Published online Jun 15, 2023. doi: 10.4251/wjgo.v15.i6.1086
Peer-review started: March 13, 2023
First decision: March 28, 2023
Revised: March 29, 2023
Accepted: May 17, 2023
Article in press: May 17, 2023
Published online: June 15, 2023
Processing time: 94 Days and 3.1 Hours
Minimally invasive or noninvasive, sensitive and accurate detection of colorectal cancer (CRC) is urgently needed in clinical practice.
To identify a noninvasive, sensitive and accurate circular free DNA marker detected by digital polymerase chain reaction (dPCR) for the early diagnosis of clinical CRC.
A total of 195 healthy control (HC) individuals and 101 CRC patients (38 in the early CRC group and 63 in the advanced CRC group) were enrolled to establish the diagnostic model. In addition, 100 HC individuals and 62 patients with CRC (30 early CRC and 32 advanced CRC groups) were included separately to validate the model. CAMK1D was dPCR. Binary logistic regression analysis was used to establish a diagnostic model including CAMK1D and CEA.
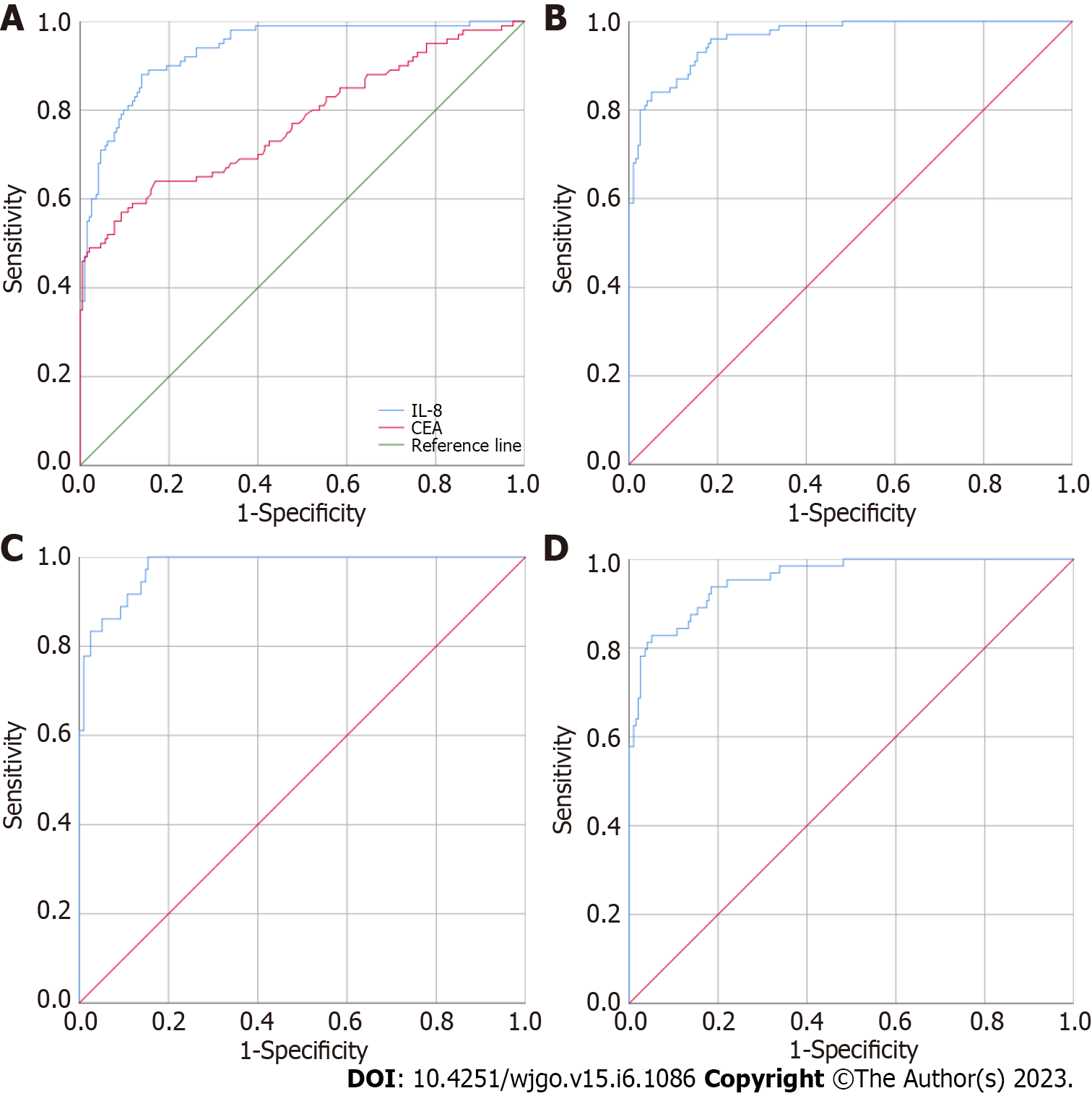
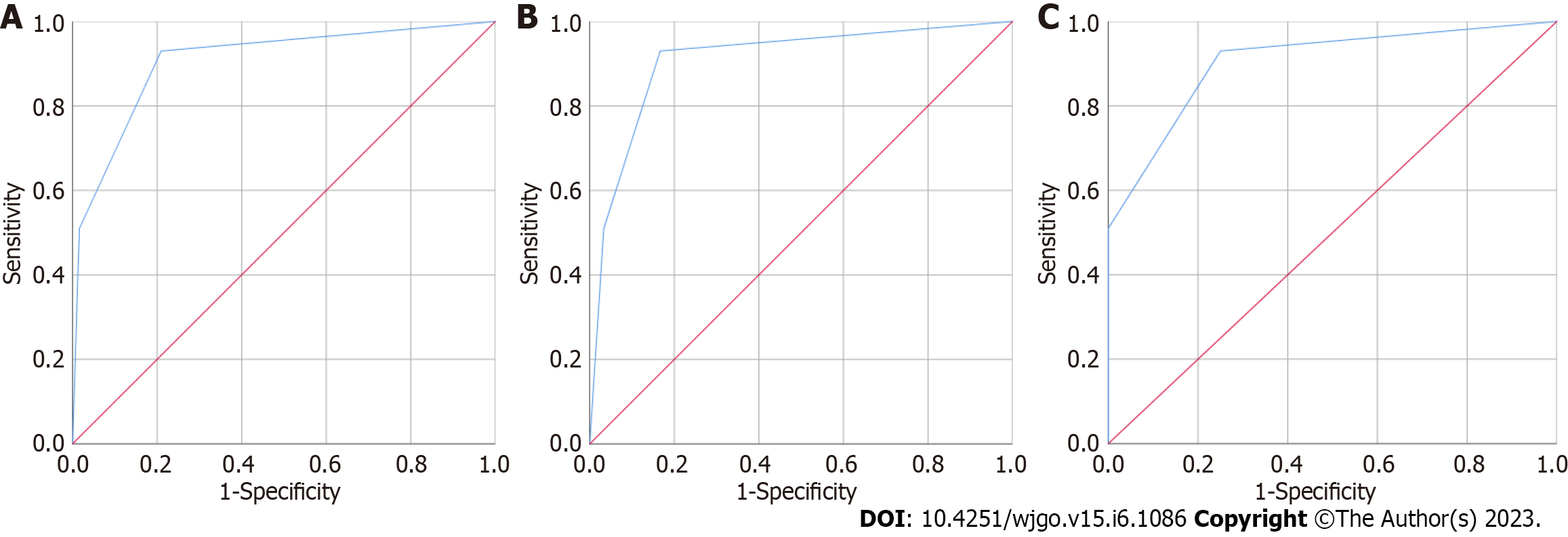
To differentiate between the 195 HCs and 101 CRC patients (38 early CRC and 63 advanced CRC patients), the common biomarkers CEA and CAMK1D were used alone or in combination to evaluate their diagnostic value. The area under the curves (AUCs) of CEA and CAMK1D were 0.773 (0.711, 0.834) and 0.935 (0.907, 0.964), respectively. When CEA and CAMK1D were analyzed together, the AUC was 0.964 (0.945, 0.982). In differentiating between the HC and early CRC