Published online Mar 15, 2023. doi: 10.4251/wjgo.v15.i3.504
Peer-review started: December 20, 2022
First decision: January 9, 2023
Revised: January 19, 2023
Accepted: March 2, 2023
Article in press: March 2, 2023
Published online: March 15, 2023
Processing time: 84 Days and 16 Hours
Hepatocellular carcinoma (HCC) is one of the most common digestive system cancers with high mortality rates worldwide. The main ingredients in Mu Ji Fang Granules (MJF) are alkaloids, flavonoids, and polysaccharides. MJF has been used in the clinical treatment of hepatitis, cirrhosis and HCC for more than 30 years. Few previous studies have focused on the mechanism of MJF on tumor immu-nology in the treatment of HCC.
To explore the mechanism of action of MJF on tumor immunology in the treatment of HCC.
The absorbable ingredients of MJF were identified using Molecule Network related to High Performance Liquid Chromatography-Electron Spray Ionization-Time of Flight- Mass Spectrometry, and hub potential anti-HCC targets were screened using network pharmacology and pathway enrichment analysis. Forty male mice were randomly divided into the Blank, Model, and MJF groups (1.8, 5.4, and 10.8 g/kg/d) following 7 d of oral administration. Average body weight gain, spleen and thymus indices were calculated, tumor tissues were stained with hematoxylin and eosin, and Interferon gamma (IFN-γ), Tumor necrosis factor α (TNF-α), Interleukin-2, aspartate aminotransferase, alanine aminotransferase, alpha-fetoprotein (AFP), Fas, and FasL were measured by Enzyme-linked Immunosorbent Assay. Relevant mRNA expression of Bax and Bcl2 was evaluated by Real Time Quantitative PCR (RT-qPCR) and protein expression of Transforming growth factor β1 (TGF-β1) and Mothers against decapentaplegic homolog (SMAD) 4 was assessed by Western blotting. The HepG2 cell line was treated with 10 mg/mL, 20 mg/mL, 30 mg/mL, 40 mg/mL of MJF, and another 3 groups were treated with TGF-β1 inhibitor (LY364947) and different doses of MJF. Relevant mRNA expression of TNF-α, IFN-γ, Bax and Bcl2 was evaluated by RT-qPCR and protein expression of TGF-β1, SMAD2, p-SMAD2, SMAD4, and SMAD7 was assessed by Western blotting.
It was shown that MJF improved body weight gain and tumor inhibition rate in H22 tumor-bearing mice, protected immune organs and liver function, reduced the HCC indicator AFP, affected immunity and apoptosis, and up-regulated the TGF-β1/SMAD signaling pathway, by increasing the relative expression of TGF-β1, SMAD2, p-SMAD2 and SMAD4 and decreasing SMAD7, reducing immune factors TNF-α and IFN-γ, decreasing apoptosis cytokines Fas, FasL and Bcl2/Bax, and inhibiting the effect of LY364947 in HepG2 cells.
MJF inhibits HCC by activating the TGF-β1/SMAD signaling pathway, and affecting immune and apoptotic cytokines, which may be due to MJF adjusting immune escape and apoptosis.
Core Tip: Mu Ji Fang Granules (MJF), a Chinese patent medicine, has been used in hepatitis, cirrhosis and hepatocellular carcinoma (HCC) for more than 30 years. MJF was identified with Molecule Network related to High Performance Liquid Chromatography-Electron Spray Ionization-Time of Flight- Mass Spectrometry, and hub potential anti-HCC targets were screened using network pharmacology and pathway enrichment analysis in H22 tumor-bearing mice and HepG2 cells. It was shown that MJF improved body weight gain and tumor inhibition rate, protected immune organs and liver function, affected immunity and apoptosis, up-regulated the Transforming growth factor β1(TGF-β1)/ Mothers against decapentaplegic homolog(SMAD) signaling pathway, and inhibited the effect of TGF-β1 inhibitor (LY364947).
- Citation: Zhang YB, Bao YR, Wang S, Li TJ, Tai H, Leng JP, Yang XX, Wang BC, Meng XS. Possible mechanisms associated with immune escape and apoptosis on anti-hepatocellular carcinoma effect of Mu Ji Fang granules. World J Gastrointest Oncol 2023; 15(3): 504-522
- URL: https://www.wjgnet.com/1948-5204/full/v15/i3/504.htm
- DOI: https://dx.doi.org/10.4251/wjgo.v15.i3.504
In the World Health Organization 2018 World Cancer Report, liver cancer is the sixth most common cancer and the fourth most common cause of cancer death worldwide, and the age-standardized rates in Asia and Africa were 2 to 3 times those in America, Europe, and Oceania[1]. In the latest report on cancer epidemiology in China, liver cancer was the third and seventh most common malignancy in males and females, with an incidence rate of 12.74% and 5.40%, and the second and third highest mortality rate of 16.36% and 9.79%, respectively[2].
Mu Ji Fang granules (MJF, also known as Fufang Mu Ji Granules), a Chinese patent medicine produced by Dandong Pharmaceutical Group Co., Ltd (Liaoning, China), derived from a folk prescription of the Manchu medicine, consists of 4 herbs including Sophorae Tonkinensis Radix[3], Cuscutae Semen[4], Juglans mandshurica Maxim[5] and Coriolus versicolor[6] (Table 1), processed into proprietary Chinese medicine, has been used in patients with hepatitis, cirrhosis and hepatocellular carcinoma (HCC) for many years and has a considerable curative effect in clinical practice[7]. Previous studies of MJF mainly focused on its clinical efficacy in hepatitis B[8,9] and fundamental pharmacological effects[10], but few reported the mechanism of action on tumor immunology in the treatment of HCC. Our research group previously established the High Performance Liquid Chromatography (HPLC) fingerprint chromatogram of MJF, combined with HPLC-Electron Spray Ionization-Time of Flight- Mass Spectrometry(HPLC-ESI-TOF-MS), and identified the main compounds in MJF to be alkaloids, flavonoids, and polysaccharides. Based on these findings we hypothesize that MJF might be able to promote tumor cell apoptosis, anti-inflammatory activity and enhance immunity.
| Herb | Latin name | Family and genus | Plant | Medicinal parts |
| Sophorae tonkinensis Radix | Sophorae tonkinensis Radix et rhizoma | Leguminosae | Sophora tonkinensis Gapnep | Root |
| Cuscutae semen | Cuscutae semen | Convolvulaceae | Cuscuta australis R. Br. or Cuscuta chinensis Lam | Seed |
| Juglans mandshurica Maxim | Juglans mandshurica Maxim | Juglandaceae | Juglans mandshurica | Bark |
| Coriolus versicolor | Coriolus versicolor | Basidiomycetes | Coriolus versicolor (L. ex Fr.) Quel | Fruit body |
In this study, we used Molecule Network (MN) coupled with UPLC-ESI-TOF-MS to analyze the compounds in mouse plasma and a chemical profile was built based on the structures of these compounds. Combined with the compounds from MN, network pharmacology was adopted to analyze the anti-HCC pharmacological mechanisms of MJF. H22 tumor-bearing mice and HepG2 cells were introduced as experimental support for verification, Enzyme Linked Immunosorbent Assay (ELISA), Real Time Quantitative PCR (RT-qPCR), and Western blotting assays were used to evaluate the expression levels of target genes in vivo and in vitro. Our data indicated that MJF had an anti-HCC effect by regulating immune escape and promoting apoptosis.
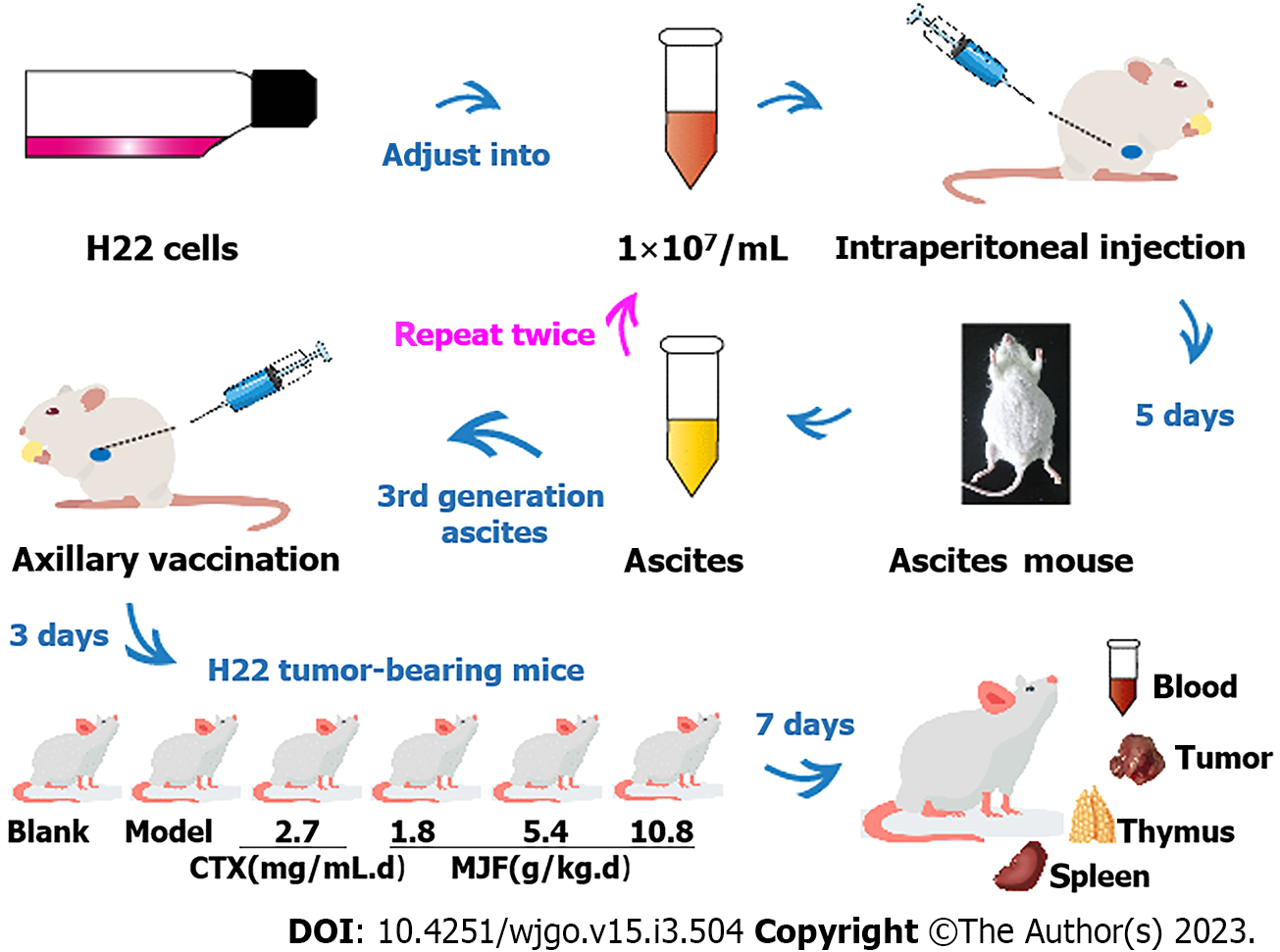
Eight-week-old Institute of Cancer Research (ICR) male mice weighing 18-22 g, (Experimental Animal Center of Liaoning Changsheng Biological Technology Co., Ltd., License No. SCXK Liao 2015-0001), were adaptively raised in a controlled-environment animal experiment room (temperature 20°C-25°C, humidity 45%-65%, light/dark cycle for 12 h/12 h) for 7 d with free access to food and tap water. All experimental procedures were performed with the approval of the animal guidelines and protocols established by the Medicine Ethics Review Committee of Animal Experiments of Liaoning University of Traditional Chinese Medicine (TCM) and The Affiliated Hospital of Liaoning University of TCM. Mouse HCC H22 cell strain (Jiangsu Chi Scientific Co., Ltd.) was cultured in RPMI-1640 culture medium (Gibco, Thermo Fisher Scientific, Waltham, MA, United States) containing 1% streptomycin and 10% fetal bovine serum (100 μg/mL penicillin, 100 μg/mL streptomycin) (Gibco Inc. United States) in an incubator at 37 °C, 5% CO2 and 95% of relative saturation humidity. H22 cells in the logarithmic growth period were adjusted to 1 × 107/mL with saline for intraperitoneal injection into mice at 0.2 mL each animal. Five days later, the mice developed ascites due to the growth of H22 cells in the abdominal cavity. We extracted abdominal ascites from the mice and adjusted them to 1 × 107/mL with saline as the first generation ascites and repeated the above intraperitoneal injection twice in healthy mice. The third generation ascites were harvested and adjusted to 1 × 107/mL with saline as the final implant cell line. An axillary vaccination was conducted in each mouse using 0.2 mL H22 diluted ascites. Three days later, nodules were observed in the right armpit of the mouse, indicating that the established model was successful.
