Published online Jul 15, 2023. doi: 10.4251/wjgo.v15.i7.1182
Peer-review started: December 7, 2022
First decision: January 18, 2023
Revised: March 30, 2023
Accepted: April 30, 2023
Article in press: April 30, 2023
Published online: July 15, 2023
Processing time: 217 Days and 1.5 Hours
Gastric cancer (GC) is a malignant tumor with high morbidity and mortality. Expression of COL5A2 is significantly elevated in GC. Abnormal expression of noncoding RNAs (ncRNAs) have been found in GC, including microRNA (miRNA) and long noncoding RNA (lncRNA). Competing endogenous RNA network plays an important regulatory role in GC. However, its specific regulatory mechanism has not been elucidated.
To gain insight into the ncRNA regulatory mechanism and immune microenvironment related to COL5A2 in GC.
RNA sequencing data and clinical information from The Cancer Genome Atlas data portal were used to analyze the expressions of COL5A2, miRNA and lncRNA related to the prognosis of GC. Cox regression analysis and Kyoto Encyclopedia of Genes and Genomes analysis were performed to assess the risk factors and relevant function of COL5A2. StarBase was used to predict the interaction of miRNA–lncRNA or miRNA–mRNA in GC. The relationship between COL5A2, miR-144-3p and ENTPD1-AS1 were verified by dual luciferase reporter assay. The association of COL5A2 with immune cell infiltration were analyzed using the Tumor Immune Estimation Resource database and single sample gene set enrichment analysis. The expression of COL5A2 and macrophages in paired GC tissues were detected by immunohistochemical staining.
We verified that the upregulation of COL5A2 expression was associated with the prognosis of GC and was an independent risk factor for GC. miR-144-3p was downregulated and correlated with the prognosis of GC. miR-144-3p regulated the expression of COL5A2 through direct interaction with COL5A2. ENTPD1-AS1 was elevated in GC and competitively bound to miR-144-3p, thus inhibiting the expression of miR-144-3p. ENTPD1-AS1 enhanced the expression of COL5A2 through sponging miR-144-3p. Compared to paired normal tissue, COL5A2 expression was upregulated at the protein level, especially in the middle and late stages of GC. The high expression of COL5A2 was positively linked to macrophage infiltration in GC.
COL5A2 regulated by ENTPD1-AS1–miR-144-3p was associated with poor prognosis and macrophage infiltration in GC. This could be a new biomarker and therapeutic target in GC.
Core Tip: Gastric cancer (GC) is a malignant tumor with high fatality rate. Competing endogenous RNA network and infiltration of immune cells play an important role in the development of GC. In this study, we verified that high expression of COL5A2 was closely related to poor prognosis and was an independent risk factor for GC. We predicted and validated that long noncoding RNA ENTPD1-AS1 regulated the expression of COL5A2 through sponging miR-144-3p. Additionally, we confirmed that upregulation of COL5A2 expression strongly correlated with immune infiltration of macrophages. ENTPD1-AS1-miR-144-3p-COL5A2 might be a new therapeutic target for GC.
- Citation: Yuan HM, Pu XF, Wu H, Wu C. ENTPD1-AS1–miR-144-3p-mediated high expression of COL5A2 correlates with poor prognosis and macrophage infiltration in gastric cancer. World J Gastrointest Oncol 2023; 15(7): 1182-1199
- URL: https://www.wjgnet.com/1948-5204/full/v15/i7/1182.htm
- DOI: https://dx.doi.org/10.4251/wjgo.v15.i7.1182
Gastric cancer (GC) remains the fourth leading cause of cancer death worldwide. About 95% of cases are gastric adenocarcinoma (GAD) subtype[1]. The survival rate of GC has improved with medical advances. However, the median survival time for advanced GC is < 12 mo[2]. Accordingly, exploring the molecular mechanism of GC is of great significance for finding better therapeutic targets and better treatment.
The extracellular matrix is mainly composed of collagen and is related to proliferation, differentiation, migration, and metabolism of cancer[3]. Recent studies have shown that COL5A2, one of the collagen genes, is upregulated in several types of cancer and associated with immune cells infiltration[4,5]. Traditionally, COL5A2 is considered tightly related to the occurrence of classical Ehlers–Danlos syndrome[6,7]. However, high expression of COL5A2 is found to be associated with worse prognosis and drug resistance[8-10]. COL5A is also found closely associated with immune cell infiltration, which may be related to the inhibitory effect of collagen on the production of CCL2[5]. In proliferative diabetic retinopathy, COL5A2 is closely related to the infiltration of M2 macrophages[11].
Noncoding RNAs (ncRNAs), such as microRNAs (miRNAs), long noncoding RNAs (lncRNAs) and circular RNAs (circRNAs), are critically involved in GC development[12]. ncRNAs could affect the proliferation, invasion, migration and metastasis of GC by regulating target miRNA genes[13]. Studies have revealed that miR-144-3p can affect the occurrence, development, and prognosis of cancer by inhibiting the expression of target genes[14,15]. ncRNAs can upregulate the expression of cancer-promoting genes by inhibiting the expression of miR-144-3p[16,17]. ENTPD1-AS1 is an antisense lncRNA that may be associated with short stature[18]. Recently, ENTPD1-AS1 is considered to be a new ncRNA that regulates the proliferation and apoptosis of cancer cells and also serves as a prognostic marker in glioblastoma multiforme[19,20]. However, the ncRNA regulatory mechanism and immune infiltration of COL5A2 are still unclear in GC.
In this study, we verified that high expression of COL5A2 in GC was closely related to poor prognosis and was an independent risk factor. We predicted and verified a new competing endogenous RNA (ceRNA) network, namely, ENTPD1-AS1 regulated COL5A2 expression through sponging miR-144-3p. We confirmed that upregulation of COL5A2 expression strongly correlated with immune infiltration of macrophages. ENTPD1-AS1–miR-144-3p regulation of COL5A2 correlated with poor prognosis and macrophage infiltration in GC.
We collected 40 paired GC and normal specimens from the Eighth Affiliated Hospital of Sun Yat-Sen University. These tissues were obtained from the patients undergoing GC surgery. All the patients signed informed consent forms. This research was approved by the Ethics Committee of the Eighth Affiliated Hospital of the Sun Yat-Sen University.
Human Embryonic Kidney cells (293T) and human GAD cell line (AGS) were cultured in dulbecco's modified eagle medium (GIBCO, Invitrogen, Carlsbad, CA, United States) containing 10% FBS (GIBCO) and 100 U/mL penicillin, and 0.1 mg/mL streptomycin (Invitrogen, Carlsbad, CA, United States).
The Gene Expression Profiling Interactive Analysis (GEPIA; http://gepia.cancer-pku.cn/) database was utilized for the expression and prognostic analysis of COL5A2 in numerous cancers.
The mRNA expression data of GAD in tumor and normal tissues (446 cases of tumor, and 45 cases of normal tissue) and clinical material were obtained from The Cancer Genome Atlas (TCGA) public database (https://portal.gdc.cancer.gov). The basic information of human miRNA was downloaded from http://www.mirbase.org. We conducted a differential analysis and survival analysis on the expression of COL5A2 in normal tissues and GC tissues. Data transformation was achieved using Perl script. R version 4.1.2 was used in several analyses.
We organized the clinical data of GAD and matched the expression of COL5A2 in the corresponding samples. Samples with incomplete or missing data were excluded from this analysis, and finally 322 specimens were obtained. Univariate and multivariate Cox proportional hazards regression models were used to assess COL5A2 for prediction of overall survival (OS).
We analyzed Kyoto Encyclopedia of Genes and Genomes (KEGG) signaling pathway enrichment of COL5A2 by gene set enrichment analysis (GSEA). We found gene markers corresponding to 22 immune cells and extracted the expression of these gene markers from the TCGA data. Using single sample GSEA (ssGSEA), we determined the infiltration of these immune cells in COL5A2 high expression and low expression groups.
We used starBase (http://starbase.sysu.edu.cn/) to predict the interaction of miRNA–mRNA or miRNA–lncRNA. StarBase is a powerful target gene prediction software, and it includes 7 functional domains for prediction. We set the screening conditions for at least two sites that considered the gene to be the target gene of COL5A2, and the gene was retained. The expression data of miR-144-3p and ENTPD1-AS1 came from the TCGA database. We conducted the expression analysis, correlation analysis, and survival analysis using R version 4.1.2.
