Published online Oct 15, 2024. doi: 10.4251/wjgo.v16.i10.4194
Revised: June 9, 2024
Accepted: June 28, 2024
Published online: October 15, 2024
Processing time: 261 Days and 21.8 Hours
The clinical effects and detailed roles of long non-coding RNA (LncRNA) steroid receptor RNA activator 1 (SRA1) in esophageal squamous cell carcinoma (ESCC) remain ambiguous. In the present study, the complementary sites between lncRNA SRA1, miRNA-363-5p, and phospholysine phosphohistidine inorganic pyrophosphate phosphatase (LHPP) predicted via bioinformatics analysis stimulated us to hypothesize that miRNA-363-5p/LHPP axis might be required for SRA1-mediated ESCC progression.
To investigate the molecular events of SRA1 in the malignant behavior in ESCC.
Thirty-eight ESCC tissues and paired adjacent normal tissues were acquired. SRA1 expression was detected in ESCC tissues and cell lines using quantitative reverse transcription-polymerase chain reaction. Cell counting Kit-8 assay, transwell invasion assay, glycolysis assay, and xenograft tumor model were performed to address the malignant biological behaviors of ESCC cells after the introduction of SRA1. The t-test and the χ2 test were used for com
SRA1 downregulation was identified in ESCC. ESCC patients exhibiting a low SRA1 expression faced shorter overall survival than those with a high SRA1 expression. The introduction of SRA1 inhibited cell proliferation, glucose uptake, and lactate production in ESCC. In vivo, the growth of ESCC was hindered by SRA1 overexpression. Then, SRA1 overexpresses the LHPP by inhibiting miRNA-363-5p. Lastly, the introduction of small interfering RNA si-LHPP or miRNA-363-5p mimic could abrogate the inhibition roles triggered by SRA1.
SRA1 inhibits the oncogenicity of ESCC via miRNA-363-5p/LHPP axis. The SRA1/miRNA-363-5p/LHPP pathway may be a therapeutic target for ESCC.
Core Tip: In this study, we identified abnormal expression of steroid receptor RNA activator 1 (SRA1) in esophageal squamous cell carcinoma (ESCC). SRA1 was significantly downregulated in ESCC tissues and cell lines, and its low expression was strongly associated with advanced tumor stage, metastasis, larger tumor size, and poor survival. Functional and rescue assays demonstrated that SRA1 could impede ESCC cell proliferation and glycolysis via miRNA-363-5p and phospholysine phosphohistidine inorganic pyrophosphate phosphatase (LHPP). Mechanistically, SRA1 elevated LHPP expression by sponging miRNA-363-5p, thereby inhibiting ESCC cell proliferation and glycolysis.
- Citation: He M, Qi Y, Zheng ZM, Sha M, Zhao X, Chen YR, Chen ZH, Qian RY, Yao J, Yang ZD. Long noncoding RNA steroid receptor RNA activator 1 inhibits proliferation and glycolysis of esophageal squamous cell carcinoma. World J Gastrointest Oncol 2024; 16(10): 4194-4208
- URL: https://www.wjgnet.com/1948-5204/full/v16/i10/4194.htm
- DOI: https://dx.doi.org/10.4251/wjgo.v16.i10.4194
There were 604100 new cases and 544076 deaths from esophageal squamous cell carcinoma (ESCC) estimated by 2020[1,2]. Because of the lack of obvious clinical symptoms and effective screening methods in the early stage, most patients with esophageal cancer are already in the middle and advanced stages when they are diagnosed[3,4].
Long non-coding RNAs (lncRNAs) are a class of biological macromolecules with a length greater than 200 nucleotides[5,6]. Tumor cells often use glycolysis to metabolize glucose[7]. Generally, increased glycolysis can meet the energy requirements of tumor cells and provide a favorable microenvironment for tumor occurrence and development[7]. Studies found that many lncRNAs are involved in the glycolysis process of tumors[8-11]. MiRNAs, in the form of oncogenes or tumor suppressor genes, play a role in promoting or suppressing tumors in ESCC[12]. Tumor suppressor protein, phospholysine phosphohistidine inorganic pyrophosphate phosphatase (LHPP), has the opposite effect of histidine kinase[13]. Studies found that low expression of LHPP can lead to up-regulation of histidine phosphorylation[14-16]. In the present study, the complementary sites between lncRNA steroid receptor RNA activator 1 (SRA1), miRNA-363-5p, and LHPP predicted via bioinformatics analysis stimulated us to hypothesize that miRNA-363-5p/LHPP axis might be required for SRA1-mediated ESCC progression.
In this study, the expression of SRA1 was verified in ESCC tissues. Then, we characterized the clinical implication as well as the biological function of SRA1 in ESCC. We found that SRA1 functions as a tumor suppressor gene in ESCC. In terms of mechanism, SRA1 inhibits the glycolysis and malignant biological behaviors of ESCC cells via functioning as a molecular sponge for miRNA-363-5p and upmodulating LHPP expression.
ESCC and paired adjacent tissues (the area of tumor margin > 5 cm) were acquired from 38 ESCC patients in the Huai’an Hospital of Huai’an City from January to December 2016. The age of the patients ranged from 54 to 77 years (median age was 65 years). There were 26 males and 12 females. The inclusion criteria for ESCC patients in this study were as follows: The patient was diagnosed as ESCC by two pathologists; The patient's personal information and pathological data were complete; The patient did not receive therapy before radical ESCC surgery; The patient completed the follow-up (36 months), and obtain complete follow-up data. The exclusion criteria for patients were as follows: The patient was re-diagnosed as non-squamous cell carcinoma; The patient's personal information and pathological data were missing; Patients sign an informed consent form for participation. Study was performed under the Ethics Committee of the Huai’an Hospital of Huai’an City (No. 2016.0112).
RNA extraction and purification were carried out following the manufacturer's guidelines using TRIzol reagent (Invitrogen, Carlsbad, CA, United States). The concentration and purity of RNA in each sample were assessed using a NanoDrop ND-1000. Additionally, the integrity of the RNA was evaluated using an Agilent 2100 system (Agilent, Santa Clara, CA, United States). After eliminating ribosomal RNAs, the remaining RNA molecules were subjected to high-temperature fragmentation, resulting in the generation of smaller fragments. Subsequently, the fragmented RNA pieces underwent reverse transcription to produce cDNA.
Candidate lncRNAs were selected using two criteria: (1) Transcripts with a length less than 200 nucleotides and reads coverage below 3 were excluded to eliminate interference from other noncoding RNAs (such as ribosomal RNA, transfer RNA, small nucleolar RNA, and small nuclear RNA); and (2) The coding ability of the transcripts was assessed using the coding-noncoding index and coding potential calculator, and transcripts with coding potential were excluded. The remaining transcripts were then categorized as lncRNAs.
Normal esophageal epithelial cells Het-1A and ESCC cells (TE1, Eca109, KYSE30, EC9706, KYSE180) were used. Cells were kept in DMEM under the condition of 5% CO2 and 37 °C. SRA1 was cloned into pcDNA3.1 vector to construct SRA1 overexpressing plasmid (pcDNA3.1-SRA1). The pcDNA3.1 vector was used as a negative control (NC).
