Published online May 15, 2024. doi: 10.4251/wjgo.v16.i5.1908
Peer-review started: November 7, 2023
First decision: December 28, 2023
Revised: January 8, 2024
Accepted: February 22, 2024
Article in press: February 22, 2024
Published online: May 15, 2024
Processing time: 184 Days and 11.3 Hours
As the primary microtubule organizing center in animal cells, centrosome abnor
To explore the role of centrosome-related genes (CRGs) in colon cancer.
CRGs were collected from public databases. Consensus clustering analysis was performed to separate the Cancer Genome Atlas cohort. Univariate Cox and least absolute shrinkage selection operator regression analyses were performed to identify candidate prognostic CRGs and construct a centrosome-related signature (CRS) to score colon cancer patients. A nomogram was developed to evaluate the CRS risk in colon cancer patients. An integrated bioinformatics analysis was conducted to explore the correlation between the CRS and tumor immune micro
A total of 726 CRGs were collected from public databases. A CRS was constructed, which consisted of the following four genes: TSC1, AXIN2, COPS7A, and MTUS1. Colon cancer patients with a high-risk signature had poor survival. Patients with a high-risk signature exhibited decreased levels of plasma cells and activated memory CD4+ T cells. Regarding treatment response, patients with a high-risk signature were resistant to immu
We constructed a centrosome-related prognostic signature that can accurately predict the prognosis of colon cancer patients, contributing to the development of individualized treatment for colon cancer.
Core Tip: Centrosome abnormalities, as the main microtubule tissue center of animal cells, are associated with human colon cancer. Our aim was to investigate the role of centrosome related genes (CRGs) in colon cancer. A total of 726 CRGs were collected from the public database. We constructed a centrosome-related signature composed of four genes: TSC1, AXIN2, COPS7A, and MTUS1. Colon cancer patients with high-risk characteristics had a low survival rate. Patients with high-risk characteristics exhibited decreased plasma cell levels and memory CD4+ T cell activation. Regarding treatment response, patients with high-risk characteristics were resistant to immunotherapy, chemotherapy, and targeted therapy. The expression of COPS7A was relatively high in endothelial cells and fibroblasts. MTUS1 was highly expressed in endothelial cells, fibroblasts, and malignant cells. We constructed a prognostic marker related to the centrosome, which can accurately predict the prognosis of colon cancer patients and contribute to the development of individualized colon cancer treatment.
- Citation: Wang HY, Diao Y, Tan PZ, Liang H. Four centrosome-related genes to predict the prognosis and drug sensitivity of patients with colon cancer. World J Gastrointest Oncol 2024; 16(5): 1908-1924
- URL: https://www.wjgnet.com/1948-5204/full/v16/i5/1908.htm
- DOI: https://dx.doi.org/10.4251/wjgo.v16.i5.1908
Colon cancer is the most commonly diagnosed cancer worldwide and the second leading cause of cancer death[1]. According to the latest online epidemiological database, there were over 1.9 million new cases of colon cancer in 2020, with 900000 deaths in the same year[2]. Colon adenocarcinoma (COAD) is the most common, accounting for 98% of colon cancer cases[3]. Even with the rapid development of cancer screening methods, many patients are still diagnosed with multiple symptoms in the late stage, such as blood bacteria or colon obstruction[2]. Unfortunately, approximately 20% of colon cancer patients are diagnosed with stage IV every year[4]. Therefore, with the improvement of surgical treatment and chemotherapy, it is also crucial to explore more diagnostic biomarkers and possible treatment targets.
The centrosome is the major microtubule nucleating organelle in animal cells. It plays a crucial role in the orientation and stabilization of the mitotic spindle[5]. Centrosome abnormalities have been detected in all major types of cancer, implicating their roles in tumorigenesis and cancer progression[6]. Centrosome amplification/overduplication, a hallmark of cancer, was detected in cancer cells driven by cytokinesis failure[7]. In colon cancer, mutations in many oncogenes and tumor suppressor genes can affect the structure and activity of the centrosome. The former include β-catenin and BRAF,etc., and the latter include APC, TP53, and others[8].
Centrosomes have been extensively studied in the context of cancer development and progression. To the best of our knowledge, few studies have reported on the centrosome-related prognostic model of colon cancer. In this study, 726 centrosome-related genes (CRGs) were collected from public databases. Univariate Cox and least absolute shrinkage selection operator (LASSO) regression analyses were then conducted to screen for prognostic CRGs in patients with colon cancer. The following four crucial genes were obtained: TSC1, AXIN2, COPS7A, and MTUS1. A centrosome-related prognostic model for patients with colon cancer was constructed based on these four genes. Patients with colon cancer were divided into high- and low-risk groups according to the centrosome-related signature (CRS) score. The high-risk group presented with a worse prognosis and a lower level of plasma cells or activated memory CD4+ T cells. In addition, the high-risk group was resistant to immunotherapy, chemotherapy, and targeted therapy.
The raw RNA sequencing (RNA-seq) dataset of COAD and clinical information data profiles were downloaded from the Cancer Genome Atlas (TCGA) database (https://portal.gdc.cancer.gov/)[3,9-14]. After removing patients with duplicate or incomplete follow-up information, 465 TCGA-COAD samples and 41 adjacent samples were included in the following study. The TCGA-COAD dataset was randomly divided into two subgroups (1:1), one as the training dataset and the other as the testing dataset. The external dataset GSE103479 was downloaded from the Gene Expression Omnibus database (https://www.ncbi.nlm.nih.gov/geo/)[15]. Somatic mutation data were downloaded from Genomic Data Commons (GDC, https://portal.gdc.cancer.gov/).
To separate the TCGA-COAD dataset, Consensus clustering (CC) was performed using the “ConsensusClusterPlus” package of the R language[16]. The CC parameter “maxK” was set as “10”, “clusterAlg” was set as “km”, and “distance” was set as “pearson”.
Gene set enrichment analysis (GSEA) was conducted to enrich the Kyoto Encyclopedia of Genes and Genomes (KEGG) pathways of highly and lowly expressed CRGs in the TCGA-COAD cohort[17-19]. In addition, the package “clusterProfiler” of the R language was used to perform enrichment analysis of the KEGG pathways and Gene Ontology (GO) terms of differentially expressed genes between CRG groups with high and low expression[20].
Univariate Cox and LASSO regression analyses were performed to identify candidate prognostic CRGs and construct a CRS to score colon cancer patients[21]. Genes with a P value < 0.05 were considered as candidates and were subjected to the LASSO Cox regression. Heat maps of the CRS were plotted using the “heatmap” R package. Efficiency of the CRS was tested using a Kaplan-Meier plotter and receiver operating characteristic (ROC) analyses[22]. ROC analyses were performed using the “timeROC” R package[23].
To evaluate the CRS risk in patients with colon cancer, we developed a nomogram including clinical features, such as gender, N, age, risk, T, and M. The nomogram plot was displayed using the “regplot” package[24,25]. A calibration map was produced (R package “rms” and “survminer”) to compare the nomogram-predicted overall survival (OS) and observed OS. A concordance index plot was generated (R package “rms” and “survminer”) to compare the cumulative hazard between the nomogram high- and low-risk groups. A decision curve analysis (DCA) was used (R package “survminer” and “ggDCA”) to predict the 1-, 3-, and 5-year OS. ROC analyses were performed using the “timeROC” R package.
To explore the correlation between CRS and immunomodulators and immune cells, the expression data of model genes were collected, and immunomodulators in each cohort were analyzed. The tumor microenvironment scores were then calculated by different algorithms, such as ESTIMATE, CIBERSORT, TIMER, and ssGSEA[26-28]. The tumor immune dysfunction and exclusion (TIDE) algorithm was applied to analyze the relationship between CRS and response to immunotherapy[29].
We used the “oncoPredict” package of the R language to evaluate the correlation between CRS and response to chemo
We downloaded the single-cell RNA-seq data for colon cancer and the corresponding paired bulk RNA-seq data from the TISCH database (http://tisch1.comp-genomics.org/). Subsequently, we analyzed each signature gene using GSE146771-Smart.
All statistical analyses were performed using R 4.2.2 (https://www.rproject.org/) and its packages. P < 0.05 was considered statistically significant.
