Published online Aug 15, 2023. doi: 10.4251/wjgo.v15.i8.1424
Peer-review started: March 24, 2023
First decision: May 19, 2023
Revised: May 20, 2023
Accepted: June 19, 2023
Article in press: June 19, 2023
Published online: August 15, 2023
Processing time: 139 Days and 5 Hours
Colorectal cancer (CRC) is a major global health burden. The current diagnostic tests have shortcomings of being invasive and low accuracy.
To explore the combination of intestinal microbiome composition and multi-target stool DNA (MT-sDNA) test in the diagnosis of CRC.
We assessed the performance of the MT-sDNA test based on a hospital clinical trial. The intestinal microbiota was tested using 16S rRNA gene sequencing. This case-control study enrolled 54 CRC patients and 51 healthy controls. We iden
MT-sDNA was associated with Bacteroides. MT-sDNA and carcinoembryonic antigen (CEA) were positively correlated with the existence of Parabacteroides, and alpha-fetoprotein (AFP) was positively associated with Faecalibacterium and
There is a positive correlation of MT-sDNA, CEA, and AFP with intestinal microbiome. Eight biomarkers including six genera of gut microbiota, MT-sDNA, and CEA showed a prominent sensitivity and specificity for CRC prediction, which could be used as a non-invasive method for improving the diagnostic accuracy for this malignancy.
Core Tip: There is a positive correlation of multi-target stool DNA (MT-sDNA), carcinoembryonic antigen (CEA), and alpha-fetoprotein with intestinal microbiome. Eight biomarkers including six genera of gut microbiota, MT-sDNA, and CEA showed a prominent sensitivity (98.1%) and specificity (92.3%) for colorectal cancer prediction, which could be used as a non-invasive method for improving the diagnostic accuracy for this malignancy.
- Citation: Fan JQ, Zhao WF, Lu QW, Zha FR, Lv LB, Ye GL, Gao HL. Fecal microbial biomarkers combined with multi-target stool DNA test improve diagnostic accuracy for colorectal cancer. World J Gastrointest Oncol 2023; 15(8): 1424-1435
- URL: https://www.wjgnet.com/1948-5204/full/v15/i8/1424.htm
- DOI: https://dx.doi.org/10.4251/wjgo.v15.i8.1424
Colorectal cancer (CRC) is the third most common cancer and the fifth-leading cause of cancer-related deaths in China[1]. Although colonoscopy is considered the gold standard in the diagnosis of CRC, it has shortcomings of being invasive and expensive[2]. Thus, non-invasive and effective diagnostic methods are needed urgently. Tumor markers such as carcinoembryonic antigen (CEA), carbohydrate antigen 199 (CA199), and alpha-fetoprotein (AFP) are not specific and sensitive in detecting CRC, which may lead to delayed treatment and reduced survival rates[3]. Fortunately, increasing evidence suggests that gut bacteria play an important role in the development of CRC[4,5]. Fecal bacteria can be used as non-invasive biomarkers for CRC diagnosis[6]. Combined tests for different intestinal bacteria in CRC had a sensitivity and specificity ranging from 54.0%-84.6%, and 63.1%-90.0%, respectively[7,8]. Due to the complexity of the gut micro
A total of 105 participants with an age range from 40 to 74 years who visited the anorectal department and health examination center of the Affiliated Hospital of Medical School of Ningbo University from January 2021 to November 2022 were recruited.
The inclusion criteria for patients with CRC were: (1) Newly diagnosed sporadic CRC patients with an expected survival > 3 mo; and (2) The patients did not undergo colorectal surgery or chemoradiotherapy. The inclusion criteria for HC were: The subjects underwent colonoscopy and the results were normal.
The exclusion criteria were: (1) Subjects with chronic diarrhea; (2) Subjects with tumor metastasis or recurrence; (3) Subjects with a family history of CRC; (4) Subjects with inflammatory bowel disease, obesity, diabetes, or other metabolic diseases; (5) Subjects with preoperative radiotherapy and chemotherapy; (6) Subjects who had taken laxatives, antibiotics, microecological preparations, and other drugs in the past 1 mo; (7) Subjects with cirrhosis; and (8) Subjects with colorectal polyps.
All participants provided informed consent and the study was approved by the Human Research and Ethics Committee of the Affiliated Hospital of Medical School of Ningbo University (approval number: KY20211104). This study conforms to the principles of the Declaration of Helsinki in 2013.
Fecal samples (4-5 g) were collected prior to bowel preparation for colonoscopy and before surgical removal of intestinal tumor tissue from CRC patients. All experimental procedures related to the MT-sDNA tests [fecal immunochemical test (FIT), NDRG4 and BMP3 methylation, KRAS mutation] were carried out using the commercial kit ColoClear® (New Horizon Health Technology, Hangzhou, China). The details regarding probes and primers, as well as the risk prediction algorithm, were the same as those described in a previous report[11]. In this MT-sDNA risk prediction model, a risk score was provided as a single output. If the risk score value was ≥ 165, the test was considered “positive”. If the risk score was < 165, the test was regarded as “negative”[12].
Fresh fecal samples (≥ 1 g) from all participants were collected prior to colonoscopy and frozen at -80 °C immediately. Total microbial genomic DNA was extracted from the fecal samples using E.Z.N.A®fecal DNA Kit (Omega, United States) according to the manufacturer’s recommendations. The sequences of the V1-V9 variable regions of the bacterial 16S rRNA gene were amplified using primers 27F (5’-AGRGTTYGATYMTGGCTCAG-3’) and 1492R (5’-RGYTACCTTGTTACGACTT-3’)[13]. The polymerase chain reaction (PCR) conditions to amplify the prokaryotic 16S fragments were consistent with those described in the previous literature[14]. The PCR amplication products were confirmed by 2% agarose gel electrophoresis and purified with AxyPrep DNA Gel Extraction Kit (Axygen Biosciences, United States). We conducted 16S rDNA sequencing on the PacBio Sequel IIe platform (Biozeron Biotechnology, Shanghai, China).