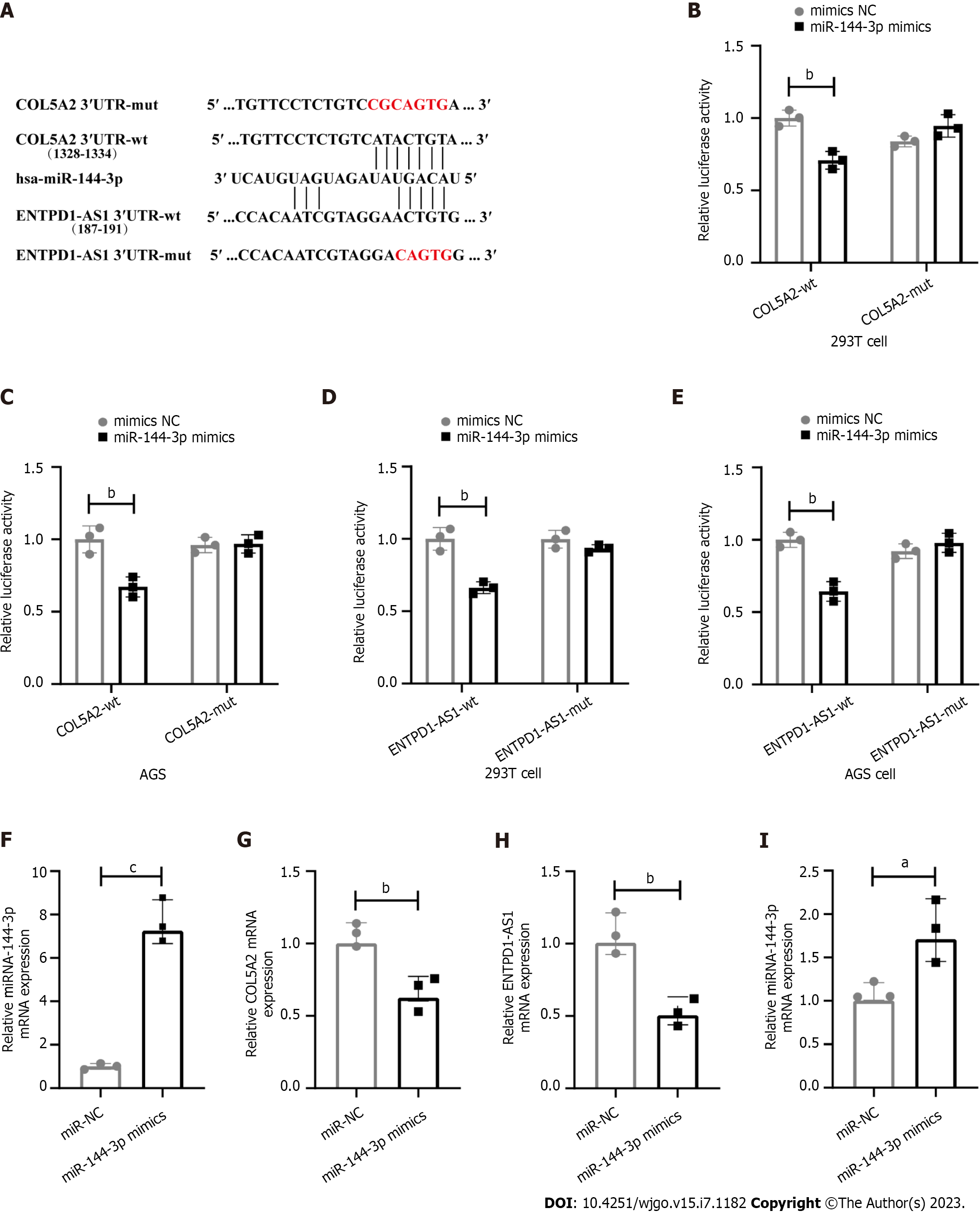
The sequences of COL5A2 or ENTPD1-AS1 that may bind to miR-144-3p were cloned into the pmirGLO vector (Sangon Biotech Co., Ltd. Shanghai, China). We constructed four kinds of double luciferase report plasmids: COL5A2 3′UTR-wt, COL5A2 3′UTR-mut, ENTPD1-AS1-wt and ENTPD1-AS1-mut.
siRNAs, miRNA mimics and their corresponding negative controls (NC) were designed by GenePharma Co., Ltd., (Shanghai, China). The sequence was as follows: MiR-144-3p mimics: (UACAGUAUAGAUGAUGUACU), mimics NC: (UUGUACUACACAAAAGUACUG), si-ENTPD1-AS1: (GGCCCGUAAUGGAGAUCGATT, UCGAUCUCCAUUACGGGCCTT), si-NC: (UUCUCCGAACGUGUCACGUTT, ACGUGACACGUUCGGAGAATT).
We seeded cells into 24-well plates at a density of 1.5 × 105 cells per well. When the cells reached 60% to 70%, dual luciferase reporter vector and miR-144-3p mimics or NC mimics were transfection into 293T cells and AGS cells in the presence of Lipofectamine™ and P3000™ (L3000001, Invitrogen, United States). In some experiments, mixture of Lipofectamine™ and miR-144-3p or si-ENTPD1-AS1 was transfection into AGS cells. After 6 h, fresh medium was replaced. 48 h later, Fluorescence intensity was detected by Dual-Luciferase® Reporter Assay System (E1910, Promega, United States).
RNAs were extracted by Trizol from AGS cells. RNA was reversely transcribed into cDNA with Evo M-MLV RT Premix (AG11701, Accurate Biotechnology, Hunan, China) and then detected gene expression with SYBR® Green Premix Pro Taq HS quantitative polymerase chain reaction (qPCR) Kit for qPCR (AG1170, Accurate Biotechnology, Hunan, China). The primers were synthesized by Sangon Biotech Co., Ltd (Shanghai, China). COL5A2: (Forward: GGATCACAGGGACCAAGAGGAGAG, Reverse: GCACCAGGTTGACCAGGAACAC), ENTP1-AS1 (Forward: CCTGCCTCTGCCTCCAAGTAG, Reverse: TTCGAGACCAGCCTGACCAAC), hsa-miR-144-3p (RT Primer: GTCGTATCCAGTGCAGGGTCCGAGGTATTCGCACTGGATACGACAGTACA, Forward: GCGCGCGTACAGTATAGATGA, Reverse: ATCCAGTGCAGGGTCCGAGG). U6 (Forward: GGAACGATACAGAGAAGATTAGC, Reverse: TGGAACGCTTCACGAATTTGCG), GAPDH (Forward: TGTGTCCGTCGTGGATCTGA, Reverse: GCAGCTGTGACACACAGTA). miRNA was detected by stem-loop. U6 or GAPDH as internal control. The relative expression of genes was calculated by 2−ΔΔCt.
Tumor IMmune Estimation Resource (TIMER) database (https://cistrome.shinyapps.io/timer/) integrated the results of multiple algorithms based on the sequencing results of TCGA. We used the TIMER and GEPIA database to analyze the correlation between COL5A2 and immune cells or immune checkpoints.
All tissue slides were dewaxed, hydrated, and antigen was retrieved. We inactivated endogenous enzymatic activity and blocked nonspecific sites. Primary and secondary antibodies were incubated and rendered with Diaminobenzidine (DAB). The concentration of primary antibodies was anti-COL5A2 (1:100, Thermo Fisher, PA5-38880, United States), anti-CD68 (1:2000, Abcam, ab955, United Kingdom). We selected the corresponding secondary antibody according to the primary antibody (1:100, Santa Cruz Biotechnology, sc-2357/sc-516102); DAB detection kit (50:1, Servicebio, G1212, China). All slides were observed and counted by a Carl Zeiss microscope (Axio Imager A2, Germany). The CD68+ cell counts were conducted by taking the average value of three high power fields (HP). Based on the number of CD68+ cells, the macrophage infiltration was categorized into weak (30–60/HP), moderate (60–90/HP), and strong (> 90/HP) subgroups.
R with the survminer, survival, Ggforest and limma packages was used for analysis. GraphPad 8 with unpaired t-test and Spearman correlations was used for some analyses. aP < 0.05 was considered statistically significant.
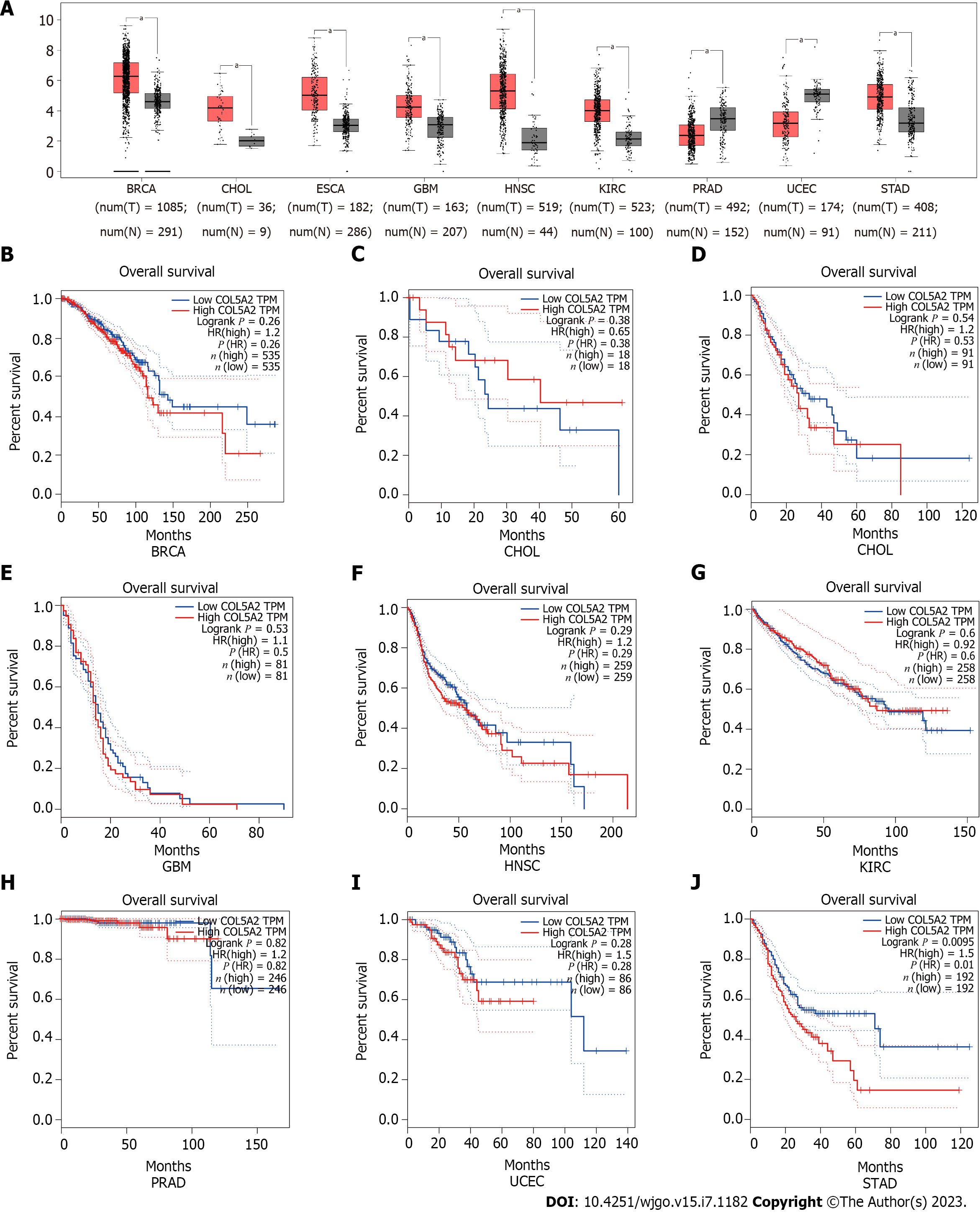
To identify the differential expression of COL5A2 in pan-cancer, the expression of COL5A2 was assessed in 18 types of cancer by GEPIA. The expression of COL5A2 was significantly increased in nine cancers compared with the normal group (Figure 1A, P < 0.05). There was no significant difference in the other nine cancers (Supplementary Figure 1). Among the cancers with significant differences in COL5A2 expression, OS analysis was conducted using GEPIA database. No significant prognostic difference in 8 out of 9 cancers was observed (Figure 1B–I). Only in GC, high expression of COL5A2 was associated with poorer prognosis (Figure 1J, P = 0.01).
