Published online Aug 15, 2023. doi: 10.4251/wjgo.v15.i8.1384
Peer-review started: March 30, 2023
First decision: April 25, 2023
Revised: May 29, 2023
Accepted: July 7, 2023
Article in press: July 7, 2023
Published online: August 15, 2023
Processing time: 133 Days and 9.7 Hours
Altered miR-188-3p expression has been observed in various human cancers.
To investigate the miR-188-3p expression, its roles, and underlying molecular events in gastric cancer.
Fifty gastric cancer and paired normal tissues were collected to analyze miR-188-3p and CBL expression. Normal and gastric cancer cells were used to manipulate miR-188-3p and CBL expression through different assays. The relationship between miR-188-3p and CBL was predicted bioinformatically and confirmed using a luciferase gene reporter assay. A Kaplan-Meier analysis was used to associate miR-188-3p or CBL expression with patient survival. A nude mouse tumor cell xenograft assay was used to confirm the in vitro data.
MiR-188-3p was found to be lower in the plasma of gastric cancer patients, tissues, and cell lines compared to their healthy counterparts. It was associated with overall survival of gastric cancer patients (P < 0.001), tumor differentiation (P < 0.001), lymph node metastasis (P = 0.033), tumor node metastasis stage (I/II vs III/IV, P = 0.024), and American Joint Committee on Cancer stage (I/II vs III/IV, P = 0.03). Transfection with miR-188-3p mimics reduced tumor cell growth and invasion while inducing apoptosis and autophagy. CBL was identified as a direct target of miR-188-3p, with its expression antagonizing the effects of miR-188-3p on gastric cancer (GC) cell proliferation by inducing tumor cell apoptosis and autophagy through the inactivation of the Akt/mTOR signaling pathway. The in vivo data confirmed antitumor activity via CBL downregulation in gastric cancer.
The current data provides ex vivo, in vitro, and in vivo evidence that miR-188-3p acts as a tumor suppressor gene or possesses antitumor activity in GC.
Core Tip: Our study provided evidence for the antitumor activity of miR-188-3p in gastric cancer. We investigated the underlying molecular mechanisms by demonstrating that miR-188-3p inhibits gastric cancer progression and malignant behavior by suppressing Akt/mTOR signaling pathway activity and targeting CBL expression.
- Citation: Lin JJ, Luo BH, Su T, Yang Q, Zhang QF, Dai WY, Liu Y, Xiang L. Antitumor activity of miR-188-3p in gastric cancer is achieved by targeting CBL expression and inactivating the AKT/mTOR signaling. World J Gastrointest Oncol 2023; 15(8): 1384-1399
- URL: https://www.wjgnet.com/1948-5204/full/v15/i8/1384.htm
- DOI: https://dx.doi.org/10.4251/wjgo.v15.i8.1384
Gastric cancer is a significant global health burden, ranking as the third most common cause of cancer-related mortality, with an estimated 783000 deaths worldwide in 2018[1-3]. The incidence of gastric cancer varies considerably by region, with exceptionally high rates in China and Japan[3]. Up to 90% of gastric adenocarcinoma cases are associated with Helicobacter pylori infection, while Epstein-Barr virus infection accounts for 10% of gastric cancer cases globally[3]. Clinically, patients diagnosed with advanced-stage gastric cancer who undergo surgery or chemotherapy have a 5-year survival rate[4,5] of only 18%. Further investigation into the molecular biology, pathogenesis, and etiology of gastric cancer could lead to the development of novel strategies and biomarkers for effective treatment, early diagnosis, prognosis prediction, and treatment outcome assessment in gastric cancer patients.
To this end, microRNA is a class of small single-stranded noncoding RNA molecules in the human genome that are 18-22 nucleotides long and regulates the expression of the target protein-coding mRNAs through RNA silencing and/or post-transcriptional gene regulation[6]. In particular, miRNAs can bind to the three prime untranslated regions (3′-UTR) of the targeting mRNA to suppress their translation into protein or promote mRNA degradation and suppress gene expression[7]. Some published studies have shown that altered mRNA expression and functions contribute to cancer development and progression[8-11]. Several of these studies have also reported altered miR-188-3p expression in various human cancers, including gastric cancer[12-22]. For example, miR-183-3p expression was reduced in non-small cell lung cancer vs. normal tissues, whereas miR-188-3p overexpression inhibited tumor cell proliferation and colony formation[14,16]. MiR-183-3p expression was also reduced in human hepatocellular carcinoma compared to normal tissues. Upregulation of miR-188-3p inhibited tumor cell growth and migration[15]. Moreover, another type of noncoding RNA, Circ_0109291, enhanced oral cancer cell resistance to cisplatin by inhibiting the miR-188-3p expression and inducing ABCB1 expression in vitro. By contrast, the miR-188-3p inhibitor could invert the suppressive effect of circ_0109291 silencing on tumor cell cisplatin resistance[17]. However, another previous genome-wide miRNA analysis showed that detecting miR-188-3p expression could predict colorectal cancer prognosis and that miR-188-3p overexpression induced colorectal cancer cell migration in vitro and metastasis in vivo[18]. In gastric cancer, a study of miR-188-3p showed that the expression thereof through Circ_0078607 silencing could suppress gastric cancer development[13]. It has been shown that the role of epigenetic regulation could be as a promising therapeutic target in cancers, e.g., the onco-miRs could epigenetically regulate colon carcinogenesis and development of resistance, while targeting of such regulation was demonstrated as a promising therapeutic potential[23]. This study further investigated the miR-188-3p expression, its roles, and underlying molecular events in gastric cancer ex vivo, in vitro, and in vivo. The results of this work provide further supporting data for miR-188-3p being used as a tumor marker and therapeutic strategy for the early detection, prediction of treatment outcome, and effective treatment of gastric cancer patients.
This study examined 50 paired gastric cancer and adjacent non-tumor tissues from the Department of Surgery of Hospital of Nanfang Hospital, Southern Medical University, China, between January 2018 and December 2018. All patients were histologically diagnosed with gastric adenocarcinoma and classified according to the WHO grading system. The tumor stage using the tumor node metastasis (TNM) classification was based on the 2011 Union for International Cancer[24]. After tumor resections, fresh patient tissue specimens were obtained from the surgical room, snap-frozen in liquid nitrogen, and stored at -80℃. In addition, plasma samples from 20 gastric cancer patients and 20 healthy controls were collected. This study was approved by the Ethics Committee of the Southern Medical University, and all participants provided written informed consent before enrolling in the study.
Total cellular RNA from tissues and cells was isolated using the Trizol reagent (Invitrogen, Carlsbad, CA, United States) and was reversely transcribed into cDNA with the M-MLV RT Kit (Promega, Madison, MI, United States) according to the manufacturers’ instructions. These cDNA samples were then subjected to amplify miR-188-3p and U6 expression using qPCR with specific RT and PCR primers (GeneCopoeia, Rockville, MD, United States) with the SYBR® Green PCR Master Mix (Toyobo, Osaka, Japan). The relative miR-188-3p level was compared with that of U6 using the 2-ΔΔCt method. The primer and probe sequences of miR-188-3p were 5'-CTCCCACATGCAGGGTTTG-'3, 5'-CTCAACTGGTGTCGTGGAGTC-3', and 5'-CACATGCAGGGTTTGCAAA -3'.
The human immortalized normal gastric epithelial GES-1 and gastric carcinoma cell lines AGS, and HGC-27 were originally obtained from American Type Culture Collection (Manassas, VA, United States), while the gastric cancer SGC-7901, BGC-823, and MGC-803 cell lines were obtained from the Beijing Institute of Cancer Research (Beijing, China) and the gastric cancer MKN-28 and MKN-45 cell lines from the Japanese Collection of Research Bioresources Cell Bank (Osaka, Japan). These cells were grown in RPMI-1640 containing 10% fetal bovine serum (FBS) at 37°C in a humidified incubator supplemented with 5% CO2.
The double-stranded Hsa-miR-188-3p mimics (5'-GUCCCACAUGCAGGGUUUGCA-3'), Hsa-miR-188-3p inhibitor (5'-UGCAAACCCUGCAUGUGGGAG-3'), and their corresponding negative controls (NC) were purchased from the Suzhou Jima Gene Co. (Suzhou, China). Gastric cancer BCG823 and AGS cells were seeded overnight and transiently transfected with the miR-188-3p mimics and their corresponding m-NC (miR-188-3p inhibitor) and i-NC controls using Lipofecta
To assess the changes in cell viability after miR-188-3p expression manipulation, we utilized the Cell Counting Kit-8 (CCK-8) kit from Dojindo Laboratories (Tokyo, Japan). In brief, the transfected cells were plated at a density of 1 × 5 cells/well in a 96-well plate and grown for up to 4 d. Every 24 h, a serum-free medium and 10 microliters of the CCK-8 solution were refreshed in each well, which were then further incubated for an additional 2 h. The optical density of each well was then measured to assess the cell viability using a microplate reader (Thermo Fisher Scientific, Waltham, MA, United States) at 450 nm. All experiments were performed in triplicate.
