Published online Apr 15, 2024. doi: 10.4251/wjgo.v16.i4.1465
Peer-review started: November 14, 2023
First decision: January 5, 2024
Revised: January 15, 2024
Accepted: February 18, 2024
Article in press: February 18, 2024
Published online: April 15, 2024
Processing time: 148 Days and 18.6 Hours
Colorectal cancer has a low 5-year survival rate and high mortality. Human β-defensin-1 (hBD-1) may play an integral function in the innate immune system, contributing to the recognition and destruction of cancer cells. Long non-coding RNAs (lncRNAs) are involved in the process of cell differentiation and growth.
To investigate the effect of hBD-1 on the mammalian target of rapamycin (mTOR) pathway and autophagy in human colon cancer SW620 cells.
CCK8 assay was utilized for the detection of cell proliferation and determination of the optimal drug concentration. Colony formation assay was employed to assess the effect of hBD-1 on SW620 cell proliferation. Bioinformatics was used to screen potentially biologically significant lncRNAs related to the mTOR pathway. Additionally, p-mTOR (Ser2448), Beclin1, and LC3II/I expression levels in SW620 cells were assessed through Western blot analysis.
hBD-1 inhibited the proliferative ability of SW620 cells, as evidenced by the reduction in the colony formation capacity of SW620 cells upon exposure to hBD-1. hBD-1 decreased the expression of p-mTOR (Ser2448) protein and increased the expression of Beclin1 and LC3II/I protein. Furthermore, bioinformatics analysis identified seven lncRNAs (2 upregulated and 5 downregulated) related to the mTOR pathway. The lncRNA TCONS_00014506 was ultimately selected. Following the inhibition of the lncRNA TCONS_00014506, exposure to hBD-1 inhibited p-mTOR (Ser2448) and promoted Beclin1 and LC3II/I protein expression.
hBD-1 inhibits the mTOR pathway and promotes autophagy by upregulating the expression of the lncRNA TCONS_00014506 in SW620 cells.
Core Tip: Colorectal cancer has a low 5-year survival rate and high mortality. Human β-defensin-1 (hBD-1) is likely to play an integral function in the innate immune system, contributing to the recognition and destruction of cancer cells. Hence, we explored the effect of hBD-1 on colon cancer SW620 cells, which could be good for the development of therapy for colorectal cancer.
- Citation: Zhao YX, Cui Y, Li XH, Yang WH, An SX, Cui JX, Zhang MY, Lu JK, Zhang X, Wang XM, Bao LL, Zhao PW. Human β-defensin-1 affects the mammalian target of rapamycin pathway and autophagy in colon cancer cells through long non-coding RNA TCONS_00014506. World J Gastrointest Oncol 2024; 16(4): 1465-1478
- URL: https://www.wjgnet.com/1948-5204/full/v16/i4/1465.htm
- DOI: https://dx.doi.org/10.4251/wjgo.v16.i4.1465
Colorectal cancer is a prevalent and highly aggressive malignancy of the gastrointestinal tract. Having the third highest incidence among all malignancies worldwide, colorectal cancer is considered one of the primary contributors to cancer-related fatalities globally[1]. According to the location of disease onset, colorectal cancer is divided into rectal cancer and colon cancer. Colon cancer often has no specific symptoms in the early stage, and changes in bowel habits, stool characteristics, abdominal pain, abdominal mass, intestinal obstruction, and systemic symptoms (anemia, weight loss, fatigue, low-grade fever, etc.) gradually appear in the late stage of disease progression[2,3]. A 5-year survival rate of only 11% was recorded in patients with colorectal cancer at TMN stage 4 or with distant metastases[4,5].
Human β-defensin-1 (hBD-1) is a small cationic host defense peptide produced by neutrophils and epithelial cells. It plays a vital role in innate immunity by effectively combating microbial pathogens or neutralizing bacterial toxins. Additionally, it contributes to adaptive immunity by acting as a chemical inducer and activator of immune cells and is a vital component of innate immune responses[6,7]. hBD-1 demonstrates varying expression patterns in diverse types of cancer. It is recognized as a tumor suppressor due to its ability to promote cancer cell apoptosis while simultaneously inhibiting the migration and invasion of cancer cells[4,8,9]. Hence, hBD-1 may play an integral function in the innate immune system, contributing to the recognition and destruction of cancer cells.
Autophagy is a highly conserved process of eukaryotic cell cycling[10]. In the context of cancer, autophagy is a general metabolic adaptation for cancer cell development, enabling the circulation of cellular components (CCs) under metabolic stress or during anticancer therapy. This allows cancer cells to survive in the challenging hypoxic and low-nutrient tumor microenvironments[11].
Despite the identification of numerous long non-coding RNAs (lncRNAs) through high-throughput RNA sequencing, a limited portion of them has undergone comprehensive functional characterization. The functional and biological relevance of most of these lncRNAs remains unelucidated. These already characterized lncRNAs are involved in the process of cell differentiation and growth. Moreover, these lncRNAs have been implicated in the onset and progression of numerous diseases, including cancer[12]. Research has confirmed that lncRNAs are vital in the onset and progression of colon cancer, and approximately 200 differentially expressed lncRNAs have been identified in colon tumors[13]. Hence, it is reasonable to speculate that lncRNAs may hold potential as biomarkers and potential targets for colon cancer therapy.
This study focused on investigating whether hBD-1 affects autophagy via lncRNAs in colon cancer SW620 cells and its impact on the expression of these lncRNAs within SW620 cells. This research will lay the groundwork for a more comprehensive examination of the role of hBD-1 in colon cancer. Furthermore, bioinformatics analysis of differentially expressed lncRNAs was performed to offer novel insights that could aid in the diagnosis and treatment of colon cancer.
The human colon cancer cell line SW620 was obtained from Wuhan Procell Life Science & Technology Co., Ltd, and hBD-1 was purchased from Sino Biological, Inc. (China). The total RNA extraction kit was purchased from TIANGEN Biotech (Beijing) Co., Ltd. The Trizol kit was acquired from Takara Bio Inc. (Japan), and chloroform was purchased from Beijing Dingguo Changsheng Biotechnology Co., Ltd. EL Transfection Reagent was purchased from TransGen Biotech Co., Ltd (Beijing). Additionally, si-lncRNA (Suzhou Gemma Biotechnology Co., Ltd.), BCA kit (Shanghai Epizyme Biotech Co., Ltd.), and Western blot reagents (Beyotime Biotech. Inc., Shanghai) were used. The antibody was purchased from Cell Signaling Technology Inc. (United States).
