Published online Jun 15, 2023. doi: 10.4251/wjgo.v15.i6.1005
Peer-review started: February 1, 2023
First decision: February 16, 2023
Revised: February 27, 2023
Accepted: April 17, 2023
Article in press: April 17, 2023
Published online: June 15, 2023
Processing time: 133 Days and 18.5 Hours
Transfer RNA (tRNA)-derived small RNAs (tsRNAs) are small fragments that form when tRNAs severe. tRNA halves (tiRNAs), a subcategory of tsRNA, are involved in the oncogenic processes of many tumors. However, their specific role in sessile serrated lesions (SSLs), a precancerous lesion often observed in the colon, has not yet been elucidated.
To identify SSL-related tiRNAs and their potential role in the development of SSLs and serrated pathway of colorectal cancer (CRC).
Small-RNA sequencing was conducted in paired SSLs and their adjacent normal control (NC) tissues. The expression levels of five SSL-related tiRNAs were validated by q-polymerase chain reaction. Cell counting kit-8 and wound healing assays were performed to detect cell proliferation and migration. The target genes and sites of tiRNA-1:33-Pro-TGG-1 (5′tiRNA-Pro-TGG) were predicted by TargetScan and miRanda algorithms. Metabolism-associated and immune-related pathways were analyzed by single-sample gene set enrichment analysis. Functional analyses were performed to establish the roles of 5′tiRNA-Pro-TGG based on the target genes.
In total, we found 52 upregulated tsRNAs and 28 downregulated tsRNAs in SSLs compared to NC. The expression levels of tiRNA-1:33-Gly-CCC-2, tiRNA-1:33-Pro-TGG-1, and tiRNA-1:34-Thr-TGT-4-M2 5′tiRNAs were higher in SSLs than those in NC, while that of 5′tiRNA-Pro-TGG was associated with the size of SSLs. It was demonstrated that 5′tiRNA-Pro-TGG promoted cell proliferation and migration of RKO cell in vitro. Then, heparanase 2 (HPSE2) was identified as a potential target gene of 5′tiRNA-Pro-TGG. Its lower expression was associated with a worse prognosis in CRC. Further, lower expression of HPSE2 was observed in SSLs compared to normal controls or conventional adenomas and in BRAF-mutant CRC compared to BRAF-wild CRC. Bioinformatics analyses revealed that its low expression was associated with a low interferon γ response and also with many metabolic pathways such as riboflavin, retinol, and cytochrome p450 drug metabolism pathways.
tiRNAs may profoundly impact the development of SSLs. 5′tiRNA-Pro-TGG potentially promotes the progression of serrated pathway CRC through metabolic and immune pathways by interacting with HPSE2 and regulating its expression in SSLs and BRAF-mutant CRC. In the future, it may be possible to use tiRNAs as novel biomarkers for early diagnosis of SSLs and as potential therapeutic targets in serrated pathway of CRC.
Core Tip: Our study identified the transfer RNA-derived small RNAs expression profile of sessile serrated lesions (SSLs) for the first time and found that tRNA halves (tiRNAs)-1:33-Pro-TGG-1, which was associated with polyp size, were highly expressed in SSLs and promoted oncogenesis in colorectal cancer cell. Furthermore, tiRNA-1:33-Pro-TGG-1 potentially promotes the progression of serrated pathway colorectal cancer (CRC) through metabolic and immune pathways by interacting with HPSE2 in SSLs and BRAF mutant CRC. In the future, tiRNA-1:33-Pro-TGG-1 may serve as a potential target for the early diagnosis of SSLs and treatment of CRC that arises from the serrated pathway.
- Citation: Wang XY, Zhou YJ, Chen HY, Chen JN, Chen SS, Chen HM, Li XB. 5’tiRNA-Pro-TGG, a novel tRNA halve, promotes oncogenesis in sessile serrated lesions and serrated pathway of colorectal cancer. World J Gastrointest Oncol 2023; 15(6): 1005-1018
- URL: https://www.wjgnet.com/1948-5204/full/v15/i6/1005.htm
- DOI: https://dx.doi.org/10.4251/wjgo.v15.i6.1005
Colorectal cancer (CRC) typically develops from colorectal polyps. there are two main categories: Conventional adenomas (ADs) and serrated lesions (SLs)[1]. SLs, precancerous lesions often observed in the colorectum, are again of four subtypes: (1) Sessile SLs with or without dysplasia (SSLs-D and SSLs, respectively); (2) Traditional serrated adenomas; (3) Hyperplastic polyps; and (4) Unclassified serrated adenomas[2].
The association between common colorectal polyps and the development of CRC has been well-studied. In recent years, a large number of studies have also confirmed the malignant potential of SLs, in particular, that of SSLs[3]. Unlike ADs, which tend to develop into microsatellite-stability or micro
Recent research has revealed the essential role of non-coding RNAs, including long non-coding RNAs, transfer (t) RNAs, and micro (mi) RNAs, in the development and progression of various diseases[10-12]. tRNAs play an important role in the translation of proteins by transporting amino acids to the ribosome[13]. However, under conditions of nutritional, physicochemical, and oxidative stress, cells selectively reduce protein synthesis to conserve energy. Under these situations, tRNAs may be enzymatically cleaved to form tRNA-derived small RNAs (tsRNAs)[14,15]. tsRNAs can be classified into two categories based on their length and enzymatic cleavage sites: tRNA halves (tiRNAs) and tRNA-related fragments (tRFs). tiRNAs comprise 31–40 nucleotides (nt). They form when mature tRNAs are cleaved in the anticodon loop region comprising 5′tiRNAs and 3′tiRNAs. tRFs are 14–30 nt in length and form from mature or precursor tRNA. tRFs have four isoforms: tRF-1, tRF-2, tRF-3, and tRF-5.
It was reported in the last decade that tsRNAs are involved in the regulation of several physiological processes. For example, 5′tiRNA-GLY can promote the proliferative and invasive capabilities of papillary thyroid cancer cells by binding to RBM17[16]. Other studies have shown that tRF-Val can attach directly to binding protein EEF1A1 to promote proliferation and inhibit apoptosis of gastric cancer cells[17]. It was also reported that tRF-Gly promotes migration of hepatocellular carcinoma cells by binding to NDFIP2 in liver cancer[18]. Thus, although the role of tsRNAs in diseases remains an interesting research topic, dysregulation of their expression may present possible biomarkers for many diseases, such as CRC and breast cancer[19-21]. However, the expression and function of tsRNAs and small tRNA-derived fragments in colorectal polyps, particularly those of SSLs, have not yet been explored.
This study aims to identify the expression profiles of tsRNAs in SSLs and paired normal control (NC) tissues using small-RNA sequencing and their potential role in the development of SSLs and serrated pathway of CRC.
Twenty paired SSL tissues and adjacent normal tissues belonging to the same patient were collected from Renji Hospital, School of Medicine, Shanghai JiaoTong University (China). The inclusion criteria were as follows: Age, ≥ 18 years; adequate bowel preparation and cecum reach; and received a colonoscopy with an assured diagnosis of SSL by experienced endoscopists. Two pathologists confirmed the diagnosis using biopsy specimens according to the 2019 World Health Organization 5th classification. Our study was approved by the Ethics Committee of Renji Hospital No. KY2021-004.
RNA sequencing (RNA-Seq) data of SSLs, ADs, and the corresponding control tissue were obtained from GSE76987 from the Gene Expression Omnibus database (https://www.ncbi.nlm.nih.gov/gds/). The gene expression data of colorectal adenocarcinoma were downloaded from The Cancer Genome Atlas Program (TCGA, https://portal.gdc.cancer.gov/) and cBioPortal for Cancer Genomics (https://www.cbioportal.org/).
Total RNA was extracted from fresh tissues stored in RNA using TRIzol (Invitrogen, CA, United States). The purity and concentration of the total RNA samples were determined with NanoDrop ND-1000 (Thermo Fisher Scientific, DE, United States).
