Published online Apr 15, 2024. doi: 10.4251/wjgo.v16.i4.1564
Peer-review started: December 22, 2023
First decision: January 9, 2024
Revised: January 26, 2024
Accepted: February 18, 2024
Article in press: February 18, 2024
Published online: April 15, 2024
Processing time: 111 Days and 0.8 Hours
Colorectal cancer (CRC) is the third most common cancer and a significant cause of cancer-related mortality globally. Resistance to chemotherapy, especially during CRC treatment, leads to reduced effectiveness of drugs and poor patient outcomes. Long noncoding RNAs (lncRNAs) have been implicated in various pathophysiological processes of tumor cells, including chemotherapy resistance, yet the roles of many lncRNAs in CRC remain unclear.
To identify and analyze the lncRNAs involved in oxaliplatin resistance in CRC and to understand the underlying molecular mechanisms influencing this resistance.
Gene Expression Omnibus datasets GSE42387 and GSE30011 were reanalyzed to identify lncRNAs and mRNAs associated with oxaliplatin resistance. Various bioinformatics tools were employed to elucidate molecular mechanisms. The expression levels of lncRNAs and mRNAs were assessed via quantitative reverse transcription-polymerase chain reaction. Functional assays, including MTT, wound healing, and Transwell, were conducted to investigate the functional implications of lncRNA alterations. Interactions between lncRNAs and trans
LncRNA prion protein testis specific (PRNT) was found to be upregulated in oxaliplatin-resistant CRC cell lines and negatively correlated with homeodomain interacting protein kinase 2 (HIPK2) expression. PRNT was demonstrated to sponge transcription factor zinc finger protein 184 (ZNF184), which in turn could regulate HIPK2 expression. Altered expression of PRNT influenced CRC cell sensitivity to oxaliplatin, with overexpression leading to decreased sensitivity and decreased expression reducing resistance. Both RIP and luciferase reporter assays indicated that ZNF184 and HIPK2 are targets of PRNT. The PRNT/ZNF184/HIPK2 axis was implicated in promoting CRC progression and oxaliplatin resistance both in vitro and in vivo.
The study concludes that PRNT is upregulated in oxaliplatin-resistant CRC cells and modulates the expression of HIPK2 by sponging ZNF184. This regulatory mechanism enhances CRC progression and resistance to oxaliplatin, positioning PRNT as a promising therapeutic target for CRC patients undergoing oxaliplatin-based chemotherapy.
Core Tip: The revelation that long noncoding RNA prion protein testis specific, which is overexpressed in oxaliplatin-resistant colorectal cancer (CRC) cells, regulates the expression of homeodomain interacting protein kinase 2 by sponging transcription factor zinc finger protein 184 unveils a promising target for enhancing the efficacy of chemotherapy. This mechanistic insight could lead to the development of novel therapeutic strategies to combat resistance in CRC treatments.
- Citation: Li SN, Yang S, Wang HQ, Hui TL, Cheng M, Zhang X, Li BK, Wang GY. Upregulated lncRNA PRNT promotes progression and oxaliplatin resistance of colorectal cancer cells by regulating HIPK2 transcription. World J Gastrointest Oncol 2024; 16(4): 1564-1577
- URL: https://www.wjgnet.com/1948-5204/full/v16/i4/1564.htm
- DOI: https://dx.doi.org/10.4251/wjgo.v16.i4.1564
Colorectal cancer (CRC) is the third most common cancer and the second leading cause of cancer-related death in the world[1]. Studies have shown that there are more than 1.85 million cases of colorectal cancer and 850000 deaths each year[2,3]. Surgery is the primary treatment for patients with nonmetastatic colorectal cancer. However, for patients with locally advanced or metastatic colorectal cancer, chemotherapy is essential[2,4]. Chemotherapy plays an irreplaceable role in the adjuvant therapy of colon cancer and neoadjuvant therapy of rectal cancer. In addition, chemotherapy is also the first line of treatment for patients with metastatic colorectal cancer[5,6]. Over the past two decades, survival for patients with metastatic colorectal cancer has improved significantly due to the use of chemotherapy drugs such as oxaliplatin and irinotecan[7]. Oxaliplatin (OXA) is one of the first-choice chemotherapy drugs for the treatment of CRC, and OXA-based chemotherapy regimens are the most commonly used treatment strategy for patients with initially unresectable or metastatic and recurrent tumours[8]. However, some patients will acquire oxaliplatin resistance during chemotherapy. This phenomenon limits the practical use of oxaliplatin and worsens the prognosis of CRC patients[9]. Thus, it is necessary to elucidate the molecular mechanism and identify biomarkers of OXA resistance to improve the prognosis of CRC patients.
Long noncoding RNAs (lncRNAs) are extended transcripts of approximately 200 nucleotides in length that do not encode proteins or have limited coding ability. It has been reported that lncRNAs are involved in the regulation of many cell biological processes[10]. Studies have shown that lncRNAs can regulate the occurrence and development of tumours by participating in transcription and posttranscriptional gene expression[11,12]. Oxaliplatin resistance is one of the major challenges in the treatment of colorectal cancer, which is regulated by complex mechanisms and is associated with abnormal activation of multiple signalling pathways. Studies have shown that lncRNAs are key factors in oxaliplatin resistance, and their dysregulation can induce the activation of AKT/mTOR, Wnt/β-catenin, JNK1 and other signalling pathways and ultimately lead to chemotherapy resistance[13-15]. Transcription factors are very important for gene transcription; they can bind to the RNA produced by gene transcription and control the transcription, localization and stability of RNA. Some studies have pointed out that some lncRNAs act as ligands and bind to transcription factors to form complexes and control gene transcription[16-18]. This phenomenon plays an important role in tumours. For instance, lncRNA HOXD-AS1 promotes FRRS1 expression through the transcription factor ELF1 and promotes the proliferation of cervical cancer cells[19]. LncRNA LINC00152 can promote the growth of oral squamous cell carcinoma by directly binding to upstream transcription factor 1 to promote the expression of mitochondrial ribosomal protein L52[20]. However, the majority of lncRNAs involved in OXA resistance in CRC remain to be elucidated.
In this study, a public microarray dataset was downloaded and reanalyzed to identify potential lncRNAs associated with oxaliplatin resistance in CRC. Prion protein testis specific (PRNT) was identified as a critical lncRNA contributing to oxaliplatin resistance in CRC. PRNT was upregulated in OXA-resistant CRC cell lines, and overexpression of PRNT increased the resistance of CRC cells to oxaliplatin. Furthermore, we verified that PRNT could sponge the transcription factor zinc finger protein 184 (ZNF184) and subsequently regulate the expression of homeodomain interacting protein kinase 2 (HIPK2), promoting the progression and oxaliplatin resistance of colorectal cancer cells. Our findings unveiled the potential of PRNT as a therapeutic target in CRC patients receiving OXA-based chemotherapy.
The datasets of OXA-resistant CRC cell lines were downloaded from the Gene Expression Omnibus (GEO, http://www.ncbi.nlm.nih.gov/geo/) database. GSE42387 contained 9 OXA-resistant samples and 9 OXA-sensitive samples, while GSE30011 contained 14 OXA-resistant samples and 14 OXA-sensitive samples (Table 1). The R software packages “sva” and “limma” were used to normalize and summarize these high-throughput sequencing data. The differentially expressed genes were identified by the R software package “limma”. The P values and fold change (FC) were adjusted by the Benjamini-Hochberg method. The protein-protein interaction (PPI) network was constructed by the STRING online database (http://string-db.org), and Cytoscape software was used for analysis of the PPI network. The catRAPID database (http://s.tartaglialab.com/page/catrapid_group) was used to predict transcription factors combined with PRNT. An animal database (http://bioinfo.life.hust.edu.cn/AnimalTFDB4/#/) was used to predict transcription factors that bind to HIPK2. The Joint Analysis of the Structural Parameters of Analytical Regulation (JASPAR) database (https://jaspar.genereg.net/) describes a matrix model of DNA-binding preferences for transcription factors and other DNA patterns, which was used to predict the binding sites of genes to transcription factors. The Comparative Toxicogenomics database (CTD; http://ctdbase.org/) provides information about interactions between chemicals and gene products and their relationships to diseases, which was used to validate the functions of genes. Gene set enrichment analysis (GSEA) is a computational method that evaluates gene expression data and provides a variety of biological pathways. The R software package “clusterprofiler” and GSEA software were used to conduct GSEA. The reference gene set for GSEA is h.all.v6.2.symbols. gmt from the Molecular Signature database (MSigDB). The criterion for significance was a nominal P value < 5% and a false-positive rate < 25%. Gene set permutations were performed 1000 times for each analysis.
| Series | Platform | Affymetrix GeneChip | Sensitive | Resistant | |
| 1 | GSE42387 | GPL16297 | Agilent-014850 Whole Human Genome Microarray 4x44K G4112F (Agilent Systematic Name, collapsed probe, version) | 9 | 9 |
| 2 | GSE30011 | GPL2006 | Human 19K oligo array | 14 | 14 |
The OXA-resistant CRC cell lines HCT116 and LoVo were purchased from Shanghai Yiyan Biotechnology Co., Ltd. The cells were stored in a humidified incubator with 5% CO2 at 37 °C. Oxaliplatin was purchased from Sigma Aldrich (Shanghai, China).
