Published online May 15, 2024. doi: 10.4251/wjgo.v16.i5.1890
Peer-review started: October 19, 2023
First decision: December 29, 2023
Revised: January 10, 2024
Accepted: March 14, 2024
Article in press: March 14, 2024
Published online: May 15, 2024
Processing time: 202 Days and 21.5 Hours
Serpin peptidase inhibitor clade H member 1 (SERPINH1) was initially recog
To investigate the effects of SERPINH1 on CRC cells and its specific mechanism.
Quantitative real-time polymerase chain reaction, western blotting analysis, The Cancer Genome Atlas data mining and immunohistochemistry were employed to examine SERPINH1 expression in CRC cell lines and tissues. A series of in-vitro assays were performed to demonstrate the function of SERPINH1 and its possible mechanisms in CRC.
SERPINH1 demonstrated elevated expression levels in both CRC cells and tissues, manifested at both mRNA and protein tiers. Elevated SERPINH1 levels correlated closely with advanced T stage, lymph node involvement, and distant metastasis, exhibiting a significant association with poorer overall survival among CRC pa
These findings imply a crucial involvement of SERPINH1 in the advancement and escalation of CRC, potentially positioning it as a novel candidate for prognostic assessment and therapeutic intervention in CRC management.
Core Tip: The expression of serpin peptidase inhibitor clade H member 1 (SERPINH1) was observed to be elevated in both colorectal cancer (CRC) cells and tissues at mRNA and protein levels. Increased SERPINH1 expression demonstrated a close association with the T stage, lymph node status, and distant metastasis in CRC patients, displaying a significant correlation with poor overall survival. Subsequent investigations revealed that the overexpression of SERPINH1 markedly enhanced the in vitro proliferation, invasion, and migration capabilities of CRC cells. Conversely, the knockdown of SERPINH1 resulted in the opposite effects. In addition, our study validated that the overexpression of SERPINH1 could stimulate the G1/S phase cell cycle transition through the activation of the phosphatidylinositol 3-kinase (PI3
- Citation: Jin XS, Chen LX, Ji TT, Li RZ. SERPINH1 promoted the proliferation and metastasis of colorectal cancer by activating PI3K/Akt/mTOR signaling pathway. World J Gastrointest Oncol 2024; 16(5): 1890-1907
- URL: https://www.wjgnet.com/1948-5204/full/v16/i5/1890.htm
- DOI: https://dx.doi.org/10.4251/wjgo.v16.i5.1890
Colorectal cancer (CRC) ranks among the prevailing malignancies globally, posing a significant global health challenge. Despite advances in early detection and therapeutic modalities, CRC continues to be a significant contributor to cancer-related morbidity and mortality. Deciphering the molecular mechanisms driving CRC progression is imperative for the advancement of innovative diagnostic and therapeutic approaches.
The serpin peptidase inhibitor, clade H [heat shock protein 47 (HSP47)], member 1, commonly referred to as serpin peptidase inhibitor clade H member 1 (SERPINH1) or HSP47, is a molecular chaperone protein primarily known for its role in collagen biosynthesis and extracellular matrix homeostasis[1]. The synthesis of HSP47 consistently mirrors that of collagen in developing tissues, various cell lines, and pathological conditions associated with collagen, such as fibrosis[2].
Emerging evidence suggests that SERPINH1 may also play a pivotal role in cancer biology. The upregulation of SERPINH1 has been observed in various cancer types, and it has been associated with tumor growth, metastasis, and poor clinical outcomes[3-5]. For instance, increased SERPINH1 expression regulated EMT and gastric cancer metastasis via the Wnt/β-catenin signaling pathway[3]. A study conducted by Pan et al[6] demonstrated that SERPINH1 holds promise as a candidate biomarker for stratifying Clear cell renal cell carcinoma patients into low- and high-risk groups. Furthermore, SERPINH1 has been shown to be associated with dasatinib responses and interstitial subtypes[7], and circCAMSAP1 promotes NPC proliferation and metastasis by binding to the 3’ untranslated region of SERPINH1 nasopharyngeal carcinoma[4]. Additionally, it has been reported that SERPINH1 (HSP47) enhances CRC cell survival by suppressing apoptosis, elevating AKT phosphorylation, and downregulating the expression of the AKT-specific phosphatase PHLPP1 in response to chemotherapy exposure[8]. However, in addition to the effect on CRC cells treated with chemotherapy, the role of SERPINH1 in CRC progression and its underlying molecular mechanisms remain poorly understood in the current scientific literature.
In this study, we observed a marked elevation of SERPINH1 expression in both CRC cell lines and tissues. Subsequent in vitro experiments revealed that the overexpression of SERPINH1 resulted in a substantial enhancement of CRC cell proliferation, invasion, migration, and tumorigenic potential. Conversely, the silencing of SERPINH1 had the opposite effect, inhibiting these cellular processes. Furthermore, we extended our inquiry to elucidate the molecular mechanisms associated with SERPINH1-mediated effects. It was evident that the upregulation of SERPINH1 accelerated the transition from G1 to S phase of the cell cycle. Of particular significance, this transition was initiated by SERPINH1 through the activation of AKT signaling, concurrent with the downregulation of mechanistic target of rapamycin (mTOR) activity and FOXO1 transcriptional competence. Moreover, we observed that the inhibition of the mTOR pathway had the capacity to induce the suppression of invasion, migration, and proliferation in cells with elevated SERPINH1 expression. The results of this study indicate that SERPINH1 may act as an oncogene in the progression of CRC and could serve as a valuable prognostic indicator for this malignancy.
