Published online Apr 15, 2024. doi: 10.4251/wjgo.v16.i4.1532
Peer-review started: December 5, 2023
First decision: December 14, 2023
Revised: December 29, 2023
Accepted: February 3, 2024
Article in press: February 3, 2024
Published online: April 15, 2024
Processing time: 127 Days and 10.4 Hours
Peutz-Jeghers syndrome (PJS) is a rare hereditary neoplastic disorder mainly associated with serine/threonine kinase 11 (STK11/LKB1) gene mutations. Preimplantation genetic testing can protect a patient’s offspring from mutated genes; however, some variations in this gene have been interpreted as variants of uncertain significance (VUS), which complicate reproductive decision-making in genetic counseling.
To identify the pathogenicity of two missense variants and provide clinical guidance.
Whole exome gene sequencing and Sanger sequencing were performed on the peripheral blood of patients with PJS treated at the Reproductive and Genetic Hospital of Citic-Xiangya. Software was employed to predict the protein structure, conservation, and pathogenicity of the two missense variation sites in patients with PJS. Additionally, plasmids were constructed and transfected into HeLa cells to observe cell growth. The differences in signal pathway expression between the variant group and the wild-type group were compared using western blot and immunohistochemistry. Statistical analysis was performed using one-way analysis of variance. P < 0.05 was considered statistically significant.
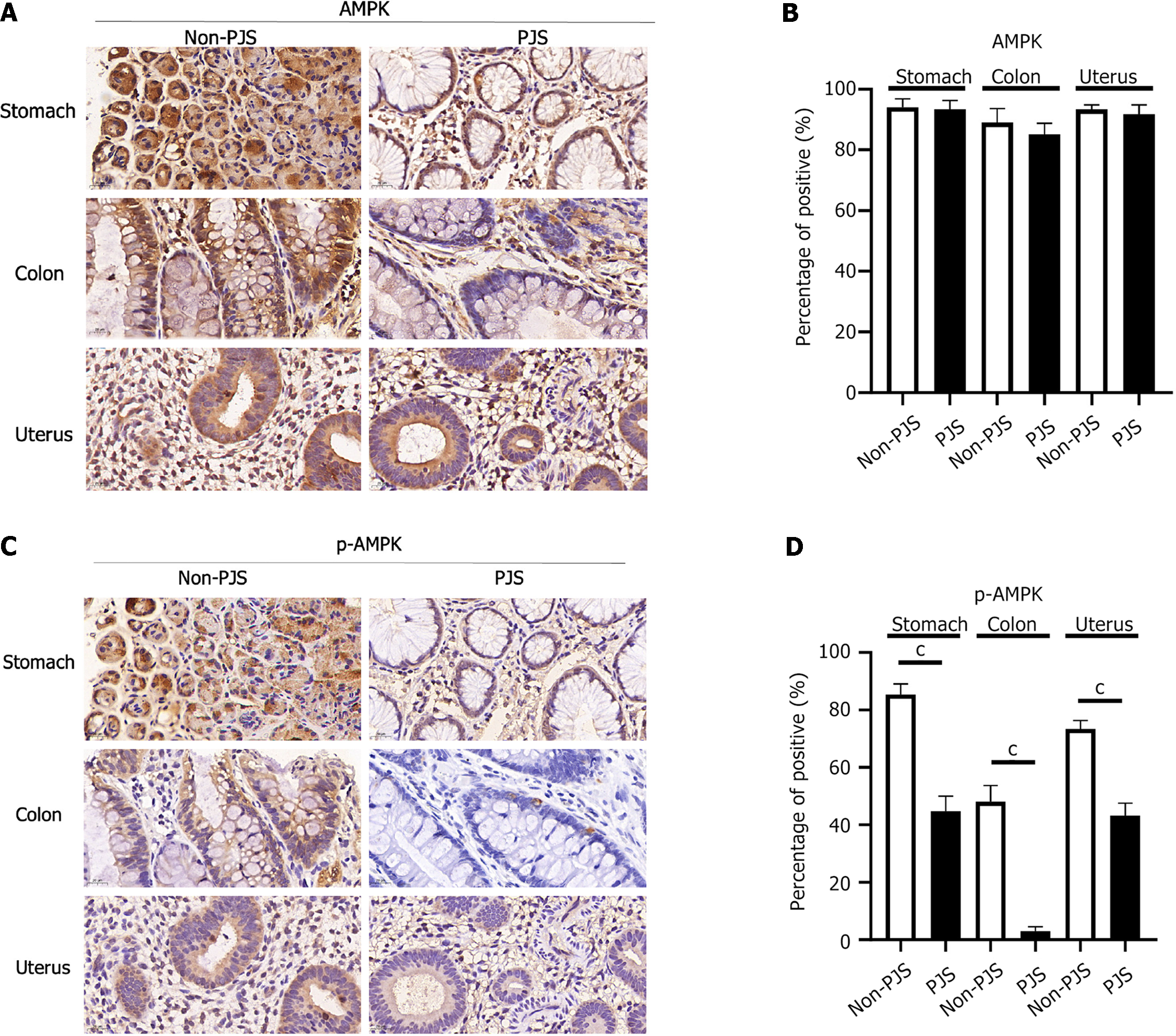
We identified two missense STK11 gene VUS [c.889A>G (p.Arg297Gly) and c.733C>T (p.Leu245Phe)] in 9 unrelated PJS families who were seeking reproductive assistance. The two missense VUS were located in the catalytic domain of serine/threonine kinase, which is a key structure of the liver kinase B1 (LKB1) protein. In vitro experiments showed that the phosphorylation levels of adenosine monophosphate-activated protein kinase (AMPK) at Thr172 and LKB1 at Ser428 were significantly higher in transfected variation-type cells than in wild-type cells. In addition, the two missense STK11 variants promoted the proliferation of HeLa cells. Subsequent immunohistochemical analysis showed that phosphorylated-AMPK (Thr172) expression was significantly lower in gastric, colonic, and uterine polyps from PJS patients with missense variations than in non-PJS patients. Our findings indicate that these two missense STK11 variants are likely pathogenic and inactivate the STK11 gene, causing it to lose its function of regulating downstream phosphorylated-AMPK (Thr172), which may lead to the development of PJS. The identification of the pathogenic mutations in these two clinically characterized PJS patients has been helpful in guiding them toward the most appropriate mode of pregnancy assistance.
These two missense variants can be interpreted as likely pathogenic variants that mediated the onset of PJS in the two patients. These findings not only offer insights for clinical decision-making, but also serve as a foundation for further research and reanalysis of missense VUS in rare diseases.
Core Tip: These two missense variants, STK11 c.889A>G (p.Arg297Gly) and STK11 c.733C>T (p.Leu245Phe), have been found to contribute to the development of Peutz-Jeghers syndrome (PJS) by impairing the STK11/adenosine monophosphate-activated protein kinase signaling pathway. It clarifies that these two germline variants, STK11 c.889A>G (p.Arg297Gly) and STK11 c.733C>T (p.Leu245Phe), are likely pathogenic mutations, providing valuable information for fertility selection in patients with PJS.
