Published online Dec 15, 2024. doi: 10.4251/wjgo.v16.i12.4548
Revised: August 5, 2024
Accepted: August 12, 2024
Published online: December 15, 2024
Processing time: 118 Days and 10.8 Hours
Survival rates following radical surgery for gastric neuroendocrine neoplasms (g-NENs) are low, with high recurrence rates. This fact impacts patient prognosis and complicates postoperative management. Traditional prognostic models, including the Cox proportional hazards (CoxPH) model, have shown limited predictive power for postoperative survival in gastrointestinal neuroectodermal tumor patients. Machine learning methods offer a unique opportunity to analyze complex relationships within datasets, providing tools and methodologies to assess large volumes of high-dimensional, multimodal data generated by biological sciences. These methods show promise in predicting outcomes across various medical disciplines. In the context of g-NENs, utilizing machine learning to predict survival outcomes holds potential for personalized postoperative management strategies. This editorial reviews a study exploring the advantages and effectiveness of the random survival forest (RSF) model, using the lymph node ratio (LNR), in predicting disease-specific survival (DSS) in postoperative g-NEN patients stratified into low-risk and high-risk groups. The findings demonstrate that the RSF model, incorporating LNR, outperformed the CoxPH model in predicting DSS and constitutes an important step towards precision medicine.
Core Tip: Liu et al’s study addresses a critical issue in determining the postoperative prognosis of gastric neuroendocrine tumors by identifying the significance of lymph node ratio. Moreover, the random survival forest model, a machine-learning approach, surpasses traditional Cox proportional hazards models by enhancing predictive accuracy, clinical utility, and overall performance. This model’s ability to stratify patient risks and personalize survival predictions can aid in formulating targeted postoperative strategies, thus realizing an important aspect of personalized “precision medicine”.
- Citation: Wang HN, An JH, Zong L. Estimating prognosis of gastric neuroendocrine neoplasms using machine learning: A step towards precision medicine. World J Gastrointest Oncol 2024; 16(12): 4548-4552
- URL: https://www.wjgnet.com/1948-5204/full/v16/i12/4548.htm
- DOI: https://dx.doi.org/10.4251/wjgo.v16.i12.4548
Liu et al[1] published a study titled “Combining lymph node ratio to develop prognostic models for postoperative gastric neuroendocrine neoplasm patients”. Their study utilized machine learning techniques to identify risk factors associated with disease-specific survival (DSS) in postoperative gastric neuroendocrine neoplasm (g-NEN) patients, and succeeded in constructing an efficient and precise prognostic model based on lymph node ratio (LNR), defined as the ratio of the number of positive lymph nodes to the number examined. It also shows off one of the most promising features of artificial intelligence or machine learning, its capacity to identify patterns from multidimensional data sets such as those found in medicine. They researched a field that is in sore need of a reliable prognostic model to guide postoperative management. g-NENs represent a rare and challenging type of gastric malignancy in oncology. These neoplasms are classified into three types: Well-differentiated neuroendocrine tumors (NETs), poorly differentiated neuroendocrine carcinomas (NEC), and mixed neuroendocrine-non-NETs. While gastric NETs typically exhibit an indolent growth pattern and are often benign, gastric NECs (g-NECs) are highly malignant, aggressive, and associated with a poor prognosis[2], showing lower postoperative survival rates and higher recurrence rates[3,4]. Currently, there are no highly effective treatment options for NENs. Various clinical characteristics significantly influence the prognosis of NEN patients[5-7]. In resected g-NECs, the presence of more than two metastatic lymph nodes, metastatic disease in over 10% of resected lymph nodes, and involvement of station 2 lymph nodes have all been demonstrated as significant prognostic indicators associated with poorer outcomes[8]. Due to its incorporation of the number of lymph nodes examined during surgery, LNR turns out to be a more advantageous parameter for prognostic estimation in such patients. Indeed, many studies have shown that the prognostic value of LNR exceeds that of the absolute number of involved lymph nodes[9] for various types of cancer. The present editorial explores the promise of machine learning as a pathway toward precision medicine, particularly in its capacity to predict postoperative outcomes for NEN patients. The advent of such artificial intelligence techniques offers unique opportunities to identify subtle patterns and factors that traditional prognostic methods might overlook[10,11].