CRC tissues were collected from the Third Affiliated Hospital of Wenzhou Medical University. The specimen was stored at -80 °C immediately after surgery. All patients were diagnosed with CRC. Approval for this study was obtained from the Ethics Committee of the Third Affiliated Hospital of Wenzhou Medical University. The clinicopathological infor
| Parameters | Group | Cases | SERPINH1 expression | ||
| Low | High | P value | |||
| Age | ≤ 53 | 24 | 10 | 14 | 0.8315 |
| > 53 | 36 | 16 | 20 | ||
| Gender | Male | 32 | 15 | 17 | 0.2837 |
| Female | 28 | 17 | 11 | ||
| T grade | T1 + T2 | 14 | 6 | 8 | 0.8032 |
| T3 + T4 | 46 | 18 | 28 | ||
| Tumor size | ≤ 5 cm | 23 | 10 | 13 | 0.2777 |
| > 5 cm | 37 | 11 | 26 | ||
| Stage | I-II | 23 | 15 | 8 | 0.0029a |
| III-IV | 37 | 10 | 27 | ||
| Distant metastasis | M0 | 34 | 24 | 10 | 0.0046a |
| M1 | 26 | 7 | 19 | ||
| Lymphatic invasion | Negative (N0) | 25 | 11 | 14 | 0.1478 |
| Positive (N1-N3) | 35 | 22 | 13 | ||
To investigate the role of SERPINH1 in CRC, HCT116 were transfected with SERPINH1 overexpression plasmids or control vectors using lipofectamine 3000 according to the manufacturer’s instructions. To knock down SERPINH1 ex
CRC cells and tissues were harvested and lysed in immunoprecipitation buffer. Protein quantification was performed using the BCA method, and 20 μg of protein samples were subjected to electrophoresis. Subsequently, membranes were exposed to primary antibodies overnight at 4 °C. Following this, horseradish peroxidase-conjugated secondary antibodies were applied and incubated for 1 h. Protein bands were visualized using the ChemiDocXRS+ System.
According to the manufacturer's instructions, the proliferation of CRC cells was detected by cell counting kit 8 (CCK-8) assay (APExBIO, Houston, United States). Ten microliters of CCK-8 solution were added to each well and incubated at 37 °C for 2 h. The absorbance at 450 nm was measured three times every 24 h using spectrophotometry.
5-ethynyl-2’-deoxyuridine (EdU) assays were conducted utilizing the cell-light EdU DNA cell proliferation kit (RiboBio, Guangzhou, China) as per the manufacturer’s instructions. The quantification of positive cells was determined based on the ratio of red to blue fluorescence staining.
A total of 1000 cells per well were seeded into a six-well plate and allowed to proliferate until reaching confluence. Following 2-3 wk of incubation, the cells were fixed with 4% paraformaldehyde for 15 min, stained with 0.1% crystal violet solution for 10 min, and the number of colonies was enumerated. The mean values were derived from three inde
Transwell chambers with 8 μm pore size were employed following the manufacturer’s protocol. CRC cell suspension (1 × 105 cells/well) in serum-free medium was seeded into the upper chamber, while medium supplemented with 10% foetal bovine serum was added to the lower compartment. Subsequently, residual cells above the chamber were wiped away using a cotton ball, and migrated cells adhering to the membrane were fixed with 4% paraformaldehyde and stained with crystal violet. The stained cells were randomly photographed and counted from three different visual fields.
Each well of the 6-well plate was inoculated with 100000 cells. The cells were collected and treated with 1 mL DNA stai
Statistical analyses were conducted using SPSS software. The data are expressed as means ± SD or standard error of the mean (SEM) derived from three independent experiments. Statistical significance was assessed using the t-test or analysis of variance (ANOVA), with a significance level set at P < 0.05.
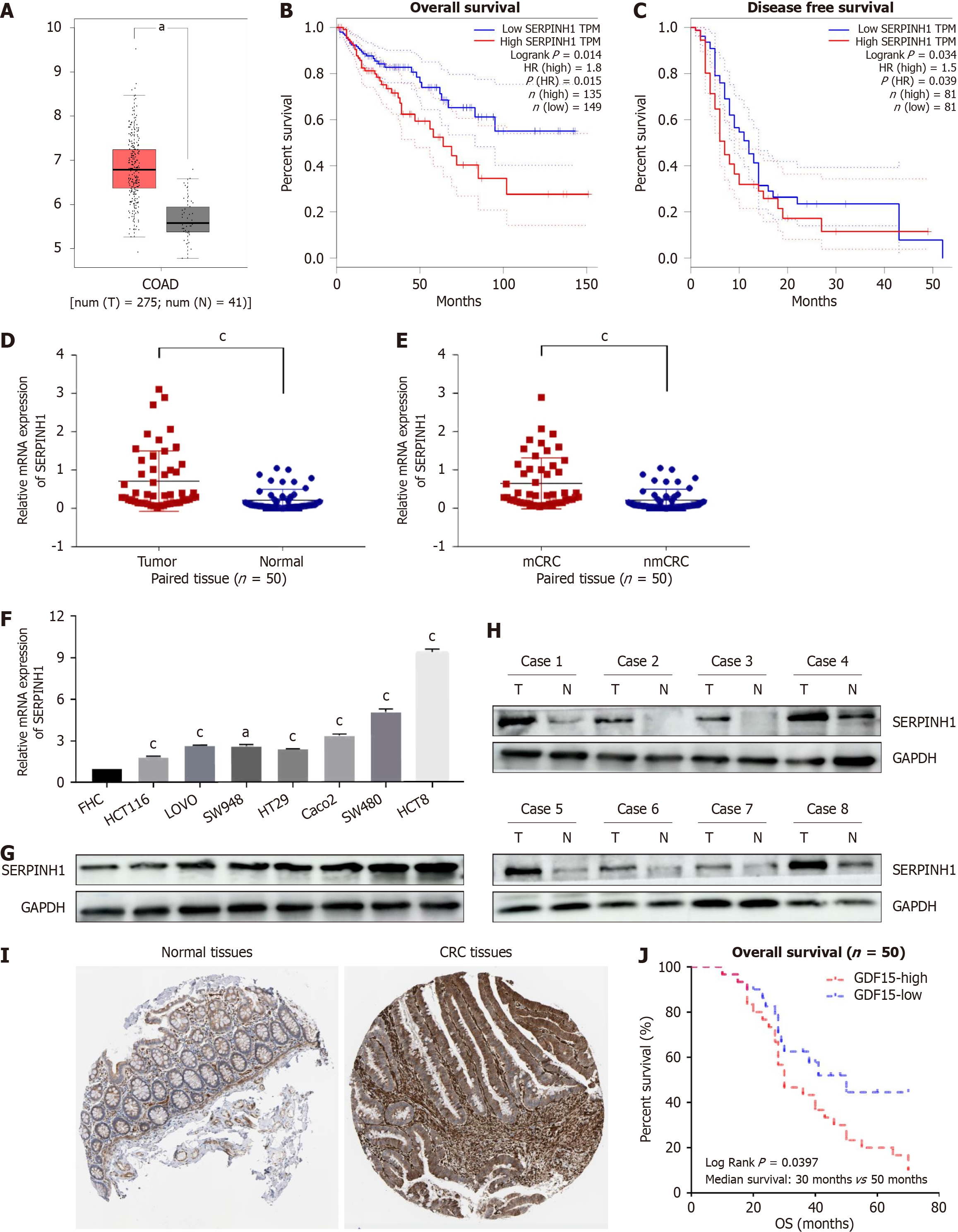
We first examined the expression of SERPINH1 in publicly available CRC datasets using The Cancer Genome Atlas (TCGA) database. The results revealed a significant up-regulation of SERPINH1 in CRC tissues compared to normal tissues (Figure 1A). Furthermore, increased expression of SERPINH1 was linked to poor overall survival [overall survival (OS), P = 0.014 and disease-free survival (DFS), P = 0.034] in COAD (Figure 1B and C). In order to confirm this finding, quantitative real-time polymerase chain reaction (qPCR) was employed to assess the expression levels of SERPINH1 mRNA in 50 freshly collected CRC tissues and their corresponding adjacent normal tissues. As illustrated in Figure 1D, the expression of SERPINH1 mRNA in CRC tissues demonstrated a substantial elevation when juxtaposed with that in adjacent non-malignant tissues (P < 0.05). Intriguingly, individuals with lymph node metastasis displayed elevated SERPINH1 mRNA levels in CRC tissues relative to those without lymph node metastasis (P < 0.05, Figure 1E). Western blotting and qPCR analysis revealed heterogeneous expression levels of SERPINH1 in FHC cells and seven CRC cell lines (Figure 1F and G). Moreover, western blotting demonstrated higher SERPINH1 protein expression levels in 8 pairs of CRC tissues compared to their corresponding normal colorectal tissues (Figure 1H). Immunohistochemistry also showed the same results (Figure 1I). This finding is consistent with previous studies that have reported elevated SERPINH1 expression in various cancer types, suggesting its potential role in tumorigenesis.