The gene-transfected cells were inoculated and cultured in a 6-well plate (200 cells/well) for 14 d. Afterward, the plate was washed with phosphate-buffered saline (PBS) three times, and cells were then fixed in ethanol for 30 s and stained with 1% crystal violet for 15 min. Cells were then rewashed using PBS. The plate was photographed, and the individual colonies containing ≥ 50 cells were counted and quantitated with the Image J software (National Institute of Health, Bethesda, MD, United States).
Tumor cell growth was further evaluated using the 5-ethynyl-2-deoxyuridine (EdU) incorporation assay with an EdU kit (RiboBio, Guangzhou, China). Briefly, tumor cells were grown and incubated overnight with 50 μmol/L of the EdU reagent for 24 h and then evaluated for cell proliferation using the Cell-Light™ EdU Cell Proliferation Detection Kit following the manufacturer’s protocol. After that, the Hoechst 33342 reagent (RiboBio, Guangzhou, China) was used to visualize tumor cell nuclei, which were reviewed and photographed under an inverted fluorescence microscope (Olympus, Tokyo, Japan) for three random horizons of cells. The number of the EdU-positive cells vs the total cells in the picture were quantitated using the Image J software (National Institute of Health, Bethesda, MD, United States).
A flow cytometric Annexin V-PE apoptosis assay kit from BD Biosciences (Franklin Lakes, NJ, United States) was obtained to assess the level of apoptosis and cell cycle distribution of gastric cancer cells after gene transfection. In brief, BGC-823 and AGS cells were grown and transfected with miR-188-3p mimics, m-NC, m-NC + Vector, miR-188-3p mimics + Vector, or miR-188-3p mimics + CBL with or without treatment with 2 µg/mL fluorouracil (5-FU) for 48 h. Cells were then harvested and washed three times with PBS and then resuspended in 500 µL of the flow buffer with the addition of 5 µL Annexin V-FITC and 10 µL PI. The samples were incubated for 15 min at room temperature, and then the level of cell apoptosis was evaluated using the BD FACS Canto II flow cytometer (BD Biosciences). The necrotic cells stained double positive for Annexin V and PI, while apoptotic cells were negative for Annexin V and PI staining and were quantified using the BD FACS Canto II software.
After gene transfection, gastric cancer cell lines were plated into six-well plates (200 cells/well) and grown in the RPMI-1640 containing 10% FBS for two weeks at 37°C. At the end of the experiment, the cells were fixed with 4% paraformaldehyde for 30 min, washed three times with PBS, and stained with 1% crystal violet solution for 10 min to visualize cell nuclei. After that, cell colonies containing ≥ 50 cells were counted and analyzed. All experiments were conducted in triplicate.
After gene transfection, tumor cells were seeded with a serum-free medium in the Transwells (Corning, Corning, NY, United States), which were pre-coated with or without 25 μg Matrigel (BD Biosciences) at a density of 2000 cells per chamber. The bottom chamber contained the growth medium supplemented with 20% FBS. The Transwells were incubated for 48 h at 37°C. After that, cells that remained on the topside of the filter were removed with cotton swabs, and the cells that migrated or invaded into the reverse side of the filter were fixed with 3.7% paraformaldehyde in PBS and stained for 10 min using 2% crystal violet. The number of cells was then counted and calculated in three typical microscopic fields (×20).
Total cellular protein was extracted using a Protein Extraction kit from Invent Biotechnologies (Plymouth, MN, United States) and quantified using the bicinchoninic acid (BCA) protein assay kit (Cell Signaling Technology, Danvers, MA, United States). The resulting protein samples with 30 μg in each lane were loaded onto 8%-12% sodium dodecyl sulfate-polyacrylamide gel electrophoresis gels for electrophoresis to separate the proteins. Subsequently, the protein in the gel was transferred onto the nitrocellulose membranes. For Western blotting, the membranes were incubated in a 5% skimmed milk powder solution at room temperature for 1 h. Next, the membranes were incubated overnight with primary antibody solutions at 4°C. Afterward, the membranes were subjected to wash with Tris-based saline-Tween 20 (TBS-T) three times and then incubated with a secondary antibody at room temperature for 2 h. After being washed with TBS-T, the positive protein signals on the membranes were detected using the Enhanced chemiluminescence reagents (Cell Signaling Technology). The antibodies used were a mouse antibody against GAPDH, β-actin, or CBL (Santa Cruz Biotechnology, Santa Cruz, CA, United States), anti-LC3 and Beclin1 (Cell Signaling Technology, Danvers, MA, United States), and a rabbit antibody against mTOR, p-mTOR, Akt (C-terminal), or p-Akt (Ser473) (Cell Signaling Technology). The dilution of each antibody followed the manufacturers’ recommendations.
To predict the mRNA target of miR-188-3p, we utilized the online tools of MiRDB (http://mirdb.org), TargetScan (http://www.targetscan.org/), miRPathDB (http://www.https://mpd.bioinf.uni-sb.de/), and TargetMiner (https://tools4mirs.org) to reveal potential targets, like CBL (Casitas B-lineage Lymphoma) and the miRNA-binding region.
A gene reporter plasmid carrying a wild type or mutant CBL 3'-UTR was purchased from Genepharma (Shanghai, China). In brief, tumor cells after miR-188-3p mimics transfection were grown and transfected with the reporter plasmid containing a luciferase gene fused to the target sequence of the wild-type (WT) 3'-UTR or mutant 3’-UTR of CBL using Lipofectamine 3000 (Invitrogen) for 36 h. Tumor cells were then lysed, and the proteins were quantified using the BCA protein assay kit (Cell Signaling Technology) and analyzed using the Luciferase Reporter Assay System (Transgen Biotech, China). All experiments were conducted in triplicate.
A lentivirus carrying miR-188-3p (UBI-MCS-SV40-EGFP-IRES-puromycin) and the red fluorescent protein gene was purchased from Gene Chem (Shanghai, China). Specifically, the plasmid carrying miR-188-3p was transfected into tumor cells and grown for 14 days to establish the puromycin-resistant cell population. Moreover, a lentivirus carrying CBL cDNA using the Ubi-MCS-3FLAG-CBh-CHERRY-IRES-puromycin vector and an Ubi-MCS-3FLAG-CBh-CHERRY-IRES-puromycin empty vector was also obtained from Gene Chem and used to establish a stably transfected cell population according to the manufacturer’s protocols. The stably transfected cell population was used for the in vivo assay in nude mice.
The in vivo study using nude mice was approved by the Experimental Animal Ethics Committee of Southern Medical University. In brief, 15 BALB/C mice of 4-6 wk of age were purchased from the Animal Research Center of Medical University and randomly divided into three groups (miR-188-3p-NC, miR-188-3p-mimics, and miR-188-3p mimics + CBL cDNA). To establish tumor cell xenografts, 1 × 106 tumor cells were implanted into each nude mouse at the right inguinal region. Tumor volume was recorded every three days, and tumor mass was calculated using the following formula: tumor volume = 1/2 × length × width2. After 30 d, the mice were sacrificed to resect gastric cancer cell xenografts.
Immunohistochemistry (IHC) analysis was conducted to evaluate CBL and Ki67 Levels in nude mouse tumor xenografts. The antibodies used were an anti-CBL (Santa Cruz Biotechnology) and anti-Ki67 (Cat. #ab15580; Abcam, Cambridge, MA, United States). In brief, nude mouse tumor xenograft tissues were processed, paraffin-embedded, and sectioned into 4 µm-thick tissue slides. Two independent investigators scored the tissue sections and quantified them with a semi-quantitative scoring system according to a previous study[25].
The data are expressed as mean ± mean standard error and analyzed statistically with the SPSS software (version 16.0; SPSS, Chicago, IL, United States). A Chi-square test or Pearson’s χ2 test was conducted to associate the miR-188-3p level with the clinical characteristics of patients. A Student’s t-test or Fisher’s exact test was used to analyze differences between the two groups. The symbol "d" indicates that the data doesn’t have a statistically significant difference (P > 0.05), whereas "a", "b", and “c” indicate that the data possesses a statistically significant difference (P < 0.05, P < 0.01. and P < 0.001, respectively).