Three serum biomarkers, i.e., CA199, CEA, and AFP, were determined by the Laboratory of the Affiliated Hospital of Ningbo University School of Medicine. Histological diagnosis and colonoscopy findings were used as the reference for determining the accuracy of the combination test for validating screening performance. All pathological diagnoses were in accordance with the diagnostic criteria of the 2010 World Health Organization Classification of Gastrointestinal Neoplasms.
The complexity of sample species diversity was analyzed by alpha diversity, and the richness and diversity of community were described by Shannon index and Simpson index. Beta diversity analysis was used to compare the samples and was evaluated by principal component analysis (PCA). Alpha diversity and beta diversity analyses were conducted using USEARCH software (version 1.1). Linear discriminant analysis (LDA) effect size (LEfSe) analysis was conducted to identify biomarkers of bacterial structure in the case-control groups[15]. Kruskal-Wallis and rank tests were used to analyze the changes and differences among the classes, and LDA was used to determine the size effects of each significant abundance group[16]. The relationship between intestinal flora, MT-sDNA, and tumor markers was analyzed by redundancy analysis (RDA). We built the random forest model to distinguish patients with CRC from HC. The analysis of disease diagnosis included the calculation of disease diagnosis index and the evaluation of diagnosis effect. Diagnostic index calculation was based on the differential genus identified in the random forest analysis to obtain the disease diagnostic index of the sample. Receiver operating characteristic (ROC) curve was used to evaluate the diagnostic effect of disease diagnostic index. The sensitivity and specificity were analyzed by ROC curve analysis with the area under the ROC curve (AUC) and 95% confidence interval calculated for the tumor markers, MT-sDNA, and intestinal flora. The analysis process was completed using vegan package, random forest package, and plotROC of R software (version 4.2, United States). t-test and chi-square test were adopted to compare the differences between different groups. P < 0.05 was considered statistically significant. Statistical analyses were performed using SPSS software (version 23.0, United States).
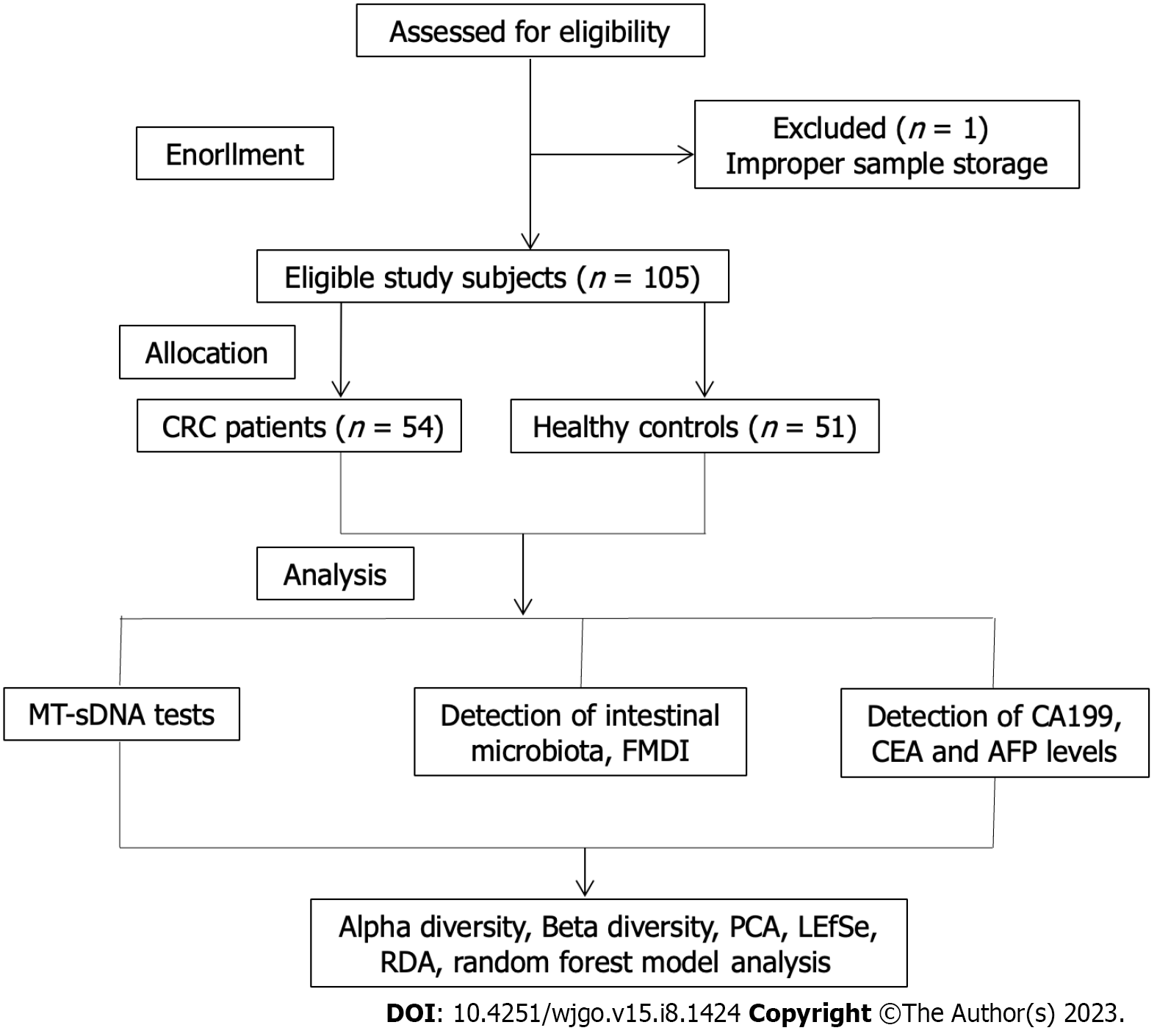
One patient was excluded from the study because of missing the sample storage standard, and 105 subjects were included eventually (Figure 1). The clinical features of the 105 participants are shown in Table 1. A total of 54 CRC patients and 51 HC were enrolled in this research, with an average age and standard deviation of 64.89 ± 9.72 and 53.94 ± 10.33, separately. The colon was the most common site (66.67%) in CRC patients. Moderate/low differentiation, ulcerative type, and Dukes stage A accounted for 92.59%, 61.11%, and 79.63% of CRC cases, respectively. There were statistically differences in terms of age, education level, and body mass index between the two groups (P < 0.05).