groups, the AUC was 0.978 (0.960, 0.995), and the sensitivity and specificity were 88.90% and 90.80%, respectively. In differentiating between the HC and advanced CRC groups, the AUC was 0.956 (0.930, 0.981), and the sensitivity and specificity were 81.30% and 95.90%, respectively. After building the diagnostic model containing CEA and CAMK1D, the AUC of the CEA and CAMK1D joint model was 0.906 (0.858, 0.954) for the validation group. In differentiating between the HC and early CRC groups, the AUC was 0.909 (0.844, 0.973), and the sensitivity and specificity were 93.00% and 83.30%, respectively. In differentiating between the HC and advanced CRC groups, the AUC was 0.904 (0.849, 0.959), and the sensitivity and specificity were 93.00% and 75.00%, respectively.
We built a diagnostic model including CEA and CAMK1D for differentiating between HC individuals and CRC patients. Compared with the common biomarker CEA alone, the diagnostic model exhibited significant improvement.
Core Tip: Minimally invasive or noninvasive, sensitive and accurate detection of colorectal cancer (CRC) is urgently needed in clinical practice. We aimed to build a joint diagnostic model based on circular free DNA for detection of colorectal cancer. We evaluated the diagnostic value of circular free CAMK1D DNA for differentiating between HC individuals and CRC patients and demonstrated that CAMK1D may represent a potential diagnostic biomarker for CRC detection. Further analysis should use the colorectal polyp group to validate the diagnostic model in future studies.
