Published online May 15, 2024. doi: 10.4251/wjgo.v16.i5.2038
Peer-review started: November 20, 2023
First decision: December 29, 2023
Revised: January 9, 2024
Accepted: March 11, 2024
Article in press: March 11, 2024
Published online: May 15, 2024
Processing time: 170 Days and 22.8 Hours
Heterogeneous ribonucleoprotein A1 (hnRNPA1) has been reported to enhance the Warburg effect and promote colon cancer (CC) cell proliferation, but the role and mechanism of the miR-490-3p/hnRNPA1-b/PKM2 axis in CC have not yet been elucidated.
To investigate the role and mechanism of a novel miR-490-3p/hnRNPA1-b/PKM2 axis in enhancing the Warburg effect and promoting CC cell proliferation through the PI3K/AKT pathway.
Paraffin-embedded pathological sections from 220 CC patients were collected and subjected to immunohistochemical analysis to determine the expression of hnRNPA1-b. The relationship between the expression values and the clinicopathological features of the patients was investigated. Differences in mRNA expression were analyzed using quantitative real-time polymerase chain reaction, while differences in protein expression were analyzed using western blot. Cell proliferation was evaluated using the cell counting kit-8 and 5-ethynyl-2’-deoxyuridine assays, and cell cycle and apoptosis were detected using flow cytometric assays. The targeted binding of miR-490-3p to hnRNPA1-b was validated using a dual luciferase reporter assay. The Warburg effect was evaluated by glucose uptake and lactic acid production assays.
The expression of hnRNPA1-b was significantly increased in CC tissues and cells compared to normal controls (P < 0.05). Immunohistochemical results demonstrated significant variations in the expression of the hnRNPA1-b antigen in different stages of CC, including stage I, II-III, and IV. Furthermore, the clinicopathologic characterization revealed a significant correlation between hnRNPA1-b expression and clinical stage as well as T classification. HnRNPA1-b was found to enhance the Warburg effect through the PI3K/AKT pathway, thereby promoting proliferation of HCT116 and SW620 cells. However, the proliferation of HCT116 and SW620 cells was inhibited when miR-490-3p targeted and bound to hnRNPA1-b, effectively blocking the Warburg effect.
These findings suggest that the novel miR-490-3p/hnRNPA1-b/PKM2 axis could provide a new strategy for the diagnosis and treatment of CC.
Core Tip: Currently, there are no ideal early diagnostic markers or specific target drugs available for the treatment of colon cancer (CC). This study confirmed the role of heterogeneous ribonucleoprotein A1 (hnRNPA1)’s selective shear monomer, hnRNPA1-b, in promoting the proliferation of CC cells and elucidated the mechanism of action of the miR-490-3p/hnRNPA1-b/PKM2 axis in modulating the proliferation of CC cells by remodeling the Warburg effect through the PI3K/AKT pathway. These findings could provide a new strategy for the diagnosis and treatment of CC.
- Citation: Wan XH, Jin GB, Yang Q, Hu JL, Liu ZL, Rao J, Wen C, Li PL, Yang XM, Huang B, Wang XZ. Novel miR-490-3p/hnRNPA1-b/PKM2 axis mediates the Warburg effect and proliferation of colon cancer cells via the PI3K/AKT pathway. World J Gastrointest Oncol 2024; 16(5): 2038-2059
- URL: https://www.wjgnet.com/1948-5204/full/v16/i5/2038.htm
- DOI: https://dx.doi.org/10.4251/wjgo.v16.i5.2038
Colon cancer (CC) is the leading malignant tumor worldwide in terms of both incidence and mortality, and there is a growing trend of younger individuals being affected[1,2]. While the 5-year survival rate for early-stage CC is approximately 90%, it drops significantly to about 14% in cases of metastatic CC, with invasive metastasis being the primary cause of death[3,4]. Unfortunately, around 60% of CC patients will eventually develop metastases[5]. Currently, there are no ideal early diagnostic markers or specific target drugs available for the treatment of CC. Therefore, it is of great significance to explore effective early diagnostic biomarkers and identify new therapeutic targets for this disease.
The phenomenon of cancer cells favoring the glycolytic metabolic pathway even under well-oxygenated conditions is known as the Warburg effect and is now considered a common hallmark of cancer. Our metabolomic study of patients with advanced CC revealed a significant association between CC and abnormalities in glycolytic/glycogenic metabolic pathways[6]. Aerobic glycolysis not only provides cancer cells with rapid energy, but also produces a large number of products to meet their growth needs and shapes an acidic microenvironment conducive to tumor cell survival[7]. Pyruvate kinase (PK) is a key rate limiting enzyme in the glycolysis process, responsible for converting phosphoenolpyruvate into pyruvate and ATP. In the original transcript of the PKM gene, alternative splicing of mutually exclusive exons 9 and 10 produces PKM1 with exon 9 and PKM2 with exon 10. PKM2 is a cancer specific splicing isomer of PK and is considered one of the key regulatory factors of the Warburg effect[8]. The heterogeneous nuclear ribonucleoprotein A1 (hnRNPA1) is the most abundant and universal expressed member of the hnRNPs[9]. hnRNPA1 also plays a major role in regulating the Warburg effect[10,11]. It is primarily involved in alternative splicing of gene precursor mRNA and mRNA transport. HnRNPA1 itself is regulated by a selective splicing mechanism and has two splice isoforms: HnRNPA1-a, formed by the exon 8 jump, and hnRNPA1-b, which retains exon 8. HnRNPA1-a is 320 amino acids long with a molecular weight of 34 KDa, while hnRNPA1-b is 372 amino acids long with a molecular weight of 39 KDa. The hnRNPA1-b isoform retains amino acids 253-303 compared to hnRNPA1-a. HnRNPA1 positively regulates vaccinia-related kinase 1, which promotes increased cyclin D1 expression and lung cancer cell proliferation, and hnRNPA1 is negatively correlated with overall survival of lung cancer patients[12]. However, the role and mechanism of hnRNPA1-b in CC is unclear.
Aerobic glycolysis in cancer is associated with several pathways, including APC/KRAS/TP53, PI3K/AKT, and WNT/β-catenin[13]. AKT and mTORC1 enhance glycolysis by reinforcing glucose uptake and phosphorylating glycolytic enzymes[14]. The sustained activation of PI3K/AKT caused by RAS gene mutations, along with epidermal growth factor receptor overexpression, is a significant factor leading to metabolic abnormalities in CC[15]. However, the role of hnRNPA1 in regulating the Warburg effect through the PI3K/AKT pathway has not been reported. We conducted mass spectrometry analysis on tissues and feces of colorectal cancer (CRC) mice, as well as HCT116 CRC cells, and found that the level of hnRNPA1 was significantly higher in the CRC group compared to the control group. After treatment with Yi Qi Dispersal Formula, the level of hnRNPA1 significantly decreased. The differential protein Kyoto Encyclopedia of Genes and Genomes analysis and PI3K/AKT signaling pathway showed a close relationship between the two groups[16].
MicroRNAs (miRNAs) are a class of single-stranded non-coding RNAs that are approximately 18-24 nucleotides in length. They interact with the 3’ untranslated regions of target genes, leading to either translation inhibition or mRNA degradation[17,18]. In the case of CRC, miR-490-3p, which is expressed at low levels, can bind to and target the frequently rearranged in advanced T-cell lymphomas (FRAT1) protein. This interaction plays a role in regulating CRC proliferation and progression through the miR-490-3p/FRAT1/β-catenin axis[19]. Additionally, miR-490-3p can also bind to transforming growth factor-beta receptor 1 to inhibit CRC cell invasion and metastasis[20]. Both national and international studies have demonstrated that hnRNPA1 can interact with mRNA, miRNA, and long non-coding RNA to regulate the proliferation, invasion, and metastasis of various cancer cells, including CRC[21-23]. HnRNPA1 is associated with cancer progression and overall patient survival[12]. TargetScan 8.0 (https://www.targetscan.org/vert_80/)[24] prediction analysis has shown that miR-490-3p has the potential to bind to the 3’ non-coding region of hnRNPA1. However, there are currently no reports on miR-490-3p targeting the hnRNPA1 isoform hnRNPA1-b to inhibit CC proliferation.
