Published online Sep 15, 2023. doi: 10.4251/wjgo.v15.i9.1567
Peer-review started: February 13, 2023
First decision: May 23, 2023
Revised: July 10, 2023
Accepted: August 6, 2023
Article in press: August 6, 2023
Published online: September 15, 2023
Processing time: 212 Days and 4.7 Hours
Cellular senescence, a state of stable growth arrest, is intertwined with human cancers. However, characterization of cellular senescence-associated phenotypes in hepatocellular carcinoma (HCC) remains unexplored.
To address this issue, we delineated cellular senescence landscape across HCC.
We enrolled two HCC datasets, TCGA-LIHC and International Cancer Genome Consortium (ICGC). Unsupervised clustering was executed to probe tumor heterogeneity based upon cellular senescence genes. Least absolute shrinkage and selection operator algorithm were utilized to define a cellular senescence-relevant scoring system. TRNP1 expression was measured in HCCs and normal tissues through immunohistochemistry, immunoblotting and quantitative real-time polymerase chain reaction. The influence of TMF-regulated nuclear protein (TRNP)1 on HCC senescence and growth was proven via a series of experiments.
TCGA-LIHC patients were classified as three cellular senescence subtypes, named C1–3. The robustness and reproducibility of these subtypes were proven in the ICGC cohort. C2 had the worst overall survival, C1 the next, and C3 the best. C2 presented the highest levels of immune checkpoints, abundance of immune cells, and immunogenetic indicators. Thus, C2 might possibly respond to immunotherapy. C2 had the lowest somatic mutation rate, while C1 presented the highest copy number variations. A cellular senescence-relevant gene signature was generated, which can predict patient survival, and chemo- or immunotherapeutic response. Experimentally, it was proven that TRNP1 presented the remarkable upregulation in HCCs. TRNP1 knockdown induced apoptosis and senescence of HCC cells and attenuated tumor growth.
These findings provide a systematic framework for assessing cellular senescence in HCC, which decode the tumor heterogeneity and tailor the pharmacological interventions to improve clinical management.
Core Tip: Cellular senescence, a state of stable growth arrest, is implicated in human cancers. Nevertheless, characterization of cellular senescence-associated phenotypes in hepatocellular carcinoma (HCC) is still indistinct. Here, we proposed a novel cellular senescence-based classification for HCC and identified TRNP1 as a novel therapeutic target.
- Citation: Wang HH, Chen WL, Cui YY, Gong HH, Li H. Cellular senescence throws new insights into patient classification and pharmacological interventions for clinical management of hepatocellular carcinoma. World J Gastrointest Oncol 2023; 15(9): 1567-1594
- URL: https://www.wjgnet.com/1948-5204/full/v15/i9/1567.htm
- DOI: https://dx.doi.org/10.4251/wjgo.v15.i9.1567
Cellular senescence is defined as an irreversible cessation of cellular division of cells with normal proliferation[1]. Human cells age due to progressive shortening of telomeres following cellular division, stress, oncogenes, etc[2]. Numerous genes have been implicated in cellular senescence as biomarkers and causal drivers[3,4]. Cellular senescence is a double-edged sword for cancer and its treatment[5,6]. The growth arrest and immunomodulatory features linked with senescence possess powerful antimalignant roles[7]. In addition, senescence bypass and secretory phenotype correlate to tumor progression and recurrence[8]. Research has unveiled the great potential for antiaging interventions as a novel antitumor strategy[9]. Nonetheless, the heterogeneity of senescence-related features makes the definition and targeting of treatment-induced senescent cells challenging[10].
Hepatocellular carcinoma (HCC) is a poorly managed malignancy with high mortality due to the lack of response to classical chemotherapy agents (doxorubicin, cisplatin, etc.) and targeted agents in the early stage[11]. For late-stage HCCs, single sorafenib or combination therapy remains the mainstay in first-line therapy, which improves overall survival by 3 mo[12]. The modest therapeutic success is largely attributable to sorafenib resistance[13]. Immunotherapy with checkpoint inhibitors (anti-PD-1/PD-L1) has displayed potent anti-HCC activity in a subset of patients[14]. The main unmet challenge in HCC immunotherapy is to discover and verify predictive biomarkers[15]. Accumulated evidence demonstrates that inducing tumor cells into senescence represents a potential anti-HCC therapy[16]. In HCCs, cellular senescence is primarily controlled by p53-dependent or -independent mechanisms[17]. Paradis et al[18] investigated replicative senescence in normal liver, chronic hepatitis C, and HCC, and demonstrated that chronic hepatitis C is a relevant model of accelerated replicative senescence and that accumulation of replicative senescent cells predispose to HCC progression[18]. Hepatic stellate cell activation and senescence also trigger the development of liver cirrhosis towards HCC[19]. Yildiz et al[20] found that cirrhosis and HCC exhibit expression patterns compatible with senescent and immortal phenotypes, respectively, while dysplasia is a transitional state. Senescence bypass exerts an essential role in hepatocellular carcinogenesis engendering systematic alteration in the transcription of genes modulating DNA repair, proliferation, differentiation, metabolism, etc[20]. Eggert et al[21] reported that while chemokines secreted by senescent hepatocytes inhibit liver cancer initiation, they enable to facilitate the growth of fully established HCC[21]. Due to the highly heterogeneous malignancy at the molecular and histological levels, characterization of cellular senescence-based classification might facilitate the personalized treatment of HCCs. Recently, cell senescence molecular subtypes have been conducted for predicting prognostic outcomes and immunotherapeutic responses of hepatitis B virus-related HCC patients[22]. A cellular-senescence-related classifier has been developed for inferring predicting prognosis, immunotherapeutic responses, and candidate agents in HCCs[23]. However, these findings are based upon retrospective analysis, and lack of experimental validation. To address these problems, our integrative analysis classified HCCs as three cellular senescence subtypes and defined a cellular senescence-relevant scoring system, which decoded the tumor heterogeneity as well as tailored the pharmacological interventions to boost clinical management of HCC.
Totally, 279 human cellular senescence genes were acquired from the CellAge database (https://genomics.senescence.info/cells/)[3,4]. Genes that induce cellular senescence present the overexpression with age in human tissue samples and are notably overrepresented in antilongevity and tumor-suppressor genes; meanwhile, genes that inhibit cellular senescence overlap with prolongevity genes and oncogenes. The detailed information is listed in Supplementary Table 1.
HCC patients were acquired from three public datasets, covering the Cancer Genome Atlas (TCGA-LIHC) database (https://portal.gdc.cancer.gov/projects/TCGA-LIHC) (n = 368), the International Cancer Genome Consortium (ICGC) portal (https://dcc.icgc.org/projects/LIRI-JP/) (n = 232), and the GSE14520 (https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE14520). Clinicopathological traits of above datasets are summarized in Supplementary Table 2. All expression data were transformed o transcripts per kilobase million, followed by log-2 conversion.
Utilizing limma package, differentially expressed cellular senescence genes were screened in HCC relative to normal liver tissues[24]. To prevent high false-positive rate, P values were adjusted via Benjamini–Hochberg approach. Adjusted P < 0.01 and |log2 fold change (FC)| > 0.58 were regarded as the criteria of differentially expressed genes.
Gene Ontology (GO) and Kyoto Encyclopedia of Genes and Genomes (KEGG) were annotated by use of clusterProfiler package[25]. GO terms and KEGG pathways with adjusted P < 0.05 were significantly enriched. The activity of fifty hallmark pathways was computed through GSVA package[26] based upon the gene sets of Molecular Signatures Database[27].
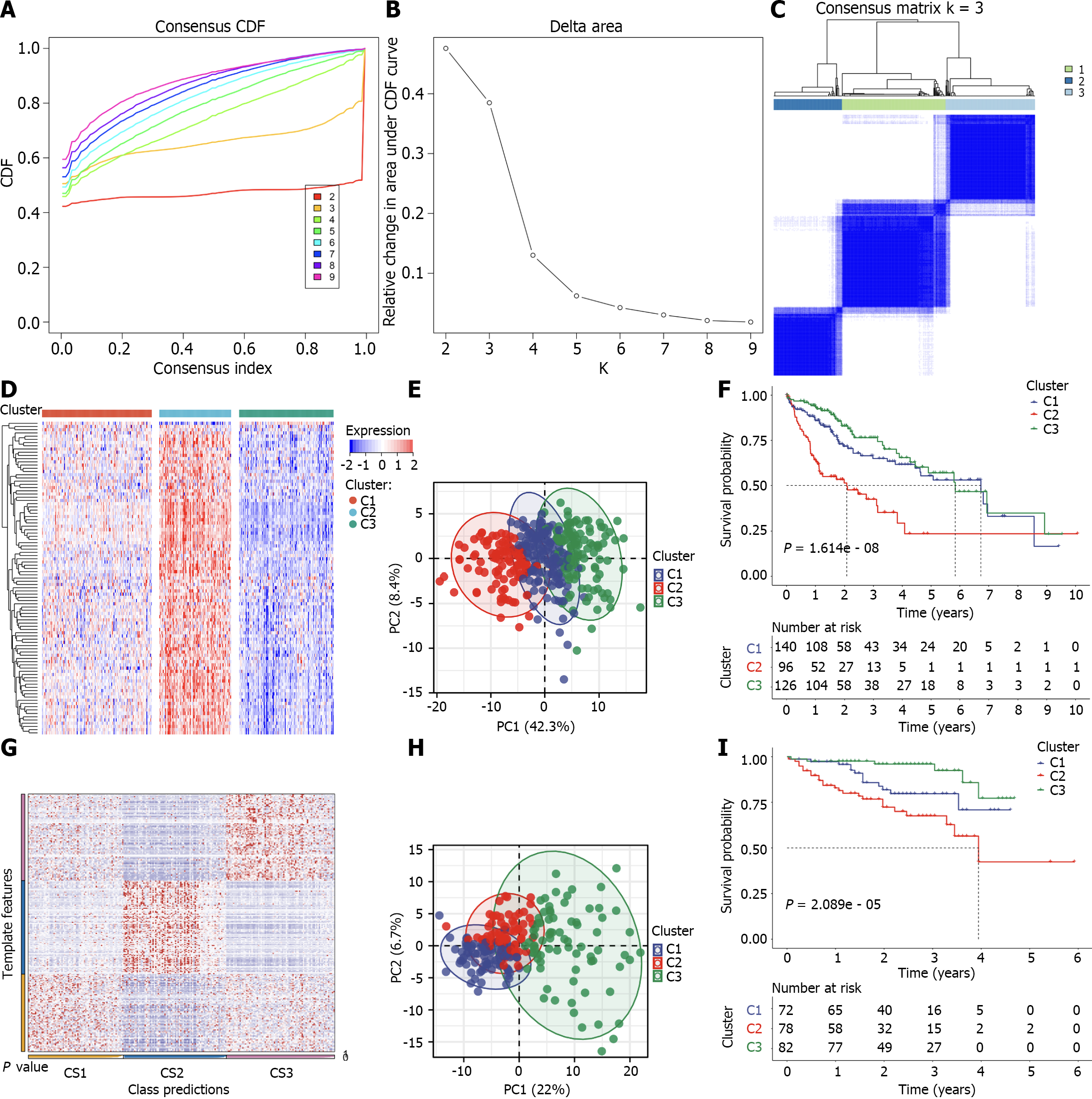
Based upon the prognostic differentially expressed cellular senescence genes derived from univariate Cox regression (P < 0.05), unsupervised clustering was implemented for TCGA-LIHC patients utilizing ConsensusClusterPlus package[28]. This process was conducted with 1000 iterations through sampling 80% of all the data for each iteration, thus ensuring clustering stability. The optimal number of clusters was identified utilizing consensus heatmap together with cumulative distribution function (CDF) curves. Principal component analysis (PCA) was utilized for recognizing and visualizing distinct subtypes.