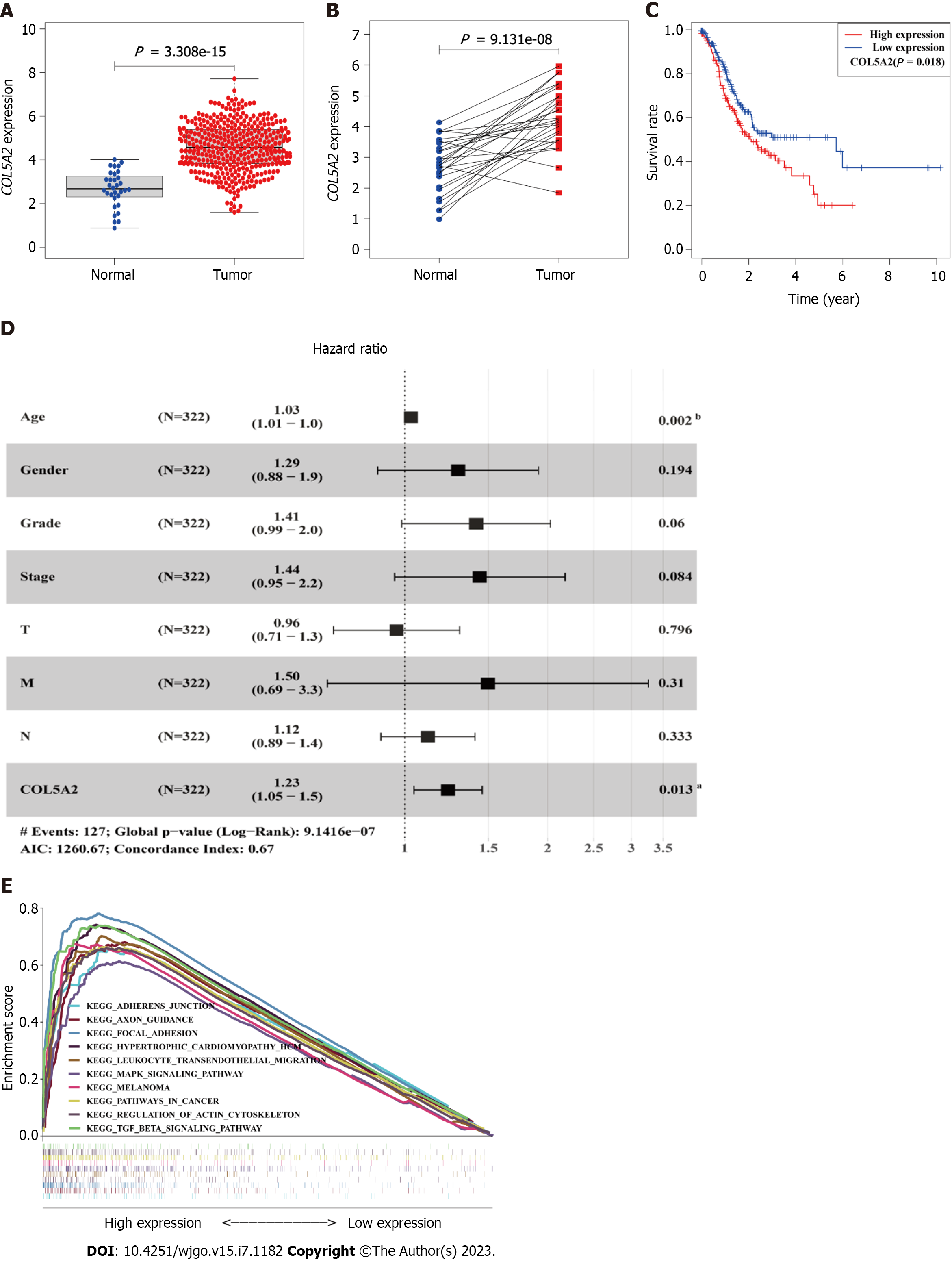
To verify the results obtained from the GEPIA database, we analyzed the expression and survival prognosis of COL5A2 using RNA-sequencing data from TCGA. In line with previous studies[21], significantly high expression of COL5A2 was found in TCGA data and 27 paired specimens (Figure 2A and B). We performed survival analysis and found a significant difference in OS between high and low COL5A2 expression groups (Figure 2C, P =0.018).
Excluding patients with missing or incomplete information, 322 of the 433 patients were included in the cox regression analysis. From clinical information, it suggested that patients with GC were mainly in the middle and late stages, with a high rate of lymph node metastasis, less distant metastasis, and a high mortality rate (Supplementary Table 1). Cancer stage, especially N stage, was the most important prognostic indicator. Age, T stage, and COL5A2 expression were risk factors for GC (Table 1). To determine whether these risk factors were independent of other factors, multivariate Cox regression was conducted. Only age [1.03 (1.01-1.1), P = 0.002] and expression of COL5A2 [1.23 (1.05-1.5), P = 0.013] were independent prognostic factors (Figure 2D).
| Parameter | Univariate analysis | P value | Multivariate analysis | P value | ||
| HR | 95%CI | HR | 95%CI | |||
| Age | 1.022 | 1.004-1.039 | 0.014a | 1.030 | 1.011-1.049 | 0.002b |
| Gender | 1.372 | 0.941-2.001 | 0.100 | 1.289 | 0.873-1.903 | 0.202 |
| Grade | 1.364 | 0.963-1.930 | 0.080 | 1.397 | 0.974-2.005 | 0.069 |
| Stae | 1.576 | 1.268-1.959 | < 0.001c | 1.446 | 0.956-2.187 | 0.081 |
| T | 1.276 | 1.022-1.593 | 0.032a | 0.954 | 0.702-1.295 | 0.761 |
| M | 1.809 | 0.973-3.364 | 0.061 | 1.475 | 0.677-3.214 | 0.328 |
| N | 1.343 | 1.147-1.573 | < 0.001c | 1.119 | 0.891-1.405 | 0.334 |
| COL5A2 | 1.278 | 1.096-1.489 | 0.002b | 2.321 | 1.215-4.434 | 0.011a |
Dividing GCs into COL5A2 high- and low-expression groups, we used KEGG enrichment to analyze which signaling pathways were associated with differential genes between the two groups. According to normalized enrichment score, nominal P value, the 10 most enriched signaling pathways in KEGG were shown in Supplementary Table 2). To be more concise, we integrated these signaling pathways into one diagram (Figure 2E). The most significantly enriched signaling pathways in the COL5A2 high expression group were the interaction between extracellular matrix and receptors, focal adhesion, and some classic cancer-related signaling pathways such as the transforming growth factor-β signaling pathway.