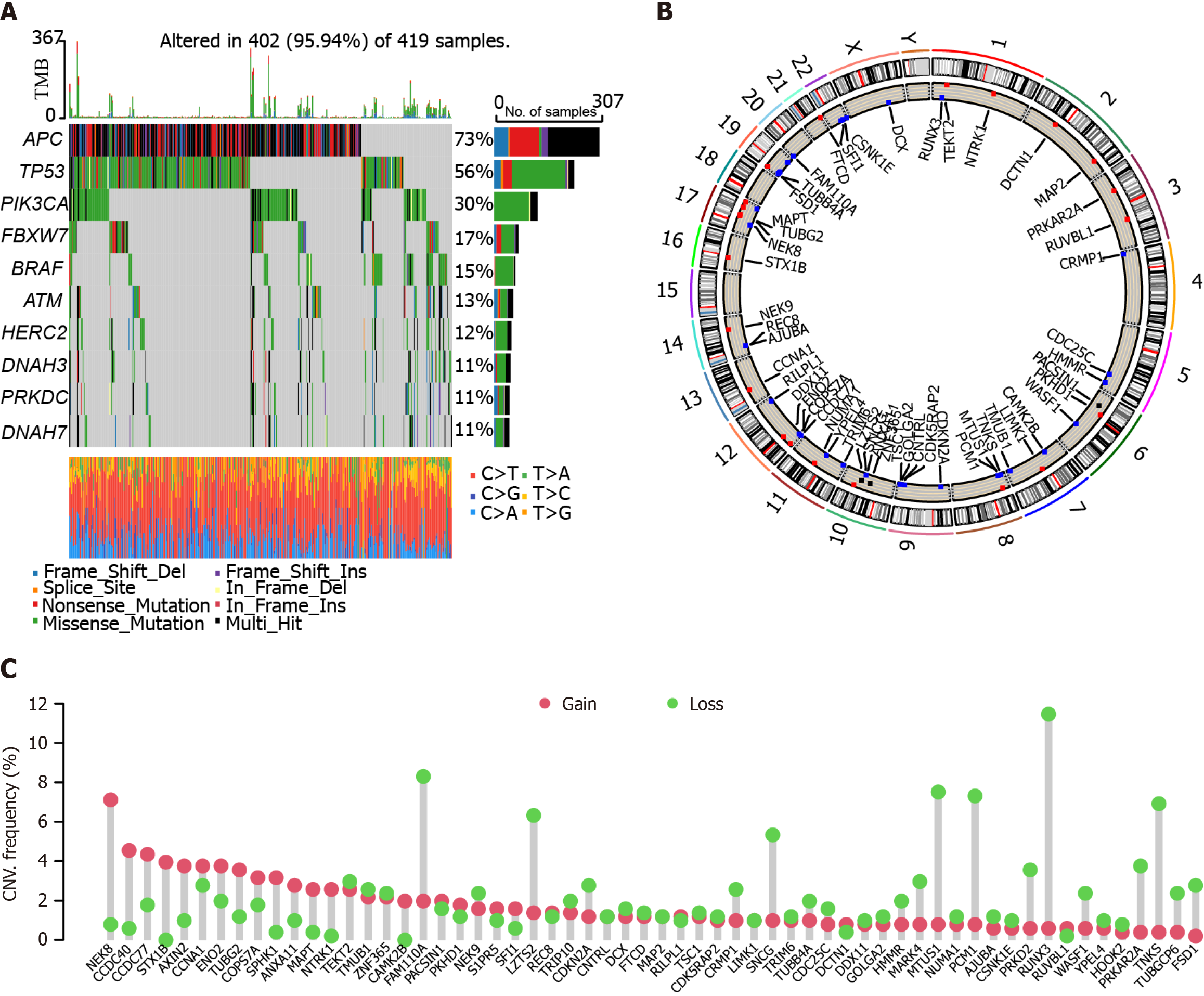
We analyzed the mutational landscapes of patients with colon cancer. Our results showed that APC, TP53, PI3KCA, FBXW7, and BRAF were the top five somatic mutational genes in these patients, with mutation frequencies of 73%, 56%, 30%, 17%, and 15%, respectively (Figure 1A). Furthermore, we plotted the circus diagram showing the distribution of mutations and genes (Figure 1B). In addition, we performed copy number variation analysis in these patients. The results showed that many genes exhibited both gain and loss, such as NEK8, CCDC40, CCDC77, and STX1B (Figure 1C).
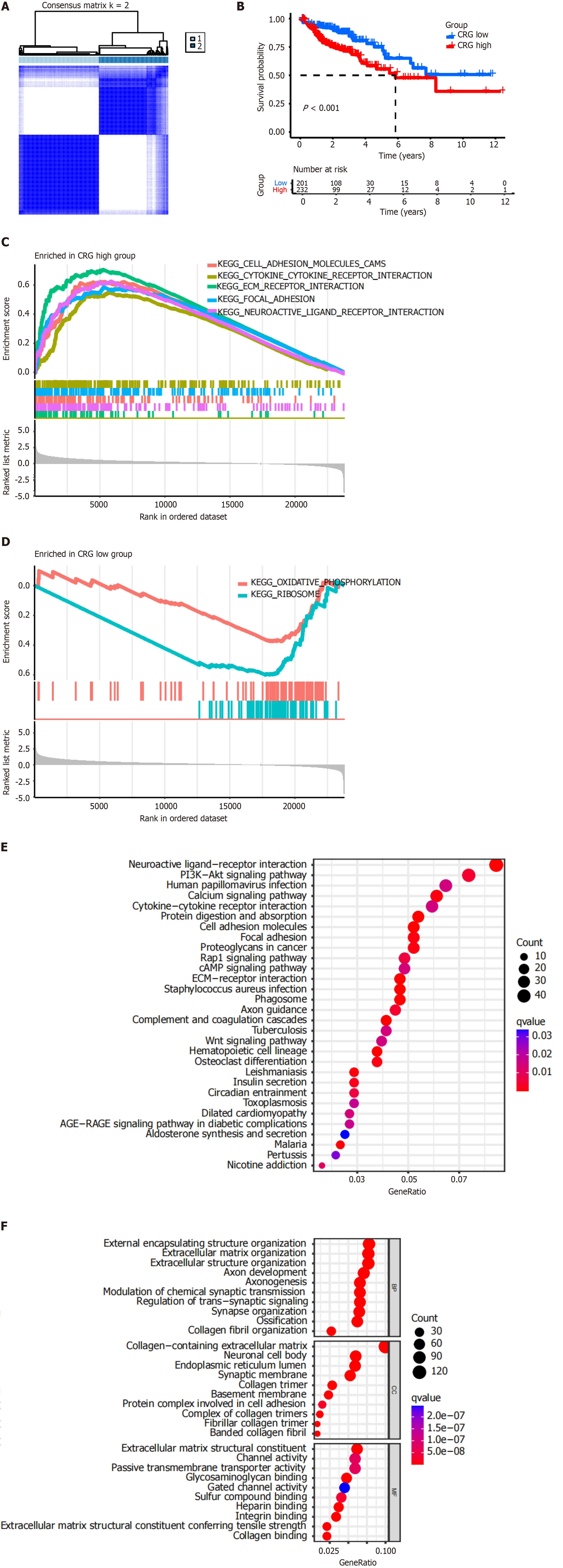
To explore unidentified subtypes of colon cancer, CC analysis was performed for genes in the TCGA-COAD cohort. The results showed that when k = 2, the patients could be divided into two clusters (Figure 2A). There was a significant difference between OS and high-risk or low-risk CRGs (P < 0.001, Figure 2B). Low-risk CRGs were associated with a favorable prognosis, whereas high-risk CRGs were associated with a poor prognosis.
The results of GSEA showed that high-risk CRGs were positively correlated with the following KEGG pathways: Cell adhesion molecules (CAM), cytokine-cytokine receptor interaction, extracellular matrix (ECM) receptor interaction, focal adhesion, and neuroactive ligand receptor interaction (Figure 2C). Low-risk CRGs were negatively correlated with oxidative phosphorylation and ribosome pathways (Figure 2D).
The results of KEGG pathway enrichment analysis showed that high-risk CRGs were enriched in the PI3K-Akt signaling pathway, cytokine-cytokine receptor interaction, CAM, focal adhesion, proteoglycans in cancer, Rap1 signaling pathway, cAMP signaling pathway, ECM-receptor interaction, and Wnt signaling pathway, etc. (Figure 2E).