Quantitative reverse transcription-polymerase chain reaction (QRT-PCR) was performed using the SYBR Green qPCR Kit (Thermo Fisher Scientific, Waltham, MA, United States) with a StepOne™ Real-Time PCR System (Applied Biosystems, Carlsbad, CA, United States). The glyceraldehyde-3-phosphate dehydrogenase (GAPDH) and U6 were used as control references for lncRNA/mRNA and miRNA. Their primers were purchased from Sangon Biotech (Shanghai, China), and the sequences as described previously are listed in Table 1. In this study, the amplification efficiencies of SRA1, miRNA-363-5p, LHPP, GAPDH and U6 were 97.8%, 96.9%, 99.8%, 98.4% and 99.2%, respectively.
| Name | Primer sequence |
| lncRNA SRA1 | F: 5′-GCTGGGCACTGGGAATGTAA-3′ |
| R: 5′-CACGACCCTACAACCCTCTG-3′ | |
| miRNA-363-5p | F: 5′-GGCGAGTTTTAATTTCTATT-3′ |
| R: 5′-ATCAACTGCTCTCGTGGA-3′ | |
| LHPP | F: 5′-GACGCAGCACTCACCCATCT-3′ |
| R: 5′-CCAGGCATTCGGTGATGTG-3′ | |
| GAPDH | F: 5′-AGAAGGCTGGGGCTCATTTG-3′ |
| R: 5′-GCAGGAGGCATTGCTGATGAT-3′ | |
| U6 | F: 5′-GCTTCGGCAGCACATATACTAAAAT-3′ |
| R: 5′-CGCTTCACGAATTTGCGTGTCAT-3′ |
After transfection, ESCC cells (2 × 103 cells/well) were plated onto 96-wells plates and cultivated for 0, 24, 48, and 72 hours, followed by addition into 10 μL CCK-8 solution (Abcam, Cambridge, MA, United States).
Glycolysis assay was performed as described previously[17-19]. After transfection, 2 × 105 ESCC cells were plated onto 6-wells plates, and the levels of glycolysis were measured (BioVision, Milpitas, CA, United States).
The membranes were incubated with LHPP (ab116175; 1:1000 dilution; Abcam, United States), glucose transport protein type 1 (GLUT1; 1:1000 dilution; ab115730, Abcam, United States), lactate dehydrogenase (LDHA; 1:1000 dilution; ab101562, Abcam, United States), GAPDH (1:3000 dilution; ab128915; Abcam, United States) antibodies.
Immunohistochemistry was performed in paraffin-embedded sections using an Immunohistochemistry kit (SV0002, BOSTER, China). Tissue samples were incubated with primary antibodies against Ki-67 (ab254123, Abcam, United States), followed by incubation with the secondary antibody.
Twelve Balb/c nude mice (3-4 weeks, male) were randomly divided into pcDNA3.1-NC group and pcDNA3.1-SRA1 group. And 1 × 106 EC9706 or TE1 cells were subcutaneously injected into nude mice. After 28 days, the mice were sacrificed. All animal experiments were conducted by the guideline of the Ethical Committees of Huai’an Hospital of Huai’an City (No. 2016.A023).
The t-test and the χ2 test were used for comparison between groups. Survival curve analysis was performed using the Kaplan–Meier method. P < 0.05 indicated statistical significance.
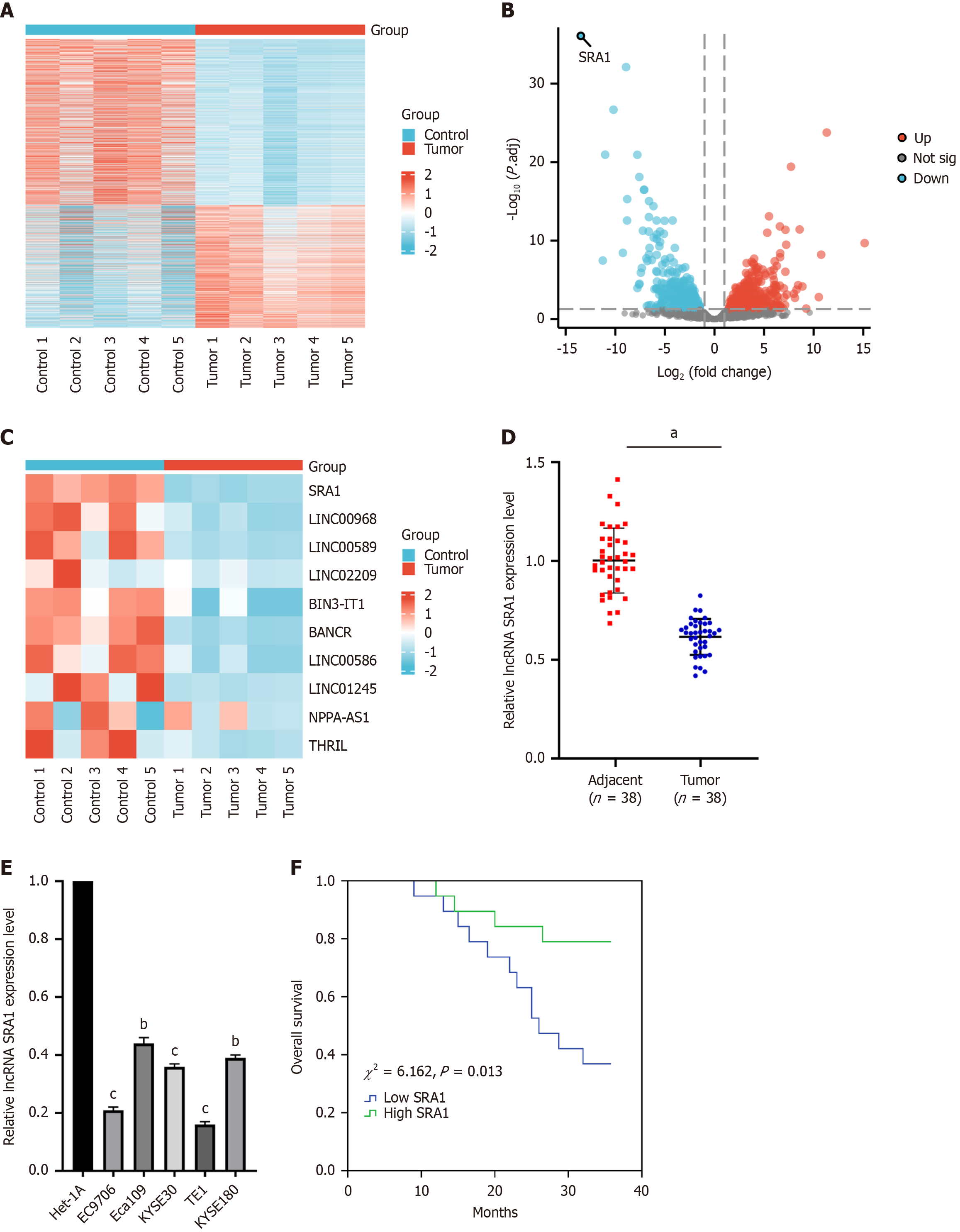
The lncRNA expression patterns were performed through hierarchical clustering between ESCC and adjacent normal tissues (Figure 1A). Changes in the lncRNA level were displayed in the volcano (Figure 1B) plot. As was observed, SRA1 had the highest fold of differential expression (Figure 1C). In clinical samples, we indicated that SRA1 expression was downregulated in ESCC tissues (Figure 1D) and cell lines (Figure 1E). Furthermore, ESCC patients with low SRA1 had shorter overall survival than patients with high SRA1 (χ2 = 6.162, P = 0.013; Figure 1F). In addition, the low SRA1 expression was correlated with lymph node metastasis, depth of tumor invasion, and TNM stage, but not with age, sex, or body mass index (Table 2).
