Published online Mar 15, 2024. doi: 10.4251/wjgo.v16.i3.659
Peer-review started: October 5, 2023
First decision: December 6, 2023
Revised: December 21, 2023
Accepted: January 18, 2024
Article in press: January 18, 2024
Published online: March 15, 2024
Processing time: 159 Days and 5.9 Hours
Pancreatic ductal adenocarcinoma (PDAC) has a poor prognosis, with a 5-year survival rate of less than 10%, owing to its late-stage diagnosis. Early detection of pancreatic cancer (PC) can significantly increase survival rates.
To identify the serum biomarker signatures associated with early-stage PDAC by serum N-glycan analysis.
An extensive patient cohort was used to determine a biomarker signature, in
The biomarker signature was created to discriminate samples derived from patients with PC from those of controls, with a receiver operating characteristic area under the curve of 0.86. In addition, the biomarker signature combined with cancer antigen 19-9 could discriminate patients with PDAC from controls, with a receiver operating characteristic area under the curve of 0.919. Glyco-model demonstrated favorable diagnostic performance in all stages of PC. The diagnostic sensitivity for stage I PDAC was 89.66%.
In a prospective validation study, this serum biomarker signature may offer a viable method for detecting early-stage PDAC.
Core Tip: This study employed a patient cohort to investigate the N-glycan signature of early-stage pancreatic cancer (PC). Serum N-glycans analysis was conducted to identify the serum biomarker signature associated with early-stage pancreatic ductal adenocarcinoma (PDAC), resulting in the identification of nine early-stage PDAC N-glycan signatures. Subsequently, utilizing these biosignatures, a diagnostic model named the “Glyco-model” was developed, demonstrating promising diagnostic performance across all stages of PC. The study revealed that the diagnostic sensitivity for stage I PDAC was determined to be 89.66%. Consequently, this diagnostic model exhibits potential as a prospective strategy for the early detection of PDAC.
- Citation: Wen YR, Lin XW, Zhou YW, Xu L, Zhang JL, Chen CY, He J. N-glycan biosignatures as a potential diagnostic biomarker for early-stage pancreatic cancer. World J Gastrointest Oncol 2024; 16(3): 659-669
- URL: https://www.wjgnet.com/1948-5204/full/v16/i3/659.htm
- DOI: https://dx.doi.org/10.4251/wjgo.v16.i3.659
Among all cancers, pancreatic cancer (PC) is the deadliest, exhibiting the lowest 5-year survival rates[1,2]. The annual incidence of PC continues to increase by approximately 1%[3]. Patients often present with non-specific symptoms such as jaundice, fatigue, changes in bowel habits, and indigestion, which makes it difficult to distinguish them from non-cancer diseases[4]. PC has a poor prognosis, primarily due to late diagnosis, with approximately 20% of patients being diag
Early diagnostic biomarkers for PC are lacking. Pancreatic ductal adenocarcinoma (PDAC) is the most common his
The tumor markers carcinoembryonic antigen (CEA), CA19-9, and CA125 play vital roles in the diagnosis and prognosis of PC[17]. These glycoprotein alterations indicate the potential involvement of abnormal glycosylation in various biological processes[18,19]. Recent advances in glycomics have led to the discovery of unique N- and O-glycans that serve as glycobiomarkers for cancer diagnosis and treatment[20]. Various tumors have altered N-linked sugar chains[21,22]. Some N-linked glycan species and other classes of glycans are associated with PC[23]. Imaging mass spectrometry has been used to evaluate the N-glycome of the human pancreas and PC in a cohort of patients with PDAC, represented by tissue microarrays and whole tissue sections, to describe the differences between PDAC and other abdominal cancers and non-cancerous pancreatic lesions[24]. Using serum N-glycome analysis, patients with PDAC can be differentiated from healthy controls on the basis of glycosylation[25]. The previous study also revealed that the serum N-glycan profile is a promising method for detecting hepatocellular carcinoma in patients with cirrhosis, based on the log ratio of the branching α-1,3 fucosylated triantennary glycan [NA3Fb, glycan peak (GP) 9] to the bigalacto core α-1,6 fucosylated bisecting biantennary glycan (NA2FB, GP7), or the log ratio of GP9 to GP4 (a single galactic core 1,6 fucosylated biantennary glycan, NG1A2F)[26]. However, there are no specific N-glycan glycobiomarkers for the early diagnosis of PC.
In this study, we designed an extensive patient cohort to identify the N-glycan signature, including patients with PDAC that were well-defined at an early stage (stages I and II). The biomarker signature was derived from a case-control study using a case-cohort design consisting of 29 patients with stage I, 22 with stage II, 4 with stage III, 16 with stage IV PDAC, and 88 controls. Serum N-glycan analysis was performed to identify the serum biomarker signatures associated with early-stage PDAC. Using a case-control study, a biomarker signature was created to discriminate between samples derived from patients with PC and those derived from controls. Moreover, the biomarker signature combined with CA19-9 could discriminate patients with PDAC from controls with a receiver operating characteristic (ROC) area under the curve (AUC) of 0.92. The N-glycan signature discriminated patients with stage I and II PDAC from controls in this independent patient cohort, with a AUC of 0.86.
