Published online Mar 15, 2023. doi: 10.4251/wjgo.v15.i3.425
Peer-review started: November 30, 2022
First decision: December 9, 2022
Revised: January 2, 2023
Accepted: February 14, 2023
Article in press: February 14, 2023
Published online: March 15, 2023
Processing time: 104 Days and 13.8 Hours
Currently, colorectal cancer (CRC) represents the third most common malignancy and the second most deadly cancer worldwide, with a higher incidence in developed countries. Like other solid tumors, CRC is a heterogeneous genomic disease in which various alterations, such as point mutations, genomic rear
Core Tip: Due to the multitude of host and microbial genetic factors, the optimization of colorectal cancer (CRC) biomarkers remains difficult. The advent of next-generation sequencing (NGS) methods has facilitated the identification of previously unrecognized CRC-related genomic alterations and the CRC relationship with gut microbial composition. Hence, we have summarized the diagnostic tools used for CRC screening in the past and the present, focusing on the revolutionary role of NGS approaches in the identification of novel genomic CRC characteristics, the advancement of understanding the CRC carcinogenesis and the screening of clinically actionable targets for personalized medicine.
- Citation: Abbes S, Baldi S, Sellami H, Amedei A, Keskes L. Molecular methods for colorectal cancer screening: Progress with next-generation sequencing evolution. World J Gastrointest Oncol 2023; 15(3): 425-442
- URL: https://www.wjgnet.com/1948-5204/full/v15/i3/425.htm
- DOI: https://dx.doi.org/10.4251/wjgo.v15.i3.425
Currently, colorectal cancer (CRC) represents the third leading cause of cancer-related deaths in men and women worldwide, and the American Cancer Society estimates that the number of new colon and rectum cancer cases in the United States in 2022 will be around 106180 and 44850, respectively[1]. Despite the great progress of modern medicine, such as the development of novel therapeutic methods and the advent of new high throughput sequencing technologies, the mortality of CRC patients remains relatively high due to the lack of specific biomarkers and therapies.
Nowadays, CRC incidence largely varies across the world, and it appears to be positively correlated with the Human Development Index. For instance, in 2020 Norway, the Netherlands and Denmark reported the highest age-standardized incidence rates (41.9, 41.0 and 40.9 cases per 100000 persons, respectively) while Guinea, Gambia and Burkina Faso showed the lowest age-standardized incidence rates (3.3, 3.7 and 3.8 cases per 100000 persons, respectively)[2]. Usually, these variations reflect differences in the availability of screening services and other factors such as geographic location, environmental factors (e.g., polluted surface water sources), economic status and dietary and lifestyle habits[3].
At present, considering the difficulties in implementing significant lifestyle changes or common primary prevention strategies, screening and early detection represent the most powerful public health tool to reduce CRC mortality[4]. In general, an acceptable screening marker can only be considered by the health community if it respects specific parameters such as simplicity, safety and accuracy and has a known and defined suitable cutoff level[5]. Colonoscopy is considered the gold standard test for detecting CRC and promoting effectiveness in reducing its incidence and mortality. However, its high cost, invasiveness and reduced availability of necessary equipment hinder the establishment of organized screening settings, especially in poor countries[6].
In recent years, massive efforts have focused on next-generation sequencing (NGS) approaches to identify genes and microorganisms that are significantly associated with the malignancy due to the emerging evidence that intestinal microbial dysbiosis constitutes a crucial environmental factor in CRC onset and development[7]. Moreover, metagenomics approaches, considered a real revolution in the screening and diagnosis of different cancers, are also useful for the identification of novel potential markers for CRC diagnosis[8]. Hence, in this review, we summarized the diagnostic tools used for CRC screening in the past and the present, focusing on recent NGS approaches.
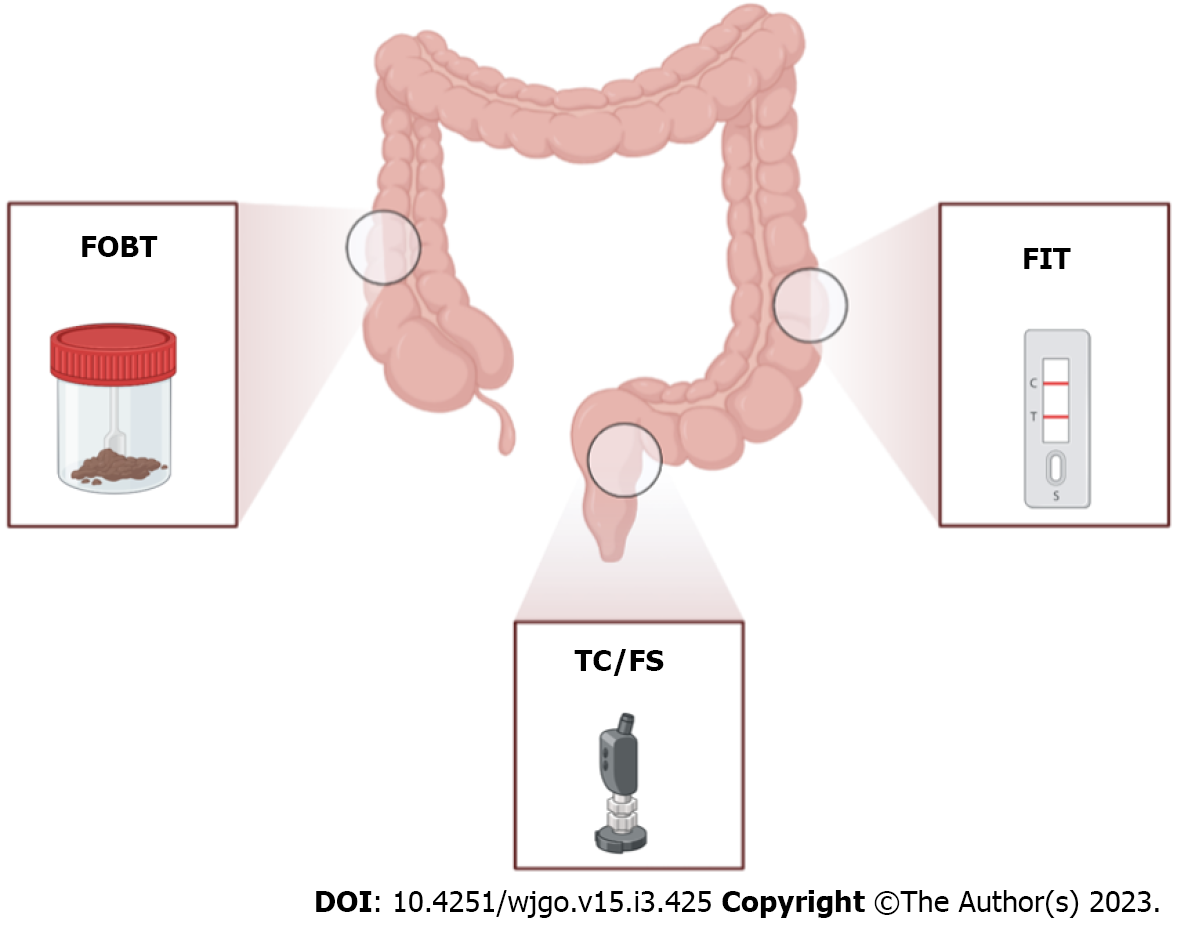