| Characteristics | Case (n = 38) | SRA1 expression | χ2 | P value | |
| High | Low | ||||
| Sex | |||||
| Male | 26 | 12 | 14 | 0.487 | 0.485 |
| Female | 12 | 7 | 5 | ||
| Age (years) | 1.310 | 0.252 | |||
| ≤ 60 | 9 | 3 | 6 | ||
| > 60 | 29 | 16 | 13 | ||
| Pathological differentiation grade | 0.186 | 0.911 | |||
| Well | 7 | 4 | 3 | ||
| Moderately | 23 | 11 | 12 | ||
| Poorly | 8 | 4 | 4 | ||
| T Stage | 5.265 | 0.022a | |||
| T1-T2 | 20 | 13 | 7 | ||
| T3-T4 | 18 | 5 | 13 | ||
| TNM Stage | 8.622 | 0.003b | |||
| I-II | 17 | 13 | 4 | ||
| III-IV | 21 | 6 | 15 | ||
| Lymph node metastasis | 8.526 | 0.004b | |||
| Negative | 19 | 14 | 5 | ||
| Positive | 19 | 5 | 14 | ||
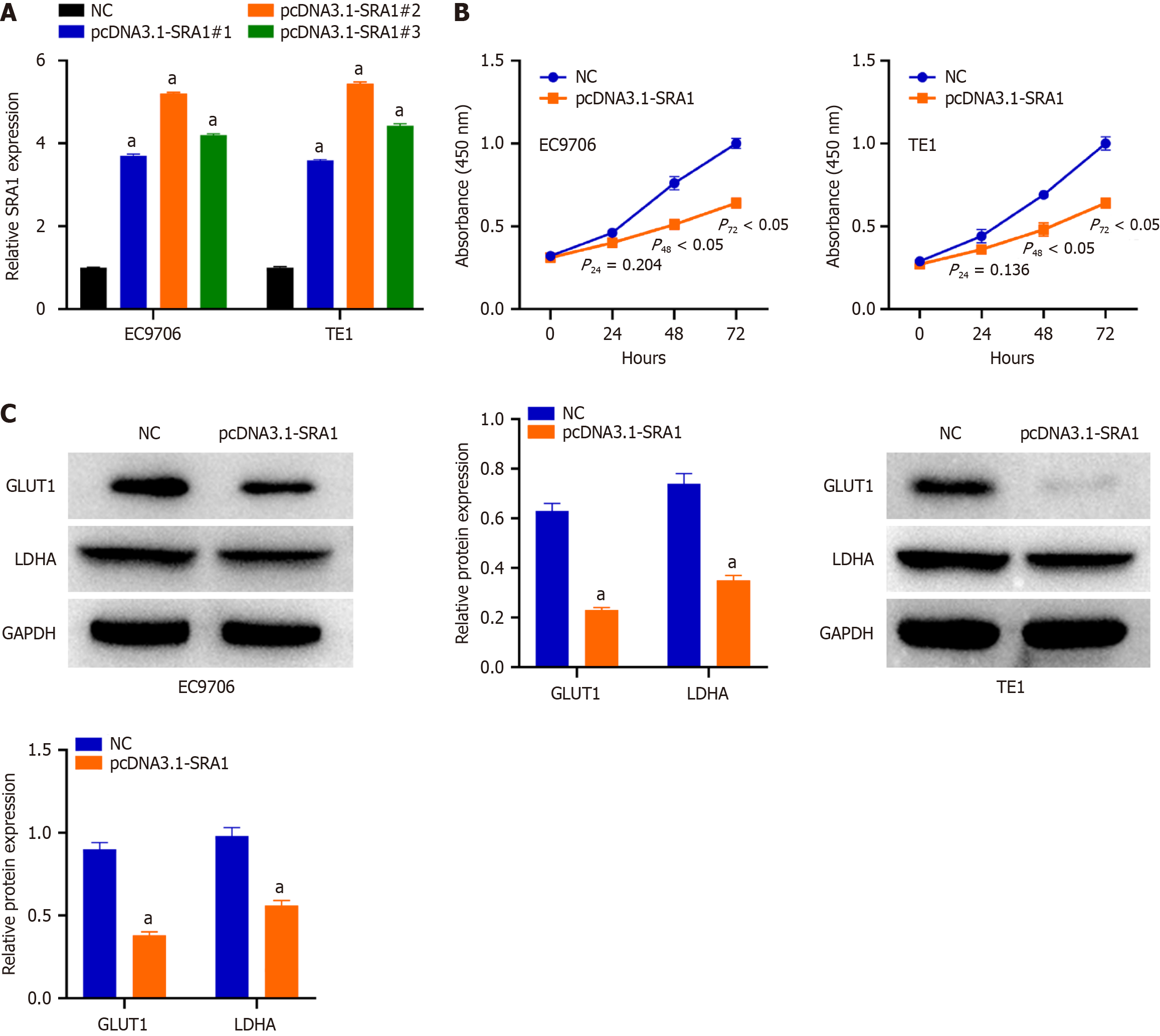
The relatively lower SRA1 was detected in the EC9706 and TE1 cells. To ascertain whether SRA1 is involved in the tumor resistance of ESCC, pcDNA3.1-SRA1 or NC was transfected into EC9706 and TE1 cells to overexpress endogenous SRA1 expression. The pcDNA3.1-SRA1#2 presented higher overexpressing efficiency; thus, the pcDNA3.1 was used (Figure 2A). Transfection with pcDNA3.1-SRA1 caused an obvious decrease in proliferation rate (Figure 2B) in EC9706 and TE1 cells. For glycolysis assay, when compared with NC group, a significant decrease in glucose consumption and lactate secretion was displayed in EC9706 and TE1 cells after pcDNA3.1-SRA1 transfection (Supplementary Figure 1). Additionally, upregulation of SRA1 triggered a decrease of GLUT1 and LDHA in EC9706 and TE1 cells than NC group (Figure 2C).