Forty mice were randomly divided into the Model group, cyclophosphamide (CTX) group (cyclophosphamide, 2.7 mg/mL), and 3 MJF groups (M-L received a low MJF dose of 1.8 g/kg, M-M received a mid MJF dose of 5.4 g/kg, M-H received a high MJF dose of 10.8 g/kg) with 8 mice in each group. A further 8 ICR male mice (weighing 18-22 g) were included in the Blank group. All mice were administered the drugs orally once a day (the Blank and Model groups received saline) for 7 d, no food was allowed 12 h before the experiment, but the animals had free access to water. Thirty minutes after the final drug administration, all mice were euthanized under deep anesthesia. Body weight was measured and blood was collected for analysis. After sacrifice, intact thymus, spleen, and tumor tissues were removed and weighed (Figure 1). The thymus index, spleen index, and tumor inhibitory rates in each mouse were calculated according to the following formulas.
Thymus index = thymus weight (mg)/ body weight (g) × 10
Spleen index = spleen weight (mg)/ body weight (g) × 10
Tumor inhibitory rate (%) = (1 - average tumor weight in the drug administration group/ average tumor weight in the Model group) × 100%
Five gram MJF granules (#20190725334, provided by Dandong Pharmaceutical Group Co., Ltd., Dandong China) were weighed and added to 30 mL anhydrous methanol (HPLC grade, Merck Co. Ltd., Darmstadt, Germany), ultrasonically extracted for 30 min, and then filtered through a micropore membrane (0.22 mm; Jinteng Corp., Tianjin, China) before use. Seventeen accurately weighed reference substances were mixed and dissolved in 10 mL of methanol to obtain a solution at the concentration of 21.00 μg/mL for protocatechuic acid, 24.54 μg/mL for ellagic acid, 18.96 μg/mL for kaempferol-3-O-rutinoside, 17.45 μg/mL for rutin, 15.09 μg/mL for hyperoside, 20.64 μg/mL for isoquercitrin, 18.44 μg/mL for astragalin, 17.42 μg/mL for kaempferol, 18.40 μg/mL for isorhamnetin, 20.62 μg/mL for marine, 23.36 μg/mL for sophocarpine, 18.43 μg/mL for gallic acid, 16.48 μg/mL for naringenin, 22.72 μg/mL for cytisine, 21.53 μg/mL for caffeate, 21.34 μg/mL for quercetin (Sichuan Victory Biological Technology Co., Ltd., Sichuan, China) and 18.92 μg/mL for ferulic acid (HPLC grade, Tianjin Kermel Chemical Co., Tianjin, China). 50 μL of thawed frozen plasma samples of MJF high dose (M-H) were placed into 1 mL centrifuge tubes, and then thoroughly mixed with 100 μL methanol and vortexed for 1 min. The solutions were centrifuged at 8000 rpm/min and 4 °C for 10 min. The supernatants were filtered through a 0.22 μm membrane for HPLC-MS analyses.
The HPLC–MS analysis was performed using an Agilent 1290 HPLC system (Agilent Technologies, Inc., CA, United States) in tandem with an Agilent 6550 quadrupole-time of flight with mass spectrometry (Agilent Co., United States). The analysis system consisted of an Agilent Proshell SB-C18 column (100 mm × 3 mm, 2.1 μm) (Agilent Technologies, Inc., CA, United States) and was used with the column temperature maintained at 35 °C; the flow rate of the mobile phase was 0.4 mL/min and the injection volume was 0.6 μL, the mobile phase consisted at 0.1% formic acid in water of A and acetonitrile of B, and the column was eluted with a linear gradient of 3%–8% B over initial to 5.0 min, 8%–15% B over 5.0-9.0 min, 15%-22% B over 9.0-12.0 min, 22%-55% B over 12-15 min, 55%-70% over 15-20 min,70%–95% B over 20.0–24.0 min, returned to 3% B for 1.0 min and then held for 2.5 min at an eluent flow rate of 0.5 mL/min. Optimal conditions for HPLC-Q-TOF-MS analysis were as follows: ion source was Dual AJS ESI, both positive and negative ion mode detection was adopted, drying gas flow of 13 L/min, drying gas temperature maintained at 350 °C, a capillary voltage of 3500 V, nebulizer pressure of 310.28 kPa, frag mentor voltage of 125 V, OCT 1RF Vpp voltage of 750 V, collision energy of 40 eV and mass spectrometric data were acquired in the mode from 1000 to 1000 m/z with an acquisition rate of 1.5 spectra/s. Data were corrected and obtained during acquisition using a correction mixed solution (Agilent Technologies, Inc., CA, United States, m/z = 112.985587, m/z = 1033.988109).
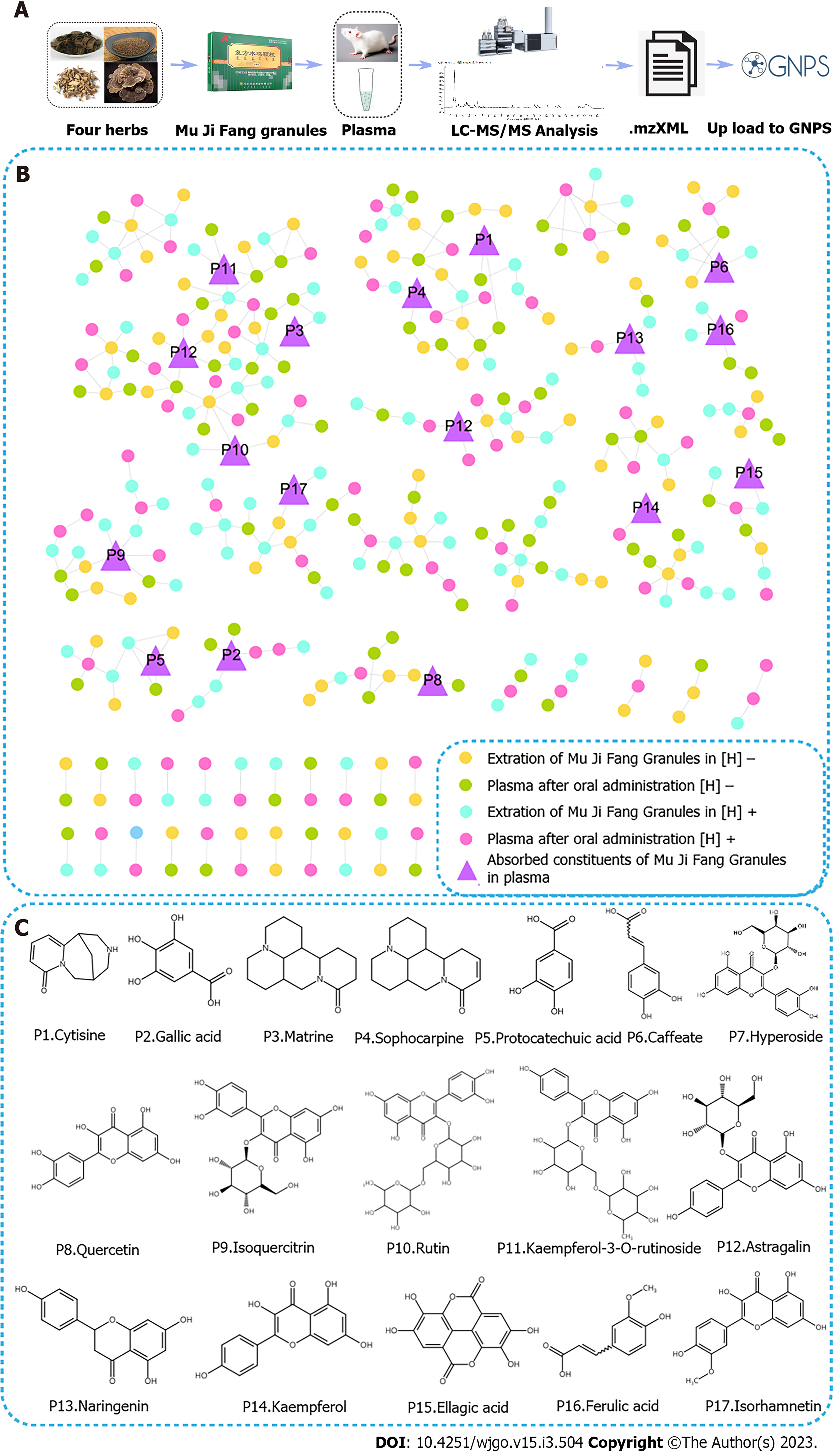
For MN data processing, MS/MS data on MJF extraction and MJF high dose were collected and converted into mzXML format using Proteo Wizard software (www.proteowizard.sourceforge.net, Proteo Wizard, Palo Alto, CA, United States) and then uploaded separately into the GNPS platform (https://gnps.ucsd.edu, UCSD, San Diego, CA, United States) (accessed on 16 November 2021) (Figure 2A). The GNPS parameters were as follows: mass error of less than 0.02 Da, matched peaks greater than 3, and cosine score greater than 0.50. After analysis, graphic format files were generated from the GNPS platform, and the files were then downloaded and imported into Cytoscape software v 3.7.0 (www.cytoscape.org, NRNB, Hill St, San Diego, CA, United States) to build the molecular network. According to Wang et al’s study[11], MN of MJF extraction and plasma after MJF oral administration were merged, and the absorbed constituents of MJF in plasma were obtained. All constituents were identified using reference substances on HPLC-Q-TOF-MS.