- Citation: Liu J, Zeng SC, Wang A, Cheng HY, Zhang QJ, Lu GX. Two missense STK11 gene variations impaired LKB1/adenosine monophosphate-activated protein kinase signaling in Peutz-Jeghers syndrome. World J Gastrointest Oncol 2024; 16(4): 1532-1546
- URL: https://www.wjgnet.com/1948-5204/full/v16/i4/1532.htm
- DOI: https://dx.doi.org/10.4251/wjgo.v16.i4.1532
Peutz-Jeghers syndrome [PJS; Mendelian Inheritance in Man (MIM), #175200, www.ncbi.nlm.gov/OMIM] is a rare hereditary neoplastic disorder characterized by mucocutaneous pigmentation and the development of multiple gastrointestinal (GI) polyps[1]. The annual incidence of PJS ranges from 1: 8300 to 1: 29000 and does not differ by sex, race, or ethnicity[2-4]. PJS has extremely high penetrance and clinical heterogeneity[5] and can lead to the formation of hamartomatous polyps in many organs, such as the jejunum, ileum, duodenum, stomach, colon, and gallbladder[6,7]. Even at a very young age, PJS patients with pathological hamartomatous polyps can experience numerous GI complications, including bleeding, anemia, abdominal pain, intussusception, obstruction, and infarction[8]. Patients with PJS are at high risk for GI and extra-GI cancer[9,10]. The disease is inherited in an autosomal dominant manner and has a 50% chance of being passed on to offspring.
The serine/threonine kinase 11 (STK11/LKB1; MIM, *602216) gene has been identified as a pathogenic factor of PJS, with germline mutations in this gene listed in the medical records of most PJS patients as a risk factor[3,7]. STK11 is a tumor suppressor gene located on chromosome 19p13.3 and includes ten exons[11]. STK11 functions mainly depend on the phosphorylation and activation of adenosine monophosphate-activated protein kinase (AMPK). As an important metabolic regulator, AMPK regulates cell generation by regulating the activity of metabolic enzymes and activating adaptive transcriptional responses[12]. STK11 is a master stress response regulator[13] and plays an important role in tumorigenesis, implicated in numerous key biological processes, including cell metabolism, cell cycle regulation, cell polarity and motility, and angiogenesis[1].
A search for STK11 gene variants in the ClinVar database revealed a total of 2481 entries with clinical significance, of which 974 (39.26%) were variants of uncertain significance (VUS), predominantly (79.98%, 779/974) missense variants until December 18, 2023. Many VUS affect diagnosis, clinical treatment, and decision-making processes because their pathogenicity cannot be defined. Some variants have been shown to be frameshift or nonsense mutations that result in abnormal protein truncation and subsequent loss of kinase activity and are therefore classified as pathogenic or potentially pathogenic. However, missense variants usually only lead to the formation of incorrect amino acid residues at the corresponding position in the translated product, leading to an abnormal protein. Whether this protein alteration affects the normal function of the gene needs to be tested experimentally. Understanding how the function of STK11 is disrupted may contribute to a better understanding of the molecular mechanisms involved in the pathogenesis and carcinogenesis of PJS. Clinical fertility counseling and reproductive assistance need to target specific variant sites, so determining the pathogenicity of VUS is urgent and important for patients.
Peripheral blood samples were collected from 9 PJS probands and 19 family members, all of whom were Chinese Han. These patients with PJS were diagnosed according to the PJS diagnostic criteria published by the European consensus in 2007[14]. When ≥ 2 individuals are affected within the same family, they are considered family cases. Patients without a family history of PJS are defined as sporadic cases. Patient information such as family history and clinical characteristics were collected from the patient’s medical records. All patients provided informed consent for this study. Clinical data collection included demographics, medical history, family history, symptoms, and surgical interventions related to PJS. Some patients underwent endoscopic or surgical polypectomy. Tissues were fixed with formalin and embedded in paraffin. The wax blocks were successively sliced and prepared for subsequent hematoxylin-eosin (HE) staining and immunohistochemical (IHC) experiments.
The peripheral blood of all probands was sent to Beijing Genomics Research Institute for Whole-exome sequencing (WES), and the tumor-related gene exons and adjacent ± 10 bp intron variations were detected. The results were verified by polymerase chain reaction (PCR)-Sanger sequencing. The identified variations were further tested in all available family members to confirm co-segregation of these variations with the disease. We used the online site primer 3 to design primers. Variation analysis was mainly based on the American Society for Medical Genetics and Genomics (ACMG) guidelines for interpreting sequence variation[15].
Amino acid conservation analysis was performed using UGENE software. Five online software programs, AlphaMissense (https://alphamissense.hegelab.org/search), PROVEAN (http://provean.jcvi.org/seq_submit.php), mutation Taster (https://www.mutationtaster.org/), PolyPhen-2 (http://genetics.bwh.harvard.edu/pph2/), and Swiss-model (http://swissmodel.expasy.org/), were used to predict the functional and structural significance of these variations.
Tissue sections were incubated with liver kinase B1 (LKB1), phosphorylated (p)-LKB1 (Ser428), AMPK, p-AMPK (Thr172) antibodies. The IHC kit used was from Leica Biosystems (catalog No.RE7280-k, Germany) and was operated according to the reagent instructions. Polyp tissue sections were stained by HE staining. An experienced pathologist reviewed these specimens and determined the histological types of these polyps. The staining of the sliced tissue with a brownish yellow color indicates positivity. Statistical analysis of IHC results was based on a direct count of the number of positive cells in five random visual fields at 70 × magnification or a visual assessment of the proportion of positive cells. Each group was repeated three times.
Five plasmids including enhanced green fluorescent protein (EGFP)-N1-vector (EGFP-Empty), PEGFP-N1-STK11 (EGFP-STK11), PEGFP-N1-STK11 c.889A>G (p.Arg297Gly), PEGFP-N1-STK11 c.733C>T (p.Leu245Phe) and PEGFP-N1-STK11 c.910C>T(p.Arg304Trp) were transfected into HeLa cells with lipofectamine 3000. Cells were collected 48 h after transfection, subsequent PCR product gel electrophoresis, Western blotting (WB), Reverse transcription quantitative PCR (RT-qPCR), and Cell Counting Kit 8 (CCK 8) experiments were performed. Transfected cells were counted and seeded in 6-well plates (1000 cells/well). Cell proliferation was detected at 12 h, 24 h, 36 h, 48 h, 60 h, and 72 h after transfection. Each group was repeated three times.
Total RNA was extracted using Trizol, and cDNA was synthesized by reverse transcription from 1 µg of total RNA using oligo (dT) primers. RT-qPCR was performed using the All-in-OneTM qPCR mix on a LightCycler 480 system (Roche, Switzerland). Primers were performed according to the primer website (https://www.ncbi.nlm.nih.gov/tools/primer). Glyceraldehyde 3-phosphate dehydrogenase (GAPDH) was used as an internal control for normalization. The level of expression endogenous, exogenous, and total mRNA of STK11 was calculated using the 2-ΔΔCt method.
Total protein was extracted using radioimmunoprecipitation assay lysis buffer and detected using bicinchoninic acid kits. Total protein was separated on 10% SDS-PAGE. After separation, the protein was elactrotransferred to a polyvinylidene difluoride membrane, and then blocked in 5% skimmed milk for 1 h at room temperature. The membranes were incubated with primary antibodies against STK11, p-STK11, AMPK, p-AMPK and GAPDH overnight at 4˚C. GAPDH served as the loading control. Membranes were subsequently incubated with secondary antibodies for 2 h at room temperature. Protein expression was detected using an enhanced chemiluminescence kit (Thermo Fisher Scientific, United States).
All statistical analyzes were performed with SPSS (version 26.0, United States) and GraphPad Prism (verison 9.0, United States). The data are expressed as mean ± SD unless otherwise noted. Comparisons among multiple groups were determined using one-way analysis of variance, followed by least significant difference or Dunnett’s T3 test. P < 0.05 was considered statistically significant.
