Published online Feb 15, 2024. doi: 10.4251/wjgo.v16.i2.475
Peer-review started: October 17, 2023
First decision: December 5, 2023
Revised: December 10, 2023
Accepted: January 8, 2024
Article in press: January 8, 2024
Published online: February 15, 2024
Processing time: 107 Days and 17.4 Hours
B56ε is a regulatory subunit of the serine/threonine protein phosphatase 2A, which is abnormally expressed in tumors and regulates various tumor cell functions. At present, the application of B56ε in pan-cancer lacks a comprehensive analysis, and its role and mechanism in hepatocellular carcinoma (HCC) are still unclear.
To analyze B56ε in pan-cancer, and explore its role and mechanism in HCC.
The Cancer Genome Atlas, Genotype-Tissue Expression, Gene Expression Profiling Interactive Analysis, and Tumor Immune Estimation Resource databases were used to analyze B56ε expression, prognostic mutations, somatic copy number alterations, and tumor immune characteristics in 33 tumors. The relationships between B56ε expression levels and drug sensitivity, immunotherapy, immune checkpoints, and human leukocyte antigen (HLA)-related genes were further analyzed. Gene Set Enrichment Analysis (GSEA) was performed to reveal the role of B56ε in HCC. The Cell Counting Kit-8, plate cloning, wound healing, and transwell assays were conducted to assess the effects of B56ε interference on the malignant behavior of HCC cells.
In most tumors, B56ε expression was upregulated, and high B56ε expression was a risk factor for adrenocortical cancer, HCC, pancreatic adenocarcinoma, and pheochromocytoma and paraganglioma (all P < 0.05). B56ε expression levels were correlated with a variety of immune cells, such as T helper 17 cells, B cells, and macro
B56ε is associated with the microenvironment, immune evasion, and immune cell infiltration of multiple tumors. B56ε plays an important role in HCC progression, supporting it as a prognostic marker and potential therapeutic target for HCC.
Core Tip: The expression of protein phosphatase 2A (PP2A) subunit B56ε is up-regulated in most tumors, and its high expression is a risk factor for adrenocortical cancer, hepatocellular carcinoma (HCC), pancreatic adenocarcinoma, and pheochromocytoma and paraganglioma. B56ε expression levels correlate with immune cells, immune checkpoint genes, human leukocyte antigen-related genes, and the sensitivity of chemotherapy drugs. In HCC, B56ε expression is related to the cancer pathway. Knockdown of B56ε expression in HCC cells can inhibit the proliferation, migration and invasion capacity of tumor cells. Our study supports PP2A subunit B56ε as a prognostic marker and potential therapeutic target for HCC.
- Citation: Wu HM, Huang YY, Xu YQ, Xiang WL, Yang C, Liu RY, Li D, Guo XF, Zhang ZB, Bei CH, Tan SK, Zhu XN. Comprehensive analysis of the protein phosphatase 2A regulatory subunit B56ε in pan-cancer and its role and mechanism in hepatocellular carcinoma. World J Gastrointest Oncol 2024; 16(2): 475-492
- URL: https://www.wjgnet.com/1948-5204/full/v16/i2/475.htm
- DOI: https://dx.doi.org/10.4251/wjgo.v16.i2.475
Hepatocellular carcinoma (HCC) is a global health challenge with a rising incidence. In the 2020 Global Cancer Statistics, HCC ranked sixth in the incidence and third in the mortality of cancers worldwide[1]. As the main histological type of liver cancer, HCC is the cause of the vast majority of liver cancer diagnoses and deaths[2,3]. The incidence of HCC varies by geography, with about 72% of cases occurring in Asia[4]. Its onset and progression are a multistep process associated with multiple risk factors, such as hepatitis B virus (HBV), hepatitis C virus, and the environment[5]. It is also controlled by genetic and epigenetic changes that inactivate tumor suppressor genes or activate oncogenes, ultimately leading to the occurrence of HCC[6]. Although after decades of exploration, we have some understanding of the molecular mechanism by which HCC occurs, the detailed pathogenesis is still poorly understood. Given the increasing rate of mortality from HCC worldwide, it is important to improve our understanding of the molecular mechanisms underlying the pathogenesis of HCC. Moreover, new diagnostic, prognostic biomarkers and therapeutic strategies are urgently needed to address this major public health issue.
Protein phosphatase 2A (PP2A) is a major serine/threonine phosphatase. It is involved in the feedback of multiple signaling pathways, affecting cell cycle progression, proliferation, transcription, and translation[7]. The PP2A holoenzyme complex contains scaffold subunit “A”, regulatory subunit “B”, and catalytic subunit “C”. The A and C subunits constitute the core enzyme, while the B subunit is responsible for regulating substrate specificity, cell loca
Recently, the B56 protein has become widely recognized and valued because of its role in the development of a variety of tumors such as melanoma[12], breast cancer[13], and prostate cancer[14]. The B56α (PPP2R5A), B56β (PPP2R5B), and B56ε (PPP2R5E) of the B56 subfamily have nuclear output signals at the C-terminus, resulting in the migration of the PP2A complex into the cytoplasm. B56γ (PPP2R5C) and B56δ (PPP2R5D) are mainly found in the nucleus because they lack the signal sequence at the C-terminus. One study shows that knockdown of B56γ could promote xenograft tumor growth and HBV-mediated migration and invasion of HCC cells in vivo[15]. Mice lacking B56δ spontaneously develop HCC, which is associated with increased carcinogenicity of c-Myc[16].
As a member of the regulatory subunit B56 subfamily, the role of B56ε in tumors has also attracted much attention. In a study of acute myeloid leukemia (AML), B56ε was shown to induce caspase-dependent apoptosis by impairing cell proliferation, affecting the activation state of AKT and reducing the colony formation capacity of leukemia cells[17]. A study also showed that B56ε can inhibit the growth of gastric cancer cells and induce cell apoptosis[18]. These results are consistent with its tumor suppressor properties in breast cancer[19] and human tongue squamous cell carcinoma[20]. Although B56ε has been studied in some cancers, the mechanism of action of B56ε in HCC is poorly understood.
To further explore the role and mechanism of B56ε in the development and progression of HCC, we conducted a combination of bioinformatics analyses and cell experiment validation in this study. The Cancer Genome Atlas (TCGA), Genotype-Tissue Expression (GTEx), Gene Expression Profiling Interactive Analysis (GEPIA), and Tumor Immune Estimation Resource (TIMER) databases were used to analyze B56ε expression, prognostic mutations, somatic copy number alterations (sCNAs), and tumor immune characteristics in 33 tumors. The relationships between B56ε expression levels and drug sensitivity, immunotherapy, immune checkpoints, and human leukocyte antigen (HLA)-related genes were further analyzed. Gene Set Enrichment Analysis (GSEA) was performed to reveal the role of B56ε in HCC. The Cell Counting Kit-8 (CCK-8), plate cloning, wound healing, and transwell assays were conducted to show the effects of B56ε interference on the malignant behaviors of HCC cells. The findings from our study will provide insight into the potential value of B56ε in the diagnosis, prognosis and treatment of HCC.
RNA sequencing (RNA-seq) data in Trusted Platform Module (TPM) format for TCGA and GTEx were downloaded from the online website UCSC XENA (https://xenabrowser.net/datapages/), which was uniformly processed by the Toil process. Data corresponding to TCGA for 33 tumors and normal tissue data corresponding to GTEx were extracted. The 33 tumors included adrenocortical carcinoma (ACC), bladder urothelial carcinoma, breast invasive carcinoma (BRCA), cervical squamous cell carcinoma (CESC), cholangiocarcinoma (CHOL), colon adenocarcinoma (COAD), lymphoid neoplasm diffuse large B cell lymphoma (DLBC), esophageal carcinoma (ESCA), glioblastoma (GBM), head and neck squamous cell carcinoma, kidney chromophobe (KICH), kidney clear cell carcinoma (KIRC), kidney renal papillary cell carcinoma (KIRP), AML, low-grade glioma (LGG), HCC, lung adenocarcinoma (LUAD), lung squamous cell carcinoma (LUSC), mesothelioma, ovarian serous cystadenoma (OV), pancreatic adenocarcinoma (PAAD), pheochromocytoma and paraganglioma (PCPG), prostate adenocarcinoma (PRAD), rectal adenocarcinoma, sarcoma, skin cutaneous melanoma (SKCM), stomach adenocarcinoma (STAD), testicular germ cell tumor (TGCT), thyroid carcinoma (THCA), thymic carcinoma (THYM), endometrial cancer (UCEC), uterine carcinosarcoma (UCS), and uveal melanoma (UVM). Statistical analyses were performed with R software (version 4.2.1).
The RNA-seq data of the STAR process of the TCGA-Liver HCC (LIHC) project was downloaded and collated from TCGA database (https://portal.gdc.cancer.gov) and extracted in TPM format. Data from the paracancerous and carcinoma samples with corresponding number pairs were extracted and statistical analysis was performed with R.
The expression of B56ε was analyzed in 33 tumors by the TIMER database (https://cistrome.shinyapps.io/timer/). RNA-seq expression data were statistically analyzed and visualized using R packages, ggplot2 (3.3.6), stats (4.2.1), and car. Expression profiles of B56ε protein levels in HCC and corresponding immunohistochemical images were obtained through an online Human Protein Atlas (HPA) (http://www.proteinatlas.org/) database.
The correlation between B56ε expression and survival in pan-carcinoma (http://dna1.bio.kyutech.ac.jp/PrognoScan/index.html) was analyzed. Univariate survival analysis was used to calculate the hazard ratio (HR) and 95% confidence interval of B56ε in 33 tumors, and the results are shown as forest plots. The prognostic value of B56ε expression in HCC was analyzed using GEPIA (http://gepia.cancer-pku.cn/). GEPIA is an interactive online platform that provides information on tumor samples from TCGA as well as normal sample information from TCGA and GTEx projects. The expression levels of B56ε in cancerous and non-cancerous tissues were divided into a B56ε high expression group and B56ε low expression group according to the median.