The results of GO term enrichment analysis showed that high-risk CRGs were enriched in the following biological processes: External encapsulating structure organization, ECM organization, extracellular structure organization, axon development, axonogenesis, modulation of chemical synaptic transmission, regulation of trans-synaptic signaling, sy
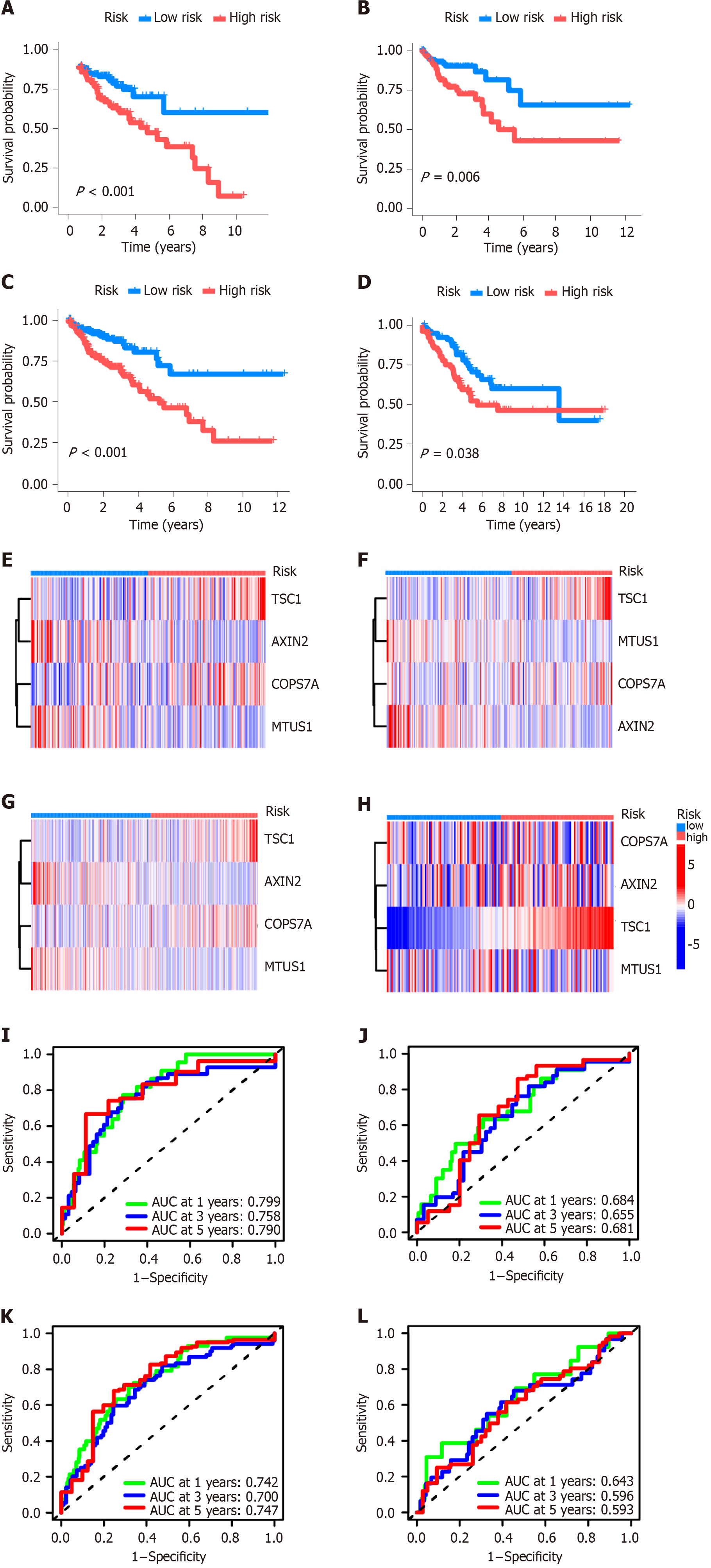
The CRS consisted of TSC1, AXIN2, COPS7A, and MTUS1 genes. Kaplan-Meier plotter analysis showed that high-risk CRS was significantly related to the prognosis of patients in the training dataset (Figure 3A) and testing datasets (Figure 3B-D). The heat maps of CRS in the training dataset and three testing datasets are shown in Figure 3E-H. In the training dataset, the area under the ROC curve for 1-, 3-, and 5-year OS was 0.799, 0.758, and 0.790, respectively (Figure 3I). In the testing dataset of the divided TCGA-COAD cohort, the area under the ROC curve for 1-, 3-, and 5-year overall OS was 0.684, 0.655, and 0.661, respectively (Figure 3J). In the testing dataset of the whole TCGA-COAD cohort, the area under the ROC curve for 1-, 3-, and 5-year OS was 0.742, 0.700, and 0.747, respectively (Figure 3K). In the GSE103479 testing dataset, the area under the ROC curve for 1-, 3-, and 5-year OS was 0.643, 0.596, and 0.593, respectively (Figure 3L).
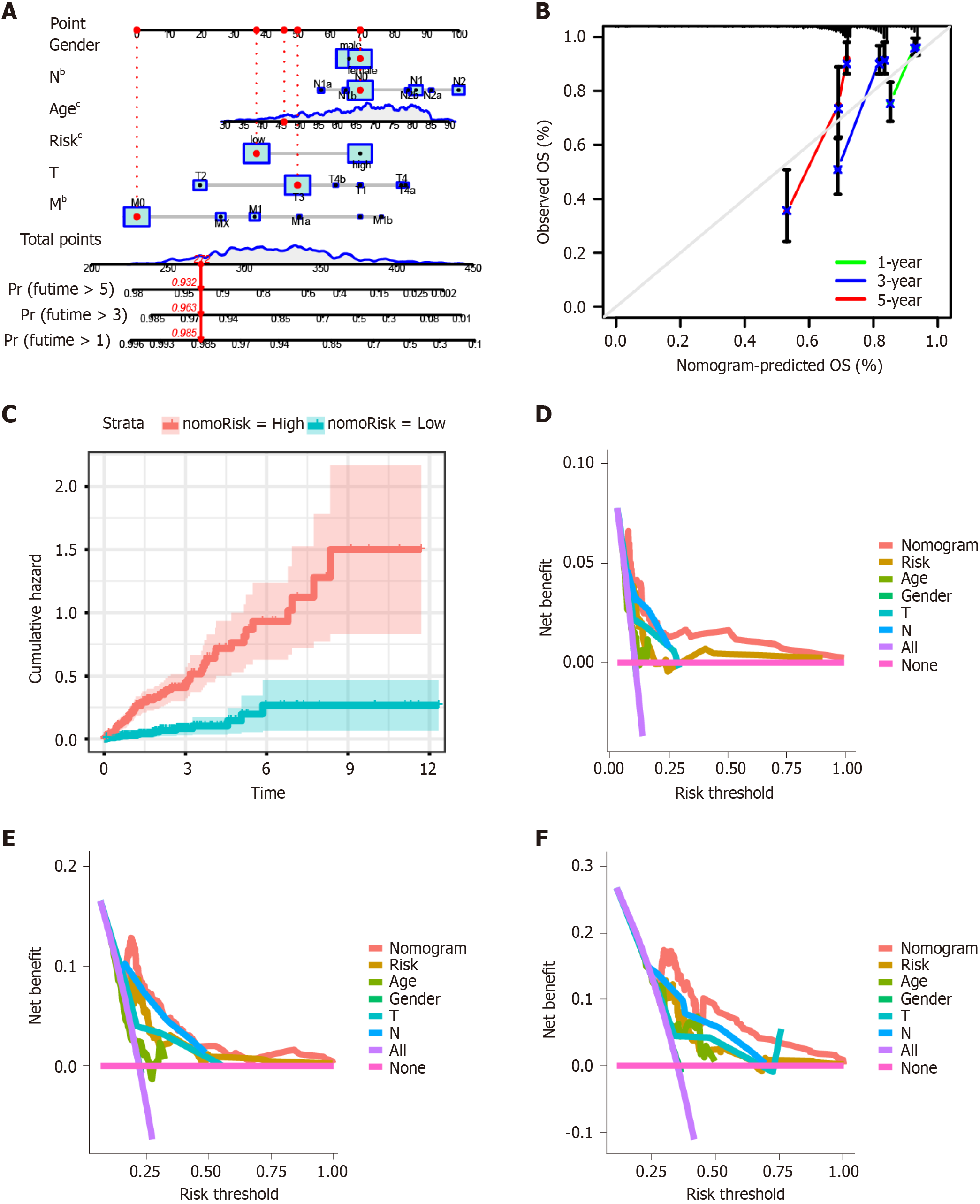
We established a nomogram model including gender, N, age, risk, T, and M using multivariate Cox and stepwise regression analyses in the TCGA-COAD cohort to estimate the 1-, 3-, and 5-year OS (Figure 4A). The calibration curves displayed the accuracy of the model in predicting the 1-, 3-, and 5-year OS (Figure 4B). The cumulative hazard index of the model showed that the cumulative hazard was significantly different between high- and low-nomorisk groups (Figure 4C). Moreover, the results of DCA showed that the accuracy of the nomogram model was the best in predicting
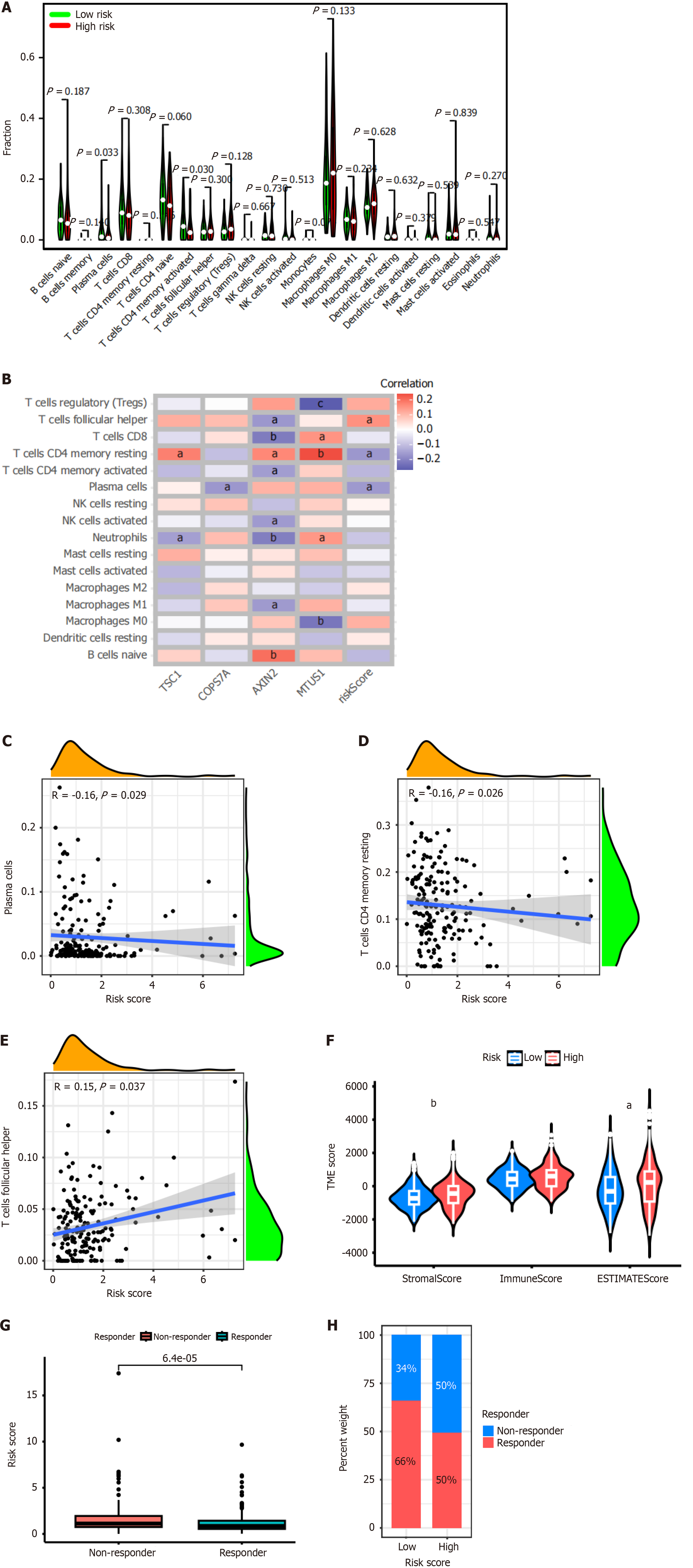
The CIBERSORT algorithm was used to analyze the distribution of 22 immune cell types in the tumor microenvironment of patients with colon cancer. The results showed that the abundance of plasma cells (P = 0.033) and activated memory CD4+ T cells (P = 0.03) was significantly lower in high-risk CRS (Figure 5A).