Since the 1970s, stool-based CRC screening was considered a successful non-invasive method with proven effectiveness given by the detection of high-risk polyps and early-stage malignancies that dramatically reduced CRC incidence and death[9] (Figure 1). The fecal occult blood test (FOBT) currently represents the early analysis for CRC screening that is recommended by the National Screening Committee[10]. This method is based on the detection of occult blood by measuring the non-protein portion of hemoglobin, the heme group, present in the stool. In particular, the heme present in a stool sample reacts with hydrogen peroxide-based developer to oxidize guaiac-infused paper, resulting in a blue color[4]. In general, FOBT has been shown to reduce both the incidence and the risk of CRC death with the advantages of ease of use and cheaper than other alternative screening approaches[11] (Table 1). Despite this, the FOBT method presents some limitations, such as the low sensitivity for colorectal adenomas that may not bleed or the specificity of the method that can be influenced by diet or drugs[12]; hence, Young et al[13] affirmed that FOBT is only suitable for limited colonoscopy resources with a need to constrain the test positivity rate[13].
| Technology | Approach | Sample types | Targeted and colorectal marker | Sensitivity/specificity for CRC | Advantages | Disadvantages |
| Chemical and immunochromatographic test | FOBT | Stool | Heme of hemoglobin | 4%-25%/95% | (1) Non-invasive; (2) Reduction of mortality (asymptomatic patients); (3) Colorimetric indicator; (4) Rapid and easy-to-carry out (self-testing); and (5) Commercially available test | (1) Low sensitivity for non-bleeding adenoma and advanced adenoma; (2) Specificity influenced by diet or drugs; (3) Must be done annually; (4) Risk of false positive results; (5) Three consecutive samples needed; (6) Only detects the blood present in the external layer of the stool; and (7) Confusing interpretation of the test results |
| FIT | Stool | Globin molecules | 62.0%-100%/94.9% | (1) Easy to use; (2) Flexible cutoff concentration; (3) Sensitive to low concentrations of globin; (4) Single sample needed; (5) Combined with FOBT inferred mortality; and (6) No dietary restriction | (1) Insensitive to digested hemoglobin; (2) Poor sensitivity for advanced adenoma; (3) Sensitivity based on threshold value of hemoglobin; and (4) Detect more distal neoplasms | |
| Visual inspection | FS | Distal colon | Polyps | 100%/100% | (1) Reduce colorectal cancer mortality and incidence; and (2) High susceptibility to detect adenomas | (1) Invasive process; (2) Not suitable for diabetic or psychotropic patients; (3) Expensive; (4) Serious harms for colonoscopy that increase with age; (5) Sigmoidoscopy was not effective for female screening (high risk for proximal colorectal cancer); and (6) Moderate-to-severe pain was reported for patients (bleeding, anxiety, etc) |
| TC | Entire colon | |||||
| Sanger sequencing methodology | Single gene sequencing | Tissue; liquid biopsy | A specific gene in human tumor DNA cells | High sensitivity (input of DNA mutated quantity < 1%) | (1) Non-invasive (blood/liquid biopsy); (2) Some mutations were prominent in colorectal cancer; (3) Bioinformatic analysis not required; (4) Simple and less time consuming; and (5) No specialized instrument in laboratory | (1) Requires high-quality DNA; (2) Heterogenous mutations genes; (3) Risk of contamination with normal tissue; and (4) Low coverage sequencing |
| ddPCR | Liquid biopsy. Tissue | Short amplicon sizes (< 100 bp) of human DNA | Very high sensitivity (input of mutated DNA quantity < 0.1% even with degraded DNA) | (1) Monitoring tumor burden in response to treatment and indicator of disease progression; (2) Precise measurement of copy number of mutated DNA and lower probability error (without standard samples); (3) Minimally invasive process; (4) Detects specific mutations; (5) Independent prognostic factor; and (6) Large target mutation | (1) No ability to detect benign lesions from plasma due to insufficient tumor burden; (2) Need an expensive instrument; (3) Limited prime-probe sets for each single nucleotide change; (4) No information in tumor-associated protein profiling; (5) Possibility of contamination with normal tissue; (6) Not strictly tumor specific; and (7) Necessity of cell search system | |
| MT-sDNA | Stool | Specific genes in human tumor DNA cells | 66%-94%/90%-96% | (1) Non-invasive test; (2) Acceptable cost; (3) Potential credibility; (4) No dietary restrictions (including food and medications); and (5) Widespread accessibility and multiple commercialized prototypes | (1) Lack of standardization or optimization of fecal DNA panels for high sensitivity and specificity; (2) Risk of contamination by microbial DNA; (3) No defined optimal interval for screening individuals; (4) Poor sensitivity for advanced adenoma; and (5) Must be repeated every 3 years | |
| Idylla system | Tissue; liquid biopsy | Specific genes in human tumor DNA cells | High sensitivity (input of DNA mutated quantity < 1%) | (1) Fully automated; (2) Real-time based-PCR molecular diagnosis system; (3) Without pre-analytical DNA extraction; (4) Lower cost and time requested for results; (5) Easily implemented in routine laboratory workflow; (6) Wide range of CRC-related mutations; and (7) Very sensitive to detect the most common CRC mutation | (1) No detection of complex genomic variants; (2) Unknown mutations were not detected; (3) Cannot detect rare and complex genomic variants not included in the reference range; and (4) Less suitable when new gene mutations appear | |