The association of B56ε expression with mutations and sCNAs in 33 tumors was first analyzed through the TIMER (http://cistrome.org/TIMER/) database. Second, the expression of B56ε was analyzed in six types of immune-infiltrating cells, namely B cells, CD4+ T cells, CD8+ T cells, neutrophils, macrophages, and dendritic cells (DCs). The relationship between B56ε expression levels and tumor purity was also determined. After a general analysis of immune cell types, the correlation between B56ε expression and multiple immunomarkers was analyzed to identify the potential subtypes of infiltrating immune cells. In addition, the relationship between B56ε expression and immune checkpoint genes/HLA-associated genes were determined. Single-sample GSEA (ssGSEA), TIMER, and ESTIMATE were used to analyze the differences between tumor infiltrating immune cells and B56ε expression levels. Drug susceptibility data were obtained from CellMiner with a screening criterion of P < 0.05 and correlation analysis was conducted using the Pearson’s test. The above data were statistically analyzed and the results were visualized in the form of box plots, heat maps, scatter plots, violin plots, or stick charts using the ggplot2 (3.3.6), stats (4.2.1), and car packages. The role of B56ε expression in HCC immunity was explored through the TISIDB (http://cis.hku.hk/TISIDB) website.
Based on “c2.cp.all.v2022.1.Hs.symbols.gmt [All Canonical Pathways] (3050)”, GSEA was performed from both Gene Ontology (GO) and Kyoto Encyclopedia of Genes and Genomes (KEGG) dimensions to explore the potential biological function of B56ε expression in HCC. The molecules of the input data were wrapped with R “org. Hs.eg.db” after ID conversion, then the “clusterProfile” package was used for GSEA analysis, and finally the “ggplot2” package was used to visualize the results.
The human hepatic cell line L02 and two types of human HCC cell lines (Hep3B and SK-Hep-1) obtained from the Cell Bank of the Chinese Academy of Sciences (Shanghai, China) were maintained in RPMI 1640, Dulbecco’s Modified Eagle Medium, and Minimum Essential Medium (Gibco, El Paso, TX, United States) with 10% fetal bovine serum (FBS) (OriCell, Guangzhou, China). Cells were cultured in a humidified incubator at 37 °C and 5% CO2.
Hep3B and SK-Hep-1 cells were digested and seeded in 6-well plates after the logarithmic growth phase was reached. When the degree of cell confluency reached 80%, transfection was performed using Lipofectamine®2000 (Invitrogen, Carlsbad, CA, United States) according to the manufacturer’s protocol.
The total RNA of cells was extracted with Trizol reagent and reverse transcribed into cDNA using MonScriptTM RTIII All-in One Mix (Monad, Shanghai, China) with dsDNase. GAPDH served as the internal reference control to quantify the relative expression of the B56ε, which was calculated using the 2-ΔΔCt method. The B56ε primer sequences were as follows: Forward primer of 5’-GACTTCATGGACACGCTAT-3’ and reverse primer of 5’-CATTCTAACTACTTCAGGGTAA-3’. The GAPDH primer sequences were as follows: forward primer of 5’-ACAACTTTGGTATCGTGGAAGG-3’ and reverse primer of 5’- GCCATCACGCCACAGTTTC-3’.
Total cell protein was extracted using lysis buffer [10 μL PMSF, 1 mL RIPA Histiocytes Lysate Buffer (strong), 10 μL protein phosphatase inhibitor cocktail]. The protein concentration was measured using a BCA protein quantitation kit (Epizyme, Shanghai, China). The following primary antibodies were used: B56ε primary antibody (1:500, PA5-118186; Invitrogen); and GAPDH primary antibody (1:10000, PR30011; Proteintech, Shanghai, China).
The CCK-8 (MedChemExpress, Shanghai, China) was used to measure cell proliferation. Cells were digested and seeded in 96-well plates. After 12 h, 24 h, 48 h, and 72 h, 10 μL CCK-8 was added to each well and cultured in the incubator for 2 h. Then, a microplate reader was used to detect the optical density value at 450 nm.
A total of 3000 cells were seeded in 6-well plate for plate cloning and cultured for 10-15 d. When the size of the cell colony was suitable, the cells were fixed in 4% neutral paraformaldehyde solution, stained with 1% crystal violet, and finally photographed for calculating.
After cell transfection for 8 h in 6-well plates, cells were scratched vertically with a 10 μL tip, washed with phosphate-buffered saline, and then covered in serum-free medium for culturing. Pictures were taken at 0 h and 48 h with a light microscope, and the documented wound-healing areas were analyzed using ImageJ software.
HCC cell migration and invasion were examined using the transwell chambers (jetbiofil, Guangzhou, China). Cells were digested 48 h after transfection, counted, and prepared in cell suspension with serum-free medium. 600 μL of 10% FBS medium was added to a 24-well plate, and then the chamber was put in the plate with 200 μL cells. After 48 h, the cells were fixed with 4% formaldehyde and stained with 1% crystal violet. Five random fields were taken under the microscope and cells passing through the chamber were calculated.
SPSS 26.0 statistical software (IBM Corp., Armonk, NY, United States) was used to analyze the data. R software (version 4.2.1) and GraphPad Prism 9 software were used for drawing graphics. The Wilcoxon test was used for immune checkpoint gene and HLA-related gene analyses. Drug sensitivity was analyzed using Pearson’s correlation coefficient. P < 0.05 was considered statistically significant.
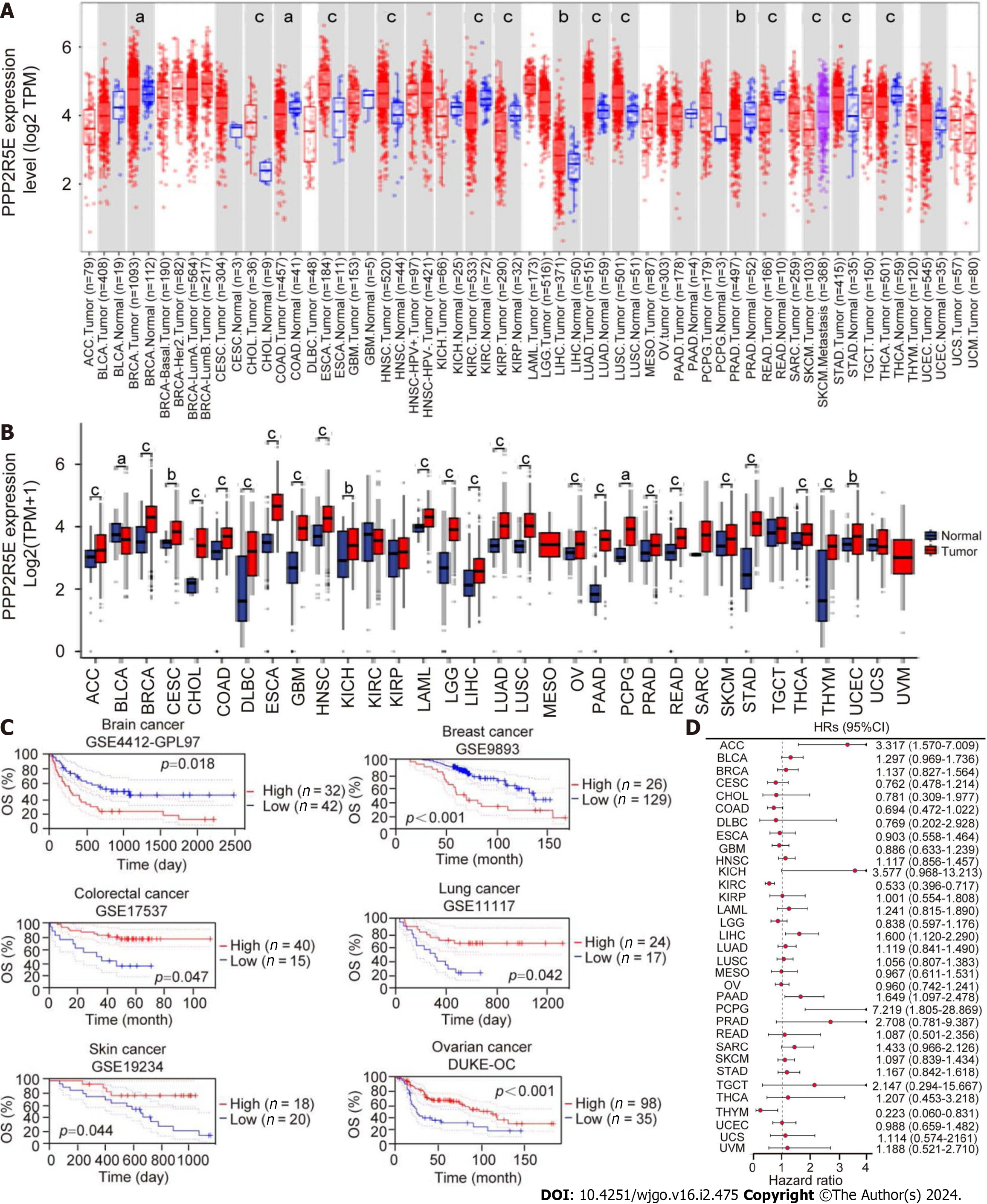
The expression of B56ε in pan-cancer was analyzed using the TIMER database. For tumors such as BRCA, CHOL, ESCA, HNSC, LIHC, LUSC, LUAD, and STAD, B56ε was expressed higher in the tumor tissues than in the normal tissues (Figure 1A). Data analysis combining TCGA and GTEx databases showed that B56ε was not only highly expressed in the above eight tumor types but also in sixteen other tumor types, including ACC, CESC, DLBC, GBM, KICH, LAML, LGG, OV, PAAD, PCPG, PRAD, READ, SKCM, THCA, THYM, and UCEC (Figure 1B). The PrognoScan online website was used to analyze the relationship between B56ε expression and prognosis of tumor patients in the Gene Expression Omnibus dataset. As shown in Figure 1C, the high expression of B56ε was associated with a poor prognosis in patients with brain or breast cancer, while the low expression of B56ε was associated with a poor prognosis in patients with colorectal, lung, skin, or ovarian cancer. Univariate analysis was used to analyze the prognostic significance of B56ε expression in 33 tumors. The results of the forest plot showed that the high expression of B56ε was a risk factor for ACC, LIHC, PAAD, and PCPG (all HR > 1, P < 0.05), but for KIRC and THYM, the high expression of B56ε was a protective factor (all HR < 1, P < 0.05) (Figure 1D). These results indicate that B56ε has a deregulated expression in tumors and its expression is correlated with the prognosis of tumor patients.