A quantitative reverse transcription-polymerase chain reaction (qRT-PCR) assay was performed to measure the expression of PRNT, ZNF184 and HIPK2. First, the total RNA in CRC cells was extracted by TRIzol reagent (Canspec Scientific Instruments Co., Ltd. Shanghai, China). Then, complementary DNA (cDNA) was generated by a RevertAid™ First Strand cDNA Synthesis Kit (Thermo Fisher Scientific, Waltham, MA, United States) according to the manufacturer’s instructions. Real-time PCR was performed on a Mastercycler5333 (Eppendorf, Hamburg, Germany) using the TB Green Premix Ex Taq™II kit (Takara, Otsu, Japan) according to the manufacturer's instructions. The relative expression of genes was calculated by the 2ΔΔCq method. The expression levels of PRNT, ZNF184 and HIPK2 in the cytoplasmic and nuclear fractions were determined by qRT-PCR. The cytoplasmic control was GAPDH, and the nuclear control was Lamin B1.
PRNT siRNAs (short interfering RNAs) and the corresponding disturbed siRNA negative control (NC) were synthesized by GenePharma (Shanghai, China). Moreover, the plasmid and negative control were synthesized by GenePharma. Cells were transiently transfected using Lipofectamine 3000 transfection reagent (Thermo Fisher Scientific) according to the manufacturer's instructions. After 24 h, qRT-PCR was used to evaluate the transfection efficiency.
An MTT assay was performed to monitor CRC cell viability. In this study, the MTT cell viability assay kit (BioAssay Systems, Xinrui Biotechnology Co., LTD, Shanghai, China) was used to determine cell viability. After transfection, CRC cells were seeded in 96-well plates (3000 cells/well) and incubated with OXA at 6 μg/ml for 48 h. Then, MTT (10 μL, 5 mg/mL) was added. After incubation for 4 h, the precipitate was dissolved in DMSO (100 μL). A microplate spectrophotometer was used to measure absorbance at 490 nm. Experiments were performed in triplicate.
A wound healing assay was performed to detect the migratory ability of CRC cells. First, CRC cells were seeded in 6-well culture plates at 5000 cells/well. Then, the CRC cells were incubated with OXA at 6 μg/mL for 24 h. A sterile pipette tip was used to create a wound. The cells were then imaged at 0 and 24 h after wounding. Experiments were performed in triplicate.
Transwell assays were performed to determine the invasion and migration abilities of CRC cells. CRC cells were seeded in 6-well plates at 1 × 105 cells/well for 24 h. CRC cells were inoculated in a Transwell chamber with 10% TBS and culture medium. Then, CRC cells were grown at 37 ℃ and 5% CO2 for 24 h. Cells in the lower chamber were fixed with 100% methanol, stained with 0.1% crystal violet, and observed under a microscope. Experiments were performed in triplicate.
The expression of ZNF184 and HIPK2 was explored by Western blot analysis. First, RIPA buffer (Solarbio Technology Co., LTD, Beijing, China) was used to lyse CRC cells, and total protein was extracted. Then, the protein was quantified by a BCA protein assay kit (Zeye Biotechnology Co., LTD, Shanghai, China). The protein in CRC cells was separated by sodium dodecyl sulfate-polyacrylamide gel electrophoresis. Then, the protein was transferred to a polyvinylidene difluoride membrane. The membrane was incubated with 5% bovine serum albumin (Thermo Fisher Scientific) for 1 h and incubated with primary antibody (dilution ratio 1:1000) overnight at 4 °C in a shaker. Then, the membrane was incubated with the corresponding two antibodies for 1 h at room temperature. The greyscale value of the images was determined by ImageJ (National Institutes of Health, United States) software. Experiments were performed in triplicate.
RNA immunoprecipitation (RIP) was performed to evaluate the interaction between lncRNAs and transcription factors. The Magna RIP™ RNA Binding Protein Immunoprecipitation Kit (Millipore, United States) was used to perform the RIP assay according to the manufacturer’s instructions. In brief, RIP lysis buffer was used to lyse CRC cells. Then, the cells were incubated with human anti-Ago2 antibody (NeoBioscience Technology Co., Ltd., Shenzhen, China) RIP buffer. Proteinase K was used to digest the protein, and qRT-PCR was used to analyse the purified RNA. Experiments were performed in triplicate.
A luciferase reporter assay was used to evaluate the interaction between mRNA and transcription factors. Sequences for mutant (Mut) and wild-type (WT) HIPK2 were subcloned and inserted into a psiCHECK-2 vector (Lianmai Biological Engineering Co., LTD, Shanghai, China). Then, the ZNF184 NC and ZNF184 mimics were cotransfected into CRC cells with luciferase reporter plasmids. After 48 h, the cells were harvested and counted. Luciferase activity was measured by a Dual Luciferase Reporter Gene Assay Kit (Biorebo Technology Co., LTD., Beijing, China) according to the manufacturer’s instructions. All assays were carried out in triplicate.
A nude mouse xenograft model was constructed to explore the effect of lncRNAs on CRC chemotherapy resistance in vivo. Animal experiments were approved by the Animal Research Committee of the Fourth Hospital of Hebei Medical University. Female BALB/c nude mice were purchased from Genet-Med Biotechnology Co., Ltd., Beijing, China. All nude mice were maintained under pathogen-free conditions. A total of 3 × 106 CRC cells were injected subcutaneously with 100 μL of PBS. They were divided into three groups: control, OV-PRNT and si-PRNT. After 1 wk, the mice were intraperitoneally injected with OXA (3 mg/kg) every 2 d. After 4 wk, the mice were killed, and the weight of the transplanted tumour was measured.
In this study, GraphPad Prism 7.0, SPSS 22.0 (SPSS Inc., Chicago, United States) and R software (version 4.0.1) were employed to conduct statistical analyses. Experimental data are expressed as the mean ± SD. Pearson’s chi-square test and two-tailed Student’s t test were used to compare the differences between different groups. Pearson correlation was used to determine the correlation. P values less than 0.05 were considered to be statistically significant.
