Published online Jun 15, 2024. doi: 10.4251/wjgo.v16.i6.2271
Revised: April 8, 2024
Accepted: April 28, 2024
Published online: June 15, 2024
Processing time: 177 Days and 15.1 Hours
The morbidity and mortality of gastrointestinal (GI) malignancies are among the highest in the world, posing a serious threat to human health. Because of the insidious onset of the cancer, it is difficult for patients to be diagnosed at an early stage, and it rapidly progresses to an advanced stage, resulting in poor treatment and prognosis. Fusobacterium nucleatum (F. nucleatum) is a gram-negative, spore-free anaerobic bacterium that primarily colonizes the oral cavity and is implicated in the development of colorectal, esophageal, gastric, and pancreatic cancers via various intricate mechanisms. Recent development in novel research suggests that F. nucleatum may function as a biomarker in GI malignancies. Detecting the abundance of F. nucleatum in stool, saliva, and serum samples of patients may aid in the diagnosis, risk assessment, and prognosis monitoring of GI malignancies. This editorial systematically describes the biological roles and mechanisms of F. nucleatum in GI malignancies focusing on the application of F. nucleatum as a biomarker in the diagnosis and prognosis of GI malignancies to promote the clinical translation of F. nucleatum and GI tumors-related research.
Core Tip: Gastrointestinal (GI) malignancies are characterized by high morbidity and mortality. Early diagnosis of GI malignancies is crucial for disease intervention and treatment. Numerous studies have shown that Fusobacterium nucleatum (F. nucleatum) is closely related to the development of various GI malignant tumors. This paper discusses the mechanism of F. nucleatum in promoting the progression of GI tumors, elaborates its clinical value as a diagnostic and prognostic biomarker, and provides ideas for the development and research of novel biomarkers for GI malignancies.
- Citation: Yu LC, Li YP, Xin YM, Mao M, Pan YX, Qu YX, Luo ZD, Zhang Y, Zhang X. Application of Fusobacterium nucleatum as a biomarker in gastrointestinal malignancies. World J Gastrointest Oncol 2024; 16(6): 2271-2283
- URL: https://www.wjgnet.com/1948-5204/full/v16/i6/2271.htm
- DOI: https://dx.doi.org/10.4251/wjgo.v16.i6.2271
Gastrointestinal (GI) malignancies, including colorectal cancer (CRC), gastric cancer (GC), esophageal cancer (EC), pancreatic cancer (PC) and so on, are a major cause of cancer-related morbidity and mortality. Globally, GI cancers account for approximately 26% of new cancer cases and 35% of cancer-related deaths[1]. GI malignancies share common risk factors, and their incidence and progression are influenced by various elements, including genetics, environment, diet, and lifestyle. The pathogenesis of GI cancers is intricate and varied[2]. However, due to the absence of specific symptoms during their early stages, it is difficult for patients to be diagnosed with GI cancer in a timely manner, resulting in expedited progression to advanced stages. As a consequence, treatment outcomes, prognosis, and five-year survival rates are poor. The 5-year survival rates for patients with advanced CRC, EC, and PC are below 20%, while the 5-year survival rate for patients with advanced GC is less than 40%[3,4]. Currently, the gold standard for diagnosing malignant GI tumors is a combination of biopsy and pathological examination. However, this method is invasive and time-consuming, making it challenging to implement for population screening purposes[5]. Furthermore, traditional tumor markers in serum like carcinoembryonic antigen (CEA), carbohydrate antigen (CA)-125, and CA19-9, which are commonly utilized for screening because of their convenience, lack the sensitivity and specificity required for consistent diagnosis of GI malignancies resulting in potential cases of missed diagnosis or misdiagnosis[6]. Therefore, it is imperative to explore a novel non-invasive biomarker for early detection, prognostic monitoring, and risk evaluation of GI malignancies.
Fusobacterium nucleatum (F. nucleatum) is a spindle-shaped, gram-negative, anaerobic bacterium devoid of spores with a large middle section and tapered ends[7]. F. nucleatum is a common opportunistic pathogen in the oral cavity that can also be found in the intestinal, urinary, and upper digestive systems. F. nucleatum has been implicated in a variety of inflammatory diseases, such as periodontitis and inflammatory bowel disease[8,9]. Through extensive research conducted on intestinal flora in recent years, it has been determined that F. nucleatum exhibits a close correlation with GI malignant tumors. F. nucleatum affected the occurrence and progression of GI malignancies by colonizing the digestive tract, contributing to the proliferation, metastasis, and chemoresistance of cancer cells. Thus, F. nucleatum may serve as a biomarker for the diagnosis and prognosis monitoring of GI malignancies[10-12].
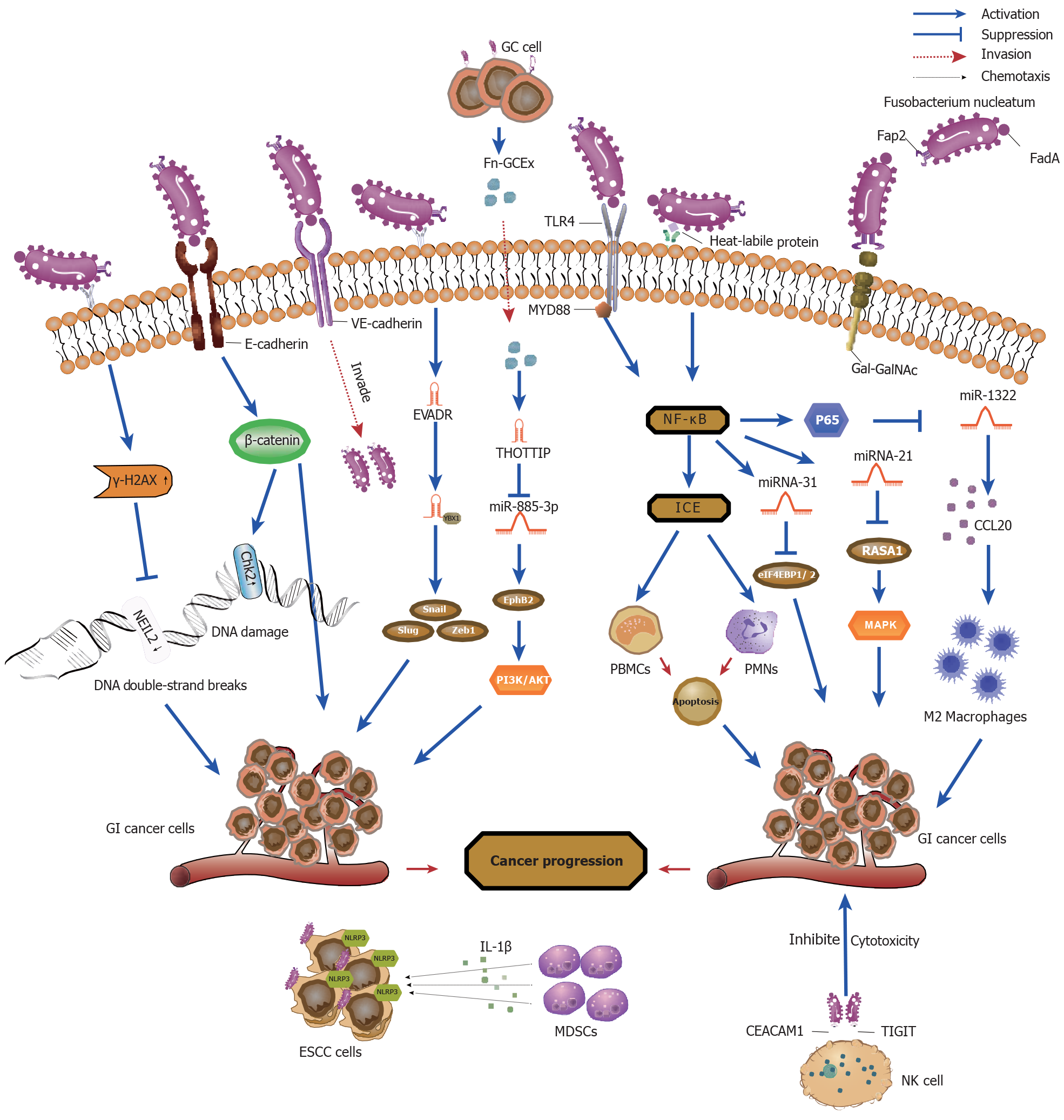
In fact, certain patterns in oral and gut microbiota have been shown to be predictive of the development of GI malignancies[13,14]. Increasing evidence suggests that F. nucleatum is more common in the saliva and stool samples of patients with GI malignancies compared to healthy controls[15]. Moreover, quantitative polymerase chain reaction (qRCR) and bacterial 16S rDNA sequence analysis revealed increased levels of F. nucleatum in GI malignancies tissues compared to adjacent normal tissues[16,17]. Notably, a high abundance of F. nucleatum was inversely associated with overall survival and exhibited high specificity and potential value as a diagnostic and prognostic marker for GI malignancies[18]. This editorial set out to summarize the latest developments regarding the potential involvement of F. nucleatum in carcinogenesis (Figure 1 and Table 1) and the significance of F. nucleatum as a diagnostic and prognostic biomarker for GI malignancies.