To further explore the clinical relevance of SERPINH1 in CRC, we analyzed the association between SERPINH1 ex
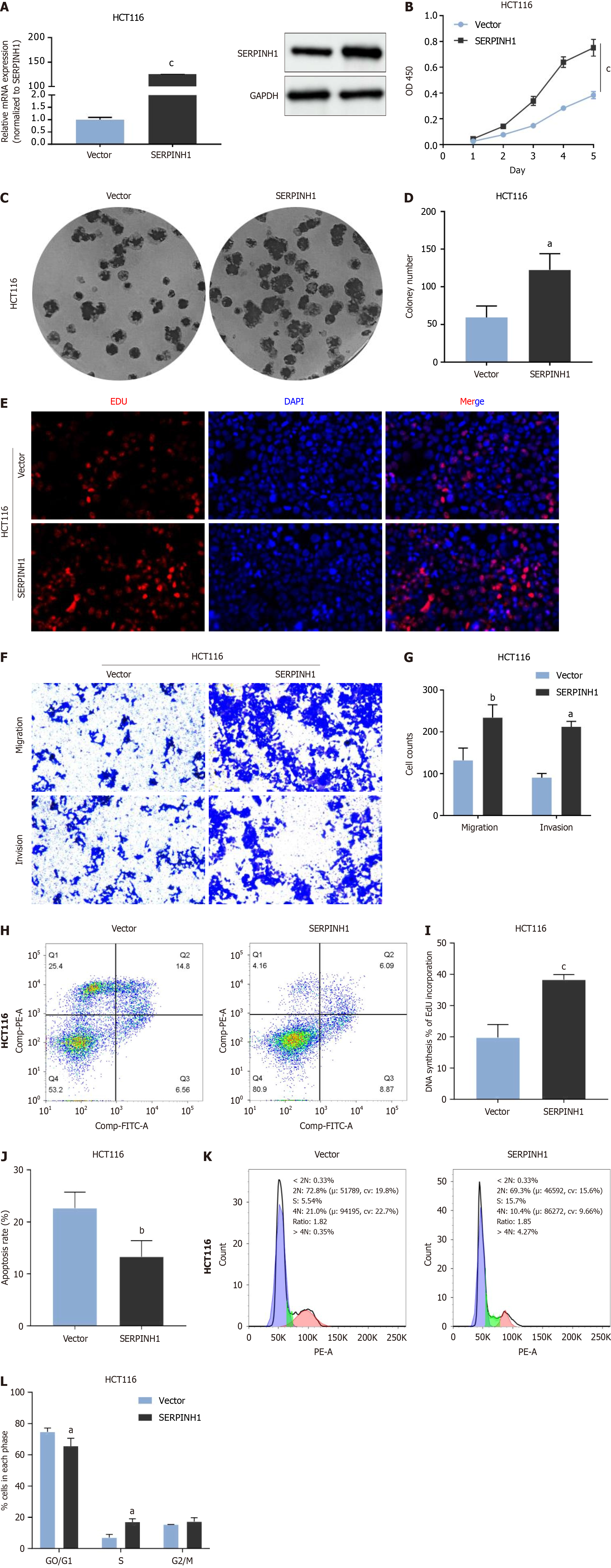
To investigate the functional role of SERPINH1 in CRC, we conducted in vitro experiments using CRC cell lines. The effi
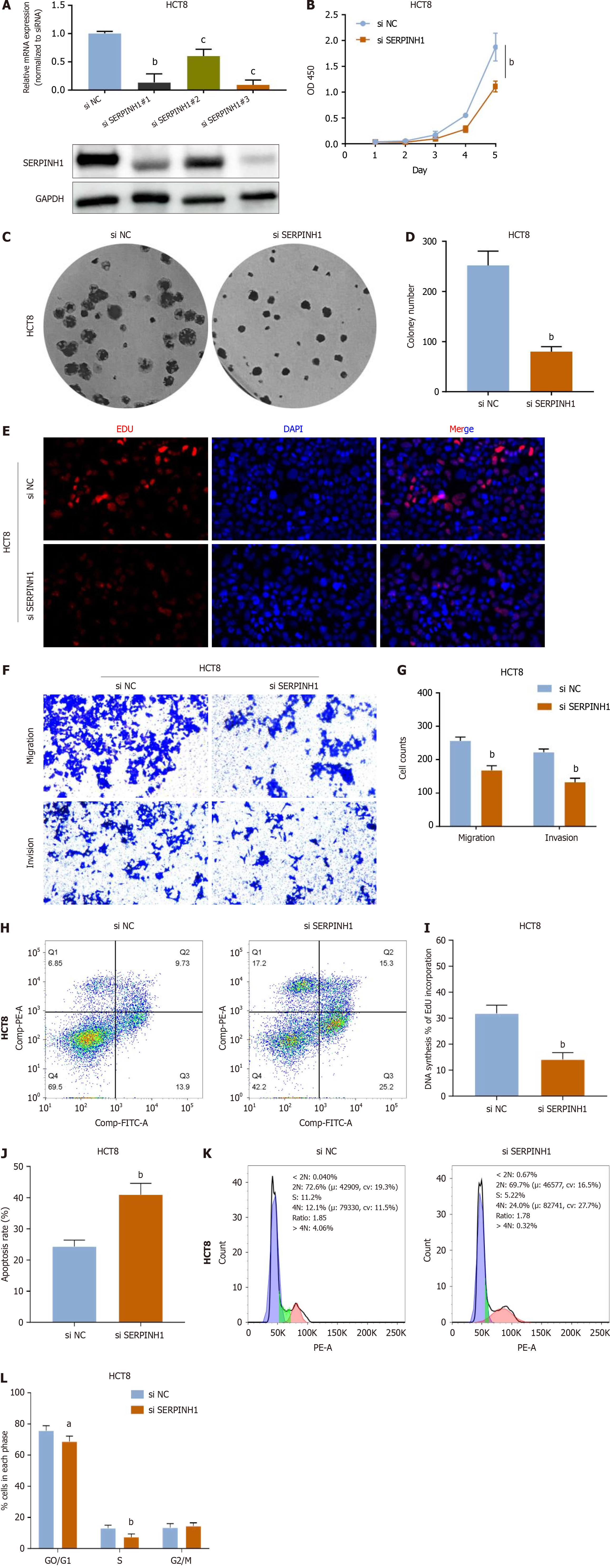
To elucidate the effect of SERPINH1 knockout in CRC cells, lentiviral vectors carrying short hairpin RNA specifically targeting SERPINH1 were used to silence endogenous SERPINH1 expression in HCT8 cells (Figure 3A). As shown in Figure 3B-E and I, knockdown of SERPINH1 significantly inhibited cell growth and proliferation. In addition, the in
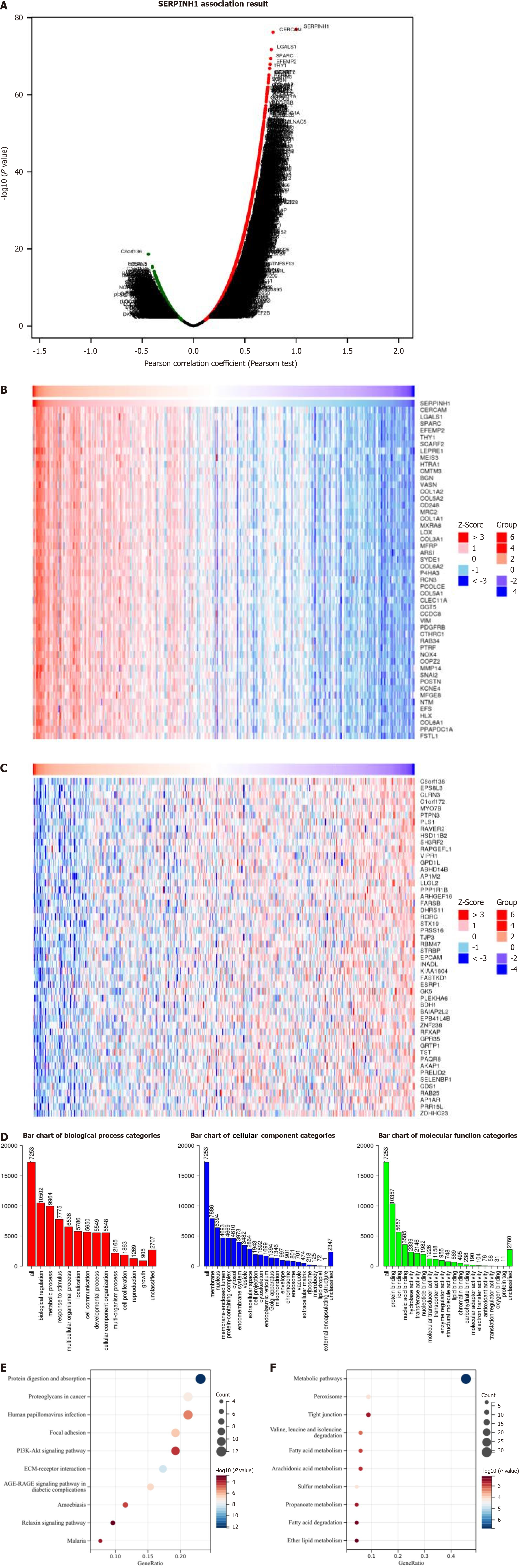
Further analyses were conducted to assess the function of SERPINH1 in colorectal adenocarcinoma (COAD). To elucidate the potential molecular mechanism of SERPINH1 in COAD, we employed the LinkedOmics database to investigate the biological role of SERPINH1. Figure 4A illustrates the genes positively and negatively correlated with SERPINH1. The top 50 genes are shown in Figure 4B and C. Figure 4D shows a summary of the biological functions of SERPINH1 co-expressed genes in the COAD cohort. In addition, we also studied the function of SERPINH1 negative gene and positive gene in COAD. Kyoto Encyclopedia of Genes and Genomes analysis showed that SERPINH1 functions were enriched in metabolism, infection, adhesion, phosphatidylinositol 3-kinase (PI3K)/AKT, etc. (Figure 4E and F). Combining the above results, it can be concluded that SERPINH1 co-expressed genes may participate in tumor progression and play a role through the PI3K/AKT pathway, which provides important clues for further study of the molecular mechanism of SERPINH1.