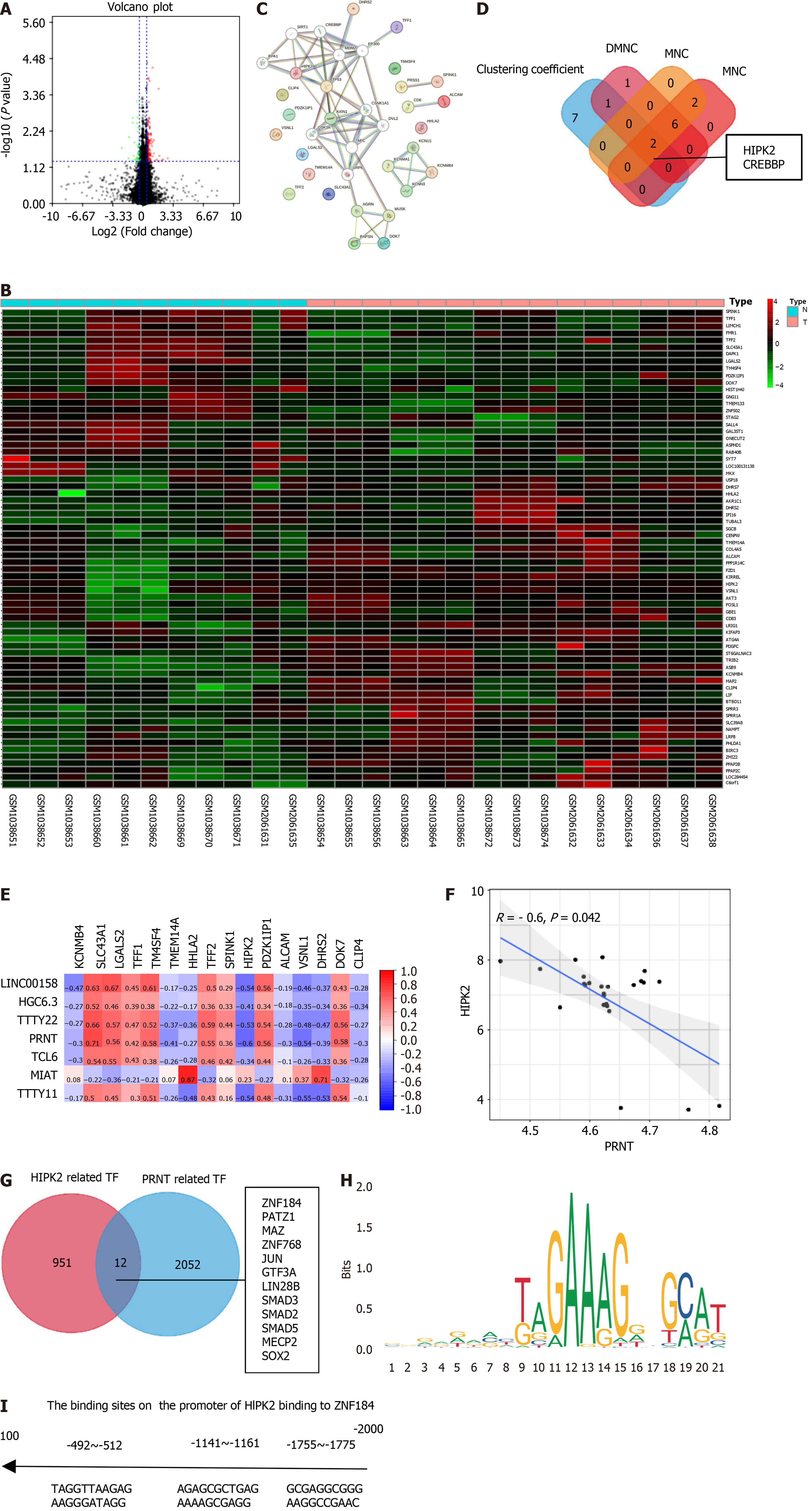
In this study, the datasets of OXA-resistant CRC cell lines GSE42387 (https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=gse42387) and GSE30011 (https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=gse30011) were downloaded and reanalyzed to identify key lncRNAs and mRNAs. These datasets contained 23 OXA-resistant samples and 23 OXA-sensitive samples. The OXA-sensitive samples were used as controls, and the volcano plot of the oxaliplatin resistance-related genes showing differential expression is shown in Figure 1A (|logFC| ≥ 1, and P < 0.05). The heatmap of the oxaliplatin resistance-related genes is shown in Figure 1B. Then, the oxaliplatin resistance-related mRNAs were input into the STRING database to generate a PPI network. The PPI network is shown in Figure 1C, which contains 37 nodes and 62 edges. Cytoscape software was used for analysis of the PPI network and to search for hub genes. Four algorithms, “clustering coefficient”, “DMNC”, “MNC”, and “EPC”, were used to find hub genes, and the intersection of the four algorithms was taken as the final result. As shown in Figure 1D, the hub genes were HIPK2 and CREBBP. The heatmap of the correlation between oxaliplatin resistance-related genes and lncRNAs is shown in Figure 1E. Among them, HIPK2 had the strongest negative correlation with PRNT, so it was included in the subsequent study. Moreover, the scatter plot of the correlation between PRNT and HIPK2 is shown in Figure 1F. The catRAPID database was used to predict transcription factors combined with PRNT. An animal database was used to predict transcription factors that bind to HIPK2. The intersection between them is shown in Figure 1G. The canonical binding motif between ZNF184 and PRNT is shown in Figure 1H. The predicted binding sites of HIPK2 and ZNF184 are shown in Figure 1I.
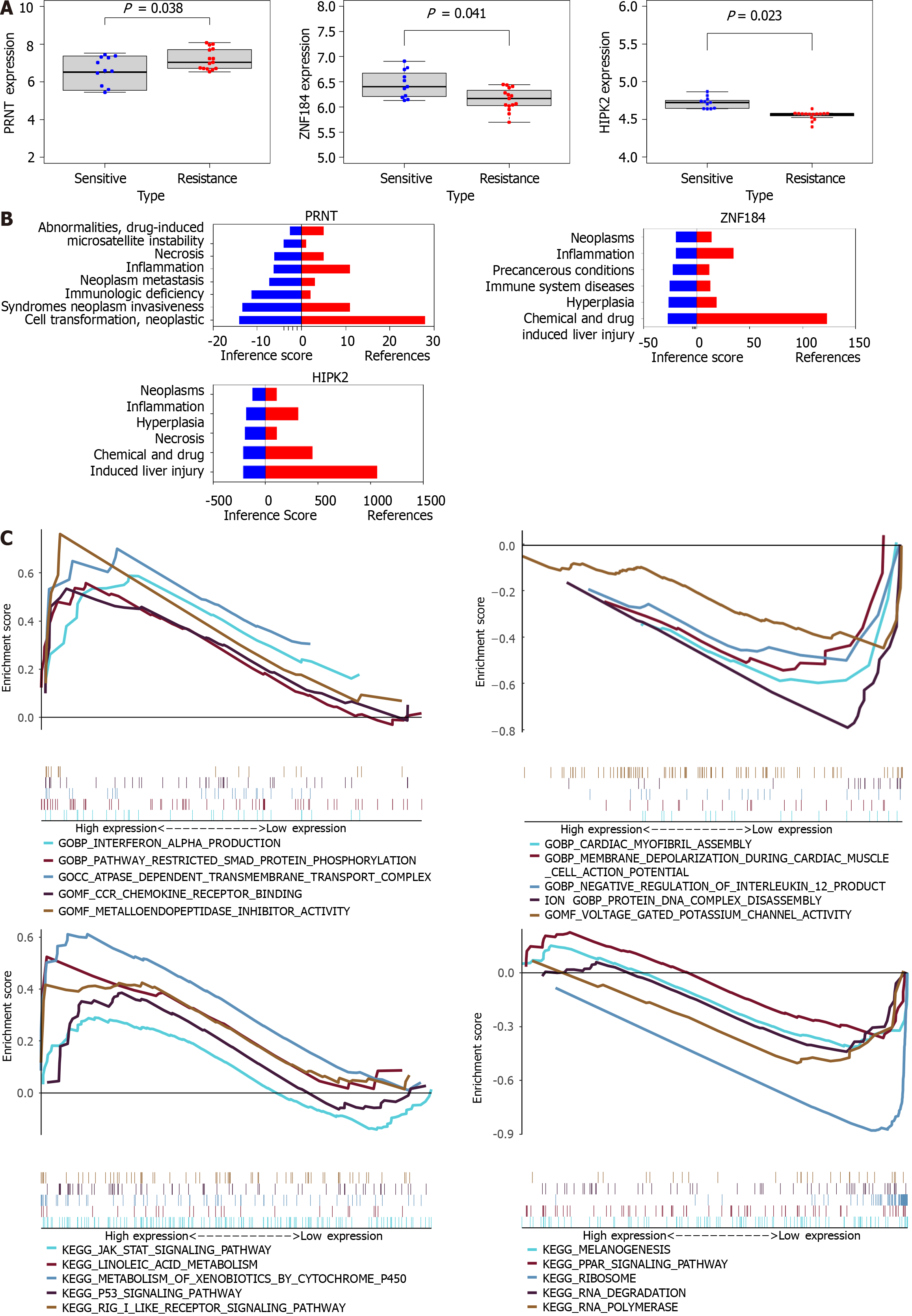
As shown in Figure 2A, PRNT was upregulated in OXA-resistant CRC datasets, and ZNF184 and HIPK2 were downregulated in OXA-resistant CRC datasets. The CTD database showed that hub genes targeted the tumour system, as shown in Figure 2B. The results of the analysis indicated 14 distinct diseases with statistical significance, which were involved in neoplasms, neoplasm invasiveness, neoplasm metastasis and so on. GSEA was performed to explore the downstream molecular mechanisms of PRNT (Figure 2C). The OXA-resistant CRC samples were divided into two groups according to the median expression level of PRNT. GO analysis based on GSEA suggested that interferon alpha production, pathway restricted smad protein phosphorylation, ATPase-dependent transmembrane transport complex and so on were upregulated, whereas cardiac myofibril assembly, membrane depolarization during cardiac muscle cell action potential, negative regulation of interleukin 12 production, and so on were downregulated. KEGG analysis indicated that the JAK STAT signalling pathway, linoleic acid metabolism, metabolism of xenobiotics by cytochrome P450 and so on were upregulated, while melanogenesis, the PPAR signalling pathway, ribosomes and so on were downregulated.