Machine learning models can analyze complex relationships within datasets and has shown promise in predicting various medical outcomes[10]. In the context of g-NENs, machine learning has the potential to predict postoperative prognosis and tailor personalized postoperative management strategies.
Traditionally, predictive models such as the Cox proportional hazards (CoxPH) model have been employed[12,13]. However, their limitations have spurred the exploration of innovative approaches[11,14]. Liu et al’s study critically compared the performance of the random survival forest (RSF) and CoxPH models in predicting DSS for patients after g-NEN surgery[1]. This greater certainty regarding outcomes allow physicians to tailor the postoperative management strategies for their patients, avoiding the pitfalls and discomfort that can be inherent to end-of-life treatment.
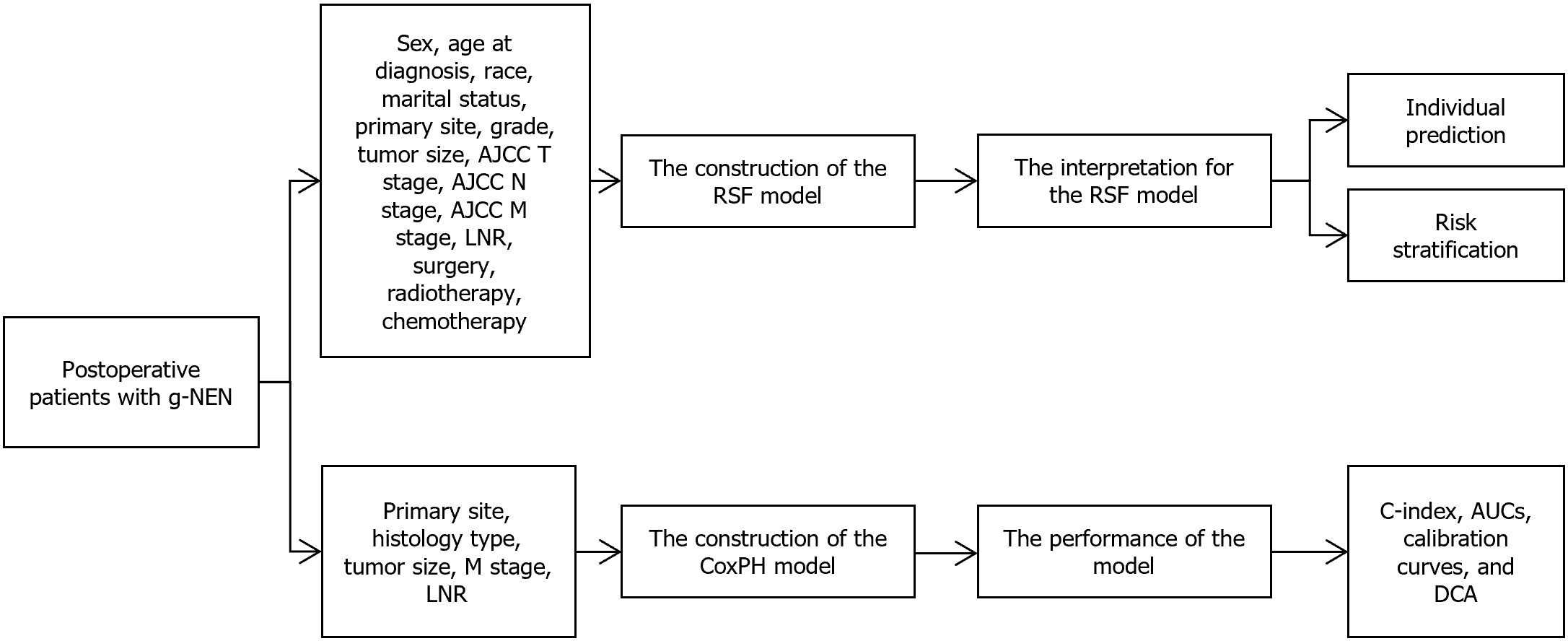
Inspired by a comprehensive cohort consisting of 286 patients from the Surveillance, Epidemiology, and End Results (SEER) database and 92 g-NEN patients from the First Affiliated Hospital of Soochow University, Liu et al[1] constructed a RSF model using 14 key features. These features encompass demographic, clinicopathologic, and tumor-specific factors. The RSF model underwent rigorous evaluation in terms of discrimination, calibration, clinical utility, and overall performance, and its performance was compared with that of traditional models.