One thousand two hundred and thirty-two HCC-related genes were extracted from DisGeNET[12] (https://www.disgenet.org) and two liver cancer databases Liverome[13] (http://Liverome.kobic.re.kr/index.php) and OncoDB. HCC[14] (http://oncodb.hcc.ibms.sinica.edu.tw). Validated and predicted targets of 17 ingredients screened from MN of MJF were collected from Swiss Target Prediction[15] (http://www.swisstargetprediction.ch), an online tool to predict the macromolecular targets (proteins from human, mouse, and rats) of small bioactive molecules. All the ingredient-related genes were mapped to HCC-related genes to obtain a total of 221 shared targets, which were then uploaded to the STRING database[16] (https://string-db.org) to acquire the protein-protein interaction (PPI) network. Cytoscape software and the Network Analyzer App were employed for topological analysis of the PPI network, and the medians of each index were calculated. Nodes of a degree over 19, betweenness centrality over 0.00283, and closeness centrality over 0.51485 were gathered as predicted targets and were uploaded to the Metascape database[17] (https://metascape.org/) for possible Gene Ontology (GO) and Kyoto Encyclopedia of Genes and Genomes (KEGG) pathways enrichment. The final results of the ingredient-pathway-target network were demonstrated by Cytoscape.
Mouse tumor tissues were immediately fixed in 4% paraformaldehyde (Beijing Solaibao Technology Co., Ltd., Beijing, China). Following paraffin embedding, the tissues were cut into 4 μm slices, stained with hematoxylin and eosin (HE), and morphological changes were observed with a microscope (× 200 high power visual field).
Plasma samples from all five groups were centrifuged at 8000 rpm/min and 4 °C for 10 min. The supernatants were then used for Interferon gamma (IFN-γ), Tumor necrosis factor α (TNF-α), Interleukin-2 (IL-2), aspartate aminotransferase (AST), alanine aminotransferase (ALT), alpha-fetoprotein (AFP), Fas, and FasL analyses by ELISA in accordance with the kit instructions (Shanghai Langton Biotechnology Co., Ltd., Shanghai, China), respectively.
The HepG2 cell strain (Jiangsu Chi Scientific Co., Ltd, Jiangsu, China) was cultured in DMEM culture medium containing 1% streptomycin and 10% fetal bovine serum (Gibco, Thermo Fisher Scientific, Waltham, MA, United States) (100 μg/mL penicillin, 100 μg/mL streptomycin) in an incubator at 37 °C, 5% CO2 and 95% of relative saturation humidity. Cells at the density of 1 × 105/mL were inoculated into 6-well plates, and after culture for 24 h the medium was removed. 4.0 g MJF was added to 20 mL 50% methanol and treated with ultrasonic extraction for 20 min (700 W, 40 kHz), and the liquid was filtered and dried. 1.0 g MJF were mixed with 0.5 mL DMSO, diluted with DMEM culture medium to 10 mg/mL, 20 mg/mL, 30 mg/mL, and 40 mg/mL, and then added to a 6-well plate with 1 mL of MJF in each well (M1, M2, M3, and M4). The Transforming growth factor β1(TGF-β1) inhibitor Ly364947 (#HY-13462, MedChem Express Co., Ltd., NJ, United States) was adjusted to 1 μM and 2.5 μM, respectively, and added to 6-well plates with 1 mL of LY364947 in each well. M2 + LY-1 and M4 + LY-2 groups contained 20 mg/mL and 40 mg/mL of MJF each and 1 μM Ly364947. 1 mL culture medium was added to the Blank group. All groups were cultured for 8 h.
Tumor tissue was placed in TRlzol reagent to obtain a homogenate, and total RNA was extracted according to the manufacturer’s instructions. The reverse transcription kit (Trans Script One-Step gDNA Removal and cDNA Synthesis Super Mix, Beyotime Biotechnology Co., Ltd., Shanghai, China) was used to reverse transcribe the extracted RNA into cDNA and processed as previously described[18]. The total RNA in HepG2 cells in each group was processed in the same way. The primer sequences for RT-qPCR of tumor tissue and HepG2 cells are listed in Tables 2 and 3.
| Name of primers | Sequences |
| Bax-F | 5’-AGGATGCGTCCACCAAGAA-3’ |
| Bax-R | 5’-CAAAGTAGAAGAGGGCAACCAC-3’ |
| Bcl-2-F | 5’-CAACACTCCCTCTTGACCTATGC-3’ |
| Bcl-2-R | 5’-GAAAATGTTCCCAAGTGAGTTAGA-3’ |
| β-actin-F | 5’-GTCCCTCACCCTCCCAAAAG-3’ |
| β-actin-R | 5’-GCTGCCTCAACACCTCAACCC-3’ |
| Name of primers | Sequences |
| TNF-α-F | 5’-ATCGGTCCCAACAAGGAGGAGAAGT-3’ |
| TNF-α-R | 5’-ACGTGGGCTACGGGCTTGTCACTC-3’ |
| IFN-γ-F | 5’-ACAACCAGGCCATCAGCAACAACATA-3’ |
| IFN-γ-R | 5’-CTGTGGGTTGTTCAGCTCGAACTT-3’ |
| Bax-F | 5’-AAACTGGTGCTCAAGGCCC-3’ |
| Bax-R | 5’-AAAGTAGGAGAGGAGGCCGTC-3’ |
| Bcl-2-F | 5’-CAGGATAACGGAGGCTGGGATG-3’ |
| Bcl-2-R | 5’-TTCACTTGTGGCCCAGATAGG-3’ |
| β-actin-F | 5’-GGACCTGACTGACTACCTC-3’ |
| β-actin-R | 5’-TACTCCTGCTTGCTGAT-3’ |
Tumor tissue proteins from the mice in each group were extracted with RIPA reagent. Proteins were separated by 10% SDS-PAGE and then transferred onto PVDF membranes (Beyotime Biotechnology Co., Ltd., Shanghai, China). The membranes were washed, blocked with BSA blocking solution for 2 h, and then incubated overnight with anti-β-actin (#20536-1-AP), anti-TGF-β1 (#21898-1-AP), anti- Mothers against decapentaplegic homolog(SMAD) 4 (#10231-1-AP), anti-SMAD7 (#25840-1-AP) (Proteintech Group, Inc., Rosemont, IL, United States), anti-SMAD2 (#5339S), and anti-phosphorylated-SMAD2 (#3108S, Cell Signaling Technology, Danvers, MA, United States) at 4 °C. After washing with TBST 3 times for 10 min each time, the membranes were incubated with secondary antibodies (Goat Anti-Rabbit IgG H + L) (#SA00001-2, Proteintech Group, Inc., Rosemont, IL, United States) for 2 h at room temperature. Immunoreactivity was determined using ECL glow (Beyotime Biotechnology Co., Ltd., Shanghai, China). Data analysis was conducted via Image J 1.8.0 software (National Institutes of Health, Bethesda, MD, United States). Protein in HepG2 cells from each group was processed using the same steps as above.
Statistical analyses were performed with SPSS 25.0 software (IBM Corporation, Armonk, NY, United States) using one-way ANOVA and data were expressed as the mean ± SD. P < 0.05 was considered statistically significant, and P < 0.01 was considered very significant. RT-qPCR data were analyzed by the 2-ΔΔ Ct algorithm[19].
A comprehensive MN based on LC-MS/MS spectra in order to reveal absorbable ingredients in MJF was performed as shown in Figure 2B. The MN map contained 933 precursor ions, including 137 clusters (nodes > 2) and 352 single nodes. A total of 17 main prototype components (P1-17) were screened out by the GNPS MS database, based on the mass measurements and fragmentation patterns confirmed by reference substances. These compounds were identified as cysteine, gallic acid, matrine, sophocarpine, protocatechuic acid, caffeate, hyperoside, quercetin, isoquercitrin, rutin, kaempferol-3-o-rutinoside, astragalin, naringenin, kaempferol, ellagic acid, ferulic acid, and isorhamnetin (Figure 2, Table 4). These compounds were of three main types: Polyphenol acids, flavonoids, and alkaloids, all of which were selected and used for the following target prediction in network pharmacology analysis.