In this study, we propose a scientific hypothesis that the novel miR-490-3p/hnRNPA1-b/PKM2 axis can remodel the Warburg effect and regulate CC proliferation through the PI3K/AKT pathway. To gain a better understanding of its mechanism of action, we aim to analyze how hnRNPA1-b enhances the Warburg effect to promote CC cell proliferation via the PI3K/AKT pathway. Additionally, we will investigate the interaction between miR-490-3p and hnRNPA1-b, aiming to inhibit the Warburg effect and CC cell proliferation. The results of this study will provide insights into the molecular mechanism of the miR-490-3p/hnRNPA1-b/PKM2 axis in remodeling the Warburg effect and regulating CC proliferation through the PI3K/AKT pathway. Furthermore, it will help identify potential biological targets for CC diagnosis and treatment.
A total of 30 fresh clinical CC tissue samples were obtained from the biospecimen bank of Jiangxi Cancer Hospital. These samples were paired with paraneoplastic tissues and normal tissues. Additionally, we obtained 220 paraffin-embedded sections of CC from the Department of Medical Pathology at Jiangxi Provincial Cancer Hospital. The 220 pathological sections were mainly obtained from patients who underwent surgery for early to mid-stage colon cancer, patients with advanced combined intestinal obstruction or active bleeding, and biopsies obtained by gastroenteroscopy. Prior to sampling, none of the patients included in the study had received chemotherapy or radiotherapy. Data on sex, age, TNM staging, and clinical staging were collected from these patients, with a total of 220 cases having complete information. The Ethics Committee of Jiangxi Cancer Hospital approved this study (No. 2023ky089), which was conducted in accordance with the Helsinki Declaration.
Total RNA was extracted using the RNA extraction kits (Shanghai Feige, RNAfast200, China) following the instructions provided. For quantitative real-time polymerase chain reaction (qPCR), the ReverTra Ace qPCR RT detection kit (Toyobo, FSQ-101, China) was used, with the SYBR Green PCR master mix (Toyobo, QPK-212, China) as the premix. β-actin and U6 were utilized as internal references for miRNA and mRNA, respectively. The primer sequences can be found in Supplementary Table 1.
Paraffin-embedded sections from stage I-IV CC patients were collected for immunohistochemical analysis. The sections were obtained from paraffin tissue blocks preserved at Jiangxi Cancer Hospital between 2006 and 2022. The sections, which were 3-μm-thick, underwent deparaffinization and rehydration. To quench endogenous peroxidase, the sections were treated with 3% hydrogen peroxide for 10 min. The primary antibody was then applied to the sections overnight at 4 °C. Subsequently, each section was incubated with 50-100 μL of secondary antibody for 50 min at room temperature. After that, the sections were restained with hematoxylin for 1 min, dehydrated, and sealed with neutral resin. Immunohistochemical images were analyzed using Image-Pro plus 6.0 software to calculate the mean optical density values.
Human CC cells (HCT116, SW480, HT29, SW620), human normal colon epithelial cells (NCM460) and human embryonic kidney 293T (HEK293T) cells were obtained from the American Type Culture Collection. NCM460 were cultured in RPMI 1640 medium supplemented with 10% fetal bovine serum (FBS) and another cell lines were maintained in DMEM supplemented with 10% FBS at a temperature of 37 °C in a 5% CO2 atmosphere. The culture medium was refreshed every 2 d.
The hnRNPA1-b target fragment was inserted into plasmid pcDNA3.1(+) to create the hnRNPA1-b overexpression vector OE-A1-b. The construct was confirmed by sequencing (Sysofer, Jiangsu, China). To silence the mRNA expression of hnRNPA1-b, the RNA interference method (siRNA) was employed (Sysofer, Jiangsu, China). The overexpression plasmid, siRNA interference sequences, and miR490-3p mimics, as well as the miR490-3p inhibitor, were transfected into human CC cell lines using Lipofectamine 3000 (Thermo Fisher Scientific, United States) to establish stable expression lines. The siRNA sequences are shown in Supplementary Table 1.
The cell counting kit-8 (CCK8) was utilized to perform the cell proliferation assay. A total of 1 × 103 cells were seeded on a 96-well plate, with each well containing 100 μL of medium. The plate was then placed in an incubator at 37 °C with 5% CO2 for 72 h. Subsequently, 10 μL of CCK-8 solution (Solarbio, CA1210, China) was added to each well, and the plate was returned to the incubator for 1-2 h. The absorbance at 450 nm was determined using an enzyme marker (Shanghai Kehua, ST-360, China). In the 5-ethynyl-2’-deoxyuridine (EdU) binding assay (Biyuntian, EdU-594, China), the nuclei of proliferating cells were stained with red fluorescence, while the nuclei of other nucleated cells were stained with blue fluorescence. The staining results were captured using a fluorescent inverted microscope (NiKon, TE200-U, Japan).
After culturing CC cells for 48 h, the cell proliferation cycle was assessed using the Propidium iodide kit and apoptosis was assessed using the Annexin V-FITC kit. Flow cytometry (Beckman Coulter, CytoFLEX S, United States) was then performed to analyze the results.
HCT116 and SW620 cells were inoculated at a density of 5 × 106 cells per dish in 60 mm culture dishes. Once attached, the cells were transfected with hnRNPA1-b overexpression or silencing vector and miR-490-3p mimics or inhibitors. Glucose uptake and lactate production were measured using a glucose assay kit (Sigma-Aldrich, 186689-07-6, United States) and a lactate assay kit (Solarbio, A019-2-1, China), respectively. Glucose uptake results were captured using a fluorescent inverted microscope (NiKon, TE200-U, Japan), while lactate production results were determined using an enzyme marker (Shanghai Kehua, ST-360, China).
Equal amounts of extracted proteins were injected into sodium dodecyl sulfate polyacrylamide gels and subsequently transferred to polyvinylidene fluoride (PVDF) membranes (Millipore, IPVH00010, United States). The membranes were then treated by washing with TBST buffer, followed by blocking with 5% skim milk powder and incubation with primary antibody. After 1 d, the membranes were incubated with a suitable secondary antibody at 37 °C for 30 min, followed by three washes with TBST for 5 min each time. Finally, ECL luminescence reagent (Novozymes, E412-01, China) was dropwise added to the front of the PVDF membrane and the developed films were fixed using an automatic film washer in a darkroom. The resulting films were scanned and archived. The antibodies used in this experiment were anti-PKM1 (Abcam, Cambridge, MA, United States), anti-PKM2 (Abcam, Cambridge, MA, United States), anti-AKT (Proteintech, 60203-2-Ig, China), anti-p-AKT (Proteintech, 28731-1-AP, China), and anti-GAPDH (Proteintech, 60004-1-Ig, China). The PI3K/AKT pathway inhibitor used was Ly294002 (MCE, HY-10108, China).
HnRNPA1-b was inserted into the pmir-GLO vector, which is a dual luciferase plasmid (hnRNPA1-b-WT). The sequence chosen for this construct was 200 bp upstream and downstream of the binding site. Additionally, mutation vectors were created for the binding site (hnRNPA1-b-MUT1, hnRNPA1-b-MUT2, hnRNPA1-b-MUT3). MiR-490-3P and its control mimic NC were synthesized. The plasmids and miRNA mimics were co-transfected into HEK293T cells. After 48 h, the luciferase activities of both firefly and sea kidney were measured. The relative luciferase intensities were calculated using sea kidney luciferase as an internal reference. Lipo2000 (Thermo Fisher, United States) was used as the staining agent. The experimental steps followed the instructions provided in the manual. Mutation sequence design is shown in Supplementary Table 2.
GraphPad Prism 9.0 was utilized for data graphing and statistical analysis, while SPSS 18.0 was used for statistical analysis of tables. The t-test was used for comparison of measurements between two groups, the One-way ANOVA was used for comparing more than two groups of measurements, and the Pearson χ2 test was utilized for count data. P < 0.05 indicated statistical significance. All experiments were independently performed three times.
