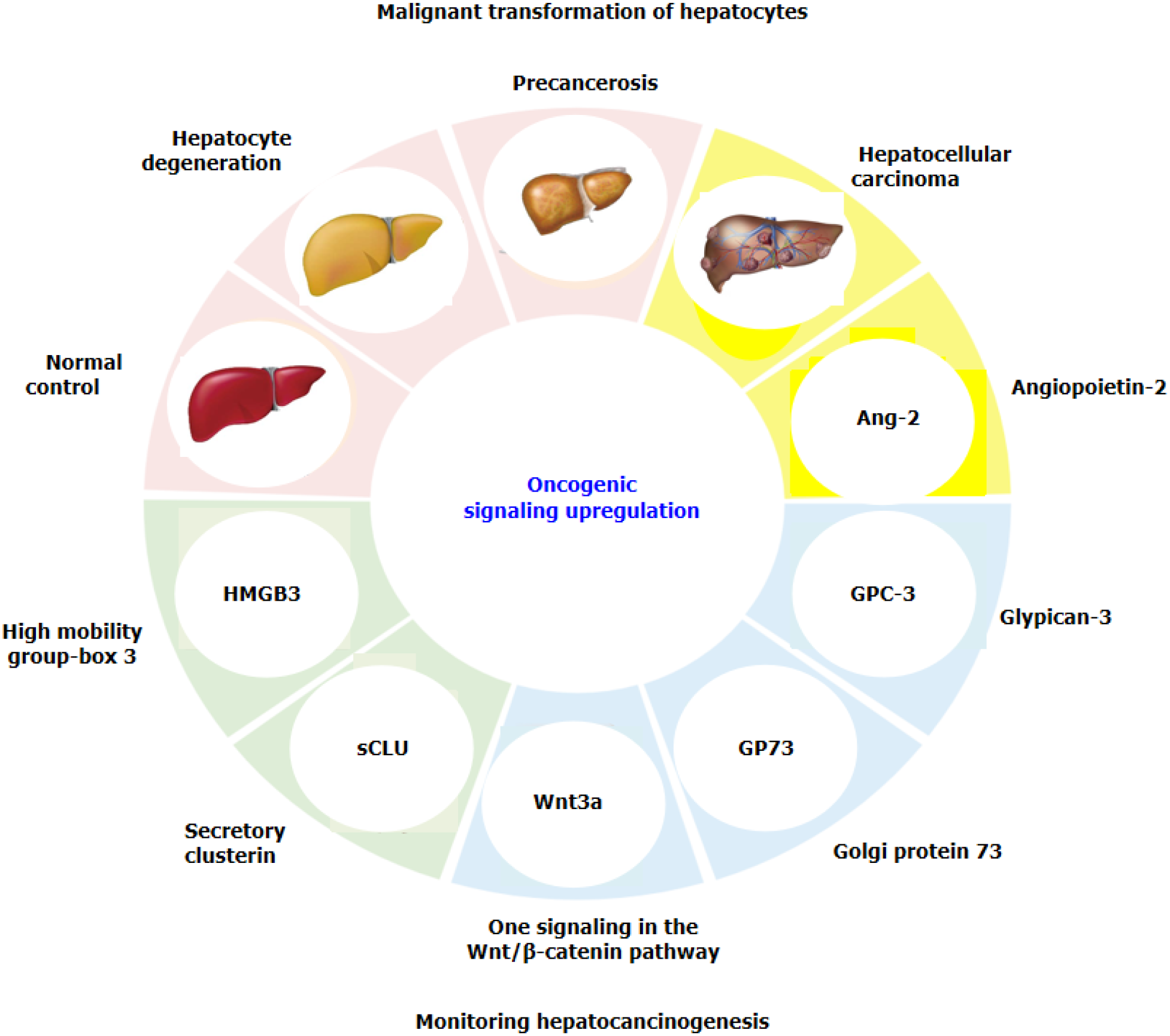
Although routine AFP detection is combined with other clinical parameters, the combined detection of multiple markers is often more sensitive or specific than the use of a single marker. A new GALAD score (sex, age, AFP, AFP-L3 and des-γ-carboxy prothrombin) has been developed[19,20] and used to detect early HCC in a population at high risk of HCC or in a cohort of patients with nonalcoholic steatohepatitis, multicentre derivation, validation and comparison[20,21]. However, identifying potential molecular mechanisms or monitoring markers with high sensitivity and specificity or identifying new therapeutic targets for HCC are urgently needed[22,23]. Recently, several specific signalling molecules related to hepatocyte malignant transformation have been reported in HCC tissues and blood, such as circulating high mobility group-Box 3 (HMGB3), Wnt3a from the Wnt/β-catenin pathway, secretory clusterin (sCLU), angiopoietin-2 (Ang-2), Golgi protein-73 (GP73), and glypican-3 (GPC3) (Figure 1). This review presents new advances in identifying promising signalling molecules for monitoring hepatocarcinogenesis in high-risk populations.
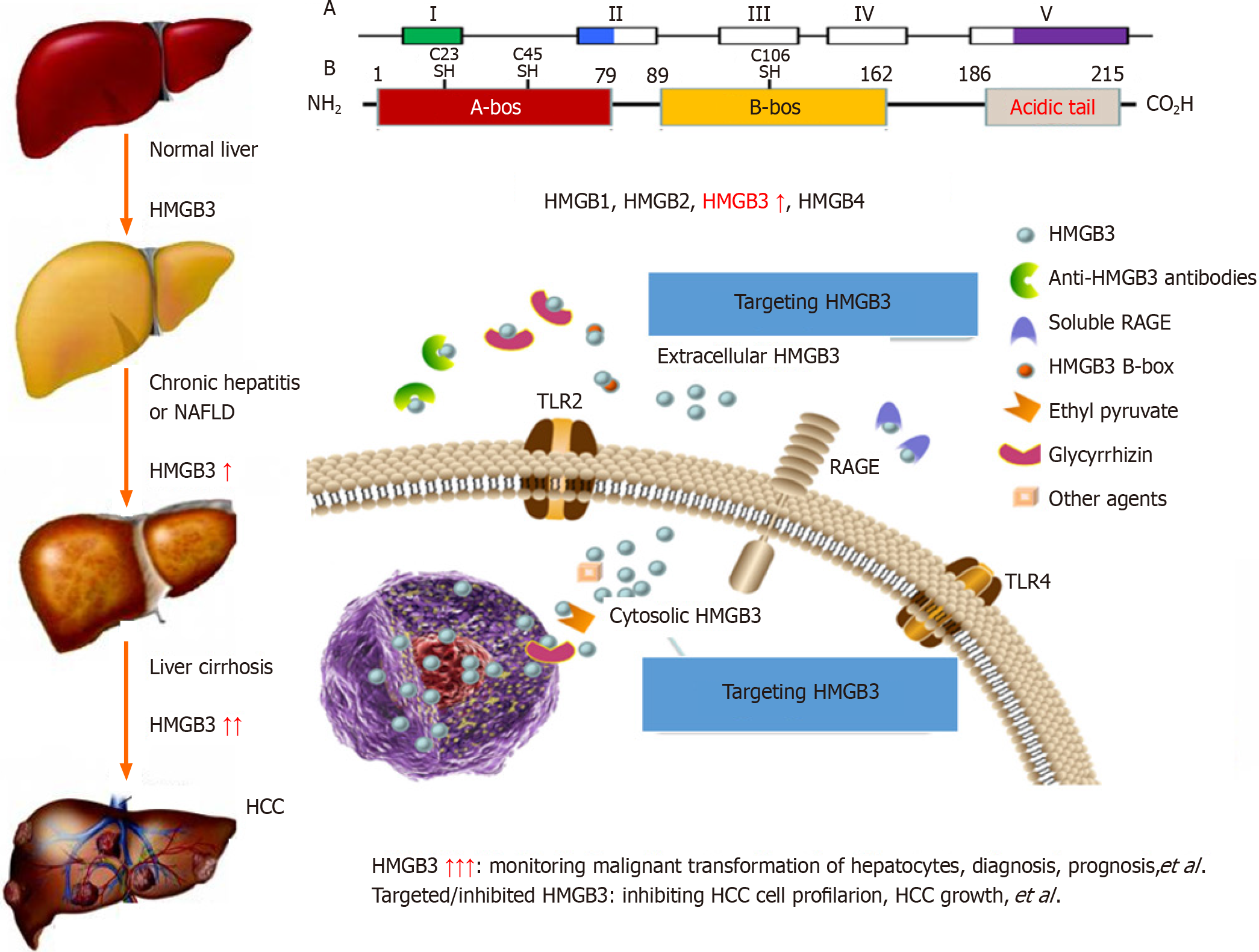
The high mobility group (HMG) protein superfamily[24] includes the HMGA[25], HMGB[26] and HMGN[27] families, which have different biological functions. The HMGB family consists of HMGB1, HMGB2, HMGB3 (Figure 2) and HMGB4 and encodes proteins that contain one or more DNA-binding motifs; these proteins are involved in multiple cellular processes, including HBV infection, cell differentiation, migration, and inflammation-related activities[28-30]. HMGB3 expression has been investigated in the cancerous tissues of patients with HCC and in the sera of patients with HBV-related chronic liver diseases[31]. HMGB3 expression in HCC samples was marked greater than that in noncancerous ones and was related to with tumour size, tumor node metastasis (TNM) stage, high recurrence rates, and there was no significant difference in patient age, sex, AFP, HBV infection or liver cirrhosis (LC). HMGB3 levels in sera of patients were analyzed in a cohort of cases with chronic liver disease, with progressive increases from benign liver disease to HCC progression. The sensitivity was 75.6 for HMGB3, 56.7 for AFP, and up to 89.0% for both combinations for HCC diagnosis. Interestingly, the percentage of HMGB3-positive patients was 55.3%, the percentage of AFP-positive patients was 39.5%, and the percentage of small-size HCC patients was 71.1%[32]. These findings indicate that HMGB3 detection is helpful for the diagnosis and differential diagnosis of benign and malignant liver diseases and might play an important role in HCC progression.