SW620 cells were grown in high-glucose DMEM with 10% fetal bovine serum and 1% penicillin/streptomycin at 37 °C in an incubator with 5% carbon dioxide.
SW620 cells (100 μL) in the logarithmic growth phase were inoculated into 96-well plates at a density of 7000 cells/well. After the cells had adhered to the plate, hBD-1 was added at final concentrations of 35, 40, 45, 50, 55, 60, 65, and 70 ng/mL, respectively. These cultures were placed at 37 °C with 5% CO2 in an incubator for 12, 24, and 48 h. Afterward, 10 μL of CCK-8 reagent was added into each well and incubated at 37 °C for 1 h. A microplate reader was utilized for assessing the optical density (OD) value at the wavelength of 450 nm. Cell inhibition rate (%) was calculated as [(OD value of control group - OD value of hBD-1 group)/(OD value of control group - OD value of blank group)] × 100%. The three assessed groups were as follows: Control group: SW620 cells + medium + CCK8 solution; hBD-1 group: SW620 cells + each concentration of hBD-1 + medium + CCK8 solution; and blank group: Medium + CCK8 solution.
SW620 cells in the logarithmic growth phase were diluted with a complete culture medium after trypsin digestion and seeded in 6-well plates at a density of 700 cells/well. The cells were categorized as a control group (without any treatment) and a hBD-1 group (exposure to 40 ng/mL hBD-1 after cell attachment). Following this process, 0.1% crystal violet was utilized to stain the cells. After 2 wk of cultivation, cell fixation was carried out, and a camera was utilized to capture the images. The cell count was subsequently determined using Image J software.
RIPA buffer was used to extract total cellular protein. After bicinchoninic acid quantification, the supernatant was mixed with 5 × loading buffer at a 4:1 ratio and placed in boiling water for 10 min. Then, 20 µg of total protein was separated by SDS-PAGE and transferred to an NC membrane. After blocking with 5% nonfat dry milk for 1 h, the primary antibody was added and incubated at 4 °C for 12-18 h. After washing with TBST thrice (10 min each), the secondary antibody was added and incubated at room temperature for 1 h, followed by three washes with TBST (10 min each). ECL chemiluminescence was evenly dropped onto the NC film. After several seconds of reaction, photographs were captured with a gel imaging system, and gray values were examined using Image J software.
The transcriptome sequencing project was completed on the Illumina sequencing platform. For this, the Illumina PE library (approximately 300 bp) was constructed and sequencing was conducted in a paired-end (PE sequencing) manner. After unloading the sequencing data, a thorough quality control process was implemented. This involved assessing the base mass distribution, base balance analysis, and repeat sequence level. Trimmomatic software was used to clean the raw data, removing the joint and low-quality reads. Subsequently, a new round of quality control was conducted on the cleaned reads to obtain comprehensive data. The clean data were then aligned with reference genomes to obtain detailed transcriptomic information. RPKM (Reads per Kilobase per Million Reads) value was used to measure gene expression, and the differentially expressed genes were identified using |logFC| > 1 and P < 0.05 as the criteria.
Differential expression analysis of lncRNAs was performed using DESeq to identify differentially expressed genes in terms of fold difference and significance level. The absolute value of logFC > 1 and P < 0.05 were used as the screening criteria. Volcano plot of differentially expressed lncRNAs was generated using the R package ggplot2.
A two-way cluster analysis of the merged set and the samples of all differentially expressed genes was performed. The process involved the use of the Pheatmap package in R, with the aim of clustering based on two criteria: The expression levels of the same lncRNA in various samples and the expression patterns of diverse lncRNAs in the same sample.
The lncRNA target genes were predicted by cis- and co-expression (differential co-expression network). The intersection of prediction results was visualized in the form of interaction networks.
Based on the KEGG database (https://www.kegg.jp/), enrichment analysis of the biological pathways, wherein the differentially expressed lncRNA target genes were located, was performed. The analysis predicted the signaling pathways in which these differentially expressed lncRNAs are involved. Through GO annotation (http://amigo.geneontology.org/amigo), gene function was annotated. The degree of enrichment was calculated according to several parameters such as rich factor, false discovery rate, and count. The top 20 most significant terms were selected after ranking for further analysis.
Target genes of lncRNAs and differentially expressed mRNAs associated with the mTOR pathway were analyzed. Specifically, the study focused on assessing lncRNA-mRNA co-expression to select the pertinent lncRNAs associated with the mTOR pathway.
The negative control (si-NC) and the small interfering RNA (si-lncRNA) of lncRNA TCONS_00014506 were transfected into SW620 cells. Subsequent experiments were carried out after an appropriate incubation period. Si-NC was utilized to ensure that any observed changes in the experiments can be attributed specifically to the presence or absence of si-lncRNA TCONS_00014506 and not to other factors. This process was executed via the EL Transfection Reagent. The si-lncRNA sequence is shown in Supplementary Table 1.
Total cellular RNA was extracted and converted into cDNA 24 h after transfection. LncRNA expression levels were detected by RT-qPCR. The sequence of primers used is shown in Supplementary Table 2.
SPSS 26.0 was utilized for statistical analyses, and the experiment was repeated more than three times. The data acquired from the experiments are presented as the mean ± SD. To assess group differences, the study employed various methods. Comparative assessment between two groups was executed via independent sample t-test, whereas comparisons among multiple groups were executed through one-way analysis of variance (ANOVA). Furthermore, the LSD t-test was utilized for pairwise comparisons. Two-sided P < 0.05 was deemed statistically significant.