| No. | RT (min) | [M-H/M+FA]- | Actual-M | Formula | Proposed compound |
| 1 | 3.412 | 189.1291 | 189.1290 | C11H14N2O | Cytisine |
| 2 | 3.585 | 169.0149 | 169.0145 | C7H6O5 | Gallic acid |
| 3 | 3.607 | 249.2185 | 249.2179 | C15H24N2O | Matrine |
| 4 | 3.615 | 247.2023 | 247.2020 | C15H22N2O | Sophocarpine |
| 5 | 4.997 | 153.0922 | 153.0914 | C7H6O4 | Protocatechuic acid |
| 6 | 7.109 | 178.0506 | 179.0349 | C15H24N2O2 | Caffeate |
| 7 | 9.003 | 463.0878 | 463.0873 | C21H20O12 | Hyperoside |
| 8 | 9.271 | 303.0512 | 303.0508 | C15H10O7 | Quercetin |
| 9 | 9.832 | 464.3002 | 464.2997 | C21H20O12 | Isoquercitrin |
| 10 | 10.183 | 610.3217 | 610.3213 | C27H30O16 | Rutin |
| 11 | 10.452 | 594.2133 | 594.2131 | C27H35O5 | Kaempferol-3-O-rutinoside |
| 12 | 10.793 | 476.3361 | 476.3364 | C21H20O11 | Astragalin |
| 13 | 12.033 | 271.0689 | 271.0681 | C15H12O5 | Naringenin |
| 14 | 15.525 | 288.3132 | 288.3128 | C15H10O6 | Kaempferol |
| 15 | 17.542 | 302.3295 | 302.3287 | C14H6O8 | Ellagic acid |
| 16 | 18.413 | 194.0819 | 194.0823 | C10H10O4 | Ferulic acid |
| 17 | 20.414 | 318.3255 | 318.3251 | C16H12O7 | Isorhamnetin |
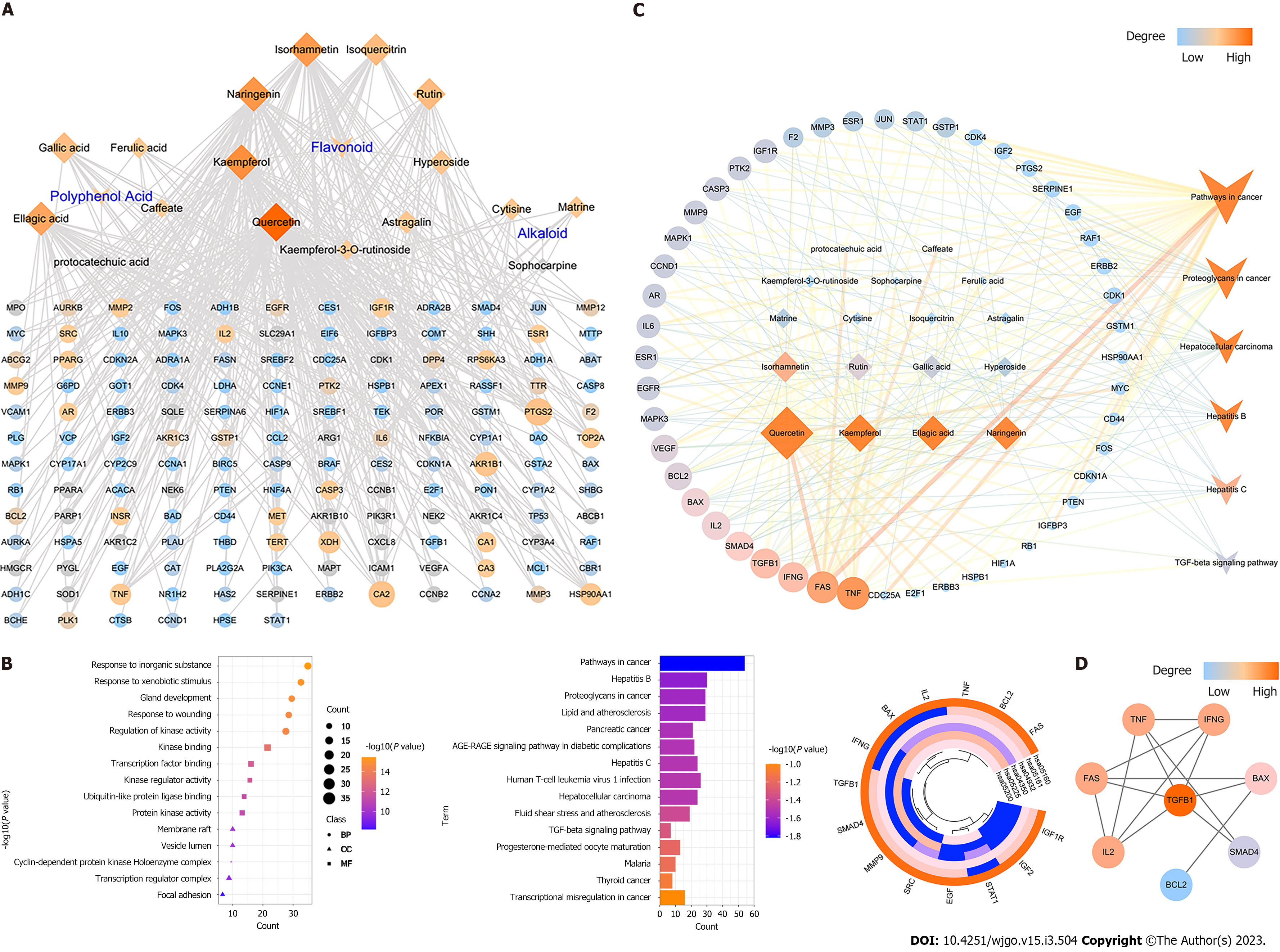
Seventeen ingredients related 1232 genes to HCC related genes were mapped, a total of 166 targets were obtained and an ingredients-anti-HCC-target network was established. The degree of each node was measured by the Network Analyzer App in Cytoscape (Figure 3A). The degree of one node indicates the number of nodes that have direct interactions with it, the higher degree one node has, the more biological processes it participates in, and the more biological importance it possesses.
From the topological analysis of 177 ingredients-anti-HCC-targets through the PPI network generated by the STRING database, 72 predicted targets were acquired. By uploading all these predicted targets into the Metascape database, we obtained the GO and KEGG pathways to enrich the analysis. Figure 3B lists the top 5 GO pathways to enrich the analysis indicating that MJF could respond to inorganic substances and xenobiotic stimulus etc. of Biological Processes (BP), kinase binding and transcription factor binding etc. of Cellular Components (CC), and membrane raft, vesicle lumen etc. of Molecular Functions (MF). Among the top 15 KEGG pathways which enriched the analysis (Figure 3B), only 6 of them were related to HCC including pathways in cancer, hepatitis B, proteoglycans in cancer, hepatitis C, HCC, and the TGF-β signaling pathway. From the heatmap in Figure 3B, 14 genes with the most significantly different abundance related to the 6 KEGG pathways of HCC were clustered, including FAS, BCL2, TNF, IL2, BAX, IFNG, TGFB1, SMAD4, MMP9 SRC, EGF, STAT1, IGF2, and IGF1R. A comprehensive predicted ingredient-anti-HCC-pathway-target network is shown in Figure 3C, where 17 potentially effective compounds related to the 6 pathways of HCC, from which, we clustered 8 hub-targets had a greater degree (above the median of 34.763) from the topological analysis of the network including TNF, BAX, BCL2, TGFB1, IFNG, FAS, IL2 and SMAD4 (Figure 3D).
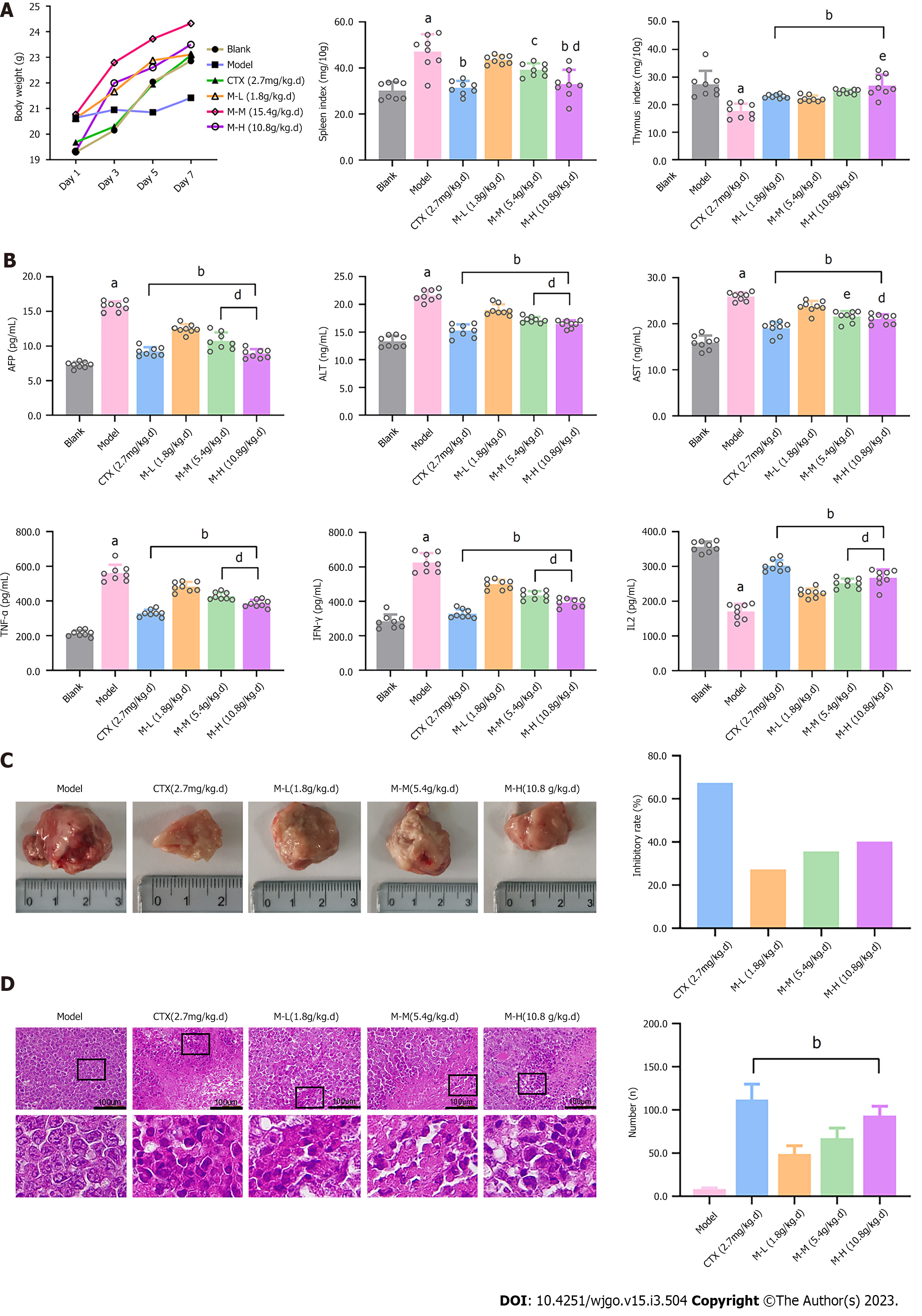
As displayed in Figure 4A, except for the Model group, the average body weight in the other groups all showed varying rapid gain. Compared with the Blank group, H22 tumor-bearing mice in the Model group had a much higher spleen index and lower thymus index (P < 0.05), due to H22 bearing tumors causing splenomegaly and thymic atrophy in mice. The CTX and MJF groups (M-L, M-M and M-H) revealed a significantly reduced spleen index and increased thymus index relative to the Model group, and in the MJF groups these levels progressed with increased dose (P < 0.05, P < 0.01). Figure 4B indicates that the concentration of AFP (HCC indicator), ALT and AST (liver function indicators), and TNF-α, IFN-γ, and IL-2 (immune factors) in the Model group markedly increased compared with the Blank group (P < 0.05), which confirmed that H22 tumors may affect immune action and inflammation in the liver and reduce liver function in mice. Compared with the Model group, CTX and MJF effectively reduced the concentration of AFP, ALT, AST, TNF-α and IFN-γ, and increased the concentration of IL2. With an increase in dose, the MJF mid and high dose groups (M-M and M-H) showed significant enhancement in adjusting the concentration of the above indicators (P < 0.05, P < 0.01).