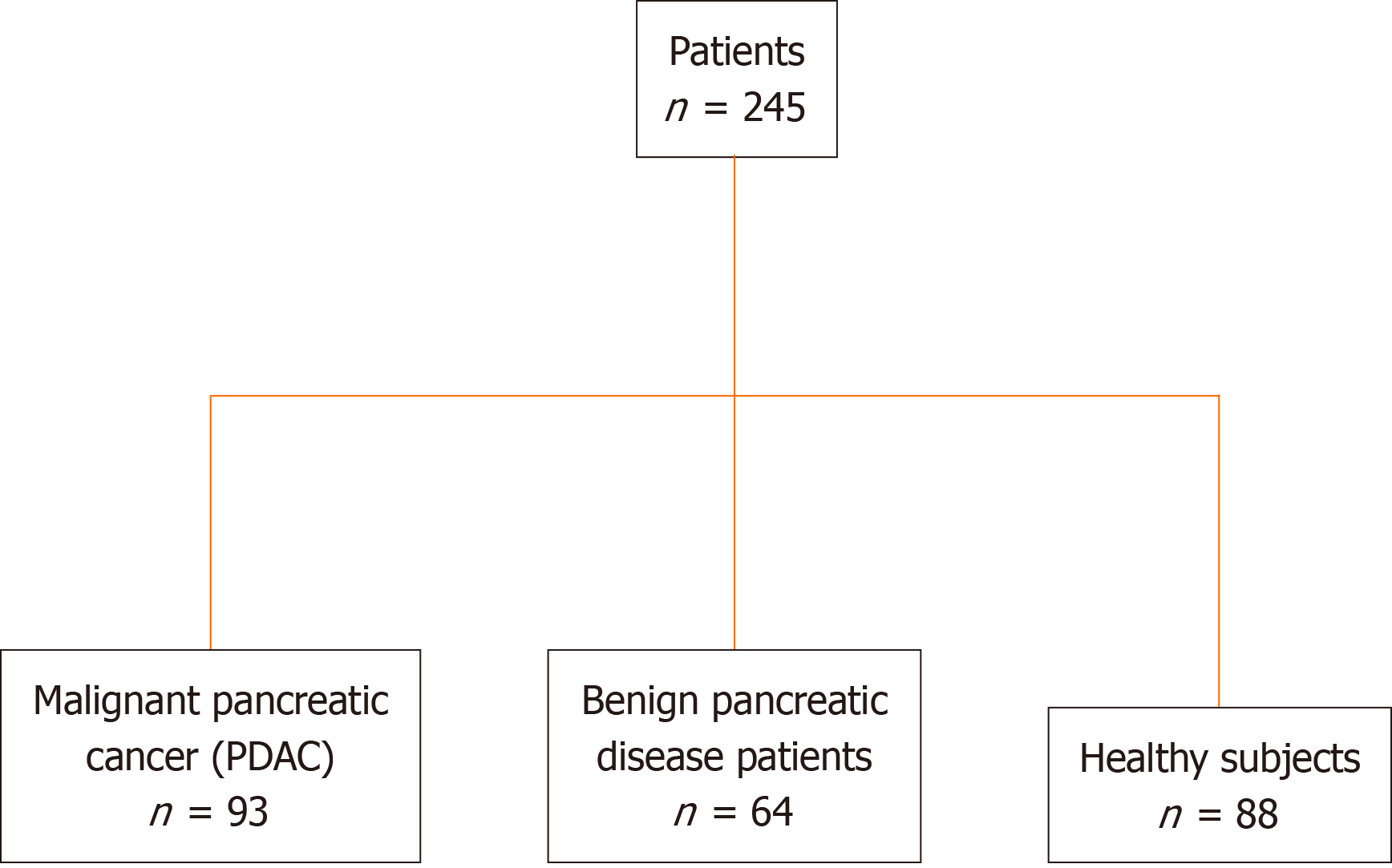
This retrospective study recruited 245 patients, including 93 patients with PDAC, 64 with benign pancreatic disease, and 88 healthy participants (Figure 1). This retrospective study performed on PDAC serum samples collected at the Nanjing Drum Tower Hospital was conducted according to the Standards for Reporting Diagnostic Accuracy Studies[27]. PDAC was staged according to the TNM staging system of the American Joint Committee on Cancer on Cancer (eighth edition).
The study cohort comprised 93 patients with PDAC, 64 with benign pancreatic disease, and 88 healthy participants (Table 1). The patients were diagnosed using computed tomography and were histologically verified. There were 29 PDAC samples from patients with stage I, 22 with stage II, 4 with stage III, 16 with stage IV, and 22 with unknown stages (Table 1). Blood samples were collected from participants who had not received any anticancer therapy. The inclusion criteria were as follows: Patients within the age range of 18-70 years, diagnosed with PDAC, had not received any prior treatment, were recruited. Serum tumor markers CEA, CA19-9, CA125, CA242, CA724, and alpha fetoprotein (AFP) were elevated.
| Characteristics | PDAC (n = 93) | Non-PDAC (n = 64) | Control (n = 88) |
| Men, n (%) | 59 (63.4) | 35 (54.7) | 38 (43.2) |
| Women, n (%) | 34 (36.6) | 29 (45.3) | 50 (56.8) |
| Age (yr) | 64 (54.74) | 54 (40.68) | 57 (43, 71) |
| CEA, n (%) | 93 (100) | 64 (100) | 82 (93.18) |
| CA19-9, n (%) | 93 (100) | 64 (100) | 69 (78.41) |
| CA125, n (%) | 93 (100) | 63 (98.44) | 42 (47.73) |
| CA242, n (%) | 93 (100) | 63 (98.33) | 0 (0.00) |
| CA724, n (%) | 93 (100) | 63 (98.44) | 0 (0.00) |
| AFP, n (%) | 93 (100) | 64 (100) | 82 (93.18) |
| Stage I, n (%) | 29 (31.18) | - | - |
| Stage II, n (%) | 22 (23.66) | - | - |
| Stage III, n (%) | 4 (4.30) | - | - |
| Stage IV, n (%) | 16 (17.20) | - | - |
| Unknown, n (%) | 22 (23.66) | - | - |
The study protocol was approved by the Ethics Committee of Drum Tower Hospital, the Affiliated Hospital of Nanjing University Medical School and conformed to the ethical guidelines of the Declaration of Helsinki. Written informed consent was obtained from all patients.
Serum was collected from whole blood using a standard protocol and centrifuged at 10000 × g for 4 min. Laboratory tests, including those for the serum tumor markers CEA, CA19-9, CA125, CA242, CA724, and AFP, were performed at local laboratories according to standard procedures.