The plasma level of miR-188-3p expression was first assayed using qRT-PCR in the plasma of gastric cancer patients. We found a significantly reduced miR-188-3p level compared to that of the plasma from healthy people (P < 0.01; Figure 1A).
Next, we assayed the miR-188-3p level in a normal GES-1 cell line and seven gastric cancer cell lines AGS, HGC-27, MKN-28, MKN-45, SGC-7901, MGC-803, and BGC-823. We found that miR-188-3p expression was lower in tumor cell lines than in GES-1 cells (Figure 1B). Furthermore, we assessed miR-188-3p expression in 50 paired noncancerous gastric mucosa (N) and gastric cancer tissues (T) using qRT-PCR. We found that 42 of 50 gastric cancer tissue samples exhibited a lower miR-188-3p expression than those of the adjacent normal tissues (Figure 1C).
To explore the effect of miR-188-3p on gastric cancer cells, we transfected the miR-188-3p mimics and inhibitors into gastric cancer cell lines and assessed the miR-188-3p level using qRT-PCR. Overall, we observed significant changes in miR-188-3p expression level in tumor cells (P < 0.01 and P < 0.001; Figure 1D and E).
Furthermore, we associated miR-188-3p expression with the survival time of patients and found that patients with a low miR-188-3p expressing gastric cancer survived much shorter than patients with high miR-188-3p expressing tumor, according to the Kaplan-Meier Plotter database data (http://kmplot.com/analysis/; P < 0.001; Figure 1F). We then associated miR-188-3p level with the clinicopathological characteristics and prognosis of gastric cancer patients. Our data showed that the miR-188-3p level was associated with gastric cancer differentiation (P < 0.001), lymph node metastasis (P = 0.033), TNM stage (I/II vs III/IV, P = 0.024), and American Joint Committee on Cancer (AJCC) stage (I/II vs III/IV, P = 0.03). However, there were no significant associations of miR-188-3p level with tumor size (10 cm or less vs 10 cm or more in size), gender, or age (60 years or younger vs 60 years or older; P > 0.05; Supplementary Table 1). The miR-188-3p level also predicted a better prognosis for gastric cancer patients (Supplementary Table 1). These data indicate that miR-188-3p may be a tumor suppressor or at least possess tumor suppressor activity in gastric cancer.
We then assessed the effects of miR-188-3p on gastric cancer AGS and BGC-823 cells using miR-188-3p mimics and inhibitors and analyzed the change in tumor cell proliferation using the EdU incorporation assay. The tumor cell proliferation rate was dramatically inhibited after miR-188-3p mimics transfection compared to control cells (m-NC cells). Conversely, tumor cell growth rate was induced after the miR-188-3p inhibitor transfection (P < 0.001; Figure 2A and B). The proliferation rate of the miR-188-3p mimics and inhibitors in gastric cancer AGS and BGC-823 cell lines was also assessed using the plate cloning experiment. The data further confirmed the EdU assay data (P < 0.001; Figure 2C and D). Moreover, the CCK-8 data revealed that the miR-188-3p mimics significantly decreased cell viability of BGC-823 and AGS cells, whereas that of the miR-188-3p inhibitor increased cell viability of BGC 823 and ACG cells (Supple
Furthermore, the gastric cancer cell invasion capacity was significantly decreased after miR-188-3p mimics transfection, whereas such a capacity was enhanced after the transfection of the miR-188-3p inhibitor (P < 0.01; Supple
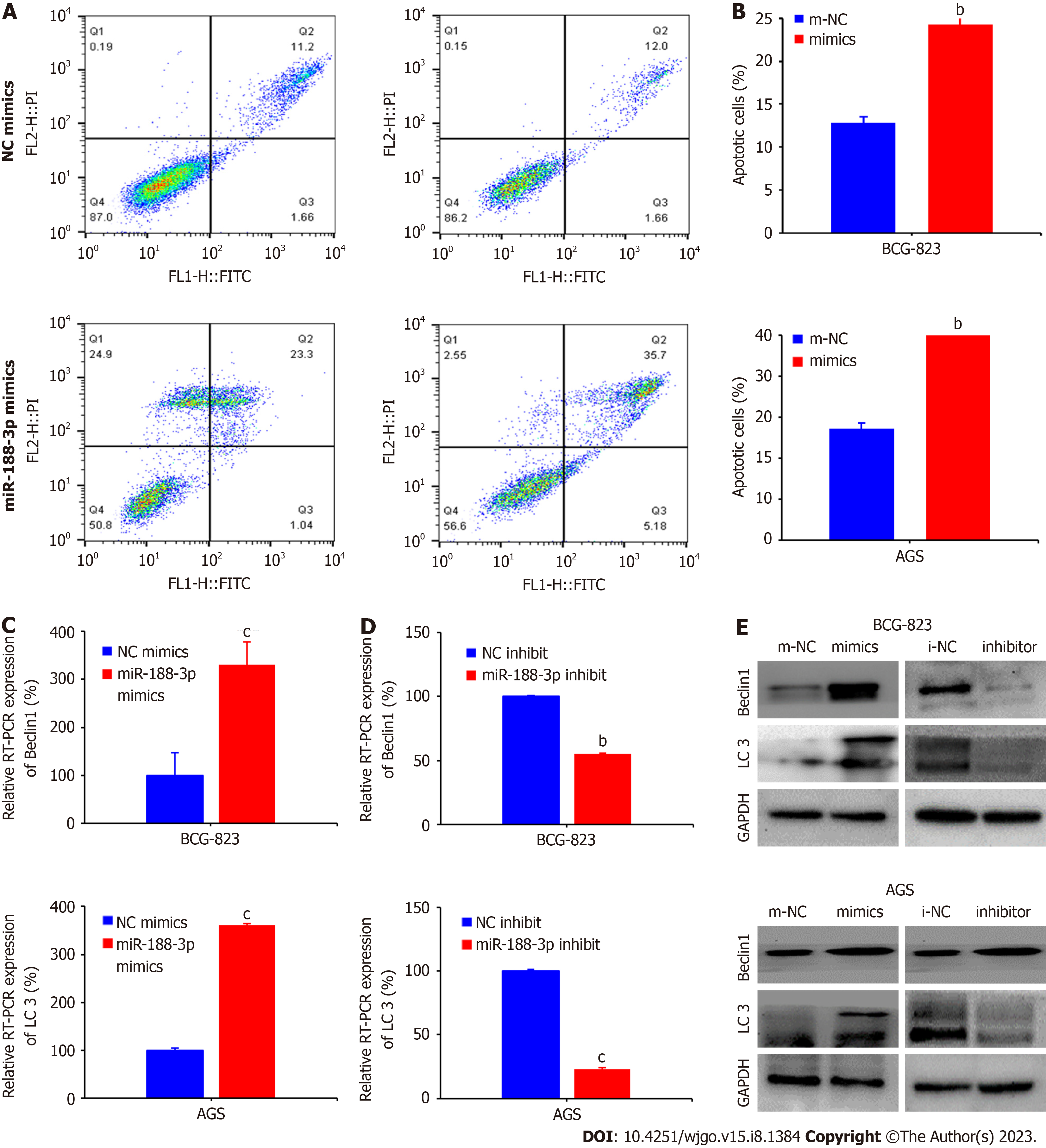
Next, we performed a flow cytometric/Annexin V-PE/7-AAD assay and found that the miR-188-3p mimics increased apoptosis of BGC-823 cells from 11.2% (cells transfected with i-NC) to 23.3% (the miR-188-3p mimics transfection), and ACG cells from 12% to 35.7% (P < 0.01; Figure 3A and B).
Moreover, miR-188-3p mimics transfection into gastric cancer AGS and BGC-823 cells increased autophagy-related gene expression (Beclin1 and LC3-II/LC3-I). In contrast, miR-188-3p inhibitor transfection into gastric cancer cells showed the opposite effect, decreasing their expression compared with i-NC (P < 0.001; Figure 3C and D). Consistent with these results, Western blot data also confirmed that miR-188-3p could induce autophagic proteins (Beclin-1 and LC3 I/II; Figure 3E).