Nearest template prediction (NTP) method is flexible for evaluating class prediction confidence for patients. Up-regulated genes were regarded as markers of each subtype with adjusted P < 0.05, which were adopted in the NTP method derived from CMScaller package[29], thus assessing the reliability and stability of subtypes.
Single-cell gene set enrichment analysis (ssGSEA), a deconvolution algorithm from GSVA package, was executed for quantifying the compositions within the tumor microenvironment (TME), comprising 22 immune cells and two stromal components (fibroblasts and endothelial cells). The ssGSEA score denoted the abundance of these TME components. The abundance of the TME components was also inferred through TIMER, CIBERSORT, CIBERSORT-ABS, QUANTISEQ, MCPCOUNTER, XCELL, together with EPIC methods.
The available mutation annotation format files from the TCGA were adopted for the analysis of somatic mutation utilizing maftools package[30]. Copy number variations (CNVs) of TCGA HCCs were stratified into three cellular senescence subtypes. Significant amplifications or deletions across the whole genome were assessed utilizing GISTIC2.0[31].
Genes with differential expression between subtypes were selected with the thresholds of adjusted P < 0.05 together with |log2FC| > 0.58. Cellular senescence subtype-relevant genes were determined following the intersection.
Univariate Cox regression analysis was utilized for picking out cellular-senescence-relevant genes with prognostic implication based upon P < 0.05. Through least absolute shrinkage and selection operator (LASSO) algorithm, the best gene subset was found out via glmnet package[32]. The cellular senescence-relevant scoring system was defined following the formula: RiskScore = Σ(coefficient (β)*Expression β), where β denoted each selected prognostic cellular senescence-relevant gene. HCCs were stratified into low- and high-RiskScore groups with the median RiskScore.
Uni- and multivariate Cox regression analyses on the cellular-senescence-relevant gene signature and conventional clinicopathological variables were executed to select independent prognostic factors in the TCGA-LIHC cohort. A nomogram based upon independent factors was generated to predict the probability of overall survival through rms package. Decision curve analysis was conducted for validating the nomogram[33].
The half-maximal inhibitory concentration (IC50) value of commonly used chemotherapy or targeted therapy drugs was inferred utilizing pRRophetic package[34]. Immunotherapy response was inferred by use of Tumor Immune Dysfunction and Exclusion (TIDE)[35].
Thirty fresh HCC tumors together with adjacent normal tissues were harvested from The Affiliated Bozhou Hospital of Anhui Medical University. No patients experienced any preoperative adjuvant treatment. HCC diagnosis was confirmed pathologically. Written informed consent was provided by each patient. This project gained the approval of the Ethics Committee of The Affiliated Bozhou Hospital of Anhui Medical University (2022-17).
TRNP1 expression in HCC or normal tissues was tested through fixing tissue sections with 4% paraformaldehyde. The sections were sealed utilizing goat serum, followed by incubation with TRNP1 antibody (1:500; ab174303; Abcam, Cambridge, MA, USA) along with secondary antibody (1/1000; ab7090). After administration with diaminobenzidine tetrahydrochloride, images were acquired under a microscope (Zeiss, Germany).
Total protein extraction was analyzed utilizing immunoblotting. Protein content was measured utilizing BCA kit (Beyotime, Shanghai, China). Proteins were separated via 12% SDS-PAGE, and transferred onto PVDF membranes that were then probed with primary antibody against TRNP1 (1/5000; ab174303; Abcam), p16 (1/500; ab151303), p21 (1/1000; ab109199) or GAPDH (1/2500; ab9485) at 4°C overnight, and secondary antibody (1/1000; ab7090) at room temperature for 2 h. Proteins were developed using ECL reagent (Beyotime).
RNA extraction was achieved utilizing RNA easy mini kit (Invitrogen, Carlsbad, CA, USA), with cDNA preparation via PrimeScript RT Master Mix (Takara, Dalian, China). Quantitative real-time polymerase chain reaction (RT-qPCR) was conducted via ChamQ SYBR qPCR Master Mix (Vazyme, Nanjing, China). The relative expression value was estimated with 2−ΔΔCt approach as well as normalized to endogenous GAPDH.
RPMI-1640 medium (Gibco) containing 10% fetal bovine serum (Gibco), and 1% penicillin–streptomycin (Gibco) was adopted for culturing SMMC-7721 and HepG2 HCC cells. All cells were maintained in an incubator at 37°C with 5% CO2. For transfection, siRNAs of TRNP1 (si-TRNP1) and negative control (si-NC) were acquired from GenePharma. Cell transfection was conducted utilizing Lipofectamine 2000 (Thermo Fisher Scientific).
Apoptotic rate was tested through flow cytometry utilizing Annexin V–fluorescein isothiocyanate (apoptosis detection kit (BD Biosciences). Cells were harvested and the assay was performed. Next, samples were assessed instantly utilizing flow cytometry (Beckman Coulter).
To investigate senescence, 104 cells were seeded onto a six-well plate. After being fixed, they were stained with senescence-associated -galactosidase activity (SA-β-gal) (Gibco).
Female BALB/c nude mice (5-wk-old, 16–18 g; Beijing Vital River Laboratory Animal Technology Co. Ltd., China) were fed under a 12-h light/dark cycle. They were divided into three groups (n = 5 per group). SMMC-7721 cells (n = 105) with si-NC, si-TRNP1#1 or si-TRNP1#2 were inoculated into the armpit. After 36 d, they were killed, with subsequent tumor excision. Tumor volume was finally calculated. This experiment gained the approval the Animal Ethics Committee of The Affiliated Bozhou Hospital of Anhui Medical University (LLSC20232071).
For between-group comparisons, unpaired Student’s t-test was adopted; Mann–Whitney U test was utilized for variables with non-normal distribution. Kaplan–Meier curves were utilized to estimate the overall survival of groups, with log-rank for testing the difference significance between groups. Survival analysis was executed utilizing survival and survminer packages. Receiver operator characteristic curves were plotted to evaluate the prediction efficacy in overall survival. Statistical analyses were implemented utilizing R packages and GraphPad Prism software. A two-sided P < 0.05 indicated statistical significance.
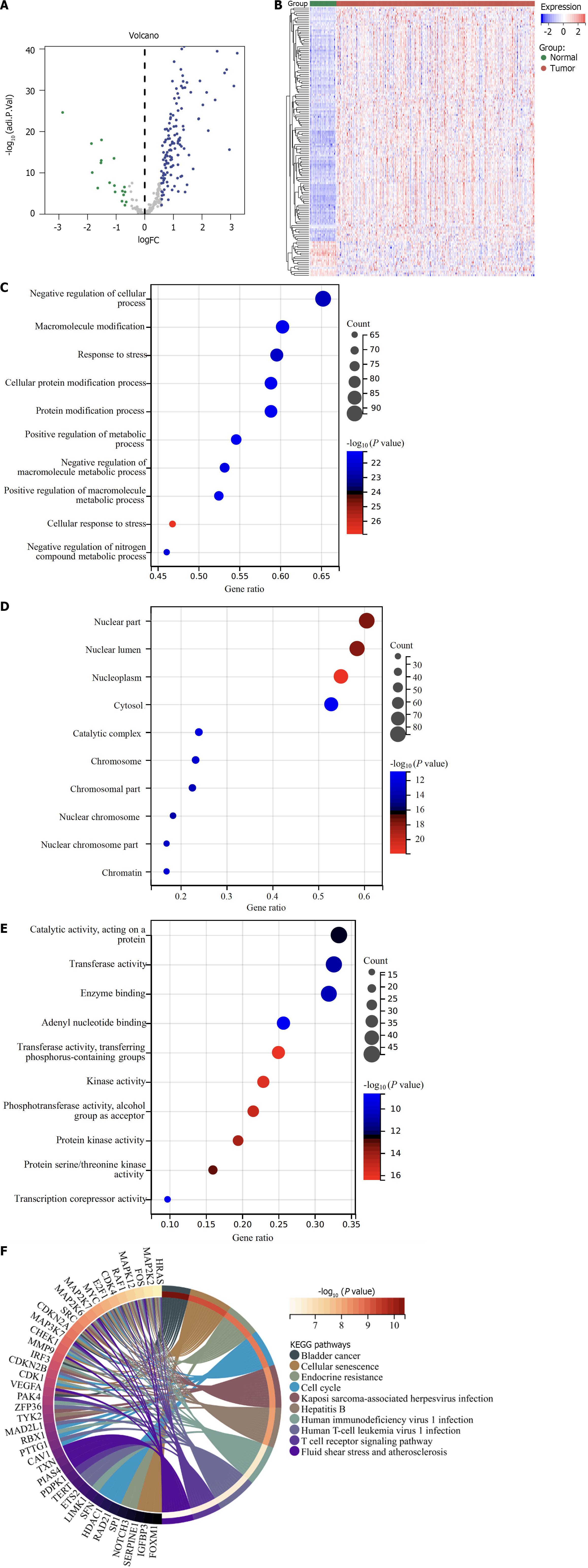
We identified 146 differentially expressed cellular senescence genes in TCGA HCCs relative to normal tissues with adjusted P < 0.01 and |log2FC| > 0.58 (Figure 1A and B; Supplementary Table 3), which might participate in HCC initiation or progression. They were linked with metabolic process, cellular senescence, cell cycle, and immunity pathways (Figure 1C–F). Their prognostic value was then assessed. Ninety-seven differentially expressed cellular senescence genes were significantly linked with HCC prognosis (Table 1).