As shown in Figure 4C, the size of the tumor in the Model group was larger and the color was darker, blood and vessels were clearly observed on the surface, while the tumors in the other groups were pale, and the size of the tumor and the average inhibitory rate in the 3 MJF groups (M-L, M-M and M-H) gradually changed with increased dose, respectively. Figure 4D shows the microscopic images and local enlarged images of HE stained sections of tumor tissue from each group. The tumor cells in the Model group were densely arranged with high cell density and large nuclei, and necrotic cells and cytoplasm were rarely seen. In the CTX group, tumor cells were loosely arranged, necrotic cells showed nucleus necrosis and rupture, and the cytoplasm was condensed and shrunken. In contrast, the three MJF groups all had different degrees of necrosis and apoptotic cell areas. The cytoplasm was highly agglutinated and condensed. In the MJF high dose group (M-H), the number of dead cells and round apoptotic bodies were more numerous, the size of the cells varied, and the connective tissue was more obvious than in the other two MJF groups (M-L and M-M).
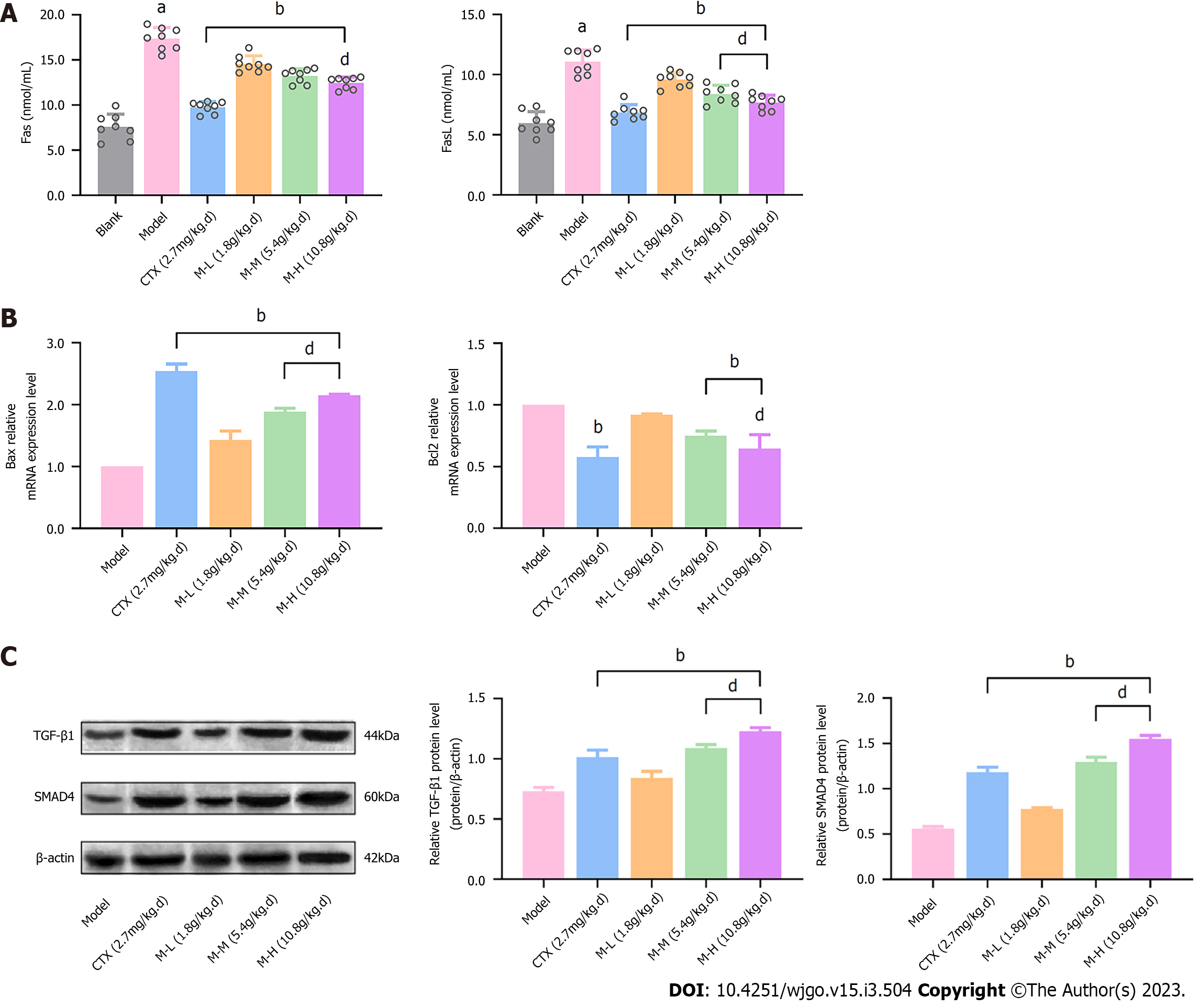
We evaluated the concentration of the cell apoptotic factors Fas and FasL, and the mRNA expression of Bax and Bcl2, two cytokines that can form homologous or heterologous dimers to regulate apoptosis, in order to determine the effect of MJF in improving the apoptosis of cancer cells. As shown in Figure 5A, the concentrations of Fas and FasL in the Model group were markedly increased relative to the Blank group, whereas, CTX and MJF (M-L, M-M and M-H) reduced the concentrations, respectively (P < 0.01). In addition, CTX and MJF also up-regulated the relative mRNA expression of Bax and down-regulated that of Bcl2 compared to the Model group (P < 0.01) (Figure 5B). Moreover, Western blot analysis was performed to determine the protein level of TGF-β1 and SMAD4, and it was demonstrated that CTX and MJF significantly increased the expression of TGF-β1 and SMAD4, and the expression levels in the MJF mid and high dose groups were even higher than that in the CTX group (P < 0.01) (Figure 5C).
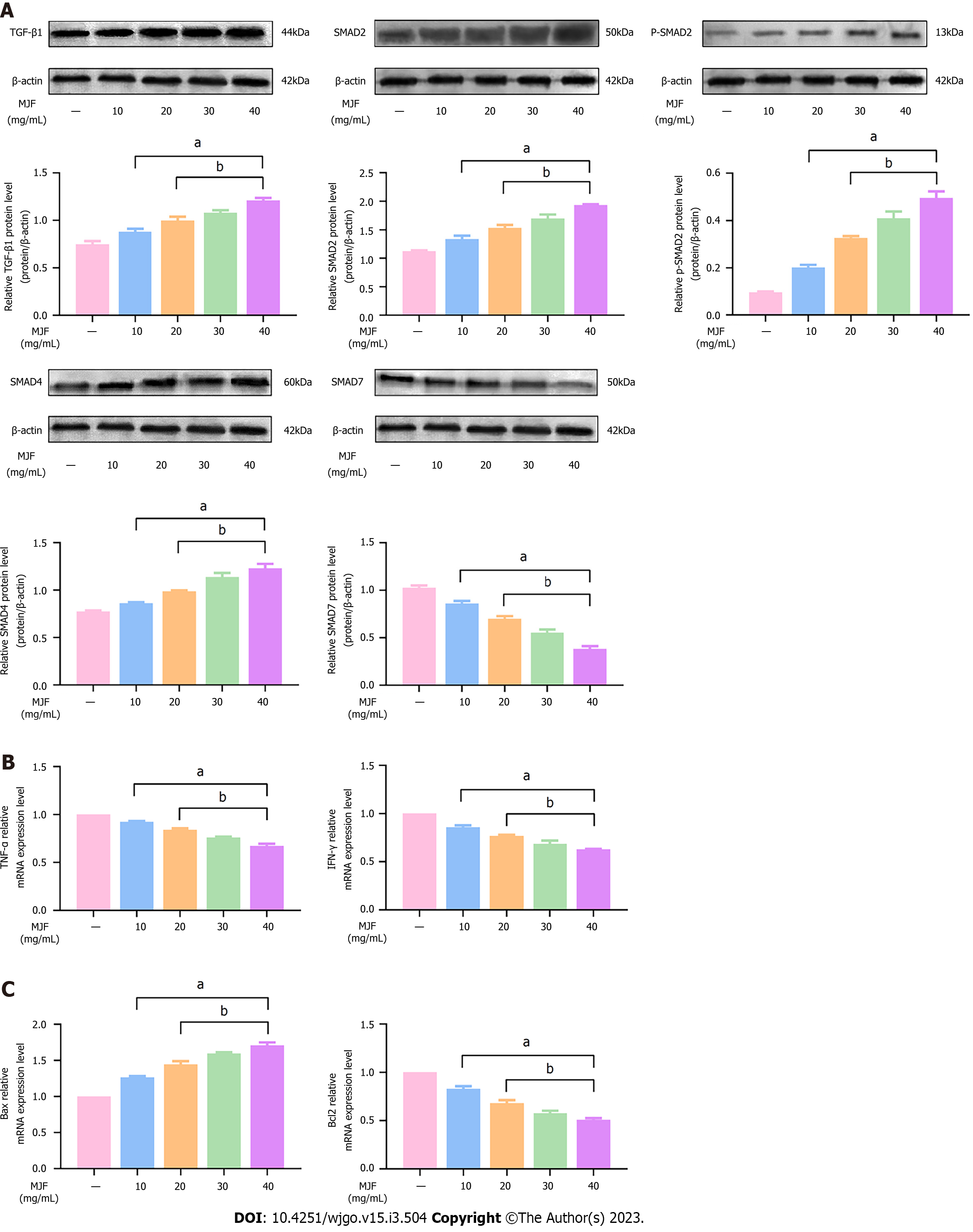
Western blot analysis was used to determine the protein expression of TGF-β1, SMAD2, p-SMAD2, SMAD4, and SMAD7, and we evaluated the effectiveness of MJF in regulating immune escape factors and promoting apoptosis in HepG2 cells, as the TGF-β1/SMAD signaling pathway plays an essential role in the development and invasion of HCC. The results showed that MJF (10 mg/mL to 40 mg/mL effectively increased the expression of TGF-β1, SMAD2, p-SMAD2 and SMAD4, and reduced the expression of SMAD7 protein relative to the levels in the Control group (MJF 0 mg/mL), respectively (P < 0.05) (Figure 6A). Furthermore, the results also showed that, compared with the Control group (MJF 0 mg/mL), mRNA expression of the immune cytokines, TNF-α and IFN-γ, in MJF (10 mg/mL to 40 mg/mL) was markedly decreased, and mRNA expression of the apoptosis cytokine, Bax in MJF (10 mg/mL to 40 mg/mL) was significantly increased and that of Bcl2 was reduced (P < 0.05) (Figure 6B and C).