| Mechanisms | Type of cancer | Potential pathway | Ref. |
| Release of virulence factors | CRC | Myloid-like FadA | [19] |
| FadA/VE-cadherin | [20] | ||
| FadA/E-adherin/β-catenin signaling | [21] | ||
| Fap2/Gal-GalNAC | [22] | ||
| Fn-DPS/ATP3/PD-L1 | [23] | ||
| Succinic acid/cGAS-interferon-β pathway | [24] | ||
| EC | Fn-DPS/CCL2/CCL7/EMT | [11] | |
| Putrescine | [70] | ||
| Induction of DNA damage | CRC | E-cadherin and chk2 | [26] |
| γ-H2AX | [27] | ||
| NEIL2 | [28] | ||
| EC | NOD1/RIPK2/NF-κB | [76] | |
| Senescence-associated secretory phenotype | [25] | ||
| Dysregulation of non-coding RNAs | CRC | TLR4/MYD88/NFκB/miRNA21/MAPK | [30] |
| miR34a/miR22/miR28 | [29] | ||
| miR-31/eukaryotic initiation factor 4f-binding protein 1/2 | [32] | ||
| miR-4717/METTL3 | [77] | ||
| miR-4474/miR-4717/CREBBP | [78] | ||
| miR-122-5p/FUT8/TGF-1/Smads axis | [36] | ||
| lncRNA EVADR/YBX1 | [34] | ||
| lncRNA ENO1-IT1 | [33] | ||
| GC | HOTTIP/miR-885-3p | [35] | |
| Modulation of tumor immune microenvironment | CRC | CD11b+ myeloid cells/arginase-1 and iNOS/CD4+T cell | [79] |
| miR-1322/polarization of M2 macrophages | [31] | ||
| CEACAM1/T cell and NK cell depletion | [38] | ||
| Fap2/TIGIT/T cell and NK cell depletion | [39] | ||
| EC | NLRP3/MDSCs | [40] | |
| KIR2DL1/CD8+ T cells | [80] | ||
| PC | CXCL1/CXCR2/MDSCs/CD8+ T cells | [73] | |
| GM-CSF, CXCL1, IL-8 and MIP-3α | [81] | ||
| AI-2/TNFSF9/CD8+ T cells | [82] | ||
| Cooperation with other intestinal microbiota | CRC | Bacteroides fragilis/PKS+E. coli | [42] |
| Enterobacteriaceae/Stenotrophomonas | [43] | ||
| Gemella morbillorum/fiber intake | [44] | ||
| RadD/Clostridioides difficile | [83] |
F. nucleatum adheres to and colonizes tumor cells by releasing adhesins, virulence factors, and metabolites, inducing the oncogenic response. Under conditions of stress and disease, it was revealed that F. nucleatum secreted adhesion FadA with amyloid properties on its surface acting as a scaffold for biofilm formation to protect F. nucleatum and promote survival in acidic environments enhancing its virulence[19]. Research has indicated that FadA bound to the vascular cell receptor VE-cadherin resulting in weakened cell-cell connections and increased endothelial cell permeability, aiding F. nucleatum to invade endothelial cells and ultimately spread throughout the body[20]. In addition to binding VE-cadherin, FadA further was shown to localize to the E-cadherin receptor, activating the β-catenin signaling pathway, mediating F. nucleatum adhesion and invasion on epithelial cells, and promoting CRC proliferation by upregulating the expression of specific oncogenes and inflammatory genes[21]. Furthermore, Abed et al[22] discovered that microbial protein Fap2 mediated the binding of F. nucleatum to overexpressed D-galactos-β (1-3)-N-acetyl-D-galactosamine (Gal-GalNAc) in CRC. This resulted in the enrichment of F. nucleatum in CRC, thus strengthening its adhesion and invasion into the tumor, and promoting immune-mediated inflammation. Furthermore, a novel F. nucleatum virulence factor, DNA hunger/stationary phase protective proteins (Fn-Dps), promoted CRC metastasis by inducing epithelial-mesenchymal transition (EMT) through upregulation of chemokine CCL2/CCL7 expression[23]. Notably, succinic acid, a metabolite of F. nucleatum, was revealed to inhibit the cyclic GMP-AMP synthase-interferon-β pathway, resulting in reduced levels of chemokines CCL5 and CXCL10 in the tumor microenvironment and attenuation of the aggregation of CD8+ T cells thereby suppressing the anti-tumor response[24].
F. nucleatum infection has been shown to promote the malignant transformation of GI tumor cells by inducing DNA damage and regulating the expression of DNA damage response genes, a crucial step in the process of carcinogenesis. It has been shown that F. nucleatum invaded chemotherapy-induced senescent esophageal squamous carcinoma (ESCC) cells and induced DNA damage and activation of DNA damage response pathways resulting in increased secretion of proinflammatory factors, exacerbating ESCC progression and chemoresistance[25]. In the C57BL/6 J-adenomatous polyposis coli Min/J mouse model, Guo et al[26] demonstrated that F. nucleatum inoculation upregulated E-cadherin and chk2 expression through FadA, resulting in DNA damage and cell proliferation of CRC cells. Moreover, it was revealed that F. nucleatum infection of CRC cells increased the level of the DNA damage marker γ-H2AX, causing DNA double-strand breaks (DSBs)[27]. In addition, the presence of F. nucleatum downregulated the expression of the DNA repair protein NEIL2, which in turn promoted DSBs and the secretion of inflammatory factors, contributing to the development of CRC[28].
Recently, an increasing number of studies have identified dysregulation of non-coding RNAs (ncRNAs), especially microRNAs (miRNAs) and long noncoding RNAs (lncRNAs), as being key to the molecular mechanisms by which F. nucleatum induces the development of GI malignancies. F. nucleatum induced pro-inflammatory cytokine secretion by recognizing Toll-like receptor (TLR) 4 and TLR2 receptors and upregulation of miR-34a and miR-135b expression[29]. Similarly, F. nucleatum infection of intestinal epithelial cells increased the proliferation and invasive ability of tumor cells through activation of the MAPK signaling pathway by inducing the expression of miRNA21 and downregulating the expression of RASA1 through the TLR4/ MYD88/NF-κB pathway[30]. Furthermore, downregulation of miRNA-1322 expression through activation of the NF-kB signaling pathway following F. nucleatum infection increased CCL20 expression to provide an immune microenvironment suitable for tumor development and metastasis[31]. Moreover, activation of the NF-kB signaling pathway induced the expression of miRNA-31, which directly acted upon eukaryotic initiation factor 4f-binding protein 1/2 to promote CRC cell proliferation by inhibiting its expression. On the other hand, miRNA-31 enhanced the inhibitory effect of F. nucleatum on autophagic flux by targeting SYNTAXIN-12 and increased the intracellular survival of F. nucleatum resulting in continuous infection of CRC cells and enhanced tumorigenicity[32].
Intriguingly, lncRNAs have emerged as important mediators of interactions between tumor cells and F. nucleatum. F. nucleatum was shown to activate lncRNA enolase1-intronic transcript 1 (ENO1-IT1) transcription by promoting the bind
Exosomes are important mediators of intercellular communication. As such, exosome content and secretion were shown to be influenced by F. nucleatum, playing a key role in tumor communication networks. Moreover, it has been reported that exosomes secreted by F. nucleatum-infected GC cells (Fn-GCEx) could enhance the proliferation, invasion, and metastasis of uninfected GC cells. LncRNA-HOTTIP, an important cancer-promoting signal in Fn-GCEx, was observed to upregulate the expression of EphB2 by sponging miRNA-885-3p resulting in activation of the PI3K/ AKT signaling pathway and promotion of the progression of GC[35]. Similarly, in F. nucleatum-infected CRC cells we observed that the RNA-binding protein hnRNPA2B1 mediated the secretion of oncogene miR-122-5P by F. nucleatum into exosomes, leading to the downregulation of its expression in CRC cells and in turn an upregulation of FUT8 expression promoting the migratory and proliferative capacities of tumor cells. Moreover, it has been further revealed that F. nucleatum activated the TGF-β1/Smads signaling pathway by regulating the miRNA-122-5p/FUT8 axis to promote EMT, resulting in tumorigenesis and development[36].
F. nucleatum has been shown to regulate the tumor immune microenvironment and suppress anti-tumor immune responses in cancer tissues through two pathways. Firstly, F. nucleatum was revealed to regulate the immune microenvironment by inhibiting the function of anti-tumor immune cells. Jewett et al[37] reported that F. nucleatum activated the NF-kB/interleukin-converting enzyme pathway through bacterial apoptosis-inducing molecules on its surface, mediated apoptosis of immune cells, such as peripheral blood mononuclear cells and polymorphonuclear cells, and suppressed their immune function. These phenomena created an immune microenvironment suitable for tumor development and metastasis. Similarly, F. nucleatum was shown to suppress the cytotoxicity of immune cells and regulate anti-tumor immunity through the interaction of Fap2 with the inhibitory receptors on natural killer cells, TIGIT and CEACAM1, which protected F. nucleatum from immune cell attack thereby promoting tumor cell metastasis[38,39].