In addition, we analyzed the correlation between the expression of each gene of CRS and 16 immune cell types (Figure 5B). The results revealed that TSC1 expression was significantly positively correlated with resting memory CD4+ T cells (P < 0.05), and significantly negatively correlated with neutrophils (P < 0.05). COPS7A expression was significantly negatively correlated with plasma cells (P < 0.05). AXIN2 expression was significantly positively correlated with memory CD4+ T cells (P < 0.05), and naïve B cells (P < 0.01). AXIN2 expression was significantly negatively correlated with T follicular helper cells (P < 0.05), CD8+ T cells (P < 0.01), activated memory CD4+ T cells (P < 0.05), activated NK cells (P < 0.05), neutrophils (P < 0.01), and M1 macrophages (P < 0.05). MTUS1 expression was significantly positively correlated with CD8+ T cells (P < 0.05), resting memory CD4+ T cells (P < 0.01), and neutrophils (P < 0.05). MTUS1 expression was significantly negatively correlated with regulating T cells (P < 0.01), and M0 macrophages (P < 0.01). In addition, the risk score of CRS was significantly positively correlated with T follicular helper cells (P < 0.05), while significantly negatively correlated with resting memory CD4+ T cells (P < 0.05), and plasma cells (P < 0.05).
Subsequently, we analyzed the correlation between the risk score of CRS and immune cell types. The results showed that the risk score was significantly negatively correlated with plasma cells (Figure 5C, R = -0.16, P = 0.029), and resting memory CD4+ T cells (Figure 5D, R = -0.16, P = 0.026). The risk score was significantly positively correlated with T follicular helper cells (Figure 5E, R = -0.15, P = 0.037).
Furthermore, we found that the stromal score and estimate score were significantly higher in the high-risk CRS group than the low-risk CRS group (P < 0.01 and P < 0.05, respectively). However, no significant difference was found between these two groups for the immune score (Figure 5F). The results of TIDE analysis showed that patients with high-risk CRS were insensitive to immunotherapy (Figure 5G and H).
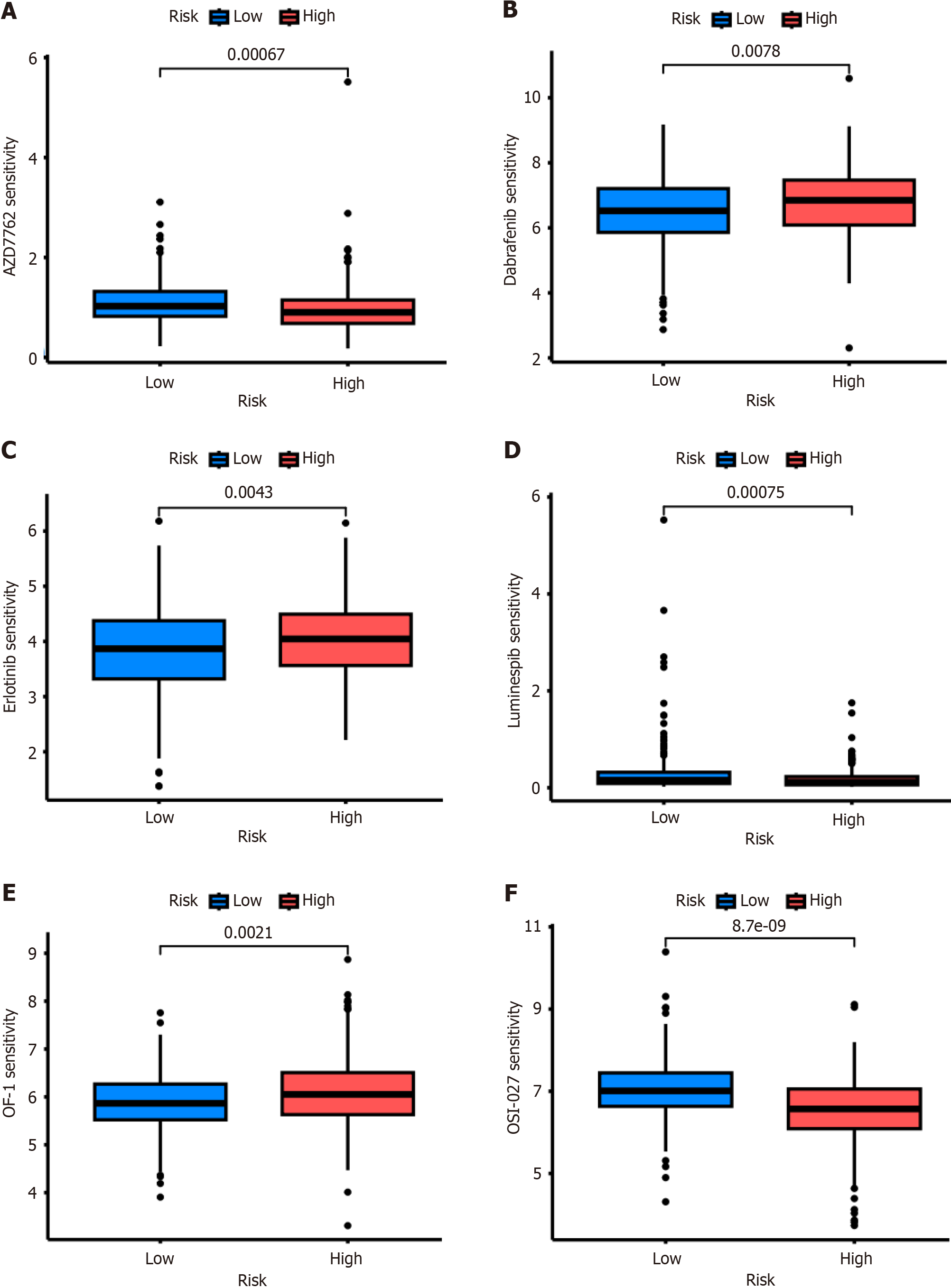
Drug sensitivity analysis showed that the patients with high-risk CRS were resistant not only to chemotherapy, such as OF-1 (Figure 6A), but also to targeted therapy, such as Dabrafenib (Figure 6B) and Erlotinib (Figure 6C). However, this high-risk group was more sensitive to AZD7762 (Figure 6D), Luminespib (Figure 6E), and OSI-027 (Figure 6F).