| Gene | HR | 95%lower | 95%upper | P value | Gene | HR | 95%lower | 95%upper | P value |
| MCRS1 | 1.6954 | 1.2504 | 2.2988 | 0.0007 | TPR | 1.2593 | 1.0274 | 1.5436 | 0.0264 |
| FASTK | 1.3714 | 1.0147 | 1.8536 | 0.0399 | HDAC1 | 1.8639 | 1.4442 | 2.4055 | < 0.0001 |
| AURKA | 1.2794 | 1.1083 | 1.4770 | 0.0008 | SPOP | 1.4186 | 1.0061 | 2.0004 | 0.0461 |
| PTTG1 | 1.3361 | 1.1782 | 1.5151 | < 0.0001 | BTG3 | 1.3304 | 1.0766 | 1.6440 | 0.0082 |
| PSMB5 | 1.8929 | 1.3286 | 2.6969 | 0.0004 | IRF5 | 1.4232 | 1.1118 | 1.8217 | 0.0051 |
| MAP2K2 | 1.5424 | 1.1846 | 2.0083 | 0.0013 | IGFBP3 | 1.1741 | 1.0435 | 1.3210 | 0.0076 |
| E2F1 | 1.2175 | 1.0804 | 1.3721 | 0.0012 | CDKN2B | 1.3951 | 1.1619 | 1.6751 | 0.0004 |
| CDK1 | 1.3097 | 1.1553 | 1.4847 | < 0.0001 | AAK1 | 1.3018 | 1.0160 | 1.6682 | 0.0371 |
| GRK6 | 1.7940 | 1.3455 | 2.3920 | < 0.0001 | ARPC1B | 1.3007 | 1.0918 | 1.5495 | 0.0032 |
| EZH2 | 1.5722 | 1.3117 | 1.8845 | < 0.0001 | LIMK1 | 1.3450 | 1.1517 | 1.5706 | 0.0002 |
| DPY30 | 1.9266 | 1.3785 | 2.6928 | 0.0001 | SFN | 1.1541 | 1.0714 | 1.2432 | 0.0002 |
| CBX8 | 1.4643 | 1.1477 | 1.8684 | 0.0022 | CSNK2A1 | 1.3825 | 1.0896 | 1.7541 | 0.0077 |
| SMARCA4 | 1.4396 | 1.1405 | 1.8170 | 0.0022 | GLB1 | 1.4265 | 1.1097 | 1.8336 | 0.0056 |
| CDKN2A | 1.1748 | 1.0571 | 1.3055 | 0.0028 | MOB3A | 1.4527 | 1.1459 | 1.8416 | 0.0020 |
| IRF3 | 1.4048 | 1.0834 | 1.8216 | 0.0103 | BRD7 | 1.3897 | 1.0401 | 1.8567 | 0.0260 |
| HRAS | 1.4678 | 1.1958 | 1.8016 | 0.0002 | PRKCD | 1.4989 | 1.2458 | 1.8035 | < 0.0001 |
| ADCK5 | 1.3095 | 1.0375 | 1.6527 | 0.0232 | CDK4 | 1.4798 | 1.2301 | 1.7800 | < 0.0001 |
| RUVBL2 | 1.7508 | 1.3029 | 2.3528 | 0.0002 | BLVRA | 1.1790 | 1.0296 | 1.3500 | 0.0172 |
| ACLY | 1.3734 | 1.1160 | 1.6901 | 0.0027 | SENP1 | 1.5189 | 1.1628 | 1.9840 | 0.0022 |
| TACC3 | 1.3618 | 1.1752 | 1.5780 | < 0.0001 | BMI1 | 1.6112 | 1.2516 | 2.0742 | 0.0002 |
| SIRT6 | 1.6515 | 1.2586 | 2.1671 | 0.0003 | DHX9 | 1.3908 | 1.1069 | 1.7477 | 0.0046 |
| SUPT5H | 1.5227 | 1.1449 | 2.0252 | 0.0039 | RSL1D1 | 1.7134 | 1.2421 | 2.3634 | 0.0010 |
| FOXM1 | 1.2772 | 1.1287 | 1.4454 | 0.0001 | PAK4 | 1.2908 | 1.0419 | 1.5990 | 0.0195 |
| PSMD14 | 1.9715 | 1.5043 | 2.5838 | < 0.0001 | PDCD10 | 1.6080 | 1.2369 | 2.0903 | 0.0004 |
| HJURP | 1.4529 | 1.2514 | 1.6869 | < 0.0001 | ASF1A | 1.7774 | 1.3749 | 2.2977 | < 0.0001 |
| TRIM28 | 1.6005 | 1.2816 | 1.9988 | < 0.0001 | PNPT1 | 1.6790 | 1.2487 | 2.2576 | 0.0006 |
| P3H1 | 1.9307 | 1.5257 | 2.4431 | < 0.0001 | MAPK12 | 1.2163 | 1.0478 | 1.4119 | 0.0101 |
| MAGOHB | 1.4601 | 1.0477 | 2.0346 | 0.0254 | UBTD1 | 1.3794 | 1.0584 | 1.7977 | 0.0173 |
| RBX1 | 1.6692 | 1.2410 | 2.2451 | 0.0007 | KDM5B | 1.3241 | 1.1029 | 1.5895 | 0.0026 |
| MAGOH | 1.6449 | 1.1977 | 2.2592 | 0.0021 | USP1 | 1.4478 | 1.1790 | 1.7778 | 0.0004 |
| CENPA | 1.4947 | 1.2958 | 1.7241 | < 0.0001 | MAP3K7 | 1.4740 | 1.1327 | 1.9180 | 0.0039 |
| EWSR1 | 1.9373 | 1.3937 | 2.693 | < 0.0001 | PKM | 1.2079 | 1.0979 | 1.3290 | 0.0001 |
| HSPA5 | 1.3234 | 1.0428 | 1.6796 | 0.0212 | NINJ1 | 1.4743 | 1.0929 | 1.9888 | 0.0110 |
| CHEK1 | 1.5484 | 1.2766 | 1.8780 | < 0.0001 | SERPINE1 | 1.1201 | 1.0226 | 1.2269 | 0.0147 |
| PIAS4 | 1.4493 | 1.0683 | 1.9663 | 0.0171 | BRCA1 | 1.4378 | 1.1653 | 1.7741 | 0.0007 |
| PRPF19 | 2.5587 | 1.7707 | 3.6974 | < 0.0001 | DEK | 1.2103 | 1.0125 | 1.4468 | 0.0360 |
| MAPKAPK5 | 2.0718 | 1.4388 | 2.9834 | < 0.0001 | NDRG1 | 1.2946 | 1.1412 | 1.4686 | < 0.0001 |
| MAD2L1 | 1.4390 | 1.2065 | 1.7164 | < 0.0001 | SRC | 1.2011 | 1.0507 | 1.3730 | 0.0073 |
| TFAP4 | 1.9651 | 1.4205 | 2.7184 | < 0.0001 | ASPH | 1.2007 | 1.0488 | 1.3746 | 0.0081 |
| G6PD | 1.3897 | 1.2500 | 1.5450 | < 0.0001 | STK32C | 1.5149 | 1.2235 | 1.8759 | 0.0001 |
| GAPDH | 1.5984 | 1.2892 | 1.9818 | < 0.0001 | CDK2AP1 | 1.2327 | 1.0010 | 1.5181 | 0.0490 |
| SMARCB1 | 1.4393 | 1.1297 | 1.8336 | 0.0032 | KDM4A | 1.3363 | 1.0476 | 1.7045 | 0.0196 |
| LEO1 | 1.4005 | 1.0359 | 1.8934 | 0.0286 | MMP9 | 1.1360 | 1.0384 | 1.2428 | 0.0054 |
| TXN | 1.3040 | 1.0667 | 1.5941 | 0.0096 | HK3 | 1.2356 | 1.0293 | 1.4832 | 0.0232 |
| HDAC4 | 1.5029 | 1.1470 | 1.9692 | 0.0031 | VEGFA | 1.2906 | 1.0657 | 1.5630 | 0.0090 |
| FXR1 | 1.6935 | 1.2509 | 2.2927 | 0.0007 | LGALS3 | 1.2041 | 1.0826 | 1.3392 | 0.0006 |
| RAD21 | 1.3480 | 1.0928 | 1.6629 | 0.0053 | AR | 0.8662 | 0.7749 | 0.9682 | 0.0115 |
| SRSF1 | 1.7610 | 1.2427 | 2.4955 | 0.0015 | BAG3 | 1.2740 | 1.0326 | 1.5718 | 0.0238 |
Prognostic differentially expressed cellular senescence genes were utilized for probing HCC heterogeneity. Utilizing unsupervised clustering, TCGA-LIHC cases were initially assigned to 2–9 clusters. Combining consensus CDF and consensus matrix, the optimal number of clusters was generated when k = 3 (Figure 2A–C). Thus, HCCs were classified as three cellular senescence subtypes, named C1–3. Prognostic differentially expressed cellular senescence genes presented the highest transcript levels in C2, followed by C1 and C3 (Figure 2D). PCA proved the extensive discrepancy in transcript levels among three subtypes (Figure 2E). Additionally, we focused on the survival difference, with C2 having the worst overall survival, C1 the next, and C3 the best (Figure 2F). Based upon up-regulated markers of each subtype (Supplementary Table 4), the robustness and reproducibility of cellular senescence subtypes were verified utilizing NTP in the ICGC cohort (Figure 2G). The discrepancy in transcript levels and overall survival among subtypes was further proven in this cohort (Figure 2H and I).
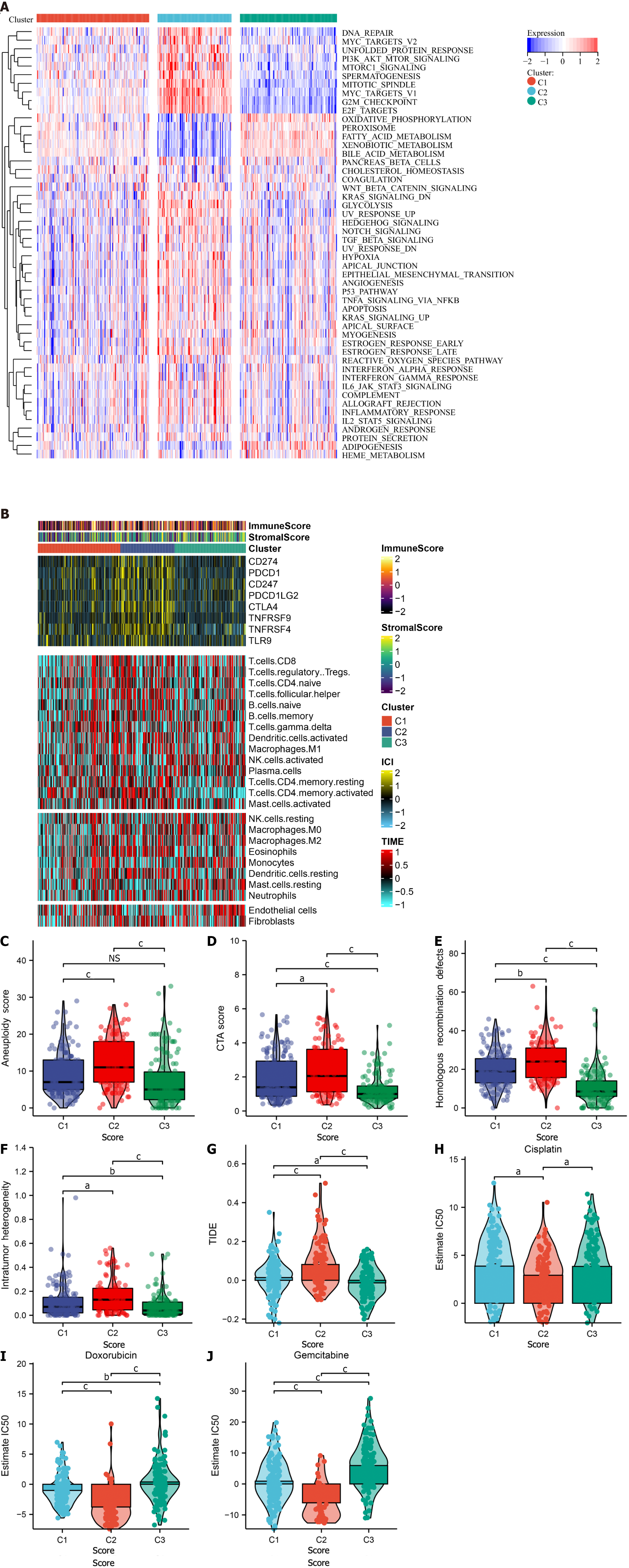
To elucidate the underlying mechanisms among the three cellular senescence subtypes, the activity of 50 hallmark pathways was inferred. Tumorigenic pathways (DNA repair, MYC, PI3K–AKT–mTOR, mTORC1, etc.) exhibited the highest activity in C2, with the lowest activity of metabolism pathways (Figure 3A). C3 presented the lowest activity of tumorigenic pathways, as well as the highest activity of metabolism pathways. Additionally, it was found that immune checkpoints displayed the highest transcript levels in C2, with the highest abundance of immune cells (Figure 3B). Immunogenetic indicators were then observed. Aneuploidy score, cancer-testicular antigen score, homologous recombination defects, and intratumor heterogeneity displayed the highest levels in C2, followed by C1 and C3 (Figure 3C–F). TIDE score was utilized to estimate the response to immune checkpoint inhibitors. Among three subtypes, C3 presented the lowest TIDE score, indicating that this subtype was most likely to respond to immune checkpoint inhibitors (Figure 3G). It was also found that cisplatin, doxorubicin and gemcitabine showed the lowest IC50 values in C2 subtype (Figure 3H–J). Thus, C2 patients were most likely to benefit from above chemotherapeutic drugs.