The common etiological causes of liver cancer progression are persistent CH and LC. It has been suggested that precancerous lesions of the liver include all stages of the disease, from dysplastic lesions and dysplastic nodules to early liver cancer[33,34]. A dynamic model of hepatocyte malignant transformation was developed in male SD rats according to the China National Invention Patent (ZL201810077848.2) by using 2-fluorenylacetamide (2-FAA) as a carcinogen. Dynamic changes in liver HMGB3 at the protein or mRNA level were analysed by immunohistochemistry (IHC) or reverse transcription-quantitative polymerase chain reaction. Pathology revealed that haematoxylin and eosin (H. After staining, the livers were divided into the hepatocyte degeneration (HD), precancerosis (PC), HCC and normal control (NC) groups. HMGB3 expression at the protein or mRNA level was not detected in the NC group, 50% in the HD group at the early stage, 100% in the PC group at the middle stage or 100% in the HCC group at the last stage, suggesting that HMGB3 expression is associated with malignant transformation of hepatocytes and could be a promising monitoring signal for hepatocarcinogenesis[35].
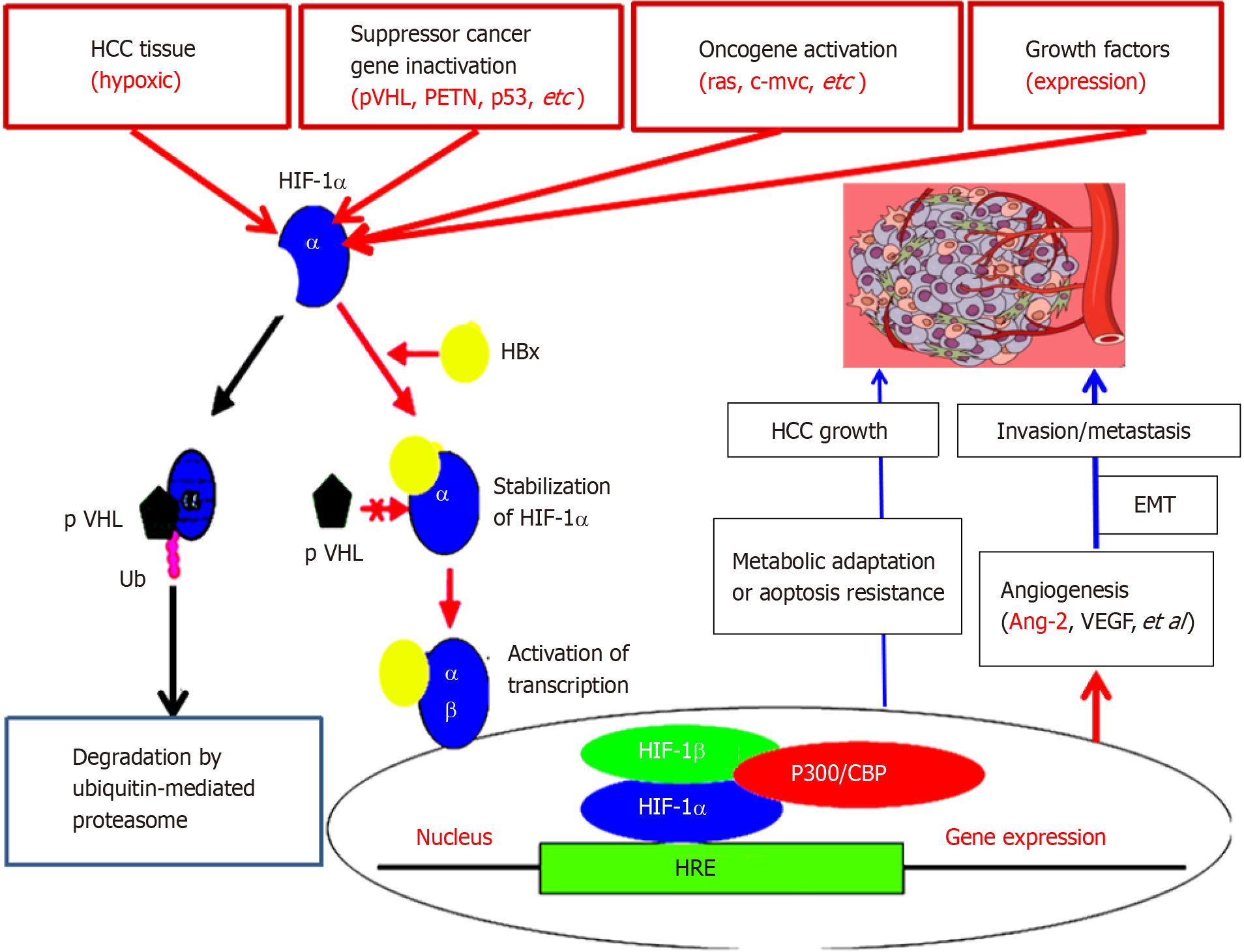
Ang-2, Ang-1, Tie2 Ligand-receptor, and vascular endothelial growth factor pathways are critical for regulating vascular maturation or stability and are associated with physiological angiogenesis or control of liver cancer[36]. Ang-2 is a key driver of HCC angiogenesis[37]. Neovascularization of HCC is a fundamental process that involves a lot of pathological processes and maintains HCC progression. The establishment of new angiogenesis depends on the complex processes of endothelial cell proliferation and organization. In addition, it may enhance the proliferation of HCC cells and the resistance to apoptosis, and promote HCC metastasis[38]. Comparative analysis revealed that the serum Ang-2 levels in the HCC, CH and LC groups were more higher (P < 0.001) than those in the NC group. The percentage of cells that were positive for Ang-2 (more than 35 μg/L) was 90.0% in HCC, 6.0% in LC, 4.0% in CH cases, and 0.0% in NC. Clinicopathological characteristics of Ang-2 in HCC patients were strongly associated (P < 0.001) with tumour size (≥ 5 cm), differentiation degree (well/moderate vs poor), gross classification (unifocal vs multifocal), AFP (≥ 50 µg/L), LC, HBV infection, portal vein invasion or lymph node metastasis and TNM stage (I-II vs III-IV). Ang-2 was positively related to vascular endothelial growth factor (VEGF) and hypoxia-inducible factor-1α (P < 0.001, Figure 3), suggesting that Ang-2 is upregulated during the progression of HCC[3,39-41].
It has been reported that ectopic expression of Ang-2 can promote the rapid development of human liver cancer and produces hemorrhage in nude mice tumor[42]. Recently, abnormal upregulation of Ang-2 was detected in a model of normal hepatocyte malignant transformation to HCC. Compared with those in the NC group, there were 70 or 93 up- or down-regulated differentially expressed genes (DEGs) at the HD stage, with Ang-2, oxidoreductase, acid mercaptoenzyme activities, peptide antigens, cofactor binding, endoplasmic reticulum and endoplasmic reticulum membrane; 1015 or 437 DEGs at the PC stage, with Ang-2, growth factors, calcium-dependent phospho-lipids, alloproteins, VEGF, insulin-like growth factor binding, oxidoreductase, acid-mercaptoenzyme, microtubule cytoskeleton in centromeric region, plasma membrane, extracellular and spindle as the main components; and 1234 or 504 DEGs at the HCC stage, with Ang-2, organic sodium transporter, microtubule movement, protein dimerization activity, alloprotein, calcium ion and cytoskeleton protein binding in the extracellular fraction, microtubule cytoskeleton, VEGF, plasma membrane and cellular components. The specific concentration (in ng/mg of liver tissue) or serum concentration (in μg/L) of Ang-2 was quantitatively investigated from the NC group to the HD group at the early stage, the PC group at the middle stage, and the HCC group at the later stage[43]. In both the liver and blood, Ang-2 levels are dynamically up-regulated in early HCC to accelerate neovascularization to meet the need for oxygen, suggesting that Ang-2 signalling is a useful biomarker for monitoring the malignant progression of chronic liver diseases or molecular targeted therapy[39,44,45].