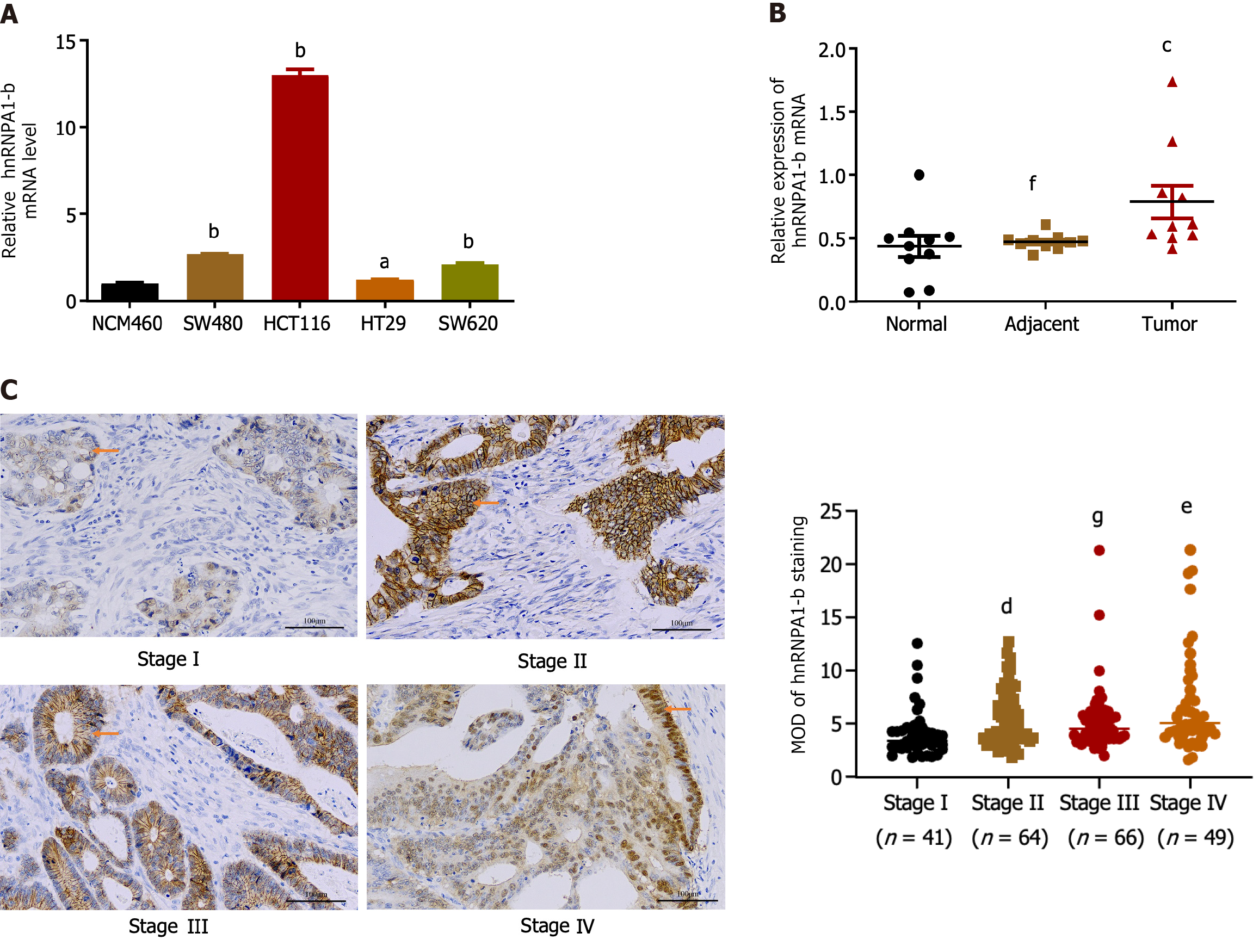
Compared to normal colon epithelial cells (NCM460), hnRNPA1-b was found to be highly expressed in CC cell lines (HCT116, SW480, HT29, SW620) (Figure 1A). For subsequent cellular experiments, HCT116 and SW620 cells, which exhibited relatively high expression, were selected. To account for individual differences, hnRNPA1-b mRNA expression was analyzed in cancerous tissues, their paired paracancerous tissues, and normal tissues from 10 CC patients. qPCR results revealed up-regulation of expression in cancerous tissues compared to normal tissues (P < 0.05) (Figure 1B). To investigate the correlation between hnRNPA1-b and the progression of CC, we conducted an immunohistochemistry analysis of the 220 paraffin-embedded sections of CC. The samples included 41 cases of stage I, 64 cases of stage II, 66 cases of stage III, and 49 cases of stage IV. The results revealed a significant difference in the expression of the hnRNPA1-b antigen among the different stages of CC (P < 0.01; Figure 1C). In addition, clinicopathologic analysis in 220 CC patients showed statistically significant differences in hnRNPA1-b expression by clinical stage and T classification (P < 0.05) (Table 1).
| Variable | hnRNPA1-b expression | χ2 | df | P value | |
| Low, no. cases | High, no. cases | ||||
| Gender | 1.654 | 1 | 0.198 | ||
| Male (125) | 75 (60) | 50 (40) | |||
| Female (95) | 65 (68.4) | 30 (31.6) | |||
| Age | 1.048 | 1 | 0.306 | ||
| ≥ 60 (120) | 80 (66.6) | 40 (33.4) | |||
| < 60 (100) | 60 (60) | 40 (40) | |||
| T classification | 15.287 | 3 | 0.004a | ||
| T1 (9) | 8 (88.9) | 1 (11.1) | |||
| T2 (35) | 30 (85.7) | 5 (14.3) | |||
| T3 (16) | 9 (56.3) | 7 (43.7) | |||
| T4 (160) | 93 (58.1) | 67 (41.9) | |||
| N classification | 1.883 | 3 | 0.757 | ||
| N0 (117) | 78 (66.7) | 39 (33.3) | |||
| N1 (57) | 33 (58.9) | 24 (41.1) | |||
| N2 (35) | 21 (60) | 14 (40) | |||
| N3 (11) | 8 (72.7) | 3 (27.3) | |||
| M classification | 3.047 | 1 | 0.081 | ||
| M0 (171) | 114 (66.7) | 57 (33.3) | |||
| M1 (49) | 26 (53.1) | 23 (46.9) | |||
| Clinical stage | 11.302 | 3 | 0.01a | ||
| 1 (41) | 35 (85.4) | 6 (14.6) | |||
| 2 (64) | 38 (59.4) | 26 (40.6) | |||
| 3 (66) | 41 (62.1) | 25 (37.9) | |||
| 4 (49) | 26 (53.1) | 23 (46.9) | |||
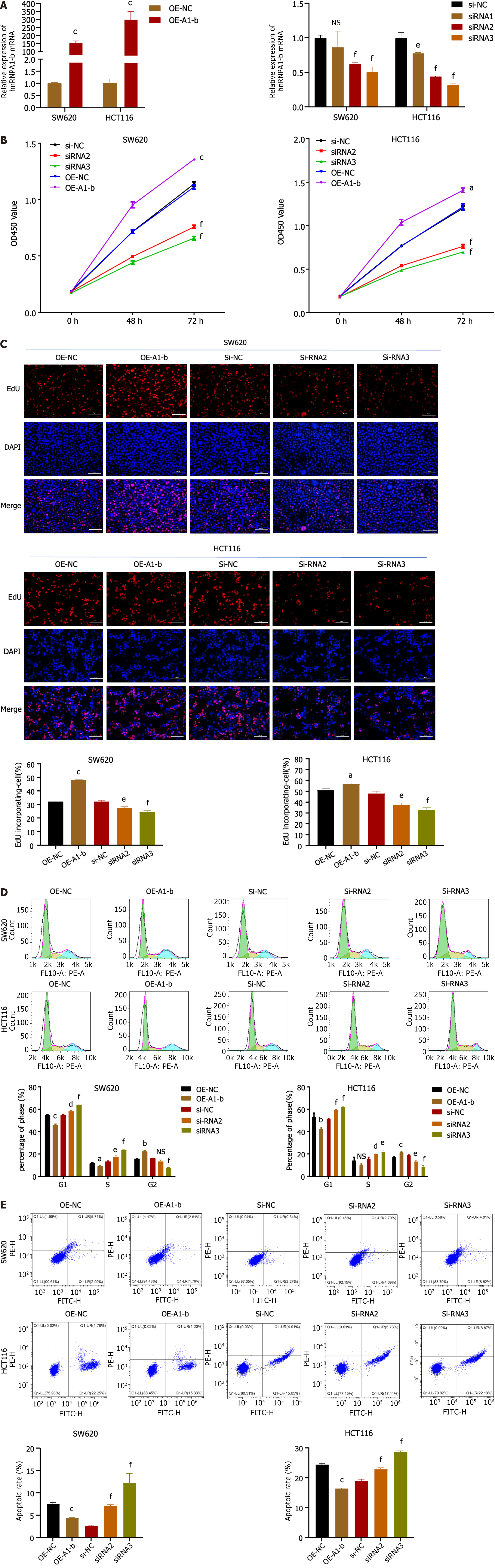
To investigate the biological function of hnRNPA1-b in CC cells, an overexpression vector (OE-A1-b) and siRNAs interfering sequences were constructed to achieve overexpression and silencing of hnRNPA1-b in HCT116 and SW620 cells. Figure 2A demonstrates that OE-A1-b significantly increased the expression of hnRNPA1-b, while siRNAs significantly decreased its expression. We selected siRNA2 and siRNA3, which exhibited a more pronounced silencing effect, for subsequent interference experiments. Both the CCK8 assay and EdU assay demonstrated that the overexpression of hnRNPA1-b significantly promoted the proliferation of HCT116 and SW620 cells, whereas the knockdown of hnRNPA1-b significantly inhibited the proliferation of HCT116 and SW620 cells (Figure 2B and C). Flow cytometry (FCM) analysis of the cell cycle revealed an increase in G1-phase cells and a decrease in G2-phase cells following the silencing of hnRNPA1-b by siRNA2 and 3, and a decrease in G1-phase cells and an increase in G2-phase cells after overexpression. The increase in cells in the G1 phase indicates slower cell division, while the increase in the G2 phase may be attributed to an increase in cells in the division phase (P < 0.05, Figure 2D). Furthermore, FCM was performed to detect apoptosis, which showed a significant increase in apoptosis after siRNA silencing and a decrease in apoptosis after overexpression (P < 0.01, Figure 2E).