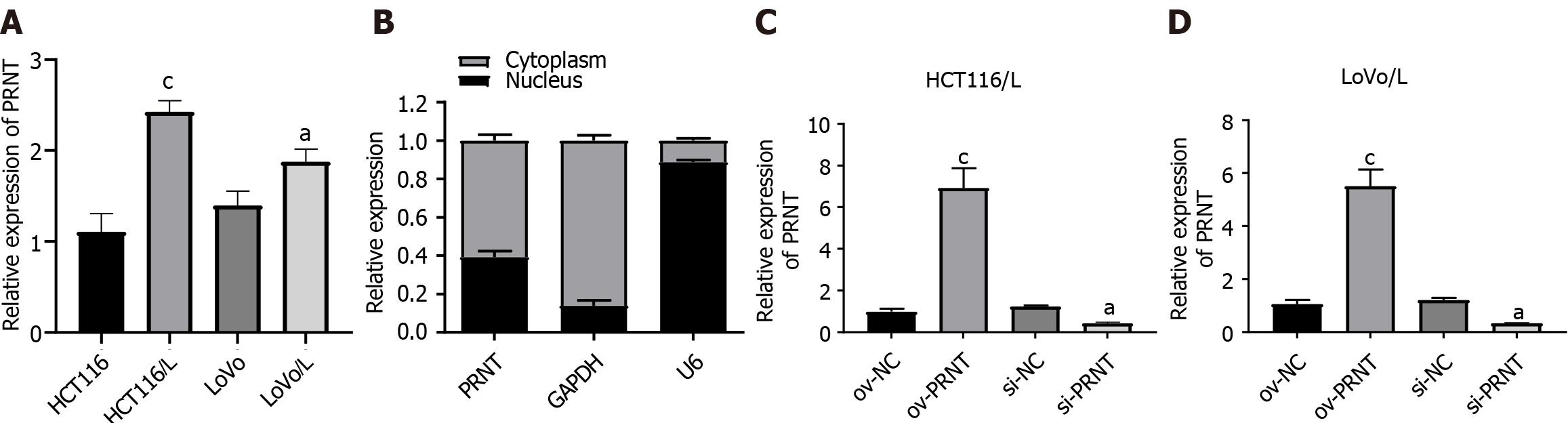
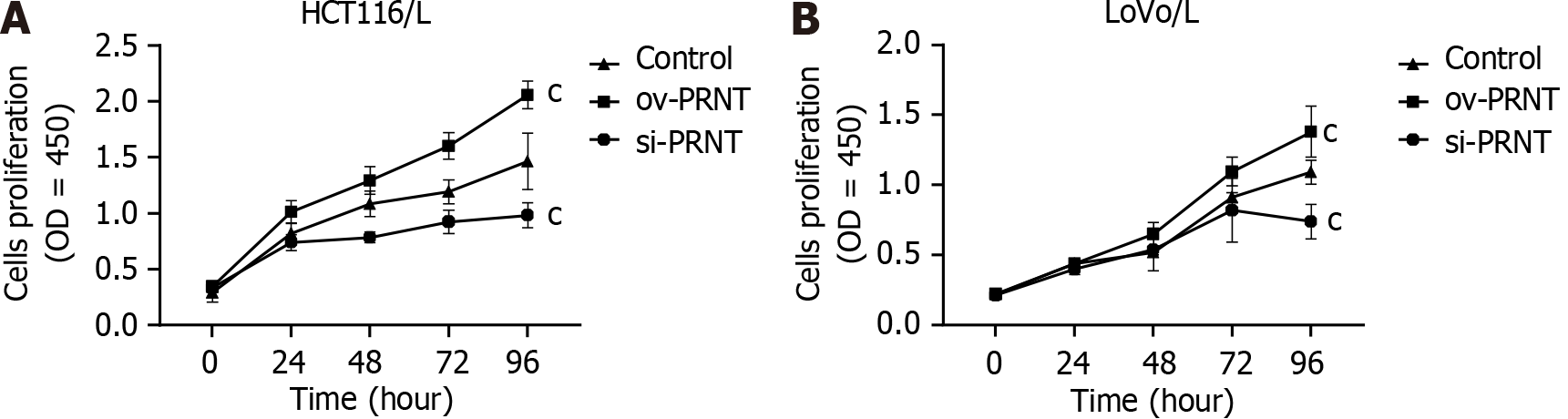
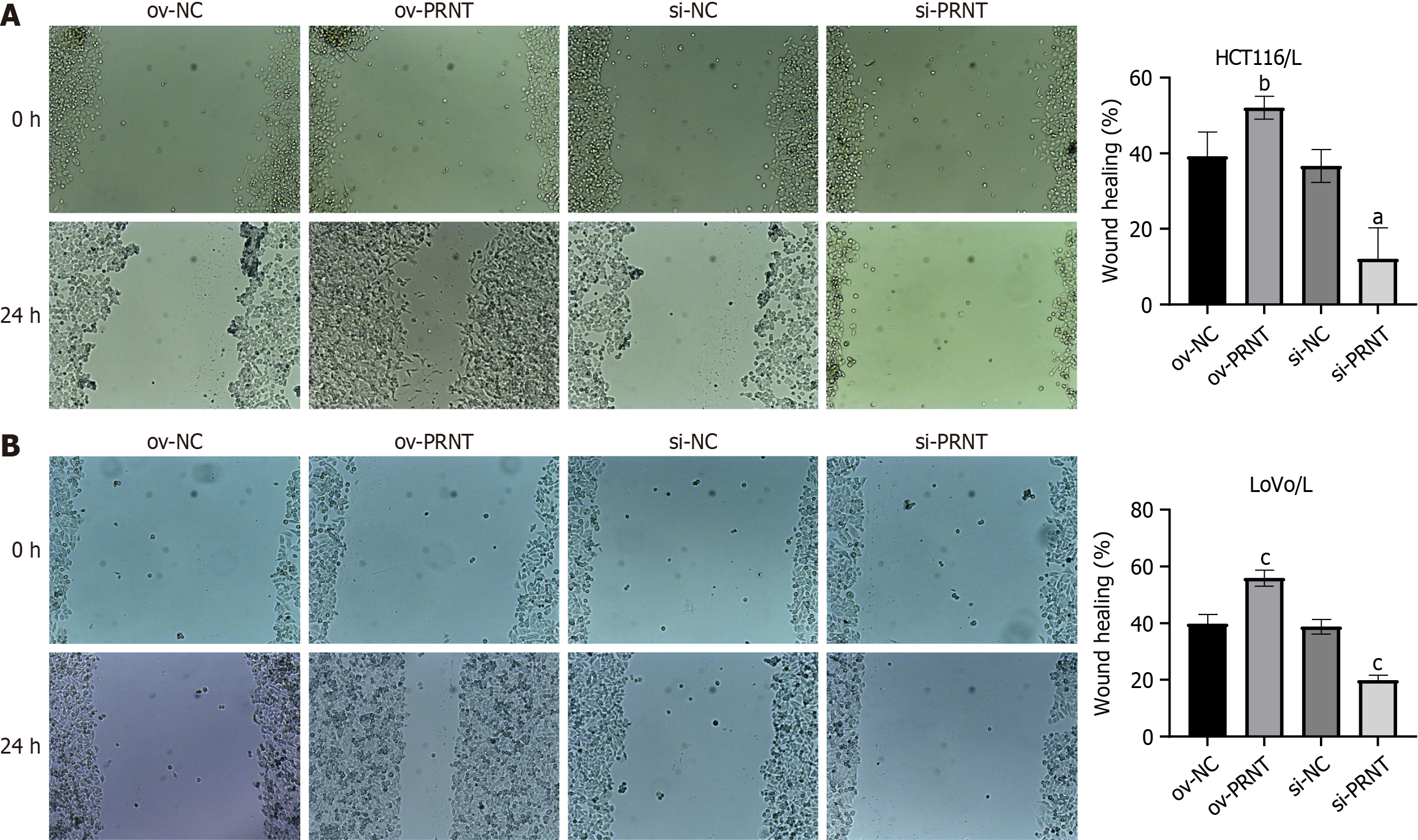
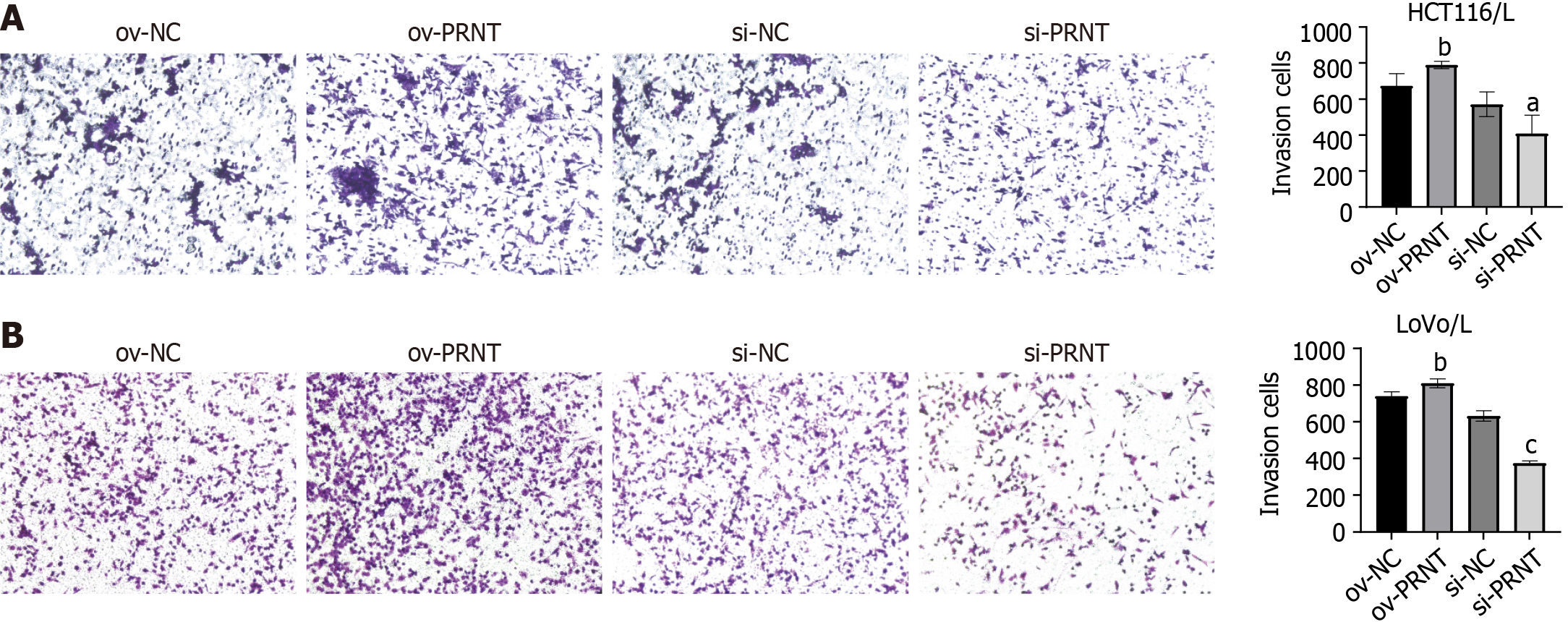
qRT-PCR was used to measure the expression of PRNT. As shown in Figure 3A, PRNT was upregulated in OXA-resistant CRC cell lines compared with OXA-sensitive CRC cell lines. Moreover, the results of the subcellular distribution assay after nuclear/cytoplasmic RNA fractionation showed that the ratio of PRNT in the nucleus to that in the cytoplasm was approximately 2:3 (Figure 3B). SiRNAs targeting PRNT (si-PRNT) and PRNT plasmid (ov-PRNT) were used to reduce or improve the expression of PRNT. As shown in Figure 3C and D, si-PRNT significantly decreased the expression and ov-PRNT significantly improved the expression of PRNT. The MTT assay showed that downregulation of PRNT significantly decreased the resistance to OXA in CRC cells, while overexpression of PRNT increased the sensitivity to OXA (Figure 4). The results of the wound healing assay suggest that PRNT knockdown could inhibit the migratory ability of CRC cells treated with OXA, and upregulated PRNT increased the migratory ability (Figure 5A and B). Transwell assays indicated that the invasion and migration abilities of CRC cells treated with OXA were improved by upregulated PRNT and reduced by downregulated PRNT (Figure 6A and B).