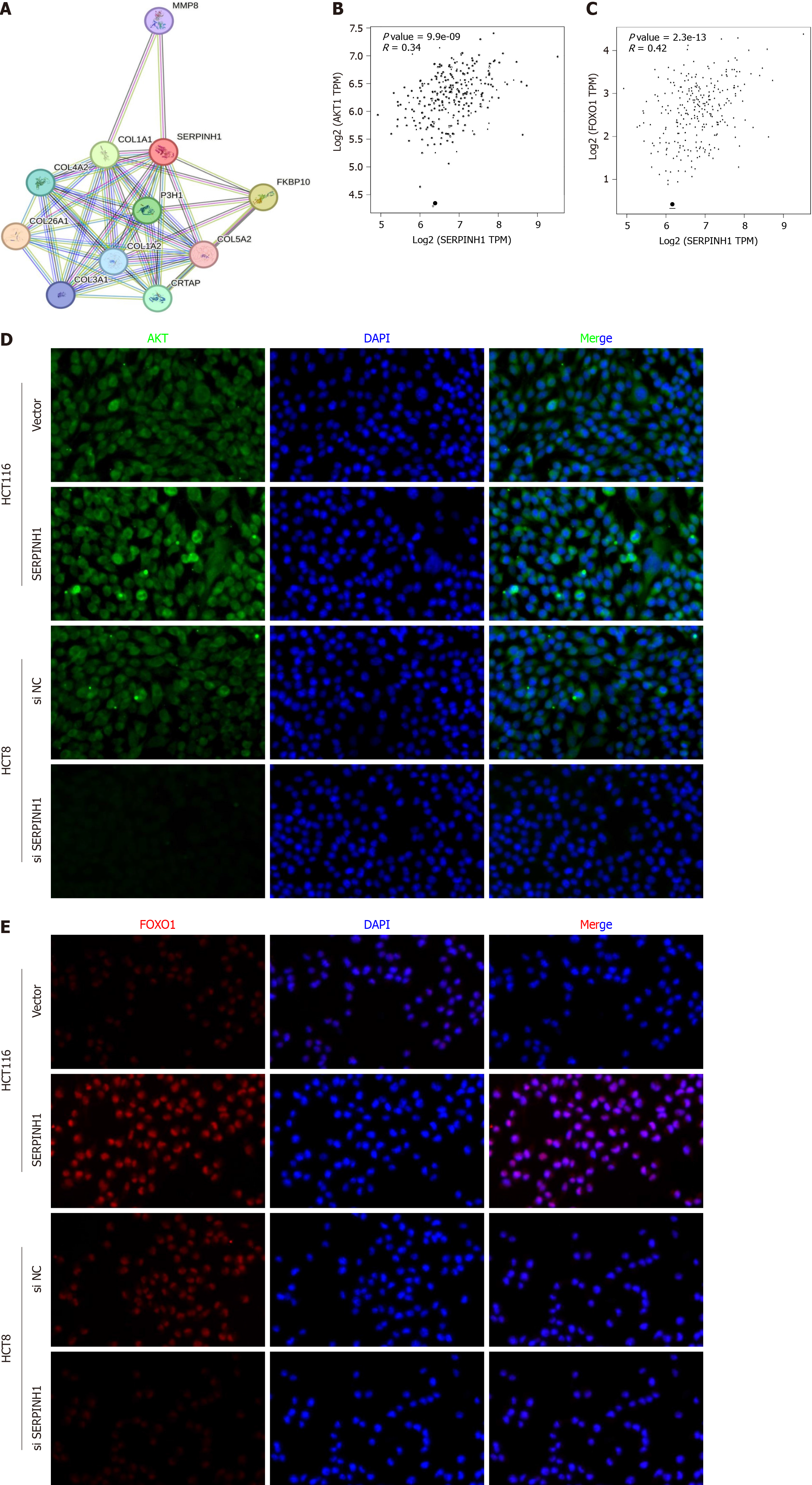
To gain a deeper understanding of the molecular mechanisms by which SERPINH1 mediates its oncogenic role in CRC, we conducted an analysis of the interaction network involving SERPINH1 (Figure 5A). Building upon the pathway enrichment analysis conducted earlier, we found that SERPINH1 may be associated with the PI3K/AKT pathway. Additionally, both mTOR and FOXO1 represent pivotal downstream effectors of the PI3K/AKT signaling pathway, and are recognized participants in cell cycle regulation and proliferation[9,10]. Therefore, we proceeded to investigate whether the pro-proliferative effects of SERPINH1 are dependent on the PI3K/AKT/mTOR and PI3K/AKT/FOXO1 signaling pathways. Initially, we analyzed the correlation between SERPINH1 and FOXO1 and AKT expression using TCGA database. The results demonstrated a positive correlation between SERPINH1 and FOXO1 and AKT expression (Figure 5B and C). This observation was further validated through immunofluorescence experiments (Figure 5D and E).
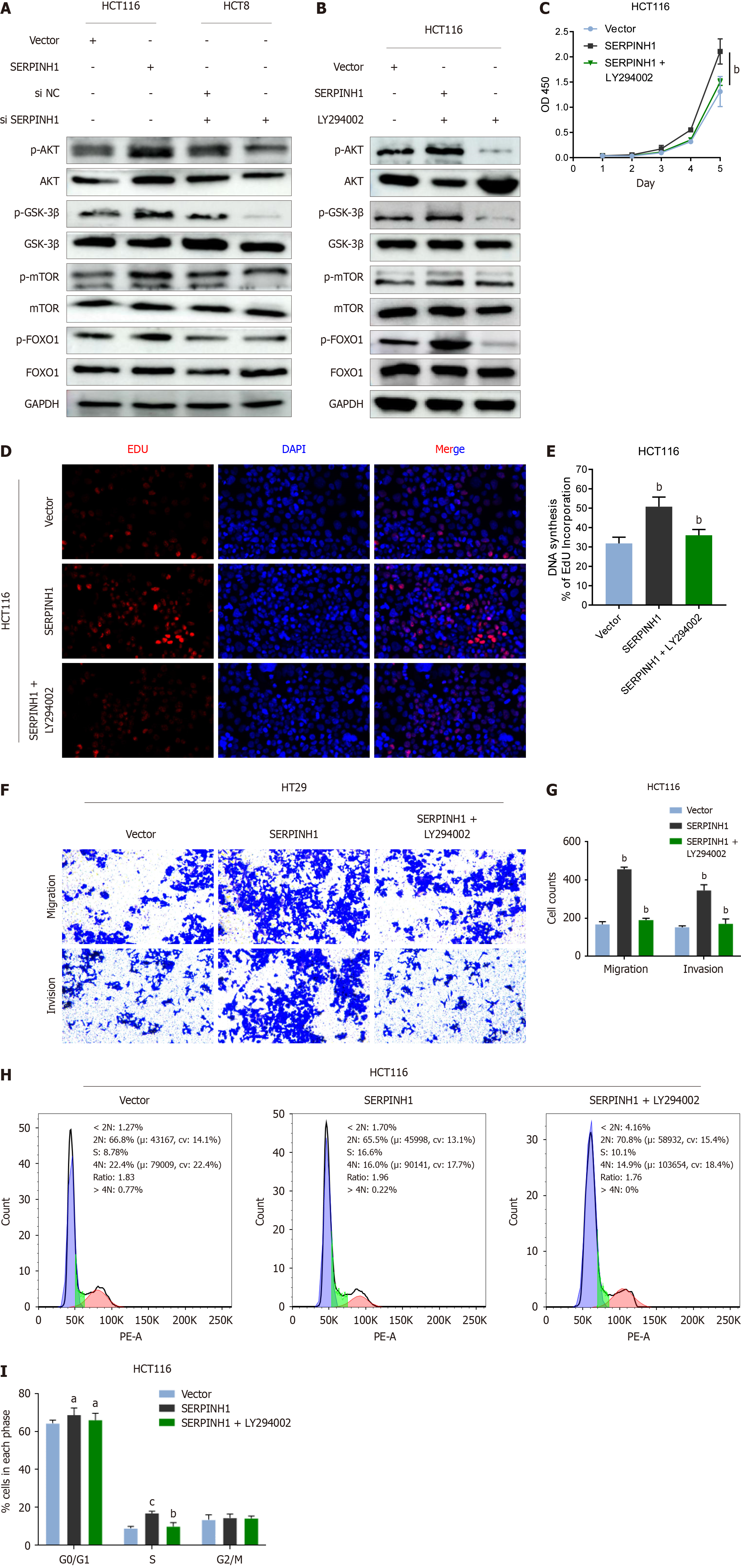
As depicted in Figure 6A, the phosphorylation of AKT [p-AKT (Ser473)], glycogen synthase kinase (GSK)-3β (p-GSK-3), mTOR (p-mTOR), and FOXO1 (p-FOXO1) proteins increased due to the overexpression of SERPINH1 and decreased upon its silencing. These findings imply a potential role for SERPINH1 in regulating the transcriptional activity of mTOR and FOXO1 through activation of the PI3K/AKT signaling pathway. To investigate this hypothesis, CRC cells overexpressing SERPINH1 were treated with an AKT inhibitor (LY294002). We observed a substantial reduction in the ex
In this study, we have demonstrated that SERPINH1 plays a crucial role in promoting the proliferation and metastasis of CRC by activating the PI3K/Akt/mTOR signaling pathway. Our findings shed light on the molecular mechanisms underlying CRC progression and provide potential targets for therapeutic intervention. One of the key findings of our study is the upregulation of SERPINH1 in CRC tissues. This is consistent with previous studies in various cancer types[3-5,11-14], which have also reported increased expression of SERPINH1, especially in CRC[8]. However, unlike the pre
Past studies have suggested the engagement of the PI3K/AKT/mTOR signaling cascade in orchestrating various cellular processes, including cell growth, adhesion, migration, and viability[9,15,16]. Activation of the AKT pathway has been demonstrated to stimulate cell proliferation and oncogenic progression through the modulation of downstream cell cycle regulators[17]. Moreover, activated AKT triggers the phosphorylation of several downstream effectors, including mTOR, FOXO1, and GSK-3β[15,18]. It has been experimentally confirmed that inhibitors targeting the mTOR result in cell cycle arrest and suppress cell proliferation in Epstein-Barr virus-related T- and natural killer-cell lymphomas[19,20]. Inte