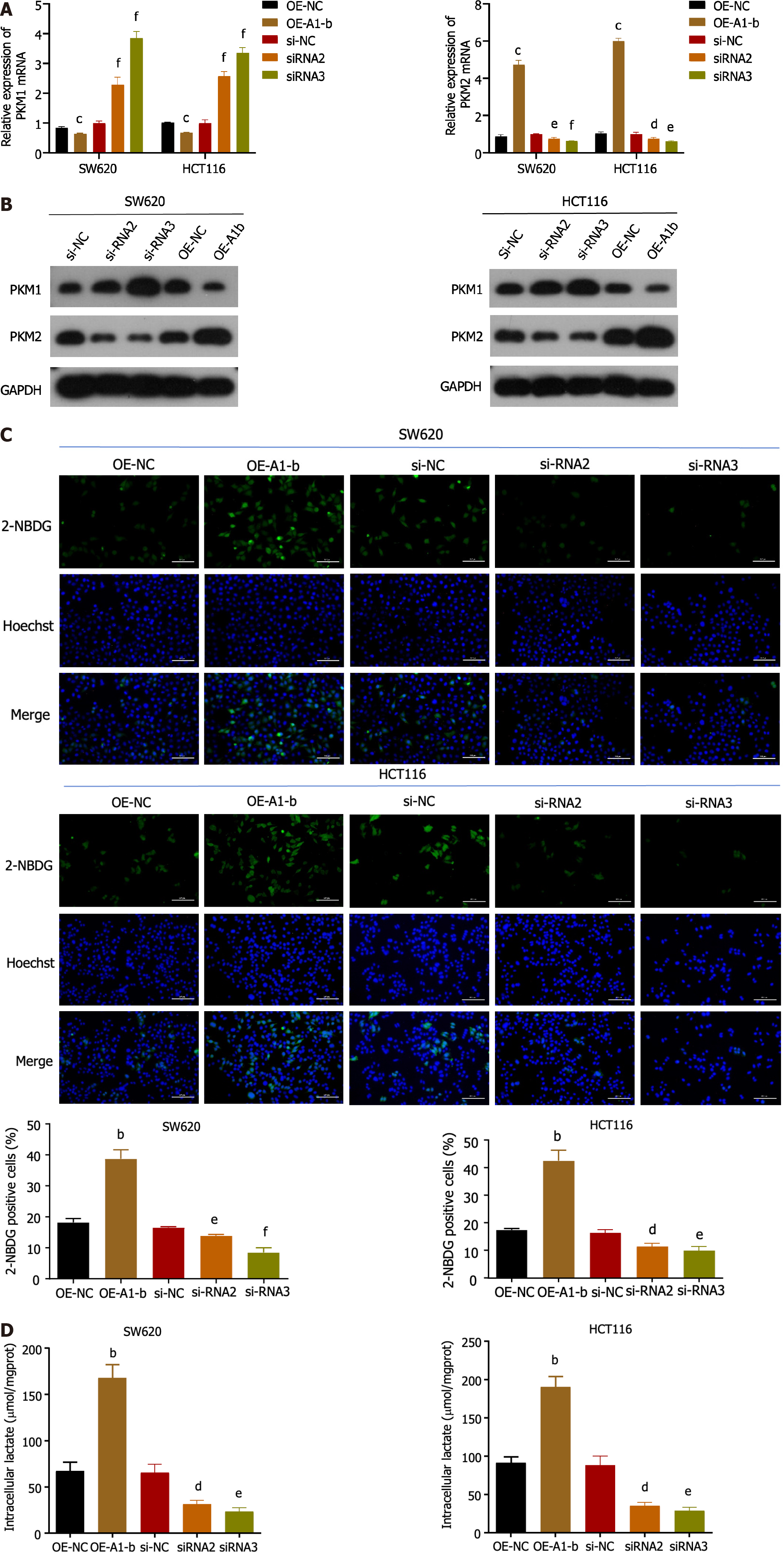
Considering the significance of PKM selective splicing in the Warburg effect, we further investigated the association between hnRNPA1-b and PKM splicing regulation. Our qPCR and western blot results demonstrated that overexpression of hnRNPA1-b led to a reduction in PKM1 expression and an increase in PKM2 expression, indicating a transition from PKM1 to PKM2. Conversely, silencing of hnRNPA1-b showed the opposite effect (Figure 3A and B). Furthermore, glucose uptake and lactate production assays revealed that overexpression of hnRNPA1-b significantly enhanced glucose uptake and promoted increased lactate production in HCT116 and SW620 cells. On the other hand, siRNAs had the opposite effect (P < 0.05; Figure 3C and D). These findings suggest that hnRNPA1-b can significantly increase the rate of glycolysis in CC cells, and according to Zhong et al[13] the lactic acid produced favors CC proliferation.