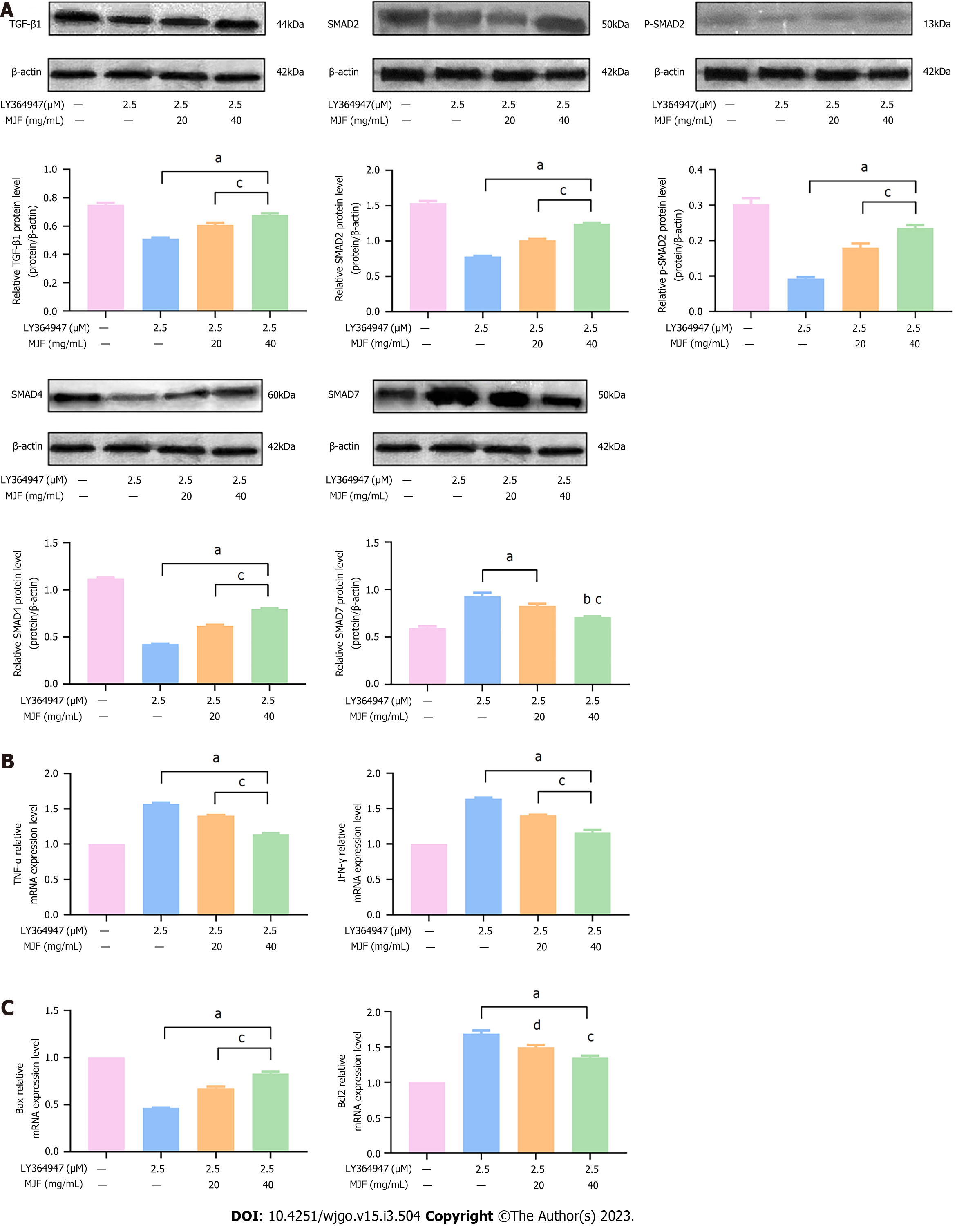
To evaluate the regulation of MJF on the TGF-β1/SMAD signaling pathway and both immune and apoptotic cytokines in HepG2 cells, LY364947, a TGF-β1 inhibitor, was added to the cell culture. As demonstrated in Figure 7A, relative to the Control group (LY364947 0 μM and MJF 0 mg/mL), LY364947 effectively inhibited the expression of TGF-β1, SMAD2, p-SMAD2 and SMAD4 protein and promoted that of SMAD7 (LY364947 2.5 μM and MJF 0 mg/mL), while, MJF (LY364947 2.5 μM, MJF 20 and 40 mg/mL) reduced the effect of LY364947 by improving the expression of TGF-β1, SMAD2, p-SMAD2 and SMAD4, and down regulating SMAD7 (P < 0.05).
With regard to the immune cytokines, TNF-α and IFN-γ, compared to the Control group (LY364947 0 μM and MJF 0 mg/mL), LY364947 significantly increased mRNA expression of TNF-α and IFN-γ (LY364947 2.5 μM and MJF 0 mg/mL), and MJF (LY364947 2.5 μM, MJF 20 and 40 mg/mL) showed a marked decrease in these two cytokines (P < 0.05) (Figure 7B). Moreover, compared with the Control group LY364947 0 μM and MJF 0 mg/mL), the expression of Bax mRNA was inhibited and that of Bcl2 was improved after treatment with LY364947 (LY364947 2.5 μM and MJF 0 mg/mL), yet, MJF (LY364947 2.5 μM, MJF 20 and 40 mg/mL) weakened this effect by increasing Bax and reducing Bcl2, respectively (P < 0.05) (Figure 7C).
HCC is a malignant tumor with high morbidity and mortality worldwide[20]. At present, the main clinical treatments for malignant liver tumors are surgery, radiation, and chemotherapy[21]. Common chemotherapy drugs have side effects such as cytotoxicity, multiple drug resistance, etc., and have different degrees of impact on liver function and the immune system[22]. TCM has a particular function in cancer treatment and prevention due to its high multi-target biological activity and low toxicity[23]. The long history and extensive study of TCM indicate its potential advantages in alleviating symptoms and improving quality of life in different patterns and stages of the disease[24]. As a Chinese patent medicine, MJF has been used in the treatment of liver cancer since the 1980s, with the exception of HCC, and has also been proved to have anti-inflammatory effects, enhances immunity, and improves liver function. Thus, in the present study, we mainly focused on exploring the mechanism of action of MJF.
In a recent study, the MN technique associated with HPLC-ESI-TOF-MS was employed in studies of natural products derived from plants, microorganisms, marine organisms, and other biological sources[25]. As a visual computational strategy, MN can be intuitively implemented based on a comparison of the theoretical MS/MS spectra to test MS/MS spectra and establish the relative network by clustering similar structures with similar mass spectra for compound identification[26]. In this study, we imported this recent approach, and established the MN of MJF solution and plasma after oral administration, and by merging the two MNs, the shared compounds were thought to be the absorbable constituents of MJF. To verify this, we also used reference substances to identify the 17 ingredients obtained from MN.
We then introduced network pharmacology to establish an image of the function and behavior of the biological network of MJF, identify potential targets and pathways of the drugs related to the disease using topology and computational methods, and explored the topology parameters of the 17 ingredients and their related targets in anti-HCC among all the ingredients-anti-HCC-pathways-targets. As a consequence, targets including TGFB1, SMAD4, TNF, IFNA1, IL2, BAX, BCL2, and FAS (TNF superfamily receptor 6) were screened out.
The TGF-β1/SMAD signaling pathway plays an important role in the development and invasion of HCC[27,28]. Cytoplasmic protein SMAD is the most critical and important signal transduction factor in the TGF β signaling pathway. When the TGF-β pathway is activated, phosphorylated SMAD2 combines with SMAD4 to form a complex, and then enters the nucleus to activate the expression of downstream target genes. SMAD 7 is a TGF-β signaling inhibitory factor, which is localized in the cell nucleus, and can regulate the activity of TGF-β[29]. IL-2 is an important cytokine which regulates immune function, promotes the proliferation and activation of T cells, and stimulates the proliferation of natural killer cells[30,31]. TNF-α and IFN-γ are essential cellular immune molecules, which affect the immune escape of cells. Clinical studies have shown that disordered TNF-α and IFN-γ levels have a significant impact on the stability of the body’s internal environment, leading to abnormal immune function in patients, and inducing the occurrence and progression of tumors[32]. Tumor cells can avoid apoptosis by up-regulating Bcl2 and down-regulating Bax[33]. Bax and Bcl2 antagonize each other in apoptosis regulation of HCC cells. With the decrease or deletion of Bax expression, apoptosis of HCC cells is gradually weakened, which may be an important reason for the rapid growth and enlargement of tumors. Fas is a cell surface death receptor and an apoptosis signaling molecule. Fas which binds to FasL activates and transmits apoptosis signals, which is an important pathway in inducing tumor cell apoptosis. Previous research showed that the apoptosis of cancer cells mediated by Fas and FasL is an important part of the body’s immune surveillance[34], and the abnormal expression of Fas and FasL can lead to disordered apoptosis, which results in massive cell proliferation due to escaping the killing effect of cytotoxic T cells[35,36].