| Variable | Colorectal cancer patients | Health controls | P value |
| Gender | 0.30 | ||
| Male | 34 (62.96) | 27 (52.94) | |
| Female | 20 (37.04) | 24 (47.06) | |
| Age | 0.00 | ||
| mean ± SD | 64.89 ± 9.72 | 53.94 ± 10.33 | |
| < 60 yr | 18 (33.33) | 35 (68.63) | |
| ≥ 60 yr | 36 (66.67) | 16 (31.37) | |
| Education level | 0.00 | ||
| Junior high school and below | 43 (79.63) | 10 (19.61) | |
| Senior high school and above | 11 (20.37) | 41 (80.39) | |
| BMI (kg/m2) | 0.00 | ||
| ≤ 18.5 | 4 (7.41) | 0 (0.00) | |
| 18.5-23.9 | 37 (68.52) | 24 (47.06) | |
| ≥ 24 | 13 (24.07) | 27 (52.94) | |
| Tumor location | |||
| Colon | 36 (66.67) | - | - |
| Rectum | 18 (33.33) | - | - |
| Pathogenic type | - | - | |
| Protrude type | 10 (18.52) | - | - |
| Infiltrating type | 11 (20.37) | - | - |
| Ulcerative type | 33 (61.11) | - | - |
| Differentiation | - | - | |
| High | 4 (7.41) | - | - |
| Medium/low | 50 (92.59) | - | - |
| Histological type | - | - | |
| Adenocarcinoma | 54 (100.00) | - | - |
| Other types | 0 (0.00) | - | - |
| Dukes stage | - | - | |
| A | 43 (79.63) | - | - |
| B | 7 (12.96) | - | - |
| C | 4 (7.41) | - | - |
| D | 0 (0.0) | - | - |
As shown in Table 2, the levels of tumor biomarkers CEA and CA199 and the DNA score were elevated in CRC patients compared with HC (P < 0.05). Fecal microbiota diagnostic index (FMDI) was -215.76 ± 539.49 and 178.47 ± 249.43 in CRC patients and HC (P < 0.05), respectively.
| Variable | Colorectal cancer patients | Health controls | P value |
| AFP | |||
| mean ± SD | 5.70 ± 17.10 | 3.61 ± 1.59 | 0.27 |
| ≤ 7 μg/L | 49 (90.74) | 49 (96.08) | |
| > 7.1 μg/L | 5 (9.26) | 2 (3.92) | |
| CEA | |||
| mean ± SD | 40.67 ± 154.34 | 2.22 ± 1.47 | 0.00 |
| ≤ 5 μg/L | 38 (70.37) | 48 (94.12) | |
| > 5.1 μg/L | 16 (29.63) | 3 (5.88) | |
| CA199 | 0.02 | ||
| mean ± SD | 59.65 ± 269.59 | 10.01 ± 9.26 | |
| ≤ 25 μg/mL | 44 (81.48) | 49 (96.08) | |
| > 25.1 μg/mL | 10 (18.52) | 2 (3.92) | |
| DNA score | |||
| mean ± SD | 787.87 ± 283.71 | 94.39 ± 41.62 | 0.00 |
| < 165 | 5 (9.26) | 50 (98.04) | |
| ≥ 165 | 49 (90.74) | 1 (1.96) |
The number of OTUs obtained in this research was 453, among which 93 unique OTUs belonged to the CRC group and 76 unique OTUs existed in the HC group. There were 284 OTUs shared between the CRC and HC groups, and details are shown in a Venn diagram (Supplementary Figure 1A).
There was no statistical difference in alpha diversity between the CRC and control groups. The Shannon index (P = 0.79) and Simpson index (P = 0.50) results are shown in Supplementary Figures 1B and C. In addition, PCA plot showed no difference in beta diversity of gut microbiota (Supplementary Figure 1D).
In order to find species with significant differences in abundance in the two groups, the cladistic diagram of LEfSe analysis was used, as shown in Figure 2. LEfSe analysis identified 23 characteristic bacterial groups (Figure 2A). An LDA score > 4 indicated a significant difference in structure between the two groups (Figure 2B). Fusobacterium, Bacteroides, Enterobacterales, Gammaproteobacteria, Proteobacteria, and Escherichia were more abundant in CRC patients than in HC. The abundance of Prevotellaceae, Lachnospiraceae, Oscillospiraceae, Eubacteriales, and Clostridia was higher in the HC group.