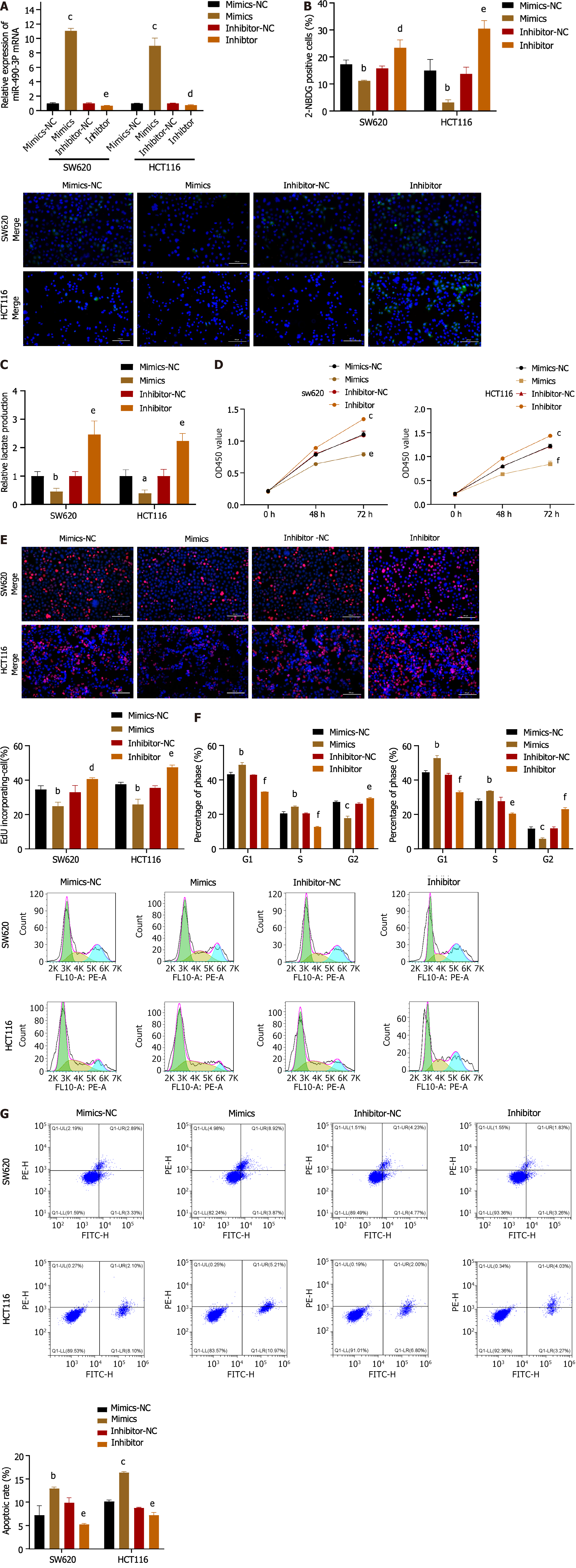
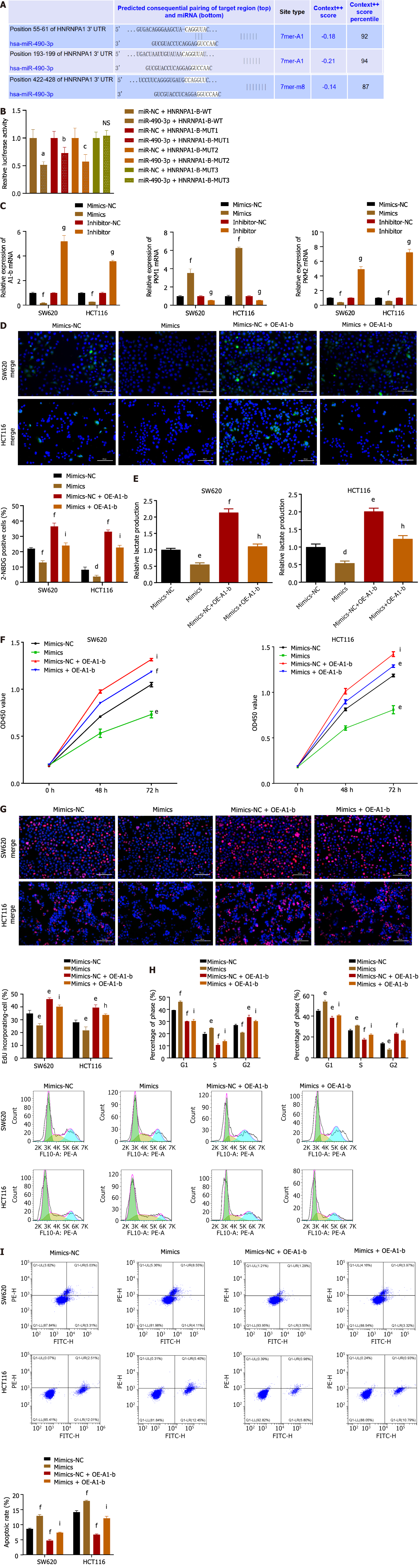
As demonstrated in Figure 4A, the introduction of miR-490-3p mimics significantly increased the mRNA expression of miR-490-3p (P < 0.001), whereas the use of inhibitors significantly decreased the mRNA expression of miR-490-3p (P < 0.01). The glucose uptake assay and lactate production assay showed that the mimics group could significantly reduce glucose uptake by HCT116 and SW620 cells and inhibit lactate production, while the inhibitor group could significantly increase glucose uptake by HCT116 and SW620 cells and promote lactate production (P < 0.05, Figure 4B and C). The results from both the CCK8 assay and EdU assay indicated that the mimics significantly inhibited the proliferation of HCT116 and SW620 cells, whereas the inhibitor promoted the proliferation of HCT116 and SW620 cells (P < 0.05; Figure 4D and E). FCM analysis of the cell cycle revealed an increase in G1 and S phase cells, as well as a decrease in G2 phase cells in the mimics group, while the inhibitor group showed a decrease in G1 and S phase cells and an increase in G2 phase cells (P < 0.05; Figure 4F). Moreover, apoptosis was assessed using FCM, which demonstrated a significant increase in apoptosis in the mimics group and a decrease in apoptosis in the inhibitor group (P < 0.01; Figure 4G). These findings suggest that miR-490-3p acts in opposition to hnRNPA1-b and exerts an inhibitory effect on CC cell proliferation.
TargetScan 8.0 predicted that miR-490-3P could bind hnRNPA1, and the binding site is shown in Figure 5A. The dual luciferase reporter assay demonstrated that miR-490-3P mimics could significantly attenuate luciferase activity by binding to wild type hnRNPA1-b (P < 0.001; Figure 5B). However, there was no significant difference in luciferase activity among the mutant groups (hRNAPA1-MUT3) (P > 0.05; Figure 5B), indicating that this site is the binding site. To further investigate the role of miR-490-3P targeting hnRNPA1-b in regulating the Warburg effect and CC cell proliferation, mimics were used to decrease the expression of hnRNPA1-b and PKM2 in HCT116 and SW620 cells, while increasing the expression of PKM1 in HCT116 and SW620 cells (P < 0.001; Figure 5C). Conversely, the inhibitor significantly increased the expression of hnRNPA1-b and PKM2 in HCT116 and SW620 cells, while decreasing the expression of PKM1 in HCT116 and SW620 cells (P < 0.001; Figure 5C). In the glucose uptake assay and lactate production assay, miR-490-3P significantly reduced the uptake of glucose by HCT116 and SW620 cells and decreased lactate production. On the other hand, OE-A1-b significantly increased the uptake of glucose by HCT116 and SW620 cells and promoted lactate production. miR-490-3P was found to attenuate the effect of OE-A1-b in promoting glucose uptake and lactate production in HCT116 and SW620 cells (P < 0.05; Figure 5D and E). Both the CCK8 assay and EdU assay demonstrated that miR-490-3P significantly reduced the proliferation of HCT116 and SW620 cells after transfection, compared to the mi-NC group. Conversely, OE-A1-b significantly increased the proliferation of HCT116 and SW620 cells after transfection. miR-490-3P was able to attenuate the role of OE-A1-b in promoting the proliferation of HCT116 and SW620 cells (P < 0.05; Figure 5F and G). The cell cycle was examined using FCM, which demonstrated that miR-490-3P increased the number of cells in the G1 phase and decreased the number of cells in the G2 phase. Conversely, OE-A1-b decreased the number of cells in the G1 phase and increased the number of cells in the G2 phase. Furthermore, miR-490-3P was able to reverse the effect of OE-A1-b (P < 0.05; Figure 5H). The FCM assay for apoptosis revealed that miR-490-3P significantly promoted apoptosis in HCT116 and SW620 cells, while OE-A1-b significantly inhibited apoptosis in these cells. Additionally, miR-490-3P was able to reverse the effect of OE-A1-b on apoptosis inhibition in HCT116 and SW620 cells (P < 0.001; Figure 5I).