- Citation: Cui Y, Zhang LJ, Li J, Xu YJ, Liu MY. Diagnostic value of circular free DNA for colorectal cancer detection. World J Gastrointest Oncol 2023; 15(6): 1086-1095
- URL: https://www.wjgnet.com/1948-5204/full/v15/i6/1086.htm
- DOI: https://dx.doi.org/10.4251/wjgo.v15.i6.1086
Colorectal cancer (CRC) is a common malignant tumor in China. The 5-year survival rate for early CRC patients after effective treatment is more than 90%. Approximately 25% of patients have local or distant metastasis at the time of initial diagnosis, and the 5-year survival rate is only 12%. Early detection, diagnosis and treatment are currently recognized methods that can effectively improve the treatment of CRC. At present, the common clinical screening tests include fecal occult blood tests and blood marker tests, but the sensitivity and specificity remain insufficient[1]. Imaging examination can effectively evaluate the scope of the lesion and the stage of the tumor, but it is of limited value in the diagnosis of early lesions. Endoscopy combined with tissue biopsy is the gold standard for the early diagnosis of CRC at present, but there are some disadvantages, such as cumbersome operation, poor compliance and the invasive nature of testing. Thus, the commonly used methods for the early diagnosis of CRC remain insufficient[2]. The identification of a minimally invasive or noninvasive, sensitive and accurate early diagnostic test is urgently needed.
Liquid biopsy technology has gained increasing interest because it is noninvasive and comprehensive and permits real-time and repeated monitoring[3]. Liquid biopsy technology primarily uses human peripheral blood, saliva, urine and other body fluid components to identify tumor heterogeneity and genetic information for the early diagnosis and individualized treatment of CRC. With the advent of precision medicine, liquid biopsy has become increasingly important. Circulating tumor cells (CTCs), circulating tumor DNA (ctDNA) and secreted proteins are the primary targets of liquid biopsy at present[4-6]. Compared with CTCs and exosomes, ctDNA is now the most widely used marker in clinical practice. A variety of tests based on ctDNA have been used in clinical practice; however, due to clearance by macrophages, the amount of circulating free DNA in body fluid is extremely low. Furthermore, ctDNA accounts for only a small portion of circulating free DNA and therefore requires high sensitivity detection equipment.
Single-stranded or double-stranded DNA is traditionally the form of ctDNA that is detected. With the development of high-throughput sequencing technology and single-cell gene amplification technology, a new type of circular free DNA has been identified[7]: Extrachromosomal circular DNA. This is a closed, circular single- or double-stranded form of DNA located in the chromatin body that can be detected in many eukaryotes, including humans[8]. Compared with free linear DNA, extrachromosomal circular DNA is not easily degraded by nucleases, and its structure is more stable[9]. Studies have also detected circular DNA in the plasma of pregnant women. Detection of the level and type of circular DNA in human plasma is expected to permit ultra-early prediction, prognosis evaluation and even targeted treatment of tumors or other physiological and pathological conditions[10,11].
Currently, detection of ctDNA is primarily achieved through polymerase chain reaction (PCR) technology and second-generation sequencing[12]. ctDNA detection based on PCR includes: (1) Amplification-refractory mutation system PCR (ARMS-PCR) technology; (2) high resolution melting curve technology (HRM); (3) digital PCR (dPCR)[13]; and (4) BEAMING technology. Compared with other PCR detection methods, dPCR has a strong reaction solution segmentation ability and has advantages of high sensitivity, high accuracy, high tolerance and absolute quantification[14]. For plasma-free DNA, the screening strategy based on second-generation sequencing technology and the sensitive detection of dPCR can realize the accurate detection of trace plasma ctDNA[15].
In our study, we aimed to provide a noninvasive, sensitive and accurate diagnostic marker detected by dPCR for the early diagnosis of CRC.
All individuals enrolled in our study provided informed consent. Our study was approved by the ethics committee. From April 2019 to July 2022, a total of 295 healthy control (HC) individuals and 163 CRC patients were enrolled in our study. The project included a colorectal polyp (CRP) group, an early CRC group and an advanced CRC group. The staging of CRC was performed in accordance with the tumor node metastasis (TNM) staging of CRC of the United States Joint Commission on Cancer and the guidelines for screening and endoscopic diagnosis and treatment of early colorectal cancer in China: (1) The inclusion criteria of the CRP group were colonoscopic diagnosis and postoperative pathological confirmation of villous/tubular adenoma, with or without mild to moderate atypical hyperplasia, or local high-grade neoplasia of villous tubular adenoma confirmed by pathology and immunohistochemistry. To be included in this group, no abnormalities could be detected on any biochemical or auxiliary examinations, patients could have no chief complaint of gastrointestinal discomfort, patients could have no clinical signs of tumor, and the adenoma (villous adenoma, mixed adenoma, or adenoma with moderate or severe dysplasia) could not exceed 1 cm in diameter; (2) To be included in the early CRC group, adenocarcinoma of the intestinal wall had to be localized in the mucosa or submucosa, and no lymphatic metastasis could be detected (i.e., stage 0-T1 tumors). Pathologically confirmed local high-grade villous tubular adenomas or adenocarcinoma of the intestinal wall localized to the mucosa or submucosa were eligible for inclusion. Patients had not received treatment by surgery, chemotherapy, radiotherapy, or other modalities prior to sample collection, and patients had not received blood transfusions in the last 3 mo; and (3) The advanced CRC group was diagnosed based on the TNM CRC staging of the United States Joint Commission on Cancer, and T2-IV stage was defined as intermediate and advanced CRC. All diagnoses were based on pathologically confirmed CRC. Patients had not undergone treatment with surgery, chemotherapy, radiotherapy, or other modalities prior to sample collection, and patients had not received any blood transfusions in the last 3 mo. CRC tissue samples and corresponding clinical examination data were available for all patients included in the study. None of the patients received chemotherapy, radiotherapy or immunotherapy before sample collection, and patients with other tumors and gastrointestinal diseases detected during the admission examination were excluded.