Some patients with PJS and a clinical phenotype seek genetic and reproductive assistance when giving birth. The demographics and clinical characteristics of 9 patients with PJS are shown in Table 1. At the time of the last follow-up, 8 patients were married, and 1 was unmarried. All patients with PJS had skin and mucosa pigmentation. The median age of the 9 PJS patients when they visited our hospital for the first time was 30 years (ranging from 23 to 45 years), of whom 5 were male (55.6%). The age at diagnosis varied widely, with some patients being diagnosed in the first few years of life and others demonstrating a later onset and milder symptoms. Eight of the 9 patients (89.0%) with PJS developed colon polyps and underwent polypectomy, and the mean age at the onset of the first GI polyp was 19.38 years (19.38 ± 4.39). The burden of polyps (cumulative number of polyps) varied from approximately 10 to several hundred. PJS polyps were distributed in the stomach in 5 patients (55.6%), in the small intestine in 7 (77.8%), and in the colorectum in 4 (44.4%). Two patients (22.2%) had uterine polyps, 1 (11.1%) had gallbladder polyps, and 1 (11.1%) had nasal polyps. Six (66.67%) were initially treated before the age of 20 years, and all patients received initial treatment before the age of 30 years. Patients with a family history of PJS were treated at a later age than those without; however, the difference was not significant (P = 0.4059). The age at first treatment differed by sex, with the first treatment occurring later in men than in women; however, the difference was not significant (P = 0.4521).
| Proband family No. | Proband gender, age (yr) | Clinical features | Age at first discovery of polyps | Previous medicine history | Nucleotide alteration, amino acid alteration | Exon/Intron, mutation type | ACMG | PJS family history | Reproductive choice | Current Clinical outcome | Pedigree analysis |
| 1 | Man, 34 | Pigmentation of skin and mucosa; multiple polyps | 19 | Intestinal polypectomy | c.180C>A (p.Tyr60Ter) | Exon1, nonsense | P | De novo | IVF-ET | The fetus had the same mutation and the pregnancy was terminated | Reject |
| 2 | Woman, 30 | Pigmentation of skin and mucosa; multiple polyps | 25 | Polypectomy was performed in the stomach, colon and uterus | c.889A>G (p.Arg297Gly) | Exon7, missense | VUS | Yes, maternal. Mother, brother | PGT | The fetus was 5 wk+ and had a spontaneous abortion | Brother carries STK11 variation but father doesn’t |
| c.1062C>G (p.Phe354Leu) | Exon 8, missense | LB | |||||||||
| 3 | Man, 27 | Pigmentation of skin and mucosa; multiple polyps | 16 | Multiple gastrointestinal polypectomy | c.733C>T (p.Leu245Phe) | Exon5, missense | VUS | De novo | PGT | The fetus was prenatally diagnosed as having no mutation and gave birth to a child | Parents had no STK11 variation |
| 4 | Man, 25 | Pigmentation of skin and mucosa; multiple polyps | 15 | Multiple gastrointestinal polypectomy | c.250A>T (p.Lys84*) | Exon1, nonsense | P | De novo | PGT | Preoperative preparation for PGT | Parents had no STK11 variation |
| 5 | Woman, 35 | Pigmentation on the face and lips; multiple polyps | 19 | Twice minimally invasive surgery for intestinal polyps and laparotomy operation | c.114_121delGCGCCGCA (p.Arg39Alafs*121) | Exon1, frame shift | P | Yes, paternal. Father, uncle, brother | PGT | The fetus was prenatally diagnosed as having no mutation and gave birth to a child | Mother and son had no STK11 variation |
| 6 | Woman, 33 | Pigmentation on the lower lip, fingers, soles of the feet, and oral mucosa; multiple polyps | 25 | Polypectomy was performed in the small intestine and uterus | c.193G>T(p.Glu65*) | Exon1, nonsense | P | De novo | PGT | Preoperative preparation for PGT | Parents had no STK11 variation |
| 7 | Man, 45 | Pigmentation on the lower lip, fingers, and oral mucosa; multiple polyps | 22 | Multiple gastrointestinal polypectomy | c.464+1G>A | Intron 3-4, splicing | LP | De novo | PGT | Preoperative preparation for PGT | Parents had no STK11 variation |
| 8 | Man, 23 | Pigmentation on the lips | / | Laser treatment for hyperpigmentation at 19 yr, no polyps found on gastrointestinal endoscopy | c.734+3A>T | Intron 5-6, splicing | LP | Yes, paternal. Father, grandfather, sister, and niece | / | Unmarried, under consideration | Father had STK11 variation |
| 9 | Woman, 24 | Pigmentation on the limbs and trunk; multiple polyps | 13 | Multiple gastrointestinal polypectomy; termination because fetal had STK11 mutation | c.734+2T>C | Intron 5-6, splicing | P | De novo | PGT | Preoperative preparation for PGT | Parents had no STK11 variation |
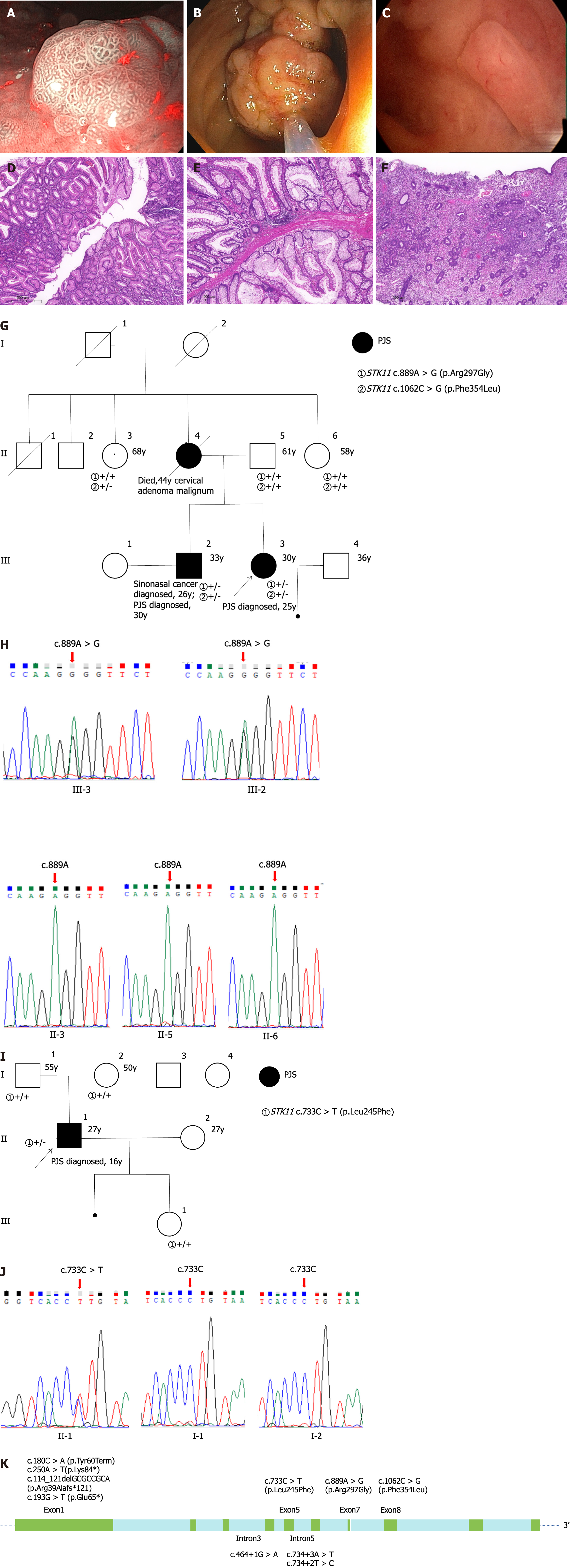
WES or STK11 variant detection was performed for the 9 unrelated PJS patients. Ten (111%) different germline variations in the STK11 gene were identified. Clinical and sociodemographic data were collected from 26 family members, 19 of whom underwent pedigree analysis for the STK11 gene (Supplementary Table 1). A total of 4 STK11 variations were identified in familial cases and 6 in sporadic cases. Of the 10 variants identified, three were nonsense variants, three were splicing variants, three were missense variants, and one was a frameshift variant. According the ACMG guidelines, the variant pathogenicity assessment indicated that 5 were pathogenic, 2 were likely pathogenic, 1 was likely benign, and 2 were VUS. Two patients with missense VUS underwent GI endoscopy at our hospital, and one had polyps removed from the stomach, colon, and uterus (Figure 1A-F). Two of the variations [c.889A>G (p.Arg297Gly) and c.733C>T (p.Leu245Phe)] were identified by Sanger sequencing (Figure 1G-J). A schematic representation of the localization of 10 STK11 variations is shown in Figure 1K; most of the variations were concentrated in the exon 1 region.