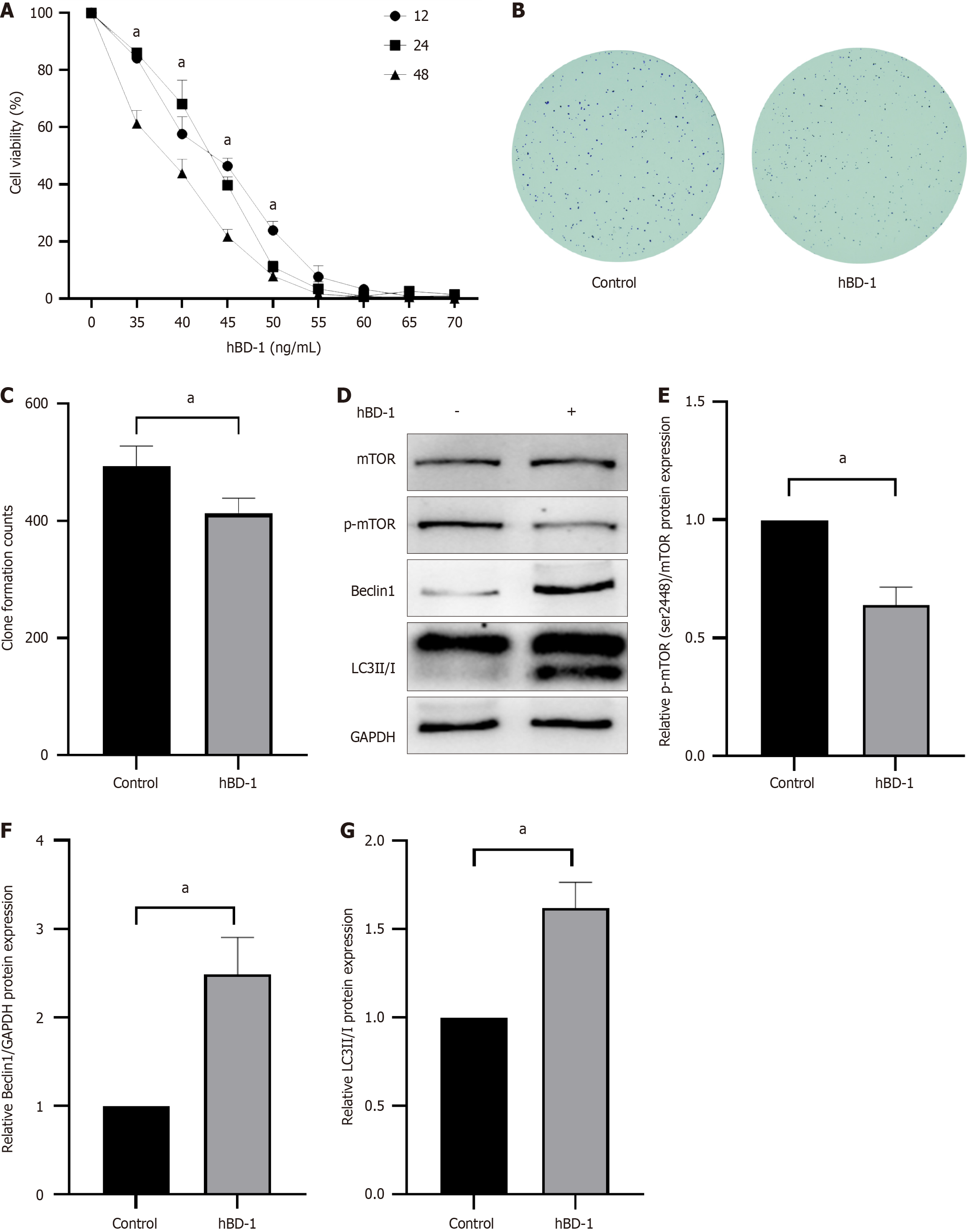
HBD-1 exerted significant effects on SW620 colon cancer cells, inhibiting their viability and clonogenic potential and promoting autophagy. The survival rate of SW620 cells decreased with increasing concentrations of hBD-1, as demonstrated via the CCK-8 assay (Figure 1A). The half maximal inhibitory concentration values of hBD-1 at 12, 24, and 48 h were measured at 43.32, 43.97, and 42.18 ng/mL, respectively. For subsequent experiments, a concentration of 40 ng/mL and a duration of 48 h were selected (P < 0.05).
The number of colonies of colon cancer SW620 cells decreased in the 40 ng/mL hBD-1 group (P < 0.05, Figure 1B and C).
To explore the effect of hBD-1 on p-mTOR, Beclin1, and LC3II/I in SW620 cells, Western blot analysis was performed. In comparison with the control cells, SW620 cells treated with 40 ng/mL hBD-1 for 48 h exhibited a decrease in the expression level of p-mTOR (P < 0.05; Figure 1D). In addition, the expression of autophagy-related proteins Beclin1 and LC3II/I was increased (P < 0.05; Figure 1D-G).