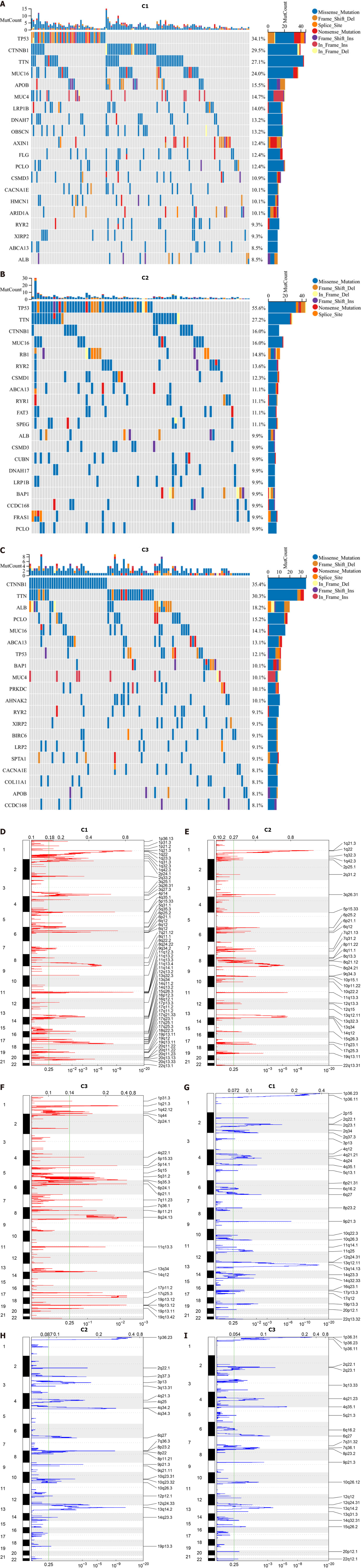
Overall, somatic mutation rate was the lowest in C2 among three cellular senescence subtypes (Figure 4A–C). Additionally, C1 presented the highest copy number amplified and deleted alterations (Figure 4D–I). Altogether, there was remarkable heterogeneity in genetic alterations among three cellular senescence subtypes.
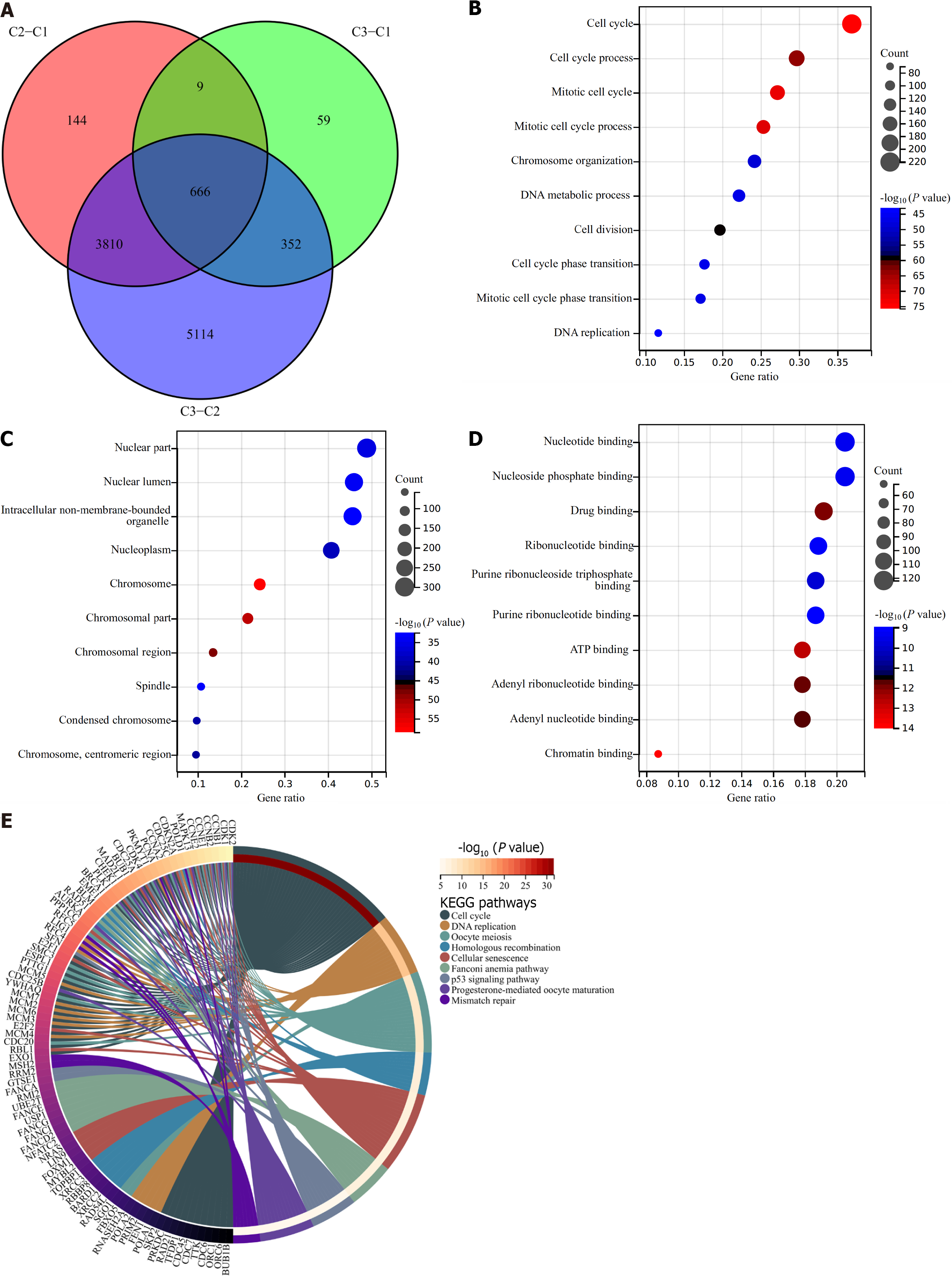
To select cellular senescence subtype-relevant genes, we assessed the genes with differential expression between cellular senescence subtypes based upon adjusted P < 0.05 together with |log2FC| > 0.58. After the intersection, 666 cellular senescence subtype-relevant genes were eventually acquired (Supplementary Table 5 and Figure 5A). We elucidated the underlying functional implications. Consequently, these cellular senescence subtype-relevant genes were remarkably linked with cell cycle, DNA replication, oocyte meiosis, homologous recombination, cellular senescence, Fanconi anemia pathway, p53 pathway, progesterone-mediated oocyte maturation, mismatch repair, etc (Figure 5B–E).
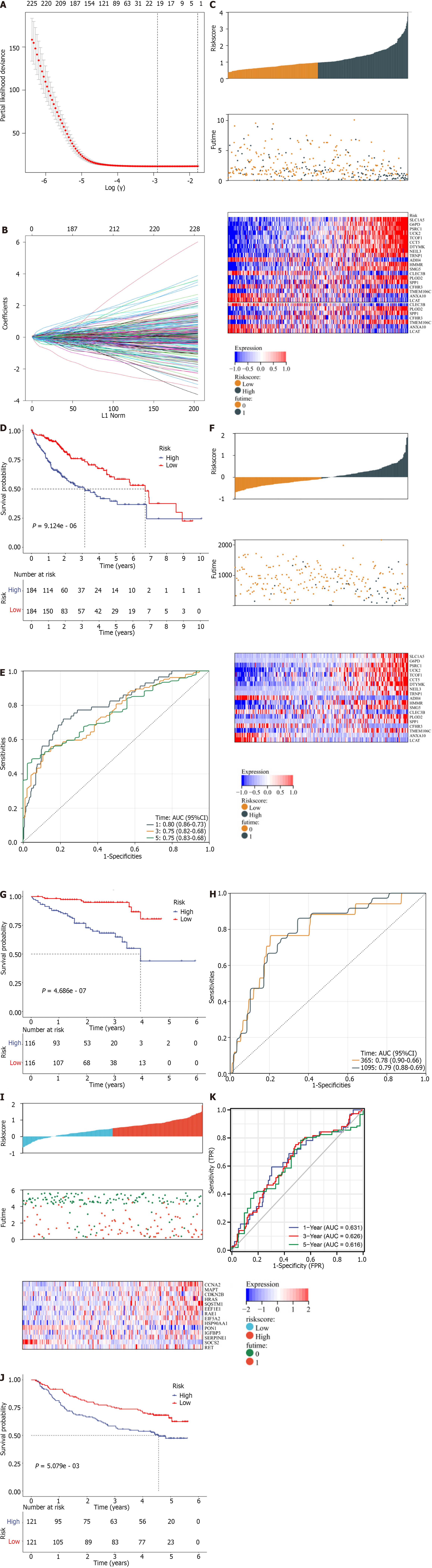
To illustrate the relationships of the cellular-senescence-relevant genes and patient survival, univariate Cox regression method was adopted. A total of 511 cellular -enescence-relevant genes presented significant correlations to TCGA-LIHC prognosis (Supplementary Table 6). These prognostic genes were entered into LASSO analysis (Figure 6A and B). A 19-gene signature was generated in accordance with the optimal λ value. The cellular senescence-relevant scoring system was computed as follows: RiskScore = 0.0610156 * transcript level of SLC1A5 + 0.049731458 * transcript level of G6PD + 0.038762092 * transcript level of PSRC1 + 0.104396819 * transcript level of UCK2 + 0.004054037 * transcript level of TCOF1 + 0.03040522 * transcript level of CCT5 + 0.002669582 * transcript level of DTYMK + 0.053080689 * transcript level of NEIL3 + 0.031271078 * transcript level of TRNP1 + (-0.037524429) * transcript level of ADH4 + 0.018409162 * transcript level of HMMR + 0.001793118 * transcript level of SMG5 + (-0.016621762) * transcript level of CLEC3B + 0.083327624 * transcript level of PLOD2 + 0.035535231 * transcript level of SPP1 + (-0.023169364) * transcript level of CFHR3 + 0.020042596 * transcript level of TMEM106C + (-0.019959706) * transcript level of ANXA10 + (-0.039577478) * transcript level of LCAT. Based upon the median RiskScore, TCGA-LIHC cases were classified as low- and high-RiskScore groups (Figure 6C). Expression levels of these selected genes exhibited the notable differences between groups. Next, K-M curves illustrated that high-RiskScore patients’ overall survival was worse (Figure 6D). AUCs at 1-, 3- and 5-year survival all exceeded 0.75, demonstrating the excellent discrimination power of the gene signature (Figure 6E).