A BD Vacutainer PPT plasma preparation tube was used to collect peripheral blood samples from patients. Within 2 h of collection, samples were centrifuged at 2000 × g for 10 minutes, and the supernatant was then divided into several aliquots. All plasma samples were kept at -80 °C until use.
Free DNA was extracted from plasma samples using a QIAamp DNA Blood Kit. ATP-dependent DNase was added to the free DNA and digested at 37 °C for 1.5 h until the final concentration was 0.4 U/μL. Linear double-stranded DNA was removed and incubated at 70 °C for 30 min to inactivate ATP-dependent DNase. The primers were designed according to the eccDNA sequence. The primer probe was designed using Primer3 software and synthesized by Invitrogen after a homologous search with BLAST. The primer and probe were diluted with deionized water, and the storage concentration was 200 μmol/L with a working concentration of 10 μmol/L. The total PCR volume was 20 μL: 10 μL 2 × ddPCRTMSuper mixture, 1.8 μL forward and reverse primers (final concentration 900 nmol/L), 0.5 μL probe (final concentration of 250 nmol/L), 4 μg template DNA, and ddH2O to a final volume of 20 μL. Then, a 20 μL reaction volume was added to the droplet generation card. All generated microdroplets were transferred to a 96-well plate for PCR amplification. The PCR conditions were as follows: 95 °C/10 min, followed by 40 cycles of 94 °C/30 s 60 °C/1 min and 98 °C/10 min. Quanta Soft 1.6 software was used to analyze the results. The system was flushed before each experiment. The sample for the 96-well plate was input, and the sample droplets were analyzed.
SPSS 22.0 was used for statistical analysis. Normally distributed data were compared using independent sample t tests. Nonnormally distributed data were compared using the rank sum test. The area under the curve (AUC), sensitivity and specificity were used to assess the diagnostic value of the indicators. P < 0.05 represents a statistically significant difference. The binary logistic regression model, which used the forward conditional method, was used to combine the indicators. The Z score test was used to compare the AUC values.
As shown in Table 1, 195 HC individuals and 101 CRC patients (38 in the early CRC group and 63 in the advanced CRC group) were enrolled for model establishment. In addition, 100 HC individuals and 62 patients with CRC (30 early CRC and 32 late CRC) were included separately to validate the model. The CRC stage was in accordance with the TNM CRC stage of the United States Joint Commission on Cancer and the guidelines for screening and endoscopic diagnosis and treatment of early CRC in China. T1 and T2 were defined as early CRC, and T3 and T4 were defined as advanced CRC. The age and sex of the patients in the model generation group and the validation group were matched. In the model generation group, 21 tumors were located in the ascending colon, 15 were located in the descending colon, 3 were located in the transverse colon, 59 were located in the sigmoid colon and 3 were located in the rectum. Twenty-one tumors were well differentiated, 57 exhibited intermediate differentiation, and 23 were poorly differentiated.
| Characteristics | CRC (training) | HC (training) | CRC (validation) | HC (validation) |
| Number | 101 | 195 | 62 | 100 |
| Age, yr | ||||
| Mean | 58 | 53 | 57 | 55 |
| Range | 29-81 | 33-57 | 33-74 | 34-67 |
| Sex | ||||
| Male | 60 | 116 | 37 | 64 |
| Female | 41 | 79 | 25 | 36 |
| TNM stage | ||||
| T1 | 11 | 11 | ||
| T2 | 27 | 21 | ||
| T3 | 44 | 7 | ||
| T4 | 19 | 23 |
As shown in Table 2, the levels of NDUFB7, CAMK1D, PIK3CD and PSEN2 were compared between the 195 HCs and 101 CRC patients. First, homogeneity of variance was tested. CAMK1D, PIK3CD and PSEN2 exhibited nonhomogeneity of variance; NDUFB7 exhibited homogeneity of variance. Three of the four indicators, CAMK1D, PIK3CD and PSEN2, exhibited statistically significant differences between the HC and CRC groups (P < 0.05). NDUFB7 exhibited no significant differences.
| Indicator | HC (n = 195) | CRC (n = 101) | F | Sig | P value |
| NDUFB7 | 1.54 (0.94, 2.31) | 2.10 (1.29, 3.08) | 0.15 | 0.70 | 0.60 |
| CAMK1D | 9.71 (6.38, 18.25) | 70.39 (35.26, 155.57) | 34.24 | < 0.01 | < 0.01 |
| PIK3CD | 297.11 (232.76, 374.69) | 333.22 (259.40, 417.90) | 10.47 | < 0.01 | 0.03 |
| PSEN2 | 5.48 (4.04, 7.21) | 8.69 (6.00, 11.67) | 5.89 | 0.02 | < 0.01 |
Based on the significant differences between the HC and CRC groups, three indicators, CAMK1D, PIK3CD and PSEN2, were used for AUC analysis. As shown in Table 3, the AUCs of CAMK1D and PIK3CD exhibited statistically significant differences (P < 0.01) between the HCs and CRC patients when the ROC curve was used to evaluate the diagnostic value. Therefore, CAMK1D and PIK3CD were selected for subsequent multiparameter diagnostic model analysis.