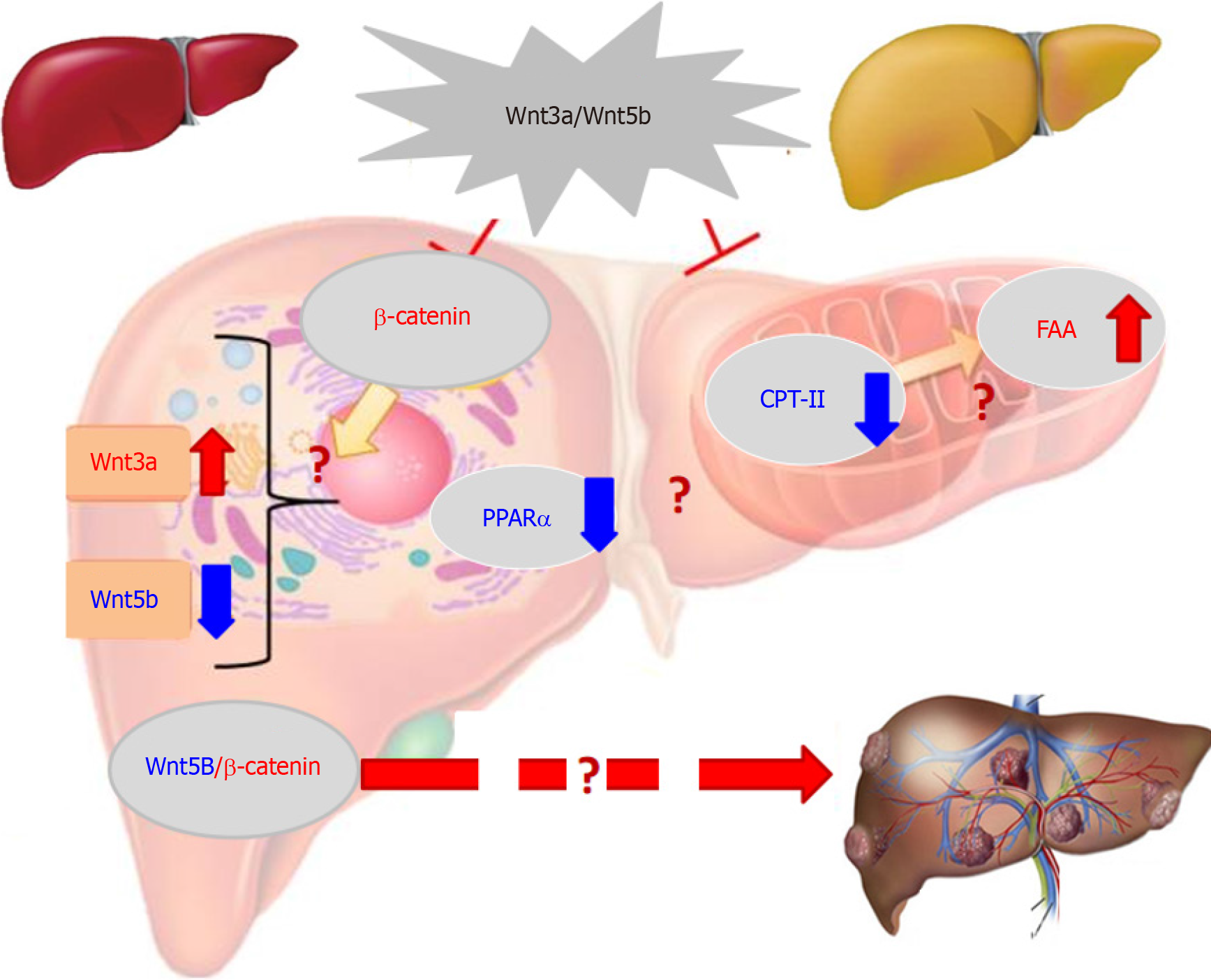
The over-expression of some members in the Wingless (Wnt)/β-catenin pathway is related to HCC occurrence or progression[46,47]. Cancer-associated Wnt molecules regulate a variety of cellular events, such as cell proliferation, apoptosis, differentiation and HCC growth, via β-catenin-dependent classical or non-classical pathways[48,49]. Wnt3a (a member of the Wnt-type MMTV integration site family) is a critical signalling molecule among 19 mammalian Wnt proteins. Located on chromosome 17 (17q21) Wnt3a is regarded as a canonical Wnt pathway an activator (Figure 4), which induces β-catenin accumulation[50,51]. HBx in HBV-related HCC can inhibit GSK-3β activity via activating Src kinase, induce the accumulation of intracellular β-catenin and activating DNA methyltransferase I. Wnt3a can bind or silence secreted frizzled-related proteins (SFP) 1 or SFP5. In HBV-related HCC, HBx can promote HCC formation by activating the Wnt pathway and reduce the inhibiting role of deacetylase 1 on β-catenin[4,52,53]. Additionally, HCV core protein during HCC progression can induce Wnt3a expression and TCF-dependent transcription, with inhibiting GSK-3β production, increasing intracellular β-catenin to nuclear transport by upregulating c-myc, WISP2, cyclin Dl, Wnt1, Wnt3a and cTGF levels for HCC growth or DNA synthesis[54,55]. These studies indicated that Wnt3a has a key role in HCC with high expression.
High Wnt3a levels in the sera or tissues of HCC patients were reported for the first time in a cohort of cases with HBV-related chronic liver diseases. The incidence of oncogenic Wnt3a expression in the cancerous group reached 96.3%, which was significantly higher (P < 0.001) than that in the surrounding group[56]. Hepatic Wnt3a in liver tissues was brown staining with gradually up-regulated expression with clinical stage, especially in advanced HCC. Clinicopathological characteristics of liver Wnt3a up-regulation in HCC were associated with differentiation degree (P < 0.001), LC (P < 0.004), HBV infection (P < 0.001), high TNM stage (P < 0.001), and poor survival rate (P < 0.001). The levels of Wnt3a were investigated in the sera of 186 patients with chronic liver disease. The incidence of circulating Wnt3a levels more than > 800 ng/L was 92.5% in patients with HCC. It was significantly associated with AFP, LC, HBV, differentiation grade, TNM, and distal metastasis. Serum Wnt3a level was obviously higher (P < 0.001) in HCC cases than that in CH, LC or NC group[57,58]. Comparative analysis between serum Wnt3 and AFP, the diagnostic specificity was 94.34% vs 69.81%, and area under the receiver operating characteristic curve was 0.994 vs 0.710 for HCC, respectively. These data indicated that up-regulated Wnt3a expression be associated with HCC progression and higher Wnt3a be a new sensitive molecular marker for HCC diagnosis or differential diagnosis[59,60].
A dynamic model of rat hepatocarcinogenesis was successfully established to investigate the alteration of Wnt3a and its monitoring value in early HCC[61]. Based on liver pathological examination with HE staining, the rats were divided into the NC, HD, PC and HCC groups. The total number of DEGs that were up- or downregulated in the liver tran
Carcinoembryonic GPC3 is a heparan sulfate proteoglycan that binds to cell membranes through glycosylphosphatidylinositol[64]. GPC3 is widely expressed in human embryos tissues, can’t been detected in livers of healthy adult persons, but GPC3 is over-expressed in hepatocarcinogenesis[65]. However, because of the deficiency of the clinical analysis method of GPC3, it is very important to detect GPC3 accurately and sensitively for the diagnosis of liver cancer. Recently, new high-sensitivity GPC3 detection technologies based on a variety of optical sensors or electro-chemical technologies have been developed, providing new perspectives for the challenges and future development of the field and also contributing to the early detection of liver cancer[16,66,67]. For HCC, the discovery of GPC3 as a promising tumor-associated antigen because GPC3 is over-expressed in cancerous tissues and is significantly associated with poor prognostic disease-free survival and overall survival[67,68]. Indeed, clinical and basic studies have demonstrated that GPC3 is a carcino- embryonic proteoglycan anchored to the membrane of HCC cells and is involved in promoting HCC growth and invasion through a variety of signaling cascades; including the Wnt pathway, which plays a well-known role in embryogenesis[69,70]. It was found that the up-regulated expression of GPC3mRNA, gene or protein in the tissues or serum of patients with liver cancer could not only specifically diagnose liver cancer, but also had a bad prognosis with the patients with liver cancer[71].
Studies of the mechanisms by which GPC3 promotes HCC progression have shown that GPC3 functions by binding to molecules such as the Wnt/β-catenin signaling or growth factors in hepatocarcinogenesis[14,72]. In addition, because of the specificity of GPC3 for HCC, serum GPC3 detection has been used in the diagnosis, prognosis and molecular targeted therapy of liver cancer[73,74]. An HCC model in SD male rats induced with 0.05% 2-FAA was confirmed by HE staining. Hepatic GPC3mRNA was analyzed by quantitatively PCR or DNA sequencing. GPC3 up-regulation has been confirmed by animal model, with brown granule-like GPC3 positive expression localized in cytoplasm by IHC, and the pathological alterations of rat hepatocytes divided into HD, PC and HCC morphological stages, with a progressive increase in total RNAs or gamma-glutamyl transferase levels in livers. The positive rates of liver GPC3mRNA, liver GPC3 and serum GPC3 expression were 83.3% or 83.3% and 38.9% in the HD group, 100% or 100% and 66.7% in the PC group, 100% or 100% and 77.8% in the rat HCC group, respectively. None in livers or blood was founded in the NC group. Significantly positive correlation was founded between hepatic liver GPC3 mRNA and total RNAs (r = 0.475), hepatic GPC3 (r = 1.0) or circulating GPC3 (r = 0.994), indicating that GPC3 is a useful marker for monitoring hepatocarcinogenesis[75].