Secondly, F. nucleatum selectively recruits immunosuppressive myeloid suppressor cells (MDSCs) and M2 macro
A multitude of studies have reported that F. nucleatum interacts with other gut microbes implicated in the occurrence and development of GI malignancies. To investigate bacterial symbiosis and its potential interactions, Tran et al[41] constructed a correlation network using the gut microbiomes of CRC patients and identified that multiple subspecies of Fusobacterium cluster in the tumor microbiome, and furthermore, that genes encoding key virulence factors (Fap2 and RadD) may undergo frequent horizontal gene transfer or recombination. Gong et al[42] postulated that in the early stages of CRC, Bacteroides fragilis-mediated inflammation induced intestinal epithelial degradation and intestinal mucosal injury, leading to an imbalance in intestinal ecology and providing the basic conditions for PKS+E. coli colonization and induction of oncogenic mutations. Cancerous intestinal epithelial cells can further recruit F. nucleatum to colonize the lesion site, and F. nucleatum in turn was shown to promote cancer cell proliferation through Fap2-mediated immune evasion. Wu et al[43] identified that in azoxymethane/dextran sulfate sodium salt mice F. nucleatum infection altered the structure of the intestinal mucosal microbial community, this was primarily manifested as an enrichment of potentially pathogenic flora and the depletion of probiotic bacteria, particularly Enterobacteriaceae and Stenotrophomonas. The structural dysregulation of the intestinal microbial community may affect the transcriptional activity of tumor-associated metabolic pathways and cause malignant transformation of tumor cells. In addition, F. nucleatum and Gemella morbi
At present, the detection of F. nucleatum is primarily reliant upon the following sample types: Tissue samples, stool samples, saliva samples and serum samples, as we illustrate in Table 2.
| Cancer | Samples | Risk assessment and detection rate of F. nucleatum | Method | Ref. |
| CRC | Tissue | 72% (proximal) and 35% (distal) | qPCR | [84] |
| CRC | Tissue | 38.8% (54/139) | ddPCR | [85] |
| CRC | Tissue | 56.30% (18/32) | qPCR | [86] |
| CRC | Tissue | 57.1% (8/14) | AP-PCR | [62] |
| Saliva | 100% (14/14) | |||
| CRC | Stool | Worse overall survival | 16S rRNA sequencing | [10] |
| Tissue | ||||
| CRC | Tissue | 50% | PCR | [87] |
| Stool | 53.5% | |||
| GC | Tissue | 26% (19/80) | qPCR | [88] |
| GC | Tissue | 44% (40/91) | qPCR | [89] |
| GC | Tissue | 28.75% (23/80) | qPCR | [66] |
| Worse overall survival | ||||
| EC | Tissue | 69.4% (68/98) | qPCR | [71] |
| PC | Tissue | 15.5% (13/84) | qRT-PCR | [73] |
| PC | Serum | 52.4% | ELISA and qPCR | [75] |
Application of F. nucleatum detection in tissues for the diagnosis and prognosis of CRC: Presently, tissue samples are the commonest sample type used in the study of F. nucleatum and CRC. Utilizing tissue, it is possible to directly analyze the relationship between F. nucleatum abundance and pathological features of the tumor as well as epigenetic alterations. Metagenomic analyses using whole-genome sequencing, transcriptome sequencing, and DNA sequencing of bacterial 16S ribosomal RNA genes revealed enrichment of Fusobacterium species in CRC compared to in adjacent normal tissues[45-47]. Moreover, it was demonstrated that the abundance of F. nucleatum in tumor tissues was correlated with the site, metastasis, and invasion depth of CRC[48,49]. Mima et al[50] measured the relative amount of F. nucleatum DNA in tumor tissues from 1069 CRC cases by qPCR and observed that in terms of epigenetics, F. nucleatum abundance was closely related to CpG island methylation phenotype, microsatellite instability and BRAF mutations in CRC.
Several studies have revealed that CRC patients with a high tissue abundance of F. nucleatum have a significantly worse prognosis than those with a low abundance of F. nucleatum. A previous clinical study suggested that high levels of F. nucleatum may be a prognostic biomarker for CRC and positively associated F. nucleatum with higher CRC-specific mortality[50]. A recent Meta-analysis revealed that high levels of F. nucleatum in tumor tissue were strongly associated with lower overall survival, disease-free survival, and cancer-specific survival in CRC patients[51]. Several studies conducted in different geographical Settings (Europe, China, and Japan) have confirmed these results[52,53]. Notably, high levels of F. nucleatum were further significantly associated with low survival in metastatic CRC[54].
In summary, the detection of F. nucleatum in tissues may be an important tool for the early detection of CRC as well as for its prognostic assessment.
Application of F. nucleatum detection in stool for the diagnosis of CRC: Recent and accumulating evidence has been reported that the ecological dysbiosis of intestinal flora is closely associated with CRC. Since bacteria account for approximately 60% of fecal dry weight, non-invasive fecal flora analysis has become widely applied in the study of intestinal flora and the diagnosis of CRC. Tunsjø et al[55] revealed markedly increased levels of F. nucleatum in fecal samples from patients with CRC in comparison to healthy controls and a polyp group, indirectly reflecting the abundance of F. nucleatum in tumor tissues. Moreover, Liang et al[56] tested the faecal samples of 203 CRC patients and 236 healthy controls by duplex qPCR and observed that F. nucleatum levels were significantly higher in CRC than in the control group, with an area under the receiver operator characteristic curve (AUC) of 0.868, 77.7% sensitivity, and 79.5% specificity. Moreover, a meta-analysis of data collected from multiple studies also revealed that the combined AUC of fecal F. nucleatum for the diagnosis of CRC was 0.8695, and the sensitivity and specificity were 81.95% and 77.95%, respectively, indicating good diagnostic value for CRC[57].
It is noteworthy that combining F. nucleatum detection with other assays has exhibited improved diagnostic performance for CRC. Currently, fecal immunochemical testing (FIT) is widely used for noninvasive screening of CRC, but it is limited by its low sensitivity for detecting advanced tumors[58]. Wong et al[59] revealed that combining FIT with F. nucleatum measurement significantly improved the detection rate of CRC with sensitivity and specificity of 92.3% and 93.0%, respectively, and of advanced adenomas with a detection rate of 38.6% and specificity of 89.0%, respectively. This combination approach was capable of detecting advanced lesions missed by FIT alone. Meanwhile, Guo et al[60] discovered that the microbial ratio of F. nucleatum to Bifidobacterium bifidum had a sensitivity of 92.3% and a specificity of 91.1% for detecting CRC, with an AUC of 0.846. Moreover, the diagnostic AUC reached 0.943 when using a combination of F. nucleatum/Bifidobacterium bifidum and F. nucleatum/Faecalibacterium prausnitzii. Overall, these results suggest that fecal quantification of F. nucleatum can contribute to the diagnosis of CRC and be used as a novel non-invasive diagnostic biomarker for CRC.
Application of F. nucleatum detection in saliva for the diagnosis and prognosis of CRC: The microbial community in the oral cavity can be anatomically linked to the microbial community in the colon through saliva. As such, fluctuations in the oral microbiome can indirectly reflect dysregulation of the intestinal microbial community. F. nucleatum is widely distributed in the oral cavity, and whole genome sequencing by Abed et al[61] identified a high level of homology between F. nucleatum in tissues and saliva from the same CRC patient, consistent with the F. nucleatum random primer multiplex PCR results of Komiya et al[62], which suggests that the F. nucleatum found in CRC originated from the oral cavity. Guven et al[63] previously demonstrated that the abundance of F. nucleatum in the saliva of CRC patients was significantly higher than that of the control group, but lacked diagnostic value. Our research group systematically analyzed F. nucleatum in the saliva of CRC patients, patients with proliferative intestinal polyps, adenoma patients and healthy controls using a multiple qPCR method, and observed that the level of F. nucleatum in the saliva of CRC patients was significantly increased. Furthermore, analysis of the receiver operator characteristic curve (ROC) revealed that F. nucleatum DNA was significantly superior to CEA, CA19-9, and their combination in the diagnosis of CRC, with an AUC of 0.841, and a sensitivity and specificity of 71.5% and 82.1%, respectively. Moreover, we identified that F. nucleatum DNA levels were negatively associated with overall survival and disease-free survival in CRC patients, serving as an independent prognostic factor[18]. Overall, salivary F. nucleatum DNA may be a noninvasive diagnostic and prognostic biomarker for CRC patients.
Application of antibody detection of F. nucleatum in serum for the diagnosis of CRC: It is often considered that some cancer-associated microorganisms could act as antigens to induce host-specific antibody production for infection diagnosis and tumor screening. As such, it has been shown that F. nucleatum induces a strong humoral response and the production of a high level of specific antibodies in CRC patients, which enables the possibility for CRC diagnosis based on serological detection of F. nucleatum antibodies[64,65]. Wang et al[64] discovered significantly higher serum levels of anti-F. nucleatum-IgA and anti-F. nucleatum-IgG in CRC patients infected with F. nucleatum compared to healthy controls and patients with benign colon disease. Remarkably, the titer of IgA was significantly higher than that of IgG, suggesting that IgA may have a more sensitive and specific diagnostic value for CRC. Additionally, they detected serum anti-F. nucleatum antibodies by ELISA and evaluated their diagnostic performance. The results revealed that anti-F. nucleatum-IgA had a sensitivity of 36.43% and a specificity of 92.71% for the diagnosis of CRC, and the diagnostic efficacy of anti-F. nucleatum-IgA was further improved by combining it with CEA (sensitivity of 53.10%, specificity of 96.41%, AUC = 0.848). The combined detection of anti-F. nucleatum-IgA with CEA and CA19-9 further improved the diagnostic efficacy for early CRC. Therefore, anti-F. nucleatum-IgA is of great value in mass screening for early CRC.