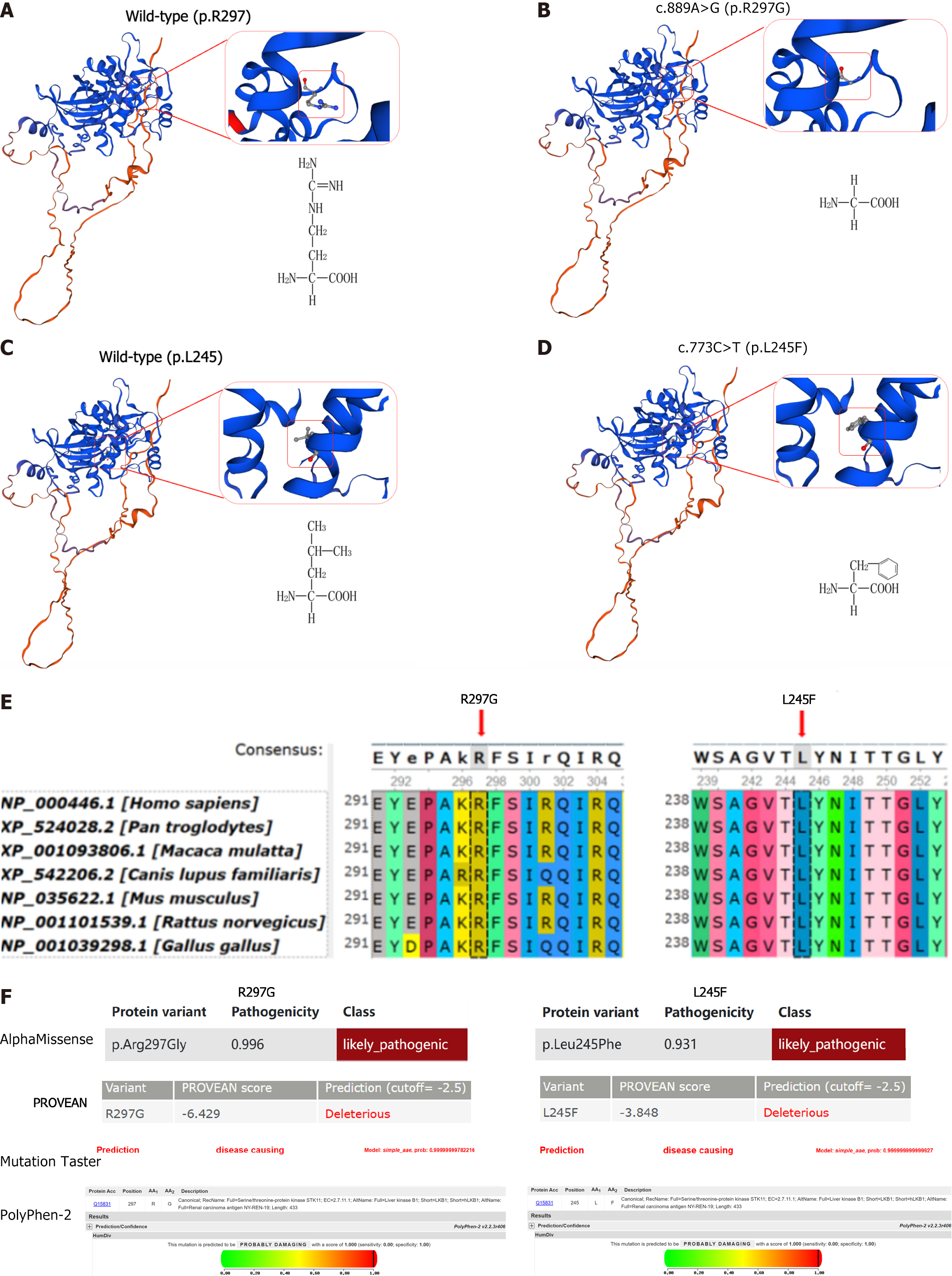
Swiss-model software was used to construct a three-dimensional (3D) model of the protein structure of STK11 (Figure 2A-D). The STK11 c.889A>G (p.Arg297Gly) variant resulted in the substitution of arginine for glycine. Arginine has a large molecular weight, electrically charged side chain, hydrophilic properties, and an alkaline R group. It plays an important role in the structure and function of proteins. Glycine has a small molecular weight, nonpolar side chain, hydrophobic properties, and a neutral R group containing only one hydrogen atom, as well as a large degree of freedom. The STK11 c.733C>T (p.Leu245Phe) variant substituted leucine with phenylalanine. Both amino acids have nonpolar side chains and are hydrophobic, but leucine is a side branched-chain amino acid, while phenylalanine has an aromatic ring in the side chain. Therefore, the variation between these two amino acids may lead to large structural changes. The two amino acids were evolutionarily conserved by UGENE (Figure 2E), while Mutation Taster, AlphaMissese, PolyPhen-2, and SIFT predicted the two missense variations (p.Arg297Gly and p.Leu245Phe) to be deleterious (Figure 2F).