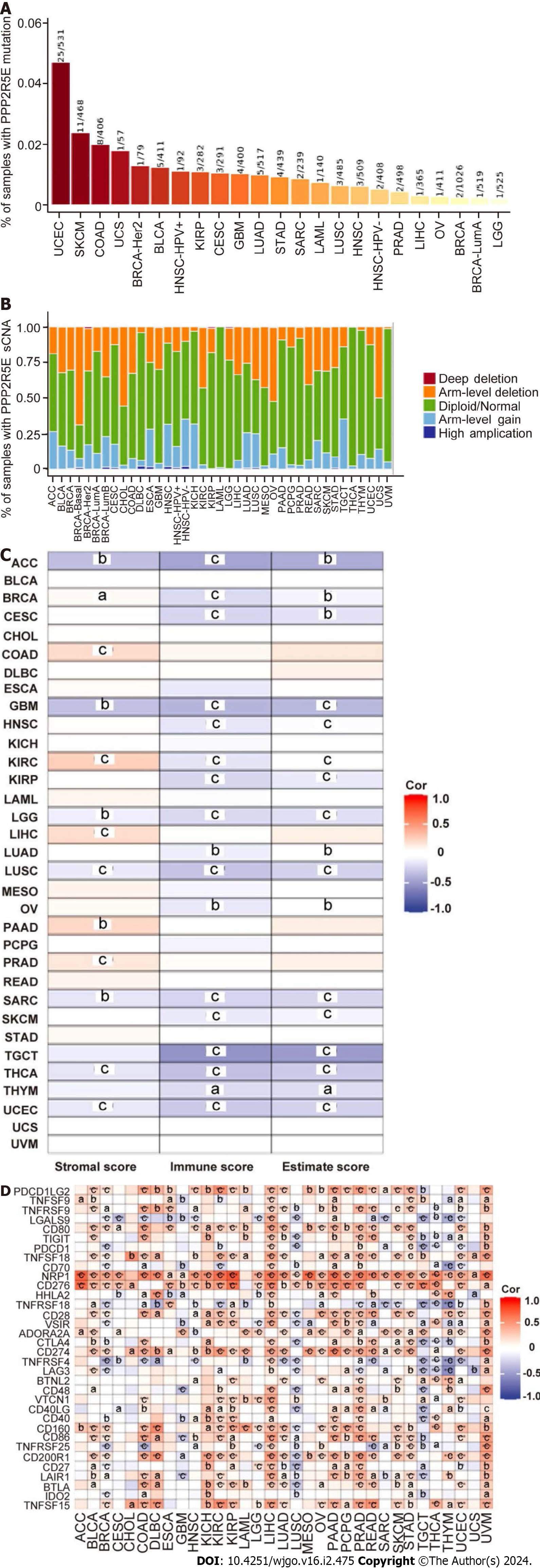
To understand the role of B56ε expression in tumors, we first analyzed the association of B56ε expression with mutations and sCNAs in 33 tumors through the TIMER database. It was found that diploid, arm-level deletion and arm-level gain were common mutation types in most cancers, accounting for a high proportion. The top four tumors with the highest mutation ratio were UCEC, SKCM, COAD, and UCS (Figures 2A and B). Second, the relationship between B56ε expression and immune status showed that B56ε expression was negatively correlated with estimated score, immune score, and stromal score (Figure 2C). In TGCT, the correlations between B56ε expression and immune score (r = -0.537) and estimated score (r = -0.494) were the strongest (all P < 0.001). In ACC, the associations between B56ε expression and stromal score (r = -0.338), immune score (r = -0.455) and estimated score (r = -0.423) were also relatively strong (all P < 0.001). Finally, more than 30 common immune checkpoint genes were collected to analyze the correlation with B56ε expression. As shown in Figure 2D, there was a positive relationship between the expression levels of B56ε and several immune checkpoint genes in some tumors such as KICH, KIRP, LIHC, PAAD, and UVM. These results suggest that B56ε expression might play a role in tumors by regulating tumor mutation, sCNAs, or immunity.
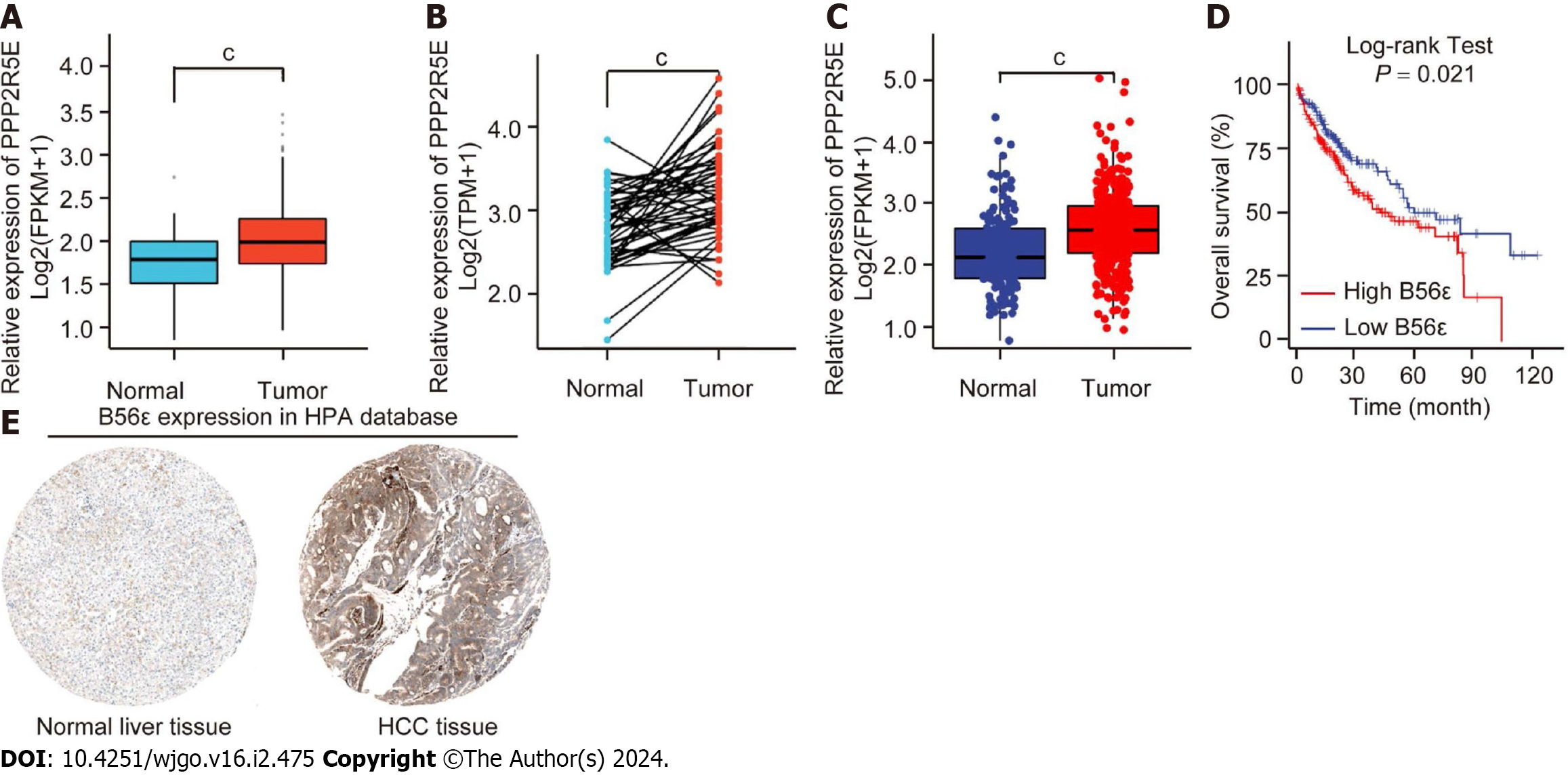
To identify the expression of B56ε in HCC tissues, we downloaded liver cancer-related data from TCGA database for bioinformatics analysis (tumor = 374, normal = 50). It was found that B56ε was highly expressed in HCC tissues compared to normal liver tissues (P < 0.001; Figures 3A and B). TCGA database combined with GTEx database was used to further analyze the expression of B56ε in HCC tissues (tumor = 371, normal = 160). As shown in Figure 3C, it was consistent with the results of TCGA database analysis. The prognostic significance of B56ε in HCC patients was analyzed using the GEPIA database. The results showed that the expression of B56ε was negatively correlated with the overall survival (OS) of patients with HCC, indicating that the higher the expression level of B56ε, the worse the prognosis of HCC patients (P < 0.05; Figure 3D).