RNA samples were extracted from four paired SSL tissues and adjacent normal tissues. The purity and concentration of the total RNA samples were determined before conducting small-RNA sequencing, as mentioned before. Next, a commercial RNA pretreatment kit (rtStar™ tRF and tiRNA Pretreatment Kit, AS-FS-005, Arraystar Inc., MD, United States) for tRF and tiRNA-seq library preparation was used, which was then used to remove some RNA modifications that interfered with small RNA-seq library construction, including 3′-aminoacyl (charged) deacylation to 3′-OH for 3′adaptor ligation, 3′-cP (2′,3′-cyclic phosphate) removal to 3′-OH for 3′adaptor ligation, 5′-OH (hydroxyl group) phosphorylation to 5′-P for 5′-adaptor ligation, m1A and m3C demethylation for reverse transcription, cDNA synthesis, and library polymerase chain reaction (PCR) amplification. The prepared RNA of each sample was ligated to 3′ and 5′ small-RNA adapters. Then, cDNA was synthesized and amplified using proprietary reverse transcription primers and amplification primers (Illumina). Subsequently, approximately 134–160 bp PCR-amplified fragments were extracted and purified using the PAGE gel. The concentration and quality of the libraries were assessed via absorbance spectrometry on Agilent BioAnalyzer 2100 (Agilent Technologies Inc., CA, United States). The libraries were denatured and diluted to a loading volume of 1.3 mL and a loading concentration of 1.8 pM. Then, they were loaded onto a reagent cartridge and forwarded to sequencing run on the Illumina NextSeq 500 system using NextSeq 500/550 V2 kit (FC-404- 2005, Illumina), according to the manufacturer′s instructions. Raw sequencing read data that passed the Illumina chastity filter were used for subsequent analysis. Trimmed reads (with 5′,3′-adaptor bases removed) were aligned to mature-tRNA and pre-tRNA reference sequences. Statistical analysis of the alignment results was applied to retain the valid sequences for subsequent tRF and tiRNA expression profiling analysis.
Sequencing quality was examined using the FastQC software (v0.11.7) and trimmed reads were aligned allowing for only one mismatch to mature-tRNA sequences. The reads that do not map are aligned allowing for only one mismatch to precursor tRNA sequences using the Bowtie software (v1.2.2, http://bowtie-bio.sourceforge.net/index.shtml). The abundance of tRF and tiRNA was evaluated using their sequencing counts and is normalized as counts per million of the total aligned reads. The differentially expressed tRFs and tiRNAs were screened based on the count value with R package edgeR. The R packages (R 4.1.2), including FactoMineR, factoextra, ggvenn, pheatmap, and ggplot2, were used for principal component analysis (PCA), Venn plots, Hierarchical clustering heatmap analysis, and Volcano plots.
Total RNA collected from 16 paired SSLs and adjacent normal tissues was extracted using TRIzol (Invitrogen, CA, United States), as stated previously. tiRNAs were reverse-transcribed into cDNA using a Bulge-Loop miRNA qRT-PCR Starter Kit (Ribobio, Guangzhou, China). Subsequently, qPCR was performed with SYBR Premix Ex Taq (Takara), as instructed by the manufacturer. The expression levels of tiRNAs were normalized to that of U6. The primers of qPCR were as follows: HPSE2-F: 5′-ATGGCCGGGCAGTAAATGG-3′; HPSE2-R: 5′-GCTGGCTCTGGAATAAATCCG -3′; ACTB-F: 5′-CACCATTGGCAATGAGCGGTTC-3′, and ACTB-R: 5′-AGGTCTTTGCGGATGTCCACGT-3′. Other primers involved in reverse transcription and qPCR were purchased from RiboBio (China).
The RKO cell line was purchased from the Typical Culture Preservation Commission Cell Bank, Chinese Academy of Sciences (Shanghai, China). The cell was cultured in the RPMI 1640 medium with 10% fetal bovine serum (Gibco, United States) at 37 °C with 5% CO2.
The RKO cell was seeded in plates (Corning Life Sciences, United States) the day before transfection. The 5′tiRNA-Pro-TGG mimic and inhibitor (50 nM), both modified with 2′-O-methyl, were purchased from GenePharma Technology (Shanghai, China) and transfected using DharmaFECT 1 siRNA transfection reagent (Thermo Fisher Scientific Dharmacon Inc., United States). The corresponding scramble sequences were used as negative controls. The RNA oligonucleotide sequences were as follows: 5′tiRNA-Pro-TGG mimic: 5′-GGCUCGUUGGUCUAGUGGUAUGAUUCUCGCUUU-3′; 5′tiRNA-Pro-TGG inhibitor: 5′-AAAGCGAGAAUCAUACCACUAGACCAACGAGCC-3′; mimic scramble control: 5′-ACGUUUGACCUGUGUCGAGUUUUCUGUUUGGCG-3′; and inhibitor scramble control: 5′-GGGAAAGCGAAUAAAUCCAAACACCCAAUCCGC-3′.
Cell proliferation was measured by CCK-8 (Dojindo, Japan). The RKO cell was seeded in 96-well plates at a density of 2 × 103 cells per well. After transfection for 48 h, 10 μL of CCK-8 solution and 100 μL of the RPMI 1640 medium per well were added to the wells after discarding the previous medium; the OD values (450 nm) were measured after 2 h. All assays were conducted three times.
The cells were inoculated in a 6-well plate. When 90% confluence was reached, a sterile 200-µL pipette tip was used to create vertical wounds. Finally, the wells were photographed under a microscope (Olympus, Japan) at × 200 magnification. The pictures were analyzed by ImageJ. All assays were conducted three times.
The ssGSEA analysis was used to investigate the expression levels of immune- and metabolism-related pathways by GSVA R package. Next, 41 metabolism pathway gene sets and 29 immune pathway gene sets were obtained from Molecular Signatures Database (MSigDB; https://www.gsea-msigdb.org/).
To investigate the potential biological function of dysregulated tiRNAs in SSLs, the target gene predictions were conducted by TargetScan (http://www.targetscan.org/vert72/) and Miranda (http://www.microrna.org/microrna/) with a context score < -0.1. KEGG pathway and GO analyses to the target gene sets were performed by using the clusterProfiler R package.
Mean and standard deviations (mean ± SD) were used to analyze all quantitative variables. Two-tailed Student′s t-tests and Wilcoxon rank test were performed. A P value < 0.05 was considered statistically significant. Spearman correlation analysis was used to determine the relationship between 5′tiRNAs and polyp size. Pearson correlation analysis was used to determine the relationship between tiRNA-1:33-Pro-TGG-1 and HPSE2. Kaplan–Meier survival analysis was performed to evaluate the association between the HPSE2-expression level and the overall survival of CRC patients. All analyses were performed by GraphPad Prism 9.3.1 (GraphPad Software, United States).