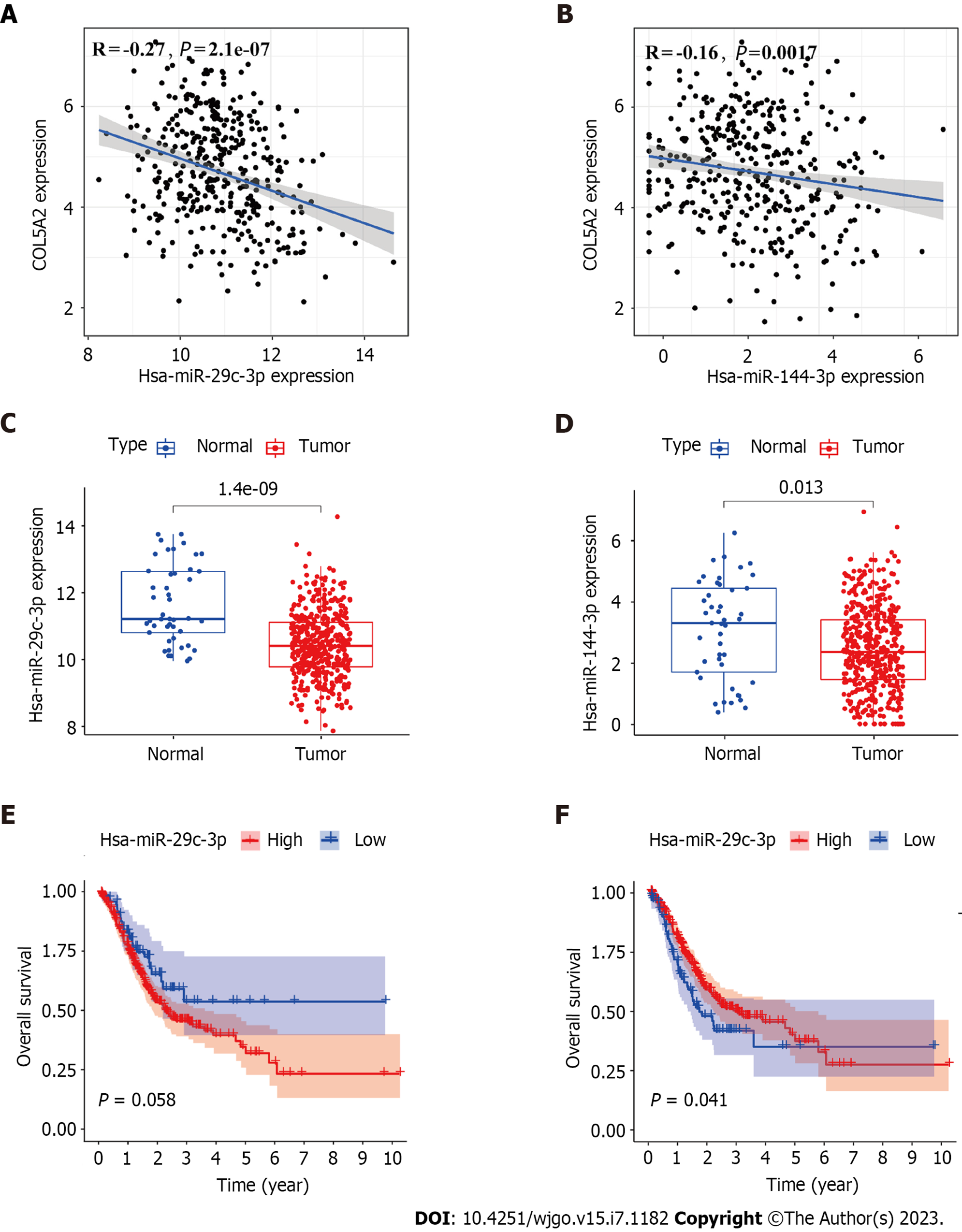
To investigate whether COL5A2 was regulated by miRNAs in GC, we used starBase to predict which miRNAs would bind to COL5A2. There were 69 miRNAs with the potential to bind to COL5A2, and it was visualized by cytoscape (Supplementary Figure 2). According to the principle of gene regulation, miRNA should negatively correlate with COL5A2. Thus, we set the screening condition that miRNA was negatively correlated with COL5A2 and P < 0.001. For COL5A2 was upregulated in GC, so the target miRNA was downregulaed in GC compared with normal tissues. miR-29c-3p and miR-144-3p were negatively correlated with COL5A2 [Figure 3A (r = -0.27, P = 2.1e-07) and Figure 3B (r = -0.16, P = 0.0017)] and markedly downregulated in GC [Figure 3C (P = 1.4e-09) and Figure 3D (P = 0.013)]. We analyzed the prognostic impact of miR-29c-3p and miR-144-3p. Only low expression of miR-144-3p had a significant effect on survival [Figure 3E (P = 0.058) and Figure 3F (P = 0.041)]. All these findings meant that miR-144-3p was the most likely upstream miRNA to regulate COL5A2 in GC.
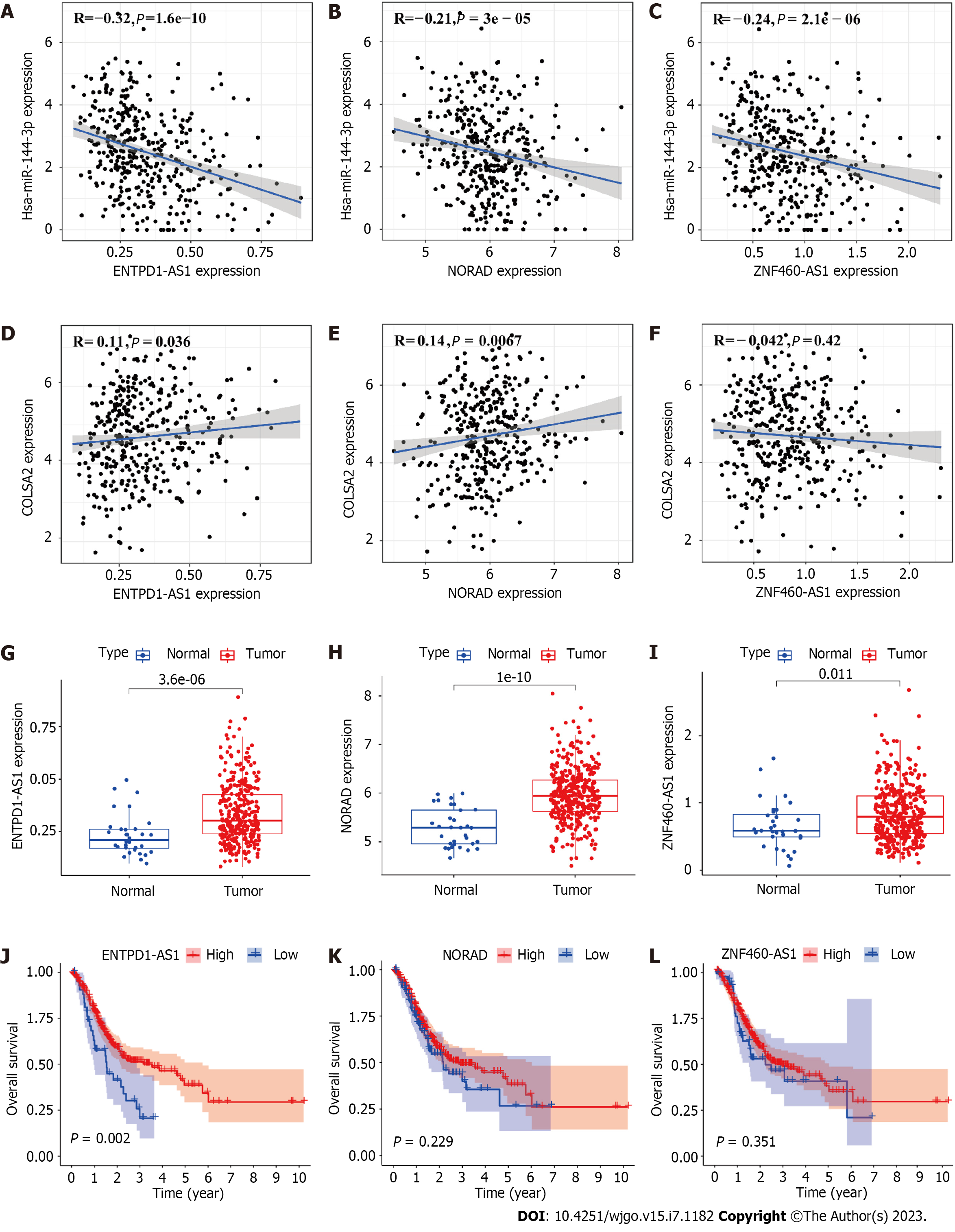
lncRNAs can competitively bind to miRNAs, which leads to upregulation of oncogenes. We predicted using starBase which lncRNAs would interact with miR-144-3p. We examined the correlation between lncRNAs, COL5A2 and miR-144-3p (Table 2). For downregulated miR-144-3p, expression of lncRNAs should be upregulated in GC. Among all lncRNAs, ENTPD1-AS1, NORAD, and ZNF460-AS1 were negatively correlated with miR-144-3p [Figure 4A (r = -0.32, P = 1.6e-10), Figure 4B (r = -0.21, P = 3e-05) and Figure 4C (r = -0.24, P = 2.1e-6)]. ENTPD1-AS1 and NORAD were positively correlated with COL5A2. ZNF460-AS1 had a negative correlation with COL5A2 [Figure 4D (r = 0.11, P = 0.036), Figure 4E (r = 0.14, P = 0.0067) and Figure 4F (r = -0.042, P = 0.42)]. We analyzed the expression of lncRNA in GC, and three lncRNAs were significantly upregulated in GC [Figure 4G (P = 3.6e-06), Figure 4H (P = 1e-10) and Figure 4I (P = 0.011)]. Kaplan–Meier analysis revealed that the higher the ENTPD1-AS1 expression, the better the OS [Figure 4J (P = 0.002), Figure 4K (P = 0.229) and Figure 4L (P = 0.351)]. By taking into account expression and prognostic analysis, our data suggested that ENTPD1-AS1 was the most likely lncRNA to regulate the miR-144-3p/COL5A2 axis in GC.