Application of F. nucleatum detection in tissues for the diagnosis and prognosis of GC: Several studies have reported that the abundance of F. nucleatum in the tumor tissues of GC patients was significantly correlated with prognosis. Boehm et al[66] detected the abundance of F. nucleatum in the tumor tissues and paracancer tissues of GC patients; the positivity rate for F. nucleatum in tumor samples was 28.75%, while in normal mucosal tissue samples it was 23.08%. Further survival analysis indicated a significantly shorter overall survival time in the positive group (524.5 d) compared to the negative group (1287 d). Overall survival was significantly longer in patients with F. nucleatum negative GC (244.5 vs 1229.5 d, P = 0.009) compared with patients with F. nucleatum positive diffuse Laurentian G. Although, no significant difference in survival was observed between intestinal and mixed-type GC. An additional study revealed that compared with healthy individuals, patients with GC had a higher abundance of F. nucleatum in gastric tissue. Furthermore, a higher F. nucleatum abundance in tumor tissues was associated with significantly poorer overall survival, however there was no significant correlation between F. nucleatum abundance in paracarcinoma tissues and overall survival[67]. Moreover, Liu et al[68] identified that the disease course of patients with F. nucleatum colonization in tumor tissues was more likely to be complicated by visceral vein thrombosis and pulmonary embolism, a finding that resulted in speculation that F. nucleatum colonization may induce pro-inflammatory and pro-thrombotic states, thus affecting the prognosis of this patient population.
Application of F. nucleatum detection in saliva for the diagnosis of GC: Considering the application of F. nucleatum in the diagnosis of GC, Chen et al[69] demonstrated that salivary F. nucleatum levels were significantly elevated in GC compared to atrophic gastritis, non-atrophic gastritis, gastric polyp, and healthy controls. ROC analysis indicated that salivary F. nucleatum abundance had diagnostic efficacy, with an AUC of 0.813, a sensitivity of 73.33%, and a specificity of 82.14% under the optimal cut-off value. The diagnostic efficacy was higher than that of traditional serum tumor markers like CEA, CA19-9, CA72-4, ferritin and sialic acid.
Application of F. nucleatum detection in tissues for the diagnosis and prognosis of EC: The abundance of F. nucleatum in ESCC tumor tissues has been revealed to be significantly higher than in adjacent normal tissues[40,70-72]. Research by Li et al[71] reported that in 98 tumor tissue samples, 69.4% (68/98) were positive for F. nucleatum, and the relative abundance of F. nucleatum in tumor tissues was significantly higher than that in paracancer tissues. Moreover, F. nucleatum was significantly enriched in patients with advanced ESCC compared to in those with early ESCC. Furth
Studies have reported that F. nucleatum infected patients were more likely to develop chemotherapy resistance, thus further affecting the survival and prognosis of ESCC patients. As such, the sensitivity of ESCC to cisplatin progressively decreased with the extension of F. nucleatum infection time[40]. Similarly, patients with high intratumoral levels of F. nucleatum exhibited greater resistance to neoadjuvant chemotherapy treatment[72]. It was therefore suggested that F. nucleatum may be a potential target for antibiotic intervention to improve treatment response rate in ESCC patients.
Application of F. nucleatum detection in tissues for the diagnosis and prognosis of PC: Hayashi et al[73] attempted to identify F. nucleatum by detecting lipopolysaccharides using immunohistochemistry and observed that lipopolysaccharides could be detected in tumor tissues of patients with PC, whereas it was not detected in normal pancreatic tissues. Moreover, it has been suggested that F. nucleatum may be associated with poor prognosis in patients with PC. In a univariate Cox regression analysis, mortality was reported to be significantly higher and survival significantly shorter in Fusobacterium positive PC cases compared with Fusobacterium negative cases[74]. Hayashi et al[73] observed that tum
Application of antibody detection of F. nucleatum in serum for the diagnosis of PC: Notably, Alkharaan et al[75] attempted to detect F. nucleatum using serum and saliva antibodies in PC patients. The results suggested that patients with intraductal papillary mucinous neoplasms (IPMN) with highly atypical hyperplasia or progression to invasive cancer had adequate antibody reactivity to oral microorganisms, and that salivary antibody reactivity to F. nucleatum and Fap2 was highly correlated. It was therefore suggested that the immunological detection of pancreatic-associated oral microorganisms may reflect the severity of IPMN, with the potential to contribute to the discovery of novel biomarkers.
In this editorial, we discuss the mechanisms of action of F. nucleatum in the development of four GI malignancies and the significance of F. nucleatum as a biomarker in the diagnosis, prognosis, and risk assessment of GI malignancies. Overall, the current literature strongly supports that F. nucleatum is involved in the development of GI malignancies primarily by releasing virulence factors, inducing DNA damage, modulating ncRNA expression, and remodeling the tumor immune microenvironment. An in-depth study of the oncogenic mechanisms of F. nucleatum may provide new strategies for the targeted treatment and prevention of GI malignancies. Importantly, the detection of F. nucleatum in feces, saliva, or serum can provide valuable new avenues for more efficacious early diagnosis and treatment of GI malignancies, greatly improving the prognosis of patients. Overall, F. nucleatum showed good predictive ability in areas such as diagnosis and prognosis determination, and is a promising diagnostic marker for GI malignancies. However, its detection efficiency is unstable, which severely limits its clinical application, and further studies are needed to improve the detection perfo
Provenance and peer review: Invited article; Externally peer reviewed.
Peer-review model: Single blind
Specialty type: Oncology
Country of origin: China
Peer-review report’s classification
Scientific Quality: Grade C
Novelty: Grade B
Creativity or Innovation: Grade B
Scientific Significance: Grade B
P-Reviewer: Bubnov R, Ukraine S-Editor: Li L L-Editor: A P-Editor: Zhao YQ
| 1. | Arnold M, Abnet CC, Neale RE, Vignat J, Giovannucci EL, McGlynn KA, Bray F. Global Burden of 5 Major Types of Gastrointestinal Cancer. Gastroenterology. 2020;159:335-349.e15. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 857] [Cited by in RCA: 1227] [Article Influence: 245.4] [Reference Citation Analysis (0)] |
| 2. | Islami F, Goding Sauer A, Miller KD, Siegel RL, Fedewa SA, Jacobs EJ, McCullough ML, Patel AV, Ma J, Soerjomataram I, Flanders WD, Brawley OW, Gapstur SM, Jemal A. Proportion and number of cancer cases and deaths attributable to potentially modifiable risk factors in the United States. CA Cancer J Clin. 2018;68:31-54. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 902] [Cited by in RCA: 1000] [Article Influence: 142.9] [Reference Citation Analysis (2)] |
| 3. | Miller KD, Nogueira L, Devasia T, Mariotto AB, Yabroff KR, Jemal A, Kramer J, Siegel RL. Cancer treatment and survivorship statistics, 2022. CA Cancer J Clin. 2022;72:409-436. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 621] [Cited by in RCA: 1551] [Article Influence: 517.0] [Reference Citation Analysis (0)] |
| 4. | Kratzer TB, Jemal A, Miller KD, Nash S, Wiggins C, Redwood D, Smith R, Siegel RL. Cancer statistics for American Indian and Alaska Native individuals, 2022: Including increasing disparities in early onset colorectal cancer. CA Cancer J Clin. 2023;73:120-146. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 65] [Cited by in RCA: 77] [Article Influence: 38.5] [Reference Citation Analysis (0)] |
| 5. | Anderson JC, Rex DK. Performing High-Quality, Safe, Cost-Effective, and Efficient Basic Colonoscopy in 2023: Advice From Two Experts. Am J Gastroenterol. 2023;118:1779-1786. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1] [Cited by in RCA: 6] [Article Influence: 3.0] [Reference Citation Analysis (0)] |