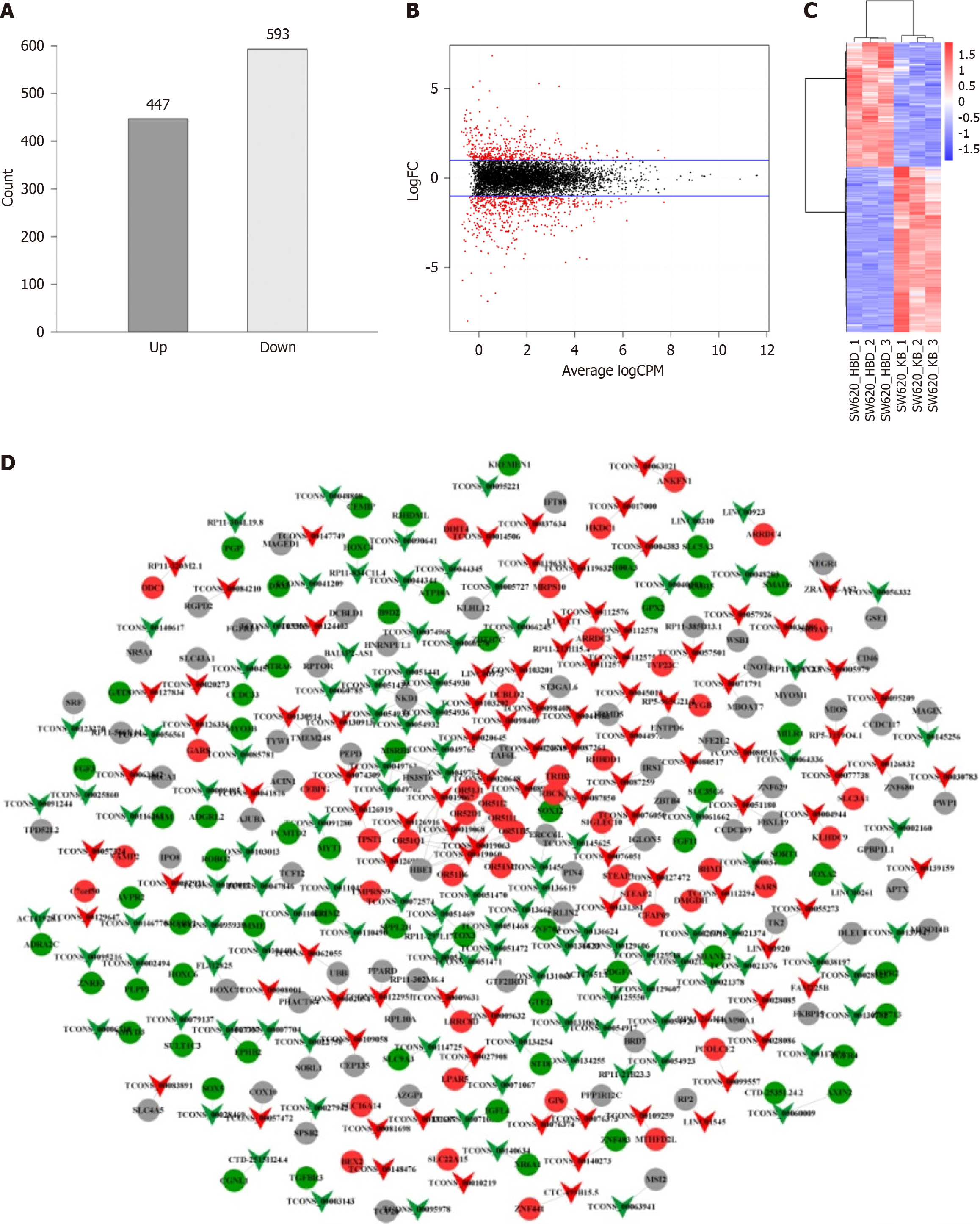
A total of 1040 differentially expressed lncRNAs (447 upregulated and 593 downregulated) were observed in the hBD-1 group (Figure 2A). Volcano plot shows the distribution of lncRNAs (Figure 2B). The overall hierarchical clustering plot of all differentially expressed lncRNAs clustered by RPKM values was generated (Figure 2C). The relationship between differentially expressed lncRNAs and their targeted mRNAs is shown in Figure 2D.
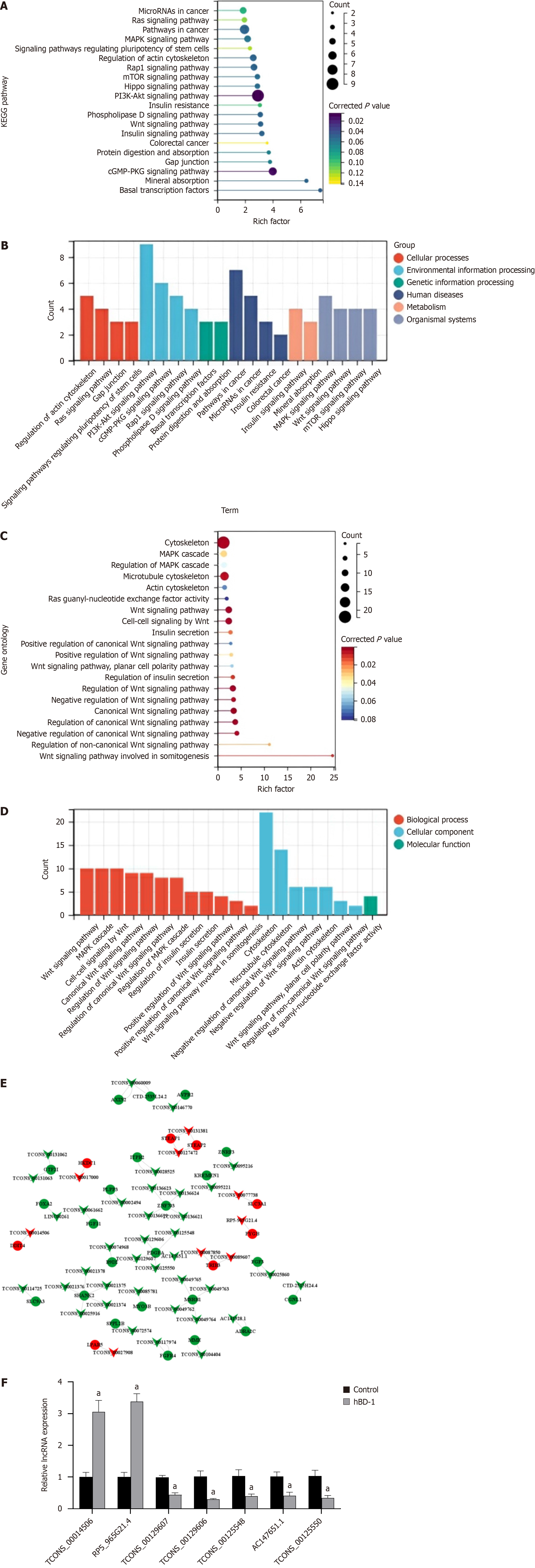
The top 20 KEGG pathways with the most significant enrichment were selected for presentation by rank order of P values (Figure 3A). Classification analysis of KEGG pathways was executed, and the resulting data are illustrated (Figure 3B). These pathways include cellular processes, genetic information processing, environmental information processing, metabolism, organismal systems, and human diseases.
The top 20 most significant GO terms were selected for plotting by rank order of P-values (Figure 3C). GO pathways were classified according to biological process (BP), CC, and molecular function (Figure 3D).
A total of 31 differentially expressed mRNAs were obtained by intersecting KEGG and GO enrichment terms, including 8 upregulated and 23 downregulated genes (Supplementary Table 3).
Differentially expressed lncRNAs and mRNAs were analyzed for lncRNA-mRNA co-expression (Figure 3E). A total of 46 lncRNAs (9 upregulated and 37 downregulated) were obtained (Supplementary Table 4). These lncRNAs hold promise as therapeutic targets for diagnosing and treating colon cancer.