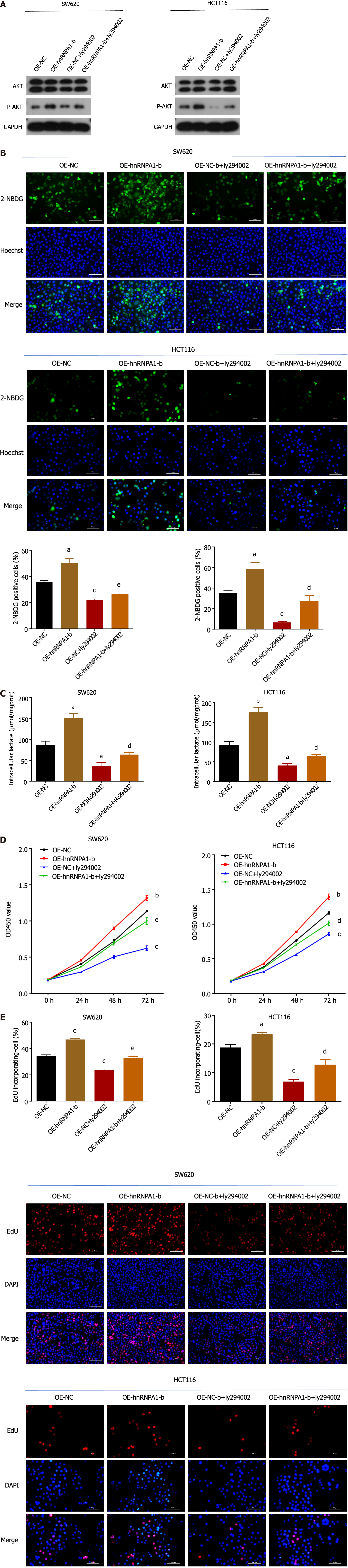
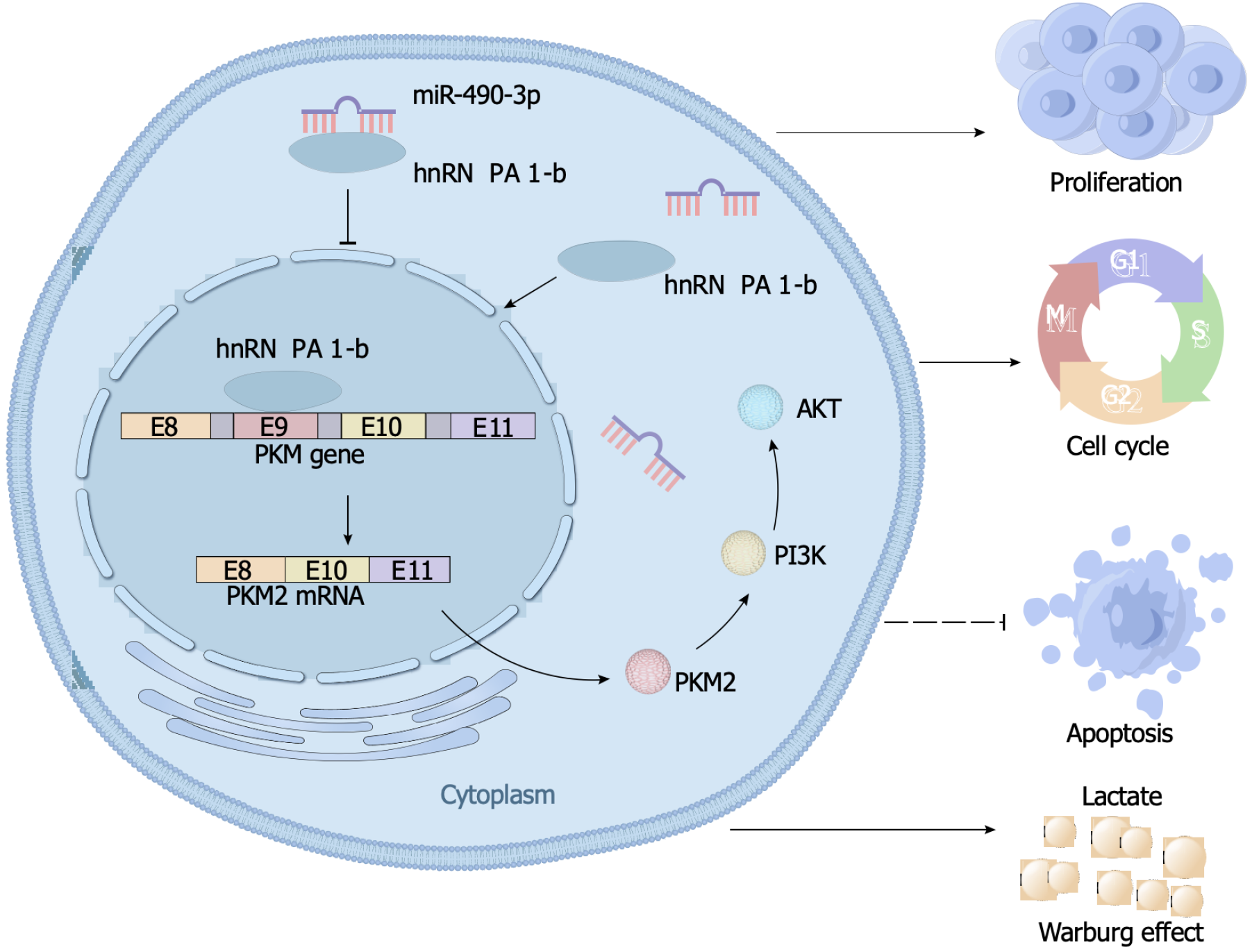
AKT is a crucial molecule in the PI3K/AKT pathway. In this study, we aimed to investigate the regulatory relationship between hnRNPA1-b and AKT to better understand the role of hnRNPA1-b in the PI3K/AKT pathway. Our Western blot results revealed an upregulation of phosphorylated AKT expression in the OE-hnRNPA1-b group compared to the control OE-NC group. Interestingly, the addition of the PI3K/AKT pathway inhibitor Ly294002 led to a downregulation of phosphorylated AKT expression in the OE-NC group, and this downregulation was partially reversed by the addition of hnRNPA1-b. However, the regulation of hnRNPA1-b and Ly294002 had no effect on non-phosphorylated AKT expression (Figure 6A). Furthermore, we conducted glucose uptake and lactate production assays to assess the impact of hnRNPA1-b on cellular metabolism. Our findings demonstrated that OE-hnRNPA1-b significantly enhanced glucose uptake and promoted increased lactate production in HCT116 and SW620 cells. Conversely, the addition of Ly294002 in the OE-NC group resulted in reduced glucose uptake and lactate production. Notably, the addition of hnRNPA1-b partially restored glucose uptake and lactate production (P < 0.05; Figure 6B and C). The CCK8 assay and EdU assay demonstrated that OE-hnRNPA1-b significantly increased the proliferation of HCT116 and SW620 cells. In the OE-NC group, the addition of Ly294002 reduced the proliferation of HCT116 and SW620 cells, which was partially restored by the addition of hnRNPA1-b (P < 0.05; Figure 6D and E). Following these experiments, it has been established that hnRNPA1-b has the ability to enhance the Warburg effect, promote the selective expression of PKM2, facilitate the proliferation and division of CC cells, and hinder the apoptosis of CC cells. However, when miR-490-3p binds to hnRNPA1-b, it exerts an inhibitory effect on the aforementioned biological processes (Figure 7).
In recent years, researchers have discovered that certain molecular abnormal splicing products can serve as effective markers for tumor diagnosis. For instance, in 2018, Scher et al[25] found that the nuclear-localized androgen receptor splice variant 7 present in tumor cells circulating in the blood could be utilized as a predictive marker for metastatic trend-resistant prostate cancer. Adult T-cell lymphoma/leukemia (ATLL) is a highly aggressive peripheral T-cell malignancy. In 2019, Japanese scholars reported that the selective spliceosome soluble CADM1 could function as an early warning marker for ATLL disease progression[26]. Additionally, in 2019, Zhou et al[27], from Tianjin, reported that the selective spliceosome of myosin 1B could potentially serve as a prognostic biomarker and therapeutic target for gliomas. Furthermore, in 2023, Jbara et al[28] discovered that the splicing factor RBFOX2 modulated alternative splicing of RHO-interacting protein to inhibit pancreatic ductal adenocarcinoma metastasis. The role and mechanism of hnRNPA1-b in CC have not been previously reported. Our study demonstrated that hnRNPA1-b is highly expressed in CC cells and tissues and significantly correlated with clinical stage and T classification (P < 0.05). Overexpression of hnRNPA1-b significantly promotes proliferation, glucose uptake, lactate production, and PKM1 to PKM2 conversion in HCT116 and SW620 cells. Conversely, silencing hnRNPA1-b expression yields opposite results. Furthermore, overexpression of hnRNPA1-b increases phosphorylated AKT expression. However, the addition of PI3K/AKT pathway inhibitors reverses the effect of hnRNPA1-b overexpression in enhancing the Warburg effect and promoting the proliferation of HCT116 and SW620 cells. These findings suggest that hnRNPA1-b enhances the Warburg effect to promote CC cell proliferation through the PI3K/AKT pathway.
Fu et al[29] suggested that hnRNPA1 has the potential to serve as a biomarker for early diagnosis and prognostic monitoring of CC. However, their study did not differentiate between hnRNPA1-b and hnRNPA1-a, and the miRNA we studied was different. In contrast, in our study we designed amplification primers and overexpression vectors that effectively distinguish hnRNPA1-b from hnRNPA1-a. We have even applied for a patent for this technology. Wu et al[30] concluded that miRNAs containing miR-490-3p are closely associated with KRAS genotype and have an important role in CRC progression. Additionally, Fu et al[29] demonstrated at the cellular level, that hnRNPA1 promotes PKM2 expression and enhances the Warburg effect. However, we have yet to determine whether miR-490-3p plays a comparable role to miR-206 in this context. In our study, we demonstrated that miR-490-3p mimics can target hnRNPA1-b and inhibit PKM2 expression, leading to a reduction in glucose uptake and lactate production in HCT116 and SW620 cells. Furthermore, we discovered that overexpression of hnRNPA1-b promotes the proliferative effects of HCT116 and SW620 cells, but this effect can be reversed by miR-490-3p. Our in vitro experiments provided evidence for a novel regulatory axis involving miR-490-3p/hnRNPA1-b/PKM2 in controlling the proliferative effects of CC.