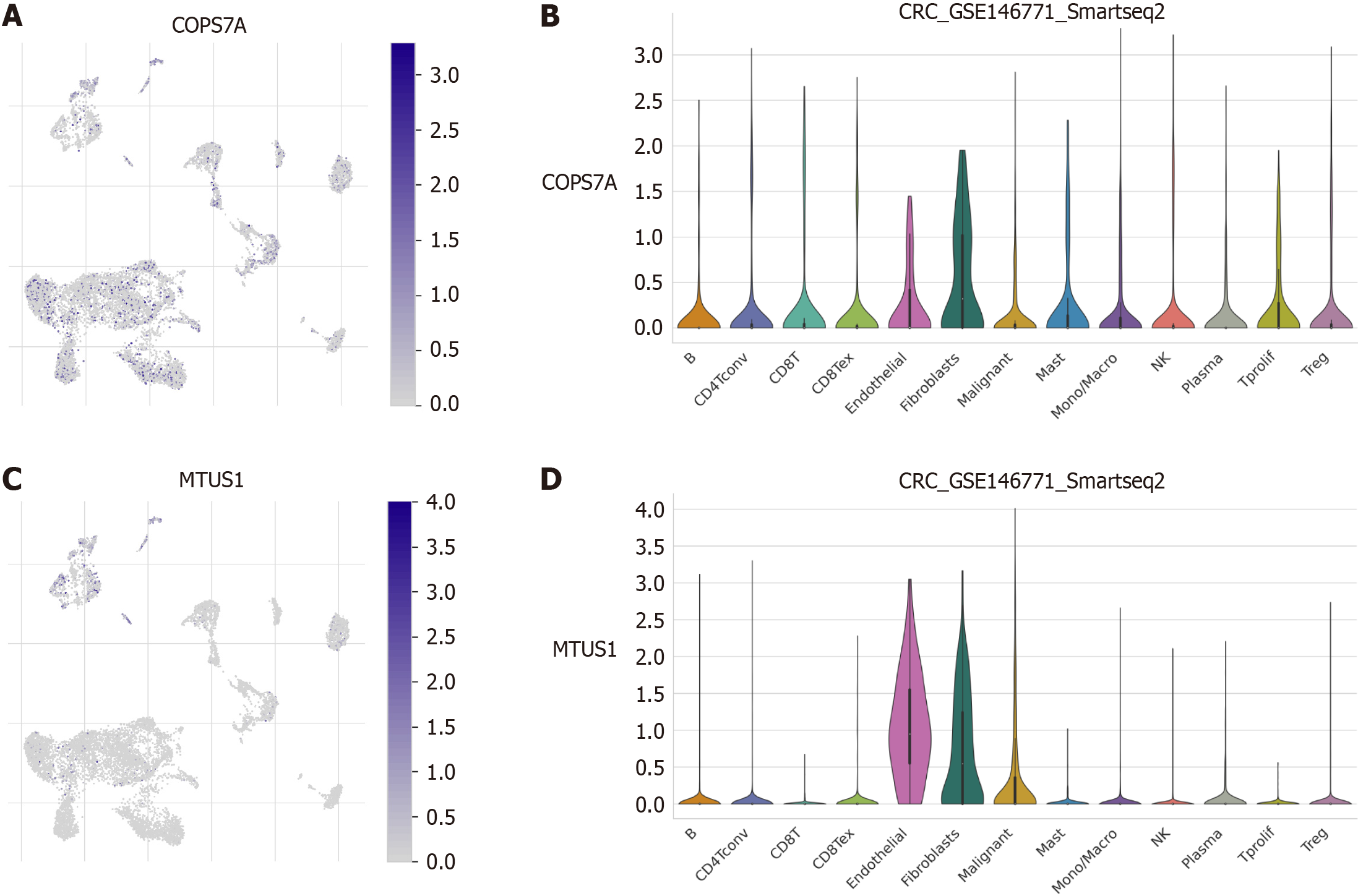
We performed single-cell transcriptome analysis to explore the immune cell landscape of the core prognostic genes of the CRS. The results revealed that COPS7A expression was relatively high in endothelial cells and fibroblasts (Figure 7A and B). MTUSI expression was high in endothelial cells, fibroblasts, and malignant cells (Figure 7C and D).
To date, most studies regarding the tools for the stratification of colon cancer patients are based on cell functions and molecular biomarkers, and few studies have focused on the role of certain subcellular structures such as centrosomes. In this study, we constructed and validated a novel prognostic CRS that independently predicts the prognosis of patients with colon cancer. The findings in this study provide a precise method predicting the prognosis and guiding treatment for patients with colon cancer.
Centrosomes are involved in the interaction between actin and tubulin, and they play a key role in the dynamics and polarity of human cells. The amplification, instability, and dysregulation of centrosomes are important factors in tumorigenesis[31].
In this study, we collected 726 CRGs from public databases. Using univariate Cox regression analysis and the LASSO algorithm, we constructed a CRS consisting of four genes (TSC1, AXIN2, COPS7A, and MTUS1) for colon cancer. TSC1 is an important component of the PI3K/AKT/MTOR signaling pathway. It plays a crucial role in cell growth, proliferation, migration, survival, autophagy, and cilia development[32-34]. Recent studies have revealed the tumor suppressor role of this gene and found that dysregulation or dysfunction of TSC1 plays a vital role in the pathogenesis of diverse human cancer types, such as liver, lung, breast, and prostate cancers[35-39]. Moreover, aberrant TSC1 is associated with poor clinical outcomes in melanoma, breast, colorectal, and gastric cancers[40-43].
AXIN2 is a crucial regulator of the Wnt-catenin signaling pathway. It is involved in cell proliferation, cytometaplasia, cell migration, and apoptosis. Although AXIN2 is a known tumor suppressor gene, recent studies have shown that it acts as an oncogene in colon cancer, liver cancer, and gastric carcinoma[44,45].
COPS7A is a member of the COP9 signalosome (CSN) complex[46]. A previous study revealed that the expression of CSN could control cell cycle progression and was associated with carcinogenesis[47]. Another study showed that the expression of COPS7A was downregulated in gastric cancer, and that COPS7A suppressed the cell proliferation of gastric cancer by inactivating the NF-κB signaling pathway[48]. A recent study reported that COPS7A expression was downregulated in breast cancer tissues[49].
MTUS1 is a tumor suppressor gene that is frequently downregulated in many human cancer types, such as pancreatic cancer, colon cancer, bladder carcinoma, head-and-neck cancer, breast cancer, gastric cancer, and lung cancer[50-59]. The low MTUS1 expression is associated with a poor prognosis in patients with various cancer types[53,55,60-63].
Subsequently, we validated the prognostic value of the signature in the TCGA training dataset and three independent testing datasets. We divided the patients with colon cancer into high- and low-risk subgroups. Kaplan-Meier plotter showed that high-risk CRS was significantly related to poor survival. The model works well, as the area under the ROC curve for 1-, 3-, and 5- year OS was 0.5-0.8 in the training dataset and three testing datasets.
To quantify the risk assessment of the signature, we constructed a nomogram based on the CRS score and several clinicopathological characteristics to estimate the 1-, 3-, and 5- year OS. The results showed that it performed well.
Furthermore, we examined whether CRS was correlated with the tumor microenvironment and response to immunotherapy. We used CIBERSORT to analyze the difference in the distribution of 22 immune cell types between high- and low-risk CRS. Our results showed that the abundance of plasma cells and activated memory CD4+ T cells was significantly lower in the high-risk group, indicating that patients in the high-risk group presented a suppressive immune microenvironment. The results of the TIDE algorithm showed that patients with a high-risk signature were immunotherapy-resistant.
In addition, our drug sensitivity analysis showed that patients with high-risk CRS were resistant not only to chem
In conclusion, we successfully constructed a novel centrosome-related prognostic signature predicting the prognosis of colon cancer patients. This model may facilitate the personalized management of colon cancer.
Centrosome abnormalities play a significant role in the development of human colon cancer, as they serve as the primary microtubule organizing center in animal cells.
The primary aim of this investigation was to explore the role of centrosome-related genes (CRGs) in the pathogenesis of colon cancer.
To examine the role of CRGs in the pathogenesis of colon cancer.
CRGs were obtained from publicly available databases. Subsequently, consensus clustering analysis was conducted to partition the Cancer Genome Atlas (TCGA)-Colon adenocarcinoma cohort. Univariate Cox and least absolute shrinkage selection operator regression analyses were employed to identify potential prognostic CRGs and establish a centrosome-related signature (CRS) for scoring patients with colon cancer. Furthermore, a nomogram was devised to assess the risk associated with the CRS in individuals diagnosed with colon cancer. A bioinformatics analysis was integrated to investigate the association between the CRS and tumor immune microenvironment, as well as the response to immunotherapy, chemotherapy, and targeted therapy. Furthermore, a single-cell transcriptome analysis was performed to examine the immune cell composition of key prognostic genes.
A cumulative count of 726 colorectal CRGs was obtained from publicly available databases. Subsequently, a colorectal cancer risk signature was developed, comprising four specific genes, namely TSC1, AXIN2, COPS7A, and MTUS1. Notably, patients with a high-risk signature exhibited unfavorable survival outcomes. Furthermore, these patients demonstrated reduced levels of plasma cells and activated memory CD4+ T cells. In terms of therapeutic response, individuals with a high-risk signature displayed resistance to immunotherapy, chemotherapy, and targeted therapy. Additionally, COPS7A expression was found to be relatively elevated in endothelial cells and fibroblasts. The expression of MTUS1 was observed to be significantly elevated in endothelial cells, fibroblasts, and malignant cells.