| 6. | Siravegna G, Mussolin B, Venesio T, Marsoni S, Seoane J, Dive C, Papadopoulos N, Kopetz S, Corcoran RB, Siu LL, Bardelli A. How liquid biopsies can change clinical practice in oncology. Ann Oncol. 2019;30:1580-1590. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 216] [Cited by in RCA: 245] [Article Influence: 40.8] [Reference Citation Analysis (0)] |
| 7. | Cho I, Blaser MJ. The human microbiome: at the interface of health and disease. Nat Rev Genet. 2012;13:260-270. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 2636] [Cited by in RCA: 2254] [Article Influence: 173.4] [Reference Citation Analysis (0)] |
| 8. | Song B, Xian W, Sun Y, Gou L, Guo Q, Zhou X, Ren B, Cheng L. Akkermansia muciniphila inhibited the periodontitis caused by Fusobacterium nucleatum. NPJ Biofilms Microbiomes. 2023;9:49. [RCA] [PubMed] [DOI] [Full Text] [Cited by in RCA: 34] [Reference Citation Analysis (0)] |
| 9. | Liu L, Liang L, Liang H, Wang M, Lu B, Xue M, Deng J, Chen Y. Fusobacterium nucleatum Aggravates the Progression of Colitis by Regulating M1 Macrophage Polarization via AKT2 Pathway. Front Immunol. 2019;10:1324. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 38] [Cited by in RCA: 79] [Article Influence: 13.2] [Reference Citation Analysis (0)] |
| 10. | Ou S, Chen H, Wang H, Ye J, Liu H, Tao Y, Ran S, Mu X, Liu F, Zhu S, Luo K, Guan Z, Jin Y, Huang R, Song Y, Liu SL. Fusobacterium nucleatum upregulates MMP7 to promote metastasis-related characteristics of colorectal cancer cell via activating MAPK(JNK)-AP1 axis. J Transl Med. 2023;21:704. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in RCA: 19] [Reference Citation Analysis (0)] |
| 11. | Li Y, Xing S, Chen F, Li Q, Dou S, Huang Y, An J, Liu W, Zhang G. Intracellular Fusobacterium nucleatum infection attenuates antitumor immunity in esophageal squamous cell carcinoma. Nat Commun. 2023;14:5788. [RCA] [PubMed] [DOI] [Full Text] [Cited by in RCA: 29] [Reference Citation Analysis (0)] |
| 12. | Chen L, Zhao R, Kang Z, Cao Z, Liu N, Shen J, Wang C, Pan F, Zhou X, Liu Z, Yang Y, Chen Q. Delivery of short chain fatty acid butyrate to overcome Fusobacterium nucleatum-induced chemoresistance. J Control Release. 2023;363:43-56. [RCA] [PubMed] [DOI] [Full Text] [Cited by in RCA: 24] [Reference Citation Analysis (0)] |
| 13. | Zhang C, Hu A, Li J, Zhang F, Zhong P, Li Y. Combined Non-Invasive Prediction and New Biomarkers of Oral and Fecal Microbiota in Patients With Gastric and Colorectal Cancer. Front Cell Infect Microbiol. 2022;12:830684. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in RCA: 24] [Reference Citation Analysis (0)] |
| 14. | Gao W, Gao X, Zhu L, Gao S, Sun R, Feng Z, Wu D, Liu Z, Zhu R, Jiao N. Multimodal metagenomic analysis reveals microbial single nucleotide variants as superior biomarkers for early detection of colorectal cancer. Gut Microbes. 2023;15:2245562. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 19] [Cited by in RCA: 1] [Article Influence: 0.5] [Reference Citation Analysis (0)] |
| 15. | Wang Y, Zhang Y, Qian Y, Xie YH, Jiang SS, Kang ZR, Chen YX, Chen ZF, Fang JY. Alterations in the oral and gut microbiome of colorectal cancer patients and association with host clinical factors. Int J Cancer. 2021;. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 24] [Cited by in RCA: 36] [Article Influence: 9.0] [Reference Citation Analysis (0)] |
| 16. | Rodriguez RM, Hernandez BY, Menor M, Deng Y, Khadka VS. The landscape of bacterial presence in tumor and adjacent normal tissue across 9 major cancer types using TCGA exome sequencing. Comput Struct Biotechnol J. 2020;18:631-641. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 34] [Cited by in RCA: 46] [Article Influence: 9.2] [Reference Citation Analysis (0)] |
| 17. | Luu K, Ye JY, Lagishetty V, Liang F, Hauer M, Sedighian F, Kwaan MR, Kazanjian KK, Hecht JR, Lin AY, Jacobs JP. Fecal and Tissue Microbiota Are Associated with Tumor T-Cell Infiltration and Mesenteric Lymph Node Involvement in Colorectal Cancer. Nutrients. 2023;15. [RCA] [PubMed] [DOI] [Full Text] [Cited by in RCA: 7] [Reference Citation Analysis (0)] |
| 18. | Zhang X, Zhang Y, Gui X, Zhang Z, Chen W, Zhang X, Wang Y, Zhang M, Shang Z, Xin Y. Salivary Fusobacterium nucleatum serves as a potential biomarker for colorectal cancer. iScience. 2022;25:104203. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in RCA: 21] [Reference Citation Analysis (0)] |
| 19. | Meng Q, Gao Q, Mehrazarin S, Tangwanichgapong K, Wang Y, Huang Y, Pan Y, Robinson S, Liu Z, Zangiabadi A, Lux R, Papapanou PN, Guo XE, Wang H, Berchowitz LE, Han YW. Fusobacterium nucleatum secretes amyloid-like FadA to enhance pathogenicity. EMBO Rep. 2021;22:e52891. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 13] [Cited by in RCA: 91] [Article Influence: 22.8] [Reference Citation Analysis (0)] |
| 20. | Fardini Y, Wang X, Témoin S, Nithianantham S, Lee D, Shoham M, Han YW. Fusobacterium nucleatum adhesin FadA binds vascular endothelial cadherin and alters endothelial integrity. Mol Microbiol. 2011;82:1468-1480. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 153] [Cited by in RCA: 208] [Article Influence: 14.9] [Reference Citation Analysis (0)] |
| 21. | Rubinstein MR, Wang X, Liu W, Hao Y, Cai G, Han YW. Fusobacterium nucleatum promotes colorectal carcinogenesis by modulating E-cadherin/β-catenin signaling via its FadA adhesin. Cell Host Microbe. 2013;14:195-206. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1511] [Cited by in RCA: 1662] [Article Influence: 138.5] [Reference Citation Analysis (1)] |
| 22. | Abed J, Emgård JE, Zamir G, Faroja M, Almogy G, Grenov A, Sol A, Naor R, Pikarsky E, Atlan KA, Mellul A, Chaushu S, Manson AL, Earl AM, Ou N, Brennan CA, Garrett WS, Bachrach G. Fap2 Mediates Fusobacterium nucleatum Colorectal Adenocarcinoma Enrichment by Binding to Tumor-Expressed Gal-GalNAc. Cell Host Microbe. 2016;20:215-225. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 326] [Cited by in RCA: 579] [Article Influence: 72.4] [Reference Citation Analysis (0)] |
| 23. | Wu Y, Guo S, Chen F, Li Y, Huang Y, Liu W, Zhang G. Fn-Dps, a novel virulence factor of Fusobacterium nucleatum, disrupts erythrocytes and promotes metastasis in colorectal cancer. PLoS Pathog. 2023;19:e1011096. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in RCA: 26] [Reference Citation Analysis (0)] |
| 24. | Jiang SS, Xie YL, Xiao XY, Kang ZR, Lin XL, Zhang L, Li CS, Qian Y, Xu PP, Leng XX, Wang LW, Tu SP, Zhong M, Zhao G, Chen JX, Wang Z, Liu Q, Hong J, Chen HY, Chen YX, Fang JY. Fusobacterium nucleatum-derived succinic acid induces tumor resistance to immunotherapy in colorectal cancer. Cell Host Microbe. 2023;31:781-797.e9. [RCA] [PubMed] [DOI] [Full Text] [Cited by in RCA: 155] [Reference Citation Analysis (0)] |
| 25. | Zhang JW, Zhang D, Yin HS, Zhang H, Hong KQ, Yuan JP, Yu BP. Fusobacterium nucleatum promotes esophageal squamous cell carcinoma progression and chemoresistance by enhancing the secretion of chemotherapy-induced senescence-associated secretory phenotype via activation of DNA damage response pathway. Gut Microbes. 2023;15:2197836. [RCA] [PubMed] [DOI] [Full Text] [Cited by in RCA: 34] [Reference Citation Analysis (0)] |
| 26. | Guo P, Tian Z, Kong X, Yang L, Shan X, Dong B, Ding X, Jing X, Jiang C, Jiang N, Yu Y. FadA promotes DNA damage and progression of Fusobacterium nucleatum-induced colorectal cancer through up-regulation of chk2. J Exp Clin Cancer Res. 2020;39:202. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 21] [Cited by in RCA: 105] [Article Influence: 21.0] [Reference Citation Analysis (0)] |
| 27. | Okita Y, Koi M, Takeda K, Ross R, Mukherjee B, Koeppe E, Stoffel EM, Galanko JA, McCoy AN, Keku TO, Okugawa Y, Kitajima T, Toiyama Y, Martens E, Carethers JM. Fusobacterium nucleatum infection correlates with two types of microsatellite alterations in colorectal cancer and triggers DNA damage. Gut Pathog. 2020;12:46. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 8] [Cited by in RCA: 31] [Article Influence: 6.2] [Reference Citation Analysis (0)] |