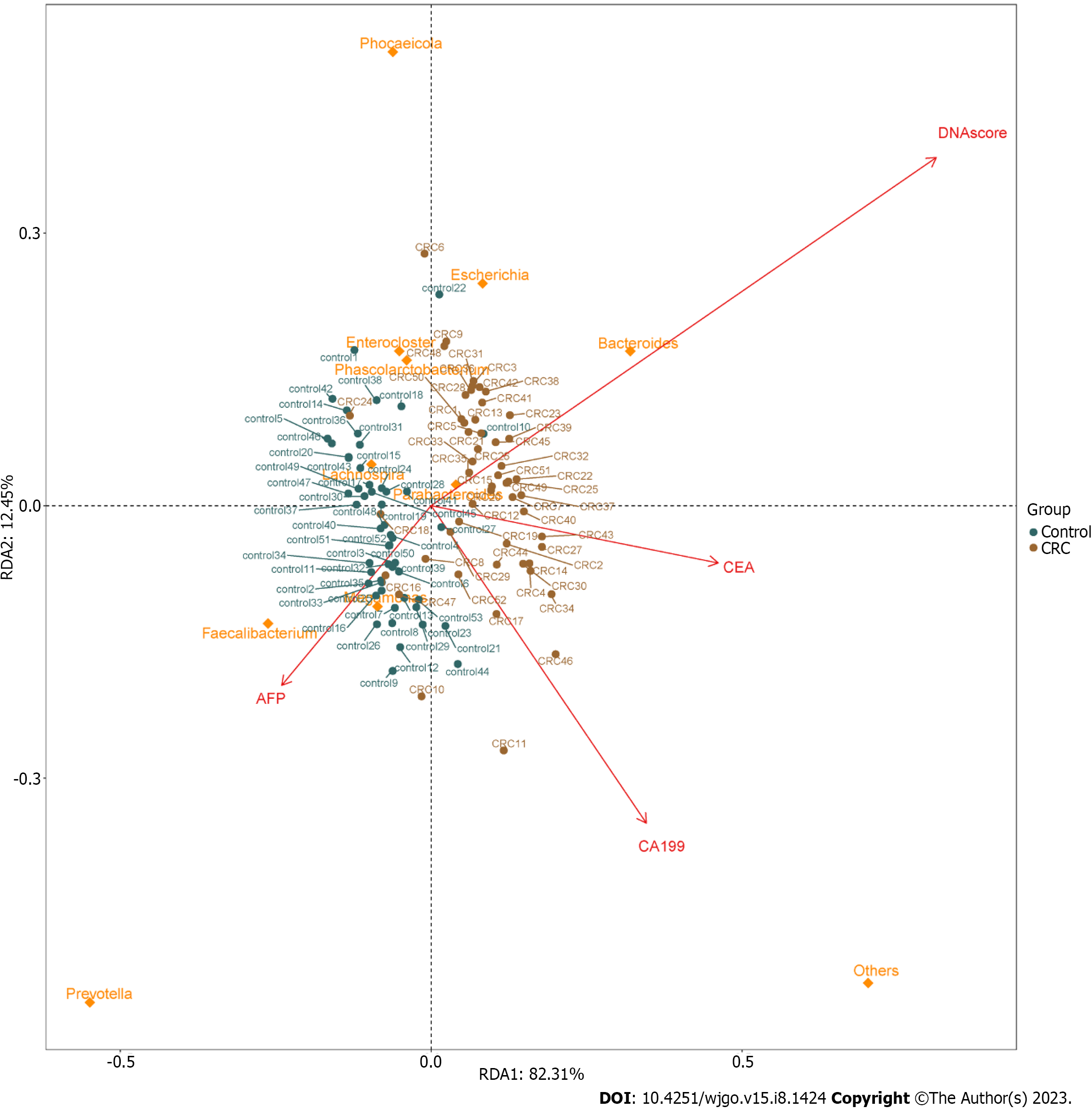
In order to analyze the relationship between different tumor markers and the relative abundance of related flora components and to observe the correlation between different flora and tumor biomarkers, RDA was performed. The results of RDA analysis showed positive correlations between MT-sDNA and Bacteroides. Parabacteroides was positively correlated with MT-sDNA and CEA, and Faecalibacterium and Megamonas was positively associated with AFP (Figure 3).
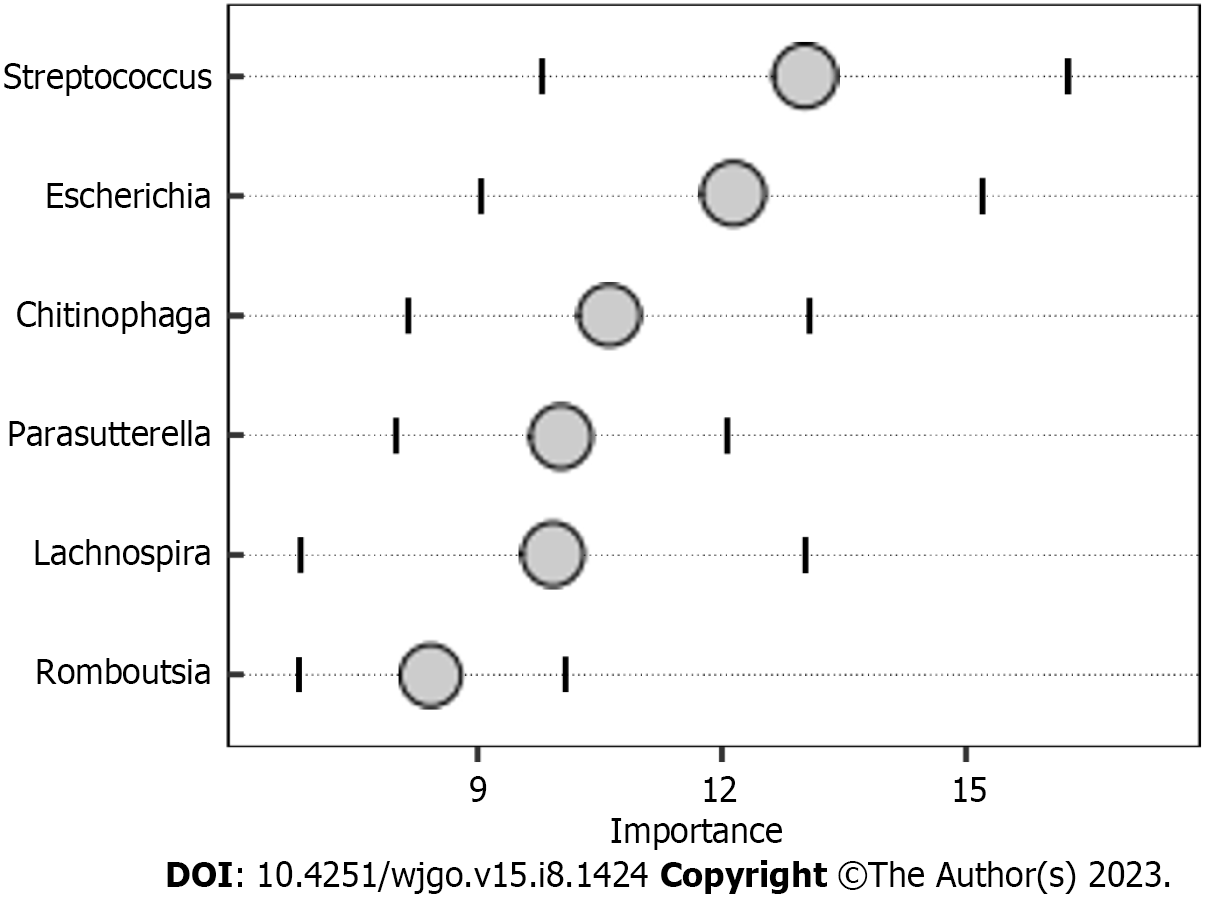
The random forest model results showed that the combination of the six genera, namely, Streptococcus, Escherichia, Chitinophaga, Parasutterella, Lachnospira, and Romboutsia (Figure 4), can distinguish CRC patients from HC. We assessed the clinical diagnostic value of intestinal microbiome, tumor markers, and MT-sDNA in CRC patients. We found that the sensitivity, specificity, and AUC of intestinal microbiome were lower than those of the combined detection of MT-sDNA and intestinal microbiome in CRC (AUC = 84.8% and 95.4%, respectively). The specificity and sensitivity of intestinal microbiome combined with MT-sDNA and CEA in the diagnosis of CRC were 92.3% and 98.1%, and the diagnostic accuracy was 97.1%, which was higher than that of either of the tumor biomarkers or MT-sDNA test alone (94.3% and 83.0%, respectively) (Figure 5).
Most of CRC cases are sporadic and follow a pattern of adenomatous to cancerous progression[17]. The development of CRC is caused by the interaction of genetic and environmental factors, and changes in intestinal microbiome are closely related to CRC.
Previous studies have confirmed changes in the bacterial composition of CRC, but no effective diagnostic model has been developed in the aspect of MT-sDNA, gut microbiota, and tumor biomarkers interactions. We performed 16S rDNA sequencing to investigate the differences in gut microbiota between patients with CRC and HC. In our study, Fusobacterium, Bacteroides, Gammaproteobacteria, Proteobacteria, Escherichia, and Enterobacterales were more abundant in CRC patients than in HC. The intestinal flora of CRC was mainly Bacteroides and Fusobacterium as found in previous studies[18-20]. Fusobacterium and Bacteroides are associated with metastasis and poor survival outcomes in CRC patients, which makes them emerging candidate biomarkers[21,22]. We speculate that the gathering of Fusobacterium may result in colonization of the colon mucosa and further promote the occurrence of CRC, suggesting that Fusobacterium may be a “driving factor” for the occurrence of CRC[23]. Bacteroides promote tumorigenesis by inducing cell proliferation and promoting tumor inflammation through