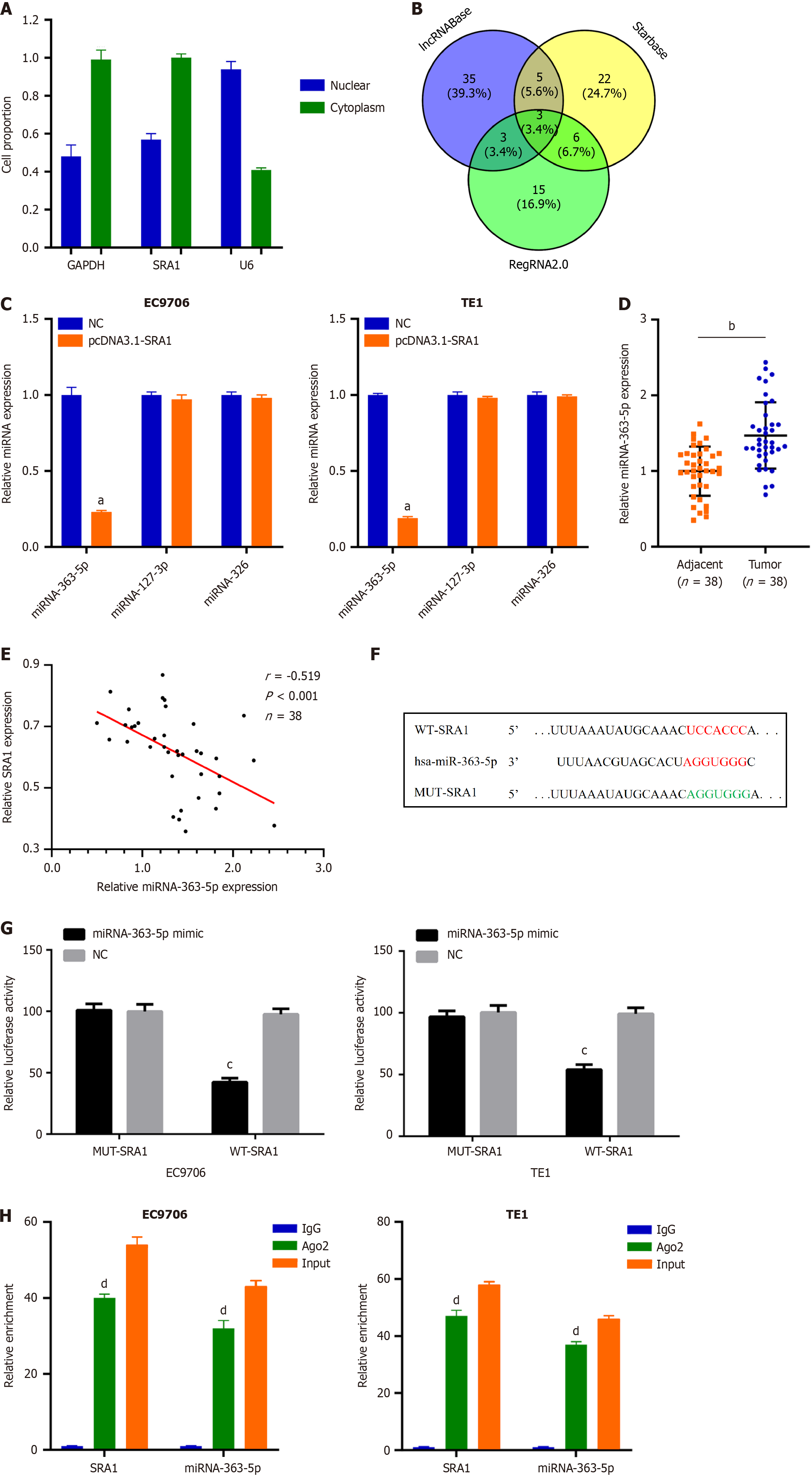
Next, SRA1 was observed in the cytoplasm (Figure 3A), which offered a theoretical basis for the ceRNA. Using lncRNABase, starBase, and RegRNA2.0, three miRNAs (miRNA-363-5p, miRNA-127-3p, and miRNA-326) were predicted to contain putative binding sites within SRA1 (Figure 3B). MiRNA-363-5p expression was inhibited in cells transfected with pcDNA3.1-SRA1 (Figure 3C). Furthermore, upregulated miRNA-363-5p was found in ESCC tissues (Figure 3D), which exhibited an inverse expression relation with SRA1 (r = -0.519, P < 0.001; Figure 3E). The Luciferase assay (Figure 3F and G) and RIP assay (Figure 3H) confirmed these findings.
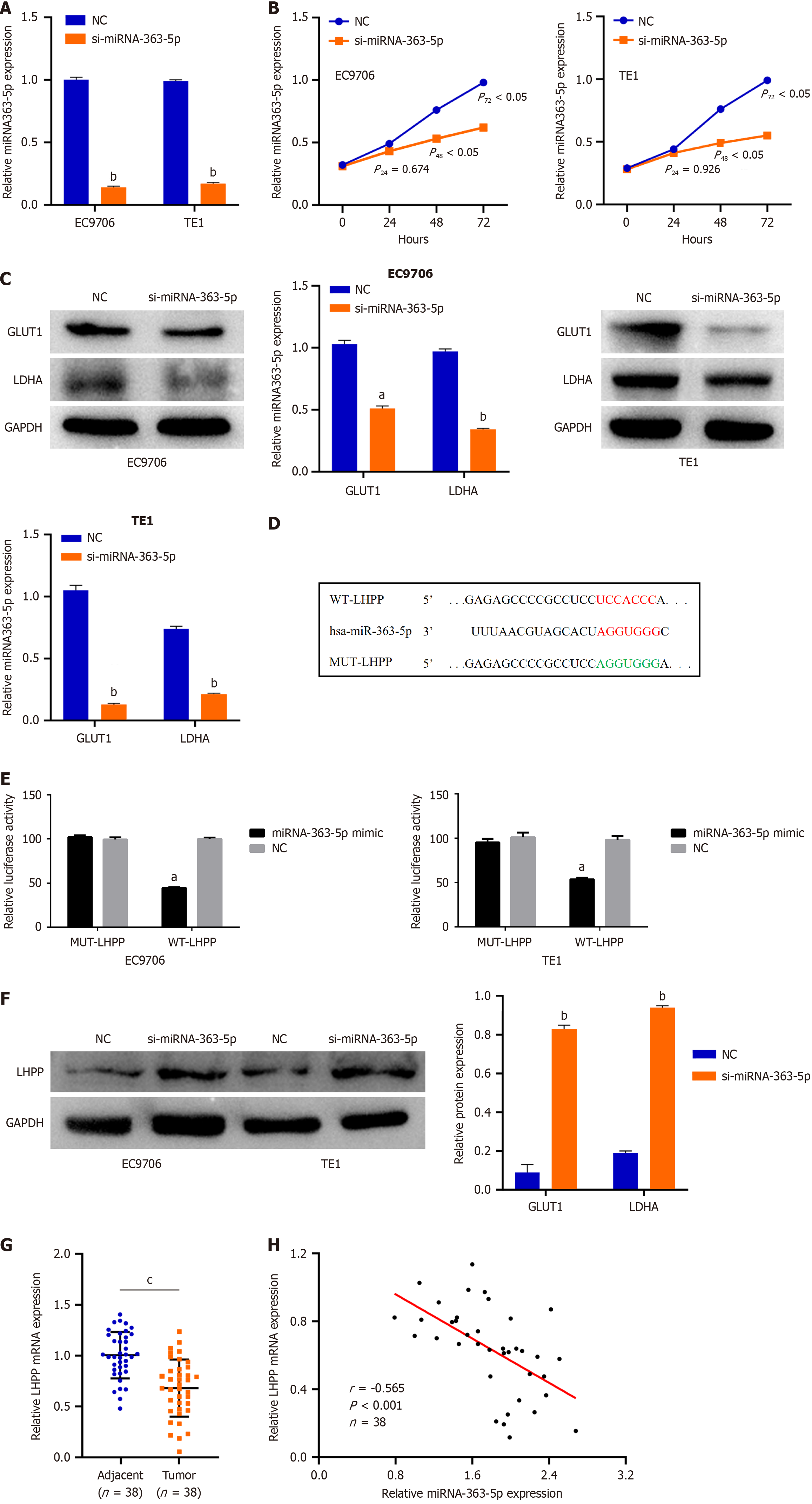
We next tried to ascertain the roles of miRNA-363-5p in ESCC cells. si-miRNA-363-5p was transfected into EC9706 and TE1 cells. MiRNA-363-5p remarkably decreased after si-miRNA-363-5p transfection (Figure 4A). Inhibition of miRNA-363-5p expression significantly attenuated proliferation (Figure 4B) and glycolysis (Figure 4C, Supplementary Figure 2) in EC9706 and TE1 cells.