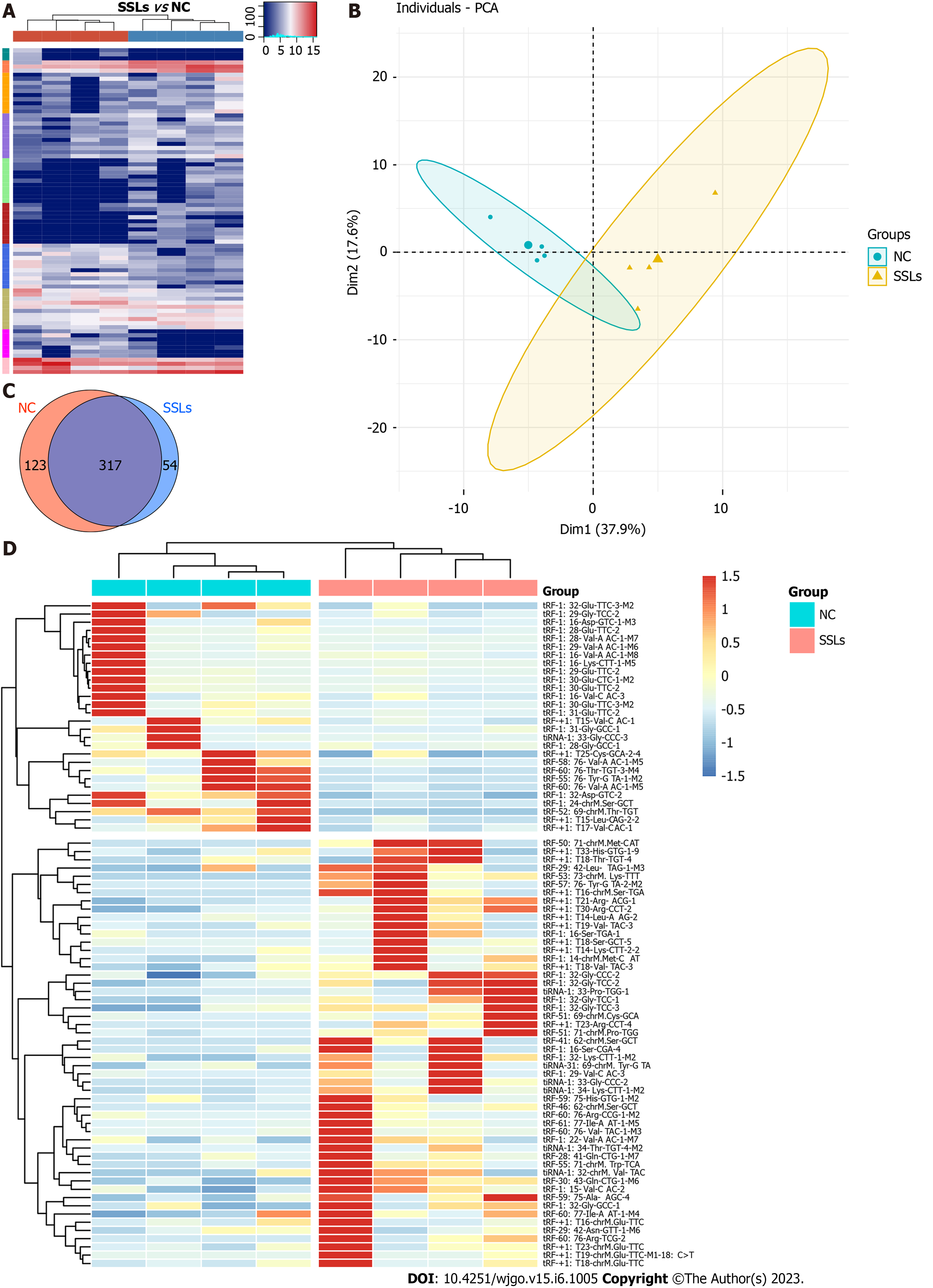
To identify the expression profiles of tRFs and tiRNAs in patients with SSLs, four pairs of SSLs and the corresponding NC tissues from different patients were collected for small-RNA sequencing analysis. Variations in all tRFs and tiRNAs expressed in SSL and NC groups are shown using a heatmap (Figure 1A). The expression levels of tRFs and tiRNAs in SSLs were different from those in NC groups, as determined by PCA (Figure 1B). We used a Venn diagram to show the tRFs and tiRNAs that were both generally and specifically expressed between the SSL and NC groups (Figure 1C). As seen in Figure 1C, 54 types of tRFs and tiRNAs were exclusively found in SSLs, while 123 types were found only in NCs. Differences in tRFs and tiRNAs found in SSLs and NCs were determined under the following conditions: absolute log2 (fold change) ≥ 1.5 and P < 0.05. Under these conditions, we identified 52 upregulated and 28 downregulated tRFs and tiRNAs and have shown them in a hierarchical cluster heatmap (Figure 1D).
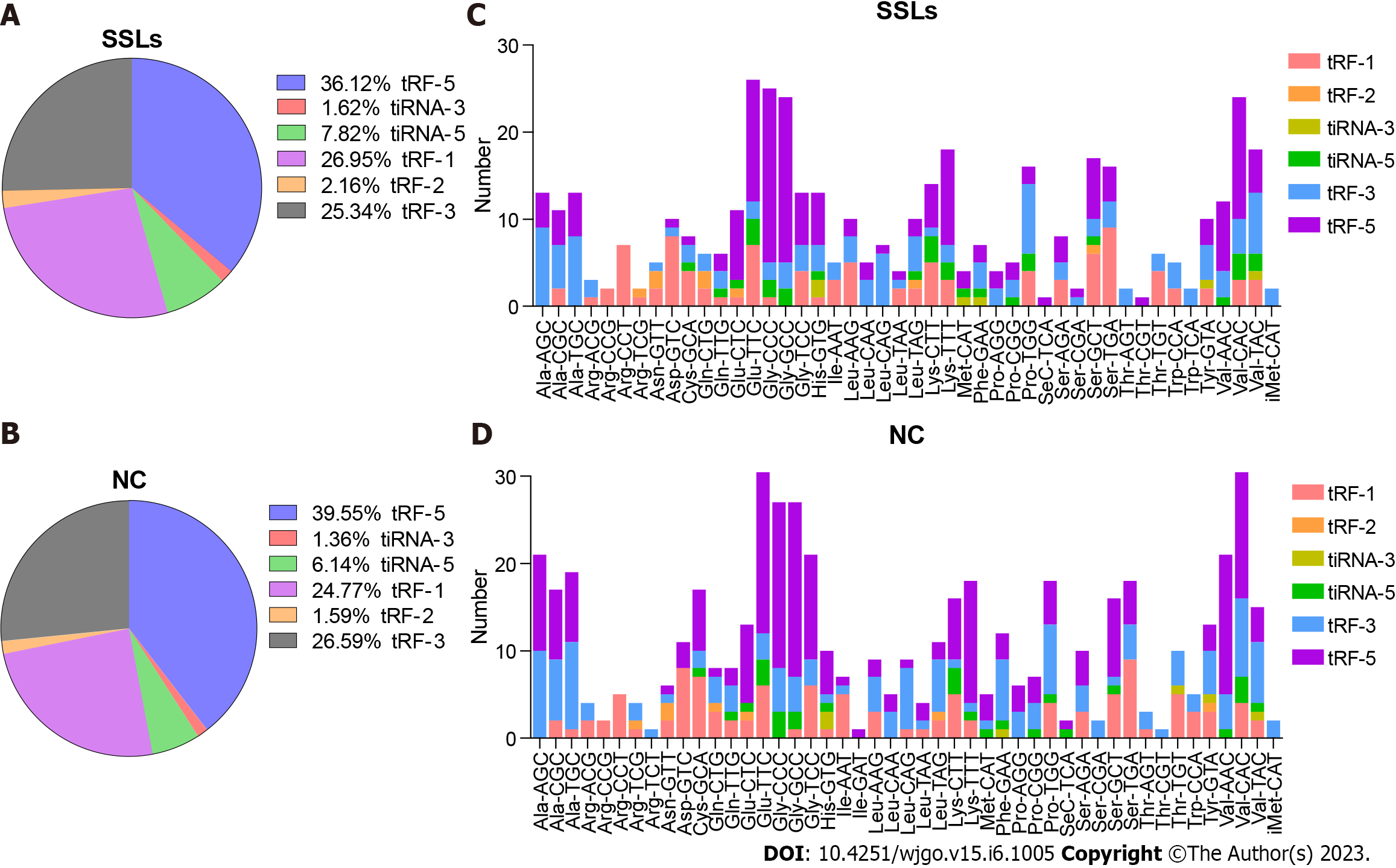
The expression of tRF-1 and 5′tiRNA increased, while that of tRF-5c and tRF-3b decreased in SSLs compared with that of NCs (Figure 2A and B). tRNAs with the same anticodon translate the same amino acid. Hence, we herein separately determined the number of different tRFs and tiRNAs with the same anticodon (Figure 2C and D).