In order to verify the interaction of ENTPD1-AS1, miR-144-3p and COL5A2, we constructed dual luciferase reporter vector (Figure 5A). The results showed that in the presence of miR-144-3p mimics, the luciferase activity of COL5A2 3′UTR-wt group was significantly reduced compared to COL5A2 3′UTR-mut group in both 293T cells and AGS cells. However, when transfected with NC mimics, there was no difference in fluorescence intensity between COL5A2 3′UTR-wt group and COL5A2 3′UTR-mut group (Figure 5B and C). Similarly, the luciferase activity was reduced when miR-144-3p mimics interacted with ENTPD1-AS1-wt instead of ENTPD1-AS1-mut. When transfected with NC mimics, the change of fluorescence intensity disappeared (Figure 5D and E). Additionally, when miR-144-3p enrichment was present, the expression of COL5A2 was decreased in AGS cells (Figure 5F and G). Compared to si-NC, si-ENTPD1-AS1 led to a decrease of miR-144-3p (Figure 5H and I). All these results suggested that ENTPD1-AS1 might promote the expression of COL5A2 by suppressing the expression of miR-144-3p.
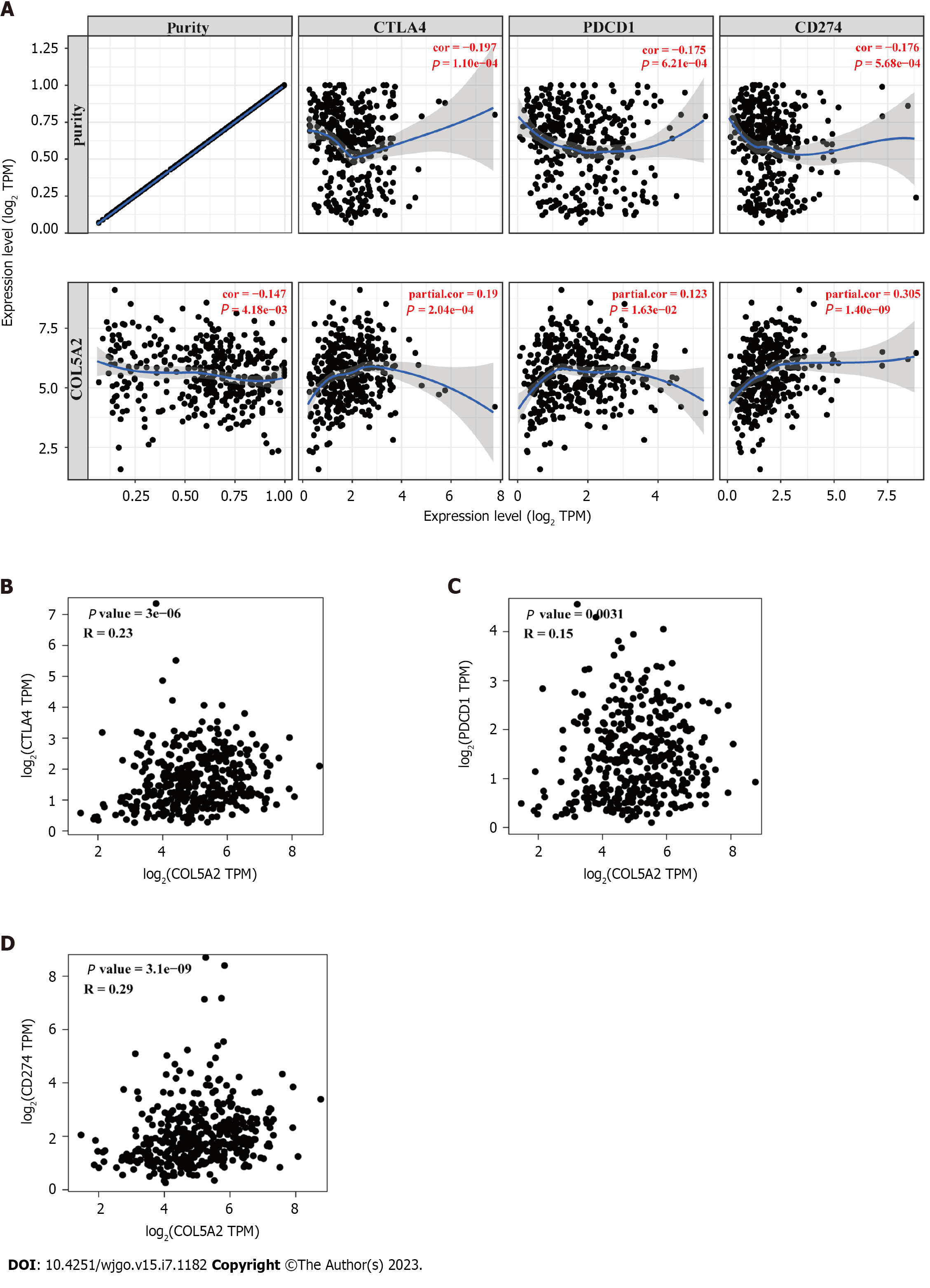
For immunotherapy having a low response in many cancers, there is a pressing need to find new target genes that could improve the efficacy of immunotherapy. Considering the cancer-promoting role of COL5A2, we analyzed the association of COL5A2 with checkpoints using the TIMER database. Our analyses revealed a high correlation between COL5A2 and CD274 and a lower correlation between COL5A2 and CTLA4 or PDCD1 (Figure 6A). We observed similar results in the GEPIA database (Figure 6B–D). These results suggested that COL5A2 mediated immune escape in GC.
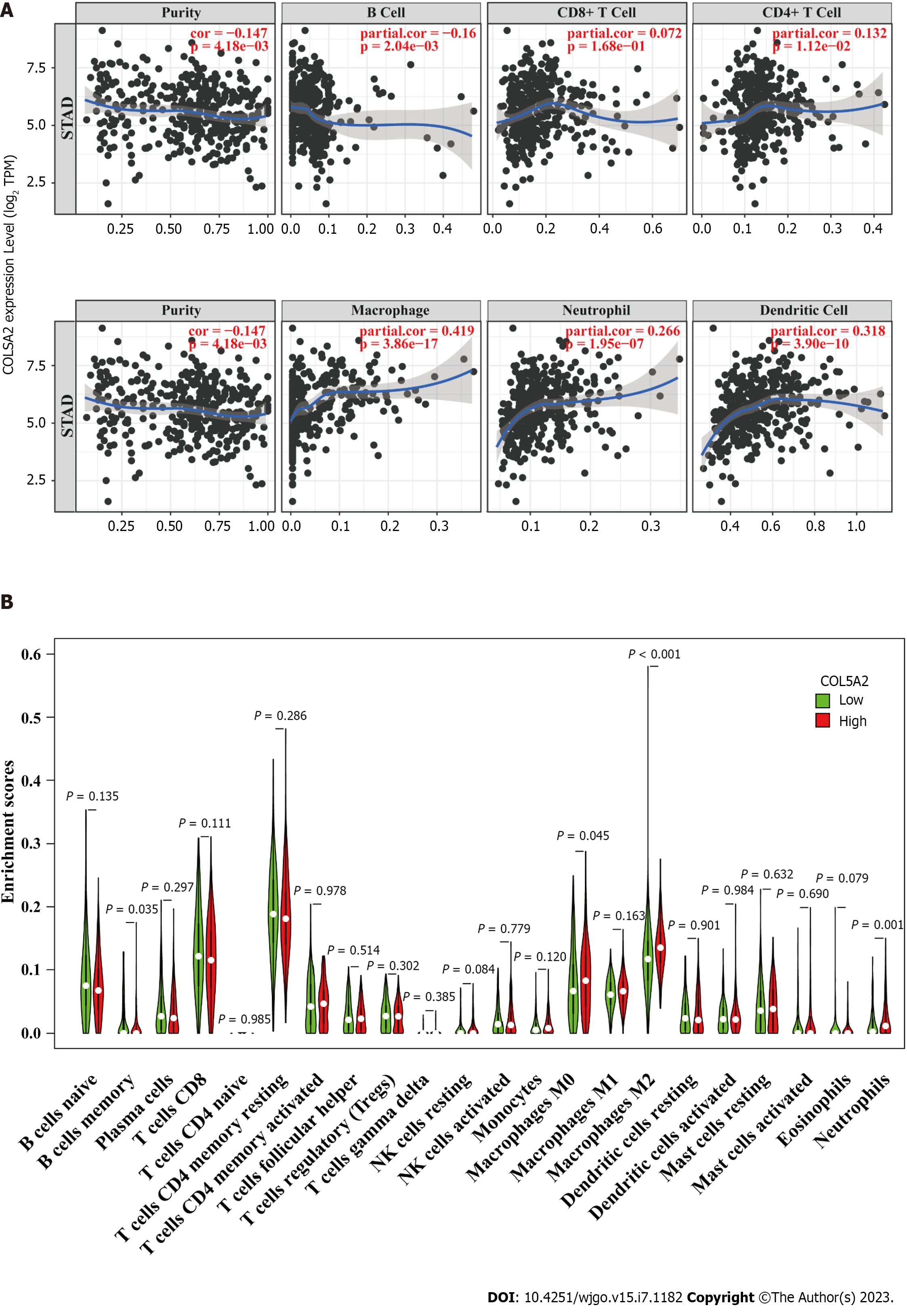
To explore the link between COL5A2 and immune cells, we determined the relationship between COL5A2 and immune cell biomarkers. We found a significant and positive association with COL5A2 and M2 macrophage biomarkers (CD163, VSIG4 and MS4A4A). The other immune cells biomarkers had a lower correlation or only some of the markers had a correlation with COL5A2 (Table 3). The TIMER database indicated that COL5A2 was significantly associated with infiltration of various immune cells, including neutrophils, macrophages and dendritic cells, but most correlated with macrophage infiltration (Figure 7A). We used ssGSEA to analyze the enrichment of 22 types of immune cells in groups with high and low COL5A2 expression. We discovered a significant difference in immune infiltration of macrophages, neutrophils and B memory cells between COL5A2 high and low expression groups. In line with our previous results, macrophage infiltration was most correlated with COL5A2 (Figure 7B, P < 0.001). These results indicated that COL5A2 was significantly positively linked to macrophage infiltration.