Patients with CC are less frequently detected in the early stages and are not suitable for surgery in the late stages, except in combination with bowel obstruction or active bleeding complications. Thus, there are more cases in stage II and III compared to stage I and IV. Furthermore, Li et al[31] investigated the relationship between hnRNPA1 monomer and chronic granulocytic leukemia (CML). The study concluded that hnRNPA1-a was predominantly expressed in healthy controls, while hnRNPA1-b was positively correlated with the progression of CML. Further research is needed to determine if the same phenomenon exists in CC, where hnRNPA1-b promotes CC while hnRNPA1-a does not. In addition, hnRNPA1-b promotes the CC metastasis mechanism, which we subsequently intend to investigate further.
In summary, this study confirmed the role of hnRNPA1’s selective shear monomer, hnRNPA1-b, in promoting the proliferation of CC cells. The study also elucidated the mechanism of action of the miR-490-3p/hnRNPA1-b/PKM2 axis in modulating the proliferation of CC cells by remodeling the Warburg effect through the PI3K/AKT pathway. These findings suggest that the novel miR-490-3p/hnRNPA1-b/PKM2 axis could provide a new strategy for the diagnosis and treatment of CC.
Studies of heterogeneous ribonucleoprotein A1 (hnRNPA1) isoforms are rare, and miR-490-3p targeting hnRNPA1-b to regulate colon cancer proliferation has not been reported.
To explore the mechanisms by which miR-490-3p and hnRNPA1-b interact with the PI3K/AKT pathway to regulate the Warburg effect and proliferation of colon cancer cells.
To investigate the role and mechanism of a novel miR-490-3p/hnRNPA1-b/PKM2 axis in enhancing the Warburg effect and promoting colon cancer cell proliferation through the PI3K/AKT pathway.
Paraffin-embedded pathological sections were obtained from 220 colon cancer patients for immunohistochemical analysis to determine the expression of hnRNPA1-b. The study investigated the relationship between the expression values and the clinicopathological features of the patients. Differences in mRNA expression were analyzed using quantitative real-time polymerase chain reaction, while differences in protein expression were analyzed using Western blot. Cell proliferation was assessed using cell counting kit-8 and 5-ethynyl-2’-deoxyuridine assays, and cell cycle and apoptosis were evaluated using flow cytometric assays. The targeted binding of miR-490-3p to hnRNPA1-b was validated using a dual luciferase reporter assay. The Warburg effect was evaluated through glucose uptake and lactic acid production assays.
The expression of hnRNPA1-b was significantly increased in colon cancer (CC) tissues and cells compared to normal controls (P < 0.05). Immunohistochemical results demonstrated significant variations in the expression of the hnRNPA1-b antigen in different stages of CC, including stage I, II-III, and IV. Furthermore, the clinicopathologic characterization revealed a significant correlation between hnRNPA1-b expression and clinical stage as well as T classification. HnRNPA1-b was found to enhance the Warburg effect through the PI3K/AKT pathway, thereby promoting proliferation of HCT116 and SW620 cells. However, the proliferation of HCT116 and SW620 cells was inhibited when miR-490-3p targeted and bound to hnRNPA1-b, effectively blocking the Warburg effect.
These findings suggest that the novel miR-1-3p/hnRNPA1-b/PKM2 axis could provide a new strategy for the diagnosis and treatment of CC.
The follow-up plan is to study the mechanism of hnRNPA1-b promoting colon cancer metastasis and drug resistance.
We would like to acknowledge the Department of Abdominal Surgery I and the biospecimen bank of Jiangxi Cancer Hospital for providing the colon cancer tissue samples. Furthermore, we express our gratitude to all the patients for their valuable contributions to this study.
Provenance and peer review: Unsolicited article; Externally peer reviewed.
Peer-review model: Single blind
Specialty type: Oncology
Country/Territory of origin: China
Peer-review report’s scientific quality classification
Grade A (Excellent): 0
Grade B (Very good): B
Grade C (Good): 0
Grade D (Fair): 0
Grade E (Poor): 0
P-Reviewer: Yoshinaga K, Japan S-Editor: Wang JJ L-Editor: A P-Editor: Zhang XD
| 1. | Siegel RL, Miller KD, Fuchs HE, Jemal A. Cancer statistics, 2022. CA Cancer J Clin. 2022;72:7-33. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 4235] [Cited by in RCA: 11419] [Article Influence: 3806.3] [Reference Citation Analysis (4)] |
| 2. | Bray F, Ferlay J, Soerjomataram I, Siegel RL, Torre LA, Jemal A. Global cancer statistics 2018: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J Clin. 2018;68:394-424. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 53206] [Cited by in RCA: 55806] [Article Influence: 7972.3] [Reference Citation Analysis (132)] |
| 3. | White A, Joseph D, Rim SH, Johnson CJ, Coleman MP, Allemani C. Colon cancer survival in the United States by race and stage (2001-2009): Findings from the CONCORD-2 study. Cancer. 2017;123 Suppl 24:5014-5036. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 99] [Cited by in RCA: 107] [Article Influence: 13.4] [Reference Citation Analysis (0)] |
| 4. | Deeb AP, Aquina CT, Monson JRT, Blumberg N, Becerra AZ, Fleming FJ. Allogeneic Leukocyte-Reduced Red Blood Cell Transfusion Is Associated with Postoperative Infectious Complications and Cancer Recurrence after Colon Cancer Resection. Dig Surg. 2020;37:163-170. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 9] [Cited by in RCA: 10] [Article Influence: 1.7] [Reference Citation Analysis (0)] |
| 5. | La Vecchia S, Sebastián C. Metabolic pathways regulating colorectal cancer initiation and progression. Semin Cell Dev Biol. 2020;98:63-70. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 98] [Cited by in RCA: 212] [Article Influence: 35.3] [Reference Citation Analysis (0)] |
| 6. | Rao J, Wan X, Tou F, He Q, Xiong A, Chen X, Cui W, Zheng Z. Molecular Characterization of Advanced Colorectal Cancer Using Serum Proteomics and Metabolomics. Front Mol Biosci. 2021;8:687229. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 12] [Cited by in RCA: 12] [Article Influence: 3.0] [Reference Citation Analysis (0)] |
| 7. | Hamanaka RB, Chandel NS. Cell biology. Warburg effect and redox balance. Science. 2011;334:1219-1220. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 89] [Cited by in RCA: 108] [Article Influence: 8.3] [Reference Citation Analysis (0)] |
| 8. | Yadav S, Bhagat SD, Gupta A, Samaiya A, Srivastava A, Shukla S. Dietary-phytochemical mediated reversion of cancer-specific splicing inhibits Warburg effect in head and neck cancer. BMC Cancer. 2019;19:1031. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 21] [Cited by in RCA: 19] [Article Influence: 3.2] [Reference Citation Analysis (0)] |
| 9. | Siculella L, Giannotti L, Di Chiara Stanca B, Spedicato F, Calcagnile M, Quarta S, Massaro M, Damiano F. A comprehensive understanding of hnRNP A1 role in cancer: new perspectives on binding with noncoding RNA. Cancer Gene Ther. 2023;30:394-403. [RCA] [PubMed] [DOI] [Full Text] [Cited by in RCA: 3] [Reference Citation Analysis (0)] |
| 10. | Yan Q, Zeng P, Zhou X, Zhao X, Chen R, Qiao J, Feng L, Zhu Z, Zhang G, Chen C. RBMX suppresses tumorigenicity and progression of bladder cancer by interacting with the hnRNP A1 protein to regulate PKM alternative splicing. Oncogene. 2021;40:2635-2650. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 24] [Cited by in RCA: 52] [Article Influence: 13.0] [Reference Citation Analysis (0)] |
