Published online Apr 15, 2024. doi: 10.4251/wjgo.v16.i4.1296
Peer-review started: October 8, 2023
First decision: January 15, 2024
Revised: January 23, 2024
Accepted: February 25, 2024
Article in press: February 25, 2024
Published online: April 15, 2024
Processing time: 185 Days and 23.1 Hours
Preoperative knowledge of mutational status of gastrointestinal stromal tumors (GISTs) is essential to guide the individualized precision therapy.
To develop a combined model that integrates clinical and contrast-enhanced computed tomography (CE-CT) features to predict gastric GISTs with specific genetic mutations, namely KIT exon 11 mutations or KIT exon 11 codons 557-558 deletions.
A total of 231 GIST patients with definitive genetic phenotypes were divided into a training dataset and a validation dataset in a 7:3 ratio. The models were constructed using selected clinical features, conventional CT features, and radiomics features extracted from abdominal CE-CT images. Three models were developed: ModelCT sign, modelCT sign + rad, and model CTsign + rad + clinic. The diagnostic performance of these models was evaluated using receiver operating characteristic (ROC) curve analysis and the Delong test.
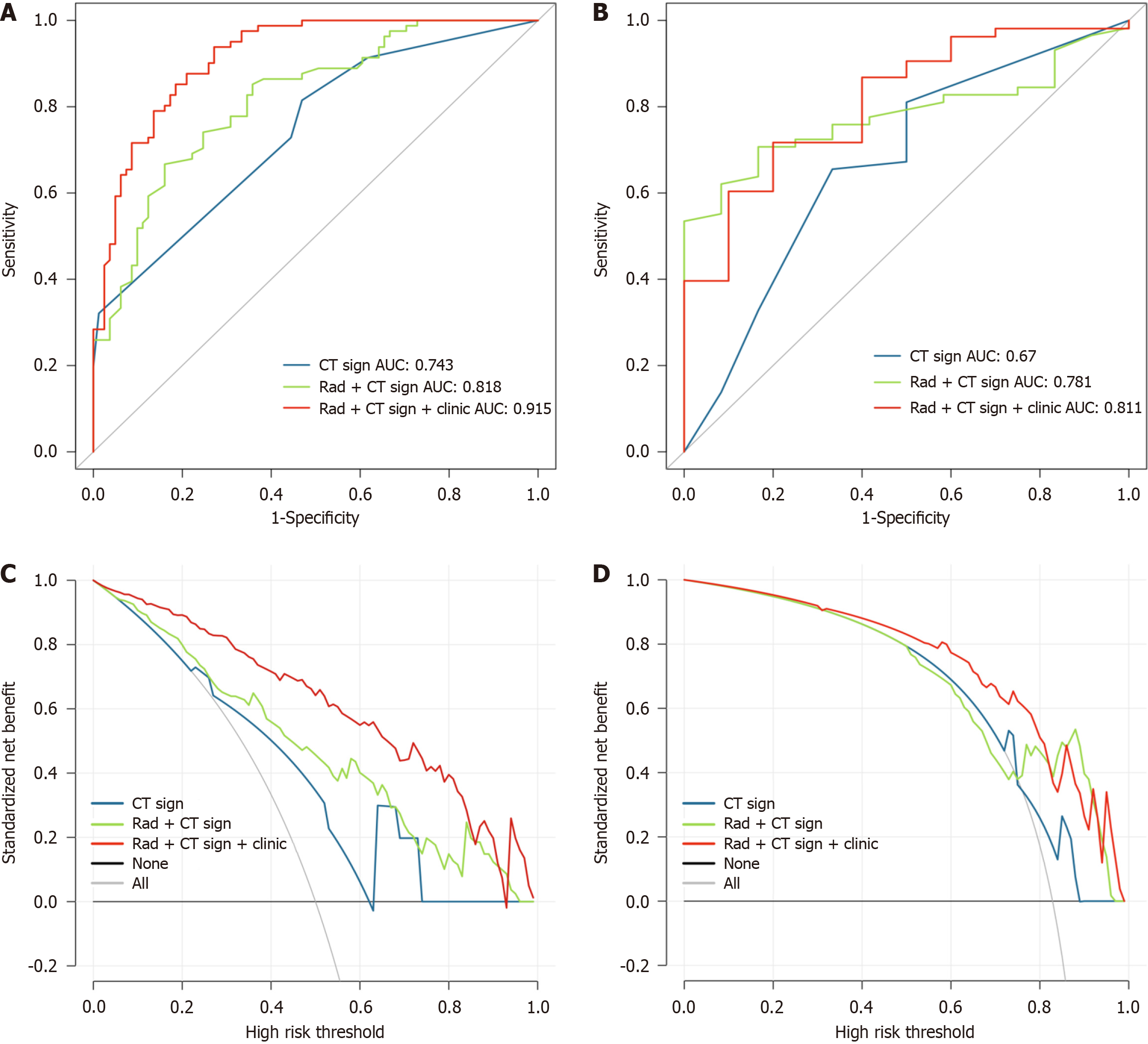
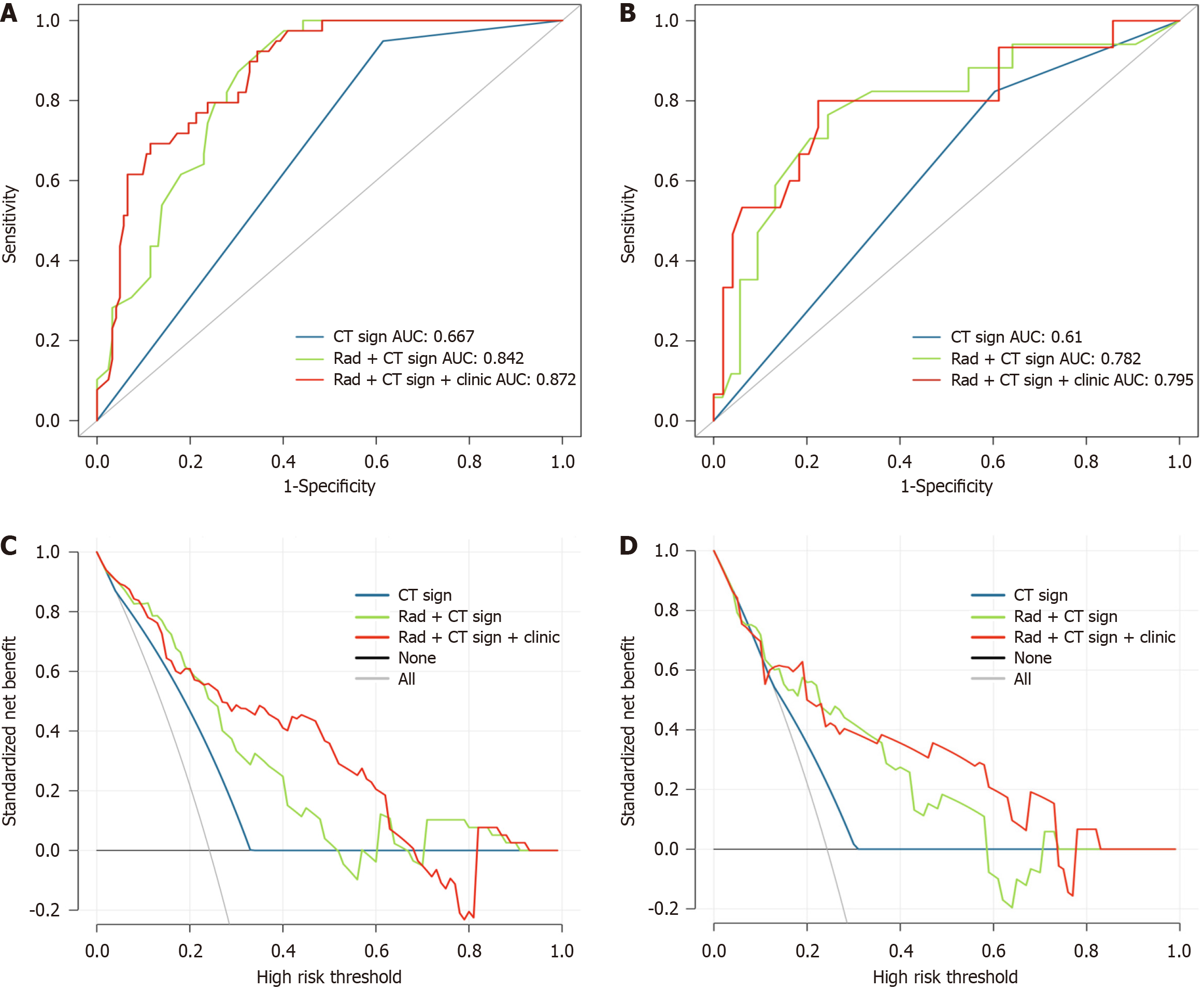
The ROC analyses revealed that in the training cohort, the area under the curve (AUC) values for modelCT sign, modelCT sign + rad, and modelCT sign + rad + clinic for predicting KIT exon 11 mutation were 0.743, 0.818, and 0.915, respectively. In the validation cohort, the AUC values for the same models were 0.670, 0.781, and 0.811, respectively. For predicting KIT exon 11 codons 557-558 deletions, the AUC values in the training cohort were 0.667, 0.842, and 0.720 for modelCT sign, modelCT sign + rad, and modelCT sign + rad + clinic, respectively. In the validation cohort, the AUC values for the same models were 0.610, 0.782, and 0.795, respectively. Based on the decision curve analysis, it was determined that the modelCT sign + rad + clinic had clinical significance and utility.
Our findings demonstrate that the combined modelCT sign + rad + clinic effectively distinguishes GISTs with KIT exon 11 mutation and KIT exon 11 codons 557-558 deletions. This combined model has the potential to be valuable in assessing the genotype of GISTs.
Core Tip: In this study, we developed and validated a radiomics model to predict the genotypes of gastric gastrointestinal stromal tumors (GISTs) using contrast-enhanced computed tomography images. Our findings demonstrated that the radiomics model exhibited a satisfactory performance in distinguishing gastric GISTs with KIT exon 11 mutations and GISTs with KIT exon 11 codons 557-558 deletions. Among the different models evaluated, the combined modelCT sign + rad + clinic demonstrated the highest predictive accuracy. This model holds promise as an effective and noninvasive approach to guide personalized treatment decisions prior to surgery.