| Indicator | AUC | SD | P value | 95%CI | |
| Lower | Upper | ||||
| CAMK1D | 0.935 | 0.015 | < 0.001 | 0.907 | 0.964 |
| PSEN2 | 0.740 | 0.031 | < 0.001 | 0.678 | 0.801 |
| PIK3CD | 0.582 | 0.036 | 0.021 | 0.511 | 0.653 |
Based on the significant differences between the HC and CRC groups and ROC curves, univariate logistic regression was performed. As shown in Tables 4 and 5, CAMK1D and PIK3CD differed significantly between the two groups (P < 0.01). Next, multivariate logistic regression analysis was performed for CAMK1D and PIK3CD, and only CAMK1D remained statistically significant (P < 0.01).
| Indicator | B | SE | Wals | P value | Exp (B) | 95%CI | |
| Lower | Upper | ||||||
| CAMK1D | 0.560 | 0.137 | 16.781 | < 0.001 | 1.751 | 1.339 | 2.289 |
| PIK3CD | 0.071 | 0.011 | 41.382 | < 0.001 | 1.074 | 1.051 | 1.097 |
| Indicator | B | SE | Wals | P value | Exp (B) | 95%CI | |
| Lower | Upper | ||||||
| CAMK1D | 0.125 | 0.039 | 10.400 | 0.001 | 1.133 | 1.050 | 1.222 |
| PIK3CD | -0.002 | 0.004 | 0.208 | 0.648 | 0.998 | 0.990 | 1.006 |
CAMK1D and the common biomarker CEA were used alone or in combination to evaluate their ability to differentiate between 195 HC individuals and 101 CRC patients (38 early CRC patients and 63 advanced CRC patients). As shown in Figure 1A, the AUCs of CEA and CAMK1D were 0.773 (0.711, 0.834) and 0.935 (0.907, 0.964), respectively. As shown in Figure 1B, the use of both CEA and CAMK1D produced an AUC of 0.964 (0.945, 0.982) by binary logistic regression analysis. Next, the diagnostic value of the CEA and CAMK1D model in the differentiation of 195 HC individuals and 38 early CRC patients was evaluated. As shown in Figure 1C, the AUC was 0.978 (0.960, 0.995), and the sensitivity and specificity were 88.90% and 90.80%, respectively. As shown in Figure 1D, when applying the model to differentiate between the 195 HC individuals and 63 advanced CRC patients, the AUC was 0.956 (0.930, 0.981), and the sensitivity and specificity were 81.30% and 95.90%, respectively.
After building the diagnostic model containing CEA and CAMK1D, 100 HC individuals and 62 patients with CRC (30 early CRC patients and 32 advanced CRC patients) were enrolled to validate the model. As shown in Figure 2A, the AUC of the CEA and CAMK1D joint model was 0.906 (0.858, 0.954). Next, the diagnostic value of the CEA and CAMK1D model in the differentiation of 100 HC individuals and 32 early CRC patients was evaluated. As shown in Figure 2B, the AUC was 0.909 (0.844, 0.973), and the sensitivity and specificity were 93.00% and 83.30%, respectively. As shown in Figure 2C, the AUC for the differentiation of the 100 HC individuals and 32 advanced CRC patients was 0.904 (0.849, 0.959), and the sensitivity and specificity were 93.00% and 75.00%, respectively.
There have been many studies exploring the use of circulating free DNA as a diagnostic and prognostic tumor biomarker[16]. Because the molecular weight of circulating free DNA is relatively large, optical microscopy can use common DNA dyes to observe extracellular DNA in M-phase cells[17]. Ultrahigh-resolution microscopy technology has been developed in recent years to aid in imaging[18]. Superresolution three-dimensional structured illumination microscopy can also be used for imaging analysis. Due to the resolution limitations of optical microscopy, it is difficult to observe and analyze fine circular DNA structures. Therefore, researchers turned to electron microscopy to solve this problem[19]. Electron microscopy has made significant contributions to structural studies. Both scanning electron microscopy and transmission electron microscopy can be used for imaging. However, due to the large sample size required and low abundance of ctDNA, this method is not commonly used at present. Transposase-accessible chromatin visualization analysis is a transposase-mediated imaging technology. This technology uses direct in situ imaging, cell sorting and depth sequencing of accessible genomes to reveal the identity of imaging elements. Single-molecule real-time sequencing technology has addressed many of the previous technological limitations. ctDNA is typically large and may contain sequences from multiple chromatin sources[20]. Therefore, it is difficult to use high-throughput sequencing to completely reconstruct the full-length sequence. ATAC-seq and DNA transposase technology were first proposed as a means of chromatin accessibility analysis in 2013[21,22]. DNA transposase can randomly insert sequences into the genome. The identification of ctDNA in plasma and serum prompted the demand for a novel detection method in plasma[14,23,24].