It has been reported that full activation of AKT requires phosphorylation at Ser473 and Thr308 sites[15,18,21]. LY294002, a small molecule, competitively and reversibly inhibits the ATP binding site of various PI3K isoforms, making it a specific inhibitor of the PI3K/AKT pathway. It suppresses tumor proliferation and prompts apoptosis in CRC cells, concomitant with decreased expression of p-AKT (Ser473)[22]. To validate the aforementioned hypothesis, we examined the levels of p-AKT Ser473 and p-mTOR. Our investigation revealed a decrease in p-AKT and p-mTOR expression in CRC cells with SERPINH1 knockdown, whereas an increase was observed in CRC cells overexpressing SERPINH1. Addi
Given that FOXO1 is one of the genes associated with cell cycle transition[10], we attempted to verify whether SERPINH1-mediated cell cycle transition relies on the PI3K/AKT/FOXO1 pathway. Similarly, we observed that p-AKT and p-FOXO1 were significantly reduced in SERPINH1-silenced CRC cells, while they increased in SERPINH1-overexpressed CRC cells. Furthermore, inhibition of AKT significantly attenuated p-AKT and phosphorylated FOXO1 levels in CRC cells overexpressing SERPINH1, resulting in the inhibition of cell growth arrest and the formation of colonies. These observations imply that the enhancement of proliferation and tumorigenesis by SERPINH1 might be linked to accelerated cell cycle progression facilitated by the activation of the PI3K/AKT/mTOR and PI3K/AKT/FOXO1 pathways. Our study suggests that the inhibition of SERPINH1 could be a potential strategy for suppressing the PI3K/Akt/mTOR signaling cascade and subsequently impeding CRC cell growth and metastasis. Small molecules or biological agents that specifically target SERPINH1 may hold promise as novel therapeutic interventions.
Enrichment analysis found that SERPINH1 was negatively correlated with fatty acid metabolism. Considering that SerpinH1 was also negatively correlated with the PI3K-AKT pathway, we found that there were studies reported that miR-145 promoted fatty acid metabolism and triacylglycerol synthesis in bovine mammary epithelial cells by inhibiting FOXO1 expression[23]. Meanwhile, the FOXO1/CD36 signaling resulted in decreased fatty acid uptake and inhibited ATP production[24]. Bastie et al[25] also demonstrated that FOXO1 activation leads to concurrent enhancements in both fatty acid uptake and oxidation. These effects are facilitated by the enrichment of CD36 on the cell membrane, which serves as a critical mediator of fatty acid-induced metastasis in gastric cancer via the AKT/GSK-3β/β-catenin signaling pathway[25]. Interestingly, our study uncovered the potential for SERPINH1 to be negatively associated with fatty acid metabolism and to promote CRC progression through activation of the PI3K/AKT/mTOR and PI3K/AKT/FOXO1 pathways. To sum up, FOXO1 and GSK-3β phosphorylation could inhibited fatty acid metabolism, partly mediated by CD36, which may be our next research direction.
It is important to note that while our study provides valuable insights into the role of SERPINH1 in CRC, there are several limitations to consider. For instance, the specific mechanisms by which SERPINH1 activates the PI3K/Akt/mTOR pathway in CRC cells require further investigation. Additionally, the in vivo and clinical relevance of our findings need to be assessed in future studies.