To clarify the mechanism of miRNA-363-5p on ESCC, bioinformatics analysis was performed to explore the target of miRNA-363-5p. LHPP was predicted to have binding sequences for miRNA-363-5p (Figure 4D). Then, luciferase reporter turned out that miRNA-363-5p mimic weakened the luciferase activity of WT-LHPP (Figure 4E). Furthermore, downregulating miRNA-363-5p enhanced the LHPP protein (Figure 4F) levels in EC9706 and TE1 cells. Besides, LHPP expression was low in ESCC tissues compared to adjacent normal tissues (Figure 4G). An inverse correlation between miRNA-363-5p and LHPP was found in ESCC (r = -0.565, P < 0.001; Figure 4H). Collectively, miRNA-363-5p directly targets LHPP in ESCC cells.
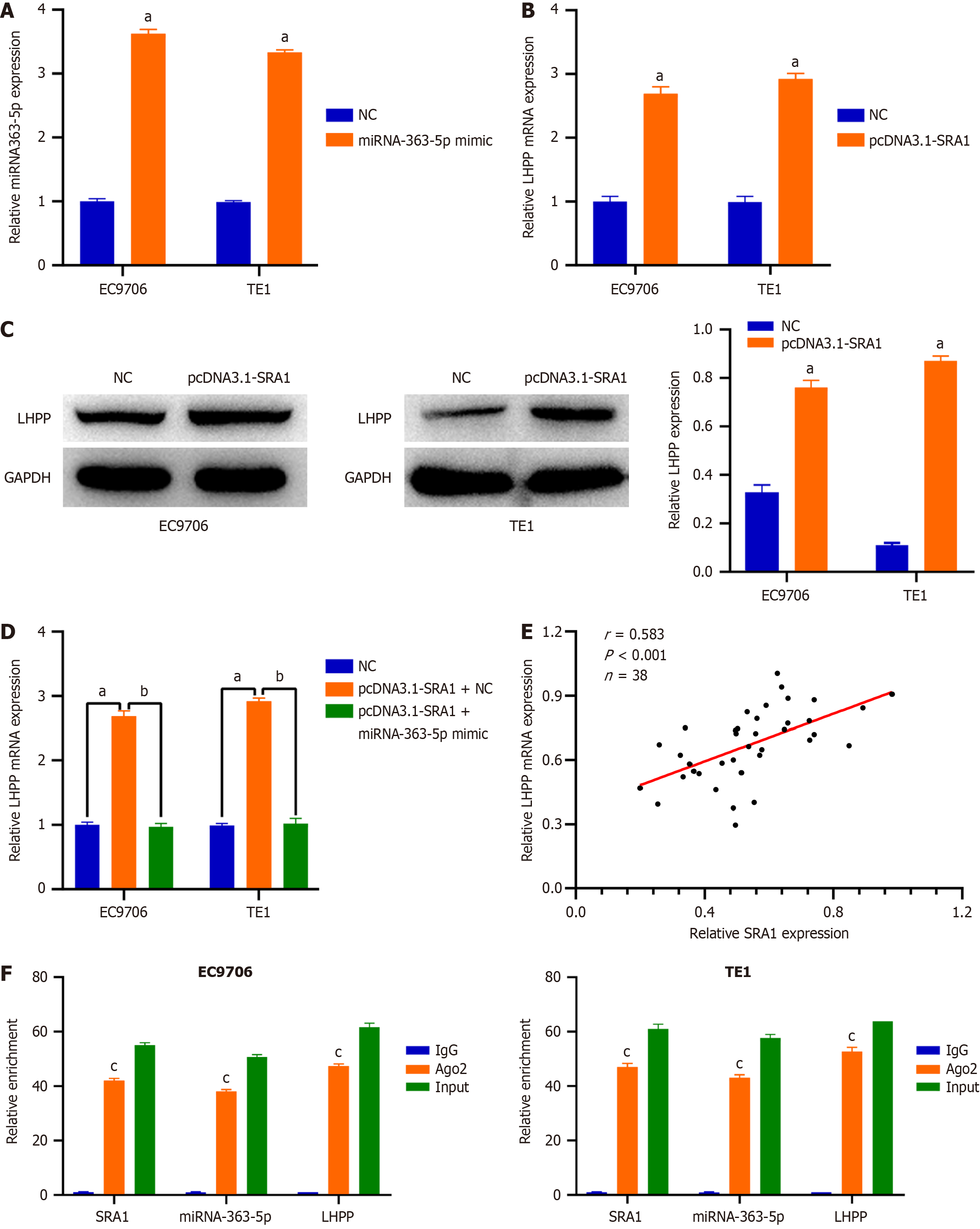
Then, EC9706 and TE1 cells were transfected with pcDNA3.1-SRA1 and miRNA-363-5p mimic either alone or in combination. The overexpression efficiency of miRNA-363-5p mimic in ESCC cells was tested by qRT-PCR. The delivery of miRNA-363-5p mimic caused a prominent miRNA-363-5p upregulation in EC9706 and TE1 cells (Figure 5A). SRA1 upregulation triggered an obvious overexpressed effect on the endogenous LHPP mRNA (Figure 5B) and protein (Figure 5C) levels in EC9706 and TE1 cells, whereas the delivery of miRNA-363-5p mimic counteracted such effect (Figure 5D). Furthermore, LHPP expression was positively correlated with SRA1 expression in ESCC tissues (r = 0.583, P < 0.001; Figure 5E). Besides, compared with the anti-IgG NC group, SRA1, miRNA-363-5p, and LHPP were preferentially concentrated in the anti-Ago2 precipitate (Figure 5F).