| Immune cell | Biomarker | R value | P value |
| B cell | CD19 | -0.053 | 3.1 × 10-1 |
| CD79A | 0.008 | 8.8 × 10-1 | |
| CD8+ T cell | CD8A | 0.078 | 1.3 × 10-1 |
| CD8B | -0.066 | 2.0 × 10-1 | |
| CD4+ T cell | CD4 | 0.312 | 8.4 × 10-10,c |
| M1 macrophage | NOS2 | 0.146 | 4.6 × 10-3,b |
| IRF5 | 0.154 | 2.7 × 10-3,b | |
| PTGS2 | 0.373 | 1.1 × 10-13,c | |
| M2 macrophage | CD163 | 0.466 | < 2.2 × 10-16,c |
| VSIG4 | 0.454 | < 2.2 × 10-16,c | |
| MS4A4A | 0.400 | < 2.2 × 10-16,c | |
| Neutrophil | CEACAM8 | 0.018 | 7.3 × 10-1 |
| ITGAM | 0.443 | < 2.2 × 10-16,c | |
| CCR7 | 0.072 | 1.7 × 10-1 | |
| Dendritic cell | HLA-DPB1 | 0.100 | 5.4 × 10-2 |
| HLA-DQB1 | 0.103 | 4.7 × 10-2,a | |
| HLA-DRA | 0.118 | 2.3 × 10-2 | |
| HLA-DPA1 | 0.096 | 6.2 × 10-2 | |
| CD1C | 0.064 | 2.2 × 10-1 | |
| NRP1 | 0.568 | < 2.2 × 10-16,c | |
| ITGAX | 0.470 | < 2.2 × 10-16,c |
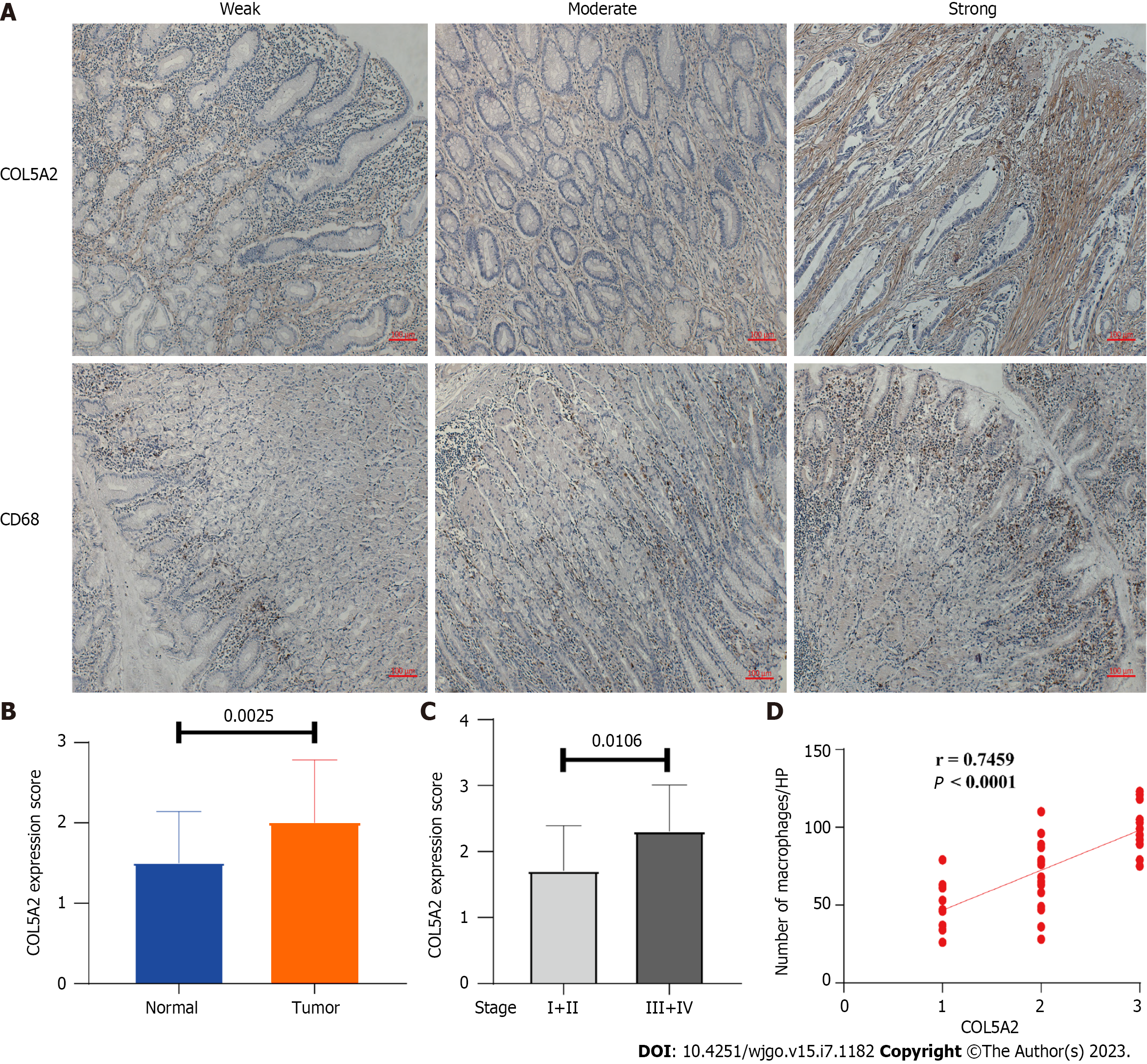
To verify the correlation between COL5A2 and macrophage infiltration in GC, we detected expression of COL5A2 and macrophages (CD68+) by immunohistochemistry in GC and paired normal tissues. According to the stain density, we divided the expression of COL5A2 into three levels, including weak, moderate, and strong staining, with corresponding scores of 1–3. According to the number of CD68+ cells, we divided macrophage infiltration into three groups: Weak (30–60/HP), moderate (60–90/HP), and strong (> 90/HP) (Figure 8A). The expression of COL5A2 at the protein level was clearly higher than in the paired peritumor tissues (Figure 8B, P = 0.025). Expression of COL5A2 between low stage GC (I and II) and high stage GC (III and IV) was assessed to determine whether COL5A2 was related to GC stage. Compared with low stage GC, the level of COL5A2 was significantly higher in high stage GC (Figure 8C, P = 0.0106). Pearson correlation analysis was performed to determine the relation between COL5A2 expression and macrophage infiltration in GC. Clearly, macrophage infiltration was consistent with COL5A2 staining intensity (Figure 8D, r = 0.7459, P < 0.0001). These results demonstrated that high expression of COL5A2 was observed in GC and COL5A2 was strongly positively correlated with macrophage infiltration.
GC is one of the tumors with high morbidity and mortality. Collagen is upregulated in advanced GC, and some collagen genes can be used as cancer biomarkers and can distinguish precancerous from cancerous lesions[22,23]. Several studies have shown that COL5A2 is upregulated in various cancers and can be used as a prognostic marker[24,25]. Here, we explored the upstream regulatory molecular mechanisms and immune function of COL5A2 in GC. We confirmed that COL5A2 was enriched and can predict poor prognosis in GC. We verified that COL5A2 was closely related to macrophage infiltration in GC. A new ceRNA network, ENTPD1-AS1–miR-144-3p–COL5A2 was identified, which may partially explain the upstream regulatory mechanism of COL5A2 in GC.
We confirmed that COL5A2 was increased in a number of tumors, but it was associated with worse survival only in GC. Cox regression analysis found that COL5A2 could be an independent prognostic factor for GC. KEGG enrichment analysis found that high expression of COL5A2 was associated with multiple signal transduction pathways. All these results revealed that COL5A2 was strongly associated with worse prognosis and has a crucial role in the development of GC. ceRNA plays a critical role in the regulation of gene expression in GC[26,27]. The mechanism of ceRNA mainly refers to that lncRNAs inhibit the negative regulation of miRNAs. For example, upregulation of circRNA KIF4A expression in GC can regulate the miRNA-144-3p–EZH2 axis to facilitate cell migration and invasion[28]. To investigate whether COL5A2 was regulated by the ceRNA network, we predicted by starBase which miRNAs could bind to COL5A2. We discovered that miR-29c-3p and miR-144-3p were most likely to regulate COL5A2 in GC. Although miR-29c reduced in GC and associated with tumor aggressiveness had been reported, our findings suggest that miR-29c-3p plays a role in GC by regulating COL5A2. Some studies have demonstrated that miR-144-3p inhibited cancer proliferation and migration, potentially serving as a biomarker in GC[16,29], which was consistent with our results that miR-144-3p was downregulated and closely related to prognosis in GC. Considering correlation analysis and expression analysis, ENTPD1-AS1, NORAD, and ZNF460–AS1 might regulate COL5A2 through a ceRNA network in GC. Three lncRNAs were negatively correlated with miR-144-3p, but only ENTPD1-AS1 and NORAD were positively correlated with COL5A2. Therefore, we focused on ENTPD1-AS1 and NORAD. Survival analysis suggested that ENTPD1-AS1 was significantly associated with GC survival. The most potentially upregulated lncRNAs was ENTPD1-AS1. Many lncRNAs have been identified as ceRNAs in GC, such as HOTAIR, MALAT1, NORAD and H19[30]. Some studies reported that lncRNA NORAD promoted GC cell growth by inhibiting expression of miR-608 or miR-433-3p[31,32]. Here, we revealed that NORAD promoted the development of GC by inhibiting expression of miR-144-3p. ENTPD1-AS1 is an antisense transcription lncRNA, which can be used as a prognostic marker in glioblastoma multiforme[19]. We confirmed that the most probable ceRNA regulatory network for COL5A2 in GC was NTPD1-AS1–miR-144-3p–COL5A2 through dual luciferase assay.