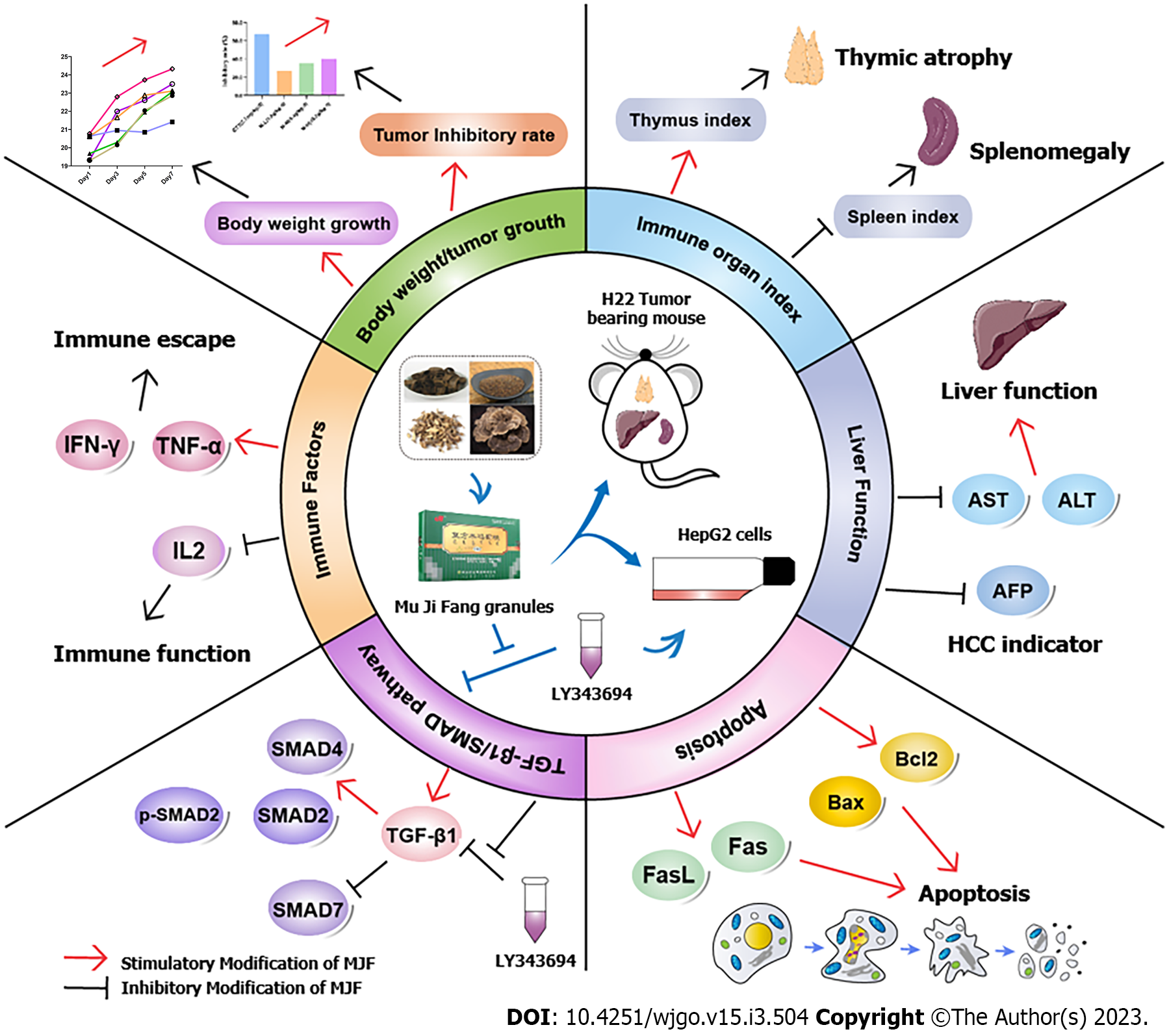
In this study, we evaluated the impact of MJF on HCC using both in vivo (H22 tumor-bearing mice) and in vitro (HepG2 cells) models (Figure 8). The results indicated that MJF could prevent HCC by improving body weight gain and tumor inhibition rate, protecting immune organs and liver function, affecting immunity and apoptosis, and up-regulating the TGF-β1/SMAD signaling pathway, by increasing the relative expression of TGF-β1, SMAD3, p-SMAD2 and SMAD4 and decreasing SMAD7, reducing immune factors TNF-α and IFN-γ, and decreasing the apoptosis cytokines Fas, FasL and Bcl2/Bax. In addition, to verify these findings, we treated HepG2 cells with the TGF-β1 inhibitor LY364947, which inhibited the TGF-β1/SMAD signaling pathway, increased Laminin Subunit Alpha 4 (LAMA4), TNF-α, IFN-γ, Fas, FasL, and Bcl2/Bax expression. However, MJF significantly inhibited the effect of LY364947. This suggested that MJF has potential effects on HCC by regulating the TGF-β1/SMAD pathway, immune and apoptotic cytokines.
Based on the identification of absorbable ingredients in mouse serum, we mainly focused on the fundamental study of potential anti-HCC targets of MJF, and intensive research on the mechanism of tumor cell apoptosis and immunology are still required. Moreover, other absorbable ingredients also need to be identified.
Our results demonstrated that MJF inhibits HCC by activating the TGF-β1/SMAD signaling pathway, immune and apoptotic cytokines, which may be due to the mechanisms of MJF in adjusting immune escape and apoptosis.
Hepatocellular carcinoma (HCC) is one of the most common digestive system cancers with high mortality rates worldwide.
Considerable effort has been expended in understanding the mechanism of Mu Ji Fang Granules (MJF) on tumor immunology in the treatment of HCC.
Our study explored the mechanism of MJF adjusting immune escape and apoptosis.
We conducted Molecule Network related to High Performance Liquid Chromatography-Electron Spray Ionization-Time of Flight- Mass Spectrometry to identify the absorbable ingredients of MJF, and hub potential anti-HCC targets were screened using network pharmacology and pathway enrichment analysis. Both in vivo and in vitro experiments were demonstrated, and multiply processes including Histopathological analysis, Immunosorbent Assay, RT-qPCR, Western blotting assays were to adopted to explore the function MJF inhibits HCC.
We found that MJF improved body weight gain and tumor inhibition rate in H22 tumor-bearing mice, protected immune organs and liver function, reduced the HCC indicator alpha-fetoprotein, affected immunity and apoptosis, and up-regulated the Transforming growth factor β1(TGF-β1)/SMAD signaling pathway, by increasing the relative expression of TGF-β1, SMAD2, p-SMAD2 and SMAD4 and decreasing SMAD7, reducing immune factors Tumor necrosis factor α and Interferon gamma, decr-easing apoptosis cytokines Fas, FasL and Bcl2/Bax, and inhibiting the effect of LY364947 in HepG2 cells.
Our findings suggested that MJF inhibits HCC by activating the TGF-β1/SMAD signaling pathway, and affecting immune and apoptotic cytokines.
Considerable effort has been expended in understanding the anti-HCC mechanism of MJF, all of which due to MJF’s particular participate in adjusting immune escape and apoptosis.
Provenance and peer review: Unsolicited article; Externally peer reviewed.
Peer-review model: Single blind
Specialty type: Oncology
Country/Territory of origin: China
Peer-review report’s scientific quality classification
Grade A (Excellent): 0
Grade B (Very good): B
Grade C (Good): C
Grade D (Fair): 0
Grade E (Poor): 0
P-Reviewer: Andersen JB, Denmark; Nardone G, Italy S-Editor: Li L L-Editor: A P-Editor: Chen YX
| 1. | International Agency for Research on Cancer. World Cancer Report: Cancer Research for Cancer Prevention. Wild CP, Weiderpass E, Stewart BW, Editor. Switzerland: WHO Press, World Health Organization, 2020: 355-367. |
| 2. | Zheng RS, Sun KX, Zhang SW, Zeng HM, Zou XN, Chen R, Gu XY, Wei WW, He J. [Report of cancer epidemiology in China, 2015]. Zhonghua Zhong Liu Za Zhi. 2019;41:19-28. [RCA] [PubMed] [DOI] [Full Text] [Cited by in RCA: 277] [Reference Citation Analysis (0)] |
| 3. | Cai L, Zou S, Liang D, Luan L. Structural characterization, antioxidant and hepatoprotective activities of polysaccharides from Sophorae tonkinensis Radix. Carbohydr Polym. 2018;184:354-365. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 109] [Cited by in RCA: 160] [Article Influence: 22.9] [Reference Citation Analysis (0)] |
| 4. | Zhang Y, Xiong H, Xu X, Xue X, Liu M, Xu S, Liu H, Gao Y, Zhang H, Li X. Compounds Identification in Semen Cuscutae by Ultra-High-Performance Liquid Chromatography (UPLCs) Coupled to Electrospray Ionization Mass Spectrometry. Molecules. 2018;23. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 23] [Cited by in RCA: 40] [Article Influence: 5.7] [Reference Citation Analysis (0)] |
| 5. | Zhou YY, Song HJ, Guo S, Wang Y, Gao HR, Zhang XJ, Sun YP, Liu Y, Yang BY, Kuang HX. A new triterpene from the green walnut husks of Juglans mandshurica Maxim. J Nat Med. 2019;73:800-804. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 7] [Cited by in RCA: 11] [Article Influence: 1.8] [Reference Citation Analysis (0)] |
| 6. | Kaplan Ö, Gökşen Tosun N, Özgür A, Erden Tayhan S, Bilgin S, Türkekul İ, Gökce İ. Microwave-assisted green synthesis of silver nanoparticles using crude extracts of Boletus edulis and Coriolus versicolor: Characterization, anticancer, antimicrobial and wound healing activities. Journal of Drug Delivery Science and Technology. 64. [RCA] [DOI] [Full Text] [Cited by in Crossref: 12] [Cited by in RCA: 8] [Article Influence: 2.0] [Reference Citation Analysis (0)] |
| 7. | Li F, Li Y, Deng ZP, Zhu XJ, Zhang ZG, Zhang XD, Tian JL, Li W, Zhao P. Traditional uses, phytochemistry, pharmacology and clinical applications of Cortex Juglandis Mandshuricae: A comprehensive review. J Ethnopharmacol. 2022;285:114887. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 9] [Cited by in RCA: 13] [Article Influence: 4.3] [Reference Citation Analysis (0)] |
| 8. | Wang L, Song YR. [Observation of Therapeutic Effects of Compound Mujj Granules in the Treatment of Chronic Hepatitis B with Hepatic Fibrosis]. Liaoning Zhongyi Zazhi. 2009;36:408-409. |
| 9. | Duan J, Jiang H, Ding Z, Xiao H, Xia W, Zhou YQ. [Clinical effect of compound muji granules in treating patients with compensatory cirrhosis induced by hepatitis B]. Zhongguo Yixue Gongcheng. 2017;(12):8-12. [DOI] [Full Text] |
| 10. | Sun J, Guo LY, Xue-Jun GU. [Effects of Muji granules on lymphocyte functions in tumor-bearing mice]. Shiyong Yixue Zazhi. 2010;26:2120-2122. [DOI] [Full Text] |
| 11. | Wang H, Hou Y, Ma X, Cui L, Bao Y, Xie Y, Li S, Meng X, Li J, Bai G. Multi-omics analysis reveals the mechanisms of action and therapeutic regimens of traditional Chinese medicine, Bufei Jianpi granules: Implication for COPD drug discovery. Phytomedicine. 2022;98:153963. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 4] [Cited by in RCA: 11] [Article Influence: 3.7] [Reference Citation Analysis (0)] |
| 12. | Piñero J, Ramírez-Anguita JM, Saüch-Pitarch J, Ronzano F, Centeno E, Sanz F, Furlong LI. The DisGeNET knowledge platform for disease genomics: 2019 update. Nucleic Acids Res. 2020;48:D845-D855. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 328] [Cited by in RCA: 1080] [Article Influence: 216.0] [Reference Citation Analysis (0)] |
| 13. | Lee L, Wang K, Li G, Xie Z, Wang Y, Xu J, Sun S, Pocalyko D, Bhak J, Kim C, Lee KH, Jang YJ, Yeom YI, Yoo HS, Hwang S. Liverome: a curated database of liver cancer-related gene signatures with self-contained context information. BMC Genomics. 2011;12 Suppl 3:S3. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 35] [Cited by in RCA: 43] [Article Influence: 3.1] [Reference Citation Analysis (0)] |