- Citation: Yin XN, Wang ZH, Zou L, Yang CW, Shen CY, Liu BK, Yin Y, Liu XJ, Zhang B. Computed tomography radiogenomics: A potential tool for prediction of molecular subtypes in gastric stromal tumor. World J Gastrointest Oncol 2024; 16(4): 1296-1308
- URL: https://www.wjgnet.com/1948-5204/full/v16/i4/1296.htm
- DOI: https://dx.doi.org/10.4251/wjgo.v16.i4.1296
Gastrointestinal stromal tumor (GIST) is the most common mesenchymal tumor of the gastrointestinal tract, with an annual incidence ranging from 6 to 22 cases per million individuals[1,2]. The stomach is the primary site of GIST onset, accounting for 60%-65% of cases[3]. Prior to the year 2000, advanced GISTs had no effective medical therapy due to their poor response to chemotherapy and radiotherapy. However, the identification of activating KIT mutations in GISTs led to the rapid development of the first tyrosine kinase inhibitor (TKI), imatinib, which significantly improved clinical outcomes for GIST patients[4,5]. In addition to KIT mutations, mutations in other genes such as PDGFRA, NF-1, BRAF, KRAS, and PIK3CA, as well as SDH deficiency, have been discovered in GISTs[1,6]. The presence of specific driver oncogenic genes in GISTs has made it a paradigm for precision medicine treatment.
The majority of GISTs harbor KIT mutations (80%) or PDGFRA mutations (5%-10%)[7,8]. Testing for KIT and PDGFRA mutations is crucial for defining GIST pathological diagnosis, predicting tumor prognosis, and guiding TKI therapy. Studies have shown that patients with PDGFRA mutations have a better prognosis compared to those with KIT mutations[9]. Among GIST patients with KIT exon 11 mutations, those with deletion or insertion-deletion mutations have a worse prognosis than those with point or repeat mutations. In addition, the presence of multiple codon deletion mutations or deletions affecting codons 557-558 on KIT exon 11 has been linked to an aggressive biological phenotype and an unfavorable prognosis[10,11]. It has been clinically observed that GISTs exhibit different response rates to imatinib depending on their mutation status. GISTs with KIT exon 11 mutations have a higher response rate and recurrence-free survival to standard imatinib therapy compared to exon 9 tumors. Most PDGFRA mutations respond to imatinib, with the exception of D842V. Therefore, predicting the mutation status of tumors is crucial for managing GISTs. However, currently, tumor mutation status can only be obtained after surgical resection or conventional invasive biopsy, making preoperative genotyping of GISTs more challenging.
Contrast-enhanced computed tomography (CE-CT) is routinely used in clinical practice for the detection and evalu
In 2021, the primary outcomes of our research were published[21]. The study revealed associations between GISTs with KIT exon 11 mutations and CE-CT images. CT radiogenomics showed promising potential in predicting the KIT exon 11 mutation status of GISTs. This study focuses specifically on gastric GISTs and aims to develop a prediction model for genotypes using CE-CT images.