Serum glycoprotein N-glycome profiling was performed on the GlyFace (Glycoprofiling by Fluorophore-Assisted Carbohydrate Electrophoresis) glycome detection technology platform provided by SysDiagno (Nanjing) Biotech Co. The results were analyzed using GeneMapper v6.0 software (Applied Biosystems). The height intensities of the nine most intense GPs were detected in all samples and normalized to the total intensity of the measured GPs.
Continuous variables are expressed as medians (interquartile ranges). Categorical variables are expressed as numbers (%). Continuous variables were compared using Student’s t-test or one-way analysis of variance. ROC curve analysis and AUC values were used to evaluate the overall diagnostic performance of single markers and diagnostic models. Sensitivities and specificities were calculated using cut-off values optimally selected upon the ROC curves. The t-test (data conforms to a normal distribution and variance homogeneity) or Wilcoxon rank-sum test (not conforms to normal distribution and homogeneity of variance) was performed to compare the two groups. The Kruskal-Wallis test was used to compare variables among multiple groups. All statistical tests were two-sided. Statistical significance was set at P < 0.05. Data were analyzed using SPSS 22.0.
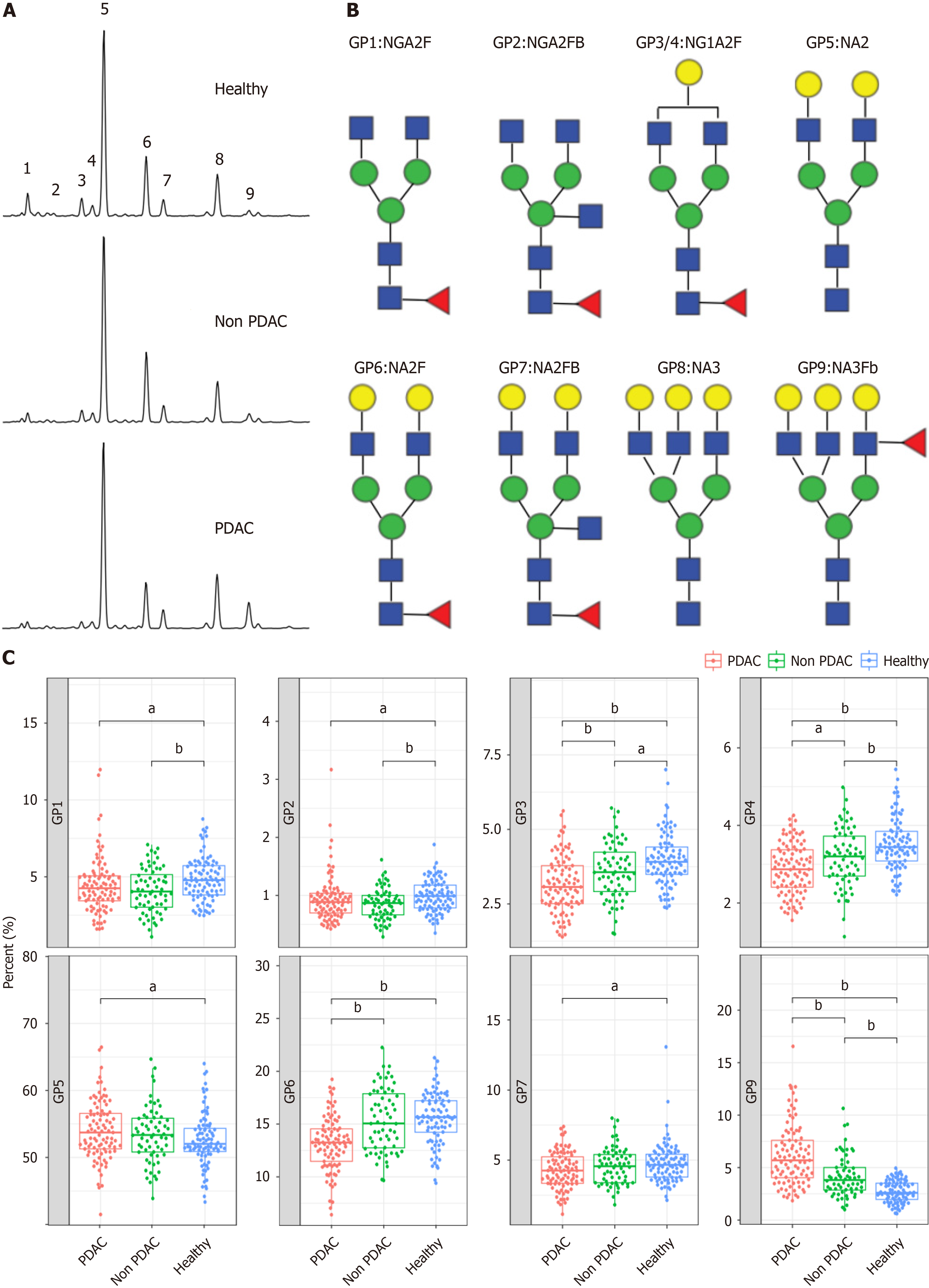
The study cohort comprised patients with PDAC, those without PDAC, and healthy controls. The clinical characteristics were comparable between the groups (Table 1 and Supplementary Figure 1). We conducted N-GP profiling in all patients. The representative profiling patterns are shown in Figure 2A. The structures of the nine N-GPs are shown in Figure 2B. Significant changes were observed in three GPs (GP3, GP6, and GP9) in the PDAC group compared to those in the non-PDAC and control groups (Figure 2C). Furthermore, tumor marker concentrations were examined for each group. Tumor markers exhibited significant differences among the three groups (Supplementary Figure 2). The differences in N-glycans among the three groups suggest the potential use of N-glycans as diagnostic markers for PDAC.