Further analysis of the differential protein expression of B56ε between normal liver tissues and HCC tissues was conducted in the HPA database. It was found that B56ε stained weakly in normal liver tissue samples, but had deeper staining in HCC tissues (Figure 3E). Univariate analysis was used to analyze the association of common pathological features with OS in HCC patients. The results showed that the differential protein expression of B56ε was statistically significant with patient OS (P < 0.05). Further inclusion of variables in the multivariate regression model analysis found that the correlation was not significant (Table 1). These results indicate that B56ε is highly expressed in HCC tissues and correlates with a poor prognosis in HCC patients.
| Variables | n | Univariate analysis | Multivariate analysis | ||
| HR (95%CI) | P value | HR (95%CI) | P value | ||
| Gender | 373 | 1.261 (0.885-1.796) | 0.204 | 1.052 (0.632-1.752) | 0.845 |
| Age | 373 | 1.205 (0.850-1.708) | 0.293 | 1.238 (0.768-1.997) | 0.381 |
| T | 370 | 2.598 (1.826-3.697) | < 0.001a | 8.023 (0.461-139.715) | 0.153 |
| N | 258 | 2.029 (0.497-8.281) | 0.375 | 4.476 (0.583-34.348) | 0.149 |
| M | 272 | 4.077 (1.281-12.973) | 0.050 | 1.865 (0.546-6.367) | 0.320 |
| Stage | 349 | 2.504 (1.727-3.631) | < 0.001a | 0.357 (0.020-6.488) | 0.487 |
| Grade | 368 | 1.091 (0.761-1.564) | 0.637 | 1.177 (0.735-1.885) | 0.498 |
| B56ε | 373 | 1.587 (1.120-2.249) | 0.009a | 1.337 (0.829-2.156) | 0.234 |
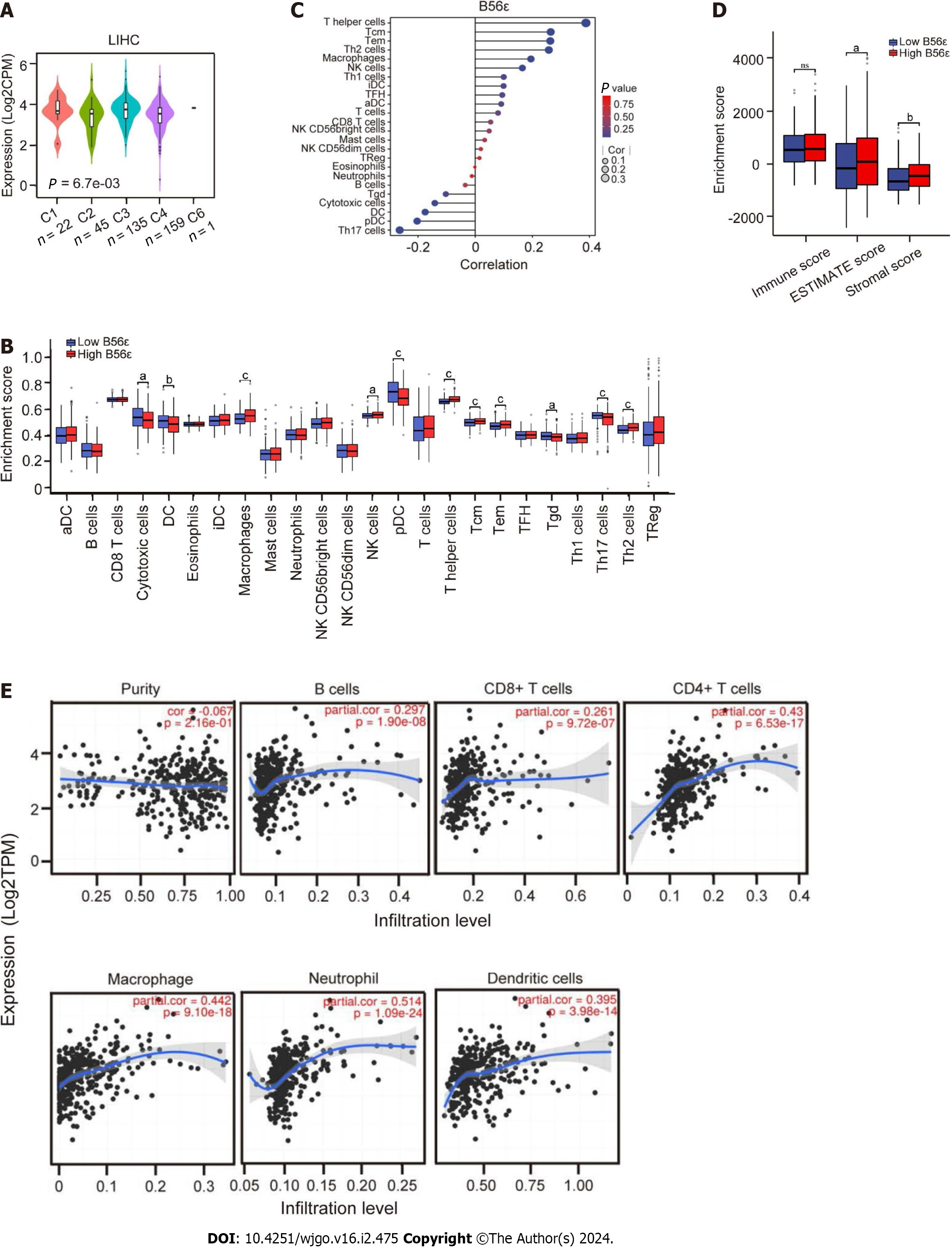
The role of B56ε expression in HCC immune subtypes was explored using the TISIDB website. Immune subtypes were divided into the following six types: C1 (wound healing), C2 (interferon gamma dominant), C3 (inflammatory), C4 (lymphocyte depleted), C5 (immunologically quiet), and C6 (transforming growth factor β dominant). As can be seen from Figure 4A, B56ε exhibited high expression in the C3 type and lowest expression in the C2 type. The immune-related function and immune infiltration of B56ε in HCC were further analyzed by the ssGSEA algorithm. The box plot in Figure 4B shows that the expression levels of macrophages, T helper (Th) cells, central memory T cells (TCM), effector memory cells (TEM), natural killer (NK) cells, and Th2 cells were significantly higher in the B56ε high expression group than in the B56ε low expression group. The opposite was true for Th17 cells, gamma delta T cells (Tgd), plasmacytoid DCs (pDCs), DCs, and cytotoxic cells. Figure 4C shows that a variety of immune cells were associated with B56ε expression including Th17 cells (r = -0.264, P < 0.001), pDCs (r = -0.203, P < 0.001), DCs (r = -0.175, P < 0.001), cytotoxic cells (r = -0.141, P < 0.01), Tgd (r = -0.103, P < 0.05), Th cells (r = 0.388, P < 0.001), TCM (r = 0.264, P < 0.001), TEM (r = 0.263, P < 0.001), Th2 cells (r = 0.258, P < 0.001), macrophages (r = 0.195, P < 0.001), and NK cells (r = 0.165, P < 0.01).
The relationship between B56ε expression and the tumor microenvironment (TME) was further analyzed by ESTIMATE, immune, and stromal score algorithms. As shown in Figure 4D, the ESTIMATE score and stromal score were significantly higher in the B56ε high expression group than in the B56ε low expression group (P < 0.05). The relationship between B56ε expression and six common immune cells were analyzed using the TIMER database. Figure 4E shows that B56ε expression was positively correlated with B cells, CD8+ T cells, CD4+ T cells, macrophages, neutrophils, and DCs (all P < 0.05). These results suggest that the expression of B56ε may have functions in HCC immunity.
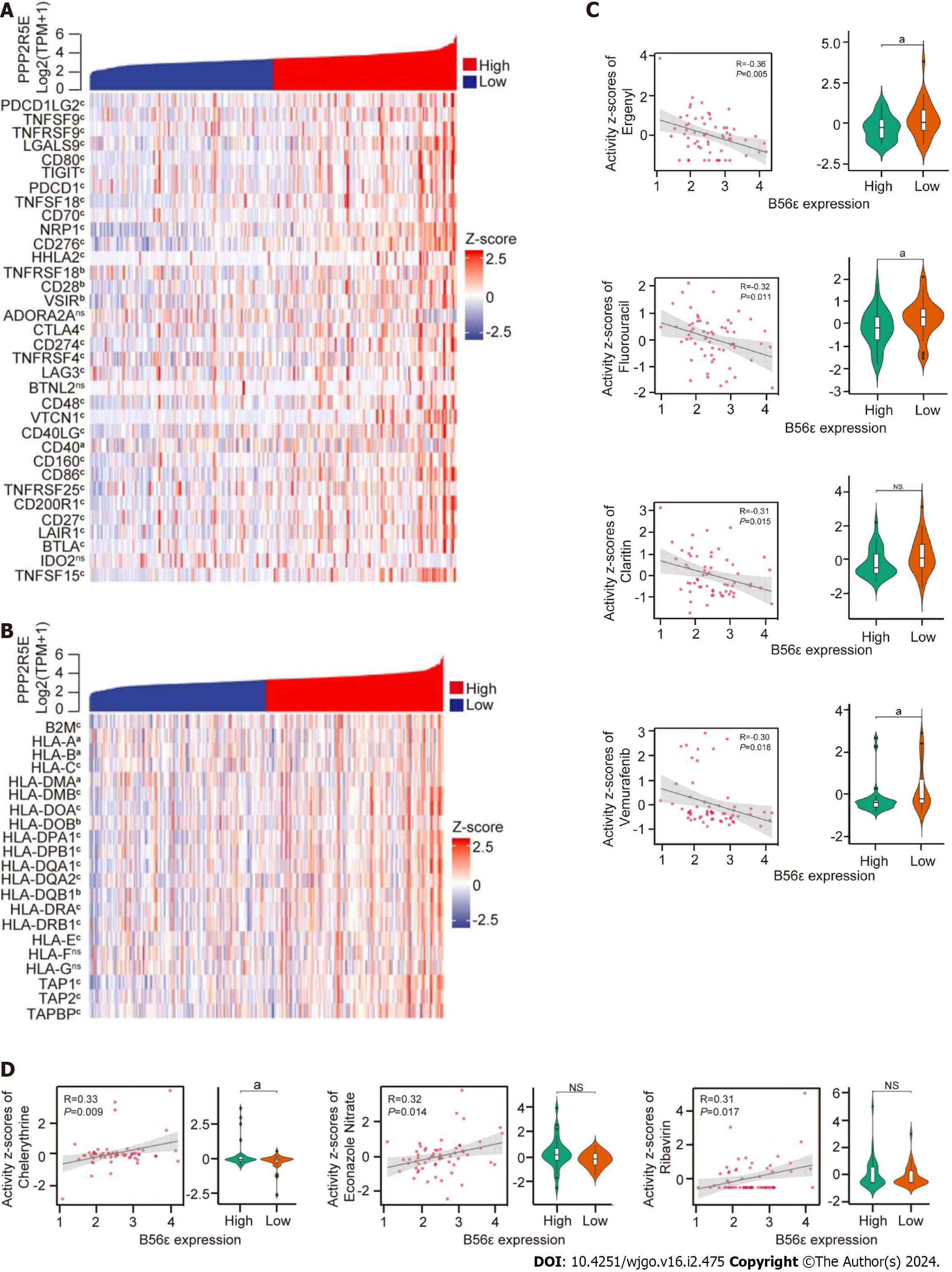
To further explore the potential mechanism of B56ε in HCC immunity, we evaluated the expression and correlations of 34 common immune checkpoints and 21 HLA-associated genes in different B56ε expression groups. Excluding the adenosine A2A receptor gene, butyrophilin-like protein 2, and indoleamine 2,3-dioxygenase 2, the remaining 31 immune checkpoints were significantly correlated with the differential expression of B56ε, and all were positively correlated (Figure 5A). As shown in Figure 5B, with the exception of the HLA-F and HLA-G genes, the remaining 19 HLA-associated genes were significantly correlated with the differential expression of 56ε, and all were positively correlated.