dPCR can precisely quantify target nucleic acids in a sample and overcomes the shortcomings of qPCR. In dPCR, the sample is first divided into many independent PCR subreactions so that each part contains either a few target sequences or no target sequences[13]. After PCR, the score of the amplification positive zone was used to quantify the concentration of the target sequence, and Poisson statistics were used to statistically define the accuracy. In addition, this approach exhibits higher tolerance for the presence of inhibitors in the sample. Each subreaction acts as a separate PCR microreactor, and the subreaction containing the amplified target sequence is detected by fluorescence. The ratio of the positive distribution to the total number of sequences can be used to determine the concentration of the target in the sample. The primary difference between dPCR and qPCR is the method of measuring the number of target sequences. Unlike qPCR, dPCR does not rely on a calibration curve for sample quantification. Therefore, dPCR avoids the limitations related to the change in reaction efficiency. dPCR is theoretically superior to qPCR because it provides an effective method to perform sample allocation and single-molecule target amplification[25]. In practice, due to its higher sensitivity, qPCR can outperform dPCR in specific applications. Calcium/calmodulin-dependent protein kinases (CAMKs) are involved in a wide range of cancer-related functions in multiple tumor types. CAMK1 may have potential prognostic value in pancreatic cancer, suggesting that CAMK1 may have a distinct role in pancreatic cancer progression[26]. In addition, CAMK1D may also be involved in immune resistance by T-cell recognition, which rapidly inhibits the terminal apoptotic cascade[27]. Other regulatory molecules, such as miRNAs and lncRNAs, may also regulate CAMK1D to participate in cancer progression. In our study, the overexpression of circular CAMK1D may also have a potential function in CRC progression[28-30].
There are still some limitations in our study. First, we only assessed the diagnostic value of our model in distinguishing between HC individuals and CRC patients; the utility of the model in the CRP group was not evaluated. Second, we only evaluated the common biomarker CEA, and other biomarkers may also possess diagnostic value. Third, the experimental protocol may affect the results.
We evaluated the diagnostic value of circular free CAMK1D in differentiating between HC individuals and CRC patients and demonstrated that CAMK1D may represent a diagnostic biomarker for CRC detection.
Endoscopy combined with tissue biopsy is currently the gold standard for the early diagnosis of colorectal cancer (CRC), but there are some disadvantages, including cumbersome operation, poor compliance and the invasive nature of testing. The commonly available methods for the early diagnosis of CRC remain insufficient.
The identification of a minimally invasive or noninvasive, sensitive and accurate early diagnostic marker for the clinical detection of CRC is urgently needed. Common biomarkers and circular free DNA may exhibit potential diagnostic value for CRC.
To evaluate the diagnostic value of circular free DNA in CRC.
A total of 195 healthy control (HC) individuals and 101 CRC patients (38 in the early CRC group and 63 in the advanced CRC group) were enrolled to generate the model. One hundred HC individuals and 62 patients with CRC (30 early CRC and 32 advanced CRC patients) were included separately to validate the model. CAMK1D was detected by digital PCR. Binary logistic regression analysis was used to establish a joint CAMK1D and CEA diagnostic model for CRC.
Inclusion of both CEA and CAMK1D in the model produced an area under the curve (AUC) of 0.964 (0.945, 0.982). For the differentiation between the HC group and early CRC group, the AUC was 0.978 (0.960, 0.995), and the sensitivity and specificity were 88.90% and 90.80%, respectively. For the differentiation between the HC group and advanced CRC group, the AUC was 0.956 (0.930, 0.981), and the sensitivity and specificity were 81.30% and 95.90%, respectively. In the validation group, the AUC of the CEA and CAMK1D joint model was 0.906 (0.858, 0.954). For differentiating between the HC group and early CRC group, the AUC was 0.909 (0.844, 0.973), and the sensitivity and specificity were 93.00% and 83.30%, respectively. For differentiating between the HC group and the advanced CRC group, the AUC was 0.904 (0.849, 0.959), and the sensitivity and specificity were 93.00% and 75.00%, respectively.
We evaluated the diagnostic value of circular free CAMK1D DNA for differentiating between HC individuals and CRC patients and demonstrated that CAMK1D may represent a potential diagnostic biomarker for CRC detection.
Further analysis should use the colorectal polyp group to validate the diagnostic model in future studies.
Provenance and peer review: Unsolicited article; Externally peer reviewed.