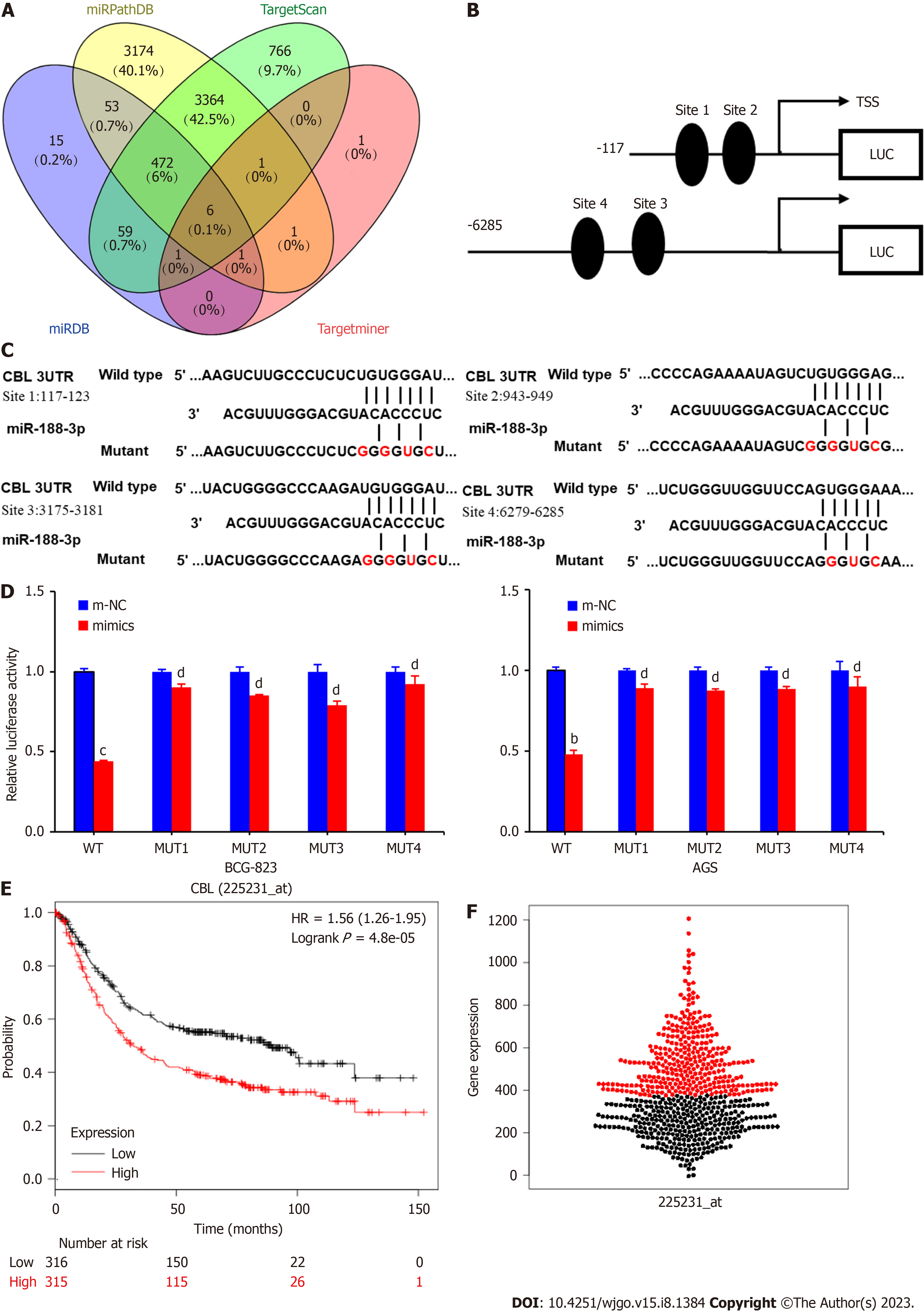
To explore the mechanism of miR-188-3p action in gastric cancer cells, we first searched the TargetScan, miRPathDB, miRDB, and the TargetMiner databases for possible target genes of miR-188-3p. We found six genes using a Wern diagram. CBL (an oncogene in gastric cancer) was selected for confirmation using the luciferase gene reporter assay. As shown in Figure 4A, four target sites show that miR-188-3p can bind to the 3'-UTR of the CBL gene (Figure 4B). The luciferase gene reporter assay confirmed this prediction, showing that miR-188-3p could bind to the 3'-UTR region of the CBL gene (WT, P < 0.01 vs MUT, P > 0.05; Figure 4C and D). We then utilized the KM-Plotter database (http://kmplot.com) and TCGA dataset (http://xena.ucsc.edu/public) to associate CBL expression with the survival of gastric cancer patients and found that CBL expression indeed was associated with survival of gastric cancer patients (Figure 4E and F). Our IHC data also showed that CBL protein was localized in the tumor cell cytoplasm and was highly expressed in tumor tissues compared to the adjacent gastric tissues (Supplementary Figure 2A). The association between miR-188-3p and CBL expression in a case-wise comparison in gastric cancer tissues was negatively correlated (Supple
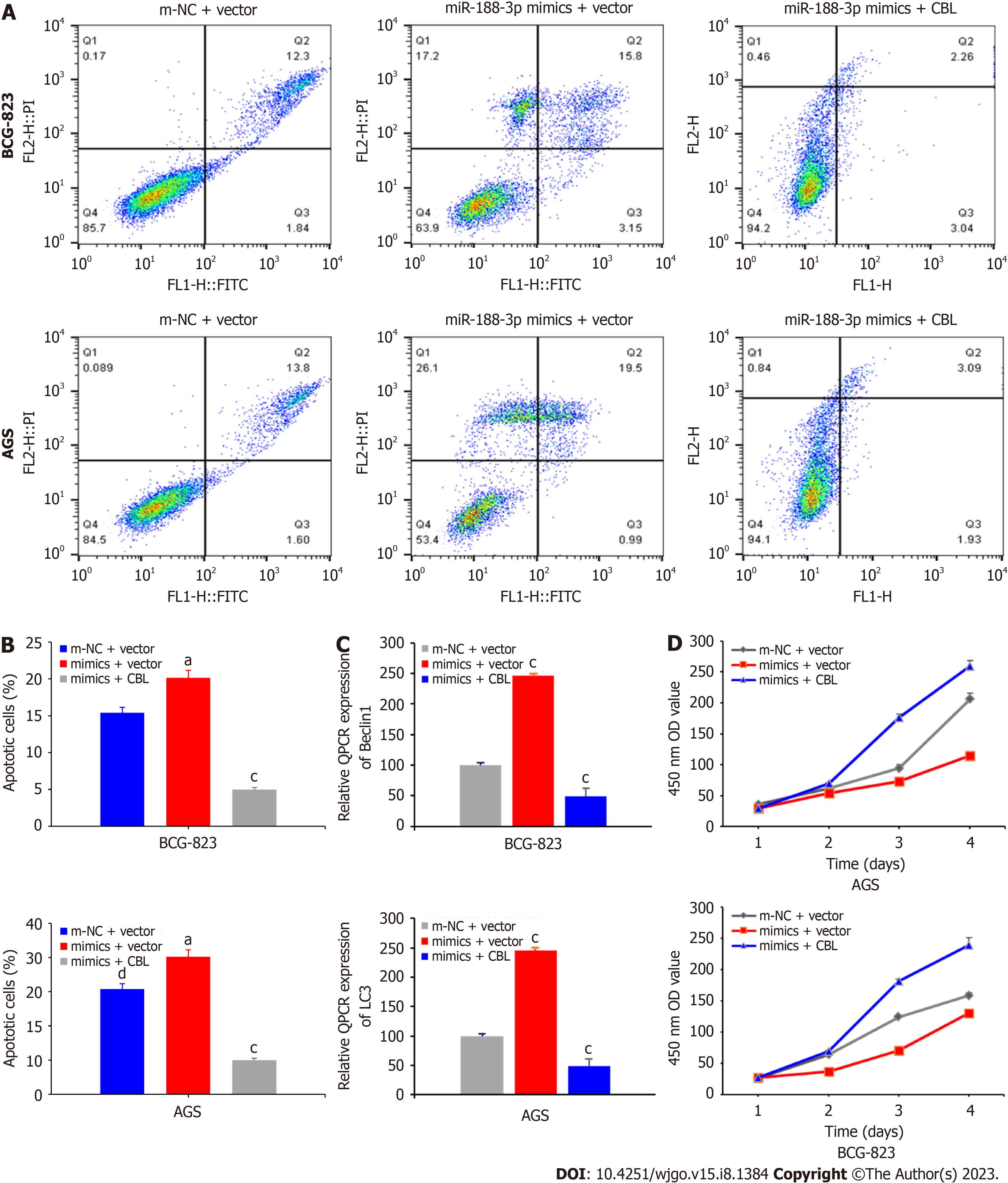
To confirm the miR-188-3p regulation of CBL in tumor cells, we analyzed the molecular effect of miR-188-3p in regulating CBL-mediated gastric cancer cell proliferation. MiR-188-3p mimics-transfected AGS and BGC-823 cells enhanced tumor cell apoptosis compared with control cells (P < 0.05 and P < 0.001; Figure 5A and B). Expression of the autophagy-related genes LC3-II/LC3-I and Beclin1 also changed accordingly (P < 0.001; Figure 5C). The proliferation rate of gastric cancer cells after miR-188-3p mimics and CBL cDNA transfection was increased significantly compared to that of miR-188-3p mimics with vector-only control transfection (Figure 5D). These results indicate that CBL transfection could antagonize the inhibiting effects of miR-188-3p in gastric cancer cells. It can be concluded that miR-188-3p could negatively regulate CBL expression to promote gastric cancer cell autophagy and apoptosis.