| 14. | Su WH, Chao CC, Yeh SH, Chen DS, Chen PJ, Jou YS. OncoDB.HCC: an integrated oncogenomic database of hepatocellular carcinoma revealed aberrant cancer target genes and loci. Nucleic Acids Res. 2007;35:D727-D731. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 63] [Cited by in RCA: 71] [Article Influence: 3.7] [Reference Citation Analysis (0)] |
| 15. | Daina A, Michielin O, Zoete V. SwissTargetPrediction: updated data and new features for efficient prediction of protein targets of small molecules. Nucleic Acids Res. 2019;47:W357-W364. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 760] [Cited by in RCA: 2250] [Article Influence: 450.0] [Reference Citation Analysis (0)] |
| 16. | Szklarczyk D, Gable AL, Nastou KC, Lyon D, Kirsch R, Pyysalo S, Doncheva NT, Legeay M, Fang T, Bork P, Jensen LJ, von Mering C. The STRING database in 2021: customizable protein-protein networks, and functional characterization of user-uploaded gene/measurement sets. Nucleic Acids Res. 2021;49:D605-D612. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 1211] [Cited by in RCA: 4827] [Article Influence: 1206.8] [Reference Citation Analysis (0)] |
| 17. | Zhou Y, Zhou B, Pache L, Chang M, Khodabakhshi AH, Tanaseichuk O, Benner C, Chanda SK. Metascape provides a biologist-oriented resource for the analysis of systems-level datasets. Nat Commun. 2019;10:1523. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 3766] [Cited by in RCA: 8788] [Article Influence: 1464.7] [Reference Citation Analysis (0)] |
| 18. | Liu Z, Luo H, Zhang L, Huang Y, Liu B, Ma K, Feng J, Xie J, Zheng J, Hu J, Zhan S, Zhu Y, Xu Q, Kong W, Wang X. Hyperhomocysteinemia exaggerates adventitial inflammation and angiotensin II-induced abdominal aortic aneurysm in mice. Circ Res. 2012;111:1261-1273. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 102] [Cited by in RCA: 132] [Article Influence: 10.2] [Reference Citation Analysis (0)] |
| 19. | Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) Method. Methods. 2001;25:402-408. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 149116] [Cited by in RCA: 133961] [Article Influence: 5581.7] [Reference Citation Analysis (1)] |
| 20. | Liu Z, Suo C, Mao X, Jiang Y, Jin L, Zhang T, Chen X. Global incidence trends in primary liver cancer by age at diagnosis, sex, region, and etiology, 1990-2017. Cancer. 2020;126:2267-2278. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 44] [Cited by in RCA: 88] [Article Influence: 17.6] [Reference Citation Analysis (0)] |
| 21. | Korean Liver Cancer Association (KLCA); National Cancer Center (NCC), Goyang, Korea. 2018 Korean Liver Cancer Association-National Cancer Center Korea Practice Guidelines for the Management of Hepatocellular Carcinoma. Korean J Radiol. 2019;20:1042-1113. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 158] [Cited by in RCA: 195] [Article Influence: 32.5] [Reference Citation Analysis (0)] |
| 22. | Marin JJG, Herraez E, Lozano E, Macias RIR, Briz O. Models for Understanding Resistance to Chemotherapy in Liver Cancer. Cancers (Basel). 2019;11. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 17] [Cited by in RCA: 29] [Article Influence: 4.8] [Reference Citation Analysis (0)] |
| 23. | Khazir J, Riley DL, Pilcher LA, De-Maayer P, Mir BA. Anticancer agents from diverse natural sources. Nat Prod Commun. 2014;9:1655-1669. [PubMed] |
| 24. | Hsiao WL, Liu L. The role of traditional Chinese herbal medicines in cancer therapy—from TCM theory to mechanistic insights. Planta Med. 2010;76:1118-1131. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 199] [Cited by in RCA: 218] [Article Influence: 14.5] [Reference Citation Analysis (0)] |
| 25. | Fox Ramos AE, Evanno L, Poupon E, Champy P, Beniddir MA. Natural products targeting strategies involving molecular networking: different manners, one goal. Nat Prod Rep. 2019;36:960-980. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 102] [Cited by in RCA: 157] [Article Influence: 26.2] [Reference Citation Analysis (0)] |
| 26. | Wang Z, Kim U, Liu J, Cheng C, Wu W, Guo S, Feng Y, Quinn RJ, Hou Y, Bai G. Comprehensive TCM molecular networking based on MS/MS in silico spectra with integration of virtual screening and affinity MS screening for discovering functional ligands from natural herbs. Anal Bioanal Chem. 2019;411:5785-5797. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 20] [Cited by in RCA: 28] [Article Influence: 4.7] [Reference Citation Analysis (0)] |
| 27. | Wakefield LM, Roberts AB. TGF-beta signaling: positive and negative effects on tumorigenesis. Curr Opin Genet Dev. 2002;12:22-29. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 650] [Cited by in RCA: 633] [Article Influence: 27.5] [Reference Citation Analysis (0)] |
| 28. | Boye A, Kan H, Wu C, Jiang Y, Yang X, He S, Yang Y. MAPK inhibitors differently modulate TGF-β/Smad signaling in HepG2 cells. Tumour Biol. 2015;36:3643-3651. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 17] [Cited by in RCA: 28] [Article Influence: 2.8] [Reference Citation Analysis (0)] |
| 29. | Syed V. TGF-β Signaling in Cancer. J Cell Biochem. 2016;117:1279-1287. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 204] [Cited by in RCA: 355] [Article Influence: 39.4] [Reference Citation Analysis (0)] |
| 30. | Waldmann TA. The biology of interleukin-2 and interleukin-15: implications for cancer therapy and vaccine design. Nat Rev Immunol. 2006;6:595-601. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 776] [Cited by in RCA: 882] [Article Influence: 46.4] [Reference Citation Analysis (0)] |
| 31. | Engel AL, Holt GE, Lu H. The pharmacokinetics of Toll-like receptor agonists and the impact on the immune system. Expert Rev Clin Pharmacol. 2011;4:275-289. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 173] [Cited by in RCA: 157] [Article Influence: 11.2] [Reference Citation Analysis (0)] |
| 32. | Cui HX, Tang L, Cheng FR, Yuan K. Antitumor Effects of Ethanol Extracts from Hyptis Rhomboidea in H(22) Tumor-bearing Mice. Pharmacogn Mag. 2017;13:571-575. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 2] [Cited by in RCA: 2] [Article Influence: 0.3] [Reference Citation Analysis (0)] |
| 33. | Croker BA, O’Donnell JA, Nowell CJ, Metcalf D, Dewson G, Campbell KJ, Rogers KL, Hu Y, Smyth GK, Zhang JG, White M, Lackovic K, Cengia LH, O’Reilly LA, Bouillet P, Cory S, Strasser A, Roberts AW. Fas-mediated neutrophil apoptosis is accelerated by Bid, Bak, and Bax and inhibited by Bcl-2 and Mcl-1. Proc Natl Acad Sci U S A. 2011;108:13135-13140. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 94] [Cited by in RCA: 99] [Article Influence: 7.1] [Reference Citation Analysis (0)] |
| 34. | Lee SH, Shin MS, Lee HS, Bae JH, Lee HK, Kim HS, Kim SY, Jang JJ, Joo M, Kang YK, Park WS, Park JY, Oh RR, Han SY, Lee JH, Kim SH, Lee JY, Yoo NJ. Expression of Fas and Fas-related molecules in human hepatocellular carcinoma. Hum Pathol. 2001;32:250-256. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 73] [Cited by in RCA: 73] [Article Influence: 3.0] [Reference Citation Analysis (0)] |
| 35. | Fukuzawa Y, Takahashi K, Furuta K, Tagaya T, Ishikawa T, Wada K, Omoto Y, Koji T, Kakumu S. Expression of fas/fas ligand (fasL) and its involvement in infiltrating lymphocytes in hepatocellular carcinoma (HCC). J Gastroenterol. 2001;36:681-688. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 38] [Cited by in RCA: 40] [Article Influence: 1.7] [Reference Citation Analysis (0)] |
| 36. | Alderson MR, Tough TW, Davis-Smith T, Braddy S, Falk B, Schooley KA, Goodwin RG, Smith CA, Ramsdell F, Lynch DH. Fas ligand mediates activation-induced cell death in human T lymphocytes. J Exp Med. 1995;181:71-77. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 679] [Cited by in RCA: 708] [Article Influence: 23.6] [Reference Citation Analysis (0)] |