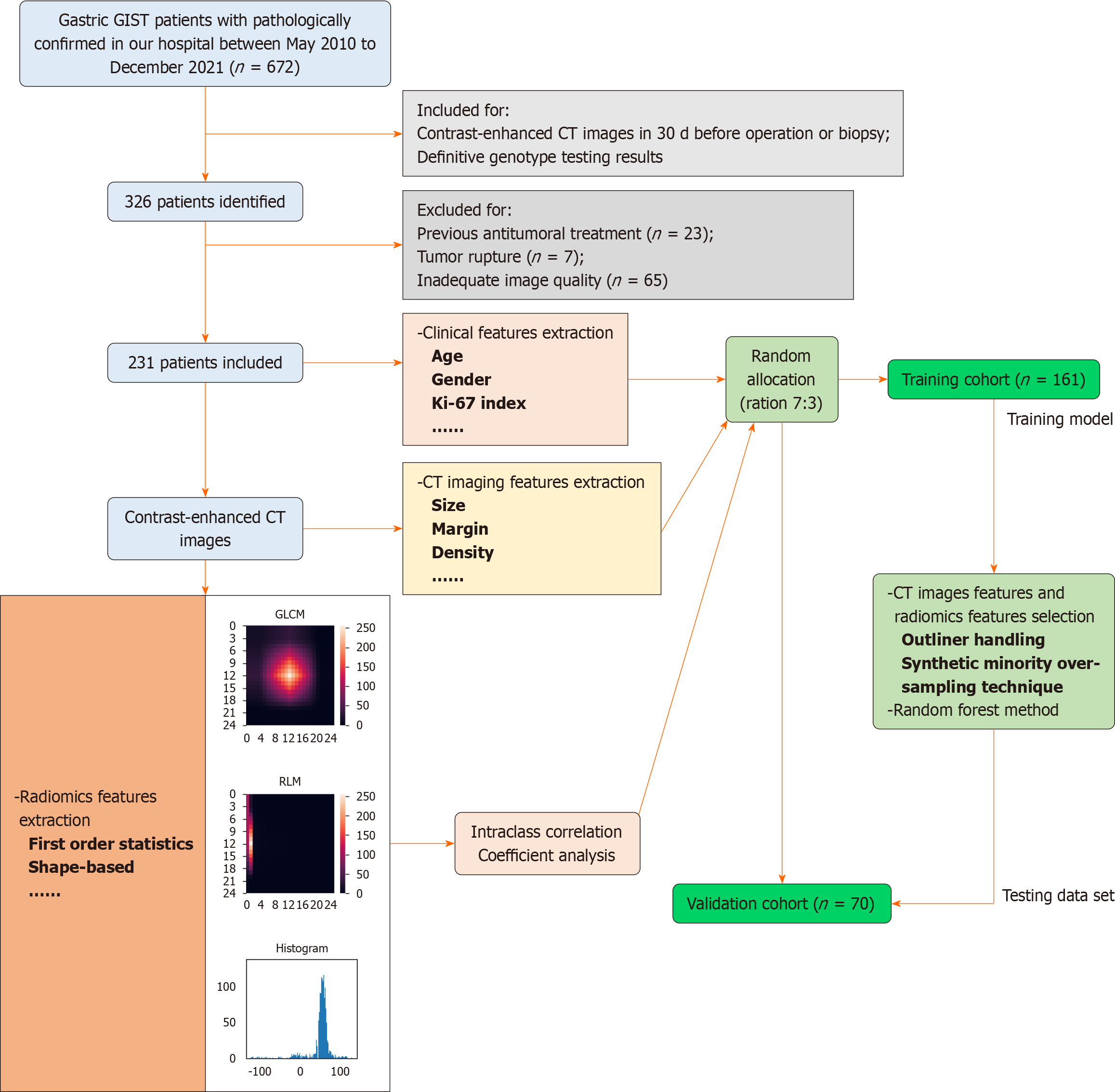
This retrospective study obtained ethical approval from the Research Ethics Board of West China Hospital, Sichuan University, China [Number: 2022(449)], and informed consent was waived due to the nature of the study. The inclusion criteria were as follows: (1) Patients who underwent CE-CT examination at our hospital within 30 d prior to surgery or biopsy; (2) patients diagnosed with primary gastric GISTs confirmed by pathological examination; and (3) patients with definitive genetic analysis results. The exclusion criteria were as follows: (1) Patients who received preoperative antitumoral treatment; (2) patients with tumor rupture; and (3) patients with inadequate CE-CT image quality, such as severe motion artifact or portal phase image thickness exceeding 5 mm. A total of 231 patients from May 2010 to December 2021 were included in the study (Figure 1). Mutation analysis was performed on the coding sequence of the KIT gene (exon 9, 11, 13, and 17) and the PDGFRA gene (exon 12, 14, and 18) using Sanger sequencing. Clinical information and pathology results were also collected.
All CT examinations were performed using three different CT scanners: A 64-slice CT scanner (Philips Medical system, Eindhoven, The Netherlands), a 128-slice CT scanner (SOMATOM Definition AS +, Siemens Healthcare, Germany), and a dual-source CT system (Somatom Definition Flash, Siemens Healthcare, Germany). Prior to the CT examination, patients were required to fast for at least 6 h and ingest 600-1000 mL of water. The CT scanning range encompassed the entire abdomen. The parameters for the CT examinations were as follows: Tube voltage of 120 kV, tube current ranging from 145 to 200 mAs, slice thickness of 2-5 mm, slice interval of 2 mm, field of view ranging from 35 cm to 50 cm, matrix size of 512 × 512, rotation time of 0.5 s, and pitch of 1.0. In all patients, an iodinated contrast agent (1.2-1.5 mL/kg) was intravenously injected using a syringe pump. Enhanced images were acquired during the arterial phase triggered at a threshold of 170 hounsfield units, and the portal venous phase was captured 30 s after the trigger.
The CT images were independently reviewed by two radiologists who were blinded to the clinicopathological data. Dis
All CE-CT images were collected and exported to the ITK-SNAP software (version 3.6.0, http://www.itk-snap.org) for manual segmentation of the ROI. For each patient, the portal vein phase images were reviewed, and the two largest cross-section slices were selected. ROIs were delineated over the solid portion of the entire lesion, excluding gas, calcification, vessels, and necrotic areas. The segmentation procedure was independently performed by two radiologists.
The Intelligence Foundry (Version 1.2, General Electric) was utilized to extract radiomics features from the lesions. A total of 554 features, comprising Original features, Co-occurrence of Local Anisotropic Gradient Orientations features, and Wavelet and local binary pattern (Wavelet-LBP) features, were extracted using PyRadiomic[23]. The reproducibility of the features was evaluated by calculating intra- and inter-class correlation coefficients (ICCs). Radiomics features that exhibited ICC values exceeding 0.75 in both intra- and inter-observer comparisons were selected for further feature analysis.
The entire dataset was randomly divided into training and internal validation datasets in a 7:3 ratio. The training dataset was exclusively used for feature selection and modeling. The feature preprocessing, feature selection, and model
Based on the features identified through the ICC analysis, features with a variance less than 1.0 were excluded. Outlier values greater than the third quartile plus twice the interquartile range were converted to the 95th percentile, while values less than the first quartile minus twice the interquartile range were converted to the 10th percentile. To address the class imbalance in the training dataset, the synthetic minority oversampling technique was employed, with 200% oversampling and 150% undersampling[24]. Subsequently, all features were normalized and standardized using the Z-Score method. The feature importance was evaluated using random forest (RF) based on the mean decrease of Gini calculated for all decision trees in the RF model. The top three important features were selected and used to construct the RF model[25,26].