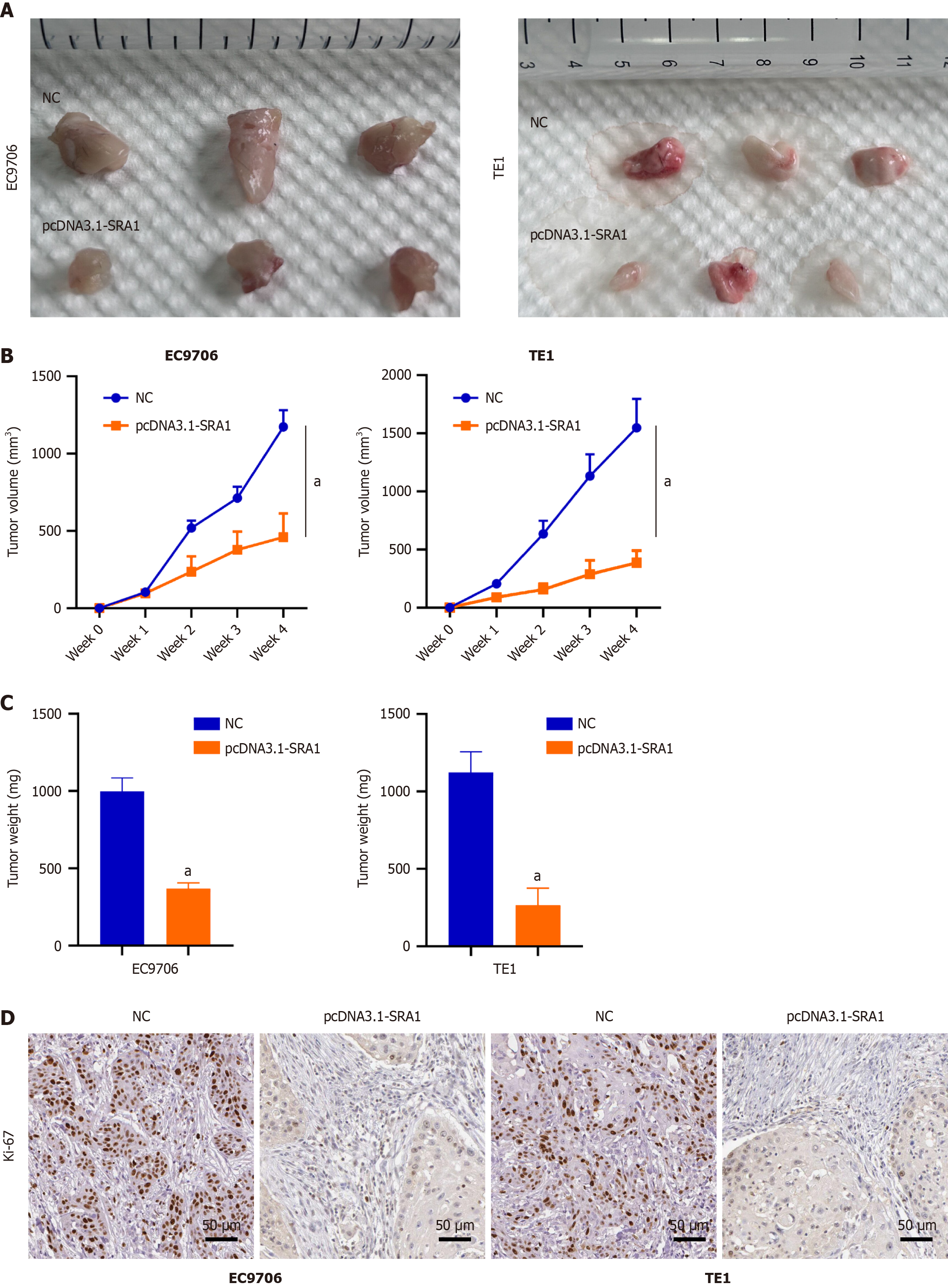
We implanted EC9706 and TE1 ESCC cells infected with pcDNA3.1-SRA1 into the nude mouse. Overexpressing SRA1 exhibited weaker tumorigenic ability (Figure 6A-C). The immunohistochemical staining found a low level of Ki-67 in the tumors with overexpressing SRA1 (Figure 6D).
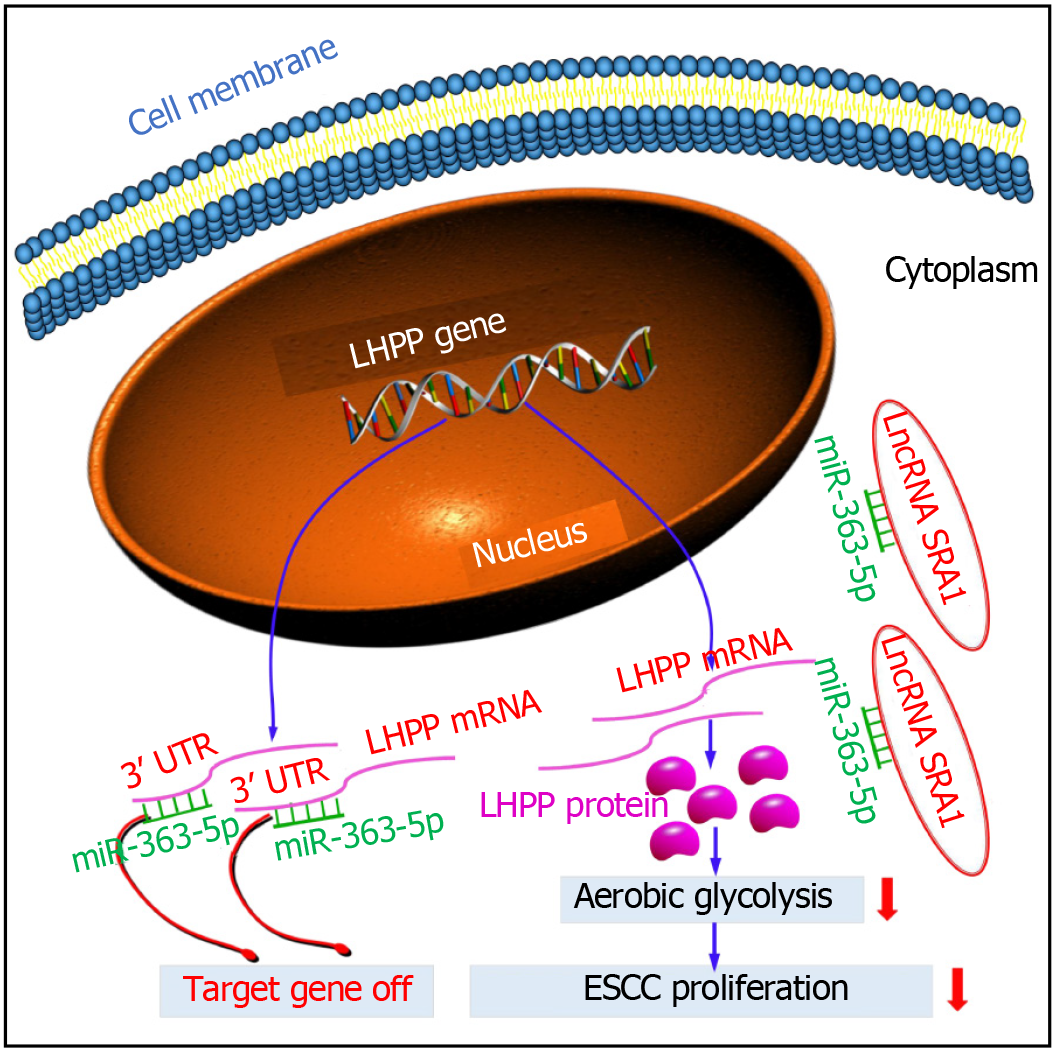
In this study, we found the abnormally expressed SRA1 in ESCC. Here, SRA1 was clearly downmodulated in ESCC tissues and cell lines, and low expression of SRA1 was significantly associated with advanced tumor stage, metastasis, larger tumor size, and poor survival. Function and rescue assays demonstrated that SRA1 could impede cell proliferation and glycolysis of ESCC cells via miRNA-363-5p and LHPP. Regarding the mechanism, SRA1 elevated LHPP expression by sponging miRNA-363-5p, leading to inhibition of ESCC cell proliferation and glycolysis (Figure 7).