| Custom panel sequencing | Tissue; liquid biopsy | Specific genes in human tumor DNA cells | 95%-100%/99%-100% | (1) Decreased sequence cost; (2) Greater sequencing depth; (3) Simple and less time consuming; (4) Robust and tissue efficient; (5) Massive parallel multigene sequencing; and (6) Provide additional information (TMB levels/relevant mutated genes/heredity cancer genes) | (1) Low coverage sequencing; (2) There is no standardized procedure; and (3) Relatively long turnaround time of 3 d | |
| Next generation sequencing | WGS/WES | Tissue; liquid biopsy | All exome and all genome in human tumor DNA cells | 95%-100%/99%-100% | (1) Detection of large-scale mutations; (2) High coverage sequencing; (3) Complete definition of the genomic landscape for WGS; and (4) Complete mutation analysis panel without the repeated testing cost and reuse of material | (1) Require bioinformatics specialists; (2) Expensive; (3) Require good quality DNA; and (4) Relatively long turnaround time of 3 d |
| Third generation sequencing | Tissue/stool/liquid biopsy | (1) Specific genes/WES/WGS in human tumor DNA cells; and (2) Microbes in stool | 95%-100%/99%-100% | (1) Identification of large-scale rearrangement; (2) Sequencing errors do not release rearrangement; (3) High coverage sequencing; and (4) Fast and real time molecular diagnosis system | (1) High percentage of somatic errors; (2) Require bioinformatic specialists for assembling and analysis in laboratory; (3) Need specialized equipment in laboratory; and (4) Cannot detect some somatic mutations | |
| Metagenomic analysis | Stool/tissue | Microbial DNA in stool by Shotgun (all the DNA) or metabarcoding DNA (16S, ITS1, ITS2, 18S, etc) | (1) Microbial flora was more abundant than human cells in stool; (2) Benign lesions do not release human cells in stool; (3) Noninvasive diagnostic test; (4) Microbiota seems to play a role major in initiation and progression of CRC; (5) Test can be potentially used on all pathogen groups; and (6) Microbiota dysbiosis induces methylation of host genes | (1) Complex bioinformatics analysis; (2) Expensive; (3) Microbiota composition depends on sample preparation, conservation, extraction protocol and many other factors; (4) Need a healthy control group; (5) Many microorganisms (virus, bacteria, fungi) have not been identified and sequenced; (6) Metabarcoding analysis provides only taxonomic affiliation based in small region; and (7) Analysis results depends on reference database |
Different immunoassay methods have been used to measure the development of antibody-globin complexes, including immunochromatography, immunoturbidimetry and enzyme-linked immunosorbent assay (Table 1)[14]. For instance, the fecal immunochemical test (FIT) is used for the detection of microscopic amounts of blood present in the stool during defecation via the utilization of antibodies targeted to globin molecules (Figure 1). The antibodies preferably target lower gastr
Randomized controlled trials showed that the visual inspection of colic mucosa through flexible sigmoidoscopy (FS) decreased CRC mortality and incidence by 22%-31% and 18%-23%, respectively (Figure 1)[19]. Overall, FS represents a safe test, but its use is limited to the distal colon and a combined strategy using FS and FOBT/FIT only increases the endoscopic workload and reduces patient participation without solving the problem. Instead, total colonoscopy allows direct visualization and polyp removal over the whole colon (Figure 1), has a very high sensitivity and specificity for CRC and is usually used as confirmatory for all other screening strategies (Table 1)[20]. Although total colonoscopy can lead to a decrease in CRC incidence (66%-90%) and mortality (31%-65%), many features (e.g., invasive, expensive and painful) dramatically reduce its acceptability as a first-line screening test; moreover, proper training programs for endoscopists as well as continuous quality assurance are necessary[21].
It is currently well established that CRC development relies upon a stepwise acquisition of several chromosome mutations. The model of the adenoma-carcinoma progression, based on the accumulation of multiple mutations and epigenetic alterations, has been widely accepted[22]. Overall, there are two types of mutational events in sporadic CRC. The first concerns about 85% of all patients and consists of frequent mutations in APC[23], TP53[24], KRAS[25], BRAF, TTN, PIK3CA[26], FBXW7[27] and SMAD4 genes[28]; the second concerns 15% of CRC-sporadic patients and is characterized by a high level of hypermethylation of the MLH1 gene, responsible for DNA mismatch repair[29]. Additionally, a different complement of mutations in somatic genes has also been described[30].