Statistical analysis was conducted using SPSS software (Version 19, Chicago, IL, United States) and R software (Version 3.6.3; http://www.Rproject.org). All statistical significance levels were two-sided, and a significance level of P < 0.05 was considered statistically significant. To compare the significant differences between different genotype groups in both the training and validation cohorts, the Mann-Whitney U test or independent sample t-test was employed. Fisher's exact test or chi-square test was utilized to identify significant differences between different groups of continuous variables. The discrimination performance of the models was evaluated using receiver operating characteristic (ROC) curves. The area under the ROC curve (AUC) was used as a comprehensive measure of performance. Specificity, sensitivity, and positive and negative predictive values were used to assess model performance at specific thresholds, which were determined by maximizing the Youden index. The Delong test was employed to compare the AUC of paired models. Internal validation was estimated by performing regular bootstrapping with 1000 bootstrap samples[27]. The goodness-of-fit of the model was assessed using the Hosmer-Lemeshow test, with a P-value greater than 0.05 indicating agreement between the observed and predicted values. Model calibration was visualized using calibration curve analysis, and the clinical net benefit of the model was evaluated using decision curve analysis (DCA).
The clinicopathological characteristics of all 231 patients included in our study are listed in Table 1. Among the 231 cases of GISTs, 192 exhibited the KIT exon 11 mutation, while 39 were characterized as wild type (23 cases), PDGFRA exon 18 mutation (12 cases), KIT exon 9 mutation (2 cases), KIT exon 17 mutation (1 case), or PDGFRA exon 14 mutation (1 case). Within the group of patients with the KIT exon 11 mutation, 56 individuals had exon 11 deletions involving codons 557-558.
| Characteristics | KIT exon 11 mutation (n = 192) | Without KIT exon 11 mutation (n = 39) | P value | KIT exon 11 mutation with deletions involving codons 557-558 (n = 56) | KIT exon 11 mutation without deletions involving codons 557-558 (n = 136) | P value |
| Gender (male) | 97 (50.5) | 24 (61.5) | 0.223 | 32 (57.1) | 65 (47.8) | 0.268 |
| Age | 55.7 ± 11.5 | 53.8 ± 13.4 | 0.360 | 53.9 ± 13.5 | 56.4 ± 10.6 | 0.222 |
| Mitosis | ||||||
| ≤ 5/50 HPF | 91 (47.4) | 24 (61.5) | 0.117 | 11 (19.6) | 80 (58.8) | < 0.010 |
| > 5/50 HPF | 101 (52.6) | 15 (38.5) | 45 (80.4) | 56 (41.2) | ||
| Risk classification | ||||||
| Very low | 2 (1.0) | 0 (0) | 0.851 | 0 (0) | 2 (1.5) | < 0.010 |
| Low | 43 (22.4) | 10 (25.7) | 4 (7.1) | 39 (28.7) | ||
| Intermediate | 59 (30.8) | 13 (33.3) | 7 (12.5) | 52 (38.2) | ||
| High | 88 (45.8) | 16 (41.0) | 45 (80.4) | 43 (31.6) |
Based on the results of the univariate analysis, gender, age, mitotic count, and risk classification did not show significant differences between the group with the KIT exon 11 mutation and the group with other types of gene mutations (P > 0.05 for all). However, a significant difference was observed in the mitotic count and risk classification between the group with KIT exon 11 codons 557-558 deletion and the group without deletions in codons 557-558 (P < 0.01).
The primary analysis of the subjective CT features is presented in Table 2. In the univariate analysis, significant diffe
| Characteristics | KIT exon 11 mutation (n = 192) | Without KIT exon 11 mutation (n = 39) | P value | KIT exon 11 mutation with deletions involving codons 557-558 (n = 56) | KIT exon 11 mutation without deletions involving codons 557-558 (n = 136) | P value |
| Size (mm) | 51 (11-224) | 45 (10-201) | 0.682 | 69 (31-224) | 42.5 (11-188) | < 0.010 |
| Shape | ||||||
| Regular | 66 (34.4) | 7 (17.9) | 0.044 | 5 (8.9) | 61 (44.9) | < 0.010 |
| Irregular | 126 (65.6) | 32 (82.1) | 51 (91.1) | 75 (55.1) | ||
| Margin | ||||||
| Well-defined | 168 (87.5) | 23 (84.6) | 0.625 | 44 (78.6) | 124 (91.2) | 0.016 |
| Ill-defined | 24 (12.5) | 6 (15.4) | 12 (21.4) | 12 (8.8) | ||
| Growth pattern | ||||||
| Endophytic | 64 (33.3) | 6 (15.4) | 0.083 | 12 (21.4) | 52 (38.2) | 0.010 |
| Exophytic | 87 (45.3) | 22 (56.4) | 25 (44.6) | 62 (45.6) | ||
| Mixed | 41 (21.4) | 11 (28.2) | 19 (33.9) | 22 (16.2) | ||
| Density | ||||||
| Hypodensity | 175 (91.1) | 34 (87.2) | 0.647 | 50 (89.3) | 125 (91.9) | 0.750 |
| Isodensity | 15 (7.8) | 4 (10.3) | 5 (8.9) | 10 (7.4) | ||
| Hyperdensity | 2 (1.0) | 1 (2.6) | 1 (1.8) | 1 (0.7) | ||
| Pattern of enhancement | ||||||
| Homogeneous | 48 (25.0) | 5 (12.8) | 0.099 | 5 (8.9) | 43 (31.6) | 0.001 |
| Heterogeneous | 144 (75.0) | 34 (87.2) | 51 (91.1) | 93 (68.4) | ||
| Degree of enhancement | ||||||
| Mild | 71 (37.0) | 11 (28.2) | 0.018 | 20 (35.7) | 51 (37.5) | 0.063 |
| Moderate | 75 (39.1) | 10 (25.6) | 28 (50.0) | 47 (34.6) | ||
| Marked | 46 (24.0) | 18 (46.2) | 8 (14.3) | 38 (27.9) | ||
| Necrosis | 121 (63.0) | 28 (71.8) | 0.296 | 43 (76.8) | 78 (57.4) | 0.011 |
| Gas | 33 (17.2) | 4 (10.3) | 0.282 | 14 (25.0) | 19 (14.0) | 0.066 |
| Cystic change | 6 (3.1) | 4 (10.3) | 0.046 | 0 (0) | 6 (4.4) | 0.110 |
| Calcification | 23 (12.0) | 2 (5.1) | 0.209 | 8 (14.3) | 15 (11.0) | 0.528 |
| Superficial ulceration | 55 (28.6) | 10 (25.6) | 0.704 | 19 (33.9) | 36 (26.5) | 0.299 |
| Intra-tumoral vessel | 86 (44.8) | 14 (35.9) | 0.307 | 34 (60.7) | 52 (38.2) | 0.004 |
| Adjacent mesangial fat infiltration | 40 (20.8) | 9 (23.1) | 0.755 | 20 (35.7) | 20 (14.7) | 0.001 |
| Adjacent organ invasion | 29 (15.1) | 7 (17.9) | 0.655 | 14 (25.0) | 15 (11.0) | 0.014 |
| Lymphadenopathy | 14 (7.3) | 3 (7.7) | 0.930 | 6 (10.7) | 8 (5.9) | 0.242 |
| Distant metastasis | 4 (2.1) | 2 (5.1) | 0.276 | 3 (5.4) | 1 (0.7) | 0.042 |
A set of 190 radiomic features, exhibiting ICC values exceeding 0.75 in intra- and inter-individual comparisons, was uti
For KIT exon 11 mutation: Three CT features (gas, growth pattern, and density in arterial phase), three radiomic features (original_firstorder_Median, original_firstorder_InterquartileRange, and original_firstorder), and six clinic features (age, size, CD34, Ki-67, mitoses, and tissue-type) were extracted to build three models: ModelCT sign, modelCT sign + rad, and modelCT sign + rad + clinic. The combined model was developed using logistic regression, incorporating the model scores generated by each independent model. In the modelCT sign + rad, the Radscore was calculated as (4.58) × rad + (1.565) × ctsign + (-2.906). In the modelCT sign + rad + clinic, the Radscore was calculated as (4.364) × rad + (1.76) × ctsign + (5.207) × clinic +