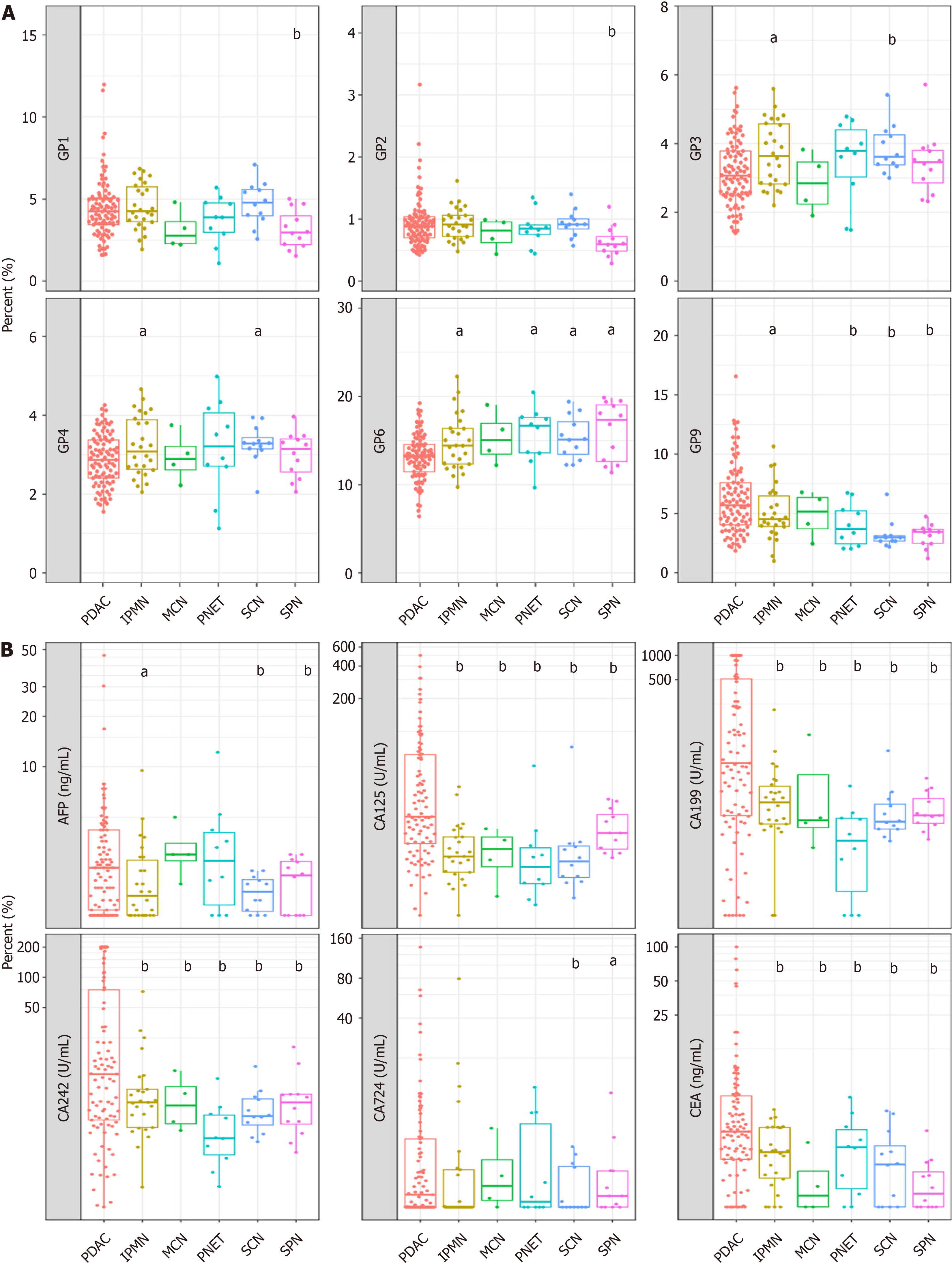
Subsequently, we assessed the discriminatory potential of the N-glycan signature in distinguishing between various clinical subgroups of patients diagnosed with PC and those diagnosed with benign pancreatic disease. Nine N-GPs were analyzed in plasma samples from 64 patients with benign pancreatic cysts (benign group) and 93 patients with pancreatic adenocarcinoma (Table 1 and Supplementary Figure 3). While GP1-GP8 did not differ significantly between patients with benign disease and patients with PDAC, patient with PDAC samples displayed a markedly higher level of the GP9 signal (Figure 3A). Furthermore, as a control, we investigated the efficacy of tumor markers in distinguishing between malignancies of pancreatic origin and benign pancreatic cysts. We found that CA125, CA19-9, and CA242 could dis
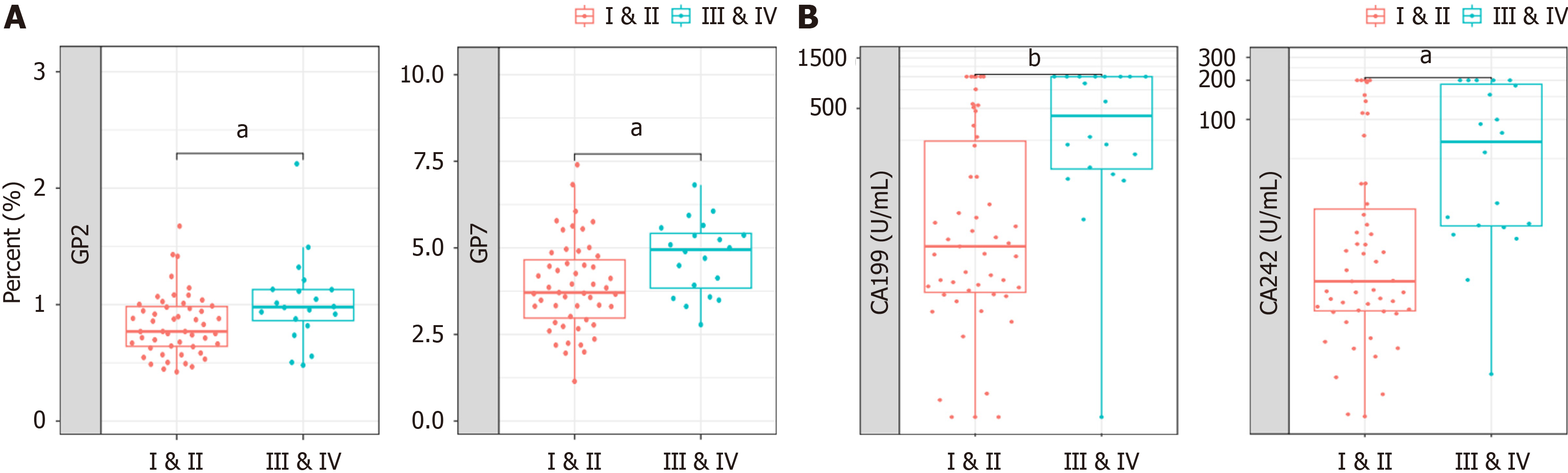
To distinguish early-stage PC, the potential of GPs as diagnostic markers in patient serum was verified in a 71-patient cohort. The participants’ clinical information regarding age and sex is shown in Supplementary Figure 4. We conducted GPs analyses in blood samples of patients with early stages I-II and advanced clinical stages III-IV to confirm the expression pattern of N-glycans. Both GP2 and GP7 distinguished patients with early- from late-stage (Figure 4A). We also compared the ability of tumor biomarkers to differentiate between patients with early-stage and advanced PDAC. The levels of CA19-9 and CA 242 were capable of distinguishing between patients with early- and late-stage PDAC (Figure 4B). This result implies that N-glycans GP2 and GP7 could be used as biomarkers to differentiate early-stage (I-II) from advanced PDAC (III-IV).