As per the KEGG pathway analysis, using │logFC│ > 1 and P < 0.05 as screening criteria, three differentially expressed mRNAs and seven differentially expressed lncRNAs associated with the autophagy pathway were enriched. These results were further confirmed by the lncRNA-mRNA co-expression analysis (Supplementary Table 5). qPCR data suggested an upregulation in the levels of TCONS_00014506 and RP5-965G21.4. Furthermore, the acquired data indicated that TCONS_00129607, TCONS_00129606, TCONS_00125548, AC147651.1, and TCONS_00125550 were downregulated (Figure 3F).
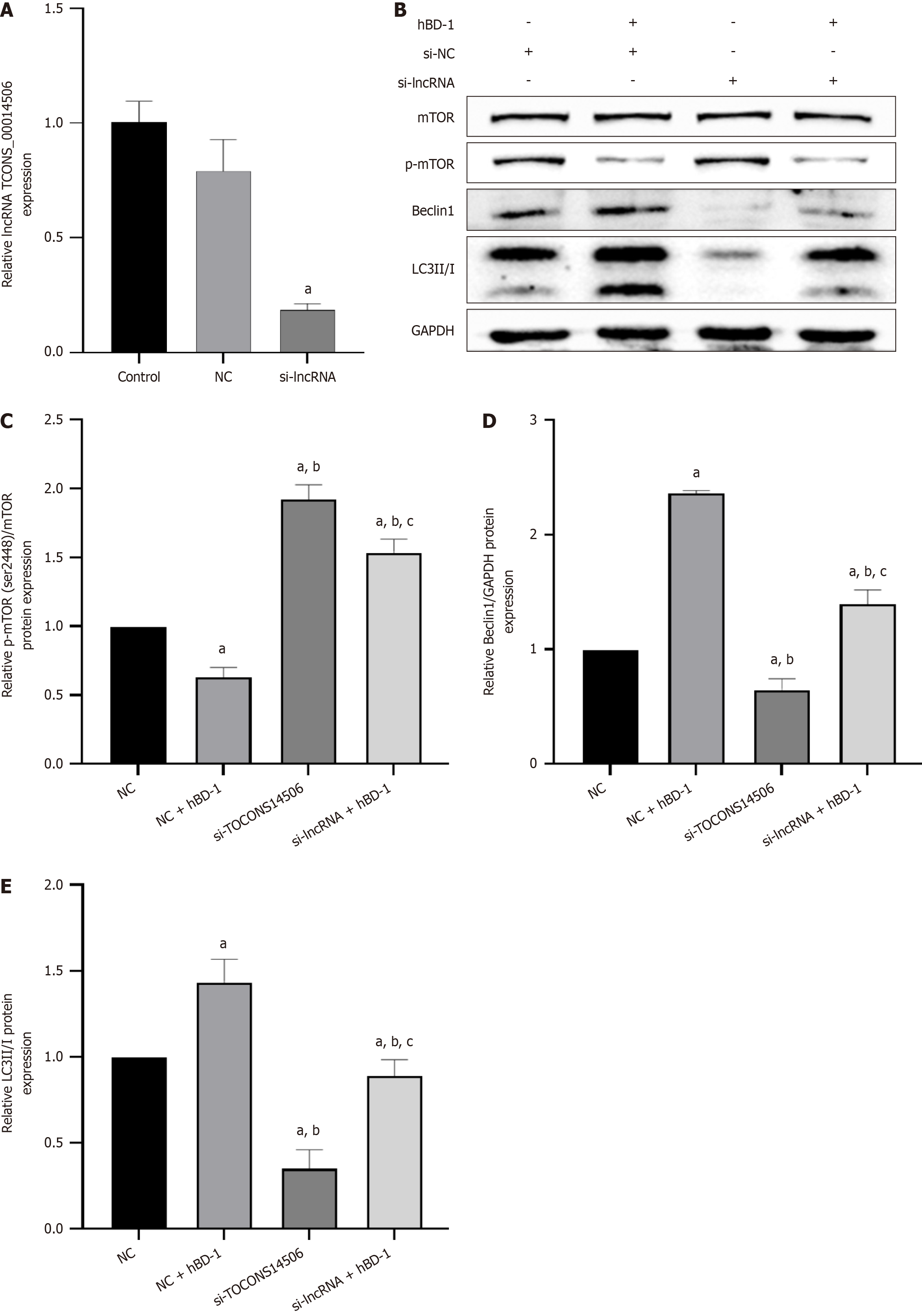
LncRNA-mRNA co-expression analysis revealed that the target gene of the lncRNA TCONS_
qPCR results showed a decrease in the expression of the lncRNA TCONS_00014506 in the si-lncRNA group in com
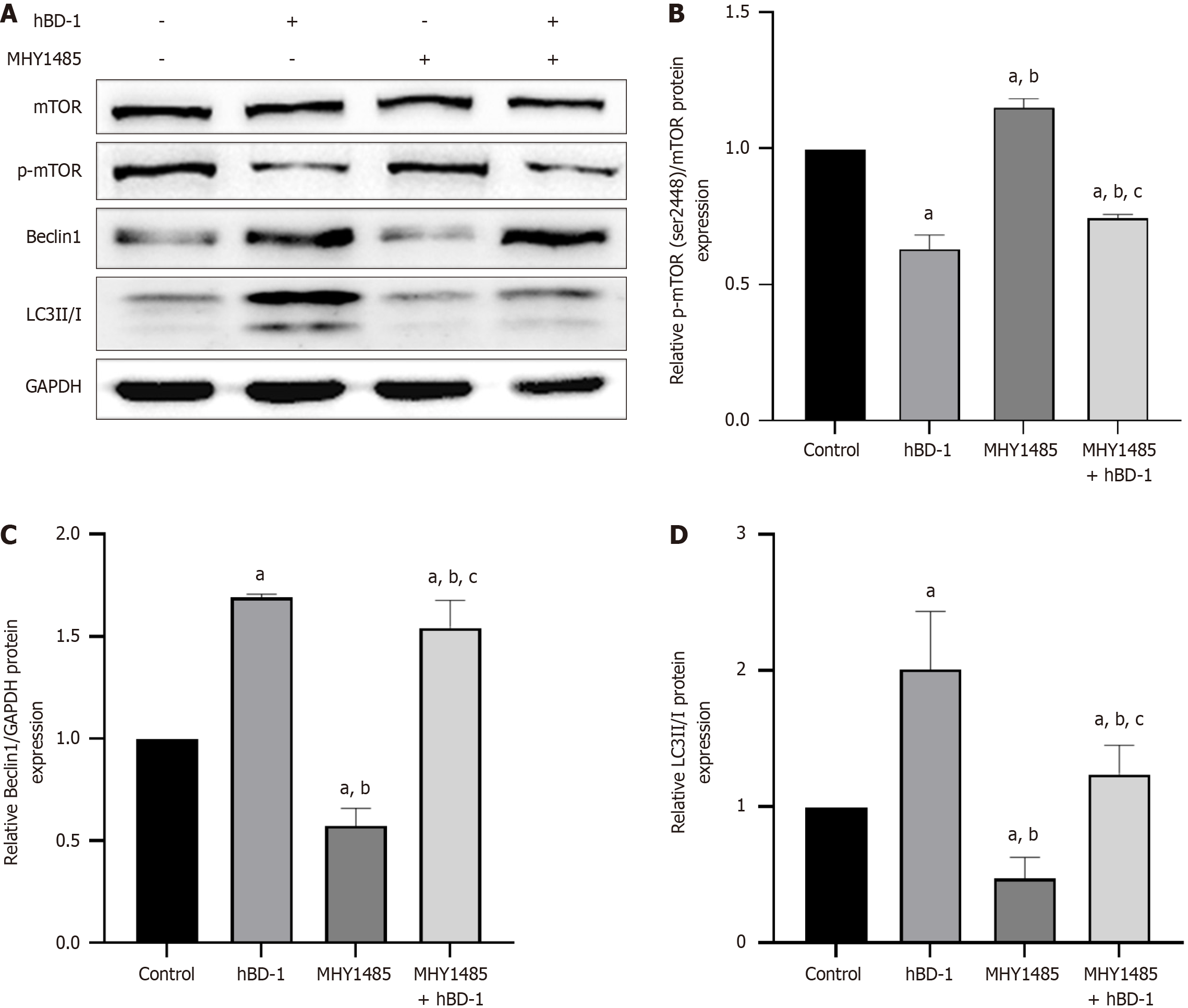
Western blot analysis revealed that MHY1485, an mTOR activator, increased the expression of p-mTOR while simultaneously reducing Beclin1 and LC3II/I expression in SW620 cells. Following exposure to hBD-1, the MHY1485-induced increase in p-mTOR was attenuated, along with an increase in Beclin1 and LC3 II/I expression (P < 0.05; Figure 5).
In China, the incidence of colon cancer has increased significantly in the past two decades. There is a tendency for colon cancer to be more common than rectal cancer[15]. The standard treatments for colorectal cancer include surgery, chemotherapy, radiotherapy, or a combination of several methods, but many patients relapse despite a series of treatments[16]. In this study, hBD-1 inhibited the viability and proliferation of SW620 cells.
hBD-1, as an antimicrobial peptide in humans, has garnered significant attention due to its involvement in cytotoxicity, cytolysis, and tumor immunity[7,8]. Notably, hBD-1 exhibits differential expression between normal tissue and tumor tissue, with varying levels along with tumor development. The significance of hBD-1 in tumorigenesis and progression is being increasingly acknowledged[17,18]. In this study, hBD-1 promoted the protein expression of Beclin1 and LC3II/I in SW620 cells, suggesting that hBD-1 promoted autophagy in SW620 cells. In cancer, autophagy is vital in the regulation of the tumor microenvironment. However, in later stages, autophagy can promote cancer cell proliferation and tumor growth, transitioning into a factor that supports cancer maintenance and contributes to treatment resistance[17-20]. Various chemotherapeutic and radiotherapeutic agents cause metabolic stress in cancer, which is accompanied by autophagy inhibition[21]. Regulation of autophagy could serve as an effective intervention strategy to prevent cancer development, limit tumor progression, and improve the efficacy of cancer treatment.