In summary, our study highlights the significance of SERPINH1 in promoting the proliferation and metastasis of CRC through the activation of the PI3K/Akt/mTOR pathway. This work contributes to our understanding of the molecular mechanisms underlying CRC progression and offers a potential avenue for the development of targeted therapies. Further research is warranted to fully elucidate the therapeutic potential of targeting SERPINH1 in CRC.
The clinical significance and biological role of serpin peptidase inhibitor clade H member 1 (SERPINH1) in colorectal cancer (CRC) remains poorly understood.
To investigate the effect of SERPINH1 on CRC cells and its specific mechanism.
To further study the specific role and mechanism of SERPINH1 in CRC and provide theoretical basis for further using SERPINH1 to improve the survival rate of CRC patients.
SERPINH1 expression in CRC cell lines and tissues was evaluated using quantitative real-time polymerase chain reaction, western blotting analysis, data mining from The Cancer Genome Atlas, and immunohistochemistry. A battery of in vitro experiments was conducted to elucidate the role of SERPINH1 and its potential mechanisms in CRC.
Our study reveals the potential for SERPINH1 to be negatively correlated with fatty acid metabolism and promote CRC progression by activating the phosphatidylinositol 3-kinase (PI3K)/AKT/mechanistic target of rapamycin (mTOR) and PI3K/AKT/FOXO1 pathways. Considering that FOXO1 and glycogen synthase kinase (GSK)-3β phosphorylation can inhibit fatty acid metabolism, which is partly mediated by CD36, this may be our next research direction.
Our study uncovered the significance of SERPINH1 in promoting CRC proliferation and metastasis by activating the PI3K/Akt/mTOR pathway. This work contributes to our understanding of the molecular mechanisms underlying CRC progression and offers a potential avenue for the development of targeted therapies.
Our study revealed the potential for SERPINH1 to be negatively correlated with fatty acid metabolism and promoted CRC progression by activating the PI3K/AKT/mTOR and PI3K/AKT/FOXO1 pathways. Considering that FOXO1 and GSK-3β phosphorylation could inhibit fatty acid metabolism, which is partly mediated by CD36, this may be our next research direction.
Provenance and peer review: Unsolicited article; Externally peer reviewed.
Peer-review model: Single blind
Specialty type: Oncology
Country/Territory of origin: China
Peer-review report’s scientific quality classification
Grade A (Excellent): 0
Grade B (Very good): B
Grade C (Good): 0
Grade D (Fair): 0
Grade E (Poor): 0
P-Reviewer: Fouad MA, Egypt S-Editor: Wang JJ L-Editor: A P-Editor: Zheng XM
| 1. | Williams RS. Heat shock protein 47 : a chaperone for the fibrous cap? Circulation. 2000;101:1227-1228. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 10] [Cited by in RCA: 9] [Article Influence: 0.4] [Reference Citation Analysis (0)] |
| 2. | Nagata K. Hsp47: a collagen-specific molecular chaperone. Trends Biochem Sci. 1996;21:22-26. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 10] [Cited by in RCA: 64] [Article Influence: 2.2] [Reference Citation Analysis (0)] |
| 3. | Tian S, Peng P, Li J, Deng H, Zhan N, Zeng Z, Dong W. SERPINH1 regulates EMT and gastric cancer metastasis via the Wnt/β-catenin signaling pathway. Aging (Albany NY). 2020;12:3574-3593. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 40] [Cited by in RCA: 139] [Article Influence: 27.8] [Reference Citation Analysis (1)] |
| 4. | Wang Y, Yan Q, Mo Y, Liu Y, Wang Y, Zhang S, Guo C, Wang F, Li G, Zeng Z, Xiong W. Splicing factor derived circular RNA circCAMSAP1 accelerates nasopharyngeal carcinoma tumorigenesis via a SERPINH1/c-Myc positive feedback loop. Mol Cancer. 2022;21:62. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 3] [Cited by in RCA: 47] [Article Influence: 15.7] [Reference Citation Analysis (0)] |
| 5. | Li J, Han T, Wang X, Wang Y, Chen X, Chen W, Yang Q. H19 may regulate the immune cell infiltration in carcinogenesis of gastric cancer through miR-378a-5p/SERPINH1 signaling. World J Surg Oncol. 2022;20:295. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in RCA: 9] [Reference Citation Analysis (0)] |
| 6. | Pan X, Huang B, Ma Q, Ren J, Liu Y, Wang C, Zhang D, Fu J, Ran L, Yu T, Li H, Wang X, Yang F, Liang C, Zhang Y, Wang S, Li W, Wang Y, Xiao B. Circular RNA circ-TNPO3 inhibits clear cell renal cell carcinoma metastasis by binding to IGF2BP2 and destabilizing SERPINH1 mRNA. Clin Transl Med. 2022;12:e994. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 9] [Cited by in RCA: 49] [Article Influence: 16.3] [Reference Citation Analysis (0)] |
| 7. | Alhalabi OT, Fletcher MNC, Hielscher T, Kessler T, Lokumcu T, Baumgartner U, Wittmann E, Schlue S, Göttmann M, Rahman S, Hai L, Hansen-Palmus L, Puccio L, Nakano I, Herold-Mende C, Day BW, Wick W, Sahm F, Phillips E, Goidts V. A novel patient stratification strategy to enhance the therapeutic efficacy of dasatinib in glioblastoma. Neuro Oncol. 2022;24:39-51. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 5] [Cited by in RCA: 25] [Article Influence: 6.3] [Reference Citation Analysis (0)] |
| 8. | Chern Y, Zhang P, Ju H, Tai IT. Heat shock protein 47 promotes tumor survival and therapy resistance by modulating AKT signaling via PHLPP1 in colorectal cancer. Cancer Biol Med. 2020;17:343-356. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 10] [Cited by in RCA: 15] [Article Influence: 3.0] [Reference Citation Analysis (0)] |
| 9. | Wang S, Cheng Z, Cui Y, Xu S, Luan Q, Jing S, Du B, Li X, Li Y. PTPRH promotes the progression of non-small cell lung cancer via glycolysis mediated by the PI3K/AKT/mTOR signaling pathway. J Transl Med. 2023;21:819. [RCA] [PubMed] [DOI] [Full Text] [Cited by in RCA: 17] [Reference Citation Analysis (0)] |