A multivariate logistic regression analysis was performed to build a diagnostic model using GP1, GP2, GP3, GP4, GP5, GP6, GP7and GP9. The diagnostic formula is as follows: Glyco-model = exp(10.696 + 11.368 × GP1 + 121.372 × GP2 - 46.884 × GP3 -66.918 × GP4 - 15.329 × GP5 - 21.862 × GP6 + 1.177 × GP7 + 47.976 × GP9)/1 + exp(10.696 + 11.368 × GP1 + 121.372 × GP2 - 46.884 × GP3 - 66.918 × GP4 - 15.329 × GP5 - 21.862 × GP6 + 1.177 × GP7 + 47.976 × GP9).
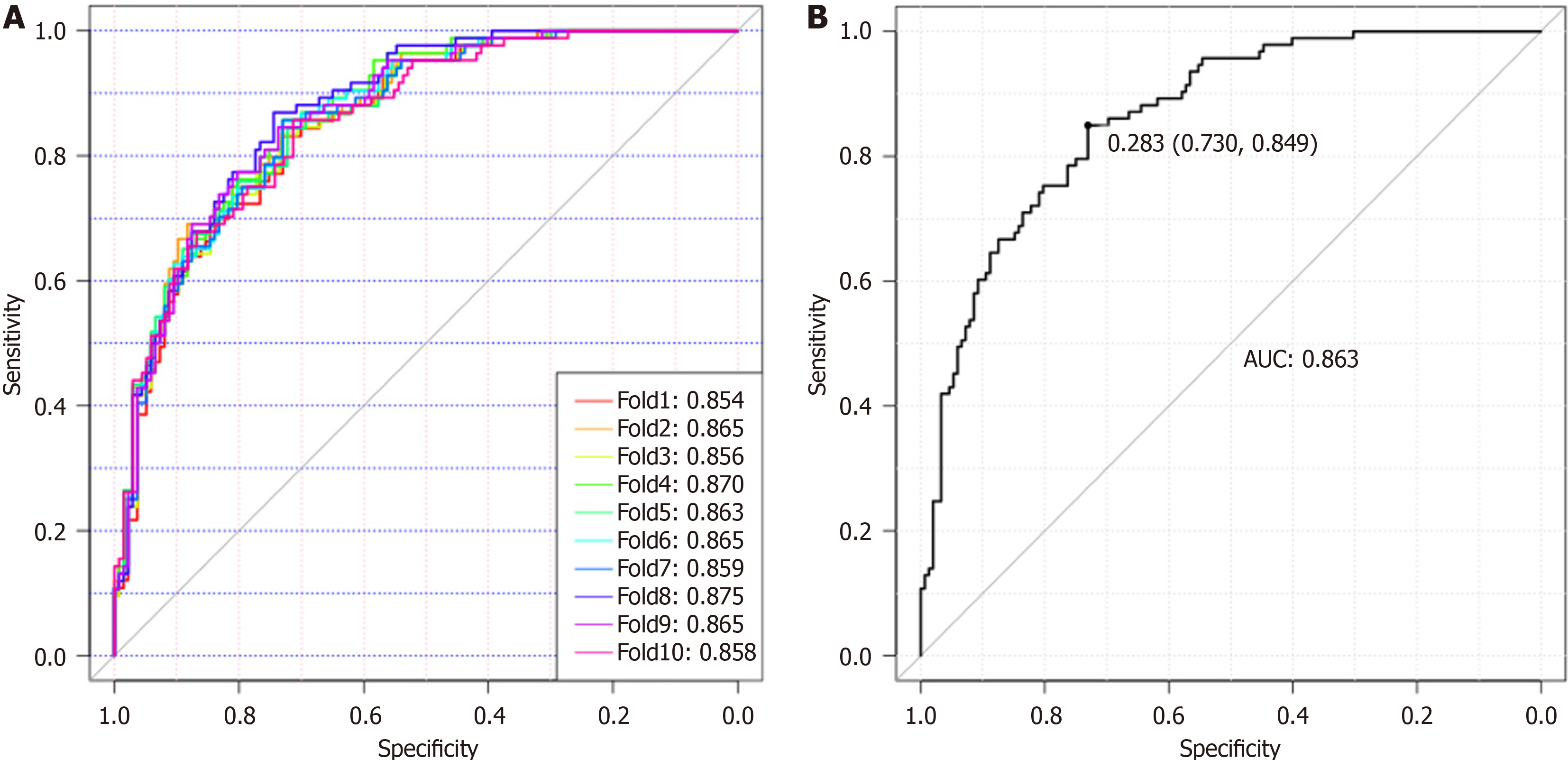
Eighty-eight normal samples, 64 non-PDAC, and 93 PDAC samples were used to construct and validate a diagnostic model for discriminating PDAC from non-PDAC and healthy individuals. To reduce the negative influence of overfitting on the PDAC predictive power, we performed a 10-fold cross-validation on the modeling training set. The training set was further divided into ten folds, of which nine were used for modeling and the remaining were used for validation. Finally, the AUC values representing the optimal diagnostic power from one of the ten-fold cross-validations, AUCROC, were 0.854-0.875 (Figure 5A). Using a cut-off value of 0.28, this model achieved good diagnostic efficiency with a validation AUROC of 0.863 (sensitivity, 84.90%; specificity, 73.00%), indicating that this Glyco-model is a promising method for detecting patients with PDAC (Figure 5B). A comparative analysis was performed to evaluate the diagnostic efficacy of the Glyco-model and tumor markers in discriminating patients with PDAC from those of non-PDAC and healthy individuals, as illustrated in Supplementary Figure 5. The results revealed that N-GP biomarkers had a significantly higher diagnostic AUROC than the tumor markers (Supplementary Figure 5 and Table 2). The concomitant utilization of N-glycans and tumor markers can substantially augment the diagnostic precision for PDAC, as shown in Supplementary Table 1. The combined use of N-glycans and CA19-9 to diagnose PC yielded an AUC value exceeding 0.919.