toxins[24]. Additionally, Proteobacteria, Escherichia, Gammaproteobacteria, and Enterobacterales were also affirmed as the CRC-associated bacteria in previous studies[25]. Proteobacteria are colonized in the gut, causing persistent intestinal inflammation[26]. Escherichia may influence the development and progression of CRC through inflammatory pathways and virulence factors[27]. Gammaproteobacteria could break leupeptin protease inhibitors and was related to colonization phenotypes[28]. Enterobacterales induced the cell death, and contained apoptosis-inducing substances[29]. Therefore, the gut microbiome has a direct pathogenic role in cell proliferation, cell death, and inflammation in CRC, suggesting that regulating the gut microbiome can be a powerful tool for CRC prevention and treatment.
Tumor markers can be used to predict the diagnosis of CRC[30]. Previous research showed that CEA and CA199 yielded an AUC of 0.74 and 0.67 to discriminate HC from CRC patients, which was similar with our results[31]. The ability of tumor markers to distinguish CRC is limited, and new biomarkers need to be found for the diagnosis of CRC. To achieve a comprehensive analysis, we first investigated the association between microbial composition and tumor markers in CRC patients. We found positive correlations between MT-sDNA and Bacteroides. Parabacteroides was positively correlated with MT-sDNA and CEA, and Faecalibacterium and Megamonas were positively associated with AFP. This suggests that gut microbiota composition affects the MT-sDNA and tumor biomarkers in CRC patients. The results of this study provide new ideas for tumor markers and intestinal flora in the drug therapy of CRC. Second, the random forest model results showed that the combination of the six genera, namely, Streptococcus, Escherichia, Chitinophaga, Parasutterella, Lachnospira, and Romboutsia, can distinguish CRC from HC. The results suggested that these six genera could be used as the potential biomarkers for CRC diagnosis. Similar to other studies, our research confirmed that the sensitivity of MT-sDNA and fecal bacteria in terms of CRC diagnosis was relatively high[32,33], indicating that MT-sDNA and gut microbiota are suitable for the diagnosis of CRC[34,35]. Previous studies mostly focused on the diagnosis accuracy of MT-sDNA and intestinal microbiome detection alone or combined with FIT, which is lacking diagnostic accuracy. There was no studies focused on the combination of MT-sDNA test, intestinal microbiome, and tumor markers in the diagnosis of CRC. We found that, in the detection of CRC, the sensitivity and accuracy of MT-sDNA combined with CEA and fecal bacteria increased, which indicated that this combination has robust advantages in distinguishing CRC patients from HC.