In light of these findings, we endeavored to develop a prognostic signature associated with CRGs that could effectively forecast the prognosis of patients with colon cancer. By doing so, we aimed to contribute to the advancement of personalized treatment strategies for individuals diagnosed with colon cancer.
This model has the potential to enhance the individualized approach to colon cancer management.
Provenance and peer review: Unsolicited article; Externally peer reviewed.
Peer-review model: Single blind
Specialty type: Oncology
Country/Territory of origin: China
Peer-review report’s scientific quality classification
Grade A (Excellent): 0
Grade B (Very good): B
Grade C (Good): 0
Grade D (Fair): 0
Grade E (Poor): 0
P-Reviewer: Pavlidis TE, Greece S-Editor: Qu XL L-Editor: Webster JR P-Editor: Zhao YQ
| 1. | Wu Z, Lu Z, Li L, Ma M, Long F, Wu R, Huang L, Chou J, Yang K, Zhang Y, Li X, Hu G, Lin C. Identification and Validation of Ferroptosis-Related LncRNA Signatures as a Novel Prognostic Model for Colon Cancer. Front Immunol. 2021;12:783362. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 19] [Cited by in RCA: 98] [Article Influence: 32.7] [Reference Citation Analysis (0)] |
| 2. | Sung H, Ferlay J, Siegel RL, Laversanne M, Soerjomataram I, Jemal A, Bray F. Global Cancer Statistics 2020: GLOBOCAN Estimates of Incidence and Mortality Worldwide for 36 Cancers in 185 Countries. CA Cancer J Clin. 2021;71:209-249. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 75126] [Cited by in RCA: 64542] [Article Influence: 16135.5] [Reference Citation Analysis (176)] |
| 3. | Chen YC, Li DB, Wang DL, Peng H. Comprehensive analysis of distal-less homeobox family gene expression in colon cancer. World J Gastrointest Oncol. 2023;15:1019-1035. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in RCA: 4] [Reference Citation Analysis (0)] |
| 4. | Zhou Y, Zang Y, Yang Y, Xiang J, Chen Z. Candidate genes involved in metastasis of colon cancer identified by integrated analysis. Cancer Med. 2019;8:2338-2347. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 9] [Cited by in RCA: 22] [Article Influence: 3.7] [Reference Citation Analysis (0)] |
| 5. | Hoffmann I. Centrosomes in mitotic spindle assembly and orientation. Curr Opin Struct Biol. 2021;66:193-198. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 12] [Cited by in RCA: 31] [Article Influence: 6.2] [Reference Citation Analysis (0)] |
| 6. | Mittal K, Kaur J, Jaczko M, Wei G, Toss MS, Rakha EA, Janssen EAM, Søiland H, Kucuk O, Reid MD, Gupta MV, Aneja R. Centrosome amplification: a quantifiable cancer cell trait with prognostic value in solid malignancies. Cancer Metastasis Rev. 2021;40:319-339. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 16] [Cited by in RCA: 27] [Article Influence: 6.8] [Reference Citation Analysis (0)] |
| 7. | Morretton JP, Simon A, Herbette A, Barbazan J, Pérez-González C, Cosson C, Mboup B, Latouche A, Popova T, Kieffer Y, Macé AS, Gestraud P, Bataillon G, Becette V, Meseure D, Nicolas A, Mariani O, Vincent-Salomon A, Stern MH, Mechta-Grigoriou F, Roman Roman S, Vignjevic DM, Rouzier R, Sastre-Garau X, Goundiam O, Basto R. A catalog of numerical centrosome defects in epithelial ovarian cancers. EMBO Mol Med. 2022;14:e15670. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 12] [Cited by in RCA: 16] [Article Influence: 5.3] [Reference Citation Analysis (0)] |
| 8. | Dang H, Schiebel E. Emerging roles of centrosome cohesion. Open Biol. 2022;12:220229. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in RCA: 15] [Reference Citation Analysis (0)] |
| 9. | Lin Z, Huang W, Yi Y, Li D, Xie Z, Li Z, Ye M. LncRNA ADAMTS9-AS2 is a Prognostic Biomarker and Correlated with Immune Infiltrates in Lung Adenocarcinoma. Int J Gen Med. 2021;14:8541-8555. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 4] [Cited by in RCA: 33] [Article Influence: 8.3] [Reference Citation Analysis (0)] |
| 10. | Yi W, Shen H, Sun D, Xu Y, Feng Y, Li D, Wang C. Low Expression of Long Noncoding RNA SLC26A4 Antisense RNA 1 Is an Independent Prognostic Biomarker and Correlate of Immune Infiltrates in Breast Cancer. Med Sci Monit. 2021;27:e934522. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Reference Citation Analysis (0)] |
| 11. | Chen J, Tang H, Li T, Jiang K, Zhong H, Wu Y, He J, Li D, Li M, Cai X. Comprehensive Analysis of the Expression, Prognosis, and Biological Significance of OVOLs in Breast Cancer. Int J Gen Med. 2021;14:3951-3960. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 5] [Cited by in RCA: 21] [Article Influence: 5.3] [Reference Citation Analysis (0)] |
| 12. | Yang D, Liu M, Jiang J, Luo Y, Wang Y, Chen H, Li D, Wang D, Yang Z. Comprehensive Analysis of DMRT3 as a Potential Biomarker Associated with the Immune Infiltration in a Pan-Cancer Analysis and Validation in Lung Adenocarcinoma. Cancers (Basel). 2022;14. [RCA] [PubMed] [DOI] [Full Text] [Cited by in RCA: 17] [Reference Citation Analysis (0)] |
| 13. | Jiao J, Jiang L, Luo Y. N6-Methyladenosine-Related RNA Signature Predicting the Prognosis of Ovarian Cancer. Recent Pat Anticancer Drug Discov. 2021;16:407-416. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1] [Cited by in RCA: 3] [Article Influence: 0.8] [Reference Citation Analysis (0)] |
| 14. | Xie L, Pan M, Zhang Z, Jiang X, Chen Y, Liu G, Zeng Y, Guan J, Lu R, Zeng L. Development and Validation of a Hypoxia-related Prognostic Model for Ovarian Cancer. Recent Pat Anticancer Drug Discov. 2022;18:161-173. [RCA] [PubMed] [DOI] [Full Text] [Reference Citation Analysis (0)] |
| 15. | Li M, Wang X, Liu J, Mao X, Li D, Wang Z, Tang Y, Wu S. Identification of Core Prognosis-Related Candidate Genes in Chinese Gastric Cancer Population Based on Integrated Bioinformatics. Biomed Res Int. 2020;2020:8859826. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 2] [Cited by in RCA: 16] [Article Influence: 3.2] [Reference Citation Analysis (0)] |
| 16. | Wilkerson MD, Hayes DN. ConsensusClusterPlus: a class discovery tool with confidence assessments and item tracking. Bioinformatics. 2010;26:1572-1573. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 1686] [Cited by in RCA: 3787] [Article Influence: 252.5] [Reference Citation Analysis (0)] |
| 17. | Yang Y, Gu X, Li Z, Zheng C, Wang Z, Zhou M, Chen Z, Li M, Li D, Xiang J. Whole-exome sequencing of rectal cancer identifies locally recurrent mutations in the Wnt pathway. Aging (Albany NY). 2021;13:23262-23283. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in RCA: 8] [Reference Citation Analysis (0)] |
| 18. | He W, Dong S, Shen J, Wu J, Zhao P, Li D, Wang D, Tang N, Zou C. Whole-genome sequencing identified novel mutations in a Chinese family with lynch syndrome. Front Oncol. 2023;13:1036356. [RCA] [PubMed] [DOI] [Full Text] [Reference Citation Analysis (0)] |
| 19. | Lyu G, Li D, Xiong H, Xiao L, Tong J, Ning C, Wang P, Li S. Quantitative Proteomic Analyses Identify STO/BBX24 -Related Proteins Induced by UV-B. Int J Mol Sci. 2020;21. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 3] [Cited by in RCA: 10] [Article Influence: 2.0] [Reference Citation Analysis (0)] |
| 20. | Wu T, Hu E, Xu S, Chen M, Guo P, Dai Z, Feng T, Zhou L, Tang W, Zhan L, Fu X, Liu S, Bo X, Yu G. clusterProfiler 4.0: A universal enrichment tool for interpreting omics data. Innovation (Camb). 2021;2:100141. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 522] [Cited by in RCA: 4752] [Article Influence: 1188.0] [Reference Citation Analysis (0)] |
| 21. | Zong Z, Hu CG, Zhou TC, Yu ZM, Tang FX, Tian HK, Li H, Wang H. Nine-long non-coding ribonucleic acid signature can improve the survival prediction of colorectal cancer. World J Gastrointest Surg. 2021;13:210-221. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in CrossRef: 1] [Cited by in RCA: 1] [Article Influence: 0.3] [Reference Citation Analysis (0)] |