Recent research has unveiled the significant role of non-coding RNAs (ncRNAs) in various BPs. Dysregulation of ncRNAs has been linked to a range of diseases, including cancer. Although there is no evidence that lncRNAs encode peptides, an increasing number of studies have elucidated the crucial functions of lncRNAs in diverse BPs. These processes encompass proliferation, differentiation, neurogenesis, embryogenesis, stem cell pluripotency, tumorigenesis, and pathogenic infection[22,23]. It has been confirmed that lncRNAs affect autophagy in a variety of cancers[24-26]. In colon cancer, a multitude of lncRNAs have been demonstrated to be crucial players in cell proliferation, apoptosis, cell metastasis and invasion, cell cycle, drug resistance, and patient prognosis[27]. In the present research, it was observed that hBD-1 stimulated the expression of the lncRNA TCONS_00014506 and concurrently inhibited p-mTOR expression. The resultant decrease in p-mTOR was accompanied by an increase in the relative protein expression of Beclin1 and LC3 II. Additionally, it was observed that inhibition of the lncRNA TCONS_00014506 resulted in increased p-mTOR expression, along with decreased Beclin1 and LC3 II protein expression. These effects were reversed upon exposure to hBD-1. It is inferred that hBD-1 inhibits mTOR phosphorylation and promotes autophagy in SW620 cells via the lncRNA TCONS_00014506.
In summary, this study verified that hBD-1 may induce autophagy in colon cancer SW620 cells by inhibiting phosphorylation of mTOR through the lncRNA TCONS00014506 at the cellular level. Additionally, associated lncRNAs hold potential as diagnostic and prognostic biomarkers, offering a novel theoretical foundation for diagnosing and treating colon cancer.
Colorectal cancer has a low 5-year survival rate and high mortality. Human β-defensin-1 (hBD-1) may play an integral function in the innate immune system, contributing to the recognition and destruction of cancer cells. Long non-coding RNAs (lncRNAs) are involved in the process of cell differentiation and growth.
There is an urgent need for innovative treatment approaches for colon cancer. Our investigation into hBD-1 has revealed its effect on autophagy in colon cancer cells. During this exploration, we hypothesized that lncRNAs might play a crucial role in influencing autophagy. Consequently, our study delves into understanding the specific functions of lncRNAs in the context of hBD-1-mediated autophagy in colon cancer cells.