| Models | ModelCT sign | ModelCT sign + rad | ModelCT sign + rad + clinic | |||
| Cohort | Training | Validation | Training | Validation | Training | Validation |
| AUC | 0.743 | 0.670 | 0.818 | 0.781 | 0.915 | 0.811 |
| Accuracy | 0.673 | 0.643 | 0.753 | 0.714 | 0.833 | 0.794 |
| Sensitivity | 0.815 | 0.672 | 0.667 | 0.690 | 0.938 | 0.830 |
| Specificity | 0.531 | 0.500 | 0.840 | 0.833 | 0.728 | 0.600 |
| NPV | 0.741 | 0.240 | 0.716 | 0.357 | 0.922 | 0.400 |
| PPV | 0.635 | 0.867 | 0.806 | 0.952 | 0.776 | 0.917 |
For deletions in KIT exon 11 codons 557-558: One CT feature (shape), three radiomic features
| Models | ModelCT sign | ModelCT sign + rad | ModelCT sign + rad + clinic | |||
| Cohort | Training | Validation | Training | Validation | Training | Validation |
| AUC | 0.667 | 0.610 | 0.842 | 0.782 | 0.872 | 0.795 |
| Accuracy | 0.522 | 0.500 | 0.689 | 0.700 | 0.720 | 0.766 |
| Sensitivity | 0.949 | 0.824 | 0.974 | 0.824 | 0.923 | 0.667 |
| Specificity | 0.385 | 0.396 | 0.598 | 0.660 | 0.656 | 0.796 |
| NPV | 0.959 | 0.875 | 0.986 | 0.927 | 0.964 | 0.886 |
| PPV | 0.330 | 0.304 | 0.437 | 0.438 | 0.462 | 0.500 |
Approximately 80% of GISTs harbor KIT mutations, while 5%-10% exhibit PDGFRA mutations. The presence and specific type of KIT and PDGFRA mutations are associated with the prognosis and clinical response to targeted therapy in GISTs[9,28]. Currently, mutation testing is typically performed on surgically resected tissue samples. However, some GIST patients are unable to undergo surgical resection at the time of initial diagnosis. For these patients, fine-needle biopsy samples provide adequate material for pathological examination but are insufficient for genetic analysis. Moreover, genetic testing is not routinely conducted in all hospitals due to its high cost. Therefore, there is an urgent need to establish a noninvasive, accurate, and cost-effective preoperative method for identifying the mutation status of GISTs.
CT is extensively employed in the detection, postoperative surveillance, and evaluation of treatment effectiveness in GISTs. Recent studies have identified several CT features associated with the differential diagnosis and high-risk categorization of GISTs, including tumor size, location, margin characteristics, hemorrhage, necrosis, heterogeneous enhancement, and adjacent organ invasion[29-31]. However, these conventional CT features rely on subjective analysis and the experience of radiologists, resulting in variability and lack of reproducibility. Radiomics, on the other hand, enables the extraction of high-throughput quantitative features from medical images using specific data characterization algorithms. This approach effectively reduces intra- and inter-observer variability. Importantly, radiomics has been widely applied in tumor diagnosis, prognosis prediction, and gene mutation analysis[32-36].
Several prior studies have reported the satisfactory performance of CT-based radiomics in the diagnosis and prediction of the malignant potential of GISTs[17,37-39]. Starmans et al[40] documented that the radiomics model achieved an AUC of 0.77 in distinguishing GISTs from non-GISTs, yielding results comparable to those of radiologists but with reduced observer dependence. Furthermore, radiomics studies in GISTs have primarily focused on predicting malignant potential and prognosis. These investigations have demonstrated the robust predictive effect and generalizability of radiomics in assessing the malignant potential of GISTs, thereby aiding clinicians in preoperative decision-making. However, there is a paucity of radiomics studies pertaining to genotype prediction. Xu et al[41] were the first to attempt differentiation of GISTs with and without KIT exon 11 mutations using CT texture analysis in a study cohort comprising 69 GISTs, with a validation group of 17 GISTs. They identified that the textural parameter standard deviation independently predicted GISTs without KIT exon 11 mutations, achieving AUC values of 0.726-0.750 in the study group and 0.904-0.962 in the validation group. Nonetheless, the relatively small sample sizes in this study may have impacted the accuracy of the findings. Starmans et al[40] also evaluated radiomics for predicting KIT mutational status in 123 patients with GISTs, reporting AUC values of 0.52 for KIT and 0.56 for KIT exon 11 mutation. These findings did not support the predictive value of the radiomics model in genetic features, likely due to study limitations. The remaining two studies both demonstrated the effective differentiation of GISTs with KIT exon 11 mutations using radiomics based on CT images[20,21]. However, the patient populations in these studies encompassed GISTs throughout the entire gastrointestinal tract, including the stomach, intestine, and colorectum, potentially introducing certain biases. It is well-known that GISTs at different sites exhibit distinct recurrence risks, with intestinal GISTs carrying a worse prognosis than gastric GISTs. Furthermore, genotypes have been closely associated with specific tumor locations, with KIT exon 11 mutations being most common in GISTs at all sites, while KIT exon 9 mutations are prevalent in intestinal GISTs, and PDGFRA exon 18 mutations are common in gastric GISTs[42].
Our study was derived from a large-scale imaging dataset and represents the first CT radiogenomics investigation specifically focused on gastric GISTs. The results revealed that the diagnostic accuracy of modelCT sign + rad + clinic for predicting KIT exon 11 mutation was significantly higher than that of modelCT sign and modelCT sign + rad, with AUC values of 0.915 in the training cohort and 0.811 in the validation cohorts. The DCA curves demonstrated that modelCT sign + rad + clinic exhibited superior predictive effectiveness compared to modelCT sign and modelCT sign + rad in the training cohorts, highlighting the clinical benefit of the combined model in distinguishing gastric GISTs with KIT exon 11 mutation.