Liver Golgi apparatus specific membrane GP73 (GOLPH2) is commonly found in epithelial cells containing GP73 mRNA transcripts (approximately 3.0 KB), the membrane consists of a short N-terminal cytoplasmic domain, a transmembrane domain and a larger C-terminal domain located on the surface of the lumen of the Golgi apparatus[76]. The bile duct epithelial cells expressed GP73 in normal liver. The overexpression of GP73 in liver has been proved to be closely related to the progression and poor prognosis of HCC[77]. Recent studies have shown that GP73 immunologically mediates chronic liver disease by enhancing sterol regulatory element binding protein (SREBP)-cleavage activating protein inte
The GP73 expression in cancerous tissue is heterogeneous and inconsistent, especially for early stage HCC[80]. Dynamic models of hepatocarcinogenesis in SD rats were generated with a diet containing 2-FAA, and the rats were divided into the NC, HD, PC and HCC groups to observe alterations in GP73 at the protein or mRNA level during the malignant transformation of rat hepatocytes. There were no increases in hepatic GP73 expression at the protein or mRNA level in NC, 66.7% or 44.4% in HD, 88.9% or 77.8% in PC, and both 100% in HCC rats. There was a positive correlation (r = 0.91, P < 0.01) between liver GP73 and serum GP73 in rat hepatocarcinogenesis, indicating that abnormal GP73 levels are a sensitive and valuable biomarker in hepatocarcinogenesis[81,82].
Liver sCLU, located on chromosome 8q21-q12, is a heterodimeric sulfated glycoprotein that has cytoprotective roles and highly conserved among species[83]. The biological function of sCLU as a small-molecule chaperone is similar to that of heat shock protein[84]. Lots of basic medicine and clinical cases studies have confirmed that liver sCLU is low expression in normal liver tissues and gradually over-expressed during the malignant transformation of hepatocytes, which is closely associated with HCC progression by contributing to angiogenesis[85,86], chemoresistance[87-89], survival and distal metastasis[90,91]. The incidence of liver sCLU expression was up to 73.3% at stage I according to IHC results[92]. The sCLU expression at mRNA or protein level increases with HCC staging, indicating that sCLU abnormality could be a useful marker to differentiate benign or malignant liver diseases[93,94].
The up-regulating expression of sCLU at early stage is considered to promote HCC progression or exacerbate patient survival, which may be related to AKT/GSK-3β phosphorylation[95]. Up to now, increasing reports have confirmed that higher sCLU expression could be a novel diagnostic or prognostic marker for HCC, and this finding should be more important role in the individualized therapy for HCC[96,97]. Regulating signal pathways by sCLU could be critical for revealing MDR in HCC. Therefore, silencing sCLU or inhibiting sCLU expression by specific inhibitors have provided new mechanistic insight into HCC targeted-therapy in injected or orthotopic models[98], indicating that sCLU could be an early monitoring biomarker or potential molecular target for HCC therapy[99,100].
In conclusion, hepatocarcinogenesis is characterized by abnormalities in key signalling pathways activated by related pathways, accompanied by complex mechanisms such as DNA methylation or the regulation of noncoding RNAs[101,102]. Monitoring abnormal signalling during hepatocarcinogenesis in high-risk populations is helpful for the early detection or diagnosis of HCC (Table 1) and can be used for timely treatment of HCC patients. On the other hand, immunotherapy targeting specific signalling pathways can be combined with other effective ways to improve or prolong survival. Despite the multiomics rapid development, advances in gene silencing, genetic engineering, molecular pharmacology or pathology, DNA splicing, transcription interference and monoclonal antibodies are expected to increase specificity and decrease side effects of immunotherapy. This technology can directly block signal molecules involved in HCC growth or molecular targets, such as radionuclides, drug carriers and immunotherapies, and might play a unique role in increasing effect of HCC treatment.
| Variables | Univariate analysis | Multivariable analysis | ||
| HR (95%CI) | P value | HR (95%CI) | P value | |
| HMGB3[32] | ||||
| Tumor diameter: ≤ 5 vs > 5 cm | 1.863 (1.300-2.671) | < 0.001 | 1.400 (0.964-2.034) | 0.077 |
| TNM stage: I/II vs III/IV | 3.870 (2.373-6.309) | < 0.001 | 2.471 (1.350-4.525) | 0.003 |
| HMGB3 expression: High vs low | 3.658 (2.208-6.061) | < 0.001 | 3.042 (1.809-5.115) | < 0.001 |
| Ang-2[39] | ||||
| Tumor diameter: ≤ 2 vs > 2 cm | 1.704 (0.994-2.921) | 0.052 | 1.120 (0.622-2.015) | 0.706 |
| TNM stage: I/II vs III/IV | 1.690 (1.283-2.228) | < 0.001 | 1.531 (1.041-2.252) | 0.030 |
| Ang-2 expression: High vs low | 3.080 (1.742-5.466) | < 0.001 | 3.144 (1.738-5.689) | < 0.001 |
| Wnt3a[56,57] | ||||
| Tumor diameter: ≤ 2 vs > 2 cm | 2.056 (1.056-4.089) | 0.030 | 2.018 (1.058-3.912) | 0.031 |
| TNM stage: I/II vs III/IV | 0.686 (0.328-1.436) | 0.318 | NA | NA |
| Wnt3a expression: High vs low | 5.656 (2.682-11.926) | < 0.001 | 3.651 (1.973-6.757) | < 0.001 |
| GPC-3[73] | ||||
| Tumor diameter: ≤ 3 vs > 3 cm | 2.011 (1.498-3.869) | 0.034 | NA | NA |
| TNM stage: I/II vs III/IV | 1.961 (1.443-2.486) | < 0.001 | NA | NA |
| GPC-3 expression: High vs low | 12.697 (3.097-52.050) | < 0.0001 | 4.259 (2.030-8.934) | < 0.001 |
| GP73[78,79] | ||||
| Tumor diameter: ≤ 2 vs > 2 cm | 0.990 (0.506-1.938) | 0.978 | NA | NA |
| TNM stage: I/II vs III/IV | 0.231 (0.121-0.440) | < 0.001 | 0.221 (0.114-0.426) | < 0.001 |
| GP73 expression: High vs low | 1.008 (1.002-1.014) | 0.005 | 0.477 (0.272-0.837) | 0.010 |
| sCLU[92,95] | ||||
| Tumor diameter: ≤ 5 vs > 5 cm | 1.758 (0.987-2.968) | 0.036 | NA | NA |
| TNM stage: I/II vs III/IV | 3.056 (1.756-5.321) | < 0.001 | 1.474 (0.589-3.690) | 0.407 |
| sCLU expression: High vs low | 2.030 (0.987-4.175) | 0.004 | NA | NA |
| 1. | Konyn P, Ahmed A, Kim D. Current epidemiology in hepatocellular carcinoma. Expert Rev Gastroenterol Hepatol. 2021;15:1295-1307. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 181] [Cited by in RCA: 167] [Article Influence: 41.8] [Reference Citation Analysis (0)] |
| 2. | Rizzo GEM, Cabibbo G, Craxì A. Hepatitis B Virus-Associated Hepatocellular Carcinoma. Viruses. 2022;14:986. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 133] [Cited by in RCA: 96] [Article Influence: 32.0] [Reference Citation Analysis (0)] |
| 3. | Radmanić L, Zidovec-Lepej S. The Role of Stem Cell Factor, Epidermal Growth Factor and Angiopoietin-2 in HBV, HCV, HCC and NAFLD. Life (Basel). 2022;12:2072. [RCA] [PubMed] [DOI] [Full Text] [Reference Citation Analysis (0)] |
| 4. | Timperi E, Barnaba V. Viral Hepatitides, Inflammation and Tumour Microenvironment. Adv Exp Med Biol. 2020;1263:25-43. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 8] [Cited by in RCA: 19] [Article Influence: 3.8] [Reference Citation Analysis (0)] |
| 5. | Zhao Y, Zhou Y, Wang D, Huang Z, Xiao X, Zheng Q, Li S, Long D, Feng L. Mitochondrial Dysfunction in Metabolic Dysfunction Fatty Liver Disease (MAFLD). Int J Mol Sci. 2023;24:17514. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in RCA: 28] [Reference Citation Analysis (0)] |
| 6. | Portincasa P. NAFLD, MAFLD, and beyond: one or several acronyms for better comprehension and patient care. Intern Emerg Med. 2023;18:993-1006. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 23] [Cited by in RCA: 24] [Article Influence: 12.0] [Reference Citation Analysis (0)] |
| 7. | Crane H, Gofton C, Sharma A, George J. MAFLD: an optimal framework for understanding liver cancer phenotypes. J Gastroenterol. 2023;58:947-964. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 15] [Cited by in RCA: 26] [Article Influence: 13.0] [Reference Citation Analysis (0)] |