To investigate the effect of hBD-1 on the mTOR pathway and autophagy in human colon cancer SW620 cells.
CCK8 assay was utilized for the detection of cell proliferation and the determination of the optimal drug concentration. Cell colony formation assay was employed to assess the effect of hBD-1 on SW620 cell proliferation. Bioinformatics was used to identify lncRNAs related to the mTOR pathway, aiming to identify those of potential biological significance. Additionally, p-mTOR (Ser2448), Beclin1, and LC3II/I expression levels in SW620 cells were assessed through Western blot analysis.
hBD-1 inhibited the proliferative ability of SW620 cells, as evidenced by the reduction in the colony formation capacity of SW620 cells upon exposure to hBD-1. hBD-1 decreased the expression of p-mTOR (Ser2448) protein and increased the expression of Beclin1 and LC3II/I protein. Furthermore, bioinformatics analysis identified seven lncRNAs (2 upregulated and 5 downregulated) related to the mTOR pathway. The lncRNA TCONS_00014506 was ultimately selected. Following the inhibition of the lncRNA TCONS_00014506, exposure to hBD-1 inhibited p-mTOR (Ser2448) and promoted Beclin1 and LC3II/I protein expression.
HBD-1 inhibits the mTOR pathway and promotes autophagy by upregulating the expression of the lncRNA TCONS_00014506 in SW620 cells.
We for the first time found that the lncRNA TCONS_00014506 can regulate mTOR during hBD-1-affected autophagy.
Provenance and peer review: Unsolicited article; Externally peer reviewed.
Peer-review model: Single blind
Specialty type: Oncology
Country/Territory of origin: China
Peer-review report’s scientific quality classification
Grade A (Excellent): 0
Grade B (Very good): 0
Grade C (Good): C
Grade D (Fair): 0
Grade E (Poor): 0
P-Reviewer: Bordonaro M, United States S-Editor: Lin C L-Editor: Wang TQ P-Editor: Zheng XM
| 1. | Sclafani F. PD-1 inhibition in metastatic dMMR/MSI-H colorectal cancer. Lancet Oncol. 2017;18:1141-1142. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 22] [Cited by in RCA: 34] [Article Influence: 4.3] [Reference Citation Analysis (0)] |
| 2. | Benson AB, Venook AP, Al-Hawary MM, Cederquist L, Chen YJ, Ciombor KK, Cohen S, Cooper HS, Deming D, Engstrom PF, Garrido-Laguna I, Grem JL, Grothey A, Hochster HS, Hoffe S, Hunt S, Kamel A, Kirilcuk N, Krishnamurthi S, Messersmith WA, Meyerhardt J, Miller ED, Mulcahy MF, Murphy JD, Nurkin S, Saltz L, Sharma S, Shibata D, Skibber JM, Sofocleous CT, Stoffel EM, Stotsky-Himelfarb E, Willett CG, Wuthrick E, Gregory KM, Freedman-Cass DA. NCCN Guidelines Insights: Colon Cancer, Version 2.2018. J Natl Compr Canc Netw. 2018;16:359-369. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 635] [Cited by in RCA: 685] [Article Influence: 97.9] [Reference Citation Analysis (1)] |
| 3. | Zhu K, Wu Y, He P, Fan Y, Zhong X, Zheng H, Luo T. PI3K/AKT/mTOR-Targeted Therapy for Breast Cancer. Cells. 2022;11. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 25] [Cited by in RCA: 88] [Article Influence: 29.3] [Reference Citation Analysis (0)] |
| 4. | Ahmad R, Singh JK, Wunnava A, Al-Obeed O, Abdulla M, Srivastava SK. Emerging trends in colorectal cancer: Dysregulated signaling pathways (Review). Int J Mol Med. 2021;47. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 29] [Cited by in RCA: 112] [Article Influence: 28.0] [Reference Citation Analysis (0)] |
| 5. | Sagaert X, Vanstapel A, Verbeek S. Tumor Heterogeneity in Colorectal Cancer: What Do We Know So Far? Pathobiology. 2018;85:72-84. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 56] [Cited by in RCA: 113] [Article Influence: 16.1] [Reference Citation Analysis (0)] |
| 6. | Fruitwala S, El-Naccache DW, Chang TL. Multifaceted immune functions of human defensins and underlying mechanisms. Semin Cell Dev Biol. 2019;88:163-172. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 70] [Cited by in RCA: 110] [Article Influence: 15.7] [Reference Citation Analysis (0)] |
| 7. | Pazgier M, Hoover DM, Yang D, Lu W, Lubkowski J. Human beta-defensins. Cell Mol Life Sci. 2006;63:1294-1313. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 393] [Cited by in RCA: 398] [Article Influence: 20.9] [Reference Citation Analysis (0)] |
| 8. | Bullard RS, Gibson W, Bose SK, Belgrave JK, Eaddy AC, Wright CJ, Hazen-Martin DJ, Lage JM, Keane TE, Ganz TA, Donald CD. Functional analysis of the host defense peptide Human Beta Defensin-1: new insight into its potential role in cancer. Mol Immunol. 2008;45:839-848. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 85] [Cited by in RCA: 76] [Article Influence: 4.5] [Reference Citation Analysis (0)] |
| 9. | Han Q, Wang R, Sun C, Jin X, Liu D, Zhao X, Wang L, Ji N, Li J, Zhou Y, Ye L, Liang X, Jiang L, Liao G, Dan H, Zeng X, Chen Q. Human beta-defensin-1 suppresses tumor migration and invasion and is an independent predictor for survival of oral squamous cell carcinoma patients. PLoS One. 2014;9:e91867. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 31] [Cited by in RCA: 42] [Article Influence: 3.8] [Reference Citation Analysis (0)] |
| 10. | Parzych KR, Klionsky DJ. An overview of autophagy: morphology, mechanism, and regulation. Antioxid Redox Signal. 2014;20:460-473. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1222] [Cited by in RCA: 1845] [Article Influence: 167.7] [Reference Citation Analysis (0)] |