| 22. | Liu M, Li L, Huang S, Pan X, Dai H, Chen ZS, Pan Y, Fang S. Prognostic and Therapeutic Values of Autophagy-related Genes in Triple-negative Breast Cancer. Recent Pat Anticancer Drug Discov. 2022;17:380-386. [RCA] [PubMed] [DOI] [Full Text] [Cited by in RCA: 6] [Reference Citation Analysis (0)] |
| 23. | Blanche P, Dartigues JF, Jacqmin-Gadda H. Estimating and comparing time-dependent areas under receiver operating characteristic curves for censored event times with competing risks. Stat Med. 2013;32:5381-5397. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 564] [Cited by in RCA: 1133] [Article Influence: 94.4] [Reference Citation Analysis (0)] |
| 24. | Han QL, Cui Z, Wang Q, Pang F, Li D, Wang D. Upregulation of OTX2-AS1 is Associated With Immune Infiltration and Predicts Prognosis of Gastric Cancer. Technol Cancer Res Treat. 2023;22:15330338231154091. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in RCA: 10] [Reference Citation Analysis (0)] |
| 25. | Liang W, Lu Y, Pan X, Zeng Y, Zheng W, Li Y, Nie Y, Li D, Wang D. Decreased Expression of a Novel lncRNA FAM181A-AS1 is Associated with Poor Prognosis and Immune Infiltration in Lung Adenocarcinoma. Pharmgenomics Pers Med. 2022;15:985-998. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1] [Cited by in RCA: 11] [Article Influence: 3.7] [Reference Citation Analysis (0)] |
| 26. | Newman AM, Liu CL, Green MR, Gentles AJ, Feng W, Xu Y, Hoang CD, Diehn M, Alizadeh AA. Robust enumeration of cell subsets from tissue expression profiles. Nat Methods. 2015;12:453-457. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 4763] [Cited by in RCA: 8891] [Article Influence: 889.1] [Reference Citation Analysis (0)] |
| 27. | Thorsson V, Gibbs DL, Brown SD, Wolf D, Bortone DS, Ou Yang TH, Porta-Pardo E, Gao GF, Plaisier CL, Eddy JA, Ziv E, Culhane AC, Paull EO, Sivakumar IKA, Gentles AJ, Malhotra R, Farshidfar F, Colaprico A, Parker JS, Mose LE, Vo NS, Liu J, Liu Y, Rader J, Dhankani V, Reynolds SM, Bowlby R, Califano A, Cherniack AD, Anastassiou D, Bedognetti D, Mokrab Y, Newman AM, Rao A, Chen K, Krasnitz A, Hu H, Malta TM, Noushmehr H, Pedamallu CS, Bullman S, Ojesina AI, Lamb A, Zhou W, Shen H, Choueiri TK, Weinstein JN, Guinney J, Saltz J, Holt RA, Rabkin CS; Cancer Genome Atlas Research Network, Lazar AJ, Serody JS, Demicco EG, Disis ML, Vincent BG, Shmulevich I. The Immune Landscape of Cancer. Immunity. 2018;48:812-830.e14. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 4007] [Cited by in RCA: 3815] [Article Influence: 545.0] [Reference Citation Analysis (0)] |
| 28. | Sturm G, Finotello F, Petitprez F, Zhang JD, Baumbach J, Fridman WH, List M, Aneichyk T. Comprehensive evaluation of transcriptome-based cell-type quantification methods for immuno-oncology. Bioinformatics. 2019;35:i436-i445. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 294] [Cited by in RCA: 609] [Article Influence: 121.8] [Reference Citation Analysis (0)] |
| 29. | Fu J, Li K, Zhang W, Wan C, Zhang J, Jiang P, Liu XS. Large-scale public data reuse to model immunotherapy response and resistance. Genome Med. 2020;12:21. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 178] [Cited by in RCA: 728] [Article Influence: 145.6] [Reference Citation Analysis (0)] |
| 30. | Maeser D, Gruener RF, Huang RS. oncoPredict: an R package for predicting in vivo or cancer patient drug response and biomarkers from cell line screening data. Brief Bioinform. 2021;22. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 46] [Cited by in RCA: 1015] [Article Influence: 253.8] [Reference Citation Analysis (0)] |
| 31. | Zhao JZ, Ye Q, Wang L, Lee SC. Centrosome amplification in cancer and cancer-associated human diseases. Biochim Biophys Acta Rev Cancer. 2021;1876:188566. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 3] [Cited by in RCA: 21] [Article Influence: 5.3] [Reference Citation Analysis (0)] |
| 32. | Xie Y, Zhao Y, Shi L, Li W, Chen K, Li M, Chen X, Zhang H, Li T, Matsuzawa-Ishimoto Y, Yao X, Shao D, Ke Z, Li J, Chen Y, Zhang X, Cui J, Cui S, Leng Q, Cadwell K, Li X, Wei H, Li H, Xiao H. Gut epithelial TSC1/mTOR controls RIPK3-dependent necroptosis in intestinal inflammation and cancer. J Clin Invest. 2020;130:2111-2128. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 59] [Cited by in RCA: 144] [Article Influence: 28.8] [Reference Citation Analysis (0)] |
| 33. | Lai M, Zou W, Han Z, Zhou L, Qiu Z, Chen J, Zhang S, Lai P, Li K, Zhang Y, Liang L, Jiang Y, Zou Z, Bai X. Tsc1 regulates tight junction independent of mTORC1. Proc Natl Acad Sci U S A. 2021;118. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 3] [Cited by in RCA: 10] [Article Influence: 2.5] [Reference Citation Analysis (0)] |
| 34. | Peixoto E, Richard S, Pant K, Biswas A, Gradilone SA. The primary cilium: Its role as a tumor suppressor organelle. Biochem Pharmacol. 2020;175:113906. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 15] [Cited by in RCA: 32] [Article Influence: 6.4] [Reference Citation Analysis (0)] |
| 35. | Dong P, Wang X, Liu L, Tang W, Ma L, Zeng W, Sun S, Zhang L, Zhang N, Shen X, Janssen HLA, Dong L, Zhang S, Chen S. Dampened VEPH1 activates mTORC1 signaling by weakening the TSC1/TSC2 association in hepatocellular carcinoma. J Hepatol. 2020;73:1446-1459. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 17] [Cited by in RCA: 29] [Article Influence: 5.8] [Reference Citation Analysis (0)] |
| 36. | Lee MC, Cai H, Murray CW, Li C, Shue YT, Andrejka L, He AL, Holzem AME, Drainas AP, Ko JH, Coles GL, Kong C, Zhu S, Zhu C, Wang J, van de Rijn M, Petrov DA, Winslow MM, Sage J. A multiplexed in vivo approach to identify driver genes in small cell lung cancer. Cell Rep. 2023;42:111990. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 14] [Cited by in RCA: 19] [Article Influence: 9.5] [Reference Citation Analysis (0)] |
| 37. | Ilozumba MN, Yao S, Llanos AAM, Omilian AR, Zhang W, Datta S, Hong CC, Davis W, Khoury T, Bandera EV, Higgins M, Ambrosone CB, Cheng TD. mTOR pathway gene expression in association with race and clinicopathological characteristics in Black and White breast cancer patients. Discov Oncol. 2022;13:34. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 2] [Cited by in RCA: 6] [Article Influence: 2.0] [Reference Citation Analysis (0)] |
| 38. | Ye F, Dewanjee S, Li Y, Jha NK, Chen ZS, Kumar A, Vishakha, Behl T, Jha SK, Tang H. Advancements in clinical aspects of targeted therapy and immunotherapy in breast cancer. Mol Cancer. 2023;22:105. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 254] [Cited by in RCA: 259] [Article Influence: 129.5] [Reference Citation Analysis (0)] |
| 39. | Hu D, Jiang L, Luo S, Zhao X, Hu H, Zhao G, Tang W. Development of an autophagy-related gene expression signature for prognosis prediction in prostate cancer patients. J Transl Med. 2020;18:160. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 41] [Cited by in RCA: 73] [Article Influence: 14.6] [Reference Citation Analysis (0)] |
| 40. | Mallela K, Kumar A. Role of TSC1 in physiology and diseases. Mol Cell Biochem. 2021;476:2269-2282. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 8] [Cited by in RCA: 28] [Article Influence: 7.0] [Reference Citation Analysis (0)] |
| 41. | Du M, Wang Y, Gu D, Guo L. Identification of vital genes and pathways associated with mucosal melanoma in Chinese. Ann Diagn Pathol. 2021;50:151648. [RCA] [PubMed] [DOI] [Full Text] [Cited by in RCA: 2] [Reference Citation Analysis (0)] |
| 42. | Wang X, Xu Y, Li T, Chen B, Yang W. Development of prognosis model for colon cancer based on autophagy-related genes. World J Surg Oncol. 2020;18:285. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 4] [Cited by in RCA: 12] [Article Influence: 2.4] [Reference Citation Analysis (0)] |