Regarding deletions in KIT exon 11 codons 557-558 of gastric GISTs, the diagnostic accuracy of modelCT sign + rad + clinic was statistically higher than that of modelCT sign and modelCT sign + rad model, with AUC values of 0.872 in the training cohort and 0.795 in the validation cohorts. The clinical benefits analysis revealed that the combined model outperformed modelCT sign and modelCT sign + rad in predicting the KIT exon 11 mutation. In the validation cohort, the sensitivity and specificity of modelCT sign + rad + clinic for predicting the KIT exon 11 mutation were 83.0% and 81.1%, respectively, surpassing the per
However, it is important to acknowledge certain limitations in our study. Firstly, it was a retrospective study, and as such, potential selection bias could not be completely eliminated. Secondly, despite the large sample size, this study was conducted at a single center, and further validation through multicenter studies is warranted. Thirdly, due to the small sample size, we did not subdivide GISTs without KIT exon 11 mutation, which is crucial for clinicians to differentiate specific types of gene mutations before surgery, such as KIT exon 9 mutation and PDGFRA exon 18 mutation, as the treatment response varies.
In conclusion, our study demonstrated that the radiomics model based on CE-CT images exhibited satisfactory performance in distinguishing gastric GISTs with KIT exon 11 mutation and GISTs with KIT exon 11 codons 557-558 deletions. The combined modelCT sign + rad + clinic demonstrated the highest predictive value, offering a potentially valuable and noninvasive approach to guide personalized treatment decisions prior to surgery.
The assessment of KIT and PDGFRA mutations plays a vital role in establishing the pathological diagnosis of gastro
Currently, tumor mutation status can only be obtained after surgical resection or conventional invasive biopsy, making preoperative genotyping of GISTs more challenging.
To develop and validate a radiomic model to predict the genotypes of gastric GISTs using contrast-enhanced computed tomography (CE-CT) images.
The models for predicting GISTs with KIT exon 11 mutations or KIT exon 11 codons 557-558 deletions were constructed using selected clinical features, conventional CT features, and radiomics features extracted from abdominal CE-CT images. Three models were developed: ModelCT sign, modelCT sign + rad, and modelCT sign + rad + clinic. The diagnostic performance of these models was evaluated using receiver operating characteristic (ROC) curve analysis and the Delong test.
The ROC analyses demonstrated the performance of different models in predicting KIT exon 11 mutation and KIT exon 11 codons 557-558 deletions. In the training cohort, the modelsCT sign, modelCT sign + rad, and modelCT sign + rad + clinic achieved area under the curve (AUC) values of 0.743, 0.818, and 0.915, respectively, for predicting KIT exon 11 mutation. In the validation cohort, the corresponding AUC values were 0.670, 0.781, and 0.811. For predicting KIT exon 11 codons 557-558 deletions, the AUC values in the training cohort were 0.667, 0.842, and 0.72 for modelCT sign, modelCT sign + rad, and modelCT sign + rad + clinic, respectively. In the validation cohort, the AUC values for the same models were 0.610, 0.782, and 0.795. Furthermore, the decision curve analysis confirmed the clinical significance and utility of the CT sign + rad + clinic model.
Our study demonstrated that the radiomics model based on CE-CT images exhibited satisfactory performance in distinguishing gastric GISTs with KIT exon 11 mutation and GISTs with KIT exon 11 codons 557-558 deletions.
This study focuses specifically on gastric GISTs and aims to develop a prediction model for genotypes using CE-CT images.
Provenance and peer review: Unsolicited article; Externally peer reviewed.
Peer-review model: Single blind
Specialty type: Gastroenterology & hepatology
Country/Territory of origin: China
Peer-review report’s scientific quality classification
Grade A (Excellent): 0
Grade B (Very good): 0
Grade C (Good): C
Grade D (Fair): 0
Grade E (Poor): 0
P-Reviewer: Dimofte GM, Romania S-Editor: Chen YL L-Editor: A P-Editor: Zheng XM
| 1. | Blay JY, Kang YK, Nishida T, von Mehren M. Gastrointestinal stromal tumours. Nat Rev Dis Primers. 2021;7:22. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 72] [Cited by in RCA: 238] [Article Influence: 59.5] [Reference Citation Analysis (0)] |
| 2. | Søreide K, Sandvik OM, Søreide JA, Giljaca V, Jureckova A, Bulusu VR. Global epidemiology of gastrointestinal stromal tumours (GIST): A systematic review of population-based cohort studies. Cancer Epidemiol. 2016;40:39-46. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 579] [Cited by in RCA: 512] [Article Influence: 56.9] [Reference Citation Analysis (1)] |
| 3. | Corless CL, Barnett CM, Heinrich MC. Gastrointestinal stromal tumours: origin and molecular oncology. Nat Rev Cancer. 2011;11:865-878. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 635] [Cited by in RCA: 620] [Article Influence: 44.3] [Reference Citation Analysis (0)] |
| 4. | Klug LR, Khosroyani HM, Kent JD, Heinrich MC. New treatment strategies for advanced-stage gastrointestinal stromal tumours. Nat Rev Clin Oncol. 2022;19:328-341. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 56] [Cited by in RCA: 78] [Article Influence: 26.0] [Reference Citation Analysis (0)] |
| 5. | Pierotti MA, Tamborini E, Negri T, Pricl S, Pilotti S. Targeted therapy in GIST: in silico modeling for prediction of resistance. Nat Rev Clin Oncol. 2011;8:161-170. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 59] [Cited by in RCA: 62] [Article Influence: 4.4] [Reference Citation Analysis (0)] |
| 6. | Dermawan JK, Rubin BP. Molecular Pathogenesis of Gastrointestinal Stromal Tumor: A Paradigm for Personalized Medicine. Annu Rev Pathol. 2022;17:323-344. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1] [Cited by in RCA: 13] [Article Influence: 3.3] [Reference Citation Analysis (0)] |
| 7. | Boikos SA, Pappo AS, Killian JK, LaQuaglia MP, Weldon CB, George S, Trent JC, von Mehren M, Wright JA, Schiffman JD, Raygada M, Pacak K, Meltzer PS, Miettinen MM, Stratakis C, Janeway KA, Helman LJ. Molecular Subtypes of KIT/PDGFRA Wild-Type Gastrointestinal Stromal Tumors: A Report From the National Institutes of Health Gastrointestinal Stromal Tumor Clinic. JAMA Oncol. 2016;2:922-928. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 188] [Cited by in RCA: 270] [Article Influence: 33.8] [Reference Citation Analysis (0)] |