| 11. | Klionsky DJ, Petroni G, Amaravadi RK, Baehrecke EH, Ballabio A, Boya P, Bravo-San Pedro JM, Cadwell K, Cecconi F, Choi AMK, Choi ME, Chu CT, Codogno P, Colombo MI, Cuervo AM, Deretic V, Dikic I, Elazar Z, Eskelinen EL, Fimia GM, Gewirtz DA, Green DR, Hansen M, Jäättelä M, Johansen T, Juhász G, Karantza V, Kraft C, Kroemer G, Ktistakis NT, Kumar S, Lopez-Otin C, Macleod KF, Madeo F, Martinez J, Meléndez A, Mizushima N, Münz C, Penninger JM, Perera RM, Piacentini M, Reggiori F, Rubinsztein DC, Ryan KM, Sadoshima J, Santambrogio L, Scorrano L, Simon HU, Simon AK, Simonsen A, Stolz A, Tavernarakis N, Tooze SA, Yoshimori T, Yuan J, Yue Z, Zhong Q, Galluzzi L, Pietrocola F. Autophagy in major human diseases. EMBO J. 2021;40:e108863. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 555] [Cited by in RCA: 981] [Article Influence: 245.3] [Reference Citation Analysis (0)] |
| 12. | Chi Y, Wang D, Wang J, Yu W, Yang J. Long Non-Coding RNA in the Pathogenesis of Cancers. Cells. 2019;8. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 581] [Cited by in RCA: 595] [Article Influence: 99.2] [Reference Citation Analysis (0)] |
| 13. | Chen S, Shen X. Long noncoding RNAs: functions and mechanisms in colon cancer. Mol Cancer. 2020;19:167. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 67] [Cited by in RCA: 207] [Article Influence: 41.4] [Reference Citation Analysis (0)] |
| 14. | Sofer A, Lei K, Johannessen CM, Ellisen LW. Regulation of mTOR and cell growth in response to energy stress by REDD1. Mol Cell Biol. 2005;25:5834-5845. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 334] [Cited by in RCA: 363] [Article Influence: 18.2] [Reference Citation Analysis (0)] |
| 15. | He Y, Sun MM, Zhang GG, Yang J, Chen KS, Xu WW, Li B. Targeting PI3K/Akt signal transduction for cancer therapy. Signal Transduct Target Ther. 2021;6:425. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 161] [Cited by in RCA: 871] [Article Influence: 217.8] [Reference Citation Analysis (0)] |
| 16. | Johdi NA, Sukor NF. Colorectal Cancer Immunotherapy: Options and Strategies. Front Immunol. 2020;11:1624. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 235] [Cited by in RCA: 297] [Article Influence: 59.4] [Reference Citation Analysis (0)] |
| 17. | Xu D, Lu W. Defensins: A Double-Edged Sword in Host Immunity. Front Immunol. 2020;11:764. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 110] [Cited by in RCA: 141] [Article Influence: 28.2] [Reference Citation Analysis (0)] |
| 18. | Bonamy C, Sechet E, Amiot A, Alam A, Mourez M, Fraisse L, Sansonetti PJ, Sperandio B. Expression of the human antimicrobial peptide β-defensin-1 is repressed by the EGFR-ERK-MYC axis in colonic epithelial cells. Sci Rep. 2018;8:18043. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 11] [Cited by in RCA: 27] [Article Influence: 3.9] [Reference Citation Analysis (0)] |
| 19. | Yun CW, Lee SH. The Roles of Autophagy in Cancer. Int J Mol Sci. 2018;19. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 377] [Cited by in RCA: 684] [Article Influence: 97.7] [Reference Citation Analysis (0)] |
| 20. | Li X, He S, Ma B. Autophagy and autophagy-related proteins in cancer. Mol Cancer. 2020;19:12. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 304] [Cited by in RCA: 1089] [Article Influence: 217.8] [Reference Citation Analysis (0)] |
| 21. | Levine B, Kroemer G. Biological Functions of Autophagy Genes: A Disease Perspective. Cell. 2019;176:11-42. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1024] [Cited by in RCA: 2063] [Article Influence: 343.8] [Reference Citation Analysis (0)] |
| 22. | Chen L, He M, Zhang M, Sun Q, Zeng S, Zhao H, Yang H, Liu M, Ren S, Meng X, Xu H. The Role of non-coding RNAs in colorectal cancer, with a focus on its autophagy. Pharmacol Ther. 2021;226:107868. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 37] [Cited by in RCA: 78] [Article Influence: 19.5] [Reference Citation Analysis (0)] |
| 23. | Tan YT, Lin JF, Li T, Li JJ, Xu RH, Ju HQ. LncRNA-mediated posttranslational modifications and reprogramming of energy metabolism in cancer. Cancer Commun (Lond). 2021;41:109-120. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 67] [Cited by in RCA: 455] [Article Influence: 91.0] [Reference Citation Analysis (0)] |
| 24. | Xu J, Deng Y, Wang Y, Sun X, Chen S, Fu G. SPAG5-AS1 inhibited autophagy and aggravated apoptosis of podocytes via SPAG5/AKT/mTOR pathway. Cell Prolif. 2020;53:e12738. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 28] [Cited by in RCA: 80] [Article Influence: 16.0] [Reference Citation Analysis (0)] |
| 25. | Luo Y, Zheng S, Wu Q, Wu J, Zhou R, Wang C, Wu Z, Rong X, Huang N, Sun L, Bin J, Liao Y, Shi M, Liao W. Long noncoding RNA (lncRNA) EIF3J-DT induces chemoresistance of gastric cancer via autophagy activation. Autophagy. 2021;17:4083-4101. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 23] [Cited by in RCA: 156] [Article Influence: 39.0] [Reference Citation Analysis (0)] |
| 26. | Jing X, Han J, Zhang J, Chen Y, Yuan J, Wang J, Neo S, Li S, Yu X, Wu J. Long non-coding RNA MEG3 promotes cisplatin-induced nephrotoxicity through regulating AKT/TSC/mTOR-mediated autophagy. Int J Biol Sci. 2021;17:3968-3980. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 18] [Cited by in RCA: 15] [Article Influence: 3.8] [Reference Citation Analysis (0)] |
| 27. | Taniue K, Akimitsu N. The Functions and Unique Features of LncRNAs in Cancer Development and Tumorigenesis. Int J Mol Sci. 2021;22. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 86] [Cited by in RCA: 146] [Article Influence: 36.5] [Reference Citation Analysis (0)] |