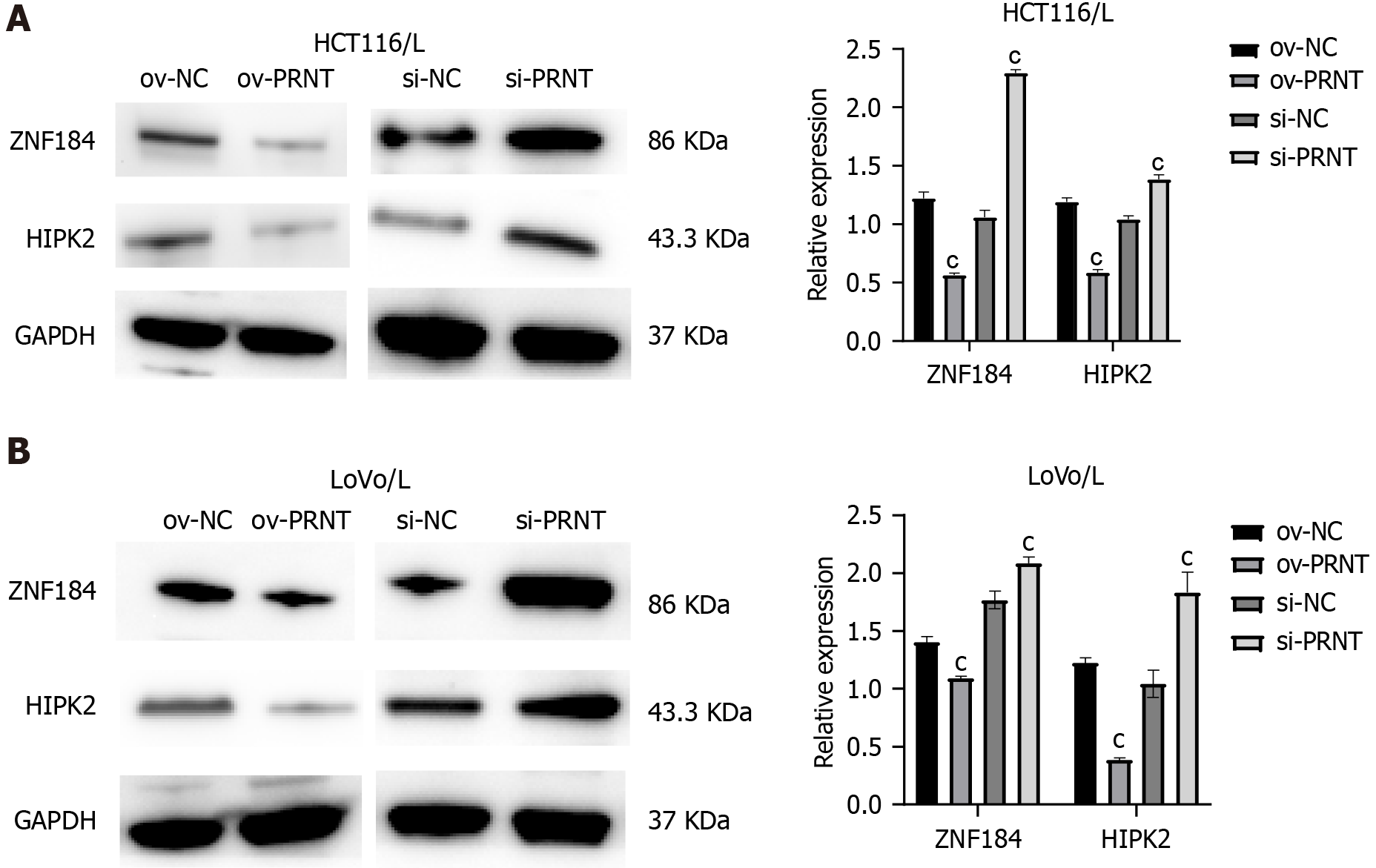
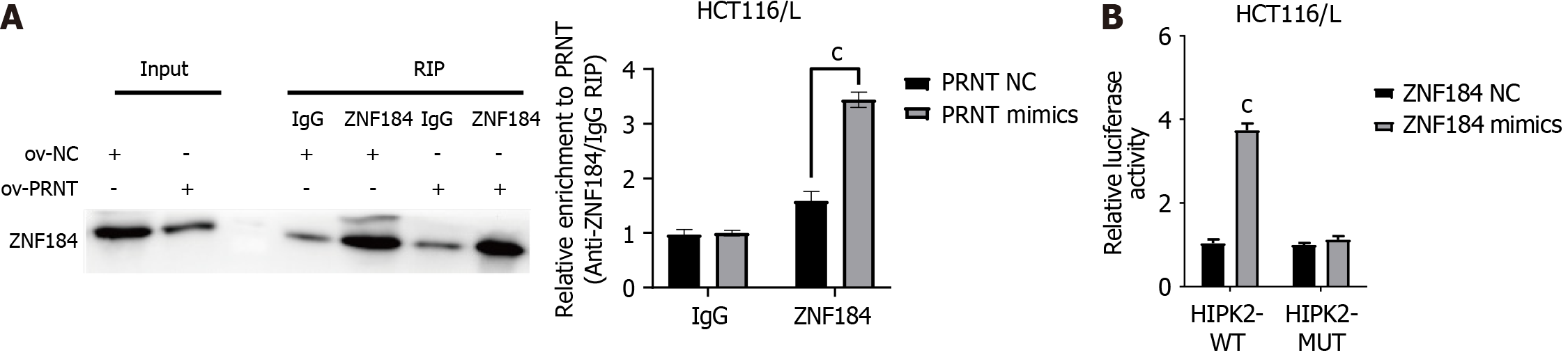
The results of bioinformatics analysis suggest that PRNT could sponge the transcription factor ZNF184 and subsequently regulate the expression of HIPK2 in CRC cells. Western blot assays were performed to validate the above results. As shown in Figure 7A and B, the expression of ZNF184 and HIPK2 was decreased by overexpression of PRNT in OXA-resistant CRC cells, whereas the expression of ZNF184 and HIPK2 was increased by downregulation of PRNT. An RNA immunoprecipitation assay was performed to evaluate the interaction between PRNT and ZNF184. As shown in Figure 8A, the expression levels of PRNT and ZNF184 were significantly higher in Ago2 pellets, suggesting that PRNT could sponge ZNF184 in CRC cells. A luciferase reporter assay was used to evaluate the interaction between HIPK2 and ZNF184. The results of the luciferase reporter assay indicated that ZNF184 mimics increased the luciferase activity in CRC cells transfected with HIPK2-WT, while the luciferase activity in CRC cells transfected with HIPK2-MUT was not increased by ZNF184 mimics (Figure 8B). The above results demonstrated that ZNF184 could directly bind to HIPK2.