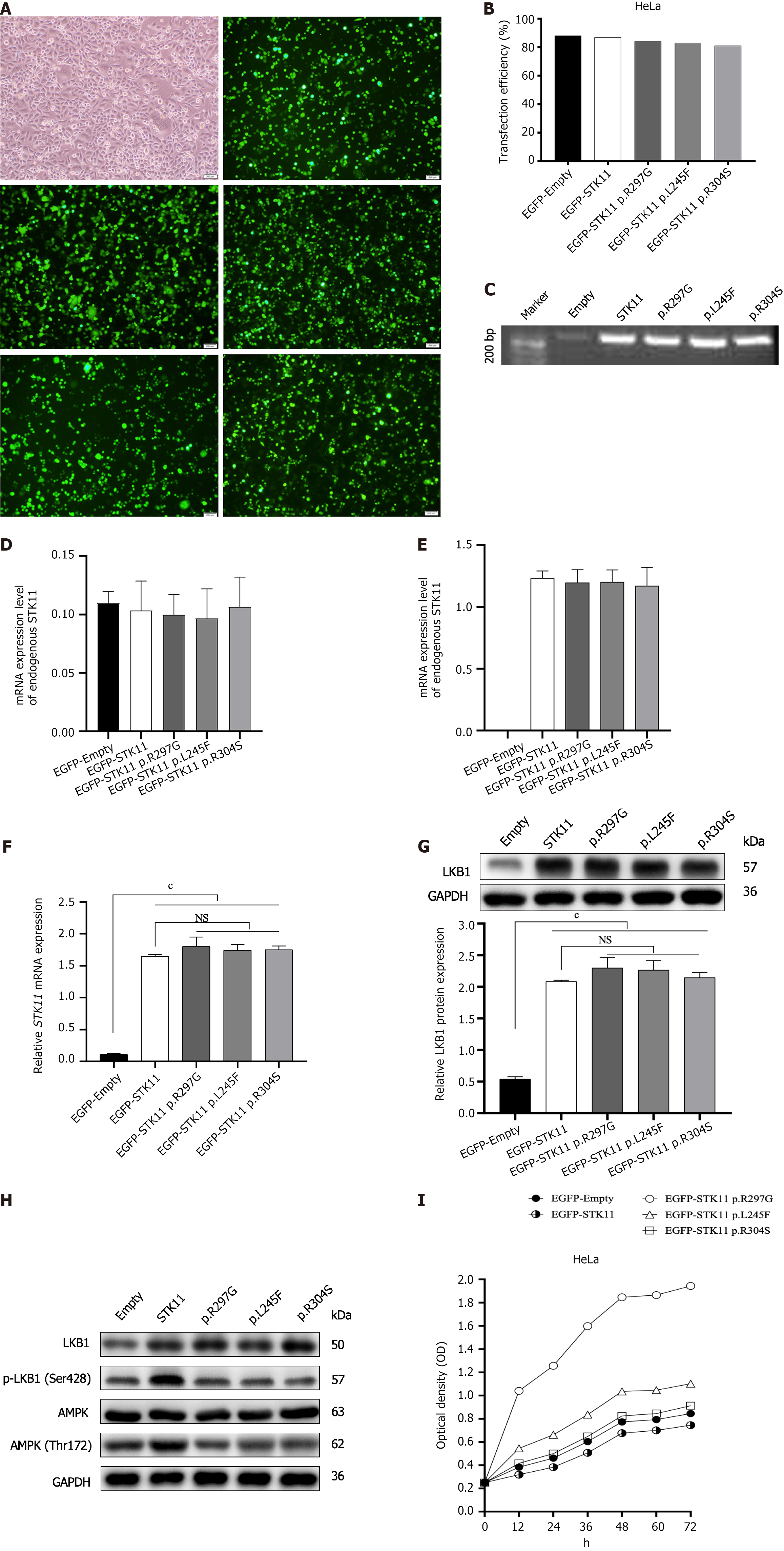
To verify whether these two missense variants affect STK11 gene transcription and translation, STK11 plasmids were established [an empty vector, wild-type vector, STK11 c.889A>G (p.Arg297Gly) variant vector, and c.733C>T (p.Leu245Phe) variant vector]. One missense variant c.910C>T (p.Arg304Trp) was interpreted as pathogenic in ClinVar; thus, a plasmid with this mutation was used as the positive control. PCR and WB were used to detect endogenous STK11 expression levels in HeLa and HEK-293T cell lines. STK11 expression levels were nearly undetectable in HeLa cells vs HEK-293T cell lines. Thus, HeLa cells were used for further studies. All plasmids were verified by gel electrophoresis and PCR product sequencing; since all carried GFP constructs, successful transfection was detected by fluorescence microscopy (Figure 3A-E). STK11 expression was examined by WB and RT-qPCR analysis 48 h after transfection. The protein and mRNA expression levels of STK11 in HeLa cells transfected with the STK11 variation and wild-type vectors were significantly increased in comparison with those in cells transfected with the empty vector. However, there was no significant difference in STK11 expression in cells transfected with the wildtype vector and the STK11 variant vectors (Figure 3F and G). These results suggest that the two missense STK11 variants have negligible effects on STK11 transcription and translation.
We next tested LKB1, p-LKB1, AMPK, and p-AMPK protein levels to detect the effects of these two missense VUS on LKB1 protein function and whether they play a role through the downstream AMPK signaling pathway. p-LKB1 and p-AMPK levels were significantly lower in all cells transfected with variant plasmids than in wild-type cells (Figure 3H). We performed CCK-8 assays to further determine whether these two STK11 VUS could effectively inhibit cell growth and proliferation. Compared with the wild-type group, the growth of cells transfected with the STK11 variant vectors was significantly accelerated (Figure 3I). These results indicate that both missense VUS disrupt the endogenous protein kinase activity of STK11 and inhibit the activation of the AMPK pathway, disrupting the inhibitory function of the STK11 gene on cell growth, leading to cell proliferation that may contribute to the development of polyps and tumors.
In summary, based on the above results, functional evidence is provided that these two missense STK11 variants are likely pathogenic according to the ACMG’s guidelines.
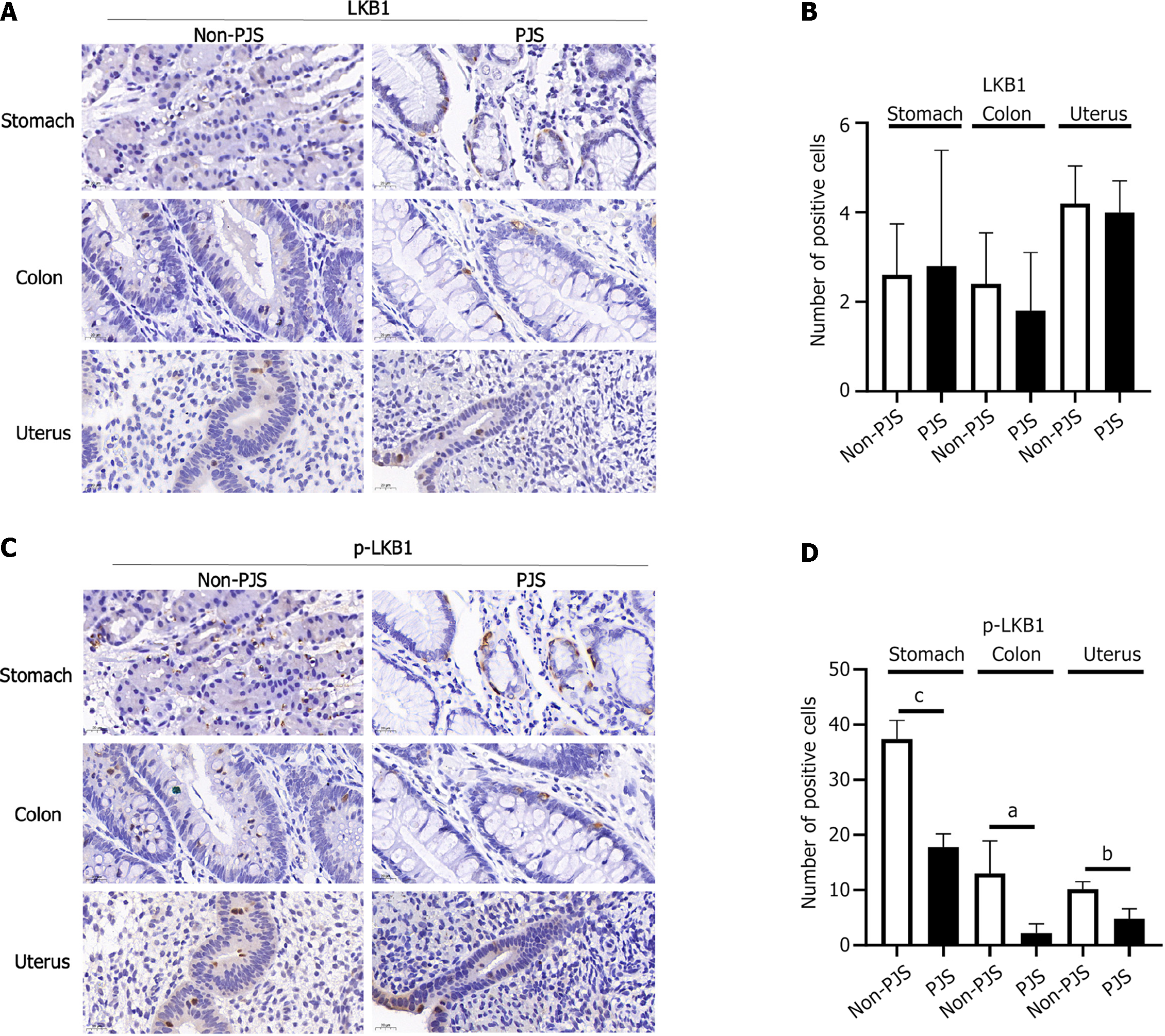
Polyp tissue from non-PJS patients with the same social factors as the PJS patients was used as the control, and the LKB1, p-LKB1, AMPK, and p-AMPK protein expression levels were compared between the two groups. The expression levels of p-STK11 and p-AMPK were decreased in the stomach, colon, and uterus in patients with PJS, and the differences were statistically significant (P < 0.05). Therefore, it is inferred that the STK11 gene may cause polyps or PJS through the LKB1/AMPK pathway (Figures 4 and 5).