| 8. | Corless CL, Schroeder A, Griffith D, Town A, McGreevey L, Harrell P, Shiraga S, Bainbridge T, Morich J, Heinrich MC. PDGFRA mutations in gastrointestinal stromal tumors: frequency, spectrum and in vitro sensitivity to imatinib. J Clin Oncol. 2005;23:5357-5364. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 579] [Cited by in RCA: 595] [Article Influence: 29.8] [Reference Citation Analysis (0)] |
| 9. | Rossi S, Gasparotto D, Miceli R, Toffolatti L, Gallina G, Scaramel E, Marzotto A, Boscato E, Messerini L, Bearzi I, Mazzoleni G, Capella C, Arrigoni G, Sonzogni A, Sidoni A, Mariani L, Amore P, Gronchi A, Casali PG, Maestro R, Dei Tos AP. KIT, PDGFRA, and BRAF mutational spectrum impacts on the natural history of imatinib-naive localized GIST: a population-based study. Am J Surg Pathol. 2015;39:922-930. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 44] [Cited by in RCA: 54] [Article Influence: 5.4] [Reference Citation Analysis (0)] |
| 10. | Joensuu H. Risk stratification of patients diagnosed with gastrointestinal stromal tumor. Hum Pathol. 2008;39:1411-1419. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 699] [Cited by in RCA: 861] [Article Influence: 50.6] [Reference Citation Analysis (0)] |
| 11. | Joensuu H, Rutkowski P, Nishida T, Steigen SE, Brabec P, Plank L, Nilsson B, Braconi C, Bordoni A, Magnusson MK, Sufliarsky J, Federico M, Jonasson JG, Hostein I, Bringuier PP, Emile JF. KIT and PDGFRA mutations and the risk of GI stromal tumor recurrence. J Clin Oncol. 2015;33:634-642. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 97] [Cited by in RCA: 126] [Article Influence: 12.6] [Reference Citation Analysis (0)] |
| 12. | Hong JH, Jung JY, Jo A, Nam Y, Pak S, Lee SY, Park H, Lee SE, Kim S. Development and Validation of a Radiomics Model for Differentiating Bone Islands and Osteoblastic Bone Metastases at Abdominal CT. Radiology. 2021;299:626-632. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 13] [Cited by in RCA: 56] [Article Influence: 14.0] [Reference Citation Analysis (0)] |
| 13. | Kirienko M, Sollini M, Corbetta M, Voulaz E, Gozzi N, Interlenghi M, Gallivanone F, Castiglioni I, Asselta R, Duga S, Soldà G, Chiti A. Radiomics and gene expression profile to characterise the disease and predict outcome in patients with lung cancer. Eur J Nucl Med Mol Imaging. 2021;48:3643-3655. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 15] [Cited by in RCA: 69] [Article Influence: 17.3] [Reference Citation Analysis (0)] |
| 14. | Li L, Kan X, Zhao Y, Liang B, Ye T, Yang L, Zheng C. Radiomics Signature: A potential biomarker for the prediction of survival in Advanced Hepatocellular Carcinoma. Int J Med Sci. 2021;18:2276-2284. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 7] [Cited by in RCA: 16] [Article Influence: 4.0] [Reference Citation Analysis (0)] |
| 15. | Schniering J, Maciukiewicz M, Gabrys HS, Brunner M, Blüthgen C, Meier C, Braga-Lagache S, Uldry AC, Heller M, Guckenberger M, Fretheim H, Nakas CT, Hoffmann-Vold AM, Distler O, Frauenfelder T, Tanadini-Lang S, Maurer B. Computed tomography-based radiomics decodes prognostic and molecular differences in interstitial lung disease related to systemic sclerosis. Eur Respir J. 2022;59. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 5] [Cited by in RCA: 32] [Article Influence: 8.0] [Reference Citation Analysis (0)] |
| 16. | Chen T, Ning Z, Xu L, Feng X, Han S, Roth HR, Xiong W, Zhao X, Hu Y, Liu H, Yu J, Zhang Y, Li Y, Xu Y, Mori K, Li G. Radiomics nomogram for predicting the malignant potential of gastrointestinal stromal tumours preoperatively. Eur Radiol. 2019;29:1074-1082. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 54] [Cited by in RCA: 54] [Article Influence: 9.0] [Reference Citation Analysis (0)] |
| 17. | Chen Z, Xu L, Zhang C, Huang C, Wang M, Feng Z, Xiong Y. CT Radiomics Model for Discriminating the Risk Stratification of Gastrointestinal Stromal Tumors: A Multi-Class Classification and Multi-Center Study. Front Oncol. 2021;11:654114. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 3] [Cited by in RCA: 12] [Article Influence: 3.0] [Reference Citation Analysis (0)] |
| 18. | Wang C, Li H, Jiaerken Y, Huang P, Sun L, Dong F, Huang Y, Dong D, Tian J, Zhang M. Building CT Radiomics-Based Models for Preoperatively Predicting Malignant Potential and Mitotic Count of Gastrointestinal Stromal Tumors. Transl Oncol. 2019;12:1229-1236. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 25] [Cited by in RCA: 39] [Article Influence: 6.5] [Reference Citation Analysis (0)] |
| 19. | Zhao Y, Feng M, Wang M, Zhang L, Li M, Huang C. CT Radiomics for the Preoperative Prediction of Ki67 Index in Gastrointestinal Stromal Tumors: A Multi-Center Study. Front Oncol. 2021;11:689136. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 3] [Cited by in RCA: 3] [Article Influence: 0.8] [Reference Citation Analysis (0)] |
| 20. | Liu B, Liu H, Zhang L, Song Y, Yang S, Zheng Z, Zhao J, Hou F, Zhang J. Value of contrast-enhanced CT based radiomic machine learning algorithm in differentiating gastrointestinal stromal tumors with KIT exon 11 mutation: a two-center study. Diagn Interv Radiol. 2022;28:29-38. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1] [Cited by in RCA: 1] [Article Influence: 0.3] [Reference Citation Analysis (0)] |
| 21. | Liu X, Yin Y, Wang X, Yang C, Wan S, Yin X, Wu T, Chen H, Xu Z, Li X, Song B, Zhang B. Gastrointestinal stromal tumors: associations between contrast-enhanced CT images and KIT exon 11 gene mutation. Ann Transl Med. 2021;9:1496. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 2] [Cited by in RCA: 1] [Article Influence: 0.3] [Reference Citation Analysis (0)] |
| 22. | Palatresi D, Fedeli F, Danti G, Pasqualini E, Castiglione F, Messerini L, Massi D, Bettarini S, Tortoli P, Busoni S, Pradella S, Miele V. Correlation of CT radiomic features for GISTs with pathological classification and molecular subtypes: preliminary and monocentric experience. Radiol Med. 2022;127:117-128. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 4] [Cited by in RCA: 24] [Article Influence: 8.0] [Reference Citation Analysis (0)] |
| 23. | van Griethuysen JJM, Fedorov A, Parmar C, Hosny A, Aucoin N, Narayan V, Beets-Tan RGH, Fillion-Robin JC, Pieper S, Aerts HJWL. Computational Radiomics System to Decode the Radiographic Phenotype. Cancer Res. 2017;77:e104-e107. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1922] [Cited by in RCA: 3837] [Article Influence: 479.6] [Reference Citation Analysis (0)] |