| 11. | Lan Z, Yao X, Sun K, Li A, Liu S, Wang X. The Interaction Between lncRNA SNHG6 and hnRNPA1 Contributes to the Growth of Colorectal Cancer by Enhancing Aerobic Glycolysis Through the Regulation of Alternative Splicing of PKM. Front Oncol. 2020;10:363. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 28] [Cited by in RCA: 67] [Article Influence: 13.4] [Reference Citation Analysis (0)] |
| 12. | Ryu HG, Jung Y, Lee N, Seo JY, Kim SW, Lee KH, Kim DY, Kim KT. HNRNP A1 Promotes Lung Cancer Cell Proliferation by Modulating VRK1 Translation. Int J Mol Sci. 2021;22. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 3] [Cited by in RCA: 17] [Article Influence: 4.3] [Reference Citation Analysis (0)] |
| 13. | Zhong X, He X, Wang Y, Hu Z, Huang H, Zhao S, Wei P, Li D. Warburg effect in colorectal cancer: the emerging roles in tumor microenvironment and therapeutic implications. J Hematol Oncol. 2022;15:160. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in RCA: 155] [Reference Citation Analysis (0)] |
| 14. | Szwed A, Kim E, Jacinto E. Regulation and metabolic functions of mTORC1 and mTORC2. Physiol Rev. 2021;101:1371-1426. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 60] [Cited by in RCA: 449] [Article Influence: 112.3] [Reference Citation Analysis (0)] |
| 15. | Hoxhaj G, Manning BD. The PI3K-AKT network at the interface of oncogenic signalling and cancer metabolism. Nat Rev Cancer. 2020;20:74-88. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1028] [Cited by in RCA: 1342] [Article Influence: 268.4] [Reference Citation Analysis (0)] |
| 16. | Wan X, Tou F, Zeng J, Chen X, Li S, Chen L, Zheng Z, Rao J. Integrative analysis and identification of key elements and pathways regulated by Traditional Chinese Medicine (Yiqi Sanjie formula) in colorectal cancer. Front Pharmacol. 2022;13:1090599. [RCA] [PubMed] [DOI] [Full Text] [Cited by in RCA: 4] [Reference Citation Analysis (0)] |
| 17. | Strubberg AM, Madison BB. MicroRNAs in the etiology of colorectal cancer: pathways and clinical implications. Dis Model Mech. 2017;10:197-214. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 79] [Cited by in RCA: 112] [Article Influence: 14.0] [Reference Citation Analysis (0)] |
| 18. | Ahadi A. The significance of microRNA deregulation in colorectal cancer development and the clinical uses as a diagnostic and prognostic biomarker and therapeutic agent. Noncoding RNA Res. 2020;5:125-134. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 21] [Cited by in RCA: 29] [Article Influence: 5.8] [Reference Citation Analysis (0)] |
| 19. | Zheng K, Zhou X, Yu J, Li Q, Wang H, Li M, Shao Z, Zhang F, Luo Y, Shen Z, Chen F, Shi F, Cui C, Zhao D, Lin Z, Zheng W, Zou Z, Huang Z, Zhao L. Corrigendum to "Epigenetic silencing of miR-490-3p promotes development of an aggressive colorectal cancer phenotype through activation of the Wnt/β-catenin signaling pathway". Cancer Letters, 376 (2016) 178-87. Cancer Lett. 2021;519:346-348. [RCA] [PubMed] [DOI] [Full Text] [Reference Citation Analysis (0)] |
| 20. | Xu X, Chen R, Li Z, Huang N, Wu X, Li S, Li Y, Wu S. MicroRNA-490-3p inhibits colorectal cancer metastasis by targeting TGFβR1. BMC Cancer. 2015;15:1023. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 32] [Cited by in RCA: 47] [Article Influence: 4.7] [Reference Citation Analysis (0)] |
| 21. | Zhu HE, Li T, Shi S, Chen DX, Chen W, Chen H. ESCO2 promotes lung adenocarcinoma progression by regulating hnRNPA1 acetylation. J Exp Clin Cancer Res. 2021;40:64. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 10] [Cited by in RCA: 35] [Article Influence: 8.8] [Reference Citation Analysis (0)] |
| 22. | Wen Z, Lian L, Ding H, Hu Y, Xiao Z, Xiong K, Yang Q. LncRNA ANCR promotes hepatocellular carcinoma metastasis through upregulating HNRNPA1 expression. RNA Biol. 2020;17:381-394. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 27] [Cited by in RCA: 54] [Article Influence: 10.8] [Reference Citation Analysis (0)] |
| 23. | Chen MS, Lin CH, Huang LY, Qiu XM. CircRNA SMARCC1 Sponges MiR-140-3p to Regulate Cell Progression in Colorectal Cancer. Cancer Manag Res. 2020;12:4899-4910. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 13] [Cited by in RCA: 14] [Article Influence: 2.8] [Reference Citation Analysis (0)] |
| 24. | McGeary SE, Lin KS, Shi CY, Pham TM, Bisaria N, Kelley GM, Bartel DP. The biochemical basis of microRNA targeting efficacy. Science. 2019;366. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 609] [Cited by in RCA: 890] [Article Influence: 148.3] [Reference Citation Analysis (0)] |
| 25. | Scher HI, Graf RP, Schreiber NA, Jayaram A, Winquist E, McLaughlin B, Lu D, Fleisher M, Orr S, Lowes L, Anderson A, Wang Y, Dittamore R, Allan AL, Attard G, Heller G. Assessment of the Validity of Nuclear-Localized Androgen Receptor Splice Variant 7 in Circulating Tumor Cells as a Predictive Biomarker for Castration-Resistant Prostate Cancer. JAMA Oncol. 2018;4:1179-1186. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 173] [Cited by in RCA: 187] [Article Influence: 26.7] [Reference Citation Analysis (0)] |
| 26. | Nakahata S, Syahrul C, Nakatake A, Sakamoto K, Yoshihama M, Nishikata I, Ukai Y, Matsuura T, Kameda T, Shide K, Kubuki Y, Hidaka T, Kitanaka A, Ito A, Takemoto S, Nakano N, Saito M, Iwanaga M, Sagara Y, Mochida K, Amano M, Maeda K, Sueoka E, Okayama A, Utsunomiya A, Shimoda K, Watanabe T, Morishita K. Clinical significance of soluble CADM1 as a novel marker for adult T-cell leukemia/lymphoma. Haematologica. 2021;106:532-542. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 4] [Cited by in RCA: 5] [Article Influence: 1.3] [Reference Citation Analysis (0)] |
| 27. | Zhou X, Wang R, Li X, Yu L, Hua D, Sun C, Shi C, Luo W, Rao C, Jiang Z, Feng Y, Wang Q, Yu S. Splicing factor SRSF1 promotes gliomagenesis via oncogenic splice-switching of MYO1B. J Clin Invest. 2019;129:676-693. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 64] [Cited by in RCA: 114] [Article Influence: 19.0] [Reference Citation Analysis (0)] |
| 28. | Jbara A, Lin KT, Stossel C, Siegfried Z, Shqerat H, Amar-Schwartz A, Elyada E, Mogilevsky M, Raitses-Gurevich M, Johnson JL, Yaron TM, Ovadia O, Jang GH, Danan-Gotthold M, Cantley LC, Levanon EY, Gallinger S, Krainer AR, Golan T, Karni R. RBFOX2 modulates a metastatic signature of alternative splicing in pancreatic cancer. Nature. 2023;617:147-153. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 33] [Cited by in RCA: 54] [Article Influence: 27.0] [Reference Citation Analysis (0)] |
| 29. | Fu R, Yang P, Amin S, Li Z. A novel miR-206/hnRNPA1/PKM2 axis reshapes the Warburg effect to suppress colon cancer growth. Biochem Biophys Res Commun. 2020;531:465-471. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 26] [Cited by in RCA: 32] [Article Influence: 6.4] [Reference Citation Analysis (0)] |
| 30. | Wu X, Li Z, Huang N, Li X, Chen R. Study of KRAS-Related miRNA Expression in Colorectal Cancer. Cancer Manag Res. 2022;14:2987-3008. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in RCA: 5] [Reference Citation Analysis (0)] |
| 31. | Li SQ, Liu J, Zhang J, Wang XL, Chen D, Wang Y, Xu YM, Huang B, Lin J, Li J, Wang XZ. Transcriptome profiling reveals the high incidence of hnRNPA1 exon 8 inclusion in chronic myeloid leukemia. J Adv Res. 2020;24:301-310. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 11] [Cited by in RCA: 27] [Article Influence: 5.4] [Reference Citation Analysis (0)] |