This study analyzed data from 7685 patients in the SEER database from 2000 to 2019, of which 286 met the inclusion criteria. Included patients had primary g-NEN, underwent curative surgery, and had complete pathological information. The exclusion criteria (n = 7399) included: (1) Cases without histopathological evidence; (2) History of other malignancies; (3) Cases lacking detailed clinical data such as differentiation grade, tumor size, or tumor node metastasis (TNM) stage; (4) Cases without information on survival duration or those who died within one month; (5) Cases that did not undergo surgery or had only local surgery; and (6) Cases without information on the number of examined lymph nodes and positive lymph nodes. Patients from the SEER database were randomly divided into a training set and an internal validation set at a ratio of 8:2. The external test cohort consisted of 92 patients from the First Affiliated Hospital of Soochow University, covering the period from 2011 to 2020.
Both RSF and CoxPH models were constructed. For the CoxPH model, univariate and multivariate regression analyses identified primary site, histologic type, size, M stage, and LNR as independent risk factors. These selected independent risk factors were then used to develop the CoxPH model, which was visualized using a nomogram. The RSF model, utilizing random forest techniques such as feature and sample bootstrapping, demonstrated faster training times and reduced estimation bias. This model was built using 14 factors: Sex, age at diagnosis, race, marital status, primary site, differentiation grade, tumor size, American Joint Committee on Cancer (AJCC) T stage, AJCC N stage, AJCC M stage, LNR, surgery at the primary site, radiotherapy, and chemotherapy. Optuna was used to determine the optimal hyperparameters for the RSF model: 330 estimators, a minimum of 5 samples per split, and a minimum of 1 sample per leaf. Shapley additive explanations (SHAP) plots were used to interpret the RSF model. Patients were then assigned risk scores and divided into low-risk, medium-risk, and high-risk groups, providing valuable insights for identifying high-risk populations and facilitating timely clinical interventions. Kaplan-Meier analysis confirmed the stratification for all cohorts (P < 0.0001) (Figure 1). Additionally, individualized survival predictions were made, allowing for a clear prediction of the impact of all admission variables on each patient’s prognosis.
The performance of the 8th edition AJCC TNM staging system, CoxPH model, and RSF model was evaluated using the C-index, areas under the receiver operating characteristic curves (AUCs), calibration curves, and decision curve analysis (DCA). The RSF model was further interpreted using SHAP values. In an external test set, the RSF model outperformed the 8th AJCC TNM staging system and the CoxPH model, with C-index values of 0.769, 0.744, and 0.723, respectively.
The 1-, 3-, and 5-year AUCs for the 8th AJCC TNM staging system were 0.690, 0.769, and 0.770, respectively. For the CoxPH model, the AUCs were 0.786, 0.834, and 0.810. The RSF model achieved AUCs of 0.803, 0.895, and 0.869 at 1, 3, and 5 years, respectively. DCA indicated that the RSF model had a higher net benefit compared to the other models (Table 1).
| Model | Cohort | C-index | AUC | ||
| 1-year | 3-year | 5-year | |||
| CoxPH | Training | 0.834 (0.789-0.879) | 0.848 (0.763-0.930) | 0.881 (0.831-0.932) | 0.875 (0.822-0.927) |
| Internal | 0.871 (0.802-0.940) | 0.843 (0.717-0.969) | 0.948 (0.892-1.000) | 0.990 (0.969-1.000) | |
| External | 0.744 (0.665-0.822) | 0.786 (0.622-0.889) | 0.834 (0.735-0.934) | 0.810 (0.688-0.931) | |
| RSF | Training | 0.940 (0.924-0.956) | 0.962 (0.938-0.989) | 0.979 (0.963-0.995) | 0.971 (0.951-0.992) |
| Internal | 0.870 (0.818-0.921) | 0.867 (0.761-0.973) | 0.955 (0.899-1.000) | 0.986 (0.960-1.000) | |
| External | 0.769 (0.691-0.846) | 0.803 (0.608-0.891) | 0.895 (0.814-0.976) | 0.869 (0.769-0.970) | |
SHAP analysis indicated that histologic type was the most significant variable in the RSF model, followed by LNR, T stage, and M stage. Elevated LNR levels were linked to worse patient outcomes.