| 28. | Sayed IM, Chakraborty A, Abd El-Hafeez AA, Sharma A, Sahan AZ, Huang WJM, Sahoo D, Ghosh P, Hazra TK, Das S. The DNA Glycosylase NEIL2 Suppresses Fusobacterium-Infection-Induced Inflammation and DNA Damage in Colonic Epithelial Cells. Cells. 2020;9. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 20] [Cited by in RCA: 38] [Article Influence: 7.6] [Reference Citation Analysis (0)] |
| 29. | Proença MA, Biselli JM, Succi M, Severino FE, Berardinelli GN, Caetano A, Reis RM, Hughes DJ, Silva AE. Relationship between Fusobacterium nucleatum, inflammatory mediators and microRNAs in colorectal carcinogenesis. World J Gastroenterol. 2018;24:5351-5365. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in CrossRef: 62] [Cited by in RCA: 86] [Article Influence: 12.3] [Reference Citation Analysis (0)] |
| 30. | Yang Y, Weng W, Peng J, Hong L, Yang L, Toiyama Y, Gao R, Liu M, Yin M, Pan C, Li H, Guo B, Zhu Q, Wei Q, Moyer MP, Wang P, Cai S, Goel A, Qin H, Ma Y. Fusobacterium nucleatum Increases Proliferation of Colorectal Cancer Cells and Tumor Development in Mice by Activating Toll-Like Receptor 4 Signaling to Nuclear Factor-κB, and Up-regulating Expression of MicroRNA-21. Gastroenterology. 2017;152:851-866.e24. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 455] [Cited by in RCA: 713] [Article Influence: 89.1] [Reference Citation Analysis (1)] |
| 31. | Xu C, Fan L, Lin Y, Shen W, Qi Y, Zhang Y, Chen Z, Wang L, Long Y, Hou T, Si J, Chen S. Fusobacterium nucleatum promotes colorectal cancer metastasis through miR-1322/CCL20 axis and M2 polarization. Gut Microbes. 2021;13:1980347. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 32] [Cited by in RCA: 180] [Article Influence: 60.0] [Reference Citation Analysis (0)] |
| 32. | Tang B, Lu X, Tong Y, Feng Y, Mao Y, Dun G, Li J, Xu Q, Tang J, Zhang T, Deng L, He X, Lan Y, Luo H, Zeng L, Xiang Y, Li Q, Zeng D, Mao X. MicroRNA-31 induced by Fusobacterium nucleatum infection promotes colorectal cancer tumorigenesis. iScience. 2023;26:106770. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in RCA: 15] [Reference Citation Analysis (0)] |
| 33. | Hong J, Guo F, Lu SY, Shen C, Ma D, Zhang X, Xie Y, Yan T, Yu T, Sun T, Qian Y, Zhong M, Chen J, Peng Y, Wang C, Zhou X, Liu J, Liu Q, Ma X, Chen YX, Chen H, Fang JY. F. nucleatum targets lncRNA ENO1-IT1 to promote glycolysis and oncogenesis in colorectal cancer. Gut. 2021;70:2123-2137. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 65] [Cited by in RCA: 172] [Article Influence: 43.0] [Reference Citation Analysis (0)] |
| 34. | Lu X, Xu Q, Tong Y, Zhang Z, Dun G, Feng Y, Tang J, Han D, Mao Y, Deng L, He X, Li Q, Xiang Y, Wang F, Zeng D, Tang B, Mao X. Long non-coding RNA EVADR induced by Fusobacterium nucleatum infection promotes colorectal cancer metastasis. Cell Rep. 2022;40:111127. [RCA] [PubMed] [DOI] [Full Text] [Cited by in RCA: 40] [Reference Citation Analysis (0)] |
| 35. | Xin Y, Li X, Zhang M, Shang Z, Luo Z, Wang Y, Gui X, Liu Q, Li T, Zeng S, Schiöth HB, Zhang X, Zhang Y. Fusobacterium nucleatum-induced exosomal HOTTIP promotes gastric cancer progression through the microRNA-885-3p/EphB2 axis. Cancer Sci. 2023;114:2360-2374. [RCA] [PubMed] [DOI] [Full Text] [Cited by in RCA: 25] [Reference Citation Analysis (0)] |
| 36. | Zhang M, Wang Y, Yu L, Zhang Y, Shang Z, Xin Y, Li X, Ning N, Zhang X. Fusobacterium nucleatum promotes colorectal cancer metastasis by excretion of miR-122-5p from cells via exosomes. iScience. 2023;26:107686. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in RCA: 12] [Reference Citation Analysis (0)] |
| 37. | Jewett A, Hume WR, Le H, Huynh TN, Han YW, Cheng G, Shi W. Induction of apoptotic cell death in peripheral blood mononuclear and polymorphonuclear cells by an oral bacterium, Fusobacterium nucleatum. Infect Immun. 2000;68:1893-1898. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 81] [Cited by in RCA: 77] [Article Influence: 3.1] [Reference Citation Analysis (0)] |
| 38. | Gur C, Maalouf N, Shhadeh A, Berhani O, Singer BB, Bachrach G, Mandelboim O. Fusobacterium nucleatum supresses anti-tumor immunity by activating CEACAM1. Oncoimmunology. 2019;8:e1581531. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 53] [Cited by in RCA: 105] [Article Influence: 17.5] [Reference Citation Analysis (0)] |
| 39. | Gur C, Ibrahim Y, Isaacson B, Yamin R, Abed J, Gamliel M, Enk J, Bar-On Y, Stanietsky-Kaynan N, Coppenhagen-Glazer S, Shussman N, Almogy G, Cuapio A, Hofer E, Mevorach D, Tabib A, Ortenberg R, Markel G, Miklić K, Jonjic S, Brennan CA, Garrett WS, Bachrach G, Mandelboim O. Binding of the Fap2 protein of Fusobacterium nucleatum to human inhibitory receptor TIGIT protects tumors from immune cell attack. Immunity. 2015;42:344-355. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1015] [Cited by in RCA: 975] [Article Influence: 97.5] [Reference Citation Analysis (0)] |
| 40. | Liang M, Liu Y, Zhang Z, Yang H, Dai N, Zhang N, Sun W, Guo Y, Kong J, Wang X, Wang M, Zhou F. Fusobacterium nucleatum induces MDSCs enrichment via activation the NLRP3 inflammosome in ESCC cells, leading to cisplatin resistance. Ann Med. 2022;54:989-1003. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 3] [Cited by in RCA: 30] [Article Influence: 10.0] [Reference Citation Analysis (0)] |
| 41. | Tran HNH, Thu TNH, Nguyen PH, Vo CN, Doan KV, Nguyen Ngoc Minh C, Nguyen NT, Ta VND, Vu KA, Hua TD, Nguyen TNT, Van TT, Pham Duc T, Duong BL, Nguyen PM, Hoang VC, Pham DT, Thwaites GE, Hall LJ, Slade DJ, Baker S, Tran VH, Chung The H. Tumour microbiomes and Fusobacterium genomics in Vietnamese colorectal cancer patients. NPJ Biofilms Microbiomes. 2022;8:87. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 11] [Cited by in RCA: 16] [Article Influence: 5.3] [Reference Citation Analysis (0)] |
| 42. | Gong D, Adomako-Bonsu AG, Wang M, Li J. Three specific gut bacteria in the occurrence and development of colorectal cancer: a concerted effort. PeerJ. 2023;11:e15777. [RCA] [PubMed] [DOI] [Full Text] [Cited by in RCA: 7] [Reference Citation Analysis (0)] |
| 43. | Wu N, Feng YQ, Lyu N, Wang D, Yu WD, Hu YF. Fusobacterium nucleatum promotes colon cancer progression by changing the mucosal microbiota and colon transcriptome in a mouse model. World J Gastroenterol. 2022;28:1981-1995. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in CrossRef: 8] [Cited by in RCA: 14] [Article Influence: 4.7] [Reference Citation Analysis (1)] |
| 44. | Shimomura Y, Zha L, Komukai S, Narii N, Sobue T, Kitamura T, Shiba S, Mizutani S, Yamada T, Sawada N, Yachida S. Mediation effect of intestinal microbiota on the relationship between fiber intake and colorectal cancer. Int J Cancer. 2023;152:1752-1762. [RCA] [PubMed] [DOI] [Full Text] [Cited by in RCA: 10] [Reference Citation Analysis (0)] |
| 45. | Marchesi JR, Dutilh BE, Hall N, Peters WH, Roelofs R, Boleij A, Tjalsma H. Towards the human colorectal cancer microbiome. PLoS One. 2011;6:e20447. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 442] [Cited by in RCA: 429] [Article Influence: 30.6] [Reference Citation Analysis (0)] |
| 46. | Kostic AD, Gevers D, Pedamallu CS, Michaud M, Duke F, Earl AM, Ojesina AI, Jung J, Bass AJ, Tabernero J, Baselga J, Liu C, Shivdasani RA, Ogino S, Birren BW, Huttenhower C, Garrett WS, Meyerson M. Genomic analysis identifies association of Fusobacterium with colorectal carcinoma. Genome Res. 2012;22:292-298. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1199] [Cited by in RCA: 1498] [Article Influence: 107.0] [Reference Citation Analysis (0)] |
| 47. | Castellarin M, Warren RL, Freeman JD, Dreolini L, Krzywinski M, Strauss J, Barnes R, Watson P, Allen-Vercoe E, Moore RA, Holt RA. Fusobacterium nucleatum infection is prevalent in human colorectal carcinoma. Genome Res. 2012;22:299-306. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1164] [Cited by in RCA: 1495] [Article Influence: 106.8] [Reference Citation Analysis (0)] |
| 48. | Li YY, Ge QX, Cao J, Zhou YJ, Du YL, Shen B, Wan YJ, Nie YQ. Association of Fusobacterium nucleatum infection with colorectal cancer in Chinese patients. World J Gastroenterol. 2016;22:3227-3233. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in CrossRef: 141] [Cited by in RCA: 142] [Article Influence: 15.8] [Reference Citation Analysis (0)] |