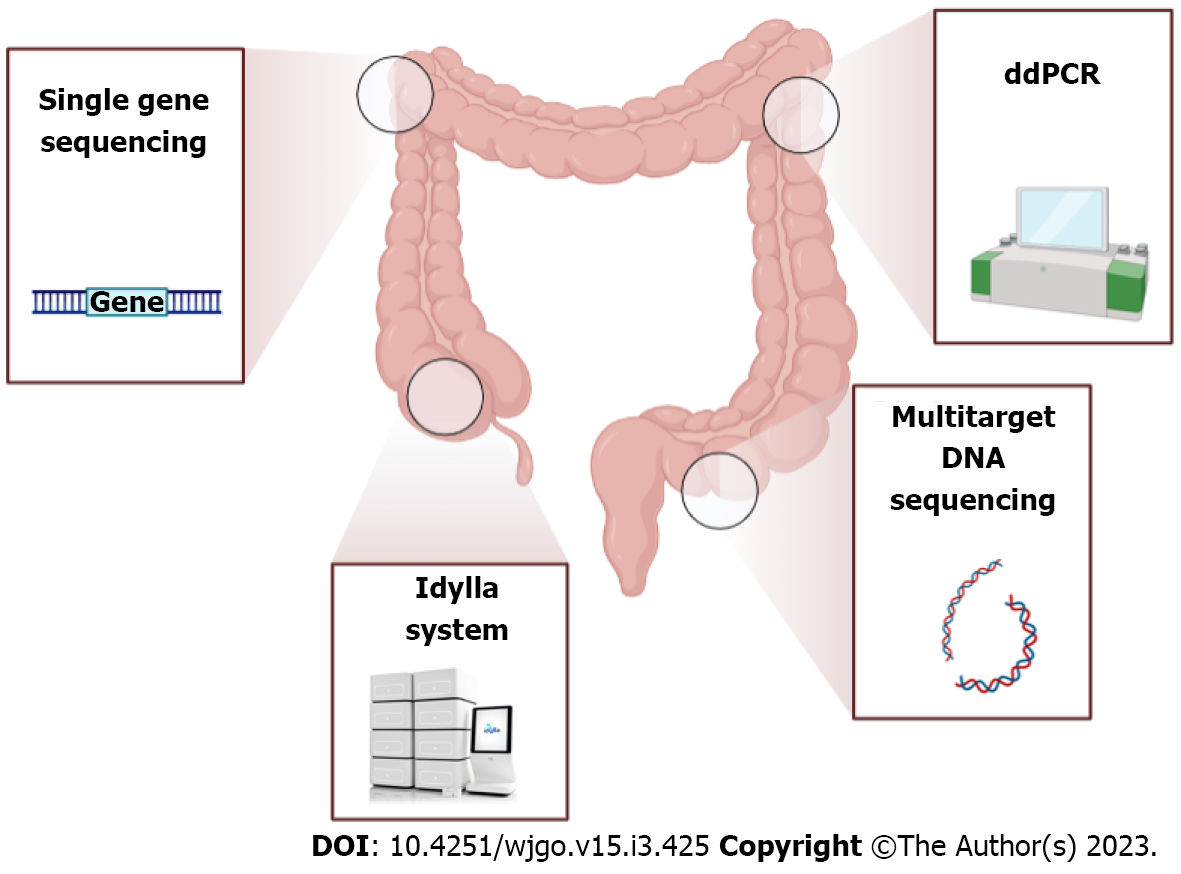
Considering their role in resistance to multiple treatment strategies, genotyping of gene mutations currently represents an important diagnostic and therapeutic tool (Figure 2). For instance, a mutation in APC, a tumor suppressor gene highly mutated in 57% of CRC cases and involved in DNA replication and repair processes, has been documented to strongly influence the chemotherapy response[31]. Also, SMAD4 gene mutations were observed in 2%-20% of CRC cases and were usually associated with poor response to cetuximab treatment[32]. In addition, several RAF mutations have been implicated in the induction of genomic instability, driving the proliferation of cancer cells[33], while heterogeneous KRAS mutations have been identified in almost 40% of CRC patients[34] (with a substitution in the G12C position as the most common detected), having a consequent association with anti-epidermal growth factor receptor treatment resistance[35].
To better represent the cancer heterogeneity using NGS technology, Ye et al[36] proposed a protocol for conducting rigorous systematic reviews and meta-analyses on the accuracy of KRAS mutation detection in CRC using non-invasive liquid biopsy samples[36]. Generally, liquid biopsies represent the collection of tumor-derived biomarkers in the blood or other body fluids, such as urine, saliva, stool or cerebrospinal fluid. Circulating tumor DNA (ctDNA), circulating tumor cells and exosomes are the most common tumor-related biomarkers assessed on liquid biopsy so far[37]. Moreover, the Food and Drug Administration recently approved a liquid biopsy test to analyze the frequency of KRAS, NRAS and BRAF hotspot mutations in ctDNA that could represent good CRC prognostic factors[38].
The multi-target stool DNA (MT-sDNA) test allows the identification of specific gene mutations in human tumor DNA cells separately from the more abundant microbial DNA in the stool (Figure 2). During the last few years, several key technological advances have led to increasingly accurate approaches to stool DNA testing including: (1) The use of a DNA preservative swab for stool collection; (2) The improvement of the target capture and amplification methods; and (3) The identification of new informative marker panels[39]. Zou et al[40] produced a methyl-binding domain protein bound to a column of nickel-agarose resin to increase the assay sensitivity for detecting methylated DNA markers in stool[40]. Subsequently, multiple prototypes of MT-sDNA test were commercialized, but only two were approved in August 2014 by the Food and Drug Administration for screening people at average risk for CRC aged over 50 years[29]. To date, both the American Cancer Society and the United Services Preventive Services Task Force affirmed that the MT-sDNA test can be repeated every 3 years to provide a decrease in CRC incidence and mortality with an acceptable cost and have approved this test for screening people of ages 45 years to 49 years[41,42].