A nude mouse xenograft model was constructed to explore the effect of PRNT on CRC OXA resistance in vivo. All nude mice were divided into three groups: control, OV-PRNT and si-PRNT. A total of 3 × 106 CRC cells were injected subcutaneously with 100 μL PBS, and after 1 wk, the mice were intraperitoneally injected with OXA (3 mg/kg) every 2 d. After 4 wk, the mice were killed, and the weight of the transplanted tumour was measured. The results of the nude mouse xenograft model indicated that the upregulation of PRNT significantly increased the resistance to OXA in CRC cells, while the downregulation of PRNT increased the sensitivity to OXA (Figure 9).
Colorectal cancer is the third most common cause of cancer and the second leading cause of cancer-related death in both men and women worldwide. Chemotherapy plays an irreplaceable role in the adjuvant treatment of colon cancer and neoadjuvant treatment of rectal cancer. Moreover, chemotherapy is the first-line therapy for patients with metastatic colorectal cancer[5,6]. Oxaliplatin is a platinum compound of diaminocyclohexane. The target site of oxaliplatin is DNA. The platinum atom forms cross-binding with DNA to antagonize its replication and transcription[21]. At present, oxaliplatin-based chemotherapy regimens play an important role in the treatment of colorectal cancer. However, due to acquired or primary resistance, oxaliplatin has unsatisfactory therapeutic effects in some patients with colorectal cancer[22,23]. Therefore, it is necessary to elucidate the molecular mechanism of OXA resistance to improve the prognosis of colorectal cancer patients. In this study, datasets of OXA-resistant CRC cell lines were downloaded and reanalyzed to identify the key lncRNAs involved in OXA resistance in CRC cells. PRNT was selected and included in subsequent studies. PRNT is a lncRNA located on chromosome 20p13. It has been reported that PRNT is abnormally methylated in breast cancer and is significantly associated with angiogenesis in cancer[24]. However, whether and how PRNT contributes to OXA resistance in CRC remain to be elucidated.
In this study, the expression of PRNT in CRC cell lines was determined. PRNT was upregulated in OXA-resistant CRC cell lines compared with OXA-sensitive CRC cell lines, indicating that PRNT was potentially correlated with OXA resistance. Subsequently, we conducted in vitro experiments to further explore the possible mechanism of PRNT. Gain- and loss-of-function experiments performed in vivo and in vitro suggested that PRNT could increase OXA resistance in CRC. To our knowledge, our study was the first to show the important role of PRNT in OXA resistance in CRC.
Studies have shown that the function of lncRNAs is closely related to their precise subcellular localization[25,26]. In this study, the subcellular location of PTAR in CRC cells was explored to determine the functional mechanisms. The results suggest that the ratio of PRNT in the nucleus to the cytoplasm is approximately 2:3. LncRNAs located in the nucleus can regulate cell functions by adsorbing transcription factors[25,26]. For instance, LncRNA CEBPA-DT is upregulated in human HCC tissues with distant metastasis after surgery and is closely associated with poorer prognosis in HCC patients. A mechanistic study showed that CEBPA-DT can bind to heteroribonucleoprotein C (hnRNPC), promote the cytoplasmic translocation of hnRNPC, enhance the interaction between hnRNPC and DDR2 mRNA, and then promote the expression of DDR2[27]. Moreover, Wang et al[28] indicated that the expression of LINC00665 is upregulated in gastric cancer tissues and is associated with poor prognosis in patients with gastric cancer. Mechanistically, LINC00665 binds to the transcription factor YBX1, which regulates the transcriptional activity of Wnt3a, and downregulation of LINC00665 blocks the activation of the Wnt/β-catenin signalling pathway[28]. In this study, the transcription factors that bind to PRNT were predicted, and ZNF184 exhibited the potential to bind to PRNT. ZNF184encodes a Kruppel C2H2-type zinc-finger protein family member. ZNF184 is overexpressed in oesophageal squamous cell carcinoma (ESCC) tissues and cells, and the expression of ZNF184 upregulates FTO, thereby enhancing MYC expression and promoting the progression of ESCC[29]. Moreover, epidemiological studies indicated that ZNF184 was significantly associated with lung cancer (P = 5.50x10-6)[30]. In CRC, the expression, biological function and clinical significance of ZNF184 have not been reported. In this study, we found that ZNF184 was downregulated in CRC cells. Moreover, a luciferase reporter assay demonstrated that ZNF184 could directly bind to the hub gene HIPK2.
The HIPK2 gene is located on human chromosome 7q34, which encodes a conserved serine/threonine kinase and interacts with homeodomain transcription factors and many other transcription factors, such as p53[31]. HIPK2 is a serine-threonine kinase that phosphorylates and regulates a large number of transcription regulators and chromatin modifiers. The function of HIPK2 in tumours remains uncertain, and relevant studies suggest that HIPK2 may play a role in tumour inhibition or cancer promotion depending on the cell environment[31]. HIPK2 plays an important role in a variety of tumours. For instance, HIPK2 is expressed at low levels in oral squamous cell carcinoma, and low expression of HIPK2 promotes the invasion of tumour cells and metastasis of cervical lymph nodes. Mechanistically, E-cadherin expression is triggered by mediating P53, which directly targets the CDH1 (encoding E-cadherin) promoter[32]. In colorectal cancer, the percentage of HIPK2-positive cells increases with tumour progression, is significantly associated with tumour lymph node metastasis stage and is associated with worse prognosis. In addition, HIPK2 is physically involved in the active RAS complex, and HIPK2 depletion impairs ERK phosphorylation and the growth of tumours derived from KRAS mutant colorectal cancer cells[33]. Moreover, Zhou et al[34] indicated that the HIPK2 protein regulates the phosphorylation of p53 in colorectal cancer cells, as well as the levels of Bax and Bcl-2 in CRC. The above studies have demonstrated the important role of HIPK2 in malignant tumours, especially colorectal cancer. In addition, HIPK2 plays an important role in tumour chemotherapy resistance. HIPK2 overexpression has been reported to negatively regulate Wip1 expression in bladder cancer cells, thereby overcoming cisplatin resistance in bladder cancer RT4-CISR cells[35]. Moreover, Alessandra Verdina et al[35] indicated that a high percentage of HIPK2+ cells is associated with treatment susceptibility to stage II CRC. In addition, HIPK2 depletion induced CRC cell resistance to 5-FU and OXA. In this study, HIPK2 was downregulated and PRNT was upregulated in OXA-resistant CRC datasets. PRNT enhances OXA resistance in CRC in vitro. Moreover, RIP and luciferase reporter assays demonstrated that PRNT could regulate the expression of HIPK2 in CRC cells by sponging the transcription factor ZNF184. Among them, HIPK2 is found in colorectal cancer, which is similar to the conclusion of the above study. This study demonstrates that HIPK2 can promote oxaliplatin resistance in colorectal cancer. Moreover, our study highlights the related upstream lncRNA-involved regulatory axis. In addition, a nude mouse xenograft model was constructed to explore the effect of PRNT on CRC OXA resistance in vivo. As expected, the downregulation of PRNT significantly increased the resistance to OXA in CRC cells, while overexpression of PRNT increased the sensitivity to OXA.