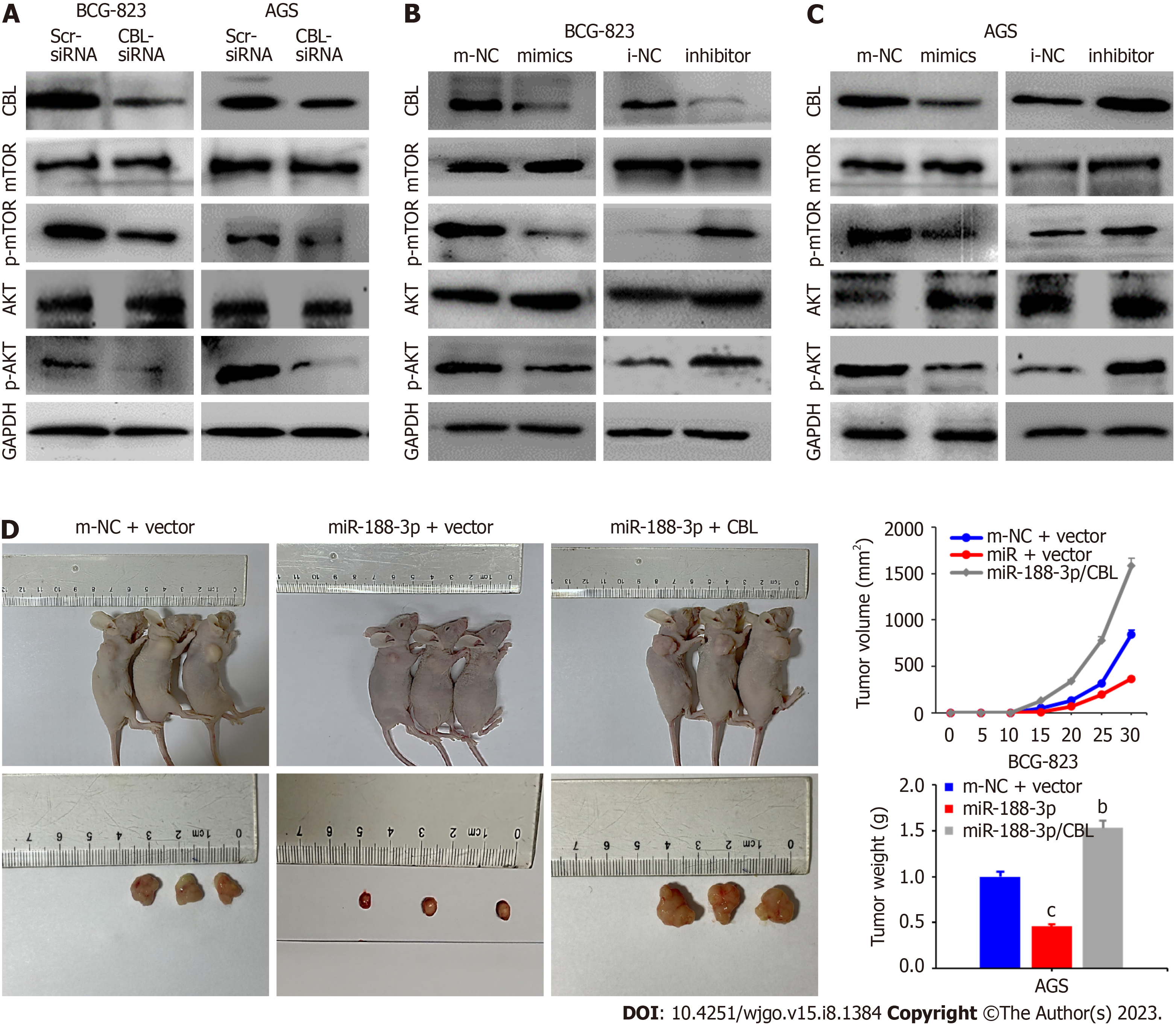
Previous studies have demonstrated that the AKT/mTOR signaling pathway was related to autophagy occurrence[26-28]. Therefore, we detected the expression of Akt and mTOR in our cell model system and found that the level of the phosphorylated proteins was lower than usual. By contrast, the level of becline1 protein was increased in CBL siRNA-transfected tumor cells (Figure 6A). In addition, miR-188-3p mimics transfection reduced the level of phosphorylated Akt and phosphorylated mTOR proteins in tumor cells in vitro. However, it increased the level of becline1 protein, whereas miR-188-3p inhibitor exhibited an inverse effect on gastric cancer cells (Figure 6B). Moreover, CBL overexpression antagonized the miR-188-3p-induced reduction of p-Akt and p-mTOR expression (Figure 6C).
To verify whether miR-188-3p inhibits gastric cancer cell growth in vivo, we implanted BGC-823 cells after miR-188-3p mimics, miR-188-3p/CBL cDNA, or m-NC transfection to establish a xenograft tumor model in nude mice. Our data revealed that miR-188-3p mimics transfection inhibited gastric cancer cell growth and that the tumor size of the miR-188-3p/CBL group was significantly greater than those of the m-NC and miR-188-3p groups (both P < 0.001; Figure 6D and E). Meanwhile, more ki67-positive and CBL-positive cells were observed in miR-188-3p/CBL cDNA-transfected gastric cancer cells (Supplementary Figure 3A and B). These findings illustrate that miR-188-3p inhibited tumor cell xenograft growth in vivo, whereas CBL expression antagonized inhibition of miR-188-3p-induced proliferation in vivo.
Gastric cancer morbidity and mortality account for the second- and third-highest death rates in China among all malignancies. Gastric cancer also has a poor clinical prognosis[5] worldwide[2]. However, the associated molecular mechanisms of this disease have not been well explained. Various high-throughput sequencing techniques have recently been used to analyze altered genome-wide miRNA gene expression in gastric cancer. These studies have revealed that aberrant expression of different miRNAs was present in gastric cancer and led to novel gastric cancer biomarker discovery and potential targets for gastric cancer treatment[29-31]. In the current study, we focused on miR-188-3p and found a significant miR-188-3p downregulation in gastric cancer samples (plasma and tissues vs. normal controls). The expression of miR-188-3p was also associated with better patient prognosis and inversely associated with gastric cancer differentiation, metastasis to the lymph node, high TNM stages, and AJCC stage. Moreover, miR-188-3p expression inhibited gastric cancer cell growth but induced apoptosis and autophagy. Autophagy in cells can be a double-edged sword for multidrug resistance (MDR) of tumors; for example, it is involved in MDR development to protect cancer cells from chemotherapy drugs but can also kill MDR cancer cells with inactivated apoptotic pathways[32]. Thus, autophagy also plays a dual role in tumorigenesis, tumor progression, and resistance to chemotherapy[33,34]. In addition, autophagy can exert anticancer effects by eliminating damaged organelles and recycling degradation products in normal cells. Paradoxically, excessive autophagy can plunge cancer cells into "autophagic cell death" or "type II programmed cell death"[35,36].
Molecularly, miR-188-3p could target CBL and downregulate CBL expression, which was associated with the antitumor activity of miR-188-3p in vitro and in a nude mouse assay. In conclusion, our current study demonstrated the tumor suppression of miR-188-3p in gastric cancer. Further study will confirm the antitumor activity of miR-188-3p in gastric cancer as a biomarker of prognosis prediction and as a novel therapeutic target in gastric cancer. Indeed, as predicted, miR-188-3p has been shown to possess antitumor activity in most human cancers analyzed to date[12-22]. However, Pichler et al[18] reported that miR-188-3p expression enhanced colonic tumor cell migration and metastasis in colorectal cancer.