The ICGC and GSE14520 datasets were utilized to externally verify this signature. The current study stratified HCCs into low- and high-RiskScore groups based upon the median RiskScore in the ICGC dataset (Figure 6F). Overall survival rate of high-RiskScore group was prominently lower (Figure 6G). AUCs at 1- and 3-year survival were > 0.75 (Figure 6H). Above data proved the high reproducibility of the signature. The similar findings were also confirmed in the GSE14520 dataset (Figure 6I–K).
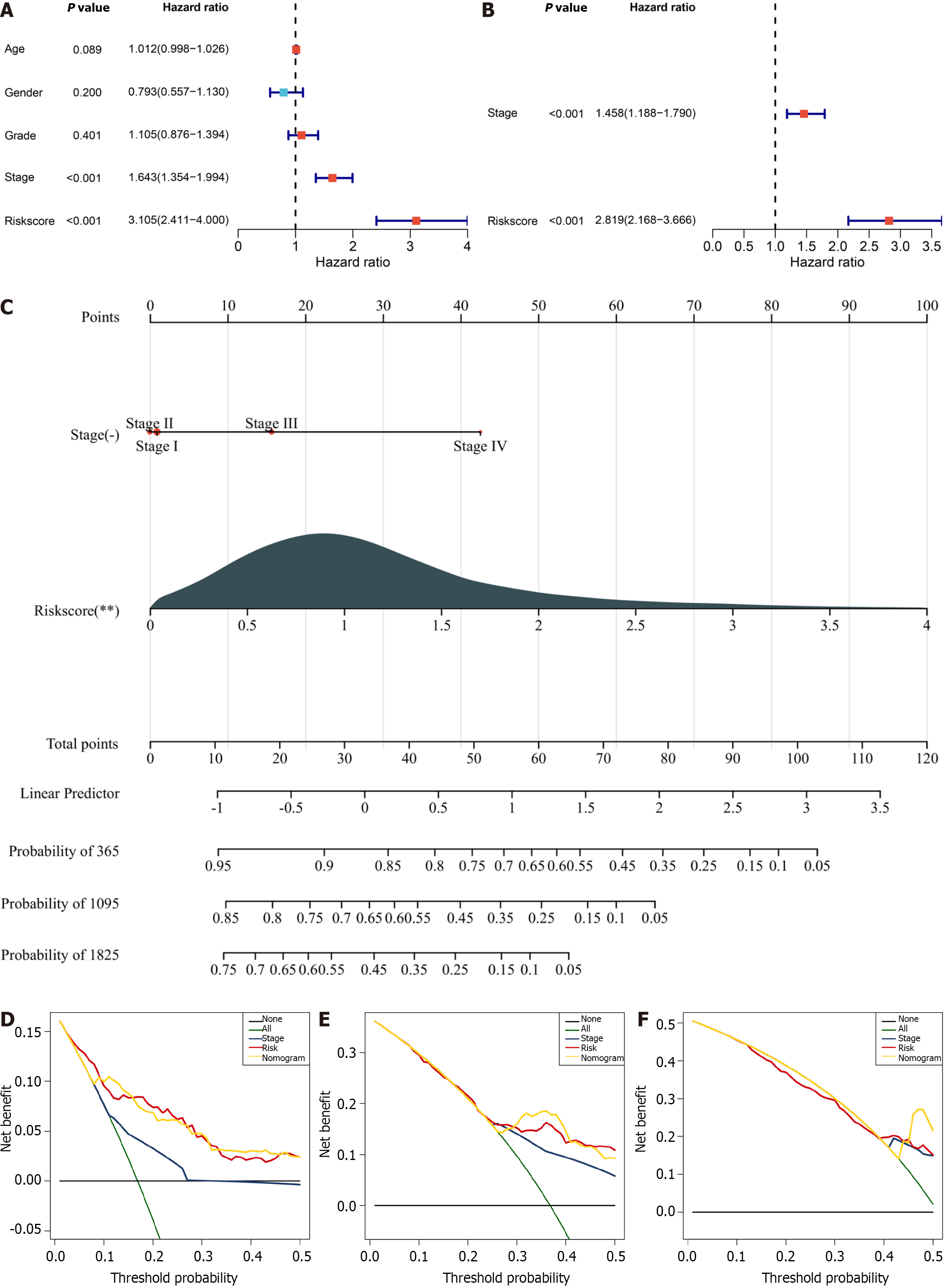
Univariate and multivariate Cox regression analyses were executed to select the independent prognostic parameters for HCCs. It was found that stage and the cellular-senescence-relevant RiskScore acted as independent risk factors of HCC prognosis (Figure 7A and B). As a visual representation of the prognostic model, a nomogram containing stage, and RiskScore was built to illustrate HCC patients’ survival more intuitively. The nomogram showed that RiskScore had the highest influence on 1-, 3- and 5-year survival of HCC patients, followed by stage (Figure 7C). Decision curve analysis demonstrated that the nomogram can accurately predict 1-, 3- and 5-year clinical outcomes (Figure 7D–F).
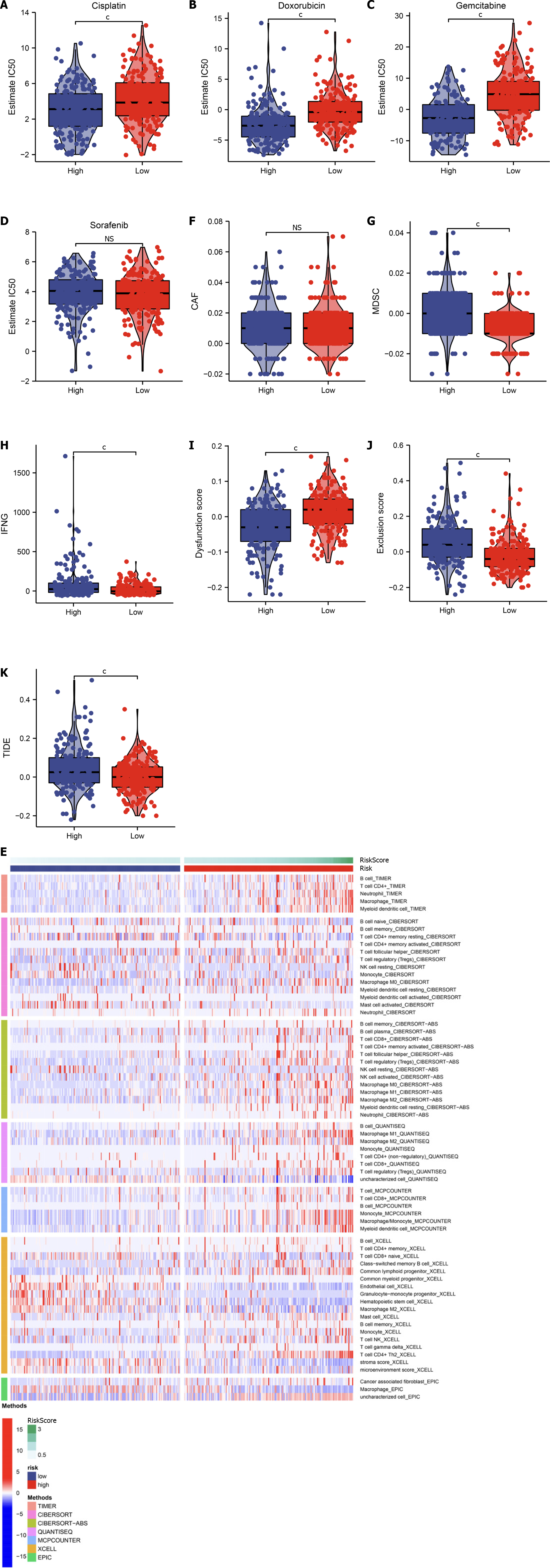
The IC50 of some chemotherapy or targeted therapy agents was estimated in TCGA-LIHC dataset. High-RiskScore HCCs presented the notably lower IC50 of cisplatin, doxorubicin, and gemcitabine relative to those with low RiskScore (Figure 8A–C). However, no significant difference in the IC50 of sorafenib was found between the two groups (Figure 8D). Accordingly, high-RiskScore HCCs more possibly responded to cisplatin, doxorubicin or gemcitabine chemotherapy.
Some reliable computational approaches were adopted to infer the abundance of the TME elements across TCGA-LIHC samples. Overall, most immune cells exhibited the higher infiltration in high-risk HCCs (Figure 8E). The TIDE method was used to predict immunotherapy response. We did not observe any difference in carcinoma-associated fibroblasts between the groups (Figure 8F). Lower myeloid-derived suppressor cells, interferon gamma, exclusion score and TIDE score as well as higher dysfunction score were found in low-RiskScore HCCs (Figure 8G–K). This indicated that low-RiskScore HCCs more possibly benefited from immunotherapy.
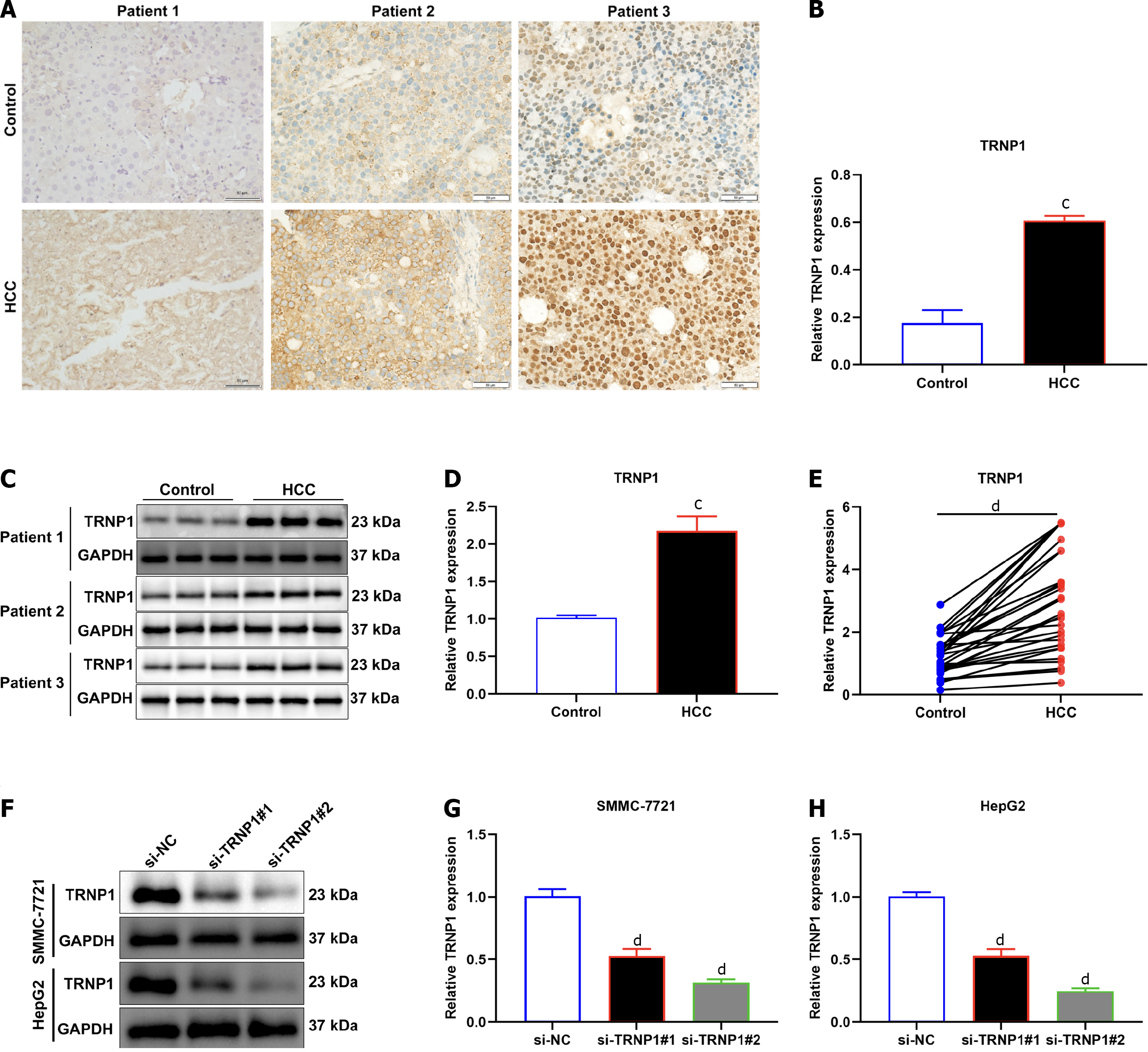
Among the genes in the cellular-senescence-relevant gene signature, the role of TRNP1 in HCC remains unclear. Therefore, we focused on TRNP1. It was proven that TRNP1 presented remarkable upregulation in HCCs relative to normal tissues in accordance with immunohistochemistry (Figure 9A and B), immunoblotting (Figure 9C and D) and RT-qPCR (Figure E). Specific siRNAs of TRNP1 were transfected into SMMC-7721 and HepG2 cells. Immunoblotting demonstrated the notable decrease in TRNP1 expression induced by siRNAs (Figure 9F–9H).