| 24. | Chawla NV, Bowyer KW, Hall LO, Kegelmeyer WP. Smote: Synthetic minority over-sampling technique. J AI Res. 2002;16:321-357. [DOI] [Full Text] |
| 25. | Breiman L. Random Forests. Machine Learning. 2001;45:5-32. [RCA] [DOI] [Full Text] [Cited by in Crossref: 56052] [Cited by in RCA: 33805] [Article Influence: 2817.1] [Reference Citation Analysis (0)] |
| 26. | Rokach L. Decision Forest: Twenty years of research. Information Fusion. 2016;27:111-125. [DOI] [Full Text] |
| 27. | Steyerberg EW, Harrell FE Jr, Borsboom GJ, Eijkemans MJ, Vergouwe Y, Habbema JD. Internal validation of predictive models: efficiency of some procedures for logistic regression analysis. J Clin Epidemiol. 2001;54:774-781. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1695] [Cited by in RCA: 1870] [Article Influence: 77.9] [Reference Citation Analysis (0)] |
| 28. | Heinrich MC, Maki RG, Corless CL, Antonescu CR, Harlow A, Griffith D, Town A, McKinley A, Ou WB, Fletcher JA, Fletcher CD, Huang X, Cohen DP, Baum CM, Demetri GD. Primary and secondary kinase genotypes correlate with the biological and clinical activity of sunitinib in imatinib-resistant gastrointestinal stromal tumor. J Clin Oncol. 2008;26:5352-5359. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 533] [Cited by in RCA: 577] [Article Influence: 33.9] [Reference Citation Analysis (0)] |
| 29. | Chen Z, Yang J, Sun J, Wang P. Gastric gastrointestinal stromal tumours (2-5 cm): Correlation of CT features with malignancy and differential diagnosis. Eur J Radiol. 2020;123:108783. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 4] [Cited by in RCA: 16] [Article Influence: 2.7] [Reference Citation Analysis (0)] |
| 30. | Xu JX, Ding QL, Lu YF, Fan SF, Rao QP, Yu RS. A scoring model for radiologic diagnosis of gastric leiomyomas (GLMs) with contrast-enhanced computed tomography (CE-CT): Differential diagnosis from gastrointestinal stromal tumors (GISTs). Eur J Radiol. 2021;134:109395. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1] [Cited by in RCA: 8] [Article Influence: 1.6] [Reference Citation Analysis (0)] |
| 31. | Zhou C, Duan X, Zhang X, Hu H, Wang D, Shen J. Predictive features of CT for risk stratifications in patients with primary gastrointestinal stromal tumour. Eur Radiol. 2016;26:3086-3093. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 74] [Cited by in RCA: 69] [Article Influence: 7.7] [Reference Citation Analysis (0)] |
| 32. | Balana C, Castañer S, Carrato C, Moran T, Lopez-Paradís A, Domenech M, Hernandez A, Puig J. Preoperative Diagnosis and Molecular Characterization of Gliomas With Liquid Biopsy and Radiogenomics. Front Neurol. 2022;13:865171. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 1] [Cited by in RCA: 16] [Article Influence: 5.3] [Reference Citation Analysis (0)] |
| 33. | Harding-Theobald E, Louissaint J, Maraj B, Cuaresma E, Townsend W, Mendiratta-Lala M, Singal AG, Su GL, Lok AS, Parikh ND. Systematic review: radiomics for the diagnosis and prognosis of hepatocellular carcinoma. Aliment Pharmacol Ther. 2021;54:890-901. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 45] [Cited by in RCA: 87] [Article Influence: 21.8] [Reference Citation Analysis (0)] |
| 34. | Lu J, Li X, Li H. A radiomics feature-based nomogram to predict telomerase reverse transcriptase promoter mutation status and the prognosis of lower-grade gliomas. Clin Radiol. 2022;77:e560-e567. [RCA] [PubMed] [DOI] [Full Text] [Cited by in RCA: 13] [Reference Citation Analysis (0)] |
| 35. | Sohn B, An C, Kim D, Ahn SS, Han K, Kim SH, Kang SG, Chang JH, Lee SK. Radiomics-based prediction of multiple gene alteration incorporating mutual genetic information in glioblastoma and grade 4 astrocytoma, IDH-mutant. J Neurooncol. 2021;155:267-276. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 1] [Cited by in RCA: 19] [Article Influence: 4.8] [Reference Citation Analysis (0)] |
| 36. | Staal FCR, van der Reijd DJ, Taghavi M, Lambregts DMJ, Beets-Tan RGH, Maas M. Radiomics for the Prediction of Treatment Outcome and Survival in Patients With Colorectal Cancer: A Systematic Review. Clin Colorectal Cancer. 2021;20:52-71. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 22] [Cited by in RCA: 74] [Article Influence: 14.8] [Reference Citation Analysis (1)] |
| 37. | Shao M, Niu Z, He L, Fang Z, He J, Xie Z, Cheng G, Wang J. Building Radiomics Models Based on Triple-Phase CT Images Combining Clinical Features for Discriminating the Risk Rating in Gastrointestinal Stromal Tumors. Front Oncol. 2021;11:737302. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 1] [Cited by in RCA: 2] [Article Influence: 0.5] [Reference Citation Analysis (0)] |
| 38. | Wang M, Feng Z, Zhou L, Zhang L, Hao X, Zhai J. Computed-Tomography-Based Radiomics Model for Predicting the Malignant Potential of Gastrointestinal Stromal Tumors Preoperatively: A Multi-Classifier and Multicenter Study. Front Oncol. 2021;11:582847. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 5] [Cited by in RCA: 18] [Article Influence: 4.5] [Reference Citation Analysis (0)] |
| 39. | Zhang QW, Zhou XX, Zhang RY, Chen SL, Liu Q, Wang J, Zhang Y, Lin J, Xu JR, Gao YJ, Ge ZZ. Comparison of malignancy-prediction efficiency between contrast and non-contract CT-based radiomics features in gastrointestinal stromal tumors: A multicenter study. Clin Transl Med. 2020;10:e291. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 4] [Cited by in RCA: 15] [Article Influence: 3.0] [Reference Citation Analysis (0)] |
| 40. | Starmans MPA, Timbergen MJM, Vos M, Renckens M, Grünhagen DJ, van Leenders GJLH, Dwarkasing RS, Willemssen FEJA, Niessen WJ, Verhoef C, Sleijfer S, Visser JJ, Klein S. Differential Diagnosis and Molecular Stratification of Gastrointestinal Stromal Tumors on CT Images Using a Radiomics Approach. J Digit Imaging. 2022;35:127-136. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 3] [Cited by in RCA: 15] [Article Influence: 5.0] [Reference Citation Analysis (0)] |
| 41. | Xu F, Ma X, Wang Y, Tian Y, Tang W, Wang M, Wei R, Zhao X. CT texture analysis can be a potential tool to differentiate gastrointestinal stromal tumors without KIT exon 11 mutation. Eur J Radiol. 2018;107:90-97. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 21] [Cited by in RCA: 30] [Article Influence: 4.3] [Reference Citation Analysis (0)] |
| 42. | Lasota J, Miettinen M. Clinical significance of oncogenic KIT and PDGFRA mutations in gastrointestinal stromal tumours. Histopathology. 2008;53:245-266. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 305] [Cited by in RCA: 285] [Article Influence: 16.8] [Reference Citation Analysis (1)] |