Data related to cancer drugs were downloaded through the CellMiner database for analysis. It was found that the expression of B56ε was negatively correlated with the sensitivity of most chemotherapy drugs, and a small number showed a positive correlation (Table 2). As shown in Figure 5C, four drugs with the strongest negative correlation and their differential expression were statistically significant in drug sensitivity, including Ergenyl (r = -0.357, P < 0.01), fluorouracil (r = -0.324, P < 0.05), Claritin (r = -0.312, P < 0.05), and vemurafenib (r = -0.303, P < 0.05). The remaining three drugs were statistically significant in the differential expression of B56ε except Claritin (all P < 0.05). Furthermore, three drugs with the strongest positive correlation with their differential expression were statistically significant in drug sensitivity (Figure 5D), including chelerythrine (r = 0.334, P < 0.01), econazole nitrate (r = 0.316, P < 0.05), and ribravirin (r = 0.307, P < 0.05). The remaining two drugs were not statistically significant in the differential expression of B56ε except chelerythrine (all P > 0.05). These results indicate that B56ε expression is positively correlated with most of immune checkpoints and negatively correlated with the sensitivity of most chemotherapy drugs.
| Drugs | Correlation | P value |
| Allopurinol | 0.255 | 0.049 |
| Fluorouracil | -0.324 | 0.011 |
| Methylprednisolone | 0.279 | 0.031 |
| Chelerythrine | 0.334 | 0.009 |
| Ergenyl | -0.357 | 0.005 |
| Ribavirin | 0.307 | 0.017 |
| Claritin | -0.312 | 0.015 |
| RAF-265 | -0.293 | 0.023 |
| Nelarabine | 0.264 | 0.042 |
| PLX-4720 | -0.271 | 0.036 |
| Econazole nitrate | 0.316 | 0.014 |
| Rabusertib | -0.276 | 0.033 |
| Vemurafenib | -0.304 | 0.018 |
| Vertex ATR inhibitor Cpd 45 | -0.265 | 0.041 |
| Dabrafenib | -0.263 | 0.042 |
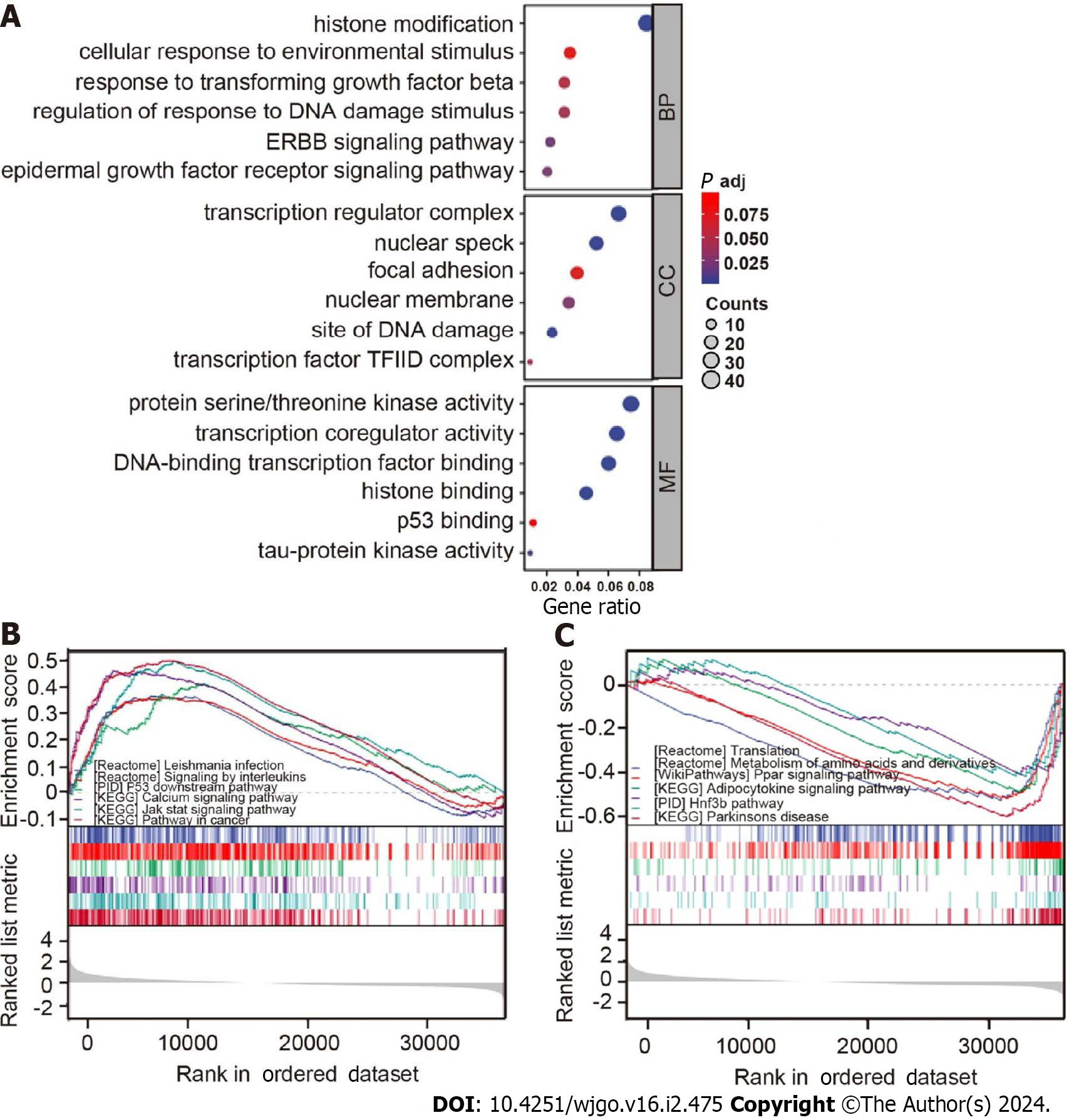
The characteristics of GO and KEGG in cells at different expression levels of B56ε were further explored by GSEA. As shown in Figure 6A, GO analysis indicated that B56ε was mainly involved in histone modification, cell response to environmental stimulation, response regulation to DNA damage stimuli, the ERBB signaling pathway, epidermal growth factor receptor signaling pathway, and other processes in the biological process category. In terms of the cellular component, it was mainly related to transcriptional regulatory complexes, nuclear spots, DNA damage sites, and transcription factor TFIID complexes. In terms of molecular function, it was mainly related to protein serine/threonine kinase activity, transcriptional co-regulatory activity, histone binding, p53 binding, and tau protein kinase activity. When the expression level of B56ε was elevated, it was mainly related to the cancer pathway, p53 downstream pathway, and interleukin-mediated signaling (Figure 6B). However, when the expression level of B56ε was decreased, it was mainly related to translation, the peroxisome proliferator-activated receptor signaling pathway, and adipocytokine signaling pathway (Figure 6C). These results show that the expression level of B56ε is correlated with different signaling pathways in HCC.