The Casitas B-lineage Lymphoma (CBL) family of genes is a class of highly conserved ubiquitin-protein ligases. A previous study showed that CBL gene mutations were implicated in human cancers, especially acute myeloid leukemia[37]. Indeed, CBL was first evaluated as an oncogene in early B-lineage lymphoma[38]. Other studies have shown that CBL is conducive to the survival and proliferation of many types of cells and plays an essential role in various human cancer developments, such as breast[39] and kidney[40] cancer. CBL alterations have also been associated with gastric cancer development[41,42], where apoptosis and migration are regulated by the Akt and mTOR signaling in gastric cancer cells[43-46]. In the current study, we demonstrated that CBL is a direct target of miR-188-3p and that miR-188-3p-inhibited gastric cancer proliferation downregulated CBL expression in vitro and in vivo. CBL overexpression reduced the miR-188-3p antitumor activity in gastric cancer. Thus, our current data indicates that miR-188-3p antitumor activity in gastric cancer could act through the downregulation of CBL expression.
Furthermore, our current study also showed that the miR-188-3p anti-gastric cancer activity inactivated the AKT/mTOR signaling pathway and induced autophagy, which is a catabolic process in various cell activities[47]. The mTOR compound is a crucial negative regulator of autophagy and regulates cell proliferation and protein synthesis[48,49]. At the gene level, mTOR can phosphorylate and suppress the kinase complex during cell autophagy[50,51]. A previous study has shown that tumor cell growth and autophagy were controlled after activation of the AKT/mTOR signaling pathway[52-55]. Our current data confirmed these previous studies, showing that miR-188-3p induced gastric cancer cell autophagy and increased levels of the p-mTOR and p-Akt proteins to activate their signaling and promote tumor cell growth and invasion capacity.
Our study is an essential addition to the literature that supports the antitumor activity of miR-188-3p in gastric cancer. We explored the underlying molecular mechanisms by illustrating the miR-188-3p-based inhibition of gastric cancer and malignant behavior (progression) by inhibiting the activity of the Akt/mTOR signaling pathway and targeting CBL expression. Our present study also contained various limitations that should be discussed. First, the number of gastric cancer cases was small, and we cannot associate miR-188-3p expression with the treatment outcome of patients. Our in vitro experiments were also preliminary, and more in-depth investigation is needed to facilitate the actual role of miR-188-3p in gastric cancer. Thus, in our future work, we will assess the detection of miR-188-3p expression as a predictive biomarker for the prognosis and treatment outcome of gastric cancer. We will also assess the use of miR-188-3p as a novel therapeutic approach against gastric cancer.
This study demonstrated the anticancer activity of miR-188-3p through the downregulation of CBL expression in gastric cancer. Our findings provide novel insights into the molecular mechanisms underlying gastric cancer development and progression and may offer a new approach to controlling this disease.
Altered miR-188-3p expression has been observed in various human cancers, while report of its role in gastric cancer needs further investigation.
Better understanding of miR-188-3p biology, functions, and molecular mechanism of action in gastric cancer could lead to develop novel strategies in control gastric cancer.
This study assessed miR-188-3p expression and functions in gastric cancer tissues and cells, respectively.
Gastric cancer and normal tissues were obtained to detect miR-188-3p expression using quantitative reverse transcriptase-polymerase chain reaction and cell lines were used to manipulate miR-188-3p expression and functions in vitro using different assays. After that, miR-188-3p regulation of CBL expression was predicted bioinformatically and confirmed using a luciferase gene reporter assay. The Kaplan-Meier analysis was used to associate miR-188-3p or CBL expression with survival of gastric cancer patients. After that, we performed a nude mouse tumor cell xenograft assay to confirm the in vitro and ex vivo data.
miR-188-3p expression was low in plasma from gastric cancer patients, tissues, and cell lines compared to controls. Downregulated miR-188-3p expression was associated with clinicopathological data from patients. Furthermore, the ex vivo, in vitro, and in vivo data confirmed miR-188-3p directly targeted CBL, while overexpression of miR-188-3p inhibited CBL autophagy through the AKT/mTOR signaling pathway to promote the proliferation of gastric cancer.
The current data provides ex vivo, in vitro, and in vivo evidence showing that miR-188-3p acts as a tumor suppressor gene or at least possesses antitumor activity in gastric cancer.
These findings provide a novel insight into the molecular mechanism underlying gastric cancer development and progression and may offer a novel approach in control of this disease in future.
We want to thank the generous support of the Guangdong Provincial Key Laboratory of Gastroenterology, Department of Gastroenterology, Nanfang Hospital, Southern Medical University, Longgang District People’s Hospital, Shenzhen.
Provenance and peer review: Unsolicited article; Externally peer reviewed.
Peer-review model: Single blind
Specialty type: Oncology
Country/Territory of origin: China
Peer-review report’s scientific quality classification
Grade A (Excellent): 0
Grade B (Very good): 0
Grade C (Good): C, C, C, C
Grade D (Fair): 0
Grade E (Poor): 0
P-Reviewer: Kotelevets SM, Russia; Liu Q, China; Sukocheva OA, Australia S-Editor: Liu JH L-Editor: A P-Editor: Xu ZH
| 1. | Ferlay J, Soerjomataram I, Dikshit R, Eser S, Mathers C, Rebelo M, Parkin DM, Forman D, Bray F. Cancer incidence and mortality worldwide: sources, methods and major patterns in GLOBOCAN 2012. Int J Cancer. 2015;136:E359-E386. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 20108] [Cited by in RCA: 20485] [Article Influence: 2048.5] [Reference Citation Analysis (19)] |
| 2. | Bray F, Ferlay J, Soerjomataram I, Siegel RL, Torre LA, Jemal A. Global cancer statistics 2018: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J Clin. 2018;68:394-424. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 53206] [Cited by in RCA: 55631] [Article Influence: 7947.3] [Reference Citation Analysis (131)] |
| 3. | Wild CP, Weiderpass E, Stewart BW. World cancer report: Cancer research for cancer prevention. Chapter 5.4. Stomach cancer, still one of the main cancer types worldwide. International agency for research on cancer, lyon, france. 2020; 333-344. |
| 4. | Rawla P, Barsouk A. Epidemiology of gastric cancer: global trends, risk factors and prevention. Prz Gastroenterol. 2019;14:26-38. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 297] [Cited by in RCA: 715] [Article Influence: 102.1] [Reference Citation Analysis (1)] |
| 5. | Chen W, Zheng R, Baade PD, Zhang S, Zeng H, Bray F, Jemal A, Yu XQ, He J. Cancer statistics in China, 2015. CA Cancer J Clin. 2016;66:115-132. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 11444] [Cited by in RCA: 13190] [Article Influence: 1465.6] [Reference Citation Analysis (3)] |
| 6. | Saliminejad K, Khorram Khorshid HR, Soleymani Fard S, Ghaffari SH. An overview of microRNAs: Biology, functions, therapeutics, and analysis methods. J Cell Physiol. 2019;234:5451-5465. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1431] [Cited by in RCA: 1310] [Article Influence: 218.3] [Reference Citation Analysis (0)] |
| 7. | Bartel DP. MicroRNAs: target recognition and regulatory functions. Cell. 2009;136:215-233. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 14460] [Cited by in RCA: 16038] [Article Influence: 1002.4] [Reference Citation Analysis (2)] |
| 8. | Acunzo M, Romano G, Wernicke D, Croce CM. MicroRNA and cancer--a brief overview. Adv Biol Regul. 2015;57:1-9. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 495] [Cited by in RCA: 495] [Article Influence: 49.5] [Reference Citation Analysis (0)] |