| 8. | Ugonabo O, Udoh US, Rajan PK, Reeves H, Arcand C, Nakafuku Y, Joshi T, Finley R, Pierre SV, Sanabria JR. The Current Status of the Liver Liquid Biopsy in MASH Related HCC: Overview and Future Directions. Biomolecules. 2023;13:1369. [RCA] [PubMed] [DOI] [Full Text] [Cited by in RCA: 1] [Reference Citation Analysis (0)] |
| 9. | Campani C, Zucman-Rossi J, Nault JC. Genetics of Hepatocellular Carcinoma: From Tumor to Circulating DNA. Cancers (Basel). 2023;15:817. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in RCA: 22] [Reference Citation Analysis (0)] |
| 10. | Liu P, Kong L, Liu Y, Li G, Xie J, Lu X. A key driver to promote HCC: Cellular crosstalk in tumor microenvironment. Front Oncol. 2023;13:1135122. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 9] [Cited by in RCA: 17] [Article Influence: 8.5] [Reference Citation Analysis (0)] |
| 11. | Yan Q, Sun YS, An R, Liu F, Fang Q, Wang Z, Xu T, Chen L, Du J. Application and progress of the detection technologies in hepatocellular carcinoma. Genes Dis. 2023;10:1857-1869. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in RCA: 14] [Reference Citation Analysis (0)] |
| 12. | Zheng Y, Gao K, Gao Q, Zhang S. Glycoproteomic contributions to hepatocellular carcinoma research: a 2023 update. Expert Rev Proteomics. 2023;20:211-220. [RCA] [PubMed] [DOI] [Full Text] [Cited by in RCA: 1] [Reference Citation Analysis (0)] |
| 13. | Hu M, Xia X, Chen L, Jin Y, Hu Z, Xia S, Yao X. Emerging biomolecules for practical theranostics of liver hepatocellular carcinoma. Ann Hepatol. 2023;28:101137. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 2] [Cited by in RCA: 3] [Article Influence: 1.5] [Reference Citation Analysis (0)] |
| 14. | Yao M, Yang JL, Wang DF, Wang L, Chen Y, Yao DF. Encouraging specific biomarkers-based therapeutic strategies for hepatocellular carcinoma. World J Clin Cases. 2022;10:3321-3333. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in CrossRef: 3] [Cited by in RCA: 1] [Article Influence: 0.3] [Reference Citation Analysis (1)] |
| 15. | Omar MA, Omran MM, Farid K, Tabll AA, Shahein YE, Emran TM, Petrovic A, Lucic NR, Smolic R, Kovac T, Smolic M. Biomarkers for Hepatocellular Carcinoma: From Origin to Clinical Diagnosis. Biomedicines. 2023;11:1852. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 23] [Cited by in RCA: 28] [Article Influence: 14.0] [Reference Citation Analysis (0)] |
| 16. | Yıldırım HÇ, Kavgaci G, Chalabiyev E, Dizdar O. Advances in the Early Detection of Hepatobiliary Cancers. Cancers (Basel). 2023;15:3880. [RCA] [PubMed] [DOI] [Full Text] [Reference Citation Analysis (0)] |
| 17. | Pan A, Truong TN, Su YH, Dao DY. Circulating Biomarkers for the Early Diagnosis and Management of Hepatocellular Carcinoma with Potential Application in Resource-Limited Settings. Diagnostics (Basel). 2023;13:676. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 9] [Cited by in RCA: 10] [Article Influence: 5.0] [Reference Citation Analysis (0)] |
| 18. | Ajuwon BI, Roper K, Richardson A, Lidbury BA. Routine blood test markers for predicting liver disease post HBV infection: precision pathology and pattern recognition. Diagnosis (Berl). 2023;10:337-347. [RCA] [PubMed] [DOI] [Full Text] [Cited by in RCA: 4] [Reference Citation Analysis (0)] |
| 19. | Truong TN, Pham TND, Hoang LB, Nguyen VT, Dao HV, Dao DVB, Alessy S, Pham HB, Pham TTT, Nguyen LDD, Nguyen K, Abaalkhail F, Manal M, Mawardi M, AlZahrani M, Alswat K, Alghamdi H, Sanai FM, Siddiqui MA, Nguyen NH, Vaidya D, Phan HT, Johnson PJ, Alqahtani SA, Dao DY. Surveillance and treatment of primary hepatocellular carcinoma (aka. STOP HCC): protocol for a prospective cohort study of high-risk patients for HCC using GALAD-score. BMC Cancer. 2023;23:875. [RCA] [PubMed] [DOI] [Full Text] [Reference Citation Analysis (0)] |
| 20. | Kim JY, Kim J, Lim YS, Gwak GY, Yeo I, Kim Y, Lee J, Shin D, Lee JH. Proteome Multimarker Panel for the Early Detection of Hepatocellular Carcinoma: Multicenter Derivation, Validation, and Comparison. ACS Omega. 2022;7:29934-29943. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 2] [Cited by in RCA: 8] [Article Influence: 2.7] [Reference Citation Analysis (0)] |
| 21. | Best J, Bechmann LP, Sowa JP, Sydor S, Dechêne A, Pflanz K, Bedreli S, Schotten C, Geier A, Berg T, Fischer J, Vogel A, Bantel H, Weinmann A, Schattenberg JM, Huber Y, Wege H, von Felden J, Schulze K, Bettinger D, Thimme R, Sinner F, Schütte K, Weiss KH, Toyoda H, Yasuda S, Kumada T, Berhane S, Wichert M, Heider D, Gerken G, Johnson P, Canbay A. GALAD Score Detects Early Hepatocellular Carcinoma in an International Cohort of Patients With Nonalcoholic Steatohepatitis. Clin Gastroenterol Hepatol. 2020;18:728-735.e4. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 94] [Cited by in RCA: 187] [Article Influence: 37.4] [Reference Citation Analysis (0)] |
| 22. | Singal AG, Reig M, Villanueva A. Emerging Tools for Hepatocellular Carcinoma Surveillance. Am J Gastroenterol. 2022;117:1948-1951. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 4] [Cited by in RCA: 15] [Article Influence: 5.0] [Reference Citation Analysis (0)] |
| 23. | Barnard Giustini A, Ioannou GN, Sirlin C, Loomba R. Review article: Available modalities for screening and imaging diagnosis of hepatocellular carcinoma-Current gaps and challenges. Aliment Pharmacol Ther. 2023;57:1056-1065. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 4] [Cited by in RCA: 4] [Article Influence: 2.0] [Reference Citation Analysis (0)] |
| 24. | Reeves R. High mobility group (HMG) proteins: Modulators of chromatin structure and DNA repair in mammalian cells. DNA Repair (Amst). 2015;36:122-136. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 56] [Cited by in RCA: 74] [Article Influence: 7.4] [Reference Citation Analysis (0)] |
| 25. | Fedele M, Fusco A. HMGA and cancer. Biochim Biophys Acta. 2010;1799:48-54. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 103] [Cited by in RCA: 119] [Article Influence: 7.9] [Reference Citation Analysis (0)] |
| 26. | Stros M. HMGB proteins: interactions with DNA and chromatin. Biochim Biophys Acta. 2010;1799:101-113. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 383] [Cited by in RCA: 465] [Article Influence: 31.0] [Reference Citation Analysis (0)] |
| 27. | Gerlitz G. HMGNs, DNA repair and cancer. Biochim Biophys Acta. 2010;1799:80-85. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 38] [Cited by in RCA: 47] [Article Influence: 2.9] [Reference Citation Analysis (0)] |
| 28. | Ueda T, Yoshida M. HMGB proteins and transcriptional regulation. Biochim Biophys Acta. 2010;1799:114-118. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 92] [Cited by in RCA: 107] [Article Influence: 7.1] [Reference Citation Analysis (0)] |
| 29. | Chen S, Dong Z, Yang P, Wang X, Jin G, Yu H, Chen L, Li L, Tang L, Bai S, Yan H, Shen F, Cong W, Wen W, Wang H. Hepatitis B virus X protein stimulates high mobility group box 1 secretion and enhances hepatocellular carcinoma metastasis. Cancer Lett. 2017;394:22-32. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 31] [Cited by in RCA: 49] [Article Influence: 6.1] [Reference Citation Analysis (0)] |
| 30. | Kwon JH, Kim J, Park JY, Hong SM, Park CW, Hong SJ, Park SY, Choi YJ, Do IG, Joh JW, Kim DS, Choi KY. Overexpression of high-mobility group box 2 is associated with tumor aggressiveness and prognosis of hepatocellular carcinoma. Clin Cancer Res. 2010;16:5511-5521. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 56] [Cited by in RCA: 72] [Article Influence: 4.8] [Reference Citation Analysis (0)] |
| 31. | Wen B, Wei YT, Zhao K. The role of high mobility group protein B3 (HMGB3) in tumor proliferation and drug resistance. Mol Cell Biochem. 2021;476:1729-1739. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 5] [Cited by in RCA: 36] [Article Influence: 9.0] [Reference Citation Analysis (0)] |
| 32. | Zheng W, Yang J, Dong Z, Wang L, Fang M, Wu W, Yao D, Yao M. High mobility group box 3 as an emerging biomarker in diagnosis and prognosis of hepatocellular carcinoma. Cancer Manag Res. 2018;10:5979-5989. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 14] [Cited by in RCA: 14] [Article Influence: 2.0] [Reference Citation Analysis (0)] |