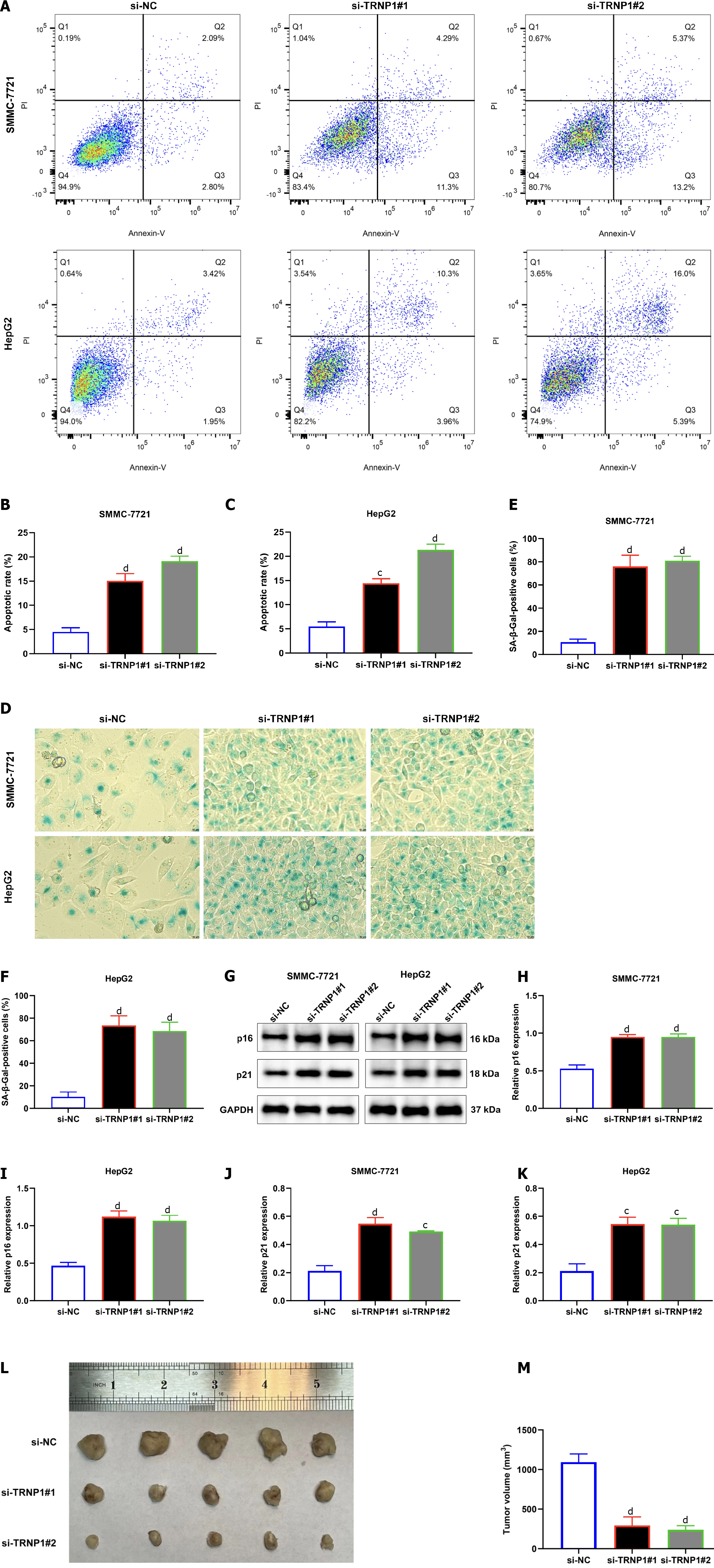
Based upon flow cytometry, apoptotic rate of SMMC-7721 and HepG2 cells was prominently elevated by TRNP1 knockdown (Figure 10A–C). SA-β-Gal staining showed that TRNP1 knockdown notably induced cellular senescence of two HCC cells (Figure 10D–F). Additionally, two cellular senescence markers: p16 and p21 were overexpressed in HCC cells with TRNP1 knockdown (Figure 10G–K), further proving the role of TRNP1 in HCC senescence. In vivo tumorigenicity models were also developed for evaluating whether TRNP1 influenced tumor growth. As a result, TRNP1 knockdown was found to decrease in vivo tumor volume (Figure 10L and M).
Cellular senescence is a permanent state of cell cycle arrest occurring in proliferating cells when face distinct stresses[36]. In cancers, senescence is usually an effective barrier against tumorigenesis because it prevents the division potential of cells[37]. Nonetheless, numerous research has demonstrated that senescent cells also have tumorigenic properties[38]. Thus, it is of significance to characterize key features of cellular senescence in HCC.
HCC is a typically fatal malignant tumor displaying genetic heterogeneity and limited therapy responses[39]. Based upon prognostic differentially expressed cellular senescence genes, we classified HCCs as three cellular senescence subtypes: C1–C3. The robustness and reproducibility of this classification were externally proven. C2 had the worst overall survival, C1 the next, and C3 the best, revealing the heterogeneity in prognostic outcomes among subtypes. Single-agent anti-PD-1 immune checkpoint blockade showed ponent efficacy in early-phase trials, but the findings were not confirmed in phase III studies[40]. In accordance with the lowest TIDE score, and immunogenetic indicators, C3 HCCs might possibly respond to immunotherapy. Additionally, C2 HCCs were most likely to benefit from chemotherapy. Thus, this classification might assist clinical decision-making. Genetic mutations associate with HCC initiation and progression[41-43]. For instance, mutant TP53 is the most frequent in HCC, affecting patient survival, and immune response[44]. CTNNB1 mutation occupies a large proportion of human HCCs, which correlates to high TMB and AFP in HCCs[45]. Among three cellular senescence subtypes, C2 presented the lowest somatic mutation rate, while C1 had the highest frequent CNVs. Accordingly, cellular senescence subtypes appear to associate with genetic mutations.
We defined a novel cellular-senescence-relevant gene signature comprising SLC1A5, G6PD, PSRC1, UCK2, TCOF1, CCT5, DTYMK, NEIL3, TRNP1, ADH4, HMMR, SMG5, CLEC3B, PLOD2, SPP1, CFHR3, TMEM106C, ANXA10, and LCAT, with the excellent power in survival prediction in HCCs. Previous research has proven the biological implications of the cellular-senescence-relevant genes in HCC. For example, SLC1A5 regulated by DDR1 contributes to HCC progression[46]. G6PD weakens ferroptosis in HCC through targeting cytochrome P450 oxidoreductase[47]. PSRC1, a hypoxia- and immune-associated gene, associates with HCC survival[48]. The nonmetabolic role of UCK2 facilitates HCC metastasis via EGFR–AKT signaling activation[49]. TCOF1 coordinates oncogenic activation and rRNA generation as well as results in HCC initiation[50]. Clinical features of HCC patients are notably associated with survival outcomes. To better optimize the cellular-senescence-relevant gene signature and improve the prediction accuracy, we incorporated stage in combination with the cellular-senescence-relevant gene signature to build a nomogram that enabled us to generate the individual survival probability in HCC patients. High-RiskScore HCCs might respond to cisplatin, doxorubicin or gemcitabine, while low-RiskScore HCCs more possibly benefit from immunotherapy, proving the potential of the cellular-senescence-relevant gene signature in inferring therapeutic efficacy. Among the cellular-senescence-relevant genes, the expression and role of TRNP1 in HCC remain indistinct. Only bioinformatics evidence demonstrated the prognostic significance of TRNP1 in HCC[51]. Our study experimentally proved that TRNP1 was upregulated in HCC, and TRNP1 knockdown induced apoptosis and senescence of HCC cells and attenuated tumor growth. Thus, TRNP1 potentially participates in HCC senescence and progression, which might be a promising therapeutic target.
Our study had some limitations. Due to the lack of HCC patients who received neoadjuvant immunotherapy, the relationship between cellular senescence subtypes and relevant gene signature with immunotherapeutic response requires further verification in the immunotherapy cohorts. Despite the external verification in the ICGC dataset, the predictive efficacy of cellular senescence-relevant gene signature needs to be proven in prospective cohorts.
Our findings showed the importance of cellular senescence in HCC classification and pharmacological interventions for clinical management. Additionally, we defined a cellular-senescence-relevant scoring system that can infer patient survival and therapeutic efficacy. Considering the clinically relevant parameters were closely linked with HCC survival, we incorporated stage in combination with the cellular-senescence-relevant gene signature to build a nomogram. Our integrated analysis provides a valuable framework for comprehending cellular senescence in HCC, which sheds light on the senescence-associated biomarker discovery as well as therapeutic targets.
Cellular senescence, a state of stable growth arrest, is intertwined with human cancers. Due to the highly heterogeneous malignancy at the molecular and histological levels, characterization of cellular-senescence-based classification might facilitate the personalized treatment of hepatocellular carcinoma (HCC).
Nonetheless, the heterogeneity of cellular-senescence-related features makes the definition and targeting of treatment-induced senescent cells challenging.
This study aimed to characterize cellular-senescence-based phenotypes in HCC, and identify a novel cellular-senescence-related therapeutic target.
We enrolled two HCC datasets, TCGA-LIHC and International Cancer Genome Consortium (ICGC). Unsupervised clustering was executed to probe tumor heterogeneity based upon cellular senescence genes. Least absolute shrinkage and selection operator algorithm was utilized to define a cellular-senescence-relevant scoring system. TRNP1 expression was measured in HCCs and normal tissues through immunohistochemistry, immunoblotting and quantitative real-time polymerase chain reaction. The influence of TRNP1 on HCC senescence and growth was proven via a series of experiments.