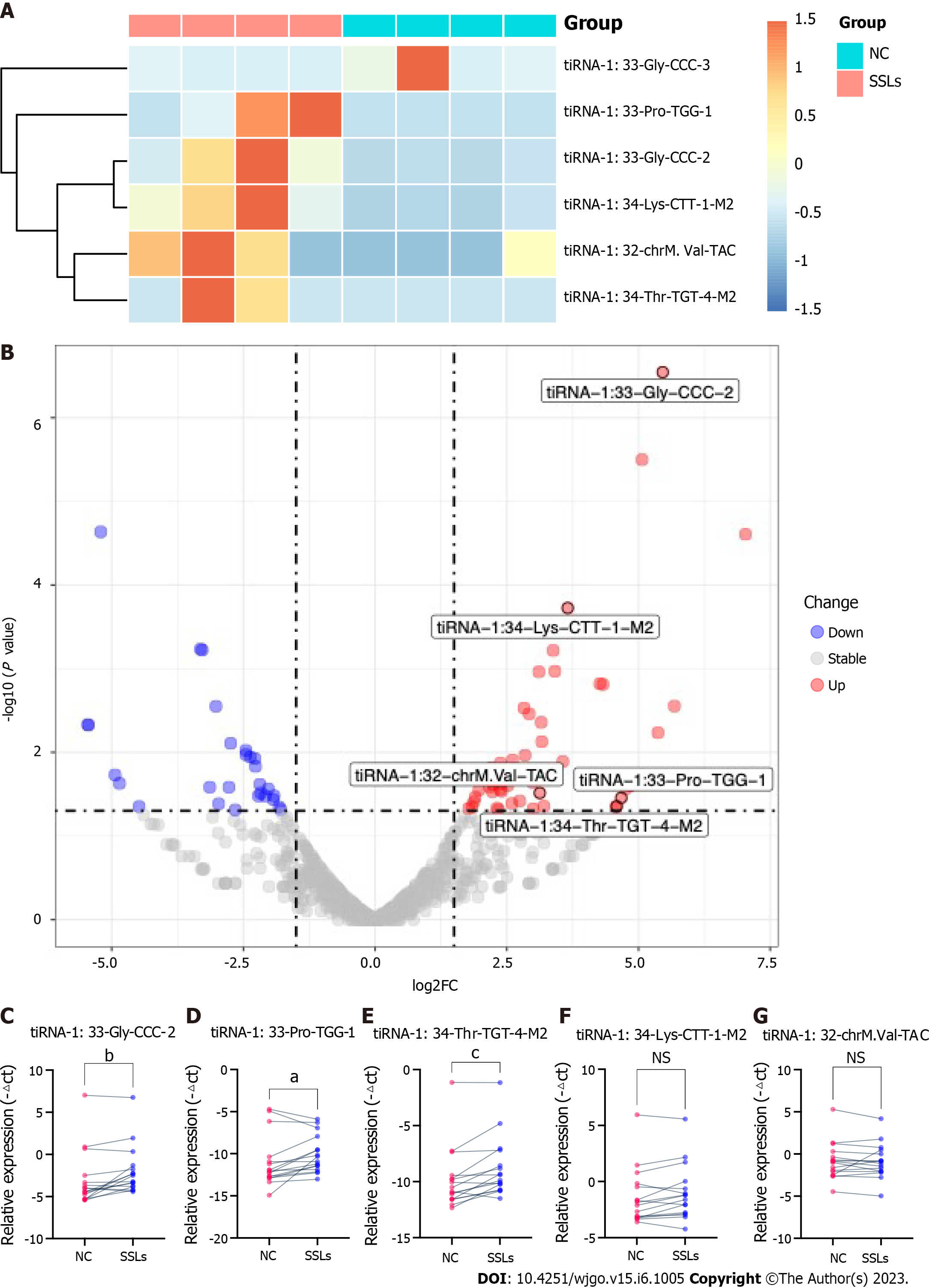
5′tiRNAs play an important role in the development of many diseases, including CRC. Because our analysis showed a significant increase in tiRNA-5 in SSLs compared to that in NCs, we further verified the expression of 5′tiRNA in SSLs. Our previous screening criteria showed that six 5′tiRNAs (five upregulated and one downregulated) with different expression levels emerged between SSLs and NCs. tiRNA-1:33-Gly-CCC-2, tiRNA-1:33-Pro-TGG-1, tiRNA-1:34-Thr-TGT-4-M2, tiRNA-1:34-Lys-CTT-1-M2, and tiRNA-1:32-chrM.Val-TAC were upregulated in SSLs with 43.99-, 25.50-, 24.00-, 12.61-, and 8.73-fold change, respectively (P < 0.05), while tiRNA-1:33-Gly-CCC-3 was downregulated with a 36.95-fold change in SSLs compared to that in NCs (P < 0.05, Figure 3A and B).
Since we previously reported increased levels of 5′tiRNA expression in SSLs (Figure 2A and B), we herein focus on the abovementioned five upregulated 5′tiRNAs in SSLs. To further validate our sequencing data, we collected 16 pairs of SSLs and the corresponding NCs to confirm the expressions of the five upregulated 5′tiRNAs using RT-PCR (Figure 3C-G). The size of all collected lesions ranged from 4 to 15 mm, with an average of 6.31 ± 3.07 mm. tiRNA-1:33-Gly-CCC-2, tiRNA-1:33-Pro-TGG-1, and tiRNA-1:34-Thr-TGT-4-M2 were significantly upregulated in SSLs compared to those in the paired NC (P = 0.0059, 0.0309, and 0.0008, respectively). The expression of tiRNA-1:34-Lys-CTT-1-M2 and tiRNA-1:32-chrM.Val-TAC did not show any statistically significant differences between SSLs and NCs (P = 0.0641 and 0.9838, respectively).
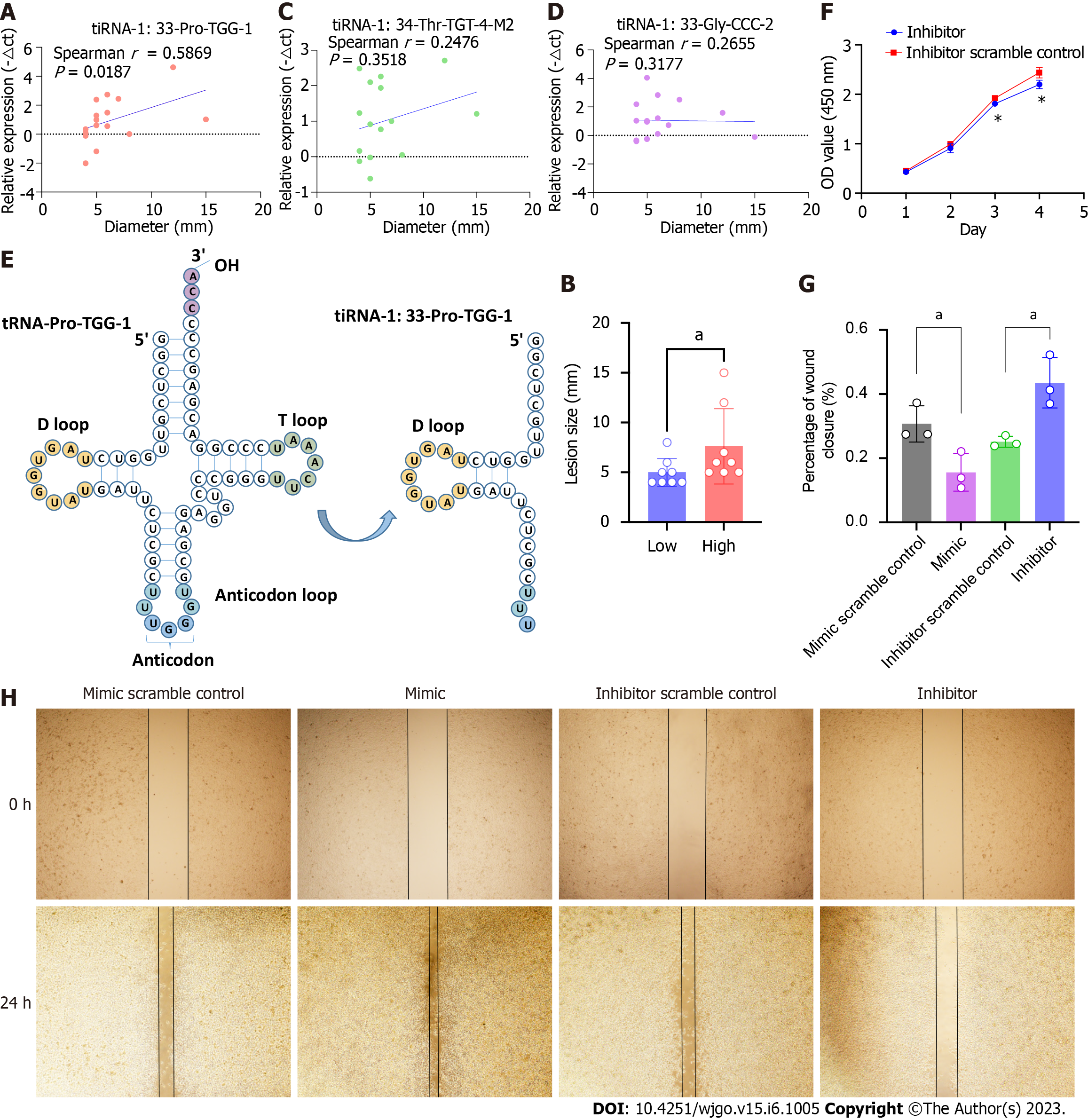
We further analyzed the correlation between the lesion size and the expression levels of the three 5′tiRNAs that had been validated as significantly highly expressed in SSLs. It was tiRNA-1:33-Pro-TGG-1 (Figure 4A and B), not tiRNA-1:33-Gly-CCC-2 or tiRNA-1:34-Thr-TGT-4-M2 (Figure 4C and D), that positively correlated with lesion size. Therefore, we focused on tiRNA-1:33-Pro-TGG-1, also known as 5′tiRNA-Pro-TGG. It comprises 33 nucleotides and is a type of 5′tiRNA that originated in tRNA-Pro-TGG-1 (Figure 4E). The inhibition of 5′tiRNA-Pro-TGG reduces the proliferation (Figure 4F) and migratory capacity of cancer cells in RKO, a colon cancer cell line carrying the BRAF V600E mutation, while its overexpression enhanced the migratory capacity of the cancer cells (Figure 4G and H).