| 49. | Eisele Y, Mallea PM, Gigic B, Stephens WZ, Warby CA, Buhrke K, Lin T, Boehm J, Schrotz-King P, Hardikar S, Huang LC, Pickron TB, Scaife CL, Viskochil R, Koelsch T, Peoples AR, Pletneva MA, Bronner M, Schneider M, Ulrich AB, Swanson EA, Toriola AT, Shibata D, Li CI, Siegel EM, Figueiredo J, Janssen KP, Hauner H, Round J, Ulrich CM, Holowatyj AN, Ose J. Fusobacterium nucleatum and Clinicopathologic Features of Colorectal Cancer: Results From the ColoCare Study. Clin Colorectal Cancer. 2021;20:e165-e172. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 5] [Cited by in RCA: 4] [Article Influence: 1.0] [Reference Citation Analysis (0)] |
| 50. | Mima K, Nishihara R, Qian ZR, Cao Y, Sukawa Y, Nowak JA, Yang J, Dou R, Masugi Y, Song M, Kostic AD, Giannakis M, Bullman S, Milner DA, Baba H, Giovannucci EL, Garraway LA, Freeman GJ, Dranoff G, Garrett WS, Huttenhower C, Meyerson M, Meyerhardt JA, Chan AT, Fuchs CS, Ogino S. Fusobacterium nucleatum in colorectal carcinoma tissue and patient prognosis. Gut. 2016;65:1973-1980. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 530] [Cited by in RCA: 754] [Article Influence: 83.8] [Reference Citation Analysis (0)] |
| 51. | Huangfu SC, Zhang WB, Zhang HR, Li Y, Zhang YR, Nie JL, Chu XD, Chen CS, Jiang HP, Pan JH. Clinicopathological and prognostic significance of Fusobacterium nucleatum infection in colorectal cancer: a meta-analysis. J Cancer. 2021;12:1583-1591. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 16] [Cited by in RCA: 15] [Article Influence: 3.8] [Reference Citation Analysis (0)] |
| 52. | Yamaoka Y, Suehiro Y, Hashimoto S, Hoshida T, Fujimoto M, Watanabe M, Imanaga D, Sakai K, Matsumoto T, Nishioka M, Takami T, Suzuki N, Hazama S, Nagano H, Sakaida I, Yamasaki T. Fusobacterium nucleatum as a prognostic marker of colorectal cancer in a Japanese population. J Gastroenterol. 2018;53:517-524. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 87] [Cited by in RCA: 122] [Article Influence: 17.4] [Reference Citation Analysis (0)] |
| 53. | Flanagan L, Schmid J, Ebert M, Soucek P, Kunicka T, Liska V, Bruha J, Neary P, Dezeeuw N, Tommasino M, Jenab M, Prehn JH, Hughes DJ. Fusobacterium nucleatum associates with stages of colorectal neoplasia development, colorectal cancer and disease outcome. Eur J Clin Microbiol Infect Dis. 2014;33:1381-1390. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 285] [Cited by in RCA: 384] [Article Influence: 34.9] [Reference Citation Analysis (0)] |
| 54. | Lee DW, Han SW, Kang JK, Bae JM, Kim HP, Won JK, Jeong SY, Park KJ, Kang GH, Kim TY. Association Between Fusobacterium nucleatum, Pathway Mutation, and Patient Prognosis in Colorectal Cancer. Ann Surg Oncol. 2018;25:3389-3395. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 45] [Cited by in RCA: 73] [Article Influence: 10.4] [Reference Citation Analysis (0)] |
| 55. | Tunsjø HS, Gundersen G, Rangnes F, Noone JC, Endres A, Bemanian V. Detection of Fusobacterium nucleatum in stool and colonic tissues from Norwegian colorectal cancer patients. Eur J Clin Microbiol Infect Dis. 2019;38:1367-1376. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 18] [Cited by in RCA: 30] [Article Influence: 5.0] [Reference Citation Analysis (0)] |
| 56. | Liang Q, Chiu J, Chen Y, Huang Y, Higashimori A, Fang J, Brim H, Ashktorab H, Ng SC, Ng SSM, Zheng S, Chan FKL, Sung JJY, Yu J. Fecal Bacteria Act as Novel Biomarkers for Noninvasive Diagnosis of Colorectal Cancer. Clin Cancer Res. 2017;23:2061-2070. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 165] [Cited by in RCA: 229] [Article Influence: 25.4] [Reference Citation Analysis (0)] |
| 57. | Peng BJ, Cao CY, Li W, Zhou YJ, Zhang Y, Nie YQ, Cao YW, Li YY. Diagnostic Performance of Intestinal Fusobacterium nucleatum in Colorectal Cancer: A Meta-Analysis. Chin Med J (Engl). 2018;131:1349-1356. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 22] [Cited by in RCA: 26] [Article Influence: 3.7] [Reference Citation Analysis (0)] |
| 58. | Shaukat A, Levin TR. Current and future colorectal cancer screening strategies. Nat Rev Gastroenterol Hepatol. 2022;19:521-531. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 8] [Cited by in RCA: 239] [Article Influence: 79.7] [Reference Citation Analysis (1)] |
| 59. | Wong SH, Kwong TNY, Chow TC, Luk AKC, Dai RZW, Nakatsu G, Lam TYT, Zhang L, Wu JCY, Chan FKL, Ng SSM, Wong MCS, Ng SC, Wu WKK, Yu J, Sung JJY. Quantitation of faecal Fusobacterium improves faecal immunochemical test in detecting advanced colorectal neoplasia. Gut. 2017;66:1441-1448. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 157] [Cited by in RCA: 224] [Article Influence: 28.0] [Reference Citation Analysis (0)] |
| 60. | Guo S, Li L, Xu B, Li M, Zeng Q, Xiao H, Xue Y, Wu Y, Wang Y, Liu W, Zhang G. A Simple and Novel Fecal Biomarker for Colorectal Cancer: Ratio of Fusobacterium Nucleatum to Probiotics Populations, Based on Their Antagonistic Effect. Clin Chem. 2018;64:1327-1337. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 67] [Cited by in RCA: 110] [Article Influence: 15.7] [Reference Citation Analysis (0)] |
| 61. | Abed J, Maalouf N, Manson AL, Earl AM, Parhi L, Emgård JEM, Klutstein M, Tayeb S, Almogy G, Atlan KA, Chaushu S, Israeli E, Mandelboim O, Garrett WS, Bachrach G. Colon Cancer-Associated Fusobacterium nucleatum May Originate From the Oral Cavity and Reach Colon Tumors via the Circulatory System. Front Cell Infect Microbiol. 2020;10:400. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 56] [Cited by in RCA: 153] [Article Influence: 30.6] [Reference Citation Analysis (0)] |
| 62. | Komiya Y, Shimomura Y, Higurashi T, Sugi Y, Arimoto J, Umezawa S, Uchiyama S, Matsumoto M, Nakajima A. Patients with colorectal cancer have identical strains of Fusobacterium nucleatum in their colorectal cancer and oral cavity. Gut. 2019;68:1335-1337. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 124] [Cited by in RCA: 206] [Article Influence: 34.3] [Reference Citation Analysis (0)] |
| 63. | Guven DC, Dizdar O, Alp A, Akdoğan Kittana FN, Karakoc D, Hamaloglu E, Lacin S, Karakas Y, Kilickap S, Hayran M, Yalcin S. Analysis of Fusobacterium nucleatum and Streptococcus gallolyticus in saliva of colorectal cancer patients. Biomark Med. 2019;13:725-735. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 26] [Cited by in RCA: 28] [Article Influence: 4.7] [Reference Citation Analysis (0)] |
| 64. | Wang HF, Li LF, Guo SH, Zeng QY, Ning F, Liu WL, Zhang G. Evaluation of antibody level against Fusobacterium nucleatum in the serological diagnosis of colorectal cancer. Sci Rep. 2016;6:33440. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 60] [Cited by in RCA: 82] [Article Influence: 9.1] [Reference Citation Analysis (0)] |
| 65. | Kurt M, Yumuk Z. Diagnostic accuracy of Fusobacterium nucleatum IgA and IgG ELISA test in colorectal cancer. Sci Rep. 2021;11:1608. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 11] [Cited by in RCA: 18] [Article Influence: 4.5] [Reference Citation Analysis (0)] |
| 66. | Boehm ET, Thon C, Kupcinskas J, Steponaitiene R, Skieceviciene J, Canbay A, Malfertheiner P, Link A. Fusobacterium nucleatum is associated with worse prognosis in Lauren's diffuse type gastric cancer patients. Sci Rep. 2020;10:16240. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 76] [Cited by in RCA: 66] [Article Influence: 13.2] [Reference Citation Analysis (0)] |
| 67. | Lehr K, Nikitina D, Vilchez-Vargas R, Steponaitiene R, Thon C, Skieceviciene J, Schanze D, Zenker M, Malfertheiner P, Kupcinskas J, Link A. Microbial composition of tumorous and adjacent gastric tissue is associated with prognosis of gastric cancer. Sci Rep. 2023;13:4640. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 27] [Cited by in RCA: 29] [Article Influence: 14.5] [Reference Citation Analysis (0)] |
| 68. | Liu C, Yang Z, Tang X, Zhao F, He M, Liu C, Zhou D, Wang L, Gu B, Yuan Y, Chen X. Colonization of Fusobacterium nucleatum is an independent predictor of poor prognosis in gastric cancer patients with venous thromboembolism: a retrospective cohort study. Thromb J. 2023;21:2. [RCA] [PubMed] [DOI] [Full Text] [Reference Citation Analysis (0)] |
| 69. | Chen WD, Zhang X, Zhang MJ, Zhang YP, Shang ZQ, Xin YW, Zhang Y. Salivary Fusobacterium nucleatum serves as a potential diagnostic biomarker for gastric cancer. World J Gastroenterol. 2022;28:4120-4132. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in CrossRef: 4] [Cited by in RCA: 19] [Article Influence: 6.3] [Reference Citation Analysis (1)] |