TCGA-LIHC patients were classified as three cellular senescence subtypes, named C1–3. The robustness and reproducibility of these subtypes were proven in the ICGC cohort. C2 had the worst overall survival, C1 the next, and C3 the best. C2 presented the highest levels of immune checkpoints, abundance of immune cells, and immunogenetic indicators. Thus, C2 might respond to immunotherapy. C2 had the lowest somatic mutation rate, while C1 presented the highest copy number variations. A cellular-senescence-relevant gene signature was generated, which can predict patient survival, and chemo- or immunotherapeutic response. Experimentally, it was proven that TRNP1 presented with remarkable upregulation in HCCs. TRNP1 knockdown induced apoptosis and senescence of HCC cells and attenuated tumor growth.
These findings provide a systematic framework for assessing cellular senescence in HCC, which decode the tumor heterogeneity and tailor the pharmacological interventions to improve clinical management.
Cellular senescence, a state of stable growth arrest, is implicated in human cancers. Nevertheless, characterization of cellular-senescence-associated phenotypes in HCC is still indistinct. Here, we proposed a novel cellular-senescence-based classification for HCC and identified TRNP1 as a novel therapeutic target.
Provenance and peer review: Unsolicited article; Externally peer reviewed.
Peer-review model: Single blind
Specialty type: Oncology
Country/Territory of origin: China
Peer-review report’s scientific quality classification
Grade A (Excellent): 0
Grade B (Very good): 0
Grade C (Good): C, C
Grade D (Fair): 0
Grade E (Poor): 0
P-Reviewer: Nagaya M, Japan; Ozturk M, Turkey S-Editor: Fan JR L-Editor: Kerr C P-Editor: Zhang XD
| 1. | Yamamoto-Imoto H, Minami S, Shioda T, Yamashita Y, Sakai S, Maeda S, Yamamoto T, Oki S, Takashima M, Yamamuro T, Yanagawa K, Edahiro R, Iwatani M, So M, Tokumura A, Abe T, Imamura R, Nonomura N, Okada Y, Ayer DE, Ogawa H, Hara E, Takabatake Y, Isaka Y, Nakamura S, Yoshimori T. Age-associated decline of MondoA drives cellular senescence through impaired autophagy and mitochondrial homeostasis. Cell Rep. 2022;38:110444. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 6] [Cited by in RCA: 43] [Article Influence: 14.3] [Reference Citation Analysis (0)] |
| 2. | Zhang D, Liu Y, Zhu Y, Zhang Q, Guan H, Liu S, Chen S, Mei C, Chen C, Liao Z, Xi Y, Ouyang S, Feng XH, Liang T, Shen L, Xu P. A non-canonical cGAS-STING-PERK pathway facilitates the translational program critical for senescence and organ fibrosis. Nat Cell Biol. 2022;24:766-782. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 8] [Cited by in RCA: 142] [Article Influence: 47.3] [Reference Citation Analysis (0)] |
| 3. | Avelar RA, Ortega JG, Tacutu R, Tyler EJ, Bennett D, Binetti P, Budovsky A, Chatsirisupachai K, Johnson E, Murray A, Shields S, Tejada-Martinez D, Thornton D, Fraifeld VE, Bishop CL, de Magalhães JP. A multidimensional systems biology analysis of cellular senescence in aging and disease. Genome Biol. 2020;21:91. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 172] [Cited by in RCA: 238] [Article Influence: 47.6] [Reference Citation Analysis (0)] |
| 4. | Chatsirisupachai K, Palmer D, Ferreira S, de Magalhães JP. A human tissue-specific transcriptomic analysis reveals a complex relationship between aging, cancer, and cellular senescence. Aging Cell. 2019;18:e13041. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 40] [Cited by in RCA: 100] [Article Influence: 16.7] [Reference Citation Analysis (0)] |
| 5. | Laphanuwat P, Gomes DCO, Akbar AN. Senescent T cells: Beneficial and detrimental roles. Immunol Rev. 2023;316:160-175. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 9] [Cited by in RCA: 23] [Article Influence: 11.5] [Reference Citation Analysis (0)] |
| 6. | Lee DA. Cellular therapy: Adoptive immunotherapy with expanded natural killer cells. Immunol Rev. 2019;290:85-99. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 52] [Cited by in RCA: 56] [Article Influence: 9.3] [Reference Citation Analysis (0)] |
| 7. | Wang X, Ma L, Pei X, Wang H, Tang X, Pei JF, Ding YN, Qu S, Wei ZY, Wang HY, Wang X, Wei GH, Liu DP, Chen HZ. Comprehensive assessment of cellular senescence in the tumor microenvironment. Brief Bioinform. 2022;23. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 21] [Cited by in RCA: 55] [Article Influence: 18.3] [Reference Citation Analysis (0)] |
| 8. | Wang B, Demaria M. The Quest to Define and Target Cellular Senescence in Cancer. Cancer Res. 2021;81:6087-6089. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 2] [Cited by in RCA: 16] [Article Influence: 4.0] [Reference Citation Analysis (0)] |
| 9. | Basu A. The interplay between apoptosis and cellular senescence: Bcl-2 family proteins as targets for cancer therapy. Pharmacol Ther. 2022;230:107943. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 21] [Cited by in RCA: 93] [Article Influence: 23.3] [Reference Citation Analysis (0)] |
| 10. | Wang L, Lankhorst L, Bernards R. Exploiting senescence for the treatment of cancer. Nat Rev Cancer. 2022;22:340-355. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 99] [Cited by in RCA: 438] [Article Influence: 146.0] [Reference Citation Analysis (0)] |
| 11. | Sung H, Ferlay J, Siegel RL, Laversanne M, Soerjomataram I, Jemal A, Bray F. Global Cancer Statistics 2020: GLOBOCAN Estimates of Incidence and Mortality Worldwide for 36 Cancers in 185 Countries. CA Cancer J Clin. 2021;71:209-249. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 75126] [Cited by in RCA: 64693] [Article Influence: 16173.3] [Reference Citation Analysis (177)] |
| 12. | Yau T, Kang YK, Kim TY, El-Khoueiry AB, Santoro A, Sangro B, Melero I, Kudo M, Hou MM, Matilla A, Tovoli F, Knox JJ, Ruth He A, El-Rayes BF, Acosta-Rivera M, Lim HY, Neely J, Shen Y, Wisniewski T, Anderson J, Hsu C. Efficacy and Safety of Nivolumab Plus Ipilimumab in Patients With Advanced Hepatocellular Carcinoma Previously Treated With Sorafenib: The CheckMate 040 Randomized Clinical Trial. JAMA Oncol. 2020;6:e204564. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 908] [Cited by in RCA: 971] [Article Influence: 194.2] [Reference Citation Analysis (0)] |
| 13. | Krstic J, Reinisch I, Schindlmaier K, Galhuber M, Riahi Z, Berger N, Kupper N, Moyschewitz E, Auer M, Michenthaler H, Nössing C, Depaoli MR, Ramadani-Muja J, Usluer S, Stryeck S, Pichler M, Rinner B, Deutsch AJA, Reinisch A, Madl T, Chiozzi RZ, Heck AJR, Huch M, Malli R, Prokesch A. Fasting improves therapeutic response in hepatocellular carcinoma through p53-dependent metabolic synergism. Sci Adv. 2022;8:eabh2635. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 28] [Cited by in RCA: 44] [Article Influence: 14.7] [Reference Citation Analysis (0)] |
| 14. | Kang YK, Chen LT, Ryu MH, Oh DY, Oh SC, Chung HC, Lee KW, Omori T, Shitara K, Sakuramoto S, Chung IJ, Yamaguchi K, Kato K, Sym SJ, Kadowaki S, Tsuji K, Chen JS, Bai LY, Oh SY, Choda Y, Yasui H, Takeuchi K, Hirashima Y, Hagihara S, Boku N. Nivolumab plus chemotherapy versus placebo plus chemotherapy in patients with HER2-negative, untreated, unresectable advanced or recurrent gastric or gastro-oesophageal junction cancer (ATTRACTION-4): a randomised, multicentre, double-blind, placebo-controlled, phase 3 trial. Lancet Oncol. 2022;23:234-247. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 36] [Cited by in RCA: 489] [Article Influence: 163.0] [Reference Citation Analysis (0)] |
| 15. | Liu X, Niu X, Qiu Z. A Five-Gene Signature Based on Stromal/Immune Scores in the Tumor Microenvironment and Its Clinical Implications for Liver Cancer. DNA Cell Biol. 2020;39:1621-1638. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 25] [Cited by in RCA: 35] [Article Influence: 7.0] [Reference Citation Analysis (0)] |
| 16. | Xiang X, Fu Y, Zhao K, Miao R, Zhang X, Ma X, Liu C, Zhang N, Qu K. Cellular senescence in hepatocellular carcinoma induced by a long non-coding RNA-encoded peptide PINT87aa by blocking FOXM1-mediated PHB2. Theranostics. 2021;11:4929-4944. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 44] [Cited by in RCA: 90] [Article Influence: 22.5] [Reference Citation Analysis (0)] |
| 17. | Liu B, Yi J, Yang X, Liu L, Lou X, Zhang Z, Qi H, Wang Z, Zou J, Zhu WG, Gu W, Luo J. MDM2-mediated degradation of WRN promotes cellular senescence in a p53-independent manner. Oncogene. 2019;38:2501-2515. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 12] [Cited by in RCA: 24] [Article Influence: 3.4] [Reference Citation Analysis (0)] |
| 18. | Paradis V, Youssef N, Dargère D, Bâ N, Bonvoust F, Deschatrette J, Bedossa P. Replicative senescence in normal liver, chronic hepatitis C, and hepatocellular carcinomas. Hum Pathol. 2001;32:327-332. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 223] [Cited by in RCA: 221] [Article Influence: 9.2] [Reference Citation Analysis (0)] |
| 19. | Liu B, Zhou Z, Jin Y, Lu J, Feng D, Peng R, Sun H, Mu X, Li C, Chen Y. Hepatic stellate cell activation and senescence induced by intrahepatic microbiota disturbances drive progression of liver cirrhosis toward hepatocellular carcinoma. J Immunother Cancer. 2022;10. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 4] [Cited by in RCA: 60] [Article Influence: 20.0] [Reference Citation Analysis (0)] |
| 20. | Yildiz G, Arslan-Ergul A, Bagislar S, Konu O, Yuzugullu H, Gursoy-Yuzugullu O, Ozturk N, Ozen C, Ozdag H, Erdal E, Karademir S, Sagol O, Mizrak D, Bozkaya H, Ilk HG, Ilk O, Bilen B, Cetin-Atalay R, Akar N, Ozturk M. Genome-wide transcriptional reorganization associated with senescence-to-immortality switch during human hepatocellular carcinogenesis. PLoS One. 2013;8:e64016. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 42] [Cited by in RCA: 48] [Article Influence: 4.0] [Reference Citation Analysis (0)] |
| 21. | Eggert T, Wolter K, Ji J, Ma C, Yevsa T, Klotz S, Medina-Echeverz J, Longerich T, Forgues M, Reisinger F, Heikenwalder M, Wang XW, Zender L, Greten TF. Distinct Functions of Senescence-Associated Immune Responses in Liver Tumor Surveillance and Tumor Progression. Cancer Cell. 2016;30:533-547. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 314] [Cited by in RCA: 448] [Article Influence: 49.8] [Reference Citation Analysis (0)] |