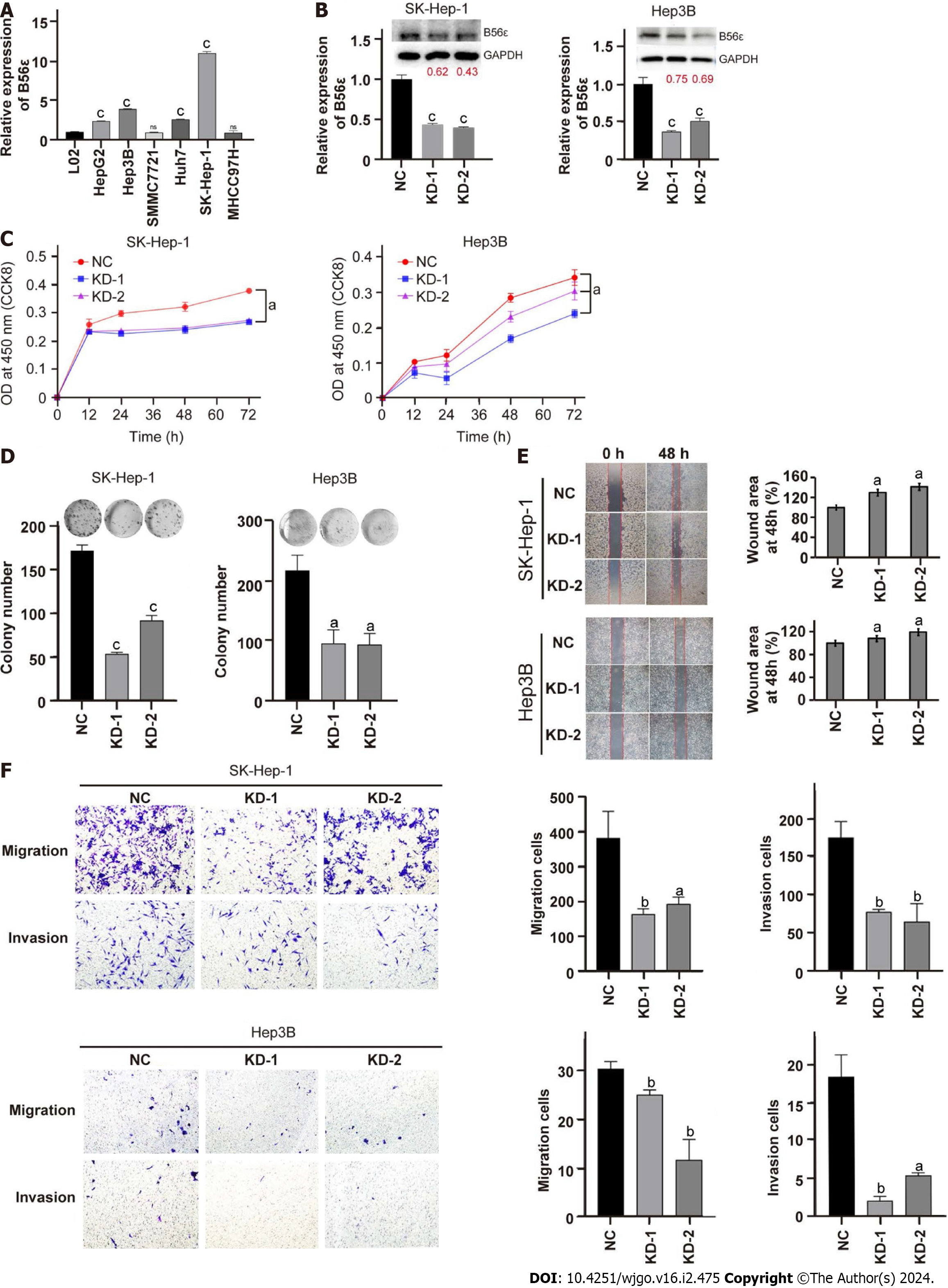
To further explore the role of B56ε in HCC progression, we knocked down B56 in HCC cells. We first detected the expression of B56ε in hepatic cells L02 and different HCC cell lines by quantitative polymerase chain reaction (qPCR). As shown in Figure 7A, B56ε was more highly expressed in SK-Hep-1 cells and Hep3B cells than in L02 cells. B56ε expression was verified by qPCR and western blotting after its knockdown in SK-Hep-1 cells and Hep3B cells (Figure 7B). Moreover, the proliferative ability of HCC cells was significantly weakened after B56ε knockdown and detected by the CCK-8 assay (Figure 7C). Plate cloning experiments also found that HCC cells proliferated more slowly after B56ε knockdown (Figure 7D). The migration and invasive ability of HCC cells after B56ε knockdown was further detected by wound healing and transwell assays. The wound-healing area was smaller in B56ε knockdown HCC cells than in control cells, indicating that the cell migration capacity was significantly decreased after B56ε knockdown (Figure 7E, P < 0.05). The same results were found in the transwell assay, which showed that HCC cell invasion and migration capacity were significantly reduced in B56ε knockdown SK-Hep-1 cells and Hep3B cells (Figure 7F). These results suggest that B56ε may promote HCC cell proliferation and metastasis.
Herein, we first verified the tumor-promoting effect of B56ε by comprehensive bioinformatics analysis and related ex
Some studies have reported that PP2A is often inactivated in human cancers and is considered a tumor suppressor[21,22]. Paradoxically, inhibition of PP2A also has the potential to be a therapeutic target for a variety of cancers. Decreased PR55α expression inhibits the migration and invasion of pancreatic cancer cells[23]. Another study showed that B55β overexpression markedly suppressed cell migration and invasion in HCC cells[24]. B56ε is one member of the PP2A B56 regulatory subunit; its role in tumors has garnered much attention and is a subject of debate. B56ε has a nuclear output signal at the C-terminus, which can lead to migration of the PP2A complex into the cytoplasm[25]. Through the TCGA, GTEx, PrognoScan, and TIMER databases, we found that B56ε expression levels were up-regulated in most tumors, and the high expression of B56ε was associated with a poor prognosis in patients with brain tumor or breast cancer. Moreover, we identified B56ε as a high-risk prognostic factor in ACC, LIHC, PAAD, and PCPG.
After determining the expression characteristics of B56ε in pan-cancer, we further investigated the key role of B56ε in HCC. The results showed that the expression level of B56ε in HCC tissues was higher than that of normal tissues, and HCC patients with high B56ε expression had a poor prognosis. Additional cell function assays showed that downregulated B56ε can inhibit HCC cell proliferation, invasion, and migration. Consistent with our results, a recent study showed that decreased B56ε can promote gastric cancer cell apoptosis to suppress cell growth[18]. Finally, GSEA was used to analyze the biological function of B56ε in HCC. GO results showed that B56ε expression was related to immune response and histone modification. When B56ε was highly expressed, it was mainly enriched in the cancer pathway, p53 downstream pathway, interleukin-mediated signaling and other related pathways, revealing the potential mechanism of B56ε in the malignant biological behavior of HCC cells.
The high incidence and mortality rate of HCC is a serious health problem worldwide[26]. Due to the insidious onset of HCC in the early stage, patients present in the intermediate to advanced stages, and the OS remains poor due to high rates of intrahepatic and extrahepatic metastasis and recurrence[27,28]. With the development of immunotherapy, the treatment of HCC has been further improved. Based on the application of cytotoxic T-lymphocyte associated protein 4 and programmed cell death protein 1/programmed death-ligand 1 monoclonal antibodies, T cell immune checkpoint inhibitors (ICIs) bring a new clinical breakthrough in tumor immunotherapy[29-33]. Especially, the immunotherapy of several solid tumor and hematological tumors has achieved satisfactory efficacy, and sheds light on immune-based HCC therapy[34-38]. ICIs have been shown to eliminate tumor cells using an efficient immune response, including non-small cell lung cancer[39], melanoma[40], and HCC.
Our study found that B56ε expression varied in different HCC immune subtypes. Moreover, the expression of B56ε was correlated with a variety of immune cells, especially tumor-infiltrating lymphocytes (TILs) and played a vital role in TME. For example, B56ε expression was significantly correlated with CD8+ T cells, macrophages, and DCs. The prognosis and immunotherapy efficacy can be predicted by TILs in the TME of cancer patients[41,42]. As a universal component of TME, macrophages have been shown to aid in immune evasion and suppression[43]. Some studies have suggested the presence of antitumor immunity in HCC patients. For example, tumor-associated antigen-specific CD8+ T-cell responses were found correlated with HCC prognosis[44]. The intratumoral density of activated cytotoxic T cells (CTLs) had a correlation with the OS of HCC patients, and the intratumoral balance between CTLs and regulatory T cells also affected the OS and disease-free survival[45]. These observations suggest that the immunogenic potential of HCC can be controlled through optimized immunotherapy.
This study had some limitations. The sample was limited to the patient information contained in TCGA dataset requires more clinical case validation. The potential mechanism of B56ε tumor-promoting and immunomodulatory effects in HCC also needs to be further verified in clinical practice.
In summary, we demonstrate that B56ε can be used as a prognostic biomarker for a variety of tumors and may modulate tumor immune cell infiltration and immune response. B56ε can promote the proliferation, invasion, and migration of HCC. Our results can provide theoretical support and new ideas for HCC treatment.
B56ε is a regulatory subunit of the protein phosphatase 2A, which is abnormally expressed in tumors and regulates various tumor cell functions.
At present, the application of B56ε in pan-carcinoma lacks a comprehensive analysis, and its role and mechanism in hepatocellular carcinoma (HCC) are still unclear.
The study aims to analyze B56ε in pan-cancer, and explore its role and mechanism in HCC.
The Cancer Genome Atlas (TCGA), Genotype-Tissue Expression, Gene Expression Profiling Interactive Analysis, and Tumor Immune Estimation Resource databases were used to analyze B56ε expression, prognostic mutations, somatic copy number alterations, and tumor immune characteristics in 33 tumors. The relationship between B56ε expression levels and drug sensitivity, immunotherapy, immune checkpoints, and human leukocyte antigen (HLA)-related genes were further analyzed. Gene Set Enrichment Analysis (GSEA) was performed to reveal the role of B56ε in HCC. Cell Counting Kit-8, plate cloning, wound healing, and transwell experiments were conducted to show the effects of B56ε interference on the malignant behaviors of HCC cells.
In most tumors, B56ε expression was upregulated, and B56ε high expression was a risk factor in adrenocortical cancer, HCC, pancreatic adenocarcinoma, and pheochromocytoma and paraganglioma (all P < 0.05). B56ε expression levels were correlated with a variety of immune cells, such as T helper 17 cells, B cells, and macrophages. There was a positive correlation between B56ε expression levels with immune checkpoint genes and HLA-related genes (all P < 0.05). The expression of B56ε was negatively correlated with the sensitivity of most chemotherapy drugs, but a small number showed a positive correlation (all P < 0.05). GSEA showed that B56ε expression was related to the cancer pathway, p53 downstream pathway, and interleukin-mediated signaling in HCC. Knockdown of B56ε expression in HCC cells inhibited the proliferation, migration, and invasion capacity of tumor cells.
B56ε may regulate the microenvironment, immune evasion, and immune cell infiltration of multiple tumors. Moreover, B56ε plays an important role in HCC progression. Our study supports B56ε as a prognostic marker and potential therapeutic target for HCC.
The patient information contained in TCGA dataset and requires more clinical case validation. In the future, the potential mechanism of B56ε tumor-promoting and immunomodulatory effects in HCC also needs to be further verified in clinical practice.
Provenance and peer review: Invited article; Externally peer reviewed.