Although our study suggests that intestinal flora combined with tumor markers can improve the diagnostic efficacy for CRC, our study have some shortcomings. First, the number of subjects included in our research was not large enough and no multicenter clinical validation was performed. A larger sample size is expected to lead to more discoveries in the future. Second, further experimental studies on mechanism investigations are needed to confirm our findings and investigate the effects of tumor markers, MT-sDNA, and intestinal flora on CRC patients. Third, environmental factors such as diet and lifestyle were not investigated in this study, and the influence of environmental factors on intestinal flora could not be analyzed. However, our findings in this study will optimize the diagnosis of CRC and provide new ideas to transform microbita-based strategies into precise diagnosis in the clinic.
Our study reveals that CRC-associated bacteria have specific differences. There are positive correlations of MT-sDNA, CEA, as well as AFP with intestinal microbiome. Fecal microbiological markers combined with MT-sDNA and CEA can enhance the sensitivity and specificity for the diagnosis of CRC, and are feasible in clinical application.
Colorectal cancer (CRC) is a major global health burden, and its incidence and mortality have increased rapidly over the past decades and resulted in massive economic burdens in China.
This case-control study enrolled 54 CRC patients and 51 healthy controls.
This research aimed to explore the characteristics of intestinal flora and its correlation with multi-target stool DNA (MT-sDNA) and tumor markers in CRC patients, and evaluate the diagnostic performance of MT-sDNA and tumor biomarkers combined with microbiota in CRC.
We evaluated the performance of the MT-sDNA test based on a hospital clinical trial. The intestinal microbiota was tested using 16S rRNA gene sequencing. We identified biomarkers of bacteria structure, analyzed the relationship between different tumor markers and the relative abundance of related flora components, and distinguished CRC patients from healthy subjects by the linear discriminant analysis effect size, redundancy analysis, and random forest analysis, respectively.
We found that MT-sDNA was closely associated with Bacteroides. MT-sDNA and carcinoembryonic antigen (CEA) were positively correlated with the existence of Parabacteroides, and alpha-fetoprotein was positively associated with Faecalibacterium and Megamonas. The random forest model results showed that the combination of the six genera, namely, Streptococcus, Escherichia, Chitinophaga, Parasutterella, Lachnospira, and Romboutsia, can distinguish CRC from health controls. The sensitivity and specificity of MT-sDNA combined with the six genera and CEA in the diagnosis of CRC were 98.1% and 92.3%, respectively, and the diagnostic accuracy was 97.1%.
MT-sDNA and tumor markers were positively correlated with intestinal flora. Intestinal flora, MT-sDNA, and tumor markers showed significant sensitivity and specificity for CRC prediction, which could be used as a non-invasive method to improve the diagnostic accuracy.
Our results will optimize the diagnosis of CRC and provide new ideas for translating microbit-based diagnostic strategies into precise diagnosis in the clinic.
Provenance and peer review: Unsolicited article; Externally peer reviewed.
Peer-review model: Single blind
Specialty type: Oncology
Country/Territory of origin: China
Peer-review report’s scientific quality classification
Grade A (Excellent): 0
Grade B (Very good): 0
Grade C (Good): C, C
Grade D (Fair): 0
Grade E (Poor): 0
P-Reviewer: Gazouli M, Greece; Jamali R, Iran S-Editor: Wang JJ L-Editor: Wang TQ P-Editor: Zhang XD
| 1. | Lin C, Li B, Tu C, Chen X, Guo M. Correlations between Intestinal Microbiota and Clinical Characteristics in Colorectal Adenoma/Carcinoma. Biomed Res Int. 2022;2022:3140070. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 9] [Cited by in RCA: 10] [Article Influence: 3.3] [Reference Citation Analysis (0)] |
| 2. | Wu W, Huang J, Yang Y, Gu K, Luu HN, Tan S, Yang C, Fu J, Bao P, Ying T, Withers M, Mao D, Chen S, Gong Y, Wong MCS, Xu W. Adherence to colonoscopy in cascade screening of colorectal cancer: A systematic review and meta-analysis. J Gastroenterol Hepatol. 2022;37:620-631. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1] [Cited by in RCA: 14] [Article Influence: 4.7] [Reference Citation Analysis (0)] |
| 3. | Olovo CV, Huang X, Zheng X, Xu M. Faecal microbial biomarkers in early diagnosis of colorectal cancer. J Cell Mol Med. 2021;25:10783-10797. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 24] [Cited by in RCA: 37] [Article Influence: 9.3] [Reference Citation Analysis (0)] |
| 4. | Sun J. Impact of bacterial infection and intestinal microbiome on colorectal cancer development. Chin Med J (Engl). 2022;135:400-408. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 6] [Cited by in RCA: 18] [Article Influence: 6.0] [Reference Citation Analysis (0)] |