| Biomarker | AUC | Cut-off | Sensitivity | Specificity |
| N-glycan | 0.86 | 0.28 | 84.90% | 73.00% |
| CA19-9 | 0.75 | 37 | 55.91% | 95.49% |
| CEA | 0.72 | 5.0 | 22.58% | 100.00% |
| CA125 | 0.76 | 35 | 32.26% | 98.10% |
| CA242 | 0.67 | 30 | 30.11% | 98.41% |
| CA724 | 0.56 | 6.9 | 20.43% | 87.30% |
The potential clinical value of the proposed model is evaluated based on its diagnostic performance. We then compared the diagnostic performance of the Glyco-model at different clinical stages of PDAC. The results showed that the Glyco-model demonstrated favorable diagnostic performance in all PC stages, with sensitivities ranging from 77.27%-90.00% (Table 3). The diagnostic sensitivity for patients with stage I PDAC was 89.66%. These results show that the Glyco-model can achieve high diagnostic ability and potentially benefit patients by enabling an early diagnosis of PC.
| Subgroup | Sensitivity |
| Overall | 84.95% (79/93) |
| Stage I | 89.66% (26/29) |
| Stage II | 77.27% (17/22) |
| Stage III & IV | 90.00% (18/20) |
| Unknown | 81.82% (18/22) |
In addition, we have conducted a comparative analysis of the diagnostic efficacy of the model across diverse non-PDAC (healthy control and patients with chronic pancreatitis) cohorts to verify the accuracy and sensitivity of the model. The N-glycan biomarker yielded a specificity of 79.6% for distinguishing patients with PC from healthy controls (Table 4).
| Subgroup | Specificity |
| Overall | 73.03% (111/152) |
| Healthy | 79.55% (70/88) |
| SPN | 83.33% (10/12) |
| PNET | 50.00% (5/10) |
| SCN | 91.67% (11/12) |
| IPMN | 50.00% (13/26) |
| MCN | 50.00% (2/4) |
We then examined the positivity rate for N-glycan (GP-9) in patients with PDAC who tested negative for various tumor markers. Patients with PDAC who tested negative for each tumor marker showed a higher positive rate of N-glycans (Table 5). The sensitivity of N-glycan features exceeded 80%. This suggests that the inclusion of N-glycans in the negative detection of tumor markers may decrease the rate of missed diagnoses in clinical settings.
| Tumor marker negative | Glyco-model positivity rate |
| CEA ≤ 5 (72) | 87.50% (63/72) |
| CA19-9 ≤ 37 (41) | 82.93% (34/41) |
| CA125 ≤ 35 (63) | 88.89% (56/63) |
| CA242 ≤ 30 (65) | 83.08% (54/65) |
| CA724 ≤ 6.9 (74) | 86.49% (64/74) |
The primary finding of this study was that N-glycome analysis can differentiate individuals with early-stage (stages I and II) PDAC from the control group. The utilization of such a test in the monitoring of: (1) High-risk patients, including those with hereditary PDAC, PC, etc.; (2) Patients with late-onset diabetes who have an elevated risk of developing PDAC within the first 3 years of diabetes; and (3) Patients with ambiguous abdominal symptoms may have clinical benefits. The World Health Organization posits that a significant number of patients with cancer can be spared from untimely mor
Differentiating PDAC from pancreatitis can pose a challenge; however, the current investigation demonstrates that the N-glycan analysis model effectively discriminates PDAC from various pancreatic inflammatory conditions. Currently, a comprehensive biological rationale for the use of N-glycans as PC biomarkers remains elusive. However, cancer pro
Caution is advised when interpreting our findings because of several limitations of the study design. First, N-glycan marker ascertainment was developed through case-control studies, thereby precluding the knowledge of its efficacy in a surveillance or therapeutic context until a prospective validation study was conducted. Second, because all patient samples were obtained at diagnosis, the behavior of N-glycan markers in patients following surgical tumor removal remains unpredictable. Furthermore, it should be noted that our study included individuals with an established disease status in both the patient and control groups. Although the obtained AUC values were elevated, it is imperative to acknowledge that this does not necessarily translate to an equivalent performance of the marker in pre-diagnostic samples.
In summary, we conducted a comprehensive case-control study of patients with PDAC, resulting in the identification and validation of an N-glycan biomarker signature. These results indicate that this biomarker signature exhibited a high level of accuracy in detecting blood samples from patients with stage I and II PDAC.
The absence of diagnostic biomarkers for pancreatic cancer (PC) poses challenges in achieving early detection.
The aim of this study is to identify novel diagnostic markers for the early detection of PC.
The identification of novel glycan markers holds the potential to differentiate early-stage PC, while the development of corresponding models can facilitate the early diagnosis of PC as well as other pancreatic ailments.