Increasing studies have demonstrated that non-coding RNAs, including miRNAs, lncRNAs and circRNAs, play a pivotal role in ESCC formation, and may be used as therapeutic targets or diagnostic and prognostic biomarkers for ESCC patients[20-26]. At present, there are many reports on SRA1 and tumor progression, but its role in different tumors reflects tumor heterogeneity. There have been many reports on the research of SRA1 in some gynecological solid tumors. These studies have found that SRA1 plays an oncogene role, such as cervical cancer[27], endometrial cancer[28], breast cancer[29]. However, a number of studies in several tumors have pointed out that SRA1 acts as a tumor suppressor gene, such as hepatocellular carcinoma[30] and prostate cancer[31]. Moreover, using online bioinformatics software and qRT-PCR analysis, we screened a few miRNAs sharing binding sites with SRA1. In the present study, our bioinformatics analyses showed that miR-363-5p has binding sites for SRA1, and further experiments showed SRA1 knockdown strongly accelerated miR-363-5p expression in ESCC cells. And miR-363-5p was found to be upmodulated, and was inversely correlated with SRA1 Levels in ESCC tissues. Importantly, using luciferase reporter and RIP assays, SRA1 still has a targeting relationship with miR-363-5p. Further, we conducted the loss-of-function and rescue experiments, and found that the suppressive effects of SRA1 overexpression on cell proliferation, glucose uptake, and lactate production of ESCC cells were rescued by miR-363-5p mimic. The partial reversal effect of miR-363-5p mimic on pcDNA3.1-SRA1 function proved that miR-363-5p was one of the downstream miRNAs of SRA1.
However, this study has some limitations. (1) Our research explored and found for the first time that SRA1 inhibits the carcinogenicity of ESCC cells by targeting the miRNA-363-5p/LHPP axis. However, the downstream regulatory molecules of LHPP need to be further studied; and (2) Finally, it is worthy of our attention that we only detected the expression of SRA1/miRNA-363-5p/LHPP in ESCC tissues. Then, by expanding the number of ESCC patients, whether the detection of serum SRA1/miRNA-363-5p/LHPP in ESCC patients can discover early noninvasive markers of ESCC patients remains to be explored.
In conclusion, we found that SRA1 inhibits the oncogenicity of ESCC via miRNA-363-5p/LHPP axis. The SRA1/miRNA-363-5p/LHPP pathway may be a therapeutic target for ESCC.
| 1. | Chu LY, Peng YH, Weng XF, Xie JJ, Xu YW. Blood-based biomarkers for early detection of esophageal squamous cell carcinoma. World J Gastroenterol. 2020;26:1708-1725. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in CrossRef: 63] [Cited by in RCA: 61] [Article Influence: 12.2] [Reference Citation Analysis (2)] |
| 2. | Sung H, Ferlay J, Siegel RL, Laversanne M, Soerjomataram I, Jemal A, Bray F. Global Cancer Statistics 2020: GLOBOCAN Estimates of Incidence and Mortality Worldwide for 36 Cancers in 185 Countries. CA Cancer J Clin. 2021;71:209-249. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 75126] [Cited by in RCA: 64445] [Article Influence: 16111.3] [Reference Citation Analysis (176)] |
| 3. | Gao R, Wang Z, Liu Q, Yang C. MicroRNA-105 plays an independent prognostic role in esophageal cancer and acts as an oncogene. Cancer Biomark. 2020;27:173-180. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1] [Cited by in RCA: 5] [Article Influence: 1.0] [Reference Citation Analysis (0)] |
| 4. | Deng W, Chen J, Xiao Z. ASO Author Reflections: Prognostic Stratification and the Value of Adjuvant Therapy in Thoracic Esophageal Squamous Cell Carcinoma Patients After Esophagectomy. Ann Surg Oncol. 2019;26:802-803. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1] [Reference Citation Analysis (0)] |
| 5. | Niu Y, Guo Y, Li Y, Shen S, Liang J, Guo W, Dong Z. LncRNA GATA2-AS1 suppresses esophageal squamous cell carcinoma progression via the mir-940/PTPN12 axis. Exp Cell Res. 2022;416:113130. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1] [Cited by in RCA: 6] [Article Influence: 2.0] [Reference Citation Analysis (0)] |
| 6. | Zhang H, Hua Y, Jiang Z, Yue J, Shi M, Zhen X, Zhang X, Yang L, Zhou R, Wu S. Cancer-associated Fibroblast-promoted LncRNA DNM3OS Confers Radioresistance by Regulating DNA Damage Response in Esophageal Squamous Cell Carcinoma. Clin Cancer Res. 2019;25:1989-2000. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 67] [Cited by in RCA: 116] [Article Influence: 16.6] [Reference Citation Analysis (0)] |
| 7. | Gao W, Zhang Y, Luo H, Niu M, Zheng X, Hu W, Cui J, Xue X, Bo Y, Dai F, Lu Y, Yang D, Guo Y, Guo H, Li H, Zhang Y, Yang T, Li L, Zhang L, Hou R, Wen S, An C, Ma T, Jin L, Xu W, Wu Y. Targeting SKA3 suppresses the proliferation and chemoresistance of laryngeal squamous cell carcinoma via impairing PLK1-AKT axis-mediated glycolysis. Cell Death Dis. 2020;11:919. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 29] [Cited by in RCA: 52] [Article Influence: 10.4] [Reference Citation Analysis (0)] |
| 8. | Xue ST, Zheng B, Cao SQ, Ding JC, Hu GS, Liu W, Chen C. Long non-coding RNA LINC00680 functions as a ceRNA to promote esophageal squamous cell carcinoma progression through the miR-423-5p/PAK6 axis. Mol Cancer. 2022;21:69. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 6] [Cited by in RCA: 86] [Article Influence: 28.7] [Reference Citation Analysis (0)] |
| 9. | Yang J, Ren B, Yang G, Wang H, Chen G, You L, Zhang T, Zhao Y. The enhancement of glycolysis regulates pancreatic cancer metastasis. Cell Mol Life Sci. 2020;77:305-321. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 197] [Cited by in RCA: 246] [Article Influence: 49.2] [Reference Citation Analysis (0)] |
| 10. | Hu T, Liu H, Liang Z, Wang F, Zhou C, Zheng X, Zhang Y, Song Y, Hu J, He X, Xiao J, King RJ, Wu X, Lan P. Tumor-intrinsic CD47 signal regulates glycolysis and promotes colorectal cancer cell growth and metastasis. Theranostics. 2020;10:4056-4072. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 35] [Cited by in RCA: 105] [Article Influence: 21.0] [Reference Citation Analysis (0)] |
| 11. | Zhang C, Xie L, Fu Y, Yang J, Cui Y. lncRNA MIAT promotes esophageal squamous cell carcinoma progression by regulating miR-1301-3p/INCENP axis and interacting with SOX2. J Cell Physiol. 2020;235:7933-7944. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 16] [Cited by in RCA: 29] [Article Influence: 5.8] [Reference Citation Analysis (0)] |
| 12. | Yu J, Chen S, Niu Y, Liu M, Zhang J, Yang Z, Gao P, Wang W, Han X, Sun G. Functional Significance and Therapeutic Potential of miRNA-20b-5p in Esophageal Squamous Cell Carcinoma. Mol Ther Nucleic Acids. 2020;21:315-331. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 9] [Cited by in RCA: 18] [Article Influence: 3.6] [Reference Citation Analysis (0)] |