| 5. | Rezasoltani S, Aghdaei HA, Jasemi S, Gazouli M, Dovrolis N, Sadeghi A, Schlüter H, Zali MR, Sechi LA, Feizabadi MM. Oral Microbiota as Novel Biomarkers for Colorectal Cancer Screening. Cancers (Basel). 2022;15. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 20] [Cited by in RCA: 22] [Article Influence: 7.3] [Reference Citation Analysis (0)] |
| 6. | Yuan B, Ma B, Yu J, Meng Q, Du T, Li H, Zhu Y, Sun Z, Ma S, Song C. Fecal Bacteria as Non-Invasive Biomarkers for Colorectal Adenocarcinoma. Front Oncol. 2021;11:664321. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 3] [Cited by in RCA: 21] [Article Influence: 5.3] [Reference Citation Analysis (0)] |
| 7. | Eklöf V, Löfgren-Burström A, Zingmark C, Edin S, Larsson P, Karling P, Alexeyev O, Rutegård J, Wikberg ML, Palmqvist R. Cancer-associated fecal microbial markers in colorectal cancer detection. Int J Cancer. 2017;141:2528-2536. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 95] [Cited by in RCA: 138] [Article Influence: 17.3] [Reference Citation Analysis (0)] |
| 8. | Zou J, Xiao Z, Wu Y, Yang J, Cui N. Noninvasive fecal testing for colorectal cancer. Clin Chim Acta. 2022;524:123-131. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 2] [Cited by in RCA: 11] [Article Influence: 2.8] [Reference Citation Analysis (0)] |
| 9. | Anand S, Liang PS. A Practical Overview of the Stool DNA Test for Colorectal Cancer Screening. Clin Transl Gastroenterol. 2022;13:e00464. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 4] [Cited by in RCA: 17] [Article Influence: 5.7] [Reference Citation Analysis (1)] |
| 10. | Tepus M, Yau TO. Non-Invasive Colorectal Cancer Screening: An Overview. Gastrointest Tumors. 2020;7:62-73. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 84] [Cited by in RCA: 67] [Article Influence: 13.4] [Reference Citation Analysis (0)] |
| 11. | Jin P, You P, Fang J, Kang Q, Gu F, Cai Y, Zhai H, Wang B, Li Y, Xu J, Wang J, He Y, Wang Y, Dai M, Sheng J. Comparison of Performance of Two Stool DNA Tests and a Fecal Immunochemical Test in Detecting Colorectal Neoplasm: A Multicenter Diagnostic Study. Cancer Epidemiol Biomarkers Prev. 2022;31:654-661. [RCA] [PubMed] [DOI] [Full Text] [Cited by in RCA: 10] [Reference Citation Analysis (1)] |
| 12. | Xu H, Chen H, Hu J, Xiong Z, Li D, Wang S, Yu J. Feasibility of quantification based on novel evaluation with stool DNA and fecal immunochemical test for colorectal cancer detection. BMC Gastroenterol. 2022;22:384. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in RCA: 7] [Reference Citation Analysis (0)] |
| 13. | Gao R, Zhu Y, Kong C, Xia K, Li H, Zhang X, Liu Y, Zhong H, Yang R, Chen C, Qin N, Qin H. Alterations, Interactions, and Diagnostic Potential of Gut Bacteria and Viruses in Colorectal Cancer. Front Cell Infect Microbiol. 2021;11:657867. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 9] [Cited by in RCA: 35] [Article Influence: 8.8] [Reference Citation Analysis (0)] |
| 14. | Shen W, Tang D, Wan P, Peng Z, Sun M, Guo X, Liu R. Identification of tissue-specific microbial profile of esophageal squamous cell carcinoma by full-length 16S rDNA sequencing. Appl Microbiol Biotechnol. 2022;106:3215-3229. [RCA] [PubMed] [DOI] [Full Text] [Cited by in RCA: 14] [Reference Citation Analysis (0)] |
| 15. | Avuthu N, Guda C. Meta-Analysis of Altered Gut Microbiota Reveals Microbial and Metabolic Biomarkers for Colorectal Cancer. Microbiol Spectr. 2022;10:e0001322. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 1] [Cited by in RCA: 58] [Article Influence: 19.3] [Reference Citation Analysis (0)] |
| 16. | Ijaz MU, Ahmed MI, Zou X, Hussain M, Zhang M, Zhao F, Xu X, Zhou G, Li C. Beef, Casein, and Soy Proteins Differentially Affect Lipid Metabolism, Triglycerides Accumulation and Gut Microbiota of High-Fat Diet-Fed C57BL/6J Mice. Front Microbiol. 2018;9:2200. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 69] [Cited by in RCA: 87] [Article Influence: 12.4] [Reference Citation Analysis (0)] |
| 17. | Liu W, Zhang R, Shu R, Yu J, Li H, Long H, Jin S, Li S, Hu Q, Yao F, Zhou C, Huang Q, Hu X, Chen M, Hu W, Wang Q, Fang S, Wu Q. Study of the Relationship between Microbiome and Colorectal Cancer Susceptibility Using 16SrRNA Sequencing. Biomed Res Int. 2020;2020:7828392. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 22] [Cited by in RCA: 68] [Article Influence: 13.6] [Reference Citation Analysis (0)] |
| 18. | Wong SH, Yu J. Gut microbiota in colorectal cancer: mechanisms of action and clinical applications. Nat Rev Gastroenterol Hepatol. 2019;16:690-704. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 384] [Cited by in RCA: 801] [Article Influence: 133.5] [Reference Citation Analysis (0)] |
| 19. | Wang WY, Zhou H, Wang Z, Zhang YH. Comparison between diagnostic performance of intestinal Fusobacterium nucleatum, Bacteroides fragilis and Escherichia coli in 5-fluorouracil resistance to colorectal cancer: A metaanalysis. Cancer Treat Res Commun. 2022;32:100536. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 2] [Reference Citation Analysis (0)] |