Peer-review model: Single blind
Specialty type: Oncology
Country/Territory of origin: China
Peer-review report’s scientific quality classification
Grade A (Excellent): 0
Grade B (Very good): B
Grade C (Good): 0
Grade D (Fair): 0
Grade E (Poor): 0
P-Reviewer: Shalaby MN, Egypt S-Editor: Wang JJ L-Editor: A P-Editor: Zhang XD
| 1. | Sung H, Ferlay J, Siegel RL, Laversanne M, Soerjomataram I, Jemal A, Bray F. Global Cancer Statistics 2020: GLOBOCAN Estimates of Incidence and Mortality Worldwide for 36 Cancers in 185 Countries. CA Cancer J Clin. 2021;71:209-249. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 75126] [Cited by in RCA: 64702] [Article Influence: 16175.5] [Reference Citation Analysis (177)] |
| 2. | Jemal A, Ward EM, Johnson CJ, Cronin KA, Ma J, Ryerson B, Mariotto A, Lake AJ, Wilson R, Sherman RL, Anderson RN, Henley SJ, Kohler BA, Penberthy L, Feuer EJ, Weir HK. Annual Report to the Nation on the Status of Cancer, 1975-2014, Featuring Survival. J Natl Cancer Inst. 2017;109. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 748] [Cited by in RCA: 1111] [Article Influence: 138.9] [Reference Citation Analysis (0)] |
| 3. | Villanueva A. Hepatocellular Carcinoma. N Engl J Med. 2019;380:1450-1462. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 2066] [Cited by in RCA: 3176] [Article Influence: 529.3] [Reference Citation Analysis (37)] |
| 4. | Singal AG, Lampertico P, Nahon P. Epidemiology and surveillance for hepatocellular carcinoma: New trends. J Hepatol. 2020;72:250-261. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 526] [Cited by in RCA: 781] [Article Influence: 156.2] [Reference Citation Analysis (0)] |
| 5. | Global Burden of Disease Liver Cancer Collaboration, Akinyemiju T, Abera S, Ahmed M, Alam N, Alemayohu MA, Allen C, Al-Raddadi R, Alvis-Guzman N, Amoako Y, Artaman A, Ayele TA, Barac A, Bensenor I, Berhane A, Bhutta Z, Castillo-Rivas J, Chitheer A, Choi JY, Cowie B, Dandona L, Dandona R, Dey S, Dicker D, Phuc H, Ekwueme DU, Zaki MS, Fischer F, Fürst T, Hancock J, Hay SI, Hotez P, Jee SH, Kasaeian A, Khader Y, Khang YH, Kumar A, Kutz M, Larson H, Lopez A, Lunevicius R, Malekzadeh R, McAlinden C, Meier T, Mendoza W, Mokdad A, Moradi-Lakeh M, Nagel G, Nguyen Q, Nguyen G, Ogbo F, Patton G, Pereira DM, Pourmalek F, Qorbani M, Radfar A, Roshandel G, Salomon JA, Sanabria J, Sartorius B, Satpathy M, Sawhney M, Sepanlou S, Shackelford K, Shore H, Sun J, Mengistu DT, Topór-Mądry R, Tran B, Ukwaja KN, Vlassov V, Vollset SE, Vos T, Wakayo T, Weiderpass E, Werdecker A, Yonemoto N, Younis M, Yu C, Zaidi Z, Zhu L, Murray CJL, Naghavi M, Fitzmaurice C. The Burden of Primary Liver Cancer and Underlying Etiologies From 1990 to 2015 at the Global, Regional, and National Level: Results From the Global Burden of Disease Study 2015. JAMA Oncol. 2017;3:1683-1691. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 1459] [Cited by in RCA: 1501] [Article Influence: 187.6] [Reference Citation Analysis (0)] |
| 6. | Chen C, Wang G. Mechanisms of hepatocellular carcinoma and challenges and opportunities for molecular targeted therapy. World J Hepatol. 2015;7:1964-1970. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 87] [Cited by in RCA: 108] [Article Influence: 10.8] [Reference Citation Analysis (4)] |
| 7. | Wlodarchak N, Xing Y. PP2A as a master regulator of the cell cycle. Crit Rev Biochem Mol Biol. 2016;51:162-184. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 187] [Cited by in RCA: 271] [Article Influence: 30.1] [Reference Citation Analysis (0)] |
| 8. | Bheri M, Pandey GK. PP2A Phosphatases Take a Giant Leap in the Post-Genomics Era. Curr Genomics. 2019;20:154-171. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 13] [Cited by in RCA: 8] [Article Influence: 1.3] [Reference Citation Analysis (0)] |
| 9. | Kong J, Li D, Zhang S, Zhang H, Fu Y, Qian B, Bei C, Tan S, Zhu X. Okadaic acid promotes epithelial-mesenchymal transition of hepatocellular carcinoma cells by inhibiting protein phosphatase 2A. J Cell Biochem. 2020;. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 4] [Cited by in RCA: 1] [Article Influence: 0.2] [Reference Citation Analysis (0)] |
| 10. | Hou CY, Ma CY, Lin YJ, Huang CL, Wang HD, Yuh CH. WNK1-OSR1 Signaling Regulates Angiogenesis-Mediated Metastasis towards Developing a Combinatorial Anti-Cancer Strategy. Int J Mol Sci. 2022;23. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in RCA: 11] [Reference Citation Analysis (0)] |
| 11. | Raman D, Pervaiz S. Redox inhibition of protein phosphatase PP2A: Potential implications in oncogenesis and its progression. Redox Biol. 2019;27:101105. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 16] [Cited by in RCA: 38] [Article Influence: 6.3] [Reference Citation Analysis (0)] |
| 12. | Mannava S, Omilian AR, Wawrzyniak JA, Fink EE, Zhuang D, Miecznikowski JC, Marshall JR, Soengas MS, Sears RC, Morrison CD, Nikiforov MA. PP2A-B56α controls oncogene-induced senescence in normal and tumor human melanocytic cells. Oncogene. 2012;31:1484-1492. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 28] [Cited by in RCA: 34] [Article Influence: 2.4] [Reference Citation Analysis (0)] |
| 13. | Eichhorn PJ, Creyghton MP, Bernards R. Protein phosphatase 2A regulatory subunits and cancer. Biochim Biophys Acta. 2009;1795:1-15. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 98] [Cited by in RCA: 275] [Article Influence: 16.2] [Reference Citation Analysis (0)] |
| 14. | Kar S, Palit S, Ball WB, Das PK. Carnosic acid modulates Akt/IKK/NF-κB signaling by PP2A and induces intrinsic and extrinsic pathway mediated apoptosis in human prostate carcinoma PC-3 cells. Apoptosis. 2012;17:735-747. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 91] [Cited by in RCA: 105] [Article Influence: 8.1] [Reference Citation Analysis (0)] |
| 15. | Che L, Du ZB, Wang WH, Wu JS, Han T, Chen YY, Han PY, Lei Z, Chen XX, He Y, Xu L, Lin X, Lin ZN, Lin YC. Intracellular antibody targeting HBx suppresses invasion and metastasis in hepatitis B virus-related hepatocarcinogenesis via protein phosphatase 2A-B56γ-mediated dephosphorylation of protein kinase B. Cell Prolif. 2022;55:e13304. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 1] [Cited by in RCA: 10] [Article Influence: 3.3] [Reference Citation Analysis (0)] |
| 16. | Lambrecht C, Ferreira GB, Omella JD, Libbrecht L, DE Vos R, Derua R, Mathieu C, Overbergh L, Waelkens E, Janssens V. Differential Proteomic Analysis of Hepatocellular Carcinomas from Ppp2r5d Knockout Mice and Normal (Knockout) Livers. Cancer Genomics Proteomics. 2020;17:669-685. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 2] [Cited by in RCA: 6] [Article Influence: 1.5] [Reference Citation Analysis (0)] |
| 17. | Cristóbal I, Cirauqui C, Castello-Cros R, Garcia-Orti L, Calasanz MJ, Odero MD. Downregulation of PPP2R5E is a common event in acute myeloid leukemia that affects the oncogenic potential of leukemic cells. Haematologica. 2013;98:e103-e104. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 21] [Cited by in RCA: 26] [Article Influence: 2.2] [Reference Citation Analysis (0)] |
| 18. | Liu X, Liu Q, Fan Y, Wang S, Liu X, Zhu L, Liu M, Tang H. Downregulation of PPP2R5E expression by miR-23a suppresses apoptosis to facilitate the growth of gastric cancer cells. FEBS Lett. 2014;588:3160-3169. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 19] [Cited by in RCA: 27] [Article Influence: 2.5] [Reference Citation Analysis (0)] |
| 19. | Dupont WD, Breyer JP, Bradley KM, Schuyler PA, Plummer WD, Sanders ME, Page DL, Smith JR. Protein phosphatase 2A subunit gene haplotypes and proliferative breast disease modify breast cancer risk. Cancer. 2010;116:8-19. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 3] [Cited by in RCA: 18] [Article Influence: 1.2] [Reference Citation Analysis (0)] |
| 20. | Tao YD, Liu X, Sun JH, Huo F, Guo HJ. miR-23a Promoting Cell Proliferation of Human Tongue Squamous Cell Carcinoma Cell through Regulating PPP2R5E. Hebei Med. 2020;11:88-92. [DOI] [Full Text] |
| 21. | Soofiyani SR, Hejazi MS, Baradaran B. The role of CIP2A in cancer: A review and update. Biomed Pharmacother. 2017;96:626-633. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 26] [Cited by in RCA: 56] [Article Influence: 7.0] [Reference Citation Analysis (0)] |
| 22. | Mazhar S, Taylor SE, Sangodkar J, Narla G. Targeting PP2A in cancer: Combination therapies. Biochim Biophys Acta Mol Cell Res. 2019;1866:51-63. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 66] [Cited by in RCA: 104] [Article Influence: 14.9] [Reference Citation Analysis (0)] |