| 33. | Liao Z, Tang C, Luo R, Gu X, Zhou J, Gao J. Current Concepts of Precancerous Lesions of Hepatocellular Carcinoma: Recent Progress in Diagnosis. Diagnostics (Basel). 2023;13:1211. [RCA] [PubMed] [DOI] [Full Text] [Reference Citation Analysis (0)] |
| 34. | Mashima E, Sawada Y, Nakamura M. Recent Advancement in Atypical Lipomatous Tumor Research. Int J Mol Sci. 2021;22:994. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 12] [Cited by in RCA: 20] [Article Influence: 5.0] [Reference Citation Analysis (0)] |
| 35. | Zheng WJ, Yao M, Fang M, Wang L, Dong ZZ, Yao DF. Abnormal expression of HMGB-3 is significantly associated with malignant transformation of hepatocytes. World J Gastroenterol. 2018;24:3650-3662. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in CrossRef: 7] [Cited by in RCA: 11] [Article Influence: 1.6] [Reference Citation Analysis (0)] |
| 36. | Tanaka S, Sugimachi K, Yamashita Y, Shirabe K, Shimada M, Wands JR. Angiogenic switch as a molecular target of malignant tumors. J Gastroenterol. 2003;38 Suppl 15:93-97. [PubMed] |
| 37. | Vanderborght B, Lefere S, Vlierberghe HV, Devisscher L. The Angiopoietin/Tie2 Pathway in Hepatocellular Carcinoma. Cells. 2020;9:2382. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 10] [Cited by in RCA: 25] [Article Influence: 5.0] [Reference Citation Analysis (0)] |
| 38. | Elgenidy A, Afifi AM, Awad AK, Jalal PK. Utility of serum angiopoietin-2 as diagnostic marker of hepatocellular carcinoma: A systematic review and diagnostic test accuracy meta-analysis. Clin Res Hepatol Gastroenterol. 2022;46:101909. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 2] [Cited by in RCA: 1] [Article Influence: 0.3] [Reference Citation Analysis (0)] |
| 39. | Li S, Yao D, Wang L, Wu W, Qiu L, Yao M, Yao N, Zhang H, Yu D, Ni Q. Expression characteristics of hypoxia-inducible factor-1α and its clinical values in diagnosis and prognosis of hepatocellular carcinoma. Hepat Mon. 2011;11:821-828. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 5] [Cited by in RCA: 30] [Article Influence: 2.1] [Reference Citation Analysis (0)] |
| 40. | Moldogazieva NT, Zavadskiy SP, Sologova SS, Mokhosoev IM, Terentiev AA. Predictive biomarkers for systemic therapy of hepatocellular carcinoma. Expert Rev Mol Diagn. 2021;21:1147-1164. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 4] [Cited by in RCA: 23] [Article Influence: 5.8] [Reference Citation Analysis (0)] |
| 41. | Li H, Liu H, Yan LJ, Ding ZN, Zhang X, Pan GQ, Han CL, Tian BW, Tan SY, Dong ZR, Wang DX, Yan YC, Li T. Performance of GALAD score and serum biomarkers for detecting NAFLD-related HCC: a systematic review and network meta-analysis. Expert Rev Gastroenterol Hepatol. 2023;17:1159-1167. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 5] [Cited by in RCA: 6] [Article Influence: 3.0] [Reference Citation Analysis (0)] |
| 42. | Lefere S, Devisscher L, Geerts A. Angiogenesis in the progression of non-alcoholic fatty liver disease. Acta Gastroenterol Belg. 2020;83:301-307. [PubMed] |
| 43. | Sun JY, Chen XY, Wang L, Ye WX, Shen SJ, Yang JL, Yao M, Yao DF. [Hypoxia-inducible factor-1α mediates and regulates angiogenesis-related factors expression in hepatocellular carcinoma]. Zhonghua Gan Zang Bing Za Zhi. 2020;28:942-948. [RCA] [PubMed] [DOI] [Full Text] [Cited by in RCA: 3] [Reference Citation Analysis (0)] |
| 44. | Dong S, Li Z, Kong J, Wu S, Gao J, Sun W. Arsenic trioxide inhibits angiogenesis of hepatocellular carcinoma after insufficient radiofrequency ablation via blocking paracrine angiopoietin-1 and angiopoietin-2. Int J Hyperthermia. 2022;39:888-896. [RCA] [PubMed] [DOI] [Full Text] [Cited by in RCA: 15] [Reference Citation Analysis (0)] |
| 45. | Orlandi P, Solini A, Banchi M, Brunetto MR, Cioni D, Ghiadoni L, Bocci G. Antiangiogenic Drugs in NASH: Evidence of a Possible New Therapeutic Approach. Pharmaceuticals (Basel). 2021;14:995. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 2] [Cited by in RCA: 14] [Article Influence: 3.5] [Reference Citation Analysis (0)] |
| 46. | Azbazdar Y, Demirci Y, Heger G, Ipekgil D, Karabicici M, Ozhan G. Comparative membrane lipidomics of hepatocellular carcinoma cells reveals diacylglycerol and ceramide as key regulators of Wnt/β-catenin signaling and tumor growth. Mol Oncol. 2023;17:2314-2336. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1] [Cited by in RCA: 11] [Article Influence: 5.5] [Reference Citation Analysis (0)] |
| 47. | Gao W, Kim H, Feng M, Phung Y, Xavier CP, Rubin JS, Ho M. Inactivation of Wnt signaling by a human antibody that recognizes the heparan sulfate chains of glypican-3 for liver cancer therapy. Hepatology. 2014;60:576-587. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 111] [Cited by in RCA: 140] [Article Influence: 12.7] [Reference Citation Analysis (0)] |
| 48. | Zeyada MS, Abdel-Rahman N, El-Karef A, Yahia S, El-Sherbiny IM, Eissa LA. Niclosamide-loaded polymeric micelles ameliorate hepatocellular carcinoma in vivo through targeting Wnt and Notch pathways. Life Sci. 2020;261:118458. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 30] [Cited by in RCA: 29] [Article Influence: 5.8] [Reference Citation Analysis (0)] |
| 49. | Yang Y, Ye YC, Chen Y, Zhao JL, Gao CC, Han H, Liu WC, Qin HY. Crosstalk between hepatic tumor cells and macrophages via Wnt/β-catenin signaling promotes M2-like macrophage polarization and reinforces tumor malignant behaviors. Cell Death Dis. 2018;9:793. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 103] [Cited by in RCA: 240] [Article Influence: 34.3] [Reference Citation Analysis (0)] |
| 50. | Li N, Wei L, Liu X, Bai H, Ye Y, Li D, Li N, Baxa U, Wang Q, Lv L, Chen Y, Feng M, Lee B, Gao W, Ho M. A Frizzled-Like Cysteine-Rich Domain in Glypican-3 Mediates Wnt Binding and Regulates Hepatocellular Carcinoma Tumor Growth in Mice. Hepatology. 2019;70:1231-1245. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 58] [Cited by in RCA: 87] [Article Influence: 14.5] [Reference Citation Analysis (0)] |
| 51. | Zhao J, Wang Y, Han M, Lu H, Chen X, Liu S, Yuan X, Han K, Liang P, Cheng J. P7TP3 inhibits tumor development, migration, invasion and adhesion of liver cancer through the Wnt/β-catenin signaling pathway. Cancer Sci. 2020;111:994-1007. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 9] [Cited by in RCA: 21] [Article Influence: 4.2] [Reference Citation Analysis (0)] |
| 52. | Nahon P, Bamba-Funck J, Layese R, Trépo E, Zucman-Rossi J, Cagnot C, Ganne-Carrié N, Chaffaut C, Guyot E, Ziol M, Sutton A, Audureau E; ANRS CO12 CirVir and CIRRAL groups. Integrating genetic variants into clinical models for hepatocellular carcinoma risk stratification in cirrhosis. J Hepatol. 2023;78:584-595. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 40] [Cited by in RCA: 40] [Article Influence: 20.0] [Reference Citation Analysis (0)] |
| 53. | Yao M, Yang JL, Wang L, Yao DF. [Carcinoembryonic type specific markers and liver cancer immunotherapy]. Zhonghua Gan Zang Bing Za Zhi. 2020;28:466-470. [RCA] [PubMed] [DOI] [Full Text] [Cited by in RCA: 4] [Reference Citation Analysis (0)] |
| 54. | Liu J, Wang Z, Tang J, Tang R, Shan X, Zhang W, Chen Q, Zhou F, Chen K, Huang A, Tang N. Hepatitis C virus core protein activates Wnt/β-catenin signaling through multiple regulation of upstream molecules in the SMMC-7721 cell line. Arch Virol. 2011;156:1013-1023. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 33] [Cited by in RCA: 43] [Article Influence: 3.1] [Reference Citation Analysis (0)] |
| 55. | Liu J, Ding X, Tang J, Cao Y, Hu P, Zhou F, Shan X, Cai X, Chen Q, Ling N, Zhang B, Bi Y, Chen K, Ren H, Huang A, He TC, Tang N. Enhancement of canonical Wnt/β-catenin signaling activity by HCV core protein promotes cell growth of hepatocellular carcinoma cells. PLoS One. 2011;6:e27496. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 88] [Cited by in RCA: 100] [Article Influence: 7.1] [Reference Citation Analysis (0)] |