The study’s limitations include selection bias from its retrospective design and the SEER database’s primary focus on the United States population, which may not generalize to Asian, especially Chinese, populations. Additionally, the lack of multi-center external validation reduces the robustness of the findings.
In the study by Song et al[12], a survival nomogram for g-NECs was constructed. Yang et al[15] developed a prognostic nomogram for g-NEN patients using computed tomography radiomic features. However, both studies failed to demonstrate that LNR is an independent risk factor. Padwal et al[16] and Jiang et al[17] employed machine learning to build prognostic models for pancreatic NEN patients, while Liu et al[1] were the first to use a random forest survival model for g-NENs. This study, through multivariable regression analysis, identified LNR as an independent risk factor, providing higher statistical power and significance.
The RSF model has become a key tool for precise postoperative prognostic estimation and optimized management of g-NENs, showing advantages over traditional models. Its capability to stratify risks and predict individual survival marks a new era of personalized prediction and optimized prognostic strategies. Artificial intelligence, particularly machine learning algorithms, holds great promise in transforming the diagnosis, treatment, and prognosis of gastric diseases by analyzing extensive medical data to identify patterns and anomalies. We predict that artificial intelligence’s role in personalized, prognosis-based management in gastric diseases will be crucial, aiding healthcare professionals in selecting the right intervention for each patient. The RSF model is expected to redefine g-NEN prognosis and guide more precise, individualized patient management.
We are grateful to those researchers who provided study data.
| 1. | Liu W, Wu HY, Lin JX, Qu ST, Gu YJ, Zhu JZ, Xu CF. Combining lymph node ratio to develop prognostic models for postoperative gastric neuroendocrine neoplasm patients. World J Gastrointest Oncol. 2024;16:3507-3520. [RCA] [DOI] [Full Text] [Full Text (PDF)] [Cited by in RCA: 2] [Reference Citation Analysis (0)] |
| 2. | Zi M, Ma Y, Chen J, Pang C, Li X, Yuan L, Liu Z, Yu P. Clinicopathological characteristics of gastric neuroendocrine neoplasms: A comprehensive analysis. Cancer Med. 2024;13:e7011. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 3] [Reference Citation Analysis (0)] |
| 3. | Iwasaki K, Barroga E, Enomoto M, Tsurui K, Shimoda Y, Matsumoto M, Miyoshi K, Ota Y, Matsubayashi J, Nagakawa Y. Long-term surgical outcomes of gastric neuroendocrine carcinoma and mixed neuroendocrine-non-neuroendocrine neoplasms. World J Surg Oncol. 2022;20:165. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in RCA: 10] [Reference Citation Analysis (0)] |
| 4. | Exarchou K, Stephens NA, Moore AR, Howes NR, Pritchard DM. New Developments in Gastric Neuroendocrine Neoplasms. Curr Oncol Rep. 2022;24:77-88. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 1] [Cited by in RCA: 7] [Article Influence: 2.3] [Reference Citation Analysis (0)] |
| 5. | Sackstein PE, O'Neil DS, Neugut AI, Chabot J, Fojo T. Epidemiologic trends in neuroendocrine tumors: An examination of incidence rates and survival of specific patient subgroups over the past 20 years. Semin Oncol. 2018;45:249-258. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 41] [Cited by in RCA: 62] [Article Influence: 8.9] [Reference Citation Analysis (0)] |
| 6. | Panzuto F, Campana D, Massironi S, Faggiano A, Rinzivillo M, Lamberti G, Sciola V, Lahner E, Manuzzi L, Colao A, Annibale B. Tumour type and size are prognostic factors in gastric neuroendocrine neoplasia: A multicentre retrospective study. Dig Liver Dis. 2019;51:1456-1460. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 39] [Cited by in RCA: 39] [Article Influence: 6.5] [Reference Citation Analysis (0)] |