| 20. | Vacante M, Ciuni R, Basile F, Biondi A. Gut Microbiota and Colorectal Cancer Development: A Closer Look to the Adenoma-Carcinoma Sequence. Biomedicines. 2020;8. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 49] [Cited by in RCA: 60] [Article Influence: 12.0] [Reference Citation Analysis (0)] |
| 21. | Peppelenbosch MP, Janmaat VT. Editorial on "A systematic review of microbial markers for risk prediction of colorectal neoplasia" by Yu and coauthors. Br J Cancer. 2022;126:1239-1240. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1] [Reference Citation Analysis (0)] |
| 22. | Yuan D, Tao Y, Wang H, Wang J, Cao Y, Cao W, Pan S, Yu Z. A comprehensive analysis of the microbiota composition and host driver gene mutations in colorectal cancer. Invest New Drugs. 2022;40:884-894. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in RCA: 14] [Reference Citation Analysis (0)] |
| 23. | Zhang S, Kong C, Yang Y, Cai S, Li X, Cai G, Ma Y. Human oral microbiome dysbiosis as a novel non-invasive biomarker in detection of colorectal cancer. Theranostics. 2020;10:11595-11606. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 21] [Cited by in RCA: 67] [Article Influence: 13.4] [Reference Citation Analysis (0)] |
| 24. | Lopez LR, Bleich RM, Arthur JC. Microbiota Effects on Carcinogenesis: Initiation, Promotion, and Progression. Annu Rev Med. 2021;72:243-261. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 17] [Cited by in RCA: 56] [Article Influence: 11.2] [Reference Citation Analysis (0)] |
| 25. | Li N, Bai C, Zhao L, Ge Y, Li X. Characterization of the fecal microbiota in gastrointestinal cancer patients and healthy people. Clin Transl Oncol. 2022;24:1134-1147. [RCA] [PubMed] [DOI] [Full Text] [Cited by in RCA: 7] [Reference Citation Analysis (0)] |
| 26. | Mirpuri J, Raetz M, Sturge CR, Wilhelm CL, Benson A, Savani RC, Hooper LV, Yarovinsky F. Proteobacteria-specific IgA regulates maturation of the intestinal microbiota. Gut Microbes. 2014;5:28-39. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 170] [Cited by in RCA: 212] [Article Influence: 19.3] [Reference Citation Analysis (0)] |
| 27. | Nouri R, Hasani A, Shirazi KM, Alivand MR, Sepehri B, Sotoodeh S, Hemmati F, Rezaee MA. Escherichia coli and Colorectal Cancer: Unfolding the Enigmatic Relationship. Curr Pharm Biotechnol. 2022;23:1257-1268. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1] [Cited by in RCA: 16] [Article Influence: 4.0] [Reference Citation Analysis (0)] |
| 28. | Li JH, Oh J, Kienesberger S, Kim NY, Clarke DJ, Zechner EL, Crawford JM. Making and Breaking Leupeptin Protease Inhibitors in Pathogenic Gammaproteobacteria. Angew Chem Int Ed Engl. 2020;59:17872-17880. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 10] [Cited by in RCA: 16] [Article Influence: 3.2] [Reference Citation Analysis (0)] |
| 29. | Arimochi H, Morita K, Nakanishi S, Kataoka K, Kuwahara T. Production of apoptosis-inducing substances from soybean protein by Clostridium butyricum: characterization of their toxic effects on human colon carcinoma cells. Cancer Lett. 2009;277:190-198. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 6] [Cited by in RCA: 10] [Article Influence: 0.6] [Reference Citation Analysis (0)] |
| 30. | Ding D, Han S, Zhang H, He Y, Li Y. Predictive biomarkers of colorectal cancer. Comput Biol Chem. 2019;83:107106. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 20] [Cited by in RCA: 36] [Article Influence: 6.0] [Reference Citation Analysis (0)] |
| 31. | Yang Y, Du L, Shi D, Kong C, Liu J, Liu G, Li X, Ma Y. Dysbiosis of human gut microbiome in young-onset colorectal cancer. Nat Commun. 2021;12:6757. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 31] [Cited by in RCA: 139] [Article Influence: 34.8] [Reference Citation Analysis (0)] |
| 32. | Agarwal A, Zhang T, Ravindran N, Thuluvath PJ, Maheshwari A. Off-Label Use of Multitarget Stool DNA Testing in Primary Care. Am J Gastroenterol. 2021;116:829-832. [RCA] [PubMed] [DOI] [Full Text] [Reference Citation Analysis (0)] |
| 33. | Yao Y, Ni H, Wang X, Xu Q, Zhang J, Jiang L, Wang B, Song S, Zhu X. A New Biomarker of Fecal Bacteria for Non-Invasive Diagnosis of Colorectal Cancer. Front Cell Infect Microbiol. 2021;11:744049. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 3] [Cited by in RCA: 25] [Article Influence: 8.3] [Reference Citation Analysis (0)] |
| 34. | Eckmann JD, Ebner DW, Kisiel JB. Multi-Target Stool DNA Testing for Colorectal Cancer Screening: Emerging Learning on Real-world Performance. Curr Treat Options Gastroenterol. 2020;. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 14] [Cited by in RCA: 22] [Article Influence: 4.4] [Reference Citation Analysis (0)] |
| 35. | Chen F, Dai X, Zhou CC, Li KX, Zhang YJ, Lou XY, Zhu YM, Sun YL, Peng BX, Cui W. Integrated analysis of the faecal metagenome and serum metabolome reveals the role of gut microbiome-associated metabolites in the detection of colorectal cancer and adenoma. Gut. 2022;71:1315-1325. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 39] [Cited by in RCA: 168] [Article Influence: 56.0] [Reference Citation Analysis (0)] |