| 13. | Hindupur SK, Colombi M, Fuhs SR, Matter MS, Guri Y, Adam K, Cornu M, Piscuoglio S, Ng CKY, Betz C, Liko D, Quagliata L, Moes S, Jenoe P, Terracciano LM, Heim MH, Hunter T, Hall MN. The protein histidine phosphatase LHPP is a tumour suppressor. Nature. 2018;555:678-682. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 98] [Cited by in RCA: 165] [Article Influence: 23.6] [Reference Citation Analysis (0)] |
| 14. | Li Z, Zhou X, Zhu H, Song X, Gao H, Niu Z, Lu J. Purpurin binding interacts with LHPP protein that inhibits PI3K/AKT phosphorylation and induces apoptosis in colon cancer cells HCT-116. J Biochem Mol Toxicol. 2021;35:e22665. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 13] [Cited by in RCA: 13] [Article Influence: 3.3] [Reference Citation Analysis (0)] |
| 15. | Sun W, Qian K, Guo K, Chen L, Xiang J, Li D, Wu Y, Ji Q, Sun T, Wang Z. LHPP inhibits cell growth and migration and triggers autophagy in papillary thyroid cancer by regulating the AKT/AMPK/mTOR signaling pathway. Acta Biochim Biophys Sin (Shanghai). 2020;52:382-389. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 15] [Cited by in RCA: 32] [Article Influence: 6.4] [Reference Citation Analysis (0)] |
| 16. | Zhang X, Kang H, Xiao J, Shi B, Li X, Chen G. LHPP Inhibits the Proliferation and Metastasis of Renal Cell Carcinoma. Biomed Res Int. 2020;2020:7020924. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 10] [Cited by in RCA: 19] [Article Influence: 3.8] [Reference Citation Analysis (0)] |
| 17. | Lu T, Ma K, Zhan C, Yang X, Shi Y, Jiang W, Wang H, Wang S, Wang Q, Tan L. Downregulation of long non-coding RNA LINP1 inhibits the malignant progression of esophageal squamous cell carcinoma. Ann Transl Med. 2020;8:675. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 2] [Cited by in RCA: 2] [Article Influence: 0.4] [Reference Citation Analysis (0)] |
| 18. | Zhang Q, Guan F, Fan T, Li S, Ma S, Zhang Y, Guo W, Liu H. LncRNA WDFY3-AS2 suppresses proliferation and invasion in oesophageal squamous cell carcinoma by regulating miR-2355-5p/SOCS2 axis. J Cell Mol Med. 2020;24:8206-8220. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 12] [Cited by in RCA: 25] [Article Influence: 5.0] [Reference Citation Analysis (0)] |
| 19. | Tang Y, Yang P, Zhu Y, Su Y. LncRNA TUG1 contributes to ESCC progression via regulating miR-148a-3p/MCL-1/Wnt/β-catenin axis in vitro. Thorac Cancer. 2020;11:82-94. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 17] [Cited by in RCA: 30] [Article Influence: 5.0] [Reference Citation Analysis (0)] |
| 20. | Wang D, You D, Pan Y, Liu P. Downregulation of lncRNA-HEIH curbs esophageal squamous cell carcinoma progression by modulating miR-4458/PBX3. Thorac Cancer. 2020;11:1963-1971. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 4] [Cited by in RCA: 10] [Article Influence: 2.0] [Reference Citation Analysis (0)] |
| 21. | Jiang B, Zhao Y, Shi M, Song L, Wang Q, Qin Q, Song X, Wu S, Fang Z, Liu X. DNAJB6 Promotes Ferroptosis in Esophageal Squamous Cell Carcinoma. Dig Dis Sci. 2020;65:1999-2008. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 14] [Cited by in RCA: 41] [Article Influence: 8.2] [Reference Citation Analysis (0)] |
| 22. | Huang X, Liu C, Li H, Dai T, Luo G, Zhang C, Li T, Lü M. Hypoxia-responsive lncRNA G077640 promotes ESCC tumorigenesis via the H2AX-HIF1α-glycolysis axis. Carcinogenesis. 2023;44:383-393. [RCA] [PubMed] [DOI] [Full Text] [Cited by in RCA: 11] [Reference Citation Analysis (0)] |
| 23. | Lu Z, Chen Z, Li Y, Wang J, Zhang Z, Che Y, Huang J, Sun S, Mao S, Lei Y, Gao Y, He J. TGF-β-induced NKILA inhibits ESCC cell migration and invasion through NF-κB/MMP14 signaling. J Mol Med (Berl). 2018;96:301-313. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 23] [Cited by in RCA: 38] [Article Influence: 5.4] [Reference Citation Analysis (0)] |
| 24. | Sang C, Chao C, Wang M, Zhang Y, Luo G, Zhang X. Identification and validation of hub microRNAs dysregulated in esophageal squamous cell carcinoma. Aging (Albany NY). 2020;12:9807-9824. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 17] [Cited by in RCA: 18] [Article Influence: 3.6] [Reference Citation Analysis (0)] |
| 25. | He Z. LINC00473/miR-497-5p Regulates Esophageal Squamous Cell Carcinoma Progression Through Targeting PRKAA1. Cancer Biother Radiopharm. 2019;34:650-659. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 16] [Cited by in RCA: 19] [Article Influence: 3.2] [Reference Citation Analysis (0)] |
| 26. | Liu T, Liang X, Yang S, Sun Y. Long noncoding RNA PTCSC1 drives esophageal squamous cell carcinoma progression through activating Akt signaling. Exp Mol Pathol. 2020;117:104543. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 2] [Cited by in RCA: 2] [Article Influence: 0.4] [Reference Citation Analysis (0)] |
| 27. | Eoh KJ, Paek J, Kim SW, Kim HJ, Lee HY, Lee SK, Kim YT. Long non-coding RNA, steroid receptor RNA activator (SRA), induces tumor proliferation and invasion through the NOTCH pathway in cervical cancer cell lines. Oncol Rep. 2017;38:3481-3488. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 8] [Cited by in RCA: 26] [Article Influence: 3.3] [Reference Citation Analysis (0)] |
| 28. | Park SA, Kim LK, Kim YT, Heo TH, Kim HJ. Long non-coding RNA steroid receptor activator promotes the progression of endometrial cancer via Wnt/ β-catenin signaling pathway. Int J Biol Sci. 2020;16:99-115. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 8] [Cited by in RCA: 21] [Article Influence: 4.2] [Reference Citation Analysis (0)] |
| 29. | Cooper C, Guo J, Yan Y, Chooniedass-Kothari S, Hube F, Hamedani MK, Murphy LC, Myal Y, Leygue E. Increasing the relative expression of endogenous non-coding Steroid Receptor RNA Activator (SRA) in human breast cancer cells using modified oligonucleotides. Nucleic Acids Res. 2009;37:4518-4531. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 74] [Cited by in RCA: 78] [Article Influence: 4.9] [Reference Citation Analysis (0)] |
| 30. | Luo P, Jing W, Zhu M, Li ND, Zhou H, Yu MX, Liang CZ, Tu JC. Decreased expression of LncRNA SRA1 in hepatocellular carcinoma and its clinical significance. Cancer Biomark. 2017;18:285-290. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 13] [Cited by in RCA: 17] [Article Influence: 2.1] [Reference Citation Analysis (0)] |
| 31. | Zhang M, Wang F, Zhang X. miRNA-627 inhibits cell proliferation and cell migration, promotes cell apoptosis in prostate cancer cells through upregulating MAP3K1, PTPRK and SRA1. Int J Clin Exp Pathol. 2018;11:255-261. [PubMed] |