| 56. | Pan L, Yao M, Zheng W, Gu J, Yang X, Qiu L, Cai Y, Wu W, Yao D. Abnormality of Wnt3a expression as novel specific biomarker for diagnosis and differentiation of hepatocellular carcinoma. Tumour Biol. 2016;37:5561-5568. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 13] [Cited by in RCA: 13] [Article Influence: 1.4] [Reference Citation Analysis (0)] |
| 57. | Pan LH, Yao M, Cai Y, Gu JJ, Yang XL, Wang L, Yao DF. Oncogenic Wnt3a expression as an estimable prognostic marker for hepatocellular carcinoma. World J Gastroenterol. 2016;22:3829-3836. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in CrossRef: 24] [Cited by in RCA: 26] [Article Influence: 2.9] [Reference Citation Analysis (0)] |
| 58. | Zheng W, Yao M, Fang M, Pan L, Wang L, Yang J, Dong Z, Yao D. Oncogenic Wnt3a: A Candidate Specific Marker and Novel Molecular Target for Hepatocellular Carcinoma. J Cancer. 2019;10:5862-5873. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 21] [Cited by in RCA: 23] [Article Influence: 3.8] [Reference Citation Analysis (0)] |
| 59. | Lu C, He Y, Duan J, Yang Y, Zhong C, Zhang J, Liao W, Huang X, Zhu R, Li M. Expression of Wnt3a in hepatocellular carcinoma and its effects on cell cycle and metastasis. Int J Oncol. 2017;51:1135-1145. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 11] [Cited by in RCA: 13] [Article Influence: 1.6] [Reference Citation Analysis (0)] |
| 60. | Lin Z, Liu J. [Corrigendum] lncRNA DQ786243 promotes hepatocellular carcinoma cell invasion and proliferation by regulating the miR‑15b‑5p/Wnt3A axis. Mol Med Rep. 2021;23:318. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 3] [Cited by in RCA: 13] [Article Influence: 3.3] [Reference Citation Analysis (0)] |
| 61. | Yao M, Wang JJ, Chen XY, Sai WL, Yang J, Wang DF, Wang L, Yao DF. Oncogenic Wnt3a is a promising sensitive biomarker for monitoring hepatocarcinogenesis. Hepatobiliary Pancreat Dis Int. 2023;22:263-269. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1] [Cited by in RCA: 5] [Article Influence: 2.5] [Reference Citation Analysis (0)] |
| 62. | Akbari S, Kunter I, Azbazdar Y, Ozhan G, Atabey N, Firtina Karagonlar Z, Erdal E. LGR5/R-Spo1/Wnt3a axis promotes stemness and aggressive phenotype in hepatoblast-like hepatocellular carcinoma cell lines. Cell Signal. 2021;82:109972. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 2] [Cited by in RCA: 18] [Article Influence: 4.5] [Reference Citation Analysis (0)] |
| 63. | Tsai HW, Cheng SW, Chen CC, Chen IW, Ho CL. A combined bioinformatics and experimental approach identifies RMI2 as a Wnt/β-catenin signaling target gene related to hepatocellular carcinoma. BMC Cancer. 2023;23:1025. [RCA] [PubMed] [DOI] [Full Text] [Reference Citation Analysis (0)] |
| 64. | Schepers EJ, Glaser K, Zwolshen HM, Hartman SJ, Bondoc AJ. Structural and Functional Impact of Posttranslational Modification of Glypican-3 on Liver Carcinogenesis. Cancer Res. 2023;83:1933-1940. [RCA] [PubMed] [DOI] [Full Text] [Cited by in RCA: 19] [Reference Citation Analysis (0)] |
| 65. | Yao M, Wang L, Fang M, Zheng W, Dong Z, Yao D. Advances in the study of oncofetal antigen glypican-3 expression in HBV-related hepatocellular carcinoma. Biosci Trends. 2016;10:337-343. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 11] [Cited by in RCA: 16] [Article Influence: 1.8] [Reference Citation Analysis (0)] |
| 66. | Tiyuri A, Baghermanesh SS, Davatgaran-Taghipour Y, Eslami SS, Shaygan N, Parsaie H, Barati M, Jafari D. Diagnostic accuracy of serum derived exosomes for hepatocellular carcinoma: a systematic review and meta-analysis. Expert Rev Mol Diagn. 2023;23:971-983. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 6] [Reference Citation Analysis (0)] |
| 67. | Xiao X, Huang Q, Lin X, Zahid KR, Huang X, Liu T, Zeng T. Current methods for the detection of glypican-3. Anal Methods. 2024;16:152-160. [RCA] [PubMed] [DOI] [Full Text] [Reference Citation Analysis (0)] |
| 68. | Pang BY, Leng Y, Wang X, Wang YQ, Jiang LH. A meta-analysis and of clinical values of 11 blood biomarkers, such as AFP, DCP, and GP73 for diagnosis of hepatocellular carcinoma. Ann Med. 2023;55:42-61. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 20] [Cited by in RCA: 27] [Article Influence: 13.5] [Reference Citation Analysis (0)] |
| 69. | Fernández-Ferreira R, Savage-Leyva R, Durán-Guerrero LF, Carranza-Sevilla MDR, Zamorano-Vazquez C, Monroy-Godínez CF, Hernández-Ramírez GR, Torres-Zazueta JM, Ceron-Ibarra E, Dorantes-Heredia R. Mandibular Metastasis as the First Manifestation of Hepatocellular Carcinoma: A Case Report. Case Rep Oncol. 2023;16:88-95. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Reference Citation Analysis (0)] |
| 70. | Wang J, Wang F, Wang N, Zhang MY, Wang HY, Huang GL. Diagnostic and Prognostic Value of Protein Post-translational Modifications in Hepatocellular Carcinoma. J Clin Transl Hepatol. 2023;11:1192-1200. [RCA] [PubMed] [DOI] [Full Text] [Cited by in RCA: 1] [Reference Citation Analysis (0)] |
| 71. | Shen J, Shen H, Ke L, Chen J, Dang X, Liu B, Hua Y. Knowledge Mapping of Immunotherapy for Hepatocellular Carcinoma: A Bibliometric Study. Front Immunol. 2022;13:815575. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 87] [Cited by in RCA: 89] [Article Influence: 29.7] [Reference Citation Analysis (0)] |
| 72. | Sun H, Xing C, Jiang S, Yu K, Dai S, Kong H, Jin Y, Shan Y, Yang W, Wang Z, Xiao J, Wang H, Wang W, Li Z, Shi K. Long term complete response of advanced hepatocellular carcinoma to glypican-3 specific chimeric antigen receptor T-Cells plus sorafenib, a case report. Front Immunol. 2022;13:963031. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in RCA: 11] [Reference Citation Analysis (0)] |
| 73. | Wang L, Pan L, Yao M, Cai Y, Dong Z, Yao D. Expression of oncofetal antigen glypican-3 associates significantly with poor prognosis in HBV-related hepatocellular carcinoma. Oncotarget. 2016;7:42150-42158. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 21] [Cited by in RCA: 25] [Article Influence: 2.8] [Reference Citation Analysis (0)] |
| 74. | Dong Z, Yao M, Wang L, Yang J, Yao D. Down-regulating glypican-3 expression: molecular-targeted therapy for hepatocellular carcinoma. Mini Rev Med Chem. 2014;14:1183-1193. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 7] [Cited by in RCA: 12] [Article Influence: 1.1] [Reference Citation Analysis (0)] |
| 75. | Yao M, Yao DF, Bian YZ, Zhang CG, Qiu LW, Wu W, Sai WL, Yang JL, Zhang HJ. Oncofetal antigen glypican-3 as a promising early diagnostic marker for hepatocellular carcinoma. Hepatobiliary Pancreat Dis Int. 2011;10:289-294. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 34] [Cited by in RCA: 37] [Article Influence: 2.6] [Reference Citation Analysis (0)] |
| 76. | Wang Y, Wan YY. Golgi protein 73, hepatocellular carcinoma and other types of cancers. Liver Res. 2020;4:161-167. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 16] [Cited by in RCA: 19] [Article Influence: 3.8] [Reference Citation Analysis (0)] |
| 77. | Butaye E, Somers N, Grossar L, Pauwels N, Lefere S, Devisscher L, Raevens S, Geerts A, Meuris L, Callewaert N, Van Vlierberghe H, Verhelst X. Systematic review: Glycomics as diagnostic markers for hepatocellular carcinoma. Aliment Pharmacol Ther. 2024;59:23-38. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1] [Cited by in RCA: 6] [Article Influence: 6.0] [Reference Citation Analysis (0)] |
| 78. | Sai W, Wang L, Zheng W, Yang J, Pan L, Cai Y, Qiu L, Zhang H, Wu W, Yao D. Abnormal Expression of Golgi Protein 73 in Clinical Values and Their Role in HBV-Related Hepatocellular Carcinoma Diagnosis and Prognosis. Hepat Mon. 2015;15:e32918. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 15] [Cited by in RCA: 16] [Article Influence: 1.6] [Reference Citation Analysis (0)] |
| 79. | Wu M, Liu Z, Li X, Zhang A, Li N. Dynamic Changes in Serum Markers and Their Utility in the Early Diagnosis of All Stages of Hepatitis B-Associated Hepatocellular Carcinoma. Onco Targets Ther. 2020;13:827-840. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 14] [Cited by in RCA: 24] [Article Influence: 4.8] [Reference Citation Analysis (0)] |