| 9. | Vishnoi A, Rani S. MiRNA Biogenesis and Regulation of Diseases: An Overview. Methods Mol Biol. 2017;1509:1-10. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 328] [Cited by in RCA: 501] [Article Influence: 71.6] [Reference Citation Analysis (0)] |
| 10. | Fabian MR, Sonenberg N, Filipowicz W. Regulation of mRNA translation and stability by microRNAs. Annu Rev Biochem. 2010;79:351-379. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 2112] [Cited by in RCA: 2433] [Article Influence: 162.2] [Reference Citation Analysis (0)] |
| 11. | Banerjee S, Karunagaran D. An integrated approach for mining precise RNA-based cervical cancer staging biomarkers. Gene. 2019;712:143961. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 13] [Cited by in RCA: 15] [Article Influence: 2.5] [Reference Citation Analysis (0)] |
| 12. | Akçakaya P, Ekelund S, Kolosenko I, Caramuta S, Ozata DM, Xie H, Lindforss U, Olivecrona H, Lui WO. miR-185 and miR-133b deregulation is associated with overall survival and metastasis in colorectal cancer. Int J Oncol. 2011;39:311-318. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 10] [Cited by in RCA: 75] [Article Influence: 5.4] [Reference Citation Analysis (0)] |
| 13. | Bian W, Liu Z, Chu Y, Xing X. Silencing of circ_0078607 prevents development of gastric cancer and inactivates the ERK1/2/AKT pathway through the miR-188-3p/RAP1B axis. Anticancer Drugs. 2021;32:909-918. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 5] [Cited by in RCA: 7] [Article Influence: 1.8] [Reference Citation Analysis (0)] |
| 14. | Yao J, Xu G, Zhu L, Zheng H. circGFRA1 Enhances NSCLC Progression by Sponging miR-188-3p. Onco Targets Ther. 2020;13:549-558. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 23] [Cited by in RCA: 40] [Article Influence: 8.0] [Reference Citation Analysis (0)] |
| 15. | Luo Z, Fan Y, Liu X, Liu S, Kong X, Ding Z, Li Y, Wei L. MiR-188-3p and miR-133b Suppress Cell Proliferation in Human Hepatocellular Carcinoma via Post-Transcriptional Suppression of NDRG1. Technol Cancer Res Treat. 2021;20:15330338211033074. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 2] [Cited by in RCA: 13] [Article Influence: 3.3] [Reference Citation Analysis (0)] |
| 16. | Zhou L, Xing C, Zhou D, Yang R, Cai M. Downregulation of lncRNA FGF12-AS2 suppresses the tumorigenesis of NSCLC via sponging miR-188-3p. Open Med (Wars). 2020;15:986-996. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 2] [Cited by in RCA: 8] [Article Influence: 1.6] [Reference Citation Analysis (0)] |
| 17. | Gao F, Han J, Wang Y, Jia L, Luo W, Zeng Y. Circ_0109291 Promotes Cisplatin Resistance of Oral Squamous Cell Carcinoma by Sponging miR-188-3p to Increase ABCB1 Expression. Cancer Biother Radiopharm. 2022;37:233-245. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 6] [Cited by in RCA: 10] [Article Influence: 2.0] [Reference Citation Analysis (0)] |
| 18. | Pichler M, Stiegelbauer V, Vychytilova-Faltejskova P, Ivan C, Ling H, Winter E, Zhang X, Goblirsch M, Wulf-Goldenberg A, Ohtsuka M, Haybaeck J, Svoboda M, Okugawa Y, Gerger A, Hoefler G, Goel A, Slaby O, Calin GA. Genome-Wide miRNA Analysis Identifies miR-188-3p as a Novel Prognostic Marker and Molecular Factor Involved in Colorectal Carcinogenesis. Clin Cancer Res. 2017;23:1323-1333. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 57] [Cited by in RCA: 63] [Article Influence: 7.9] [Reference Citation Analysis (0)] |
| 19. | Pei J, Zhang S, Yang X, Han C, Pan Y, Li J, Wang Z, Sun C, Zhang J. Long non-coding RNA RP11-283G6.5 confines breast cancer development through modulating miR-188-3p/TMED3/Wnt/β-catenin signalling. RNA Biol. 2021;18:287-302. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 3] [Cited by in RCA: 14] [Article Influence: 3.5] [Reference Citation Analysis (0)] |
| 20. | Shi W, Zhang C, Ning Z, Hua Y, Li Y, Chen L, Liu L, Chen Z, Meng Z. Long non-coding RNA LINC00346 promotes pancreatic cancer growth and gemcitabine resistance by sponging miR-188-3p to derepress BRD4 expression. J Exp Clin Cancer Res. 2019;38:60. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 45] [Cited by in RCA: 61] [Article Influence: 10.2] [Reference Citation Analysis (0)] |
| 21. | Meng F, Zhang S, Song R, Liu Y, Wang J, Liang Y, Han J, Song X, Lu Z, Yang G, Pan S, Li X, Zhou F, Wang Y, Cui Y, Zhang B, Ma K, Zhang C, Sun Y, Xin M, Liu L. NCAPG2 overexpression promotes hepatocellular carcinoma proliferation and metastasis through activating the STAT3 and NF-κB/miR-188-3p pathways. EBioMedicine. 2019;44:237-249. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 23] [Cited by in RCA: 49] [Article Influence: 8.2] [Reference Citation Analysis (0)] |
| 22. | Pei J, Zhang J, Yang X, Wu Z, Sun C, Wang Z, Wang B. TMED3 promotes cell proliferation and motility in breast cancer and is negatively modulated by miR-188-3p. Cancer Cell Int. 2019;19:75. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 35] [Cited by in RCA: 40] [Article Influence: 6.7] [Reference Citation Analysis (0)] |
| 23. | Sukocheva OA, Liu J, Neganova ME, Beeraka NM, Aleksandrova YR, Manogaran P, Grigorevskikh EM, Chubarev VN, Fan R. Perspectives of using microRNA-loaded nanocarriers for epigenetic reprogramming of drug resistant colorectal cancers. Semin Cancer Biol. 2022;86:358-375. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 31] [Cited by in RCA: 24] [Article Influence: 8.0] [Reference Citation Analysis (0)] |
| 24. | Warneke VS, Behrens HM, Hartmann JT, Held H, Becker T, Schwarz NT, Röcken C. Cohort study based on the seventh edition of the TNM classification for gastric cancer: proposal of a new staging system. J Clin Oncol. 2011;29:2364-2371. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 80] [Cited by in RCA: 83] [Article Influence: 5.9] [Reference Citation Analysis (0)] |
| 25. | Lin J, Zhu H, Hong L, Tang W, Wang J, Hu H, Wu X, Chen Y, Liu G, Yang Q, Li J, Wang Y, Lin Z, Xiao Y, Dai W, Huang M, Li G, Li A, Xiang L, Liu S. Coexpression of HOXA6 and PBX2 promotes metastasis in gastric cancer. Aging (Albany NY). 2021;13:6606-6624. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 7] [Cited by in RCA: 12] [Article Influence: 3.0] [Reference Citation Analysis (0)] |
| 26. | Chen JF, Wu P, Xia R, Yang J, Huo XY, Gu DY, Tang CJ, De W, Yang F. STAT3-induced lncRNA HAGLROS overexpression contributes to the malignant progression of gastric cancer cells via mTOR signal-mediated inhibition of autophagy. Mol Cancer. 2018;17:6. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 130] [Cited by in RCA: 191] [Article Influence: 27.3] [Reference Citation Analysis (0)] |
| 27. | Ahumada-Castro U, Silva-Pavez E, Lovy A, Pardo E, Molgό J, Cárdenas C. MTOR-independent autophagy induced by interrupted endoplasmic reticulum-mitochondrial Ca(2+) communication: a dead end in cancer cells. Autophagy. 2019;15:358-361. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 49] [Cited by in RCA: 48] [Article Influence: 8.0] [Reference Citation Analysis (0)] |
| 28. | Zhang X, Wang S, Wang H, Cao J, Huang X, Chen Z, Xu P, Sun G, Xu J, Lv J, Xu Z. Circular RNA circNRIP1 acts as a microRNA-149-5p sponge to promote gastric cancer progression via the AKT1/mTOR pathway. Mol Cancer. 2019;18:20. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 508] [Cited by in RCA: 605] [Article Influence: 100.8] [Reference Citation Analysis (0)] |
| 29. | Wu JG, Wang JJ, Jiang X, Lan JP, He XJ, Wang HJ, Ma YY, Xia YJ, Ru GQ, Ma J, Zhao ZS, Zhou R. MiR-125b promotes cell migration and invasion by targeting PPP1CA-Rb signal pathways in gastric cancer, resulting in a poor prognosis. Gastric Cancer. 2015;18:729-739. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 41] [Cited by in RCA: 51] [Article Influence: 5.1] [Reference Citation Analysis (0)] |
| 30. | Jia J, Zhan D, Li J, Li Z, Li H, Qian J. The contrary functions of lncRNA HOTAIR/miR-17-5p/PTEN axis and Shenqifuzheng injection on chemosensitivity of gastric cancer cells. J Cell Mol Med. 2019;23:656-669. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 30] [Cited by in RCA: 50] [Article Influence: 7.1] [Reference Citation Analysis (0)] |