| 70. | Ding N, Cheng Y, Liu H, Wu Y, Weng Y, Cui H, Cheng C, Zhang W, Cui Y. Fusobacterium nucleatum Infection Induces Malignant Proliferation of Esophageal Squamous Cell Carcinoma Cell by Putrescine Production. Microbiol Spectr. 2023;11:e0275922. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 10] [Cited by in RCA: 14] [Article Influence: 7.0] [Reference Citation Analysis (0)] |
| 71. | Li Z, Shi C, Zheng J, Guo Y, Fan T, Zhao H, Jian D, Cheng X, Tang H, Ma J. Fusobacterium nucleatum predicts a high risk of metastasis for esophageal squamous cell carcinoma. BMC Microbiol. 2021;21:301. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 3] [Cited by in RCA: 48] [Article Influence: 12.0] [Reference Citation Analysis (0)] |
| 72. | Yamamura K, Izumi D, Kandimalla R, Sonohara F, Baba Y, Yoshida N, Kodera Y, Baba H, Goel A. Intratumoral Fusobacterium Nucleatum Levels Predict Therapeutic Response to Neoadjuvant Chemotherapy in Esophageal Squamous Cell Carcinoma. Clin Cancer Res. 2019;25:6170-6179. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 137] [Cited by in RCA: 141] [Article Influence: 23.5] [Reference Citation Analysis (0)] |
| 73. | Hayashi M, Ikenaga N, Nakata K, Luo H, Zhong P, Date S, Oyama K, Higashijima N, Kubo A, Iwamoto C, Torata N, Abe T, Yamada Y, Ohuchida K, Oda Y, Nakamura M. Intratumor Fusobacterium nucleatum promotes the progression of pancreatic cancer via the CXCL1-CXCR2 axis. Cancer Sci. 2023;114:3666-3678. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in RCA: 29] [Reference Citation Analysis (0)] |
| 74. | Mitsuhashi K, Nosho K, Sukawa Y, Matsunaga Y, Ito M, Kurihara H, Kanno S, Igarashi H, Naito T, Adachi Y, Tachibana M, Tanuma T, Maguchi H, Shinohara T, Hasegawa T, Imamura M, Kimura Y, Hirata K, Maruyama R, Suzuki H, Imai K, Yamamoto H, Shinomura Y. Association of Fusobacterium species in pancreatic cancer tissues with molecular features and prognosis. Oncotarget. 2015;6:7209-7220. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 301] [Cited by in RCA: 288] [Article Influence: 28.8] [Reference Citation Analysis (0)] |
| 75. | Alkharaan H, Lu L, Gabarrini G, Halimi A, Ateeb Z, Sobkowiak MJ, Davanian H, Fernández Moro C, Jansson L, Del Chiaro M, Özenci V, Sällberg Chen M. Circulating and Salivary Antibodies to Fusobacterium nucleatum Are Associated With Cystic Pancreatic Neoplasm Malignancy. Front Immunol. 2020;11:2003. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 20] [Cited by in RCA: 25] [Article Influence: 5.0] [Reference Citation Analysis (0)] |
| 76. | Nomoto D, Baba Y, Liu Y, Tsutsuki H, Okadome K, Harada K, Ishimoto T, Iwatsuki M, Iwagami S, Miyamoto Y, Yoshida N, Watanabe M, Moroishi T, Komohara Y, Sawa T, Baba H. Fusobacterium nucleatum promotes esophageal squamous cell carcinoma progression via the NOD1/RIPK2/NF-κB pathway. Cancer Lett. 2022;530:59-67. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 7] [Cited by in RCA: 67] [Article Influence: 22.3] [Reference Citation Analysis (0)] |
| 77. | Xu Q, Lu X, Li J, Feng Y, Tang J, Zhang T, Mao Y, Lan Y, Luo H, Zeng L, Xiang Y, Hu L, Zhang Y, Li Q, Deng L, He X, Tang B, Mao X, Zeng D. Fusobacterium nucleatum induces excess methyltransferase-like 3-mediated microRNA-4717-3p maturation to promote colorectal cancer cell proliferation. Cancer Sci. 2022;113:3787-3800. [RCA] [PubMed] [DOI] [Full Text] [Cited by in RCA: 19] [Reference Citation Analysis (0)] |
| 78. | Feng YY, Zeng DZ, Tong YN, Lu XX, Dun GD, Tang B, Zhang ZJ, Ye XL, Li Q, Xie JP, Mao XH. Alteration of microRNA-4474/4717 expression and CREB-binding protein in human colorectal cancer tissues infected with Fusobacterium nucleatum. PLoS One. 2019;14:e0215088. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 17] [Cited by in RCA: 31] [Article Influence: 5.2] [Reference Citation Analysis (0)] |
| 79. | Kostic AD, Chun E, Robertson L, Glickman JN, Gallini CA, Michaud M, Clancy TE, Chung DC, Lochhead P, Hold GL, El-Omar EM, Brenner D, Fuchs CS, Meyerson M, Garrett WS. Fusobacterium nucleatum potentiates intestinal tumorigenesis and modulates the tumor-immune microenvironment. Cell Host Microbe. 2013;14:207-215. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1659] [Cited by in RCA: 1912] [Article Influence: 159.3] [Reference Citation Analysis (0)] |
| 80. | Wang X, Liu Y, Lu Y, Chen S, Xing Y, Yang H, Wang X, Zhang Y, Pan T, Li J, Wang M, Zhang N, Liang M, Zhou F. Clinical impact of Fn-induced high expression of KIR2DL1 in CD8 T lymphocytes in oesophageal squamous cell carcinoma. Ann Med. 2022;54:51-62. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 4] [Cited by in RCA: 9] [Article Influence: 3.0] [Reference Citation Analysis (0)] |
| 81. | Udayasuryan B, Ahmad RN, Nguyen TTD, Umaña A, Monét Roberts L, Sobol P, Jones SD, Munson JM, Slade DJ, Verbridge SS. Fusobacterium nucleatum induces proliferation and migration in pancreatic cancer cells through host autocrine and paracrine signaling. Sci Signal. 2022;15:eabn4948. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1] [Cited by in RCA: 77] [Article Influence: 25.7] [Reference Citation Analysis (0)] |
| 82. | Wu J, Wang Y, Jiang Z. Immune induction identified by TMT proteomics analysis in Fusobacterium nucleatum autoinducer-2 treated macrophages. Expert Rev Proteomics. 2020;17:175-185. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 7] [Cited by in RCA: 19] [Article Influence: 3.8] [Reference Citation Analysis (0)] |
| 83. | Engevik MA, Danhof HA, Auchtung J, Endres BT, Ruan W, Bassères E, Engevik AC, Wu Q, Nicholson M, Luna RA, Garey KW, Crawford SE, Estes MK, Lux R, Yacyshyn MB, Yacyshyn B, Savidge T, Britton RA, Versalovic J. Fusobacteriumnucleatum Adheres to Clostridioides difficile via the RadD Adhesin to Enhance Biofilm Formation in Intestinal Mucus. Gastroenterology. 2021;160:1301-1314.e8. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 24] [Cited by in RCA: 74] [Article Influence: 18.5] [Reference Citation Analysis (0)] |
| 84. | Tunsjø HS, Gundersen G, Rangnes F, Noone JC, Endres A, Bemanian V. Correction to: Detection of Fusobacterium nucleatum in stool and colonic tissues from Norwegian colorectal cancer patients. Eur J Clin Microbiol Infect Dis. 2020;39:213. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 3] [Cited by in RCA: 6] [Article Influence: 1.0] [Reference Citation Analysis (0)] |
| 85. | Datorre JG, de Carvalho AC, Dos Reis MB, Dos Reis M, Matsushita M, Santos F, Guimarães DP, Reis RM. Accuracy and Clinical Relevance of Intra-Tumoral Fusobacterium nucleatum Detection in Formalin-Fixed Paraffin-Embedded (FFPE) Tissue by Droplet Digital PCR (ddPCR) in Colorectal Cancer. Diagnostics (Basel). 2022;12. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 5] [Reference Citation Analysis (0)] |
| 86. | Dregelies T, Haumaier F, Sterlacci W, Backert S, Vieth M. Detection of Fusobacterium nucleatum in Patients with Colitis-Associated Colorectal Cancer. Curr Microbiol. 2023;80:293. [RCA] [PubMed] [DOI] [Full Text] [Reference Citation Analysis (0)] |
| 87. | Bi D, Zhu Y, Gao Y, Li H, Zhu X, Wei R, Xie R, Wei Q, Qin H. A newly developed PCR-based method revealed distinct Fusobacterium nucleatum subspecies infection patterns in colorectal cancer. Microb Biotechnol. 2021;14:2176-2186. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 22] [Cited by in RCA: 19] [Article Influence: 4.8] [Reference Citation Analysis (0)] |
| 88. | Nascimento Araujo CD, Amorim AT, Barbosa MS, Alexandre JCPL, Campos GB, Macedo CL, Marques LM, Timenetsky J. Evaluating the presence of Mycoplasma hyorhinis, Fusobacterium nucleatum, and Helicobacter pylori in biopsies of patients with gastric cancer. Infect Agent Cancer. 2021;16:70. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 3] [Cited by in RCA: 19] [Article Influence: 4.8] [Reference Citation Analysis (0)] |
| 89. | Liu C, Zhang H, Li T, Jiang Z, Yuan Y, Chen X. Fusobacterium nucleatum Promotes Megakaryocyte Maturation in Patients with Gastric Cancer via Inducing the Production of Extracellular Vesicles Containing 14-3-3ε. Infect Immun. 2023;91:e0010223. [RCA] [PubMed] [DOI] [Full Text] [Reference Citation Analysis (0)] |