Serum N-glycan analysis performed to identify the serum biomarker signatures associated with early-stage pancreatic ductal adenocarcinoma (PDAC). A multivariate logistic regression analysis was performed to build a diagnostic model.
The biomarker signature was created to discriminate samples derived from patients with PC from those of controls. Glyco-model demonstrated favorable diagnostic performance in all stages of PC.
The serum N-glycan biosignatures and the “Glyco-model” offer a viable method for detecting early-stage PDAC.
There is a desire to develop additional biomarkers that exhibit heightened sensitivity and specificity for PDAC, in order to facilitate early detection.
Provenance and peer review: Unsolicited article; Externally peer reviewed.
Peer-review model: Single blind
Specialty type: Oncology
Country/Territory of origin: China
Peer-review report’s scientific quality classification
Grade A (Excellent): 0
Grade B (Very good): 0
Grade C (Good): C
Grade D (Fair): 0
Grade E (Poor): 0
P-Reviewer: Sanyal D, India S-Editor: Wang JJ L-Editor: A P-Editor: Xu ZH
| 1. | Ying H, Dey P, Yao W, Kimmelman AC, Draetta GF, Maitra A, DePinho RA. Genetics and biology of pancreatic ductal adenocarcinoma. Genes Dev. 2016;30:355-385. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 301] [Cited by in RCA: 397] [Article Influence: 44.1] [Reference Citation Analysis (0)] |
| 2. | Siegel RL, Miller KD, Fuchs HE, Jemal A. Cancer statistics, 2022. CA Cancer J Clin. 2022;72:7-33. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 4235] [Cited by in RCA: 11365] [Article Influence: 3788.3] [Reference Citation Analysis (4)] |
| 3. | Siegel RL, Miller KD, Wagle NS, Jemal A. Cancer statistics, 2023. CA Cancer J Clin. 2023;73:17-48. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 116] [Cited by in RCA: 9819] [Article Influence: 4909.5] [Reference Citation Analysis (1)] |
| 4. | Walter FM, Mills K, Mendonça SC, Abel GA, Basu B, Carroll N, Ballard S, Lancaster J, Hamilton W, Rubin GP, Emery JD. Symptoms and patient factors associated with diagnostic intervals for pancreatic cancer (SYMPTOM pancreatic study): a prospective cohort study. Lancet Gastroenterol Hepatol. 2016;1:298-306. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 61] [Cited by in RCA: 107] [Article Influence: 11.9] [Reference Citation Analysis (0)] |
| 5. | Huang B, Huang H, Zhang S, Zhang D, Shi Q, Liu J, Guo J. Artificial intelligence in pancreatic cancer. Theranostics. 2022;12:6931-6954. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 46] [Cited by in RCA: 52] [Article Influence: 17.3] [Reference Citation Analysis (0)] |
| 6. | Kaur S, Smith LM, Patel A, Menning M, Watley DC, Malik SS, Krishn SR, Mallya K, Aithal A, Sasson AR, Johansson SL, Jain M, Singh S, Guha S, Are C, Raimondo M, Hollingsworth MA, Brand RE, Batra SK. A Combination of MUC5AC and CA19-9 Improves the Diagnosis of Pancreatic Cancer: A Multicenter Study. Am J Gastroenterol. 2017;112:172-183. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 119] [Cited by in RCA: 118] [Article Influence: 14.8] [Reference Citation Analysis (0)] |
| 7. | Gerdtsson AS, Wingren C, Persson H, Delfani P, Nordström M, Ren H, Wen X, Ringdahl U, Borrebaeck CA, Hao J. Plasma protein profiling in a stage defined pancreatic cancer cohort - Implications for early diagnosis. Mol Oncol. 2016;10:1305-1316. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 19] [Cited by in RCA: 24] [Article Influence: 2.7] [Reference Citation Analysis (0)] |
| 8. | Okano K, Suzuki Y. Strategies for early detection of resectable pancreatic cancer. World J Gastroenterol. 2014;20:11230-11240. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in CrossRef: 32] [Cited by in RCA: 36] [Article Influence: 3.3] [Reference Citation Analysis (0)] |
| 9. | Locker GY, Hamilton S, Harris J, Jessup JM, Kemeny N, Macdonald JS, Somerfield MR, Hayes DF, Bast RC Jr; ASCO. ASCO 2006 update of recommendations for the use of tumor markers in gastrointestinal cancer. J Clin Oncol. 2006;24:5313-5327. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1057] [Cited by in RCA: 1110] [Article Influence: 58.4] [Reference Citation Analysis (0)] |
| 10. | Galli C, Basso D, Plebani M. CA 19-9: handle with care. Clin Chem Lab Med. 2013;51:1369-1383. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 56] [Cited by in RCA: 66] [Article Influence: 6.0] [Reference Citation Analysis (0)] |
| 11. | Borrebaeck CA. Precision diagnostics: moving towards protein biomarker signatures of clinical utility in cancer. Nat Rev Cancer. 2017;17:199-204. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 253] [Cited by in RCA: 288] [Article Influence: 36.0] [Reference Citation Analysis (0)] |
| 12. | Hanash SM, Pitteri SJ, Faca VM. Mining the plasma proteome for cancer biomarkers. Nature. 2008;452:571-579. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 638] [Cited by in RCA: 650] [Article Influence: 38.2] [Reference Citation Analysis (0)] |
| 13. | Brand RE, Nolen BM, Zeh HJ, Allen PJ, Eloubeidi MA, Goldberg M, Elton E, Arnoletti JP, Christein JD, Vickers SM, Langmead CJ, Landsittel DP, Whitcomb DC, Grizzle WE, Lokshin AE. Serum biomarker panels for the detection of pancreatic cancer. Clin Cancer Res. 2011;17:805-816. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 192] [Cited by in RCA: 190] [Article Influence: 13.6] [Reference Citation Analysis (0)] |