| 10. | van Boxtel R, Gomez-Puerto C, Mokry M, Eijkelenboom A, van der Vos KE, Nieuwenhuis EE, Burgering BM, Lam EW, Coffer PJ. FOXP1 acts through a negative feedback loop to suppress FOXO-induced apoptosis. Cell Death Differ. 2013;20:1219-1229. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 42] [Cited by in RCA: 45] [Article Influence: 3.8] [Reference Citation Analysis (0)] |
| 11. | Kamikawaji K, Seki N, Watanabe M, Mataki H, Kumamoto T, Takagi K, Mizuno K, Inoue H. Regulation of LOXL2 and SERPINH1 by antitumor microRNA-29a in lung cancer with idiopathic pulmonary fibrosis. J Hum Genet. 2016;61:985-993. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 41] [Cited by in RCA: 54] [Article Influence: 6.0] [Reference Citation Analysis (0)] |
| 12. | Kawagoe K, Wada M, Idichi T, Okada R, Yamada Y, Moriya S, Okubo K, Matsushita D, Arigami T, Kurahara H, Maemura K, Natsugoe S, Seki N. Regulation of aberrantly expressed SERPINH1 by antitumor miR-148a-5p inhibits cancer cell aggressiveness in gastric cancer. J Hum Genet. 2020;65:647-656. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 20] [Cited by in RCA: 17] [Article Influence: 3.4] [Reference Citation Analysis (0)] |
| 13. | Wang Y, Gu W, Wen W, Zhang X. SERPINH1 is a Potential Prognostic Biomarker and Correlated With Immune Infiltration: A Pan-Cancer Analysis. Front Genet. 2021;12:756094. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 5] [Cited by in RCA: 29] [Article Influence: 9.7] [Reference Citation Analysis (0)] |
| 14. | Zhong H, Wang Z, Wei X, Liu Y, Huang X, Mo X, Tang W. Prognostic and immunological role of SERPINH1 in pan-cancer. Front Genet. 2022;13:900495. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 6] [Reference Citation Analysis (0)] |
| 15. | Glaviano A, Foo ASC, Lam HY, Yap KCH, Jacot W, Jones RH, Eng H, Nair MG, Makvandi P, Geoerger B, Kulke MH, Baird RD, Prabhu JS, Carbone D, Pecoraro C, Teh DBL, Sethi G, Cavalieri V, Lin KH, Javidi-Sharifi NR, Toska E, Davids MS, Brown JR, Diana P, Stebbing J, Fruman DA, Kumar AP. PI3K/AKT/mTOR signaling transduction pathway and targeted therapies in cancer. Mol Cancer. 2023;22:138. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 278] [Cited by in RCA: 778] [Article Influence: 389.0] [Reference Citation Analysis (0)] |
| 16. | Vasconcelos de Matos L, Volovat S, Debiasi M, Cardoso F. Unfolding the role of the PI3K/AKT/MTOR pathway in male breast cancer: A pragmatic appraisal. Breast. 2023;72:103576. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1] [Reference Citation Analysis (0)] |
| 17. | Chen G, Tian TT, Wang FQ, Pan CS, Sun K, Wang XY, Yang B, Yang Z, Tang DX, Han JY. Chanling Gao suppresses colorectal cancer via PI3K/Akt/mTOR pathway modulation and enhances quality of survival. Environ Toxicol. 2024;39:1107-1118. [RCA] [PubMed] [DOI] [Full Text] [Cited by in RCA: 3] [Reference Citation Analysis (0)] |
| 18. | Chang L, Graham PH, Hao J, Ni J, Bucci J, Cozzi PJ, Kearsley JH, Li Y. PI3K/Akt/mTOR pathway inhibitors enhance radiosensitivity in radioresistant prostate cancer cells through inducing apoptosis, reducing autophagy, suppressing NHEJ and HR repair pathways. Cell Death Dis. 2014;5:e1437. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 189] [Cited by in RCA: 255] [Article Influence: 23.2] [Reference Citation Analysis (0)] |
| 19. | Kawada J, Ito Y, Iwata S, Suzuki M, Kawano Y, Kanazawa T, Siddiquey MN, Kimura H. mTOR inhibitors induce cell-cycle arrest and inhibit tumor growth in Epstein-Barr virus-associated T and natural killer cell lymphoma cells. Clin Cancer Res. 2014;20:5412-5422. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 31] [Cited by in RCA: 44] [Article Influence: 4.0] [Reference Citation Analysis (0)] |
| 20. | Lees J, Hay J, Moles MW, Michie AM. The discrete roles of individual FOXO transcription factor family members in B-cell malignancies. Front Immunol. 2023;14:1179101. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1] [Cited by in RCA: 9] [Article Influence: 4.5] [Reference Citation Analysis (0)] |
| 21. | Tong H, Li K, Zhou M, Wu R, Yang H, Peng Z, Zhao Q, Luo KQ. Coculture of cancer cells with platelets increases their survival and metastasis by activating the TGFβ/Smad/PAI-1 and PI3K/AKT pathways. Int J Biol Sci. 2023;19:4259-4277. [RCA] [PubMed] [DOI] [Full Text] [Cited by in RCA: 8] [Reference Citation Analysis (0)] |
| 22. | Li R, Zheng Y, Zhang J, Zhou Y, Fan X. Gomisin N attenuated cerebral ischemia-reperfusion injury through inhibition of autophagy by activating the PI3K/AKT/mTOR pathway. Phytomedicine. 2023;110:154644. [RCA] [PubMed] [DOI] [Full Text] [Cited by in RCA: 33] [Reference Citation Analysis (0)] |
| 23. | Lyu M, Li F, Wang X, Xu K, Sun S. miR-145 Modulates Fatty Acid Metabolism by Targeting FOXO1 to Affect SERBP1 Activity in Bovine Mammary Epithelial Cells. J Agric Food Chem. 2023;71:7440-7450. [RCA] [PubMed] [DOI] [Full Text] [Reference Citation Analysis (0)] |
| 24. | Ying F, Liu H, Ching Tang EH, Lakhani I, Liu N, Xia Z, Liu S. Prostaglandin E receptor subtype 4 protects against diabetic cardiomyopathy by modulating cardiac fatty acid metabolism via FOXO1/CD36 signalling. Biochem Biophys Res Commun. 2021;548:196-203. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 3] [Cited by in RCA: 9] [Article Influence: 2.3] [Reference Citation Analysis (0)] |
| 25. | Bastie CC, Nahlé Z, McLoughlin T, Esser K, Zhang W, Unterman T, Abumrad NA. FoxO1 stimulates fatty acid uptake and oxidation in muscle cells through CD36-dependent and -independent mechanisms. J Biol Chem. 2005;280: 14222-14229. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 108] [Cited by in RCA: 118] [Article Influence: 5.9] [Reference Citation Analysis (0)] |