Recent studies have shown that fibrillar collagen can facilitate immune cell infiltration with bioinformatics analysis[33]. Genes associated with M2 infiltration of GC have been described in some previous studies, including COL1A1, COL4A1, COL12A1 and PDGFRB[34,35]. Wei et al[36] identified that stromal-relevant gene clusters could be used as prognostic genes and were associated with macrophage infiltration in GC. However, most of the results were obtained through database analysis and lacked experimental validation. Similar to previous studies, we found that COL5A2 was markedly positively associated with macrophage infiltration, using the TIMER database and ssGSEA. We confirmed by immunohistochemical staining that COL5A2 was significantly highly expressed, especially in the high stage of GC and was significantly positively correlated with macrophage infiltration at the protein level by IHC staining. These results give us a more complete view that macrophage infiltration may partially explain the carcinogenesis mediated by COL5A2 in GC.
There are some limitations to this study. First, although we proved the direct interaction of miR-144-3p and COL5A2 or miR-144-3p and ENTPD1-AS1 by Dual-Luciferase Reporter Assay and siRNA transfection, further experimental verification and confirmation were needed, such as RNA Binding Protein Immunoprecipitation (RIP) and in vivo experiments. Second, COL5A2 was found to be associated with poor prognosis of GC and macrophage infiltration by in vitro experiments, the sample size was small, these results thus should be verified in COL5A2 knockout mice, and tested in big samples.
We verify that COL5A2 is an independent risk factor and can be as a biomarker for GC. Our results demonstrate that COL5A2 exerts a tumor-promoting effect by promoting immune cell infiltration, especially macrophage infiltration. We have identified a novel ceRNA network that facilitates COL5A2 expression in GC, namely, lncRNA ENTPD1-AS1 upregulates the expression of COL5A2 by inhibiting the expression of miR-144-3p. These results partly explained the upstream regulatory mechanism and immune mechanism of COL5A2 in GC. COL5A2–miR-144-3p–ENTPD1-AS1 has the potential to be a novel therapeutic target for GC.
Gastric cancer (GC) is a malignant tumor with high morbidity and mortality. Expression of COL5A2 is significantly elevated in GC. However, its specific regulatory mechanism has not been elucidated.
Abnormal expression of noncoding RNAs (ncRNAs) has been found in GC, including microRNA (miRNA) and long noncoding RNA (lncRNA). The ncRNA regulatory mechanism and immune microenvironment related to COL5A2 in GC are not well understand.
To explore the competing endogenous RNA regulatory mechanism and immune mechanism of COL5A2 in GC.
StarBase was used to predict the interaction of miRNA–lncRNA or miRNA–mRNA in GC. The direct interaction between COL5A2, miR-144-3p and ENTPD1-AS1 were verified by dual luciferase reporter assay. The correlation between of COL5A2 and macrophages infiltration was analyzed through bioinformatics and validated in paired GC tissues by immunohistochemical staining.
miR-144-3p interacted directly with COL5A2 and negatively regulated the expression of COL5A2. ENTPD1-AS1 was elevated in GC and competitively bound to miR-144-3p, thus inhibiting the expression of miR-144-3p. Compared to paired normal tissue, COL5A2 expression was upregulated at the protein level, especially in the middle and late stages of GC. The high expression of COL5A2 was positively linked to macrophage infiltration in GC.
COL5A2 regulated by ENTPD1-AS1–miR-144-3p is associated with poor prognosis and macrophage infiltration in GC.
ENTPD1-AS1-miR-144-3p-COL5A2 might be a new therapeutic target for GC.
Provenance and peer review: Unsolicited article; Externally peer reviewed.
Peer-review model: Single blind
Specialty type: Oncology
Country/Territory of origin: China
Peer-review report’s scientific quality classification
Grade A (Excellent): 0
Grade B (Very good): B, B
Grade C (Good): C
Grade D (Fair): 0
Grade E (Poor): 0
P-Reviewer: de Melo FF, Brazil; Li DH, China; Liu YQ, United States S-Editor: Fan JR L-Editor: A P-Editor: Zhang XD
| 1. | Smyth EC, Nilsson M, Grabsch HI, van Grieken NC, Lordick F. Gastric cancer. Lancet. 2020;396:635-648. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1150] [Cited by in RCA: 2856] [Article Influence: 571.2] [Reference Citation Analysis (5)] |
| 2. | Sexton RE, Al Hallak MN, Diab M, Azmi AS. Gastric cancer: a comprehensive review of current and future treatment strategies. Cancer Metastasis Rev. 2020;39:1179-1203. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 459] [Cited by in RCA: 460] [Article Influence: 92.0] [Reference Citation Analysis (0)] |
| 3. | Fischer H, Stenling R, Rubio C, Lindblom A. Colorectal carcinogenesis is associated with stromal expression of COL11A1 and COL5A2. Carcinogenesis. 2001;22:875-878. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 115] [Cited by in RCA: 133] [Article Influence: 5.5] [Reference Citation Analysis (0)] |
| 4. | Lü XZ, Chen WT, Zhang CP. [Investigation of mRNA expression of collagen genes in oral squamous cell carcinoma and paired normal tissue]. Nan Fang Yi Ke Da Xue Xue Bao. 2011;31:1197-1199. [PubMed] |
| 5. | Mohan H, Krumbholz M, Sharma R, Eisele S, Junker A, Sixt M, Newcombe J, Wekerle H, Hohlfeld R, Lassmann H, Meinl E. Extracellular matrix in multiple sclerosis lesions: Fibrillar collagens, biglycan and decorin are upregulated and associated with infiltrating immune cells. Brain Pathol. 2010;20:966-975. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 28] [Cited by in RCA: 89] [Article Influence: 5.9] [Reference Citation Analysis (0)] |
| 6. | Ma N, Zhu Z, Liu J, Peng Y, Zhao X, Tang W, Jia Z, Xi H, Gao B, Wang H, Du J. Clinical and genetic analysis of classical Ehlers-Danlos syndrome patient caused by synonymous mutation in COL5A2. Mol Genet Genomic Med. 2021;9:e1632. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 1] [Cited by in RCA: 4] [Article Influence: 1.0] [Reference Citation Analysis (0)] |
| 7. | Jacinto JGP, Häfliger IM, Veiga IMB, Letko A, Benazzi C, Bolcato M, Drögemüller C. A Heterozygous Missense Variant in the COL5A2 in Holstein Cattle Resembling the Classical Ehlers-Danlos Syndrome. Animals (Basel). 2020;10. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 3] [Cited by in RCA: 7] [Article Influence: 1.4] [Reference Citation Analysis (0)] |
| 8. | Januchowski R, Zawierucha P, Ruciński M, Zabel M. Microarray-based detection and expression analysis of extracellular matrix proteins in drugresistant ovarian cancer cell lines. Oncol Rep. 2014;32:1981-1990. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 38] [Cited by in RCA: 54] [Article Influence: 4.9] [Reference Citation Analysis (0)] |
| 9. | Cao L, Chen Y, Zhang M, Xu DQ, Liu Y, Liu T, Liu SX, Wang P. Identification of hub genes and potential molecular mechanisms in gastric cancer by integrated bioinformatics analysis. PeerJ. 2018;6:e5180. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 42] [Cited by in RCA: 78] [Article Influence: 11.1] [Reference Citation Analysis (0)] |
| 10. | Meng XY, Shi MJ, Zeng ZH, Chen C, Liu TZ, Wu QJ, Li S. The Role of COL5A2 in Patients With Muscle-Invasive Bladder Cancer: A Bioinformatics Analysis of Public Datasets Involving 787 Subjects and 29 Cell Lines. Front Oncol. 2018;8:659. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 16] [Cited by in RCA: 36] [Article Influence: 6.0] [Reference Citation Analysis (0)] |
| 11. | Meng Z, Chen Y, Wu W, Yan B, Meng Y, Liang Y, Yao X, Luo J. Exploring the Immune Infiltration Landscape and M2 Macrophage-Related Biomarkers of Proliferative Diabetic Retinopathy. Front Endocrinol (Lausanne). 2022;13:841813. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in RCA: 32] [Reference Citation Analysis (0)] |
| 12. | Wei L, Sun J, Zhang N, Zheng Y, Wang X, Lv L, Liu J, Xu Y, Shen Y, Yang M. Noncoding RNAs in gastric cancer: implications for drug resistance. Mol Cancer. 2020;19:62. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 279] [Cited by in RCA: 336] [Article Influence: 67.2] [Reference Citation Analysis (0)] |