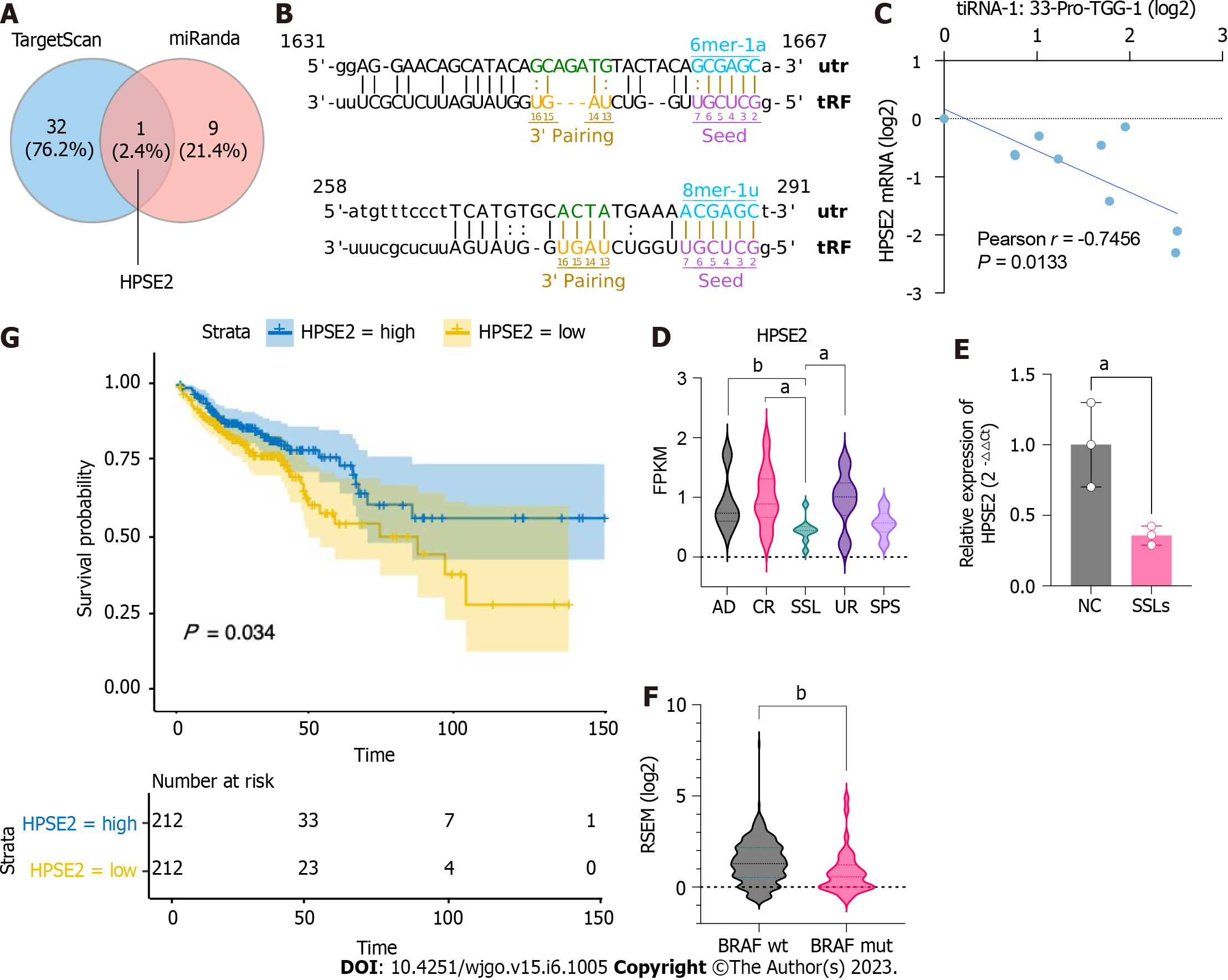
To further investigate the potential functions of upregulated 5′tiRNAs in the progression of SSLs, we identified potential target genes that might bind to 5′tiRNA-Pro-TGG using TargetScan and miRanda algorithms and could predict 502 target genes. When context plus score < -0.5 and structure score > 300, the filtering parameters of the TargetScan and miRanda algorithms were predicted to be 33 and 10 target genes, respectively, with the intersection at HPSE2 (Figure 5A). Two possible binding sites were predicted within the 3′UTR of HPSE2 for the seed regions of 5′tiRNA-Pro-TGG (Figure 5B). HPSE2 encodes heparinase II. A mutation in HPSE2 is responsible for the urofacial syndrome and has been progressively identified as a tumor suppressor. We examined the expression levels of 5′tiRNA-Pro-TGG and HPSE2 and found a significant negative correlation between their expression levels (Figure 5C). We also found that the expression level of HPSE2 in SSLs was significantly lower than that in the uninvolved right colon, control right colon tissue, and common adenoma (P < 0.05; Figure 5D). The qPCR results also confirmed that the expression level of HPSE2 in SSLs was lower than that in NC (P < 0.05; Figure 5E). Not coincidentally, the HPSE2 expression level was lower in CRC lesions carrying BRAF mutations than those with BRAF wild-type CRC (Figure 5F). An analysis of survival outcomes in CRC patients demonstrated that the lower level of HPSE2 was associated with poorer prognosis (Figure 5G).