Moreover, Heigh et al[43] performed a targeted single assay test with aberrant methylation of BMP3 alone and detected sessile serrated polyps with a sensitivity of 66% and a specificity of 91%[43]. Although additional biomarkers can be used by including multiple targets that reach the 21-target MT-sDNA test, no increase in the sensitivity or specificity was observed[44]. In general, most studies agree that MT-sDNA is effective to detect CRC with only a few exceptions. In fact, Imperiale et al[16] detected 60 out of 65 colon cancers by MT-sDNA test with an estimated sensitivity of 92.3% and a specificity of 90%, confirming that the MT-sDNA test is more sensitive than FIT, especially for the detection of lesions with high-grade dysplasia or sessile serrated polyps (≥ 1 cm). Overall, the method sensitivity varied from 62% to 91% for cancer and from 27% to 82% for advanced adenomas, with a specificity of 93% to 96% in people with normal findings on colonoscopy[45].
The advancement of the genetic knowledge in CRC and their related mutational events would improve the efficiency and the sensitivity of MT-sDNA tests by increasing the target DNA genes. Nowadays, MT-sDNA tests include quantitative molecular assays for KRAS mutations, NDRG4 and BMP3 methylation and β-actin and include eleven different DNA sequences commonly seen in colon polyps/cancers[46]. Therefore, as confirmed by a retrospective study conducted by Weiser et al[47] on 368494 subjects, the MT-sDNA test represents the most recommended CRC screening tool because of its widespread accessibility and higher sensitivity compared with other previously described methods such as FIT and FOBT (Table 1)[47].
Droplet digital polymerase chain reaction (ddPCR) is recognized as an established and trustworthy approach for clinical cancer research due to its high sensitivity (almost 74% for CRC) in comparison to traditional standard procedures, even in degraded samples[48] (Figure 2). This method consists of an enrichment strategy that allows the detection of low-level mutations by amplification of single DNA molecules without the need for standard reference curves. It is considered much easier, faster and less error-prone than real-time quantitative PCR[49]. Nowadays, ddPCR is commonly used for detecting rare alleles as molecular markers in plasma samples of pre- and postoperative CRC patients not only because of its high sensitivity for detecting tumor DNA (even with a very small fraction or degraded DNA) but also to monitor disease progression and the emergence of drug resistance[50]. Through this method, Taly et al[51] documented seven common mutations in codons 12 and 13 of the KRAS oncogene from plasma samples of CRC patients, demonstrating the clinical utility of multiplex ddPCR to screen multiple mutations with a sensitivity sufficient to detect mutations in circulating DNA obtained by non-invasive blood collection[51].
In the same context, ddPCR platforms using OncoBEAM technology demonstrated high sensitivity for plasma detection of KRAS mutations[52], and overall ddPCR has been largely applied to the detection and quantification of mutated genes including KRAS[53], BAT26[54], ITGA6 and ITGA6A[55] and hypermethylated GRIA4, VIPR2[56] and VIM[57] from both ctDNA or fecal DNA of CRC patients. Recently, Garrigou et al[58] proposed the screening of modifications in methylated ctDNA as a biomarker to monitor tumor evolution of CRC patients at different stages and concluded that it could be a universal approach to follow tumor burden of CRC patients as compared with mutated ctDNA, which requires previous tumor mutation identification[58]. To summarize, although there are many advantages of ddPCR including the high sensitivity and the large range of target mutations, its major limitation is represented by the lower availability of primer/probe sets (Table 1)[59].
The Idylla system (Biocartis, Mechelen, Belgium) consists of a cartridge-based fully automated medical device able to perform an innovative technology that consists of a conventional TaqMan reporter system and novel chemistry known as PlexPCR (amplicons containing a small region with a sequence different from that of target DNA) simultaneously with a PlexZyme (specific amplicon sequence-matched reporter probe) that allows multiplexing of numerous gene targets in one assay[60] (Figure 2). Hence, due to its ability to easily detect a wide range of CRC-related mutations, the Idylla approach can be easily implemented in pathology laboratories to reduce turnaround time[61]. It currently represents a feasible and validated test for KRAS, NRAS and epidermal growth factor receptor mutations in formalin-fixed paraffin-embedded tissues[62] and for BRAF hotspot mutations in plasma samples[63].
In addition, the Idylla system can be used to confirm uncertain outcomes of doubtful NGS results and/or in case of scarce tissue material within a few hours. For instance, Zwaenepoel et al[64] evaluated the clinical performance of the Idylla method in 330 CRC samples and demonstrated that this technology was able to give results in less than 2.5 h with only two invalid results. Many authors tested the full panel of CRC gene targets (BRAF, KRAS and NRAS) and found that the concordance between Idylla and NGS was 100% for BRAF and KRAS mutations and 94% for NRAS[65]. Therefore, this methodology is highly accurate for detecting frequent mutations and minimizing the contamination risk, in addition to reducing cost per test when compared with NGS or some conventional PCR assays. However, rare and/or complex genomic variants, which are not included in the reference ranges, cannot be detected by the Idylla system, and continuous improvement of its biomarker panel is necessary to guarantee efficient diagnosis[66].
Since the 2000s, and in coincidence with the emergence and development of new high-throughput sequencing technologies, many analyses have been undertaken to examine genetic susceptibility to diseases through genome-wide association studies. Zanke et al[67], using a multistage genetic association approach comprising 7480 CRC patients and 7779 controls, recognized a wide association of markers in chromosomal region 8q24, the same site where the SMAD7 gene is located[67]. In addition, a genome-wide association study performed by Broderick et al[68], consisting of the genotyping of 550163 tag single nucleotide polymorphisms in 940 individuals with familial CRC and 965 controls, identified three single nucleotide polymorphisms in the SMAD7 gene[68]. Subsequently, Tomlinson et al[69] confirmed these results and elucidated other markers in chromosomal regions of 8q23.3 and 10p14 at which common variants can influence the risk of CRC development[69].