| 43. | Byeon SJ, Han N, Choi J, Kim MA, Kim WH. Prognostic implication of TSC1 and mTOR expression in gastric carcinoma. J Surg Oncol. 2014;109:812-817. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 14] [Cited by in RCA: 19] [Article Influence: 1.7] [Reference Citation Analysis (0)] |
| 44. | Leclerc J, Beaumont M, Vibert R, Pinson S, Vermaut C, Flament C, Lovecchio T, Delattre L, Demay C, Coulet F, Guillerm E, Hamzaoui N, Benusiglio PR, Brahimi A, Cornelis F, Delhomelle H, Fert-Ferrer S, Fournier BPJ, Hovnanian A, Legrand C, Lortholary A, Malka D, Petit F, Saurin JC, Lejeune S, Colas C, Buisine MP. AXIN2 germline testing in a French cohort validates pathogenic variants as a rare cause of predisposition to colorectal polyposis and cancer. Genes Chromosomes Cancer. 2023;62:210-222. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in RCA: 7] [Reference Citation Analysis (0)] |
| 45. | Wang W, Liu P, Lavrijsen M, Li S, Zhang R, van de Geer WS, van de Werken HJG, Peppelenbosch MP, Smits R. Evaluation of AXIN1 and AXIN2 as targets of tankyrase inhibition in hepatocellular carcinoma cell lines. Sci Rep. 2021;11:7470. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 2] [Cited by in RCA: 23] [Article Influence: 5.8] [Reference Citation Analysis (1)] |
| 46. | Davidsson J, Johansson B. Methylation and expression analyses of Pallister-Killian syndrome reveal partial dosage compensation of tetrasomy 12p and hypomethylation of gene-poor regions on 12p. Epigenetics. 2016;11:194-204. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 4] [Cited by in RCA: 3] [Article Influence: 0.3] [Reference Citation Analysis (0)] |
| 47. | Yoshida A, Yoneda-Kato N, Panattoni M, Pardi R, Kato JY. CSN5/Jab1 controls multiple events in the mammalian cell cycle. FEBS Lett. 2010;584:4545-4552. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 24] [Cited by in RCA: 22] [Article Influence: 1.5] [Reference Citation Analysis (0)] |
| 48. | Zheng J, Zhang H, Ma R, Liu H, Gao P. Long non-coding RNA KRT19P3 suppresses proliferation and metastasis through COPS7A-mediated NF-κB pathway in gastric cancer. Oncogene. 2019;38:7073-7088. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 33] [Cited by in RCA: 53] [Article Influence: 8.8] [Reference Citation Analysis (0)] |
| 49. | He X, Xiao H, Yang R, Chen H, Wang B. lncRNA LOC339524 inhibits the proliferation of bladder cancer cells by targeting the miR-875-5p/COPS7A signaling axis. Exp Ther Med. 2021;22:1202. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in RCA: 1] [Reference Citation Analysis (0)] |
| 50. | Seibold S, Rudroff C, Weber M, Galle J, Wanner C, Marx M. Identification of a new tumor suppressor gene located at chromosome 8p21.3-22. FASEB J. 2003;17:1180-1182. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 80] [Cited by in RCA: 86] [Article Influence: 3.9] [Reference Citation Analysis (0)] |
| 51. | Ozcan O, Kara M, Yumrutas O, Bozgeyik E, Bozgeyik I, Celik OI. MTUS1 and its targeting miRNAs in colorectal carcinoma: significant associations. Tumour Biol. 2016;37:6637-6645. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 22] [Cited by in RCA: 26] [Article Influence: 2.6] [Reference Citation Analysis (0)] |
| 52. | Loo LW, Tiirikainen M, Cheng I, Lum-Jones A, Seifried A, Church JM, Gryfe R, Weisenberger DJ, Lindor NM, Gallinger S, Haile RW, Duggan DJ, Thibodeau SN, Casey G, Le Marchand L. Integrated analysis of genome-wide copy number alterations and gene expression in microsatellite stable, CpG island methylator phenotype-negative colon cancer. Genes Chromosomes Cancer. 2013;52:450-466. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 44] [Cited by in RCA: 55] [Article Influence: 4.6] [Reference Citation Analysis (0)] |
| 53. | Rogler A, Hoja S, Giedl J, Ekici AB, Wach S, Taubert H, Goebell PJ, Wullich B, Stöckle M, Lehmann J, Petsch S, Hartmann A, Stoehr R. Loss of MTUS1/ATIP expression is associated with adverse outcome in advanced bladder carcinomas: data from a retrospective study. BMC Cancer. 2014;14:214. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 14] [Cited by in RCA: 15] [Article Influence: 1.4] [Reference Citation Analysis (0)] |
| 54. | Ding X, Zhang N, Cai Y, Li S, Zheng C, Jin Y, Yu T, Wang A, Zhou X. Down-regulation of tumor suppressor MTUS1/ATIP is associated with enhanced proliferation, poor differentiation and poor prognosis in oral tongue squamous cell carcinoma. Mol Oncol. 2012;6:73-80. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 42] [Cited by in RCA: 46] [Article Influence: 3.5] [Reference Citation Analysis (0)] |
| 55. | Zhao T, Ding X, Chang B, Zhou X, Wang A. MTUS1/ATIP3a down-regulation is associated with enhanced migration, invasion and poor prognosis in salivary adenoid cystic carcinoma. BMC Cancer. 2015;15:203. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 20] [Cited by in RCA: 31] [Article Influence: 3.1] [Reference Citation Analysis (0)] |
| 56. | Molina A, Velot L, Ghouinem L, Abdelkarim M, Bouchet BP, Luissint AC, Bouhlel I, Morel M, Sapharikas E, Di Tommaso A, Honoré S, Braguer D, Gruel N, Vincent-Salomon A, Delattre O, Sigal-Zafrani B, André F, Terris B, Akhmanova A, Di Benedetto M, Nahmias C, Rodrigues-Ferreira S. ATIP3, a novel prognostic marker of breast cancer patient survival, limits cancer cell migration and slows metastatic progression by regulating microtubule dynamics. Cancer Res. 2013;73:2905-2915. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 41] [Cited by in RCA: 43] [Article Influence: 3.6] [Reference Citation Analysis (0)] |
| 57. | Frank B, Bermejo JL, Hemminki K, Sutter C, Wappenschmidt B, Meindl A, Kiechle-Bahat M, Bugert P, Schmutzler RK, Bartram CR, Burwinkel B. Copy number variant in the candidate tumor suppressor gene MTUS1 and familial breast cancer risk. Carcinogenesis. 2007;28:1442-1445. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 68] [Cited by in RCA: 75] [Article Influence: 4.2] [Reference Citation Analysis (0)] |
| 58. | Li X, Liu H, Yu T, Dong Z, Tang L, Sun X. Loss of MTUS1 in gastric cancer promotes tumor growth and metastasis. Neoplasma. 2014;61:128-135. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 20] [Cited by in RCA: 21] [Article Influence: 1.9] [Reference Citation Analysis (0)] |
| 59. | Westcott PM, Halliwill KD, To MD, Rashid M, Rust AG, Keane TM, Delrosario R, Jen KY, Gurley KE, Kemp CJ, Fredlund E, Quigley DA, Adams DJ, Balmain A. The mutational landscapes of genetic and chemical models of Kras-driven lung cancer. Nature. 2015;517:489-492. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 242] [Cited by in RCA: 267] [Article Influence: 24.3] [Reference Citation Analysis (0)] |
| 60. | Park H, Jee S, Son H, Cha H, Bang S, Ahn BK, Myung J, Paik S, Kim H. Low MTUS1 Protein Expression Is Associated with Poor Survival in Patients with Colorectal Adenocarcinoma. Diagnostics (Basel). 2023;13. [RCA] [PubMed] [DOI] [Full Text] [Reference Citation Analysis (0)] |
| 61. | Sim J, Wi YC, Park HY, Park SY, Yoon YE, Bang S, Kim Y, Jang K, Paik SS, Shin SJ. Clinicopathological Significance of MTUS1 Expression in Patients With Renal Cell Carcinoma. Anticancer Res. 2020;40:2961-2967. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 3] [Cited by in RCA: 6] [Article Influence: 1.2] [Reference Citation Analysis (0)] |
| 62. | Jee S, Kim H, Bang S, Kim Y, Park HY, Paik SS, Sim J, Jang K. Low-Level Expression of MTUS1 Is Associated with Poor Survival in Patients with Lung Adenocarcinoma. Diagnostics (Basel). 2021;11. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 2] [Cited by in RCA: 5] [Article Influence: 1.3] [Reference Citation Analysis (0)] |
| 63. | Sim J, Kim Y, Kim H, Bang S, Jee S, Park S, Shin SJ, Jang K. Loss of MTUS1 Expression Is Associated With Poor Prognosis in Patients With Gallbladder Carcinoma. In Vivo. 2020;34:125-132. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 4] [Cited by in RCA: 4] [Article Influence: 0.8] [Reference Citation Analysis (0)] |