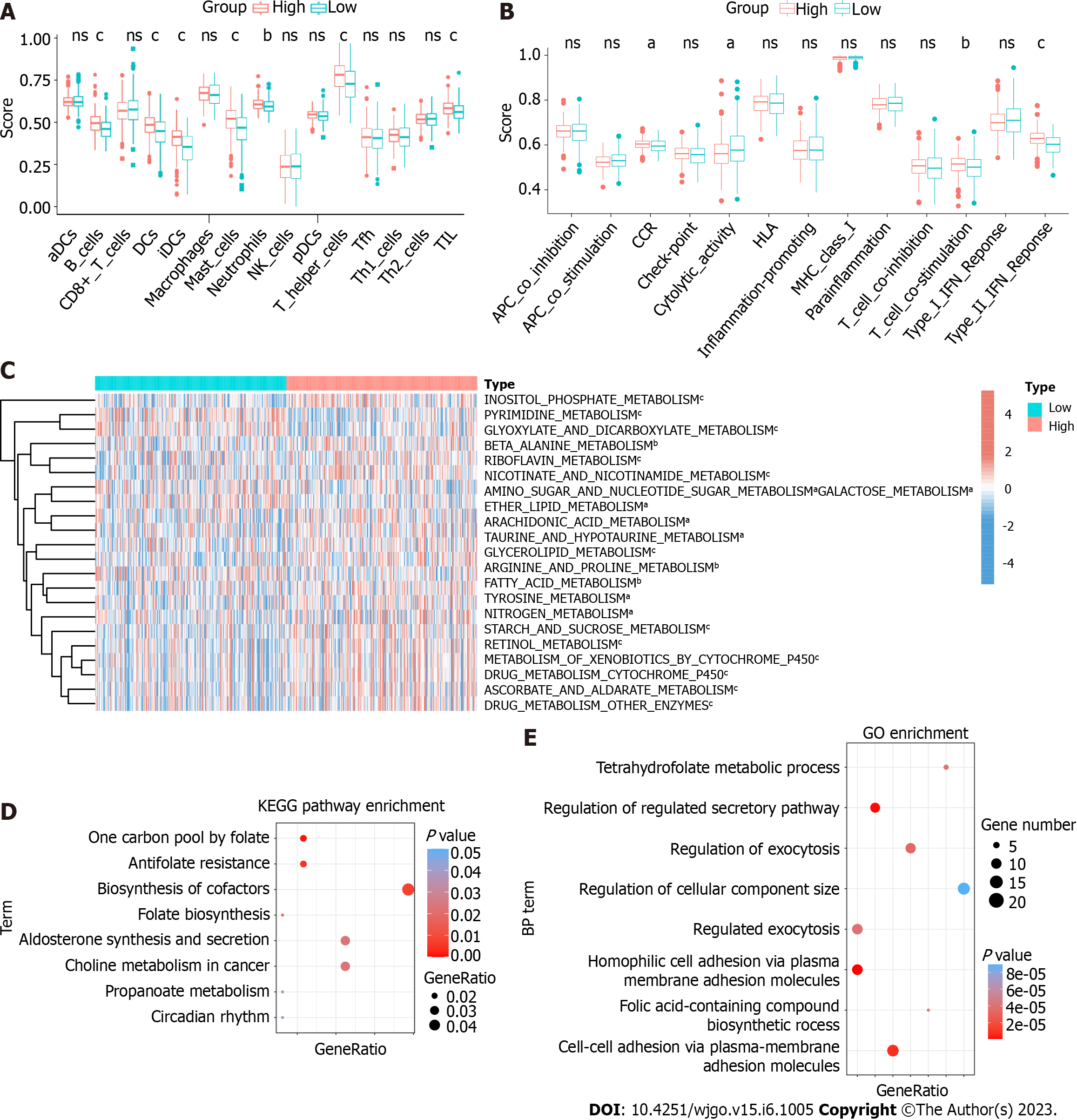
To explore the function of HPSE2 in the development of serrated pathway of CRC, we grouped CRC patients into HPSE2 high (HPSE2-high) and low (HPSE2-low) expression groups. We then scored both groups for the immune cell type (Figure 6A), immune cell function (Figure 6B), and metabolic pathways (Figure 6C) using the ssGSEA algorithm. In the HPSE2-low group, a part of the immune cell scores were lower, including that for tumor infiltration lymphocyte (TIL), dendritic cells, T helper cells, B cells, and mast cells. The scores for response to interferon γ (IFNγ) and T-cell co-stimulation were lower in the HPSE2-low group than they were in the HPSE2-high group. Notably, many metabolic pathways scored lower in the HPSE2-low-expression group, implying the downregulation of these pathways, including that of riboflavin, retinol, and cytochrome P450 drug metabolism pathways. However, the metabolism of glyoxylate, dicarboxylate, and pyrimidine upregulated in the HPSE2-low group.
We next performed KEGG and GO enrichment analyses using the potential target genes of 5′tiRNA-Pro-TGG. KEGG enrichment analysis revealed that these target genes could be involved in pathways such as the biosynthesis of cofactors, antifolate resistance, and choline metabolism in cancer (Figure 6D). GO enrichment analysis demonstrated the possible target genes involved in cell-to-cell adhesion and regulation of the secretory pathway and exocytosis (Figure 6E).
Formation of SSLs and the process by which they progress to CRC is known as the serrated neoplastic pathway. However, the mechanisms and processes involved in this pathway are still not fully understood. SSLs that progress to CRC have a prevalence of 10%–15% in the general population. The main pathology of this type of cancer involves a structurally distorted serrated crypt, with a BRAF mutation (BRAFmut), MSI-H, and CpG island methylator phenotype-high[22]. SSLs presenting with BRAFmut often develop into CRC with a worse prognosis[23]. Therefore, early diagnosis of SSLs can help in reducing the incidence of BRAFmut CRC. In addition, understanding the pathogenesis of SSLs can help identify new targets for intervention in the early and precise treatment of BRAFmut CRC.
Studies conducted in the last decade reported that tsRNAs can play an important role as a biomarker in colon cancer[24], and that 5′-tiRNA-Pro-TGG levels can be used as an independent prognostic marker in CRC for predicting its recurrence[20]. Another study showed that in CRC, higher levels of tRF-phe-GAA-031 and tRF-VAL-TCA-002 expression were associated with reduced survival. Hence, they could also be used as prognostic predictors of CRC[19]. Therefore, the present study investigated the expression levels of tsRNAs, specifically that of 5′tiRNAs, in SSLs and their potential biological roles. We sequenced small RNAs from SSLs and their corresponding NCs, and found that among the 80 dysregulated tsRNAs, 5′tiRNAs, 3′tiRNAs, and tRFs-1 were more highly expressed in the SSLs, which suggests that tiRNAs may play an important role in these lesions.
Further analysis confirmed tiRNA-1:33-Gly-CCC-2, tiRNA-1:33-Pro-TGG-1, and tiRNA-1:34-Thr-TGT-4-M2 to be significantly upregulated in SSLs with a 2.92-, 3.69-, and 2.37-fold change, respectively. The expression level of 5′tiRNA-Pro-TGG was positively correlated with the size of SSLs. In addition, 5′tiRNA-Pro-TGG promoted carcinogenic processes in the colon cancer cell. We further screened HPSE2 as the potential target gene. Interestingly, we found that HPSE2 appeared to be specifically hypo-expressed in SSLs, as well as in BRAF-mutant CRC, and its low expression predicted lower survival. 5′tiRNA-Pro-TGG is associated with poor prognosis in CRC[20]. HPSE2, a novel tumor-suppressor gene, has been reported to have reduced expression levels and poor prognosis in colon and breast cancers[25,26]. Our study identifies for the first time the specific low expression of HPSE2 in SSLs and BRAFmut CRC and reveals that it may play an essential role in the serrated pathway but not in other colorectal carcinogenesis pathways. To the best of our knowledge, this study is the first to report the high expression of 5′tiRNA-Pro-TGG in SSLs and its potential regulatory relationship with HPSE2.
Analysis of immune cells and their functions suggested that HPSE2 is involved in regulating the functions of various immune cells in CRC, including TIL, particularly in response to IFNγ. IFNγ promotes antigen presentation and tumor killing[27]. Our finding that patients with low HPSE2 had lower IFNγ-response scores suggested that it might be involved in regulating the tumor immune escape. Metabolic analysis revealed that HPSE2 could downregulate various metabolic pathways, such as riboflavin and retinol metabolism. Riboflavin may reduce the risk of CRC in women[28]. In addition, a negative association between the plasma retinol concentration and the risk of proximal colon cancer has also been reported[29]. Recently, a lack of retinoic acid synthesis was found to promote the accumulation of myeloid-derived suppressor cells (MDSC) in CRC, thereby mediating the immune escape, while exogenous retinoic acid supplementation in an in vitro model attenuated the polymorphonuclear MDSC production[30].