This study also have some limitations. Firstly, these findings are based on in vitro and xenograft models that may not fully replicate the complexity of human tumors. In addition, the study focused on a specific subset of CRC resistance mechanisms, and broader clinical relevance needs to be verified in larger patient cohorts. The regulatory axis including PRNT, HIPK2, and ZNF184 may be part of a broader network, and other factors affecting OXA resistance may be overlooked. Moreover, Another limitation lies in the excessive focus on PRNT-mediated regulation of HIPK2 in OXA resistance, which may neglect other contributing factors in CRC drug resistance. The scope of the study may not capture all the molecular events involved in the complex chemoresistance landscape. In addition, generalizations of findings may be influenced by the specific context of the study and the need for further exploration in different patient populations. Addressing these limitations will enhance the robustness and applicability of the findings.
In conclusion, this study showed that PRNT was downregulated in OXA-resistant CRC datasets and cell lines. PRNT sponges the transcription factor ZNF184 and subsequently regulates the expression of HIPK2. Through the PRNT/ZNF184/HIPK2 axis, PRNT could inhibit the progression and oxaliplatin resistance of colorectal cancer cells. In conclusion, our study highlights the potential of PRNT as a therapeutic target in CRC patients receiving OXA-based chemotherapy.
Colorectal cancer ranks as the third most prevalent form of cancer and is the second leading cause of cancer-related mortality worldwide. The development of chemotherapy resistance, especially to the drug oxaliplatin, remains a significant hurdle in the treatment of colorectal cancer, leading to reduced efficacy of anticancer drugs and worsening patient outcomes.
This study is driven by the recognition that long noncoding RNAs (lncRNAs) play a crucial role in the pathophysiology of cancer, including the development of chemotherapy resistance. However, the specific lncRNAs contributing to resistance in colorectal cancer treatment, particularly involving oxaliplatin, are not fully identified or understood.
The primary aim of this research is to delve into the role of lncRNAs in the resistance against oxaliplatin in colorectal cancer cells and to specifically pinpoint and characterize the function of the lncRNA prion protein testis specific (PRNT) in the modulation of this resistance and the overall progression of colorectal cancer.
The study approaches its objectives through the analysis of datasets from the Gene Expression Omnibus database, identifying potential lncRNAs and mRNAs that are involved in oxaliplatin resistance. A series of methodologies, including quantitative real-time polymerase chain reaction, MTT assays, wound healing, and Transwell assays, in addition to Western blotting and xenograft mouse modeling, were employed to investigate the interactions and effects within the PRNT/zinc finger protein 184 (ZNF184)/homeodomain interacting protein kinase 2 (HIPK2) regulatory axis.
It was found that the expression of lncRNA PRNT is higher in oxaliplatin-resistant colorectal cancer cell lines. PRNT appears to function by sponging the transcription factor ZNF184, which in turn regulates the expression of HIPK2, a gene inversely correlated with oxaliplatin resistance. The manipulation of PRNT levels was shown to alter the sensitivity of colorectal cancer cells to oxaliplatin, both in laboratory and animal models.
The study concludes that PRNT significantly contributes to the progression of colorectal cancer and to the resistance against oxaliplatin by interacting with ZNF184 and regulating HIPK2 expression. These findings illuminate a novel pathway of chemotherapy resistance in colorectal cancer and suggest that PRNT could be a promising target for therapeutic intervention in CRC patients undergoing oxaliplatin-based chemotherapy.
Looking ahead, the research suggests that future work should explore the potential for clinical application of PRNT-targeted therapies in colorectal cancer. There is also a need for further research to fully understand the impact of the PRNT/ZNF184/HIPK2 axis on colorectal cancer and potentially other cancers, which may open new avenues for overcoming chemotherapy resistance.
Provenance and peer review: Unsolicited article; Externally peer reviewed.
Peer-review model: Single blind
Specialty type: Gastroenterology & hepatology
Country/Territory of origin: China
Peer-review report’s scientific quality classification
Grade A (Excellent): 0
Grade B (Very good): 0
Grade C (Good): C
Grade D (Fair): 0
Grade E (Poor): 0
P-Reviewer: Cardella F, Italy S-Editor: Gong ZM L-Editor: A P-Editor: Zhao S
| 1. | Baidoun F, Elshiwy K, Elkeraie Y, Merjaneh Z, Khoudari G, Sarmini MT, Gad M, Al-Husseini M, Saad A. Colorectal Cancer Epidemiology: Recent Trends and Impact on Outcomes. Curr Drug Targets. 2021;22:998-1009. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 26] [Cited by in RCA: 152] [Article Influence: 38.0] [Reference Citation Analysis (2)] |
| 2. | Biller LH, Schrag D. Diagnosis and Treatment of Metastatic Colorectal Cancer: A Review. JAMA. 2021;325:669-685. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 398] [Cited by in RCA: 1402] [Article Influence: 350.5] [Reference Citation Analysis (0)] |
| 3. | Zhao H, Ming T, Tang S, Ren S, Yang H, Liu M, Tao Q, Xu H. Wnt signaling in colorectal cancer: pathogenic role and therapeutic target. Mol Cancer. 2022;21:144. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 2] [Cited by in RCA: 449] [Article Influence: 149.7] [Reference Citation Analysis (1)] |
| 4. | Paty PB, Garcia-Aguilar J. Colorectal cancer. J Surg Oncol. 2022;126:881-887. [RCA] [PubMed] [DOI] [Full Text] [Cited by in RCA: 5] [Reference Citation Analysis (0)] |
| 5. | Loughrey MB. Neoadjuvant immunotherapy and colorectal cancer treatment: Implications for the primary role of surgery. Colorectal Dis. 2022;24:1460-1461. [RCA] [PubMed] [DOI] [Full Text] [Cited by in RCA: 1] [Reference Citation Analysis (0)] |
| 6. | Damato A, Ghidini M, Dottorini L, Tomasello G, Iaculli A, Ghidini A, Luciani A, Petrelli F. Chemotherapy Duration for Various Indications in Colorectal Cancer: a Review. Curr Oncol Rep. 2023;25:341-352. [RCA] [PubMed] [DOI] [Full Text] [Cited by in RCA: 2] [Reference Citation Analysis (0)] |
| 7. | Abdel-Rahman O, Koski S, Mulder K. Real-world patterns of chemotherapy administration and attrition among patients with metastatic colorectal cancer. Int J Colorectal Dis. 2021;36:493-499. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 6] [Cited by in RCA: 11] [Article Influence: 2.8] [Reference Citation Analysis (0)] |
| 8. | Mauri G, Gori V, Bonazzina E, Amatu A, Tosi F, Bencardino K, Ruggieri L, Patelli G, Arena S, Bardelli A, Siena S, Sartore-Bianchi A. Oxaliplatin retreatment in metastatic colorectal cancer: Systematic review and future research opportunities. Cancer Treat Rev. 2020;91:102112. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 16] [Cited by in RCA: 38] [Article Influence: 7.6] [Reference Citation Analysis (1)] |
| 9. | Lin JF, Hu PS, Wang YY, Tan YT, Yu K, Liao K, Wu QN, Li T, Meng Q, Lin JZ, Liu ZX, Pu HY, Ju HQ, Xu RH, Qiu MZ. Phosphorylated NFS1 weakens oxaliplatin-based chemosensitivity of colorectal cancer by preventing PANoptosis. Signal Transduct Target Ther. 2022;7:54. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 6] [Cited by in RCA: 187] [Article Influence: 62.3] [Reference Citation Analysis (0)] |
| 10. | Bridges MC, Daulagala AC, Kourtidis A. LNCcation: lncRNA localization and function. J Cell Biol. 2021;220. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 528] [Cited by in RCA: 954] [Article Influence: 238.5] [Reference Citation Analysis (0)] |
| 11. | McCabe EM, Rasmussen TP. lncRNA involvement in cancer stem cell function and epithelial-mesenchymal transitions. Semin Cancer Biol. 2021;75:38-48. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 35] [Cited by in RCA: 194] [Article Influence: 38.8] [Reference Citation Analysis (0)] |