| 23. | Hein AL, Seshacharyulu P, Rachagani S, Sheinin YM, Ouellette MM, Ponnusamy MP, Mumby MC, Batra SK, Yan Y. PR55α Subunit of Protein Phosphatase 2A Supports the Tumorigenic and Metastatic Potential of Pancreatic Cancer Cells by Sustaining Hyperactive Oncogenic Signaling. Cancer Res. 2016;76:2243-2253. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 39] [Cited by in RCA: 55] [Article Influence: 6.1] [Reference Citation Analysis (0)] |
| 24. | Li J, Zhou JK, Mu X, Shen S, Xu X, Luo Y, Ming Y, Wu Y, Peng Y. Regulation of XPO5 phosphorylation by PP2A in hepatocellular carcinoma. MedComm (2020). 2022;3:e125. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 9] [Cited by in RCA: 8] [Article Influence: 2.7] [Reference Citation Analysis (0)] |
| 25. | Slupe AM, Merrill RA, Strack S. Determinants for Substrate Specificity of Protein Phosphatase 2A. Enzyme Res. 2011;2011:398751. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 61] [Cited by in RCA: 76] [Article Influence: 5.4] [Reference Citation Analysis (0)] |
| 26. | Bray F, Ferlay J, Soerjomataram I, Siegel RL, Torre LA, Jemal A. Global cancer statistics 2018: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J Clin. 2018;68:394-424. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 53206] [Cited by in RCA: 55853] [Article Influence: 7979.0] [Reference Citation Analysis (132)] |
| 27. | European Association for the Study of the Liver. EASL Clinical Practice Guidelines: Management of hepatocellular carcinoma. J Hepatol. 2018;69:182-236. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 5593] [Cited by in RCA: 6065] [Article Influence: 866.4] [Reference Citation Analysis (3)] |
| 28. | Zheng Z, Liang W, Wang D, Schroder PM, Ju W, Wu L, Zheng Z, Shang Y, Guo Z, He X. Adjuvant chemotherapy for patients with primary hepatocellular carcinoma: a meta-analysis. Int J Cancer. 2015;136:E751-E759. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 22] [Cited by in RCA: 34] [Article Influence: 3.1] [Reference Citation Analysis (0)] |
| 29. | Fehrenbacher L, Spira A, Ballinger M, Kowanetz M, Vansteenkiste J, Mazieres J, Park K, Smith D, Artal-Cortes A, Lewanski C, Braiteh F, Waterkamp D, He P, Zou W, Chen DS, Yi J, Sandler A, Rittmeyer A; POPLAR Study Group. Atezolizumab versus docetaxel for patients with previously treated non-small-cell lung cancer (POPLAR): a multicentre, open-label, phase 2 randomised controlled trial. Lancet. 2016;387:1837-1846. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1779] [Cited by in RCA: 2205] [Article Influence: 245.0] [Reference Citation Analysis (0)] |
| 30. | Herbst RS, Baas P, Kim DW, Felip E, Pérez-Gracia JL, Han JY, Molina J, Kim JH, Arvis CD, Ahn MJ, Majem M, Fidler MJ, de Castro G Jr, Garrido M, Lubiniecki GM, Shentu Y, Im E, Dolled-Filhart M, Garon EB. Pembrolizumab versus docetaxel for previously treated, PD-L1-positive, advanced non-small-cell lung cancer (KEYNOTE-010): a randomised controlled trial. Lancet. 2016;387:1540-1550. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 4116] [Cited by in RCA: 5054] [Article Influence: 561.6] [Reference Citation Analysis (0)] |
| 31. | Sharon E, Streicher H, Goncalves P, Chen HX. Immune checkpoint inhibitors in clinical trials. Chin J Cancer. 2014;33:434-444. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 83] [Cited by in RCA: 97] [Article Influence: 8.8] [Reference Citation Analysis (0)] |
| 32. | Carlino MS, Larkin J, Long GV. Immune checkpoint inhibitors in melanoma. Lancet. 2021;398:1002-1014. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 206] [Cited by in RCA: 704] [Article Influence: 176.0] [Reference Citation Analysis (0)] |
| 33. | Lipson EJ, Drake CG. Ipilimumab: an anti-CTLA-4 antibody for metastatic melanoma. Clin Cancer Res. 2011;17:6958-6962. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 307] [Cited by in RCA: 421] [Article Influence: 30.1] [Reference Citation Analysis (0)] |
| 34. | Waldman AD, Fritz JM, Lenardo MJ. A guide to cancer immunotherapy: from T cell basic science to clinical practice. Nat Rev Immunol. 2020;20:651-668. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 2409] [Cited by in RCA: 2667] [Article Influence: 533.4] [Reference Citation Analysis (0)] |
| 35. | Qin S, Ren Z, Meng Z, Chen Z, Chai X, Xiong J, Bai Y, Yang L, Zhu H, Fang W, Lin X, Chen X, Li E, Wang L, Chen C, Zou J. Camrelizumab in patients with previously treated advanced hepatocellular carcinoma: a multicentre, open-label, parallel-group, randomised, phase 2 trial. Lancet Oncol. 2020;21:571-580. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 453] [Cited by in RCA: 426] [Article Influence: 85.2] [Reference Citation Analysis (0)] |
| 36. | El-Khoueiry AB, Sangro B, Yau T, Crocenzi TS, Kudo M, Hsu C, Kim TY, Choo SP, Trojan J, Welling TH Rd, Meyer T, Kang YK, Yeo W, Chopra A, Anderson J, Dela Cruz C, Lang L, Neely J, Tang H, Dastani HB, Melero I. Nivolumab in patients with advanced hepatocellular carcinoma (CheckMate 040): an open-label, non-comparative, phase 1/2 dose escalation and expansion trial. Lancet. 2017;389:2492-2502. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 3278] [Cited by in RCA: 3315] [Article Influence: 414.4] [Reference Citation Analysis (1)] |
| 37. | Zhu AX, Finn RS, Edeline J, Cattan S, Ogasawara S, Palmer D, Verslype C, Zagonel V, Fartoux L, Vogel A, Sarker D, Verset G, Chan SL, Knox J, Daniele B, Webber AL, Ebbinghaus SW, Ma J, Siegel AB, Cheng AL, Kudo M; KEYNOTE-224 investigators. Pembrolizumab in patients with advanced hepatocellular carcinoma previously treated with sorafenib (KEYNOTE-224): a non-randomised, open-label phase 2 trial. Lancet Oncol. 2018;19:940-952. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1184] [Cited by in RCA: 1902] [Article Influence: 271.7] [Reference Citation Analysis (0)] |
| 38. | Finn RS, Ryoo BY, Merle P, Kudo M, Bouattour M, Lim HY, Breder VV, Edeline J, Chao Y, Ogasawara S, Yau T, Garrido M, Chan SL, Knox JJ, Daniele B, Ebbinghaus S, Chen E, Siegel AB, Zhu A, Cheng AL. Results of KEYNOTE-240: phase 3 study of pembrolizumab (Pembro) vs best supportive care (BSC) for second line therapy in advanced hepatocellular carcinoma (HCC). J Clin Oncol. 2019;37:4004. [RCA] [DOI] [Full Text] [Cited by in Crossref: 127] [Cited by in RCA: 129] [Article Influence: 21.5] [Reference Citation Analysis (0)] |
| 39. | Borghaei H, Paz-Ares L, Horn L, Spigel DR, Steins M, Ready NE, Chow LQ, Vokes EE, Felip E, Holgado E, Barlesi F, Kohlhäufl M, Arrieta O, Burgio MA, Fayette J, Lena H, Poddubskaya E, Gerber DE, Gettinger SN, Rudin CM, Rizvi N, Crinò L, Blumenschein GR Jr, Antonia SJ, Dorange C, Harbison CT, Graf Finckenstein F, Brahmer JR. Nivolumab versus Docetaxel in Advanced Nonsquamous Non-Small-Cell Lung Cancer. N Engl J Med. 2015;373:1627-1639. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 6945] [Cited by in RCA: 7537] [Article Influence: 753.7] [Reference Citation Analysis (0)] |
| 40. | Pasquali S, Hadjinicolaou AV, Chiarion Sileni V, Rossi CR, Mocellin S. Systemic treatments for metastatic cutaneous melanoma. Cochrane Database Syst Rev. 2018;2:CD011123. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 69] [Cited by in RCA: 104] [Article Influence: 14.9] [Reference Citation Analysis (0)] |
| 41. | Azimi F, Scolyer RA, Rumcheva P, Moncrieff M, Murali R, McCarthy SW, Saw RP, Thompson JF. Tumor-infiltrating lymphocyte grade is an independent predictor of sentinel lymph node status and survival in patients with cutaneous melanoma. J Clin Oncol. 2012;30:2678-2683. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 486] [Cited by in RCA: 614] [Article Influence: 47.2] [Reference Citation Analysis (0)] |
| 42. | Ohtani H. Focus on TILs: prognostic significance of tumor infiltrating lymphocytes in human colorectal cancer. Cancer Immun. 2007;7:4. [PubMed] |
| 43. | Gajewski TF, Schreiber H, Fu YX. Innate and adaptive immune cells in the tumor microenvironment. Nat Immunol. 2013;14:1014-1022. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 2228] [Cited by in RCA: 3065] [Article Influence: 255.4] [Reference Citation Analysis (0)] |
| 44. | Flecken T, Schmidt N, Hild S, Gostick E, Drognitz O, Zeiser R, Schemmer P, Bruns H, Eiermann T, Price DA, Blum HE, Neumann-Haefelin C, Thimme R. Immunodominance and functional alterations of tumor-associated antigen-specific CD8+ T-cell responses in hepatocellular carcinoma. Hepatology. 2014;59:1415-1426. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 288] [Cited by in RCA: 298] [Article Influence: 27.1] [Reference Citation Analysis (0)] |
| 45. | Gao Q, Qiu SJ, Fan J, Zhou J, Wang XY, Xiao YS, Xu Y, Li YW, Tang ZY. Intratumoral balance of regulatory and cytotoxic T cells is associated with prognosis of hepatocellular carcinoma after resection. J Clin Oncol. 2007;25:2586-2593. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 787] [Cited by in RCA: 892] [Article Influence: 49.6] [Reference Citation Analysis (0)] |