Our study has some limitations. Firstly, more in vitro and in vivo experiments are needed to validate the function of the tiRNAs discussed. Secondly, because of the low prevalence of SSLs and their frequent neglect by endoscopists, it became difficult to collect a large number of samples. In addition, the expression level of 5′tiRNA-Pro-TGG and its association with recurrence and prognosis in SSL patients require further studies in large samples.
Our study is the first to identify the tsRNA expression profile of SSLs. It also reported that tiRNA-1:33-Gly-CCC-2, tiRNA-1:33-Pro-TGG-1, and tiRNA-1:34-Thr-TGT-4-M2 were highly expressed in SSLs. tiRNA-1:33-Pro-TGG-1 potentially promotes the serrated pathway for CRC progression through metabolic and immune pathways by interacting with HPSE2 in SSLs and BRAFmut CRC. Our results showed that tiRNA-1:33-Pro-TGG-1 may serve as a potential target for early diagnosis of SSLs and treatment of CRC arising from the serrated pathway.
tRNA halves (tiRNAs), a subcategory of tRNA-derived small RNAs (tsRNA), are involved in the oncogenic processes of many tumors, yet their specific role in sessile serrated lesions (SSLs) has not yet been elucidated.
The motivation for this study is to identify SSL-related tiRNAs and their potential role in the development of SSLs and serrated pathways in colorectal cancer (CRC).
Endoscopic detection of SSLs is very difficult and we do not know much about the exact mechanisms through which SSLs develop and how they progress to CRC in present.
Small-RNA sequencing was conducted in paired SSLs and their adjacent normal control (NC) tissues. The expression levels of five SSL-related tiRNAs were validated by q-polymerase chain reaction. Cell counting kit and wound healing assays were performed to detect cell proliferation and migration. The target genes and sites of tiRNA-1:33-Pro-TGG-1 (5′tiRNA-Pro-TGG) were predicted by TargetScan and miRanda algorithms. Metabolism-associated and immune-related pathways were analyzed by single-sample gene set enrichment analysis. Functional analyses were performed to establish the roles of 5′tiRNA-Pro-TGG based on the target genes.
The expression levels of tiRNA-1:33-Gly-CCC-2, tiRNA-1:33-Pro-TGG-1, and tiRNA-1:34-Thr-TGT-4-M2 5′tiRNAs were higher in SSLs than those in NC, while that of 5′tiRNA-Pro-TGG was associated with the size of SSLs. It was demonstrated that 5′tiRNA-Pro-TGG promoted cell proliferation and migration of RKO cell in vitro. Then, heparanase 2 (HPSE2) was identified as a potential target gene of 5′tiRNA-Pro-TGG. Its lower expression was associated with a worse prognosis in CRC. Further, lower expression of HPSE2 was observed in SSLs compared to NC or adenomas and in BRAF-mutant CRC compared to BRAF-wild CRC. Bioinformatics analyses revealed that its low expression was associated with a low interferon γ response and also with many metabolic pathways such as riboflavin, retinol, and cytochrome p450 drug metabolism pathways.
5′tiRNA-Pro-TGG potentially promotes the progression of serrated pathway CRC through metabolic and immune pathways by interacting with HPSE2 and regulating its expression in SSLs and BRAF-mutant CRC.
In the future, it may be possible to use 5′tiRNA-Pro-TGG as a novel biomarker for early diagnosis of SSLs and as a potential therapeutic target in serrated pathway of CRC.
The authors are grateful to all the individuals who participated in the study.
Provenance and peer review: Unsolicited article; Externally peer reviewed.
Peer-review model: Single blind
Specialty type: Oncology
Country/Territory of origin: China
Peer-review report’s scientific quality classification
Grade A (Excellent): A
Grade B (Very good): B
Grade C (Good): 0
Grade D (Fair): 0
Grade E (Poor): 0
P-Reviewer: Elpek GO, Turkey; Zhang Z, China S-Editor: Li L L-Editor: A P-Editor: Zhao S
| 1. | Crockett SD, Nagtegaal ID. Terminology, Molecular Features, Epidemiology, and Management of Serrated Colorectal Neoplasia. Gastroenterology. 2019;157:949-966.e4. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 142] [Cited by in RCA: 237] [Article Influence: 39.5] [Reference Citation Analysis (0)] |
| 2. | Ahadi M, Sokolova A, Brown I, Chou A, Gill AJ. The 2019 World Health Organization Classification of appendiceal, colorectal and anal canal tumours: an update and critical assessment. Pathology. 2021;53:454-461. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 16] [Cited by in RCA: 73] [Article Influence: 18.3] [Reference Citation Analysis (0)] |
| 3. | Meester RGS, Ladabaum U. Sessile serrated polyps and colorectal cancer mortality. Lancet Gastroenterol Hepatol. 2020;5:516-517. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 4] [Cited by in RCA: 4] [Article Influence: 0.8] [Reference Citation Analysis (0)] |
| 4. | Bell PD, Anderson JC, Srivastava A. The Frontiers of Serrated Polyps. Am J Surg Pathol. 2022;46:e64-e70. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1] [Cited by in RCA: 7] [Article Influence: 1.8] [Reference Citation Analysis (0)] |
| 5. | Yang HM, Mitchell JM, Sepulveda JL, Sepulveda AR. Molecular and histologic considerations in the assessment of serrated polyps. Arch Pathol Lab Med. 2015;139:730-741. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 34] [Cited by in RCA: 38] [Article Influence: 3.8] [Reference Citation Analysis (0)] |
| 6. | Hazewinkel Y, López-Cerón M, East JE, Rastogi A, Pellisé M, Nakajima T, van Eeden S, Tytgat KM, Fockens P, Dekker E. Endoscopic features of sessile serrated adenomas: validation by international experts using high-resolution white-light endoscopy and narrow-band imaging. Gastrointest Endosc. 2013;77:916-924. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 135] [Cited by in RCA: 154] [Article Influence: 12.8] [Reference Citation Analysis (0)] |
| 7. | Erichsen R, Baron JA, Hamilton-Dutoit SJ, Snover DC, Torlakovic EE, Pedersen L, Frøslev T, Vyberg M, Hamilton SR, Sørensen HT. Increased Risk of Colorectal Cancer Development Among Patients With Serrated Polyps. Gastroenterology. 2016;150:895-902.e5. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 150] [Cited by in RCA: 180] [Article Influence: 20.0] [Reference Citation Analysis (0)] |
| 8. | He X, Hang D, Wu K, Nayor J, Drew DA, Giovannucci EL, Ogino S, Chan AT, Song M. Long-term Risk of Colorectal Cancer After Removal of Conventional Adenomas and Serrated Polyps. Gastroenterology. 2020;158:852-861.e4. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 157] [Cited by in RCA: 168] [Article Influence: 33.6] [Reference Citation Analysis (0)] |
| 9. | Lu FI, van Niekerk de W, Owen D, Tha SP, Turbin DA, Webber DL. Longitudinal outcome study of sessile serrated adenomas of the colorectum: an increased risk for subsequent right-sided colorectal carcinoma. Am J Surg Pathol. 2010;34:927-934. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 143] [Cited by in RCA: 140] [Article Influence: 9.3] [Reference Citation Analysis (0)] |
| 10. | Fabbri M, Girnita L, Varani G, Calin GA. Decrypting noncoding RNA interactions, structures, and functional networks. Genome Res. 2019;29:1377-1388. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 95] [Cited by in RCA: 97] [Article Influence: 16.2] [Reference Citation Analysis (0)] |