| 14. | Kim J, Bamlet WR, Oberg AL, Chaffee KG, Donahue G, Cao XJ, Chari S, Garcia BA, Petersen GM, Zaret KS. Detection of early pancreatic ductal adenocarcinoma with thrombospondin-2 and CA19-9 blood markers. Sci Transl Med. 2017;9. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 147] [Cited by in RCA: 187] [Article Influence: 26.7] [Reference Citation Analysis (0)] |
| 15. | Wingren C, Sandström A, Segersvärd R, Carlsson A, Andersson R, Löhr M, Borrebaeck CA. Identification of serum biomarker signatures associated with pancreatic cancer. Cancer Res. 2012;72:2481-2490. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 92] [Cited by in RCA: 84] [Article Influence: 6.5] [Reference Citation Analysis (0)] |
| 16. | Omiya K, Oba A, Inoue Y, Kobayashi K, Wu YHA, Ono Y, Sato T, Sasaki T, Ozaka M, Sasahira N, Ito H, Saiura A, Takahashi Y. Serum DUPAN-2 could be an Alternative Biological Marker for CA19-9 Nonsecretors with Pancreatic Cancer. Ann Surg. 2023;277:e1278-e1283. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 9] [Cited by in RCA: 25] [Article Influence: 12.5] [Reference Citation Analysis (0)] |
| 17. | Zhang Y, Yang C, Cheng H, Fan Z, Huang Q, Lu Y, Fan K, Luo G, Jin K, Wang Z, Liu C, Yu X. Novel agents for pancreatic ductal adenocarcinoma: emerging therapeutics and future directions. J Hematol Oncol. 2018;11:14. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 30] [Cited by in RCA: 31] [Article Influence: 4.4] [Reference Citation Analysis (0)] |
| 18. | Agrawal P, Fontanals-Cirera B, Sokolova E, Jacob S, Vaiana CA, Argibay D, Davalos V, McDermott M, Nayak S, Darvishian F, Castillo M, Ueberheide B, Osman I, Fenyö D, Mahal LK, Hernando E. A Systems Biology Approach Identifies FUT8 as a Driver of Melanoma Metastasis. Cancer Cell. 2017;31:804-819.e7. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 179] [Cited by in RCA: 240] [Article Influence: 30.0] [Reference Citation Analysis (0)] |
| 19. | Song M, Bai H, Zhang P, Zhou X, Ying B. Promising applications of human-derived saliva biomarker testing in clinical diagnostics. Int J Oral Sci. 2023;15:2. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in RCA: 90] [Reference Citation Analysis (0)] |
| 20. | Vajaria BN, Patel PS. Glycosylation: a hallmark of cancer? Glycoconj J. 2017;34:147-156. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 151] [Cited by in RCA: 211] [Article Influence: 23.4] [Reference Citation Analysis (0)] |
| 21. | Kyselova Z, Mechref Y, Al Bataineh MM, Dobrolecki LE, Hickey RJ, Vinson J, Sweeney CJ, Novotny MV. Alterations in the serum glycome due to metastatic prostate cancer. J Proteome Res. 2007;6:1822-1832. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 184] [Cited by in RCA: 192] [Article Influence: 10.7] [Reference Citation Analysis (0)] |
| 22. | Abbott KL, Nairn AV, Hall EM, Horton MB, McDonald JF, Moremen KW, Dinulescu DM, Pierce M. Focused glycomic analysis of the N-linked glycan biosynthetic pathway in ovarian cancer. Proteomics. 2008;8:3210-3220. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 86] [Cited by in RCA: 91] [Article Influence: 5.4] [Reference Citation Analysis (0)] |
| 23. | Drake RR. Glycosylation and cancer: moving glycomics to the forefront. Adv Cancer Res. 2015;126:1-10. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 47] [Cited by in RCA: 59] [Article Influence: 5.9] [Reference Citation Analysis (0)] |
| 24. | McDowell CT, Klamer Z, Hall J, West CA, Wisniewski L, Powers TW, Angel PM, Mehta AS, Lewin DN, Haab BB, Drake RR. Imaging Mass Spectrometry and Lectin Analysis of N-Linked Glycans in Carbohydrate Antigen-Defined Pancreatic Cancer Tissues. Mol Cell Proteomics. 2021;20:100012. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 53] [Cited by in RCA: 67] [Article Influence: 13.4] [Reference Citation Analysis (0)] |
| 25. | Vreeker GCM, Hanna-Sawires RG, Mohammed Y, Bladergroen MR, Nicolardi S, Dotz V, Nouta J, Bonsing BA, Mesker WE, van der Burgt YEM, Wuhrer M, Tollenaar RAEM. Serum N-Glycome analysis reveals pancreatic cancer disease signatures. Cancer Med. 2020;9:8519-8529. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 12] [Cited by in RCA: 28] [Article Influence: 5.6] [Reference Citation Analysis (0)] |
| 26. | Cong M, Ou X, Huang J, Long J, Li T, Liu X, Wang Y, Wu X, Zhou J, Sun Y, Shang Q, Chen G, Ma H, Xie W, Piao H, Yang Y, Gao Z, Xu X, Tan Z, Chen C, Zeng N, Wu S, Kong Y, Liu T, Wang P, You H, Jia J, Zhuang H. A Predictive Model Using N-Glycan Biosignatures for Clinical Diagnosis of Early Hepatocellular Carcinoma Related to Hepatitis B Virus. OMICS. 2020;24:415-423. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 5] [Cited by in RCA: 9] [Article Influence: 1.8] [Reference Citation Analysis (0)] |
| 27. | Bossuyt PM, Reitsma JB, Bruns DE, Gatsonis CA, Glasziou PP, Irwig L, Lijmer JG, Moher D, Rennie D, de Vet HC, Kressel HY, Rifai N, Golub RM, Altman DG, Hooft L, Korevaar DA, Cohen JF; STARD Group. STARD 2015: an updated list of essential items for reporting diagnostic accuracy studies. BMJ. 2015;351:h5527. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1702] [Cited by in RCA: 2089] [Article Influence: 208.9] [Reference Citation Analysis (0)] |