NGS-based diagnostic assays are increasingly adopted especially with decreasing sequencing costs. In the early stage, sequencing technologies were used to target driver genes known to contribute to CRC, but recently larger chromosomal regions have been targeted exploiting the potential of these technologies in multigene sequencing by using a very low amount of biological material from liquid or tissue biopsy samples. In this step, many efforts have been made to standardize sequencing procedures and data analyses and to generate databases that store the sequencing information. Clinicians and research communities can use this information to provide better quality care[70].
Early in 2010, The Cancer Genome Atlas project conducted a genome-scale analysis of samples obtained from 276 CRC patients, analyzed exome sequences, DNA copy number, promoter methylation and messenger RNA and microRNA expression and concluded that 16% of CRC samples were found to be hypermutated, 77% of patients displayed one or both breakpoints leading to translocation in an intergenic region and 7% of patients reported a translocation involving the TTC28 gene (an inhibitor of tumor cell growth) located on chromosome 22[71]. Furthermore, the Pan-Cancer Analysis of Whole Genomes, the International Cancer Genome Consortium and The Cancer Genome Atlas projects recently described 2658 whole genomes of tumor samples and their matching normal tissues, not only of CRC but of 38 different cancer types, providing insights into the nature and timing of the many mutational processes that shape large and small-scale somatic variation in the cancer genome[65].
According to the improvement of NGS approaches, different sequencing platforms have been developed (Illumina, Ion Torrent, SOLiD, PacBio and Nanopore) that are classified in terms of maximum output, reads per run, accuracy, run time, amount of nucleic acids necessary for analysis and reads length. In particular, they can generate short (e.g., SOLiD, Ion Torrent, Illumina) or long reads (e.g., PacBio, Nanopore). While short reads sequencing does not exceed 300 base pairs and is more suitable for CRC diagnosis, long reads sequencing determines a better coverage of the genome and is more adaptable for large deletion/insertion determination or chromosomal rearrangement[72]. Considering that both short-read and long-read sequencing have their benefits and flaws depending on the experimental aim, it is important to remark that when somatic alterations in oncogenes and tumor suppressor genes are stable throughout the tumor clonal evolution, chromosomal alterations and copy number variation (CNV) could be lost during cancer progression[73].
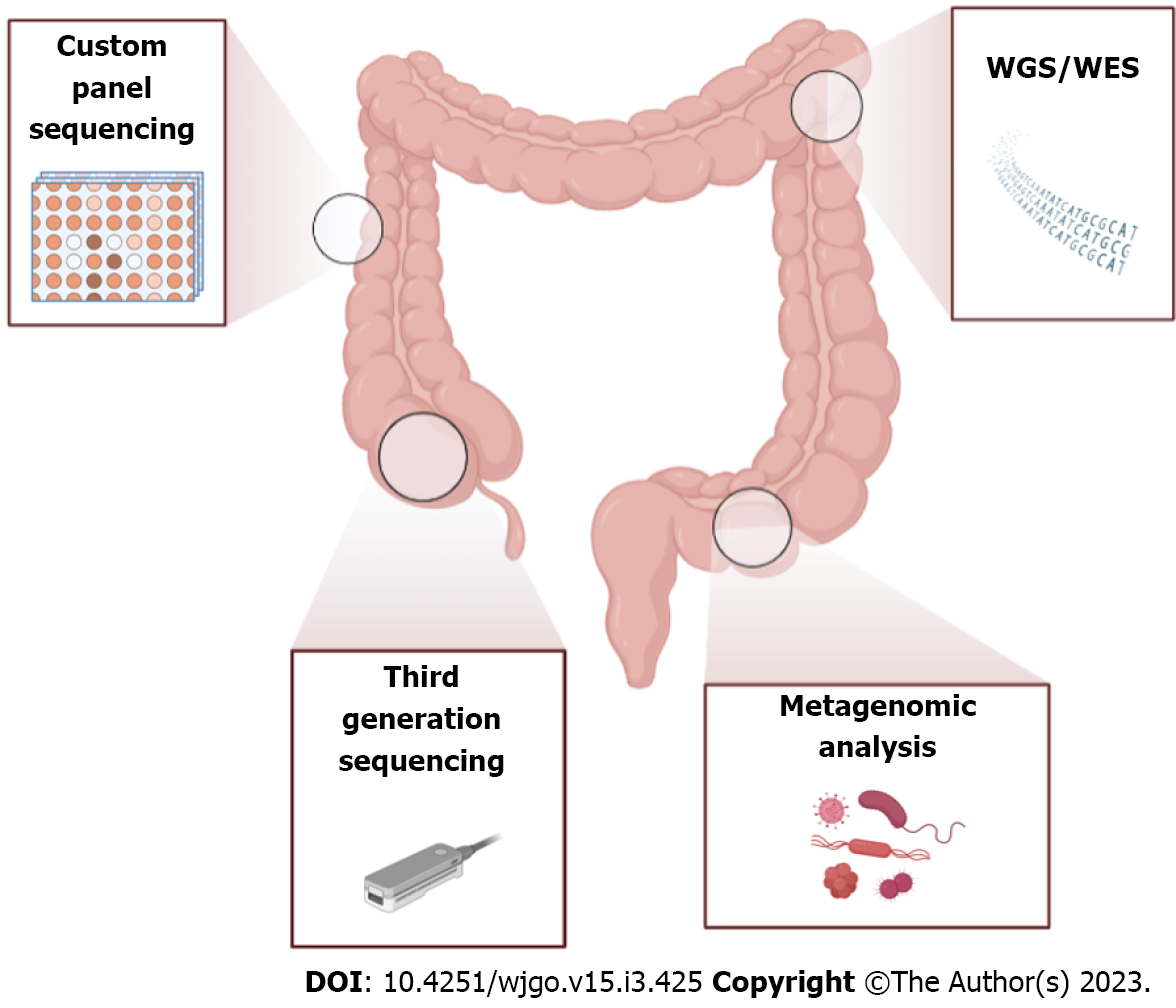
In addition, CRC represents one of the most interesting fields of NGS application because of its great quantity of activating mutations; in fact, next-gen techniques enable the identification of novel mutations/altered genes or genomic rearrangements allowing the discovery of new possible treatments[74]. In general, there are three more common NGS-based methods used for CRC studies: Custom panel; whole genome sequencing (WGS); or whole exome (WES) sequencing and third-generation sequencing approaches. In general, large-scale mutations were identified by WGS of tumor DNA, while point mutations were identified by targeted sequencing (Table 1).