| 11. | Hulshoff MS, Del Monte-Nieto G, Kovacic J, Krenning G. Non-coding RNA in endothelial-to-mesenchymal transition. Cardiovasc Res. 2019;115:1716-1731. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 65] [Cited by in RCA: 64] [Article Influence: 10.7] [Reference Citation Analysis (0)] |
| 12. | Zang J, Lu D, Xu A. The interaction of circRNAs and RNA binding proteins: An important part of circRNA maintenance and function. J Neurosci Res. 2020;98:87-97. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 191] [Cited by in RCA: 443] [Article Influence: 63.3] [Reference Citation Analysis (0)] |
| 13. | Phizicky EM, Hopper AK. tRNA biology charges to the front. Genes Dev. 2010;24:1832-1860. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 577] [Cited by in RCA: 631] [Article Influence: 42.1] [Reference Citation Analysis (0)] |
| 14. | Tao EW, Cheng WY, Li WL, Yu J, Gao QY. tiRNAs: A novel class of small noncoding RNAs that helps cells respond to stressors and plays roles in cancer progression. J Cell Physiol. 2020;235:683-690. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 27] [Cited by in RCA: 58] [Article Influence: 9.7] [Reference Citation Analysis (0)] |
| 15. | Park J, Ahn SH, Shin MG, Kim HK, Chang S. tRNA-Derived Small RNAs: Novel Epigenetic Regulators. Cancers (Basel). 2020;12. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 11] [Cited by in RCA: 51] [Article Influence: 10.2] [Reference Citation Analysis (0)] |
| 16. | Han L, Lai H, Yang Y, Hu J, Li Z, Ma B, Xu W, Liu W, Wei W, Li D, Wang Y, Zhai Q, Ji Q, Liao T. A 5'-tRNA halve, tiRNA-Gly promotes cell proliferation and migration via binding to RBM17 and inducing alternative splicing in papillary thyroid cancer. J Exp Clin Cancer Res. 2021;40:222. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 12] [Cited by in RCA: 51] [Article Influence: 12.8] [Reference Citation Analysis (0)] |
| 17. | Cui H, Li H, Wu H, Du F, Xie X, Zeng S, Zhang Z, Dong K, Shang L, Jing C, Li L. A novel 3'tRNA-derived fragment tRF-Val promotes proliferation and inhibits apoptosis by targeting EEF1A1 in gastric cancer. Cell Death Dis. 2022;13:471. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 6] [Cited by in RCA: 69] [Article Influence: 23.0] [Reference Citation Analysis (0)] |
| 18. | Zhou Y, Hu J, Liu L, Yan M, Zhang Q, Song X, Lin Y, Zhu D, Wei Y, Fu Z, Hu L, Chen Y, Li X. Gly-tRF enhances LCSC-like properties and promotes HCC cells migration by targeting NDFIP2. Cancer Cell Int. 2021;21:502. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 4] [Cited by in RCA: 26] [Article Influence: 6.5] [Reference Citation Analysis (0)] |
| 19. | Chen H, Xu Z, Cai H, Peng Y, Yang L, Wang Z. Identifying Differentially Expressed tRNA-Derived Small Fragments as a Biomarker for the Progression and Metastasis of Colorectal Cancer. Dis Markers. 2022;2022:2646173. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 2] [Cited by in RCA: 10] [Article Influence: 3.3] [Reference Citation Analysis (0)] |
| 20. | Tsiakanikas P, Adamopoulos PG, Tsirba D, Artemaki PI, Papadopoulos IN, Kontos CK, Scorilas A. High Expression of a tRNA(Pro) Derivative Associates with Poor Survival and Independently Predicts Colorectal Cancer Recurrence. Biomedicines. 2022;10. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 3] [Cited by in RCA: 21] [Article Influence: 7.0] [Reference Citation Analysis (0)] |
| 21. | Wang J, Ma G, Li M, Han X, Xu J, Liang M, Mao X, Chen X, Xia T, Liu X, Wang S. Plasma tRNA Fragments Derived from 5' Ends as Novel Diagnostic Biomarkers for Early-Stage Breast Cancer. Mol Ther Nucleic Acids. 2020;21:954-964. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 22] [Cited by in RCA: 50] [Article Influence: 10.0] [Reference Citation Analysis (0)] |
| 22. | De Palma FDE, D'Argenio V, Pol J, Kroemer G, Maiuri MC, Salvatore F. The Molecular Hallmarks of the Serrated Pathway in Colorectal Cancer. Cancers (Basel). 2019;11. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 65] [Cited by in RCA: 118] [Article Influence: 19.7] [Reference Citation Analysis (0)] |
| 23. | Angerilli V, Sabella G, Centonze G, Lonardi S, Bergamo F, Mangogna A, Pietrantonio F, Fassan M, Milione M. BRAF-mutated colorectal adenocarcinomas: Pathological heterogeneity and clinical implications. Crit Rev Oncol Hematol. 2022;172:103647. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 15] [Cited by in RCA: 9] [Article Influence: 3.0] [Reference Citation Analysis (0)] |
| 24. | Li S, Shi X, Chen M, Xu N, Sun D, Bai R, Chen H, Ding K, Sheng J, Xu Z. Angiogenin promotes colorectal cancer metastasis via tiRNA production. Int J Cancer. 2019;145:1395-1407. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 47] [Cited by in RCA: 77] [Article Influence: 12.8] [Reference Citation Analysis (0)] |
| 25. | Zhang H, Xu C, Shi C, Zhang J, Qian T, Wang Z, Ma R, Wu J, Jiang F, Feng J. Hypermethylation of heparanase 2 promotes colorectal cancer proliferation and is associated with poor prognosis. J Transl Med. 2021;19:98. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 7] [Cited by in RCA: 18] [Article Influence: 4.5] [Reference Citation Analysis (0)] |
| 26. | Wu B, Liu G, Jin Y, Yang T, Zhang D, Ding L, Zhou F, Pan Y, Wei Y. miR-15b-5p Promotes Growth and Metastasis in Breast Cancer by Targeting HPSE2. Front Oncol. 2020;10:108. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 53] [Cited by in RCA: 52] [Article Influence: 10.4] [Reference Citation Analysis (0)] |
| 27. | Alspach E, Lussier DM, Schreiber RD. Interferon γ and Its Important Roles in Promoting and Inhibiting Spontaneous and Therapeutic Cancer Immunity. Cold Spring Harb Perspect Biol. 2019;11. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 269] [Cited by in RCA: 378] [Article Influence: 63.0] [Reference Citation Analysis (0)] |
| 28. | de Vogel S, Dindore V, van Engeland M, Goldbohm RA, van den Brandt PA, Weijenberg MP. Dietary folate, methionine, riboflavin, and vitamin B-6 and risk of sporadic colorectal cancer. J Nutr. 2008;138:2372-2378. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 66] [Cited by in RCA: 74] [Article Influence: 4.4] [Reference Citation Analysis (0)] |
| 29. | Leenders M, Leufkens AM, Siersema PD, van Duijnhoven FJ, Vrieling A, Hulshof PJ, van Gils CH, Overvad K, Roswall N, Kyrø C, Boutron-Ruault MC, Fagerhazzi G, Cadeau C, Kühn T, Johnson T, Boeing H, Aleksandrova K, Trichopoulou A, Klinaki E, Androulidaki A, Palli D, Grioni S, Sacerdote C, Tumino R, Panico S, Bakker MF, Skeie G, Weiderpass E, Jakszyn P, Barricarte A, María Huerta J, Molina-Montes E, Argüelles M, Johansson I, Ljuslinder I, Key TJ, Bradbury KE, Khaw KT, Wareham NJ, Ferrari P, Duarte-Salles T, Jenab M, Gunter MJ, Vergnaud AC, Wark PA, Bueno-de-Mesquita HB. Plasma and dietary carotenoids and vitamins A, C and E and risk of colon and rectal cancer in the European Prospective Investigation into Cancer and Nutrition. Int J Cancer. 2014;135:2930-2939. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 49] [Cited by in RCA: 53] [Article Influence: 4.8] [Reference Citation Analysis (0)] |
| 30. | Sun HW, Chen J, Wu WC, Yang YY, Xu YT, Yu XJ, Chen HT, Wang Z, Wu XJ, Zheng L. Retinoic Acid Synthesis Deficiency Fosters the Generation of Polymorphonuclear Myeloid-Derived Suppressor Cells in Colorectal Cancer. Cancer Immunol Res. 2021;9:20-33. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 15] [Cited by in RCA: 19] [Article Influence: 4.8] [Reference Citation Analysis (0)] |