| 7. | Song W, Tian C. The Effect of Marital Status on Survival of Patients with Gastrointestinal Stromal Tumors: A SEER Database Analysis. Gastroenterol Res Pract. 2018;2018:5740823. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 5] [Cited by in RCA: 10] [Article Influence: 1.4] [Reference Citation Analysis (0)] |
| 8. | Tang X, Chen Y, Guo L, Zhang J, Wang C. Prognostic significance of metastatic lymph node number, ratio and station in gastric neuroendocrine carcinoma. J Gastrointest Surg. 2015;19:234-241. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 9] [Cited by in RCA: 15] [Article Influence: 1.5] [Reference Citation Analysis (0)] |
| 9. | Widschwendter P, Polasik A, Janni W, de Gregorio A, Friedl TWP, de Gregorio N. Lymph Node Ratio Can Better Predict Prognosis than Absolute Number of Positive Lymph Nodes in Operable Cervical Carcinoma. Oncol Res Treat. 2020;43:87-95. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 4] [Cited by in RCA: 5] [Article Influence: 1.0] [Reference Citation Analysis (0)] |
| 10. | Ngiam KY, Khor IW. Big data and machine learning algorithms for health-care delivery. Lancet Oncol. 2019;20:e262-e273. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 371] [Cited by in RCA: 657] [Article Influence: 131.4] [Reference Citation Analysis (0)] |
| 11. | Christou CD, Tsoulfas G. Challenges and opportunities in the application of artificial intelligence in gastroenterology and hepatology. World J Gastroenterol. 2021;27:6191-6223. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in CrossRef: 34] [Cited by in RCA: 17] [Article Influence: 4.3] [Reference Citation Analysis (8)] |
| 12. | Song X, Xie Y, Lou Y. A novel nomogram and risk stratification system predicting the cancer-specific survival of patients with gastric neuroendocrine carcinoma: a study based on SEER database and external validation. BMC Gastroenterol. 2023;23:238. [RCA] [PubMed] [DOI] [Full Text] [Cited by in RCA: 6] [Reference Citation Analysis (0)] |
| 13. | Hu P, Bai J, Liu M, Xue J, Chen T, Li R, Kuai X, Zhao H, Li X, Tian Y, Sun W, Xiong Y, Tang Q. Trends of incidence and prognosis of gastric neuroendocrine neoplasms: a study based on SEER and our multicenter research. Gastric Cancer. 2020;23:591-599. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 22] [Cited by in RCA: 55] [Article Influence: 11.0] [Reference Citation Analysis (0)] |
| 14. | Pickett KL, Suresh K, Campbell KR, Davis S, Juarez-Colunga E. Random survival forests for dynamic predictions of a time-to-event outcome using a longitudinal biomarker. BMC Med Res Methodol. 2021;21:216. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 21] [Cited by in RCA: 35] [Article Influence: 8.8] [Reference Citation Analysis (0)] |
| 15. | Yang ZH, Han YJ, Cheng M, Wang R, Li J, Zhao HP, Gao JB. Prognostic value of computed tomography radiomics features in patients with gastric neuroendocrine neoplasm. Front Oncol. 2023;13:1143291. [RCA] [PubMed] [DOI] [Full Text] [Cited by in RCA: 4] [Reference Citation Analysis (0)] |
| 16. | Padwal MK, Basu S, Basu B. Application of Machine Learning in Predicting Hepatic Metastasis or Primary Site in Gastroenteropancreatic Neuroendocrine Tumors. Curr Oncol. 2023;30:9244-9261. [RCA] [PubMed] [DOI] [Full Text] [Cited by in RCA: 3] [Reference Citation Analysis (0)] |
| 17. | Jiang C, Wang K, Yan L, Yao H, Shi H, Lin R. Predicting the survival of patients with pancreatic neuroendocrine neoplasms using deep learning: A study based on Surveillance, Epidemiology, and End Results database. Cancer Med. 2023;12:12413-12424. [RCA] [PubMed] [DOI] [Full Text] [Cited by in RCA: 7] [Reference Citation Analysis (0)] |