| 22. | Yu X, Chen P, Yi W, Ruan W, Xiong X. Identification of cell senescence molecular subtypes in prediction of the prognosis and immunotherapy of hepatitis B virus-related hepatocellular carcinoma. Front Immunol. 2022;13:1029872. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in RCA: 5] [Reference Citation Analysis (0)] |
| 23. | Luo Y, Liu H, Fu H, Ding GS, Teng F. A cellular senescence-related classifier based on a tumorigenesis- and immune infiltration-guided strategy can predict prognosis, immunotherapy response, and candidate drugs in hepatocellular carcinoma. Front Immunol. 2022;13:974377. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 2] [Cited by in RCA: 4] [Article Influence: 1.3] [Reference Citation Analysis (0)] |
| 24. | Wang Z, Yao J, Dong T, Niu X. Definition of a Novel Cuproptosis-Relevant lncRNA Signature for Uncovering Distinct Survival, Genomic Alterations, and Treatment Implications in Lung Adenocarcinoma. J Immunol Res. 2022;2022:2756611. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 2] [Cited by in RCA: 22] [Article Influence: 7.3] [Reference Citation Analysis (0)] |
| 25. | Yu G, Wang LG, Han Y, He QY. clusterProfiler: an R package for comparing biological themes among gene clusters. OMICS. 2012;16:284-287. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 11591] [Cited by in RCA: 22258] [Article Influence: 1712.2] [Reference Citation Analysis (0)] |
| 26. | Hänzelmann S, Castelo R, Guinney J. GSVA: gene set variation analysis for microarray and RNA-seq data. BMC Bioinformatics. 2013;14:7. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 7222] [Cited by in RCA: 9293] [Article Influence: 774.4] [Reference Citation Analysis (0)] |
| 27. | Liberzon A, Birger C, Thorvaldsdóttir H, Ghandi M, Mesirov JP, Tamayo P. The Molecular Signatures Database (MSigDB) hallmark gene set collection. Cell Syst. 2015;1:417-425. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 4238] [Cited by in RCA: 8407] [Article Influence: 840.7] [Reference Citation Analysis (0)] |
| 28. | Wilkerson MD, Hayes DN. ConsensusClusterPlus: a class discovery tool with confidence assessments and item tracking. Bioinformatics. 2010;26:1572-1573. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 1686] [Cited by in RCA: 3791] [Article Influence: 252.7] [Reference Citation Analysis (0)] |
| 29. | Eide PW, Bruun J, Lothe RA, Sveen A. CMScaller: an R package for consensus molecular subtyping of colorectal cancer pre-clinical models. Sci Rep. 2017;7:16618. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 124] [Cited by in RCA: 230] [Article Influence: 28.8] [Reference Citation Analysis (0)] |
| 30. | Mayakonda A, Lin DC, Assenov Y, Plass C, Koeffler HP. Maftools: efficient and comprehensive analysis of somatic variants in cancer. Genome Res. 2018;28:1747-1756. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 1228] [Cited by in RCA: 3114] [Article Influence: 444.9] [Reference Citation Analysis (0)] |
| 31. | Mermel CH, Schumacher SE, Hill B, Meyerson ML, Beroukhim R, Getz G. GISTIC2.0 facilitates sensitive and confident localization of the targets of focal somatic copy-number alteration in human cancers. Genome Biol. 2011;12:R41. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 1825] [Cited by in RCA: 2511] [Article Influence: 179.4] [Reference Citation Analysis (0)] |
| 32. | Engebretsen S, Bohlin J. Statistical predictions with glmnet. Clin Epigenetics. 2019;11:123. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 163] [Cited by in RCA: 663] [Article Influence: 110.5] [Reference Citation Analysis (0)] |
| 33. | Chen L, Niu X, Qiao X, Liu S, Ma H, Shi X, He X, Zhong M. Characterization of Interplay Between Autophagy and Ferroptosis and Their Synergistical Roles on Manipulating Immunological Tumor Microenvironment in Squamous Cell Carcinomas. Front Immunol. 2021;12:739039. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 12] [Cited by in RCA: 39] [Article Influence: 13.0] [Reference Citation Analysis (0)] |
| 34. | Geeleher P, Cox N, Huang RS. pRRophetic: an R package for prediction of clinical chemotherapeutic response from tumor gene expression levels. PLoS One. 2014;9:e107468. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 495] [Cited by in RCA: 1665] [Article Influence: 151.4] [Reference Citation Analysis (0)] |
| 35. | Jiang P, Gu S, Pan D, Fu J, Sahu A, Hu X, Li Z, Traugh N, Bu X, Li B, Liu J, Freeman GJ, Brown MA, Wucherpfennig KW, Liu XS. Signatures of T cell dysfunction and exclusion predict cancer immunotherapy response. Nat Med. 2018;24:1550-1558. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1184] [Cited by in RCA: 3401] [Article Influence: 485.9] [Reference Citation Analysis (0)] |
| 36. | Tao L, Zhang W, Zhang Y, Zhang M, Niu X, Zhao Q, Liu Z, Li Y, Diao A. Caffeine promotes the expression of telomerase reverse transcriptase to regulate cellular senescence and aging. Food Funct. 2021;12:2914-2924. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 4] [Cited by in RCA: 24] [Article Influence: 6.0] [Reference Citation Analysis (0)] |
| 37. | Chand V, Liao X, Guzman G, Benevolenskaya E, Raychaudhuri P. Hepatocellular carcinoma evades RB1-induced senescence by activating the FOXM1-FOXO1 axis. Oncogene. 2022;41:3778-3790. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 20] [Cited by in RCA: 17] [Article Influence: 5.7] [Reference Citation Analysis (0)] |
| 38. | McGettigan SE, Debes GF. Immunoregulation by antibody secreting cells in inflammation, infection, and cancer. Immunol Rev. 2021;303:103-118. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 12] [Cited by in RCA: 18] [Article Influence: 4.5] [Reference Citation Analysis (0)] |
| 39. | Tian Y, Xiao H, Yang Y, Zhang P, Yuan J, Zhang W, Chen L, Fan Y, Zhang J, Cheng H, Deng T, Yang L, Wang W, Chen G, Wang P, Gong P, Niu X, Zhang X. Crosstalk between 5-methylcytosine and N(6)-methyladenosine machinery defines disease progression, therapeutic response and pharmacogenomic landscape in hepatocellular carcinoma. Mol Cancer. 2023;22:5. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in RCA: 50] [Reference Citation Analysis (0)] |
| 40. | Sperandio RC, Pestana RC, Miyamura BV, Kaseb AO. Hepatocellular Carcinoma Immunotherapy. Annu Rev Med. 2022;73:267-278. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 11] [Cited by in RCA: 124] [Article Influence: 31.0] [Reference Citation Analysis (0)] |
| 41. | Boyault S, Rickman DS, de Reyniès A, Balabaud C, Rebouissou S, Jeannot E, Hérault A, Saric J, Belghiti J, Franco D, Bioulac-Sage P, Laurent-Puig P, Zucman-Rossi J. Transcriptome classification of HCC is related to gene alterations and to new therapeutic targets. Hepatology. 2007;45:42-52. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 827] [Cited by in RCA: 927] [Article Influence: 51.5] [Reference Citation Analysis (0)] |
| 42. | Liu M, Jiang L, Guan XY. The genetic and epigenetic alterations in human hepatocellular carcinoma: a recent update. Protein Cell. 2014;5:673-691. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 116] [Cited by in RCA: 133] [Article Influence: 12.1] [Reference Citation Analysis (0)] |
| 43. | Ren F, Li W, Xiang A, Wang L, Li M, Guo Y. Distribution and difference of APOBEC-induced mutations in the TpCpW context of HBV DNA between HCC and non-HCC. J Med Virol. 2020;92:53-61. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 3] [Cited by in RCA: 3] [Article Influence: 0.5] [Reference Citation Analysis (0)] |
| 44. | Long J, Wang A, Bai Y, Lin J, Yang X, Wang D, Jiang Y, Zhao H. Development and validation of a TP53-associated immune prognostic model for hepatocellular carcinoma. EBioMedicine. 2019;42:363-374. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 128] [Cited by in RCA: 258] [Article Influence: 43.0] [Reference Citation Analysis (0)] |
| 45. | Wang S, Shi H, Liu T, Li M, Zhou S, Qiu X, Wang Z, Hu W, Guo W, Chen X, Guo H, Shi X, Shi J, Zang Y, Cao J, Wu L. Mutation profile and its correlation with clinicopathology in Chinese hepatocellular carcinoma patients. Hepatobiliary Surg Nutr. 2021;10:172-179. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 6] [Cited by in RCA: 33] [Article Influence: 8.3] [Reference Citation Analysis (0)] |
| 46. | Pan Y, Han M, Zhang X, He Y, Yuan C, Xiong Y, Li X, Zeng C, Lu K, Zhu H, Lu X, Liu Q, Liang H, Liao Z, Ding Z, Zhang Z, Chen X, Zhang W, Zhang B. Discoidin domain receptor 1 promotes hepatocellular carcinoma progression through modulation of SLC1A5 and the mTORC1 signaling pathway. Cell Oncol (Dordr). 2022;45:163-178. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 21] [Cited by in RCA: 21] [Article Influence: 7.0] [Reference Citation Analysis (0)] |
| 47. | Cao F, Luo A, Yang C. G6PD inhibits ferroptosis in hepatocellular carcinoma by targeting cytochrome P450 oxidoreductase. Cell Signal. 2021;87:110098. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 9] [Cited by in RCA: 78] [Article Influence: 19.5] [Reference Citation Analysis (0)] |
| 48. | Hu B, Yang XB, Sang XT. Development and Verification of the Hypoxia-Related and Immune-Associated Prognosis Signature for Hepatocellular Carcinoma. J Hepatocell Carcinoma. 2020;7:315-330. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 16] [Cited by in RCA: 32] [Article Influence: 6.4] [Reference Citation Analysis (0)] |
| 49. | Cai J, Sun X, Guo H, Qu X, Huang H, Yu C, Wu H, Gao Y, Kong X, Xia Q. Non-metabolic role of UCK2 links EGFR-AKT pathway activation to metastasis enhancement in hepatocellular carcinoma. Oncogenesis. 2020;9:103. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 4] [Cited by in RCA: 25] [Article Influence: 5.0] [Reference Citation Analysis (0)] |
| 50. | Wu C, Xia D, Wang D, Wang S, Sun Z, Xu B, Zhang D. TCOF1 coordinates oncogenic activation and rRNA production and promotes tumorigenesis in HCC. Cancer Sci. 2022;113:553-564. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 4] [Cited by in RCA: 16] [Article Influence: 4.0] [Reference Citation Analysis (0)] |
| 51. | Liu J, Zhang SQ, Chen J, Li ZB, Chen JX, Lu QQ, Han YS, Dai W, Xie C, Li JC. Identifying Prognostic Significance of RCL1 and Four-Gene Signature as Novel Potential Biomarkers in HCC Patients. J Oncol. 2021;2021:5574150. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 3] [Cited by in RCA: 3] [Article Influence: 0.8] [Reference Citation Analysis (0)] |