| 12. | Xing C, Sun SG, Yue ZQ, Bai F. Role of lncRNA LUCAT1 in cancer. Biomed Pharmacother. 2021;134:111158. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 55] [Cited by in RCA: 215] [Article Influence: 43.0] [Reference Citation Analysis (0)] |
| 13. | Yue B, Liu C, Sun H, Liu M, Song C, Cui R, Qiu S, Zhong M. A Positive Feed-Forward Loop between LncRNA-CYTOR and Wnt/β-Catenin Signaling Promotes Metastasis of Colon Cancer. Mol Ther. 2018;26:1287-1298. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 91] [Cited by in RCA: 159] [Article Influence: 22.7] [Reference Citation Analysis (0)] |
| 14. | Wei Z, Zhou J, Yu H, Pu Y, Cheng Y, Zhang Y, Ji Q, Zhu H. Zuo Jin Wan Reverses the Resistance of Colorectal Cancer to Oxaliplatin by Regulating the MALAT1/miR-200s/JNK Signaling Pathway. Evid Based Complement Alternat Med. 2022;2022:3032407. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Reference Citation Analysis (0)] |
| 15. | Huang G, Li L, Liang C, Yu F, Teng C, Pang Y, Wei T, Song J, Wang H, Liao X, Li Y, Yang J. Upregulated UCA1 contributes to oxaliplatin resistance of hepatocellular carcinoma through inhibition of miR-138-5p and activation of AKT/mTOR signaling pathway. Pharmacol Res Perspect. 2021;9:e00720. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 8] [Cited by in RCA: 6] [Article Influence: 1.5] [Reference Citation Analysis (0)] |
| 16. | Vance KW, Sansom SN, Lee S, Chalei V, Kong L, Cooper SE, Oliver PL, Ponting CP. The long non-coding RNA Paupar regulates the expression of both local and distal genes. EMBO J. 2014;33:296-311. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 163] [Cited by in RCA: 178] [Article Influence: 16.2] [Reference Citation Analysis (0)] |
| 17. | Ali T, Grote P. Beyond the RNA-dependent function of LncRNA genes. Elife. 2020;9. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 54] [Cited by in RCA: 195] [Article Influence: 39.0] [Reference Citation Analysis (0)] |
| 18. | Herman AB, Tsitsipatis D, Gorospe M. Integrated lncRNA function upon genomic and epigenomic regulation. Mol Cell. 2022;82:2252-2266. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 6] [Cited by in RCA: 361] [Article Influence: 120.3] [Reference Citation Analysis (0)] |
| 19. | Liu H, Liu L, Liu Q, He F, Zhu H. LncRNA HOXD-AS1 affects proliferation and apoptosis of cervical cancer cells by promoting FRRS1 expression via transcription factor ELF1. Cell Cycle. 2022;21:416-426. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 2] [Cited by in RCA: 15] [Article Influence: 5.0] [Reference Citation Analysis (0)] |
| 20. | Zou X, Hu X, He F, Zhang M, Kong X, Rui S, Liu Y, Wang L, Zheng X, Liu J, Li Z, Luo H. LncRNA LINC00152 promotes oral squamous cell carcinoma growth via enhancing Upstream Transcription Factor 1 mediated Mitochondrial Ribosomal Protein L52 transcription. J Oral Pathol Med. 2022;51:454-463. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 3] [Cited by in RCA: 11] [Article Influence: 2.8] [Reference Citation Analysis (0)] |
| 21. | Zhang C, Xu C, Gao X, Yao Q. Platinum-based drugs for cancer therapy and anti-tumor strategies. Theranostics. 2022;12:2115-2132. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 315] [Cited by in RCA: 430] [Article Influence: 143.3] [Reference Citation Analysis (0)] |
| 22. | Li Y, Gan Y, Liu J, Li J, Zhou Z, Tian R, Sun R, Xiao Q, Li Y, Lu P, Peng Y, Shu G, Yin G. Downregulation of MEIS1 mediated by ELFN1-AS1/EZH2/DNMT3a axis promotes tumorigenesis and oxaliplatin resistance in colorectal cancer. Signal Transduct Target Ther. 2022;7:87. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 2] [Cited by in RCA: 100] [Article Influence: 33.3] [Reference Citation Analysis (0)] |
| 23. | Fakhr E, Zare F, Azadmanesh K, Teimoori-Toolabi L. LEF1 silencing sensitizes colorectal cancer cells to oxaliplatin, 5-FU, and irinotecan. Biomed Pharmacother. 2021;143:112091. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 4] [Cited by in RCA: 19] [Article Influence: 4.8] [Reference Citation Analysis (0)] |
| 24. | Sun Y, Wang R. A Risk Score System Based on the Methylation Levels of 15 RNAs in Breast Cancer. Cancer Biother Radiopharm. 2022;37:697-707. [RCA] [PubMed] [DOI] [Full Text] [Reference Citation Analysis (0)] |
| 25. | Chen LL. Linking Long Noncoding RNA Localization and Function. Trends Biochem Sci. 2016;41:761-772. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 587] [Cited by in RCA: 782] [Article Influence: 86.9] [Reference Citation Analysis (0)] |
| 26. | Guo CJ, Xu G, Chen LL. Mechanisms of Long Noncoding RNA Nuclear Retention. Trends Biochem Sci. 2020;45:947-960. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 32] [Cited by in RCA: 70] [Article Influence: 14.0] [Reference Citation Analysis (0)] |
| 27. | Cai Y, Lyu T, Li H, Liu C, Xie K, Xu L, Li W, Liu H, Zhu J, Lyu Y, Feng X, Lan T, Yang J, Wu H. LncRNA CEBPA-DT promotes liver cancer metastasis through DDR2/β-catenin activation via interacting with hnRNPC. J Exp Clin Cancer Res. 2022;41:335. [RCA] [PubMed] [DOI] [Full Text] [Cited by in RCA: 36] [Reference Citation Analysis (0)] |
| 28. | Wang J, Shen D, Li S, Li Q, Zuo Q, Lu J, Tang D, Feng Y, Yin P, Chen C, Chen T. LINC00665 activating Wnt3a/β-catenin signaling by bond with YBX1 promotes gastric cancer proliferation and metastasis. Cancer Gene Ther. 2023;30:1530-1542. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1] [Cited by in RCA: 9] [Article Influence: 4.5] [Reference Citation Analysis (0)] |
| 29. | Ke S, Wang J, Lu J, Fang M, Li R. Long intergenic non-protein coding RNA 00858 participates in the occurrence and development of esophageal squamous cell carcinoma through the activation of the FTO-m6A-MYC axis by recruiting ZNF184. Genomics. 2023;115:110593. [RCA] [PubMed] [DOI] [Full Text] [Cited by in RCA: 4] [Reference Citation Analysis (0)] |
| 30. | Zuber V, Marconett CN, Shi J, Hua X, Wheeler W, Yang C, Song L, Dale AM, Laplana M, Risch A, Witoelar A, Thompson WK, Schork AJ, Bettella F, Wang Y, Djurovic S, Zhou B, Borok Z, van der Heijden HF, de Graaf J, Swinkels D, Aben KK, McKay J, Hung RJ, Bikeböller H, Stevens VL, Albanes D, Caporaso NE, Han Y, Wei Y, Panadero MA, Mayordomo JI, Christiani DC, Kiemeney L, Andreassen OA, Houlston R, Amos CI, Chatterjee N, Laird-Offringa IA, Mills IG, Landi MT. Pleiotropic Analysis of Lung Cancer and Blood Triglycerides. J Natl Cancer Inst. 2016;108. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 12] [Cited by in RCA: 14] [Article Influence: 1.6] [Reference Citation Analysis (0)] |
| 31. | Conte A, Valente V, Paladino S, Pierantoni GM. HIPK2 in cancer biology and therapy: Recent findings and future perspectives. Cell Signal. 2023;101:110491. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1] [Cited by in RCA: 12] [Article Influence: 6.0] [Reference Citation Analysis (0)] |
| 32. | Zheng X, Pan Y, Chen X, Xia S, Hu Y, Zhou Y, Zhang J. Inactivation of homeodomain-interacting protein kinase 2 promotes oral squamous cell carcinoma metastasis through inhibition of P53-dependent E-cadherin expression. Cancer Sci. 2021;112:117-132. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 2] [Cited by in RCA: 3] [Article Influence: 0.6] [Reference Citation Analysis (0)] |
| 33. | Di Segni M, Virdia I, Verdina A, Amoreo CA, Baldari S, Toietta G, Diodoro MG, Mottolese M, Sperduti I, Moretti F, Buglioni S, Soddu S, Di Rocco G. HIPK2 Cooperates with KRAS Signaling and Associates with Colorectal Cancer Progression. Mol Cancer Res. 2022;20:686-698. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 7] [Cited by in RCA: 4] [Article Influence: 1.3] [Reference Citation Analysis (0)] |
| 34. | Zhou L, Feng Y, Jin Y, Liu X, Sui H, Chai N, Chen X, Liu N, Ji Q, Wang Y, Li Q. Verbascoside promotes apoptosis by regulating HIPK2-p53 signaling in human colorectal cancer. BMC Cancer. 2014;14:747. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 37] [Cited by in RCA: 46] [Article Influence: 4.2] [Reference Citation Analysis (0)] |
| 35. | Lin J, Zhang Q, Lu Y, Xue W, Xu Y, Zhu Y, Hu X. Downregulation of HIPK2 increases resistance of bladder cancer cell to cisplatin by regulating Wip1. PLoS One. 2014;9:e98418. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 14] [Cited by in RCA: 20] [Article Influence: 1.8] [Reference Citation Analysis (0)] |