STK11 plays a crucial role in regulating cell damage repair, energy metabolism, and tumor immune responses[16]. The LKB1 protein comprises three major domains: The N-terminal non-catalytic domain (encoded by amino acids 1-49), the catalytic kinase domain (encoded by amino acids 49-309), and the C-terminal non-catalytic regulatory domain (encoded by amino acids 309-433)[1,17]. Variations in patients with PJS are predominantly located in the catalytic domain region, leading to kinase activity dysfunction and the disruption of STK11 function[18]. Germline STK11 mutation has been confirmed to be a significant cause of PJS[19].
We conducted a review of PJS patients with STK11 gene variations at our hospital. A total of 10 variations were found, primarily in exon 1, consistent with Li et al[20]. While all PJS patients exhibited STK11 gene variations, some were VUS[21,22], and numerous STK11 gene VUS exist in the ClinVar database, with the majority being missense variations[23]. Understanding the relationship between these variations and PJS is challenging, complicating clinical decision-making, reproduction decisions, and genetic counseling for the patient[24,25], which may lead to increased psychological stress[26]. Therefore, it is crucial to determine the pathogenicity of these VUS.
Most patients with PJS have STK11 germline mutations, and their offspring face an elevated risk of inheriting the same mutation and experiencing PJS due to the autosomal dominant inheritance pattern[27]. PJS patients affected by the disease naturally do not want their offspring to experience the same effects[28]. Therefore, some patients opt for preimplantation genetic testing (PGT)[29] to prevent the transmission of disease-causing mutations to future generations[5,30]. Some PJS patients choose to pursue a natural pregnancy[31]. In cases where the female patient or the male patient’s partner becomes pregnant, prenatal diagnosis is performed to predict whether the fetus carries STK11 or other genetic mutations[32,33]. Parents can terminate the pregnancy if the fetus is a carrier of the STK11 mutation[34]; therefore, this method also allows healthy children to be born. In our study, one PJS patient chose in vitro fertilization-embryo transfer. Unfortunately, prenatal diagnosis at 18 wk of gestation revealed that the fetus carried the STK11 gene mutation; thus, the pregnancy was terminated. Seven patients (78%) chose PGT to block the transmission of the mutated genes to their offspring. It is worth mentioning that after the two missense VUS were indicated to likely be pathogenic through experiments, both patients chose PGT to assist their pregnancies, and one patient then gave birth to a healthy girl. The other suffered a spontaneous miscarriage in the fifth week of pregnancy. Therefore, identifying the pathogenicity of genetic variations may affect the patient’s reproduction decisions.
The two missense variants found in this study are VUS, complicating clinical decision-making. Clinical geneticists and reproductive specialists cannot provide help and guidance to patients with VUS since it is unclear whether the variant is related to the disease. Here, the two missense VUS were predicted by software to be harmful and affect protein structure; however, because the variants were VUS, the patients faced great difficulties in clinical fertility selection. Now that experiments have supported the idea that these variants can be interpreted as being likely pathogenic, reproductive, and genetic physicians will be able to provide better fertility counseling to patients. The findings can also be used for evidence-based genetic and reproductive counseling for PJS families. Finally, we provided valuable insights into the molecular mechanisms of PJS pathogenesis. Two patients with PJS had a child by PGT, and no pathogenic gene mutation was found in the fetus by prenatal diagnosis, suggesting that the combination of molecular and prenatal diagnosis is an effective way to prevent the recurrence or transmission of inherited diseases in these families. However, determining the genetic cause before conception is crucial in this process.
Several limitations should be noted in our study. First, because of the rarity of PJS, the number of patients included is relatively small, which may reduce the statistical analysis power. Second, the follow-up period was short; the longest follow-up was 8 years. Because the patients with PJS were generally young when they visited the doctor, no malignant tumors were found till the date of the final follow-up; thus, the correlation between PJS and cancer development could not be discussed. Third, because our hospital mainly caters to patients who need assisted reproduction and genetic counseling, there may be some bias in the selection of patients included; that is, patients visit the doctor for assisted reproduction, leading to a high proportion of patients choosing PGT.
We identified two missense variants in the STK11 gene that were experimentally evaluated for their functional significance. Our results identify the two missense variations as being likely pathogenic, providing new guidance for genetic counseling, fertility counseling, and prenatal diagnosis in patients with STK11 gene variants.
Peutz-Jeghers syndrome (PJS) is a rare hereditary tumor disease with autosomal dominant inheritance that primarily results from mutations in the STK11 gene. While certain missense variations of the STK11 gene have been identified and classified as pathogenic or likely pathogenic, numerous missense variations still remain as variants of uncertain significance (VUS). This ambiguity makes it challenging to establish the association between these variations and PJS, which poses significant challenges for clinical treatment management and fertility selection.
Among the 9 PJS families evaluated at our hospital, 2 patients with PJS were identified as having missense variations through whole-exome sequencing and were classified as VUS. Understanding how the function of STK11 is disrupted may contribute to a better understanding of the pathogenesis of PJS and the molecular mechanisms involved in the carcinogenesis process. Clarifying the pathogenicity of variations can offer patients the most appropriate reproductive guidance.
To clarify the pathogenicity of these two missense variations and to better provide patients with health management and fertility guidance.
This study used whole-exome gene sequencing and Sanger sequencing methods to identify gene variants, combining bioinformatics analysis, quantitative polymerase chain reaction, immunoblotting, immunohistochemistry, and a variety of in vitro and in vivo functional assays to investigate pathogenicity.
Bioinformatics software analysis indicated that these two missense variants are deleterious. The phosphorylation levels of adenosine monophosphate-activated protein kinase (AMPK) and liver kinase B1 in the variant group were significantly lower than those in the wild-type group. Both missense STK11 variants promoted the proliferation of HeLa cells. The expression of phosphorylated-AMPK was significantly lower in PJS patients with a missense variant in gastric, colon, and uterine polyps compared to non-PJS patients. These missense variants inactivate the STK11 gene, disrupting the regulation of downstream AMPK proteins, thereby mediating the occurrence of PJS. As a result, they are interpreted as likely pathogenic, providing valuable information for patients in reproductive decision-making.