During the last decade, several pipelines based on NGS approaches have been developed, and additional somatic mutations and chromosomal aberrations were detected in CRC samples (Figure 3). To simplify routine adoption of NGS tools, Zheng et al[75] considered a custom-designed panel of genes of only 2.2 Mb (exons and partial introns of cancer driver of more than 600 genes) and deduced a 9-loci model for detecting microsatellite instability (MSI) with 100% sensitivity and specificity compared with MSI and 84.3% overall concordance with immunohistochemistry staining[75]. Many authors have undertaken the simultaneous sequencing of many driver genes including low allele frequencies using NGS technologies and have emphasized the importance of the fine classification of mutational status as some cancers were associated with poor prognosis treatment[76]. In this regard, the comprehension of the wide heterogeneity of CRC lesions seems to be an extremely important point for tracing the therapeutic approach of the patient and developing effective strategies for early CRC detection and prevention.
Liquid biopsy samples have been investigated more than tumor tissue samples because of their non-invasiveness and their better representation of cancer heterogeneity[77]. In this context, Myint et al[78] developed a multiregional NGS approach from circulating cell-free DNA using a customized targeted CRC panel consisting of all coding exons of 116 genes, 22 genes recurrently amplified/deleted, 51 copy number regions, 121 MSI regions and 2 gene fusions (RSPO2 and RSPO3) and confirmed the widespread genetic heterogeneity in six adenoma samples, which affected the driver genes MMR, APC, PIK3CA, TP53 and SMAD4[78]. Additionally, based on an NGS analysis of a panel of 324 CRC-associated genes, Stahler et al[79] documented frequent single nucleotide variations in the TP53, APC, KRAS, PIK3CA, BRAF, SMAD4 and FBXW7 genes, and copy number alterations in the MYC and FLT3 genes[79].
Furthermore, Leary et al[80] developed a “personalized analysis of rearranged ends” approach, which can identify translocations and copy number alterations in CRC and other solid tumors. In addition, personalized analysis of rearranged ends can detect 57 regions containing putative somatic rearrangements, with an average of 14 rearrangements per sample[80]. Moreover, targeted sequencing strategies based on short reads and CNV determination could represent a good strategy for CRC studies. In fact, Gould et al[81] confirmed that an NGS approach using short fragments presented a sensitivity > 96% and a specificity > 99% for detecting samples with CNVs in the terminal five exons of PMS2[81].
Additionally, Corti et al[82] developed multiple DNA NGS approaches coupled with the computational and bioinformatics algorithm “IDEA” to target a WES of about 30 Mb, a custom panel of genes of about 603 Kb (frequently mutated genes) and another of 918 Kb (intron-exon junction to precisely identify the genomic breakpoint)[82]. Currently, IDEA represents a flexible and comprehensive pipeline for the management of CRC patients and is suitable for identifying several genetic alterations from a non-invasive sample (ctDNA) such as single nucleotide variants, insertions and deletions, gene copy-number alterations and chromosomal rearrangements in the KRAS, BRAF, PIK3CA and ERBB2 genes (usually involved in drug resistance). In general, sequencing of smaller target regions provides greater sequencing depth which allows for better recognition of low gene frequency variation. Hence a customized gene approach is more suitable for clinical oncology laboratories for many advantages such as the simplicity, low cost and fast of the method and the non-need for bioinformatics specialists in the laboratories (Table 1).
The contribution of MSI to the tumor mutational burden (TMB) due to a defective mismatch repair system is considered important in about 15% of CRC patients. According to the phenotype, MSI tumors can be divided into two distinct MSI phenotypes: MSI-high and MSI-low[83]. Recently, considering that the defective mismatch repair phenotype is crucial to define the efficacy of immune checkpoint inhibitor treatment, Xiao et al[84] used WES to evaluate the immune microenvironment and 2539 microsatellite loci in a group of 98 CRC patients. They concluded that the microenvironment of TMB-high was significantly more immune-responsive than TMB-low[84]. On the other hand, Gurjao et al[85] demonstrated the presence of a novel alkylating mutational signature, identified through the WES of 900 CRC patients and predicted that KRAS p.G12D, KRAS p.G13D and PIK3CA p.E545K driver mutations were mainly targeted by the alkylating signature in non-hypermutated patients[85].
Moreover, Chang et al[86] performed the WES of DNA obtained from tumor tissues of 32 surgical CRC patients and identified the well-known recurrent mutations in the APC, TP53, KRAS and FBXW7 genes and unreported mutations in additional 14 genes[86]. Furthermore, many authors confirmed that WGS largely contributed to determining the significant role of non-coding regions such as enhancers, transcription factor binding sites, promoters and 3’untranslated regions in CRC carcinogenesis[87]. In addition, WGS was used to demonstrate that metastatic lesions were enriched in gene mutations affecting PI3K-AKt signaling, cell adhesion and extracellular matrix processes[88]. Finally, Dashti et al[89] conceived a new technique based on a novel concept called ‘gene-motif,’ which identified seven CRC subtypes that can be effectively used to develop a personalized treatment[89].