| 31. | YiRen H, YingCong Y, Sunwu Y, Keqin L, Xiaochun T, Senrui C, Ende C, XiZhou L, Yanfan C. Long noncoding RNA MALAT1 regulates autophagy associated chemoresistance via miR-23b-3p sequestration in gastric cancer. Mol Cancer. 2017;16:174. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 189] [Cited by in RCA: 276] [Article Influence: 34.5] [Reference Citation Analysis (0)] |
| 32. | Li YJ, Lei YH, Yao N, Wang CR, Hu N, Ye WC, Zhang DM, Chen ZS. Autophagy and multidrug resistance in cancer. Chin J Cancer. 2017;36:52. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 303] [Cited by in RCA: 514] [Article Influence: 64.3] [Reference Citation Analysis (0)] |
| 33. | Amaravadi RK, Kimmelman AC, Debnath J. Targeting Autophagy in Cancer: Recent Advances and Future Directions. Cancer Discov. 2019;9:1167-1181. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 337] [Cited by in RCA: 662] [Article Influence: 110.3] [Reference Citation Analysis (0)] |
| 34. | Mathew R, Karantza-Wadsworth V, White E. Role of autophagy in cancer. Nat Rev Cancer. 2007;7:961-967. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 1483] [Cited by in RCA: 1452] [Article Influence: 80.7] [Reference Citation Analysis (0)] |
| 35. | Shimizu S. Autophagic Cell Death and Cancer Chemotherapeutics. In: K. Nakao, N. Minato and S. Uemoto. Innovative Medicine: Basic Research and Development. Tokyo, 2015. [DOI] [Full Text] |
| 36. | Russell RC, Guan KL. The multifaceted role of autophagy in cancer. EMBO J. 2022;41:e110031. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1] [Cited by in RCA: 112] [Article Influence: 37.3] [Reference Citation Analysis (0)] |
| 37. | Naramura M, Nadeau S, Mohapatra B, Ahmad G, Mukhopadhyay C, Sattler M, Raja SM, Natarajan A, Band V, Band H. Mutant Cbl proteins as oncogenic drivers in myeloproliferative disorders. Oncotarget. 2011;2:245-250. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 40] [Cited by in RCA: 37] [Article Influence: 2.6] [Reference Citation Analysis (0)] |
| 38. | Langdon WY, Hartley JW, Klinken SP, Ruscetti SK, Morse HC 3rd. v-cbl, an oncogene from a dual-recombinant murine retrovirus that induces early B-lineage lymphomas. Proc Natl Acad Sci USA. 1989;86:1168-1172. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 243] [Cited by in RCA: 245] [Article Influence: 6.8] [Reference Citation Analysis (0)] |
| 39. | Wang Y, Chen L, Wu Z, Wang M, Jin F, Wang N, Hu X, Liu Z, Zhang CY, Zen K, Chen J, Liang H, Zhang Y, Chen X. miR-124-3p functions as a tumor suppressor in breast cancer by targeting CBL. BMC Cancer. 2016;16:826. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 85] [Cited by in RCA: 82] [Article Influence: 9.1] [Reference Citation Analysis (0)] |
| 40. | Ding M, Sun X, Zhong J, Zhang C, Tian Y, Ge J, Zhang CY, Zen K, Wang JJ, Wang C. Decreased miR-200a-3p is a key regulator of renal carcinoma growth and migration by directly targeting CBL. J Cell Biochem. 2018;119:9974-9985. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 12] [Cited by in RCA: 20] [Article Influence: 2.9] [Reference Citation Analysis (0)] |
| 41. | Dong Q, Liu YP, Qu XJ, Hou KZ, Li LL. Expression of c-Cbl, Cbl-b, and epidermal growth factor receptor in gastric carcinoma and their clinical significance. Chin J Cancer. 2010;29:59-64. [PubMed] |
| 42. | Ito R, Nakayama H, Yoshida K, Matsumura S, Oda N, Yasui W. Expression of Cbl linking with the epidermal growth factor receptor system is associated with tumor progression and poor prognosis of human gastric carcinoma. Virchows Arch. 2004;444:324-331. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 20] [Cited by in RCA: 23] [Article Influence: 1.1] [Reference Citation Analysis (0)] |
| 43. | Qi HY, Qu XJ, Liu J, Hou KZ, Fan YB, Che XF, Liu YP. Bufalin induces protective autophagy by Cbl-b regulating mTOR and ERK signaling pathways in gastric cancer cells. Cell Biol Int. 2019;43:33-43. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 10] [Cited by in RCA: 24] [Article Influence: 3.4] [Reference Citation Analysis (0)] |
| 44. | Chen S, Wu J, Jiao K, Wu Q, Ma J, Chen D, Kang J, Zhao G, Shi Y, Fan D. MicroRNA-495-3p inhibits multidrug resistance by modulating autophagy through GRP78/mTOR axis in gastric cancer. Cell Death Dis. 2018;9:1070. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 57] [Cited by in RCA: 80] [Article Influence: 11.4] [Reference Citation Analysis (0)] |
| 45. | Hibdon ES, Razumilava N, Keeley TM, Wong G, Solanki S, Shah YM, Samuelson LC. Notch and mTOR Signaling Pathways Promote Human Gastric Cancer Cell Proliferation. Neoplasia. 2019;21:702-712. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 27] [Cited by in RCA: 62] [Article Influence: 10.3] [Reference Citation Analysis (0)] |
| 46. | Wang J, Feng W, Dong Y, Mao X, Guo F, Luo F. MicroRNA495 regulates human gastric cancer cell apoptosis and migration through Akt and mTOR signaling. Oncol Rep. 2018;40:3654-3662. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 3] [Cited by in RCA: 6] [Article Influence: 0.9] [Reference Citation Analysis (0)] |
| 47. | Xu Z, Han X, Ou D, Liu T, Li Z, Jiang G, Liu J, Zhang J. Targeting PI3K/AKT/mTOR-mediated autophagy for tumor therapy. Appl Microbiol Biotechnol. 2020;104:575-587. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 166] [Cited by in RCA: 417] [Article Influence: 69.5] [Reference Citation Analysis (0)] |
| 48. | Sun Y, Jiang Y, Huang J, Chen H, Liao Y, Yang Z. CISD2 enhances the chemosensitivity of gastric cancer through the enhancement of 5-FU-induced apoptosis and the inhibition of autophagy by AKT/mTOR pathway. Cancer Med. 2017;6:2331-2346. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 18] [Cited by in RCA: 20] [Article Influence: 2.5] [Reference Citation Analysis (0)] |
| 49. | Tao W, Li Y, Zhu M, Li C, Li P. LncRNA NORAD promotes effort and inhibits apoptosis of gastric cancer by regulating mir-214 / akt/mtor axis. Onco Targets Ther 2019; 12. [RCA] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 35] [Cited by in RCA: 40] [Article Influence: 6.7] [Reference Citation Analysis (0)] |
| 50. | Paquette M, El-Houjeiri L, Pause A. mTOR Pathways in Cancer and Autophagy. Cancers (Basel). 2018;10. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 175] [Cited by in RCA: 221] [Article Influence: 31.6] [Reference Citation Analysis (0)] |
| 51. | Kim YC, Guan KL. mTOR: a pharmacologic target for autophagy regulation. J Clin Invest. 2015;125:25-32. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1352] [Cited by in RCA: 1554] [Article Influence: 155.4] [Reference Citation Analysis (0)] |
| 52. | Lee HJ, Venkatarame Gowda Saralamma V, Kim SM, Ha SE, Raha S, Lee WS, Kim EH, Lee SJ, Heo JD, Kim GS. Pectolinarigenin Induced Cell Cycle Arrest, Autophagy, and Apoptosis in Gastric Cancer Cell via PI3K/AKT/mTOR Signaling Pathway. Nutrients. 2018;10. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 67] [Cited by in RCA: 97] [Article Influence: 13.9] [Reference Citation Analysis (0)] |
| 53. | Lu R, Yang Z, Xu G, Yu S. miR-338 modulates proliferation and autophagy by PI3K/AKT/mTOR signaling pathway in cervical cancer. Biomed Pharmacother. 2018;105:633-644. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 22] [Cited by in RCA: 36] [Article Influence: 5.1] [Reference Citation Analysis (0)] |
| 54. | Wang SS, Chen YH, Chen N, Wang LJ, Chen DX, Weng HL, Dooley S, Ding HG. Hydrogen sulfide promotes autophagy of hepatocellular carcinoma cells through the PI3K/Akt/mTOR signaling pathway. Cell Death Dis. 2017;8:e2688. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 130] [Cited by in RCA: 133] [Article Influence: 16.6] [Reference Citation Analysis (0)] |
| 55. | Fan XJ, Wang Y, Wang L, Zhu M. Salidroside induces apoptosis and autophagy in human colorectal cancer cells through inhibition of PI3K/Akt/mTOR pathway. Oncol Rep. 2016;36:3559-3567. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 66] [Cited by in RCA: 86] [Article Influence: 9.6] [Reference Citation Analysis (0)] |