These findings provide a basis for further research on and reanalysis of clinical decision making related to rare disease missense VUS.
Further investigation into the role of STK11 gene in mediating PJS is required. Identifying the pathogenicity of more missense VUS is crucial to offer more accurate medical services for patients.
We thank the patients, their families, and the referring clinicians for their valuable contributions to this study.
Provenance and peer review: Unsolicited article; Externally peer reviewed.
Peer-review model: Single blind
Specialty type: Genetics & heredity
Country/Territory of origin: China
Peer-review report’s scientific quality classification
Grade A (Excellent): 0
Grade B (Very good): B
Grade C (Good): 0
Grade D (Fair): 0
Grade E (Poor): 0
P-Reviewer: Gragnaniello V, Italy S-Editor: Chen YL L-Editor: A P-Editor: Zhang XD
| 1. | Forte G, Cariola F, De Marco K, Manghisi A, Guglielmi FA, Armentano R, Lippolis G, Giorgio P, Simone C, Disciglio V. A novel STK11 gene mutation (c.388dupG, p.Glu130Glyfs*33) in a Peutz-Jeghers family and evidence of higher gastric cancer susceptibility associated with alterations in STK11 region aa 107-170. Genes Dis. 2022;9:288-291. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 2] [Cited by in RCA: 4] [Article Influence: 1.3] [Reference Citation Analysis (0)] |
| 2. | Kirakosyan E, Lokhmatov M. High-Tech Diagnostic Methods and Enteroscopic Treatment of Children with Peutz-Jeghers Syndrome. Eur J Pediatr Surg. 2020;30:529-535. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 4] [Cited by in RCA: 4] [Article Influence: 0.8] [Reference Citation Analysis (0)] |
| 3. | Klimkowski S, Ibrahim M, Ibarra Rovira JJ, Elshikh M, Javadi S, Klekers AR, Abusaif AA, Moawad AW, Ali K, Elsayes KM. Peutz-Jeghers Syndrome and the Role of Imaging: Pathophysiology, Diagnosis, and Associated Cancers. Cancers (Basel). 2021;13. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 4] [Cited by in RCA: 30] [Article Influence: 7.5] [Reference Citation Analysis (0)] |
| 4. | Monahan KJ, Bradshaw N, Dolwani S, Desouza B, Dunlop MG, East JE, Ilyas M, Kaur A, Lalloo F, Latchford A, Rutter MD, Tomlinson I, Thomas HJW, Hill J; Hereditary CRC guidelines eDelphi consensus group. Guidelines for the management of hereditary colorectal cancer from the British Society of Gastroenterology (BSG)/Association of Coloproctology of Great Britain and Ireland (ACPGBI)/United Kingdom Cancer Genetics Group (UKCGG). Gut. 2020;69:411-444. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 242] [Cited by in RCA: 293] [Article Influence: 58.6] [Reference Citation Analysis (0)] |
| 5. | Kariv R, Dahary D, Yaron Y, Petel-Galil Y, Malcov M, Rosner G. Whole Genome Sequencing Applied in Familial Hamartomatous Polyposis Identifies Novel Structural Variations. Genes (Basel). 2022;13. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in RCA: 3] [Reference Citation Analysis (0)] |
| 6. | McGarrity TJ, Amos CI, Baker MJ. Peutz-Jeghers Syndrome. 2001 Feb 23. In: GeneReviews® [Internet]. Seattle (WA): University of Washington, Seattle; 1993–. [PubMed] |
| 7. | Wang Z, Liang L, Liu L, Wang Z, Wang Y, Yu Z, Wu B, Chen Y. Changes in the Gut Microbiome Associated with Intussusception in Patients with Peutz-Jeghers Syndrome. Microbiol Spectr. 2023;11:e0281922. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 8] [Cited by in RCA: 5] [Article Influence: 2.5] [Reference Citation Analysis (0)] |
| 8. | Wagner A, Aretz S, Auranen A, Bruno MJ, Cavestro GM, Crosbie EJ, Goverde A, Jelsig AM, Latchford A, Leerdam MEV, Lepisto A, Puzzono M, Winship I, Zuber V, Möslein G. The Management of Peutz-Jeghers Syndrome: European Hereditary Tumour Group (EHTG) Guideline. J Clin Med. 2021;10. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 21] [Cited by in RCA: 77] [Article Influence: 19.3] [Reference Citation Analysis (0)] |
| 9. | Liu S, Ma Y, You W, Li J, Li JN, Qian JM. Hamartomatous polyposis syndrome associated malignancies: Risk, pathogenesis and endoscopic surveillance. J Dig Dis. 2021;22:444-451. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 2] [Cited by in RCA: 4] [Article Influence: 1.0] [Reference Citation Analysis (0)] |
| 10. | Tian LL, Guo JZ, Yin YQ, Dang XH, Huo LJ. Analysis of a pedigree of Peutz-Jeghers syndrome and RET proto-oncogene mutation: one case report and literature review. Transl Cancer Res. 2020;9:3007-3011. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Reference Citation Analysis (0)] |
| 11. | Altamish M, Dahiya R, Singh AK, Mishra A, Aljabali AAA, Satija S, Mehta M, Dureja H, Prasher P, Negi P, Kapoor DN, Goyal R, Tambuwala MM, Chellappan DK, Dua K, Gupta G. Role of the Serine/Threonine Kinase 11 (STK11) or Liver Kinase B1 (LKB1) Gene in Peutz-Jeghers Syndrome. Crit Rev Eukaryot Gene Expr. 2020;30:245-252. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 5] [Cited by in RCA: 14] [Article Influence: 3.5] [Reference Citation Analysis (0)] |
| 12. | Li L, Yao Y, Zhao J, Cao J, Ma H. Dehydroepiandrosterone protects against hepatic glycolipid metabolic disorder and insulin resistance induced by high fat via activation of AMPK-PGC-1α-NRF-1 and IRS1-AKT-GLUT2 signaling pathways. Int J Obes (Lond). 2020;44:1075-1086. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 13] [Cited by in RCA: 25] [Article Influence: 5.0] [Reference Citation Analysis (0)] |
| 13. | Zyla RE, Hahn E, Hodgson A. Gene of the month: STK11. J Clin Pathol. 2021;74:681-685. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 2] [Cited by in RCA: 11] [Article Influence: 2.8] [Reference Citation Analysis (0)] |
| 14. | Beggs AD, Latchford AR, Vasen HF, Moslein G, Alonso A, Aretz S, Bertario L, Blanco I, Bülow S, Burn J, Capella G, Colas C, Friedl W, Møller P, Hes FJ, Järvinen H, Mecklin JP, Nagengast FM, Parc Y, Phillips RK, Hyer W, Ponz de Leon M, Renkonen-Sinisalo L, Sampson JR, Stormorken A, Tejpar S, Thomas HJ, Wijnen JT, Clark SK, Hodgson SV. Peutz-Jeghers syndrome: a systematic review and recommendations for management. Gut. 2010;59:975-986. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 516] [Cited by in RCA: 458] [Article Influence: 30.5] [Reference Citation Analysis (0)] |