Peer-review model: Single blind
Specialty type: Gastroenterology and hepatology
Country/Territory of origin: China
Peer-review report’s scientific quality classification
Grade A (Excellent): 0
Grade B (Very good): B, B
Grade C (Good): 0
Grade D (Fair): 0
Grade E (Poor): 0
P-Reviewer: Bianciardi E, Italy; Quaresima S, Italy S-Editor: Yan JP L-Editor: A P-Editor: Yan JP
| 1. | Kuipers EJ, Grady WM, Lieberman D, Seufferlein T, Sung JJ, Boelens PG, van de Velde CJ, Watanabe T. Colorectal cancer. Nat Rev Dis Primers. 2015;1:15065. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1014] [Cited by in RCA: 1122] [Article Influence: 112.2] [Reference Citation Analysis (0)] |
| 2. | Goldstein MJ, Mitchell EP. Carcinoembryonic antigen in the staging and follow-up of patients with colorectal cancer. Cancer Invest. 2005;23:338-351. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 265] [Cited by in RCA: 295] [Article Influence: 18.4] [Reference Citation Analysis (0)] |
| 4. | Alix-Panabières C, Pantel K. Liquid Biopsy: From Discovery to Clinical Application. Cancer Discov. 2021;11:858-873. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 146] [Cited by in RCA: 584] [Article Influence: 146.0] [Reference Citation Analysis (0)] |
| 5. | Junqueira-Neto S, Batista IA, Costa JL, Melo SA. Liquid Biopsy beyond Circulating Tumor Cells and Cell-Free DNA. Acta Cytol. 2019;63:479-488. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 32] [Cited by in RCA: 45] [Article Influence: 7.5] [Reference Citation Analysis (0)] |
| 6. | Chen M, Zhao H. Next-generation sequencing in liquid biopsy: cancer screening and early detection. Hum Genomics. 2019;13:34. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 162] [Cited by in RCA: 331] [Article Influence: 55.2] [Reference Citation Analysis (0)] |
| 7. | Wang M, Chen X, Yu F, Ding H, Zhang Y, Wang K. Extrachromosomal Circular DNAs: Origin, formation and emerging function in Cancer. Int J Biol Sci. 2021;17:1010-1025. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 12] [Cited by in RCA: 34] [Article Influence: 8.5] [Reference Citation Analysis (0)] |
| 8. | Sun Z, Ji N, Zhao R, Liang J, Jiang J, Tian H. Extrachromosomal circular DNAs are common and functional in esophageal squamous cell carcinoma. Ann Transl Med. 2021;9:1464. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 29] [Cited by in RCA: 27] [Article Influence: 6.8] [Reference Citation Analysis (0)] |
| 9. | Zuo S, Yi Y, Wang C, Li X, Zhou M, Peng Q, Zhou J, Yang Y, He Q. Extrachromosomal Circular DNA (eccDNA): From Chaos to Function. Front Cell Dev Biol. 2021;9:792555. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 8] [Cited by in RCA: 36] [Article Influence: 12.0] [Reference Citation Analysis (0)] |
| 10. | Wang T, Zhang H, Zhou Y, Shi J. Extrachromosomal circular DNA: a new potential role in cancer progression. J Transl Med. 2021;19:257. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 12] [Cited by in RCA: 45] [Article Influence: 11.3] [Reference Citation Analysis (0)] |
| 11. | Yu W, Hurley J, Roberts D, Chakrabortty SK, Enderle D, Noerholm M, Breakefield XO, Skog JK. Exosome-based liquid biopsies in cancer: opportunities and challenges. Ann Oncol. 2021;32:466-477. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 328] [Cited by in RCA: 557] [Article Influence: 139.3] [Reference Citation Analysis (0)] |
| 12. | Zhu H, Zhang H, Xu Y, Laššáková S, Korabečná M, Neužil P. PCR past, present and future. Biotechniques. 2020;69:317-325. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 128] [Cited by in RCA: 185] [Article Influence: 37.0] [Reference Citation Analysis (0)] |
| 13. | Váňová B, Malicherova B, Burjanivová T, Liskova A, Janikova K, Jasek K, Lasabová Z, Tatár M, Plank L. Droplet digital PCR as a novel dia-gnostic tool. Klin Onkol. 2021;34:33-39. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1] [Cited by in RCA: 6] [Article Influence: 1.5] [Reference Citation Analysis (0)] |
| 14. | Feng Z, Shu Y. An Overview of Digital PCR. Bing Du Xue Bao. 2017;33:103-107. [PubMed] |
| 15. | Lin J, Su G, Su W, Zhou C. [Progress in digital PCR technology and application]. Sheng Wu Gong Cheng Xue Bao. 2017;33:170-177. [RCA] [PubMed] [DOI] [Full Text] [Cited by in RCA: 3] [Reference Citation Analysis (0)] |
| 16. | Wu S, Turner KM, Nguyen N, Raviram R, Erb M, Santini J, Luebeck J, Rajkumar U, Diao Y, Li B, Zhang W, Jameson N, Corces MR, Granja JM, Chen X, Coruh C, Abnousi A, Houston J, Ye Z, Hu R, Yu M, Kim H, Law JA, Verhaak RGW, Hu M, Furnari FB, Chang HY, Ren B, Bafna V, Mischel PS. Circular ecDNA promotes accessible chromatin and high oncogene expression. Nature. 2019;575:699-703. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 342] [Cited by in RCA: 382] [Article Influence: 63.7] [Reference Citation Analysis (0)] |