| 80. | Eissa M, Awad S, Barakat S, Saleh A, Rozaik S. Serum Golgi protein 73 as a sensitive biomarker for early detection of hepatocellular carcinoma among Egyptian patients with hepatitis C virus-related cirrhosis. Med J Armed Forces India. 2021;77:331-336. [RCA] [PubMed] [DOI] [Full Text] [Cited by in RCA: 9] [Reference Citation Analysis (0)] |
| 81. | Sai WL, Yao M, Shen SJ, Zheng WJ, Sun JY, Wu MN, Wang L, Yao DF. Dynamic expression of hepatic GP73 mRNA and protein and circulating GP73 during hepatocytes malignant transformation. Hepatobiliary Pancreat Dis Int. 2020;19:449-454. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 13] [Cited by in RCA: 12] [Article Influence: 2.4] [Reference Citation Analysis (0)] |
| 82. | Ye JZ, Yan SM, Yuan CL, Wu HN, Zhang JY, Liu ZH, Li YQ, Luo XL, Lin Y, Liang R. GP73 level determines chemotherapeutic resistance in human hepatocellular carcinoma cells. J Cancer. 2018;9:415-423. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 10] [Cited by in RCA: 20] [Article Influence: 2.9] [Reference Citation Analysis (0)] |
| 83. | Xiu P, Dong XF, Li XP, Li J. Clusterin: Review of research progress and looking ahead to direction in hepatocellular carcinoma. World J Gastroenterol. 2015;21:8262-8270. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in CrossRef: 16] [Cited by in RCA: 22] [Article Influence: 2.2] [Reference Citation Analysis (0)] |
| 84. | Wang C, Zhang Y, Guo K, Wang N, Jin H, Liu Y, Qin W. Heat shock proteins in hepatocellular carcinoma: Molecular mechanism and therapeutic potential. Int J Cancer. 2016;138:1824-1834. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 79] [Cited by in RCA: 80] [Article Influence: 8.9] [Reference Citation Analysis (0)] |
| 85. | Hwang S, Lee DH, Lee IK, Park YM, Jo I. Far-infrared radiation inhibits proliferation, migration, and angiogenesis of human umbilical vein endothelial cells by suppressing secretory clusterin levels. Cancer Lett. 2014;346:74-83. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 20] [Cited by in RCA: 22] [Article Influence: 2.0] [Reference Citation Analysis (0)] |
| 86. | Xiu P, Dong X, Xu Z, Zhu H, Liu F, Wei Z, Zhai B, Kanwar JR, Jiang H, Li J, Sun X. Secretory clusterin contributes to oxaliplatin resistance by activating Akt pathway in hepatocellular carcinoma. Cancer Sci. 2013;104:375-382. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 48] [Cited by in RCA: 59] [Article Influence: 4.9] [Reference Citation Analysis (0)] |
| 87. | Xiu P, Xu Z, Liu F, Li Z, Li T, Zou F, Sun X, Li J. Downregulating sCLU enhances the sensitivity of hepatocellular carcinoma cells to gemcitabine by activating the intrinsic apoptosis pathway. Dig Dis Sci. 2014;59:1798-1809. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 13] [Cited by in RCA: 15] [Article Influence: 1.4] [Reference Citation Analysis (0)] |
| 88. | Wang X, Zou F, Zhong J, Yue L, Wang F, Wei H, Yang G, Jin T, Dong X, Li J, Xiu P. Secretory Clusterin Mediates Oxaliplatin Resistance via the Gadd45a/PI3K/Akt Signaling Pathway in Hepatocellular Carcinoma. J Cancer. 2018;9:1403-1413. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 12] [Cited by in RCA: 28] [Article Influence: 4.0] [Reference Citation Analysis (0)] |
| 89. | Zhang J, Wu M, Xu Y, Song Q, Zheng W. Secretory Clusterin: A Promising Target for Chemoresistance of Hepatocellular Carcinoma. Mini Rev Med Chem. 2020;20:1153-1165. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 2] [Cited by in RCA: 3] [Article Influence: 0.8] [Reference Citation Analysis (0)] |
| 90. | Wang C, Jiang K, Gao D, Kang X, Sun C, Zhang Q, Li Y, Sun L, Zhang S, Guo K, Liu Y. Clusterin protects hepatocellular carcinoma cells from endoplasmic reticulum stress induced apoptosis through GRP78. PLoS One. 2013;8:e55981. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 48] [Cited by in RCA: 58] [Article Influence: 4.8] [Reference Citation Analysis (0)] |
| 91. | Wang C, Jin G, Jin H, Wang N, Luo Q, Zhang Y, Gao D, Jiang K, Gu D, Shen Q, Huo X, Hu F, Ge T, Zhao F, Chu W, Shu H, Yao M, Cong W, Qin W. Clusterin facilitates metastasis by EIF3I/Akt/MMP13 signaling in hepatocellular carcinoma. Oncotarget. 2015;6:2903-2916. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 39] [Cited by in RCA: 53] [Article Influence: 5.3] [Reference Citation Analysis (0)] |
| 92. | Zheng W, Yao M, Sai W, Qian Q, Pan L, Qiu L, Huang J, Wu W, Yao D. Diagnostic and prognostic significance of secretory clusterin expression in patients with hepatocellular carcinoma. Tumour Biol. 2016;37:999-1008. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 15] [Cited by in RCA: 11] [Article Influence: 1.2] [Reference Citation Analysis (0)] |
| 93. | Beheshti Namdar A, Kabiri M, Mosanan Mozaffari H, Aminifar E, Mehrad-Majd H. Circulating Clusterin Levels and Cancer Risk: A Systematic Review and Meta-Analysis. Cancer Control. 2022;29:10732748211038437. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in RCA: 6] [Reference Citation Analysis (0)] |
| 94. | Rasmy HS, Mohammed HA, Mohammed ES, Ahmed ASM, Isaac A. Serum clusterin as a promising diagnostic and prognostic marker for hepatocellular carcinoma after locoregional treatment. Egypt J Immunol. 2022;29:26-40. [PubMed] |
| 95. | Yao M, Fang M, Zheng W, Dong Z, Yao D. Role of secretory clusterin in hepatocarcinogenesis. Transl Gastroenterol Hepatol. 2018;3:48. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 12] [Cited by in RCA: 18] [Article Influence: 2.6] [Reference Citation Analysis (0)] |
| 96. | Zheng W, Yao M, Wu M, Yang J, Yao D, Wang L. Secretory clusterin promotes hepatocellular carcinoma progression by facilitating cancer stem cell properties via AKT/GSK-3β/β-catenin axis. J Transl Med. 2020;18:81. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 36] [Cited by in RCA: 40] [Article Influence: 8.0] [Reference Citation Analysis (0)] |
| 97. | Zhong J, Yu X, Dong X, Lu H, Zhou W, Li L, Li Z, Sun P, Shi X. Downregulation of secreted clusterin potentiates the lethality of sorafenib in hepatocellular carcinoma in association with the inhibition of ERK1/2 signals. Int J Mol Med. 2018;41:2893-2900. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 5] [Cited by in RCA: 8] [Article Influence: 1.1] [Reference Citation Analysis (0)] |
| 98. | Narahara S, Watanabe T, Nagaoka K, Fujimoto N, Furuta Y, Tanaka K, Tokunaga T, Kawasaki T, Yoshimaru Y, Setoyama H, Oniki K, Saruwatari J, Tateyama M, Naoe H, Tanaka M, Tanaka Y, Sasaki Y. Clusterin and Related Scoring Index as Potential Early Predictors of Response to Sorafenib in Hepatocellular Carcinoma. Hepatol Commun. 2022;6:1198-1212. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 4] [Cited by in RCA: 2] [Article Influence: 0.7] [Reference Citation Analysis (0)] |
| 99. | Zheng W, Yao M, Qian Q, Sai W, Qiu L, Yang J, Wu W, Dong Z, Yao D. Oncogenic secretory clusterin in hepatocellular carcinoma: Expression at early staging and emerging molecular target. Oncotarget. 2017;8:52321-52332. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 12] [Cited by in RCA: 19] [Article Influence: 2.4] [Reference Citation Analysis (0)] |
| 100. | Yao M, Sai W, Zheng W, Wang L, Dong Z, Yao D. Secretory Clusterin as a Novel Molecular-targeted Therapy for Inhibiting Hepatocellular Carcinoma Growth. Curr Med Chem. 2020;27:3290-3301. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 6] [Cited by in RCA: 4] [Article Influence: 0.8] [Reference Citation Analysis (0)] |
| 101. | Lv Y, Sun X. Role of miRNA in pathogenesis, diagnosis, and prognosis in hepatocellular carcinoma. Chem Biol Drug Des. 2024;103:e14352. [RCA] [PubMed] [DOI] [Full Text] [Cited by in RCA: 11] [Reference Citation Analysis (1)] |
| 102. | Zhao F, Tan F, Tang L, Du Z, Chen X, Yang Y, Zhou G, Yuan C. Long Non-coding RNA DLGAP1-AS1 and DLGAP1-AS2: Two Novel Oncogenes in Multiple Cancers. Curr Med Chem. 2023;30:2822-2834. [RCA] [PubMed] [DOI] [Full Text] [Reference Citation Analysis (0)] |