| 15. | Richards S, Aziz N, Bale S, Bick D, Das S, Gastier-Foster J, Grody WW, Hegde M, Lyon E, Spector E, Voelkerding K, Rehm HL; ACMG Laboratory Quality Assurance Committee. Standards and guidelines for the interpretation of sequence variants: a joint consensus recommendation of the American College of Medical Genetics and Genomics and the Association for Molecular Pathology. Genet Med. 2015;17:405-424. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 19696] [Cited by in RCA: 22475] [Article Influence: 2247.5] [Reference Citation Analysis (0)] |
| 16. | Long LL, Ma SC, Guo ZQ, Zhang YP, Fan Z, Liu LJ, Liu L, Han DD, Leng MX, Wang J, Guo XJ, Tan JL, Cai XT, Lin Y, Pan X, Wu DH, Bai X, Dong ZY. PARP Inhibition Induces Synthetic Lethality and Adaptive Immunity in LKB1-Mutant Lung Cancer. Cancer Res. 2023;83:568-581. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 17] [Cited by in RCA: 11] [Article Influence: 5.5] [Reference Citation Analysis (0)] |
| 17. | Hanks SK, Quinn AM, Hunter T. The protein kinase family: conserved features and deduced phylogeny of the catalytic domains. Science. 1988;241:42-52. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 3605] [Cited by in RCA: 3822] [Article Influence: 103.3] [Reference Citation Analysis (0)] |
| 18. | Islam MJ, Khan AM, Parves MR, Hossain MN, Halim MA. Prediction of Deleterious Non-synonymous SNPs of Human STK11 Gene by Combining Algorithms, Molecular Docking, and Molecular Dynamics Simulation. Sci Rep. 2019;9:16426. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 39] [Cited by in RCA: 56] [Article Influence: 9.3] [Reference Citation Analysis (0)] |
| 19. | Huber-Keener KJ. Cancer genetics and breast cancer. Best Pract Res Clin Obstet Gynaecol. 2022;82:3-11. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1] [Cited by in RCA: 1] [Article Influence: 0.3] [Reference Citation Analysis (0)] |
| 20. | Li YX, LV XS, Xia JH. Mutation characteristic of STK11 gene in Chinese with Peutz-Jeghers syndrome. Zhonghua Yixue Yichuanxue Zazhi. 2001;18:4-7. [RCA] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 4] [Cited by in RCA: 6] [Article Influence: 0.4] [Reference Citation Analysis (0)] |
| 21. | Frebourg T. The challenge for the next generation of medical geneticists. Hum Mutat. 2014;35:909-911. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 37] [Cited by in RCA: 37] [Article Influence: 3.4] [Reference Citation Analysis (0)] |
| 22. | Brito S, Póvoas M, Dupont J, Lopes AI. Peutz-Jeghers syndrome: early clinical expression of a new STK11 gene variant. BMJ Case Rep. 2015;2015. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1] [Cited by in RCA: 1] [Article Influence: 0.1] [Reference Citation Analysis (0)] |
| 23. | Landrum MJ, Chitipiralla S, Brown GR, Chen C, Gu B, Hart J, Hoffman D, Jang W, Kaur K, Liu C, Lyoshin V, Maddipatla Z, Maiti R, Mitchell J, O'Leary N, Riley GR, Shi W, Zhou G, Schneider V, Maglott D, Holmes JB, Kattman BL. ClinVar: improvements to accessing data. Nucleic Acids Res. 2020;48:D835-D844. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 229] [Cited by in RCA: 625] [Article Influence: 125.0] [Reference Citation Analysis (0)] |
| 24. | Ataei-Kachouei M, Nadaf J, Akbari MT, Atri M, Majewski J, Riazalhosseini Y, Garshasbi M. Double Heterozygosity of BRCA2 and STK11 in Familial Breast Cancer Detected by Exome Sequencing. Iran J Public Health. 2015;44:1348-1352. [PubMed] |
| 25. | Gunawardena K, Sirisena ND, Anandagoda G, Neththikumara N, Dissanayake VHW. Germline variants of uncertain significance, their frequency, and clinico-pathological features in a cohort of Sri Lankan patients with hereditary breast cancer. BMC Res Notes. 2023;16:95. [RCA] [PubMed] [DOI] [Full Text] [Reference Citation Analysis (0)] |
| 26. | van Lier MG, Mathus-Vliegen EM, van Leerdam ME, Kuipers EJ, Looman CW, Wagner A, Vanheusden K. Quality of life and psychological distress in patients with Peutz-Jeghers syndrome. Clin Genet. 2010;78:219-226. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 8] [Cited by in RCA: 13] [Article Influence: 0.9] [Reference Citation Analysis (0)] |
| 27. | Jelsig AM, van Overeem Hansen T, Gede LB, Qvist N, Christensen LL, Lautrup CK, Frederiksen JH, Sunde L, Ousager LB, Ljungmann K, Bertelsen B, Karstensen JG. Survival, surveillance, and genetics in patients with Peutz-Jeghers syndrome: A nationwide study. Clin Genet. 2023;104:81-89. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 7] [Cited by in RCA: 7] [Article Influence: 3.5] [Reference Citation Analysis (0)] |
| 28. | Wang S, Huang G, Wang JX, Tian L, Zuo XL, Li YQ, Yu YB. Altered Gut Microbiota in Patients With Peutz-Jeghers Syndrome. Front Microbiol. 2022;13:881508. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 12] [Cited by in RCA: 10] [Article Influence: 3.3] [Reference Citation Analysis (0)] |
| 29. | Villy MC, Frydman N, Moutou C, Thierry G, Raad J, Colas C, Steffann J, Metras J, Chabbert-Buffet N, Parc Y, Richard S, Benusiglio PR. Preimplantation genetic testing in patients with genetic susceptibility to cancer. Fam Cancer. 2023;22:119-125. [RCA] [PubMed] [DOI] [Full Text] [Reference Citation Analysis (0)] |
| 30. | Xu X, Song R, Hu K, Li Y, Jin H, Chen B, Song W, Zhang Y, Xu J, Sun Y. Multidisciplinary management for Peutz-Jeghers syndrome and prevention of vertical transmission to offspring using preimplantation genetic testing. Orphanet J Rare Dis. 2022;17:64. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 2] [Cited by in RCA: 4] [Article Influence: 1.3] [Reference Citation Analysis (0)] |
| 31. | Wang Z, Liu S, Wang Y, Chen J, Wu B. Prenatal diagnosis in a hereditary Peutz-Jeghers syndrome family with high cancer risk. BMC Med Genet. 2018;19:66. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 3] [Cited by in RCA: 6] [Article Influence: 0.9] [Reference Citation Analysis (0)] |
| 32. | Leaver M, Wells D. Non-invasive preimplantation genetic testing (niPGT): the next revolution in reproductive genetics? Hum Reprod Update. 2020;26:16-42. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 59] [Cited by in RCA: 109] [Article Influence: 21.8] [Reference Citation Analysis (0)] |
| 33. | Sullivan-Pyke C, Dokras A. Preimplantation Genetic Screening and Preimplantation Genetic Diagnosis. Obstet Gynecol Clin North Am. 2018;45:113-125. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 34] [Cited by in RCA: 51] [Article Influence: 7.3] [Reference Citation Analysis (0)] |
| 34. | Byrjalsen A, Roos L, Diemer T, Karstensen JG, Løssl K, Jelsig AM. Preimplantation genetic testing in two Danish couples affected by Peutz-Jeghers syndrome. Scand J Gastroenterol. 2023;58:314-318. [RCA] [PubMed] [DOI] [Full Text] [Cited by in RCA: 3] [Reference Citation Analysis (0)] |