| 17. | Paulsen T, Shibata Y, Kumar P, Dillon L, Dutta A. Small extrachromosomal circular DNAs, microDNA, produce short regulatory RNAs that suppress gene expression independent of canonical promoters. Nucleic Acids Res. 2019;47:4586-4596. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 99] [Cited by in RCA: 110] [Article Influence: 18.3] [Reference Citation Analysis (0)] |
| 18. | Paulsen T, Kumar P, Koseoglu MM, Dutta A. Discoveries of Extrachromosomal Circles of DNA in Normal and Tumor Cells. Trends Genet. 2018;34:270-278. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 154] [Cited by in RCA: 172] [Article Influence: 24.6] [Reference Citation Analysis (0)] |
| 19. | Hull RM, Houseley J. The adaptive potential of circular DNA accumulation in ageing cells. Curr Genet. 2020;66:889-894. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 11] [Cited by in RCA: 32] [Article Influence: 6.4] [Reference Citation Analysis (0)] |
| 20. | Zhu J, Zhang F, Du M, Zhang P, Fu S, Wang L. Molecular characterization of cell-free eccDNAs in human plasma. Sci Rep. 2017;7:10968. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 46] [Cited by in RCA: 83] [Article Influence: 10.4] [Reference Citation Analysis (0)] |
| 21. | Kumar P, Kiran S, Saha S, Su Z, Paulsen T, Chatrath A, Shibata Y, Shibata E, Dutta A. ATAC-seq identifies thousands of extrachromosomal circular DNA in cancer and cell lines. Sci Adv. 2020;6:eaba2489. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 98] [Cited by in RCA: 114] [Article Influence: 22.8] [Reference Citation Analysis (0)] |
| 22. | Su Z, Saha S, Paulsen T, Kumar P, Dutta A. ATAC-Seq-based Identification of Extrachromosomal Circular DNA in Mammalian Cells and Its Validation Using Inverse PCR and FISH. Bio Protoc. 2021;11:e4003. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 3] [Cited by in RCA: 13] [Article Influence: 3.3] [Reference Citation Analysis (0)] |
| 23. | Feng XJ, Yi HM, Ren XX, Ren JL, Ge JR, Wang FG. [Digital PCR and its application in biological detection]. Yi Chuan. 2020;42:363-373. [RCA] [PubMed] [DOI] [Full Text] [Cited by in RCA: 3] [Reference Citation Analysis (0)] |
| 24. | Mann L, Seibt KM, Weber B, Heitkam T. ECCsplorer: a pipeline to detect extrachromosomal circular DNA (eccDNA) from next-generation sequencing data. BMC Bioinformatics. 2022;23:40. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 6] [Cited by in RCA: 38] [Article Influence: 12.7] [Reference Citation Analysis (0)] |
| 25. | Quan PL, Sauzade M, Brouzes E. dPCR: A Technology Review. Sensors (Basel). 2018;18. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 374] [Cited by in RCA: 392] [Article Influence: 56.0] [Reference Citation Analysis (0)] |
| 26. | Lei Y, Yu T, Li C, Li J, Liang Y, Wang X, Chen Y. Expression of CAMK1 and its association with clinicopathologic characteristics in pancreatic cancer. J Cell Mol Med. 2021;25:1198-1206. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 8] [Cited by in RCA: 28] [Article Influence: 5.6] [Reference Citation Analysis (0)] |
| 27. | Volpin V, Michels T, Sorrentino A, Menevse AN, Knoll G, Ditz M, Milenkovic VM, Chen CY, Rathinasamy A, Griewank K, Boutros M, Haferkamp S, Berneburg M, Wetzel CH, Seckinger A, Hose D, Goldschmidt H, Ehrenschwender M, Witzens-Harig M, Szoor A, Vereb G, Khandelwal N, Beckhove P. CAMK1D Triggers Immune Resistance of Human Tumor Cells Refractory to Anti-PD-L1 Treatment. Cancer Immunol Res. 2020;8:1163-1179. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 9] [Cited by in RCA: 22] [Article Influence: 4.4] [Reference Citation Analysis (0)] |
| 28. | Dimitrova N, Gocheva V, Bhutkar A, Resnick R, Jong RM, Miller KM, Bendor J, Jacks T. Stromal Expression of miR-143/145 Promotes Neoangiogenesis in Lung Cancer Development. Cancer Discov. 2016;6:188-201. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 108] [Cited by in RCA: 118] [Article Influence: 13.1] [Reference Citation Analysis (0)] |
| 29. | Sui MH, Zhang WW, Geng DM, Sun DJ. CircPRKCI regulates proliferation, migration and cycle of lung adenocarcinoma cells by targeting miR-219a-5p-regulated CAMK1D. Eur Rev Med Pharmacol Sci. 2021;25:1899-1909. [RCA] [PubMed] [DOI] [Full Text] [Cited by in RCA: 1] [Reference Citation Analysis (0)] |
| 30. | Wang L, Lin Y, Meng H, Liu C, Xue J, Zhang Q, Li C, Zhang P, Cui F, Chen W, Jiang A. Long non-coding RNA LOC283070 mediates the transition of LNCaP cells into androgen-independent cells possibly via CAMK1D. Am J Transl Res. 2016;8:5219-5234. [PubMed] |