| 13. | Shin VY, Chu KM. MiRNA as potential biomarkers and therapeutic targets for gastric cancer. World J Gastroenterol. 2014;20:10432-10439. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in CrossRef: 223] [Cited by in RCA: 282] [Article Influence: 25.6] [Reference Citation Analysis (1)] |
| 14. | Xue X, Pan J, Zhang H, Lu Y, Mao Q, Ma K. Baihe Dihuang (Lilium Henryi Baker and Rehmannia Glutinosa) decoction attenuates somatostatin interneurons deficits in prefrontal cortex of depression via miRNA-144-3p mediated GABA synthesis and release. J Ethnopharmacol. 2022;292:115218. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 7] [Cited by in RCA: 18] [Article Influence: 6.0] [Reference Citation Analysis (0)] |
| 15. | Xing B, Shen C, Yang Q, Wang Z, Tan W. miR-144-3p represses hepatocellular carcinoma progression by affecting cell aerobic glycolysis via FOXK1. Int J Exp Pathol. 2023;. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 4] [Reference Citation Analysis (0)] |
| 16. | Wang B, Xu L, Zhang J, Cheng X, Xu Q, Wang J, Mao F. LncRNA NORAD accelerates the progression and doxorubicin resistance of neuroblastoma through up-regulating HDAC8 via sponging miR-144-3p. Biomed Pharmacother. 2020;129:110268. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 20] [Cited by in RCA: 36] [Article Influence: 7.2] [Reference Citation Analysis (0)] |
| 17. | Cao H, Xu X, Wang K, Li C. Circ_0047835 Combines with miR-144-3p to Promote the Proliferation, Invasion, Migration, and Fibrosis of TGF-β1-Treated Human Tenon's Capsule Fibroblasts by Upregulating SP1. Curr Eye Res. 2023;48:371-381. [RCA] [PubMed] [DOI] [Full Text] [Reference Citation Analysis (0)] |
| 18. | Zhang X, Liu Q, Zhang H, Zhao S, Huang J, Sovannary T, Bunnath L, Aun HS, Samnom H, Su B, Chen H. The distinct morphological phenotypes of Southeast Asian aborigines are shaped by novel mechanisms for adaptation to tropical rainforests. Natl Sci Rev. 2022;9:nwab072. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 2] [Cited by in RCA: 7] [Article Influence: 1.8] [Reference Citation Analysis (0)] |
| 19. | Zhou M, Zhang Z, Zhao H, Bao S, Cheng L, Sun J. An Immune-Related Six-lncRNA Signature to Improve Prognosis Prediction of Glioblastoma Multiforme. Mol Neurobiol. 2018;55:3684-3697. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 84] [Cited by in RCA: 134] [Article Influence: 16.8] [Reference Citation Analysis (0)] |
| 20. | Tonge DP, Darling D, Farzaneh F, Williams GT. Whole-genome-scale identification of novel non-protein-coding RNAs controlling cell proliferation and survival through a functional forward genetics strategy. Sci Rep. 2022;12:182. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 2] [Cited by in RCA: 1] [Article Influence: 0.3] [Reference Citation Analysis (0)] |
| 21. | Li Z, Liu Z, Shao Z, Li C, Li Y, Liu Q, Zhang Y, Tan B, Liu Y. Identifying multiple collagen gene family members as potential gastric cancer biomarkers using integrated bioinformatics analysis. PeerJ. 2020;8:e9123. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 12] [Cited by in RCA: 12] [Article Influence: 2.4] [Reference Citation Analysis (0)] |
| 22. | Januchowski R, Świerczewska M, Sterzyńska K, Wojtowicz K, Nowicki M, Zabel M. Increased Expression of Several Collagen Genes is Associated with Drug Resistance in Ovarian Cancer Cell Lines. J Cancer. 2016;7:1295-1310. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 97] [Cited by in RCA: 135] [Article Influence: 15.0] [Reference Citation Analysis (0)] |
| 23. | Weng K, Huang Y, Deng H, Wang R, Luo S, Wu H, Chen J, Long M, Hao W. Collagen family genes and related genes might be associated with prognosis of patients with gastric cancer: an integrated bioinformatics analysis and experimental validation. Transl Cancer Res. 2020;9:6246-6262. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in RCA: 8] [Reference Citation Analysis (0)] |
| 24. | Ren X, Chen X, Fang K, Zhang X, Wei X, Zhang T, Li G, Lu Z, Song N, Wang S, Qin C. COL5A2 Promotes Proliferation and Invasion in Prostate Cancer and Is One of Seven Gleason-Related Genes That Predict Recurrence-Free Survival. Front Oncol. 2021;11:583083. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 13] [Cited by in RCA: 33] [Article Influence: 8.3] [Reference Citation Analysis (0)] |
| 25. | Yin W, Zhu H, Tan J, Xin Z, Zhou Q, Cao Y, Wu Z, Wang L, Zhao M, Jiang X, Ren C, Tang G. Identification of collagen genes related to immune infiltration and epithelial-mesenchymal transition in glioma. Cancer Cell Int. 2021;21:276. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 11] [Cited by in RCA: 43] [Article Influence: 10.8] [Reference Citation Analysis (0)] |
| 26. | Ghafouri-Fard S, Vafaee R, Shoorei H, Taheri M. MicroRNAs in gastric cancer: Biomarkers and therapeutic targets. Gene. 2020;757:144937. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 16] [Cited by in RCA: 41] [Article Influence: 8.2] [Reference Citation Analysis (0)] |
| 27. | Bure IV, Nemtsova MV. Methylation and Noncoding RNAs in Gastric Cancer: Everything Is Connected. Int J Mol Sci. 2021;22. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 5] [Cited by in RCA: 17] [Article Influence: 4.3] [Reference Citation Analysis (0)] |
| 28. | Yan H, Han L, He N, Li R, He S. Upregulated Circular RNA KIF4A Promotes Cell Migration and Invasion by Regulating MicroRNA-144-3p/EZH2 Axis in Gastric Cancer. J Oncol. 2022;2022:3985621. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in RCA: 5] [Reference Citation Analysis (0)] |
| 29. | Lario S, Brunet-Vega A, Quílez ME, Ramírez-Lázaro MJ, Lozano JJ, García-Martínez L, Pericay C, Miquel M, Junquera F, Campo R, Calvet X. Expression profile of circulating microRNAs in the Correa pathway of progression to gastric cancer. United European Gastroenterol J. 2018;6:691-701. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 8] [Cited by in RCA: 10] [Article Influence: 1.4] [Reference Citation Analysis (0)] |
| 30. | Zhao Z, Liu H, Zhou X, Fang D, Ou X, Ye J, Peng J, Xu J. Necroptosis-Related lncRNAs: Predicting Prognosis and the Distinction between the Cold and Hot Tumors in Gastric Cancer. J Oncol. 2021;2021:6718443. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 65] [Cited by in RCA: 141] [Article Influence: 35.3] [Reference Citation Analysis (0)] |
| 31. | Miao Z, Guo X, Tian L. The long noncoding RNA NORAD promotes the growth of gastric cancer cells by sponging miR-608. Gene. 2019;687:116-124. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 35] [Cited by in RCA: 51] [Article Influence: 8.5] [Reference Citation Analysis (0)] |
| 32. | Wang J, Sun Y, Zhang X, Cai H, Zhang C, Qu H, Liu L, Zhang M, Fu J, Zhang J, Wang J, Zhang G. Oxidative stress activates NORAD expression by H3K27ac and promotes oxaliplatin resistance in gastric cancer by enhancing autophagy flux via targeting the miR-433-3p. Cell Death Dis. 2021;12:90. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 18] [Cited by in RCA: 55] [Article Influence: 13.8] [Reference Citation Analysis (0)] |
| 33. | Zhang H, Ding C, Li Y, Xing C, Wang S, Yu Z, Chen L, Li P, Dai M. Data mining-based study of collagen type III alpha 1 (COL3A1) prognostic value and immune exploration in pan-cancer. Bioengineered. 2021;12:3634-3646. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 3] [Cited by in RCA: 28] [Article Influence: 7.0] [Reference Citation Analysis (0)] |
| 34. | Liu B, Ma X, Ha W. Identification of Potential Prognostic Biomarkers Associated With Macrophage M2 Infiltration in Gastric Cancer. Front Genet. 2021;12:827444. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Reference Citation Analysis (0)] |
| 35. | Chivu-Economescu M, Necula LG, Matei L, Dragu D, Bleotu C, Sorop A, Herlea V, Dima S, Popescu I, Diaconu CC. Collagen Family and Other Matrix Remodeling Proteins Identified by Bioinformatics Analysis as Hub Genes Involved in Gastric Cancer Progression and Prognosis. Int J Mol Sci. 2022;23. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 25] [Cited by in RCA: 25] [Article Influence: 8.3] [Reference Citation Analysis (0)] |
| 36. | Wei S, Lu J, Lou J, Shi C, Mo S, Shao Y, Ni J, Zhang W, Cheng X. Gastric Cancer Tumor Microenvironment Characterization Reveals Stromal-Related Gene Signatures Associated With Macrophage Infiltration. Front Genet. 2020;11:663. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 11] [Cited by in RCA: 34] [Article Influence: 6.8] [Reference Citation Analysis (0)] |