In comparison to WES, the WGS approach has the advantage of increasing the overall variant accuracy and poor coverage but is more expensive and requires fresh-frozen tumor material to perform analysis of the highest quality (Figure 3 and Table 1).
Third-generation sequencing of long reads has been developed and represents the most suitable approach for the identification of deletion/duplication breakpoints and complex structural variants and CNV-neutral rearrangements such as inversions and large intronic insertions[90] (Figure 3). Indeed, many studies affirmed that long-read sequencing technologies have potential advantages over existing alternatives especially when pathogenic variants are in complex genomic regions, such as the recurrent PMS2 insertion-deletion variant. Using a locus-specific amplicon template, Watson et al[91] undertook Nanopore long-read sequencing to assess the CRC diagnostic accuracy of this platform. Pairwise comparison between sequencing results derived from short-read NGS and unidirectional Sanger sequencing and the consensus Nanopore dataset revealed 100% sequence identity[91]. Furthermore, reads produced by Nanopore oxford technology were able to identify both the 5’ and 3’ junctions and revealed detailed insertion sequence information[92].
Genetic factors that concern somatic mutations in KRAS, APC, p53, mismatch repair genes and other chromosomal aberrations explain less than 35% of all diagnosed CRCs, and many environmental exposures seem to modulate the cancer risk[93]. For instance, metagenomics studies based on 16S rRNA sequencing that has been recently conducted have documented the presence of more than a thousand microbial species in the human gastrointestinal tract carrying more than 100 times as many genes as the human genome[94] (Figure 3).
Therefore, considering the high microbial diversity in humans and their contribution to host health and pathological or malignant conditions, it was suggested that about 20% of the global cancer burden can be linked to microbial agents[95]. However, in addition to the several factors that can considerably modify the gut microbiota (GM) composition (e.g., age, sex, nationality, dietary and lifestyle habits, drugs or alcohol abuse)[96], multiple experimental challenges can influence the results of GM studies such as sampling methods and consistency[97], storage sample conditions[98], DNA extraction methods[99], type of primers used and pipelines adopted for data analyses[100]. For all these reasons, it is very hard to define a baseline microbial community for healthy people, especially due to the impossibility of obtaining biopsy samples from healthy controls. Therefore, tumor-adjacent tissue has been regarded as the healthy control, but many efforts have recently been expended to standardize the experimental and analytical methods[101]. The two most common metagenomics approaches for GM characterization are shotgun sequencing and metabarcoding.
These NGS-based approaches both contain three basic steps: Library preparation; sequencing; and data analysis. Sequencing libraries are typically created by fragmenting DNA and adding specialized adapters to both ends to allow the DNA fragments to bind to the sequencer flow cell. Due to unique barcodes added to each library that are used to distinguish between the libraries during data analysis, multiple libraries can be pooled together and sequenced in the same run (a process known as multiplexing). During the next sequencing step of the NGS workflow, the sequencer amplifies the DNA fragments, resulting in millions of copies of single-stranded DNA. In detail, chemically modified nucleotides bind to the DNA template strand through natural complementarity, and each nucleotide contains a fluorescent tag and a reversible terminator that blocks the incorporation of the next base. The fluorescent signal indicates which nucleotide has been added, and the terminator is cleaved so the next base can bind. After reading the forward DNA strand, the reads are washed away, and the process repeats for the reverse strand. After sequencing, the instrument software identifies nucleotides (a process called base calling) and the predicted accuracy of those base calls. Finally, data analysis can be performed with standard tools or with customized techniques[102].
While shotgun sequencing results in a very complicated data output (i.e. a huge amount of information that can be up to 1.5 terabases per run) because it simultaneously provides functional and taxonomic information about bacteria, fungi, viruses and a variety of other microorganisms, metabarcoding has a less complex data output and provides only taxonomic information about the bacterial (16S region sequencing) or fungal (ITS sequencing) composition of the sample (Table 1)[103]. Thus, metagenome-wide association studies have identified a correlation between many microbial species/gene markers and CRC, promoting the development of an affordable diagnostic test using both stool and tissue samples[104,105].
Much evidence has documented GM involvement in different diseases, including CRC. In particular, recent reports have demonstrated a bacterial driver-passenger model for CRC initiation and progression and showed that the first epithelial transformations can be supported by certain intestinal bacteria[106]. In 2012, Tjalsma et al[107] proposed a bacterial driver-passenger model for CRC in which pathogenic driver bacteria interact transiently with host cells to initiate CRC development and are then replaced by other passenger bacteria species that were unable to colonize healthy colon tissue but benefitted from altered metabolism of tumors cells[107]. To date, Wang et al[108] have identified Bacillus spp., Bradyrhizobium spp., Methylobacterium spp. and Streptomyces spp. as potential driver bacteria and Fusobacterium spp. and Campylobacter spp. as certain and abundant passenger bacteria[108].
Moreover, Luan et al[109] characterized the mucosa-adherent fungal microbiota of paired biopsy samples of adenomas and adjacent healthy tissue from 27 subjects using barcoded high-throughput sequencing that targeted the ITS region and reported a different fungal composition in patients with different adenoma stages and identified the phylum Glomeromycota as a possible powerful CRC marker[109]. Consistently, recent findings obtained through the WGS approach demonstrated that the Ascomycota/Basidiomycota ratio could represent a potential novel marker for early CRC detection[110]. Furthermore, Coker et al[111] used a shotgun metagenomics approach to evaluate the role of the archeome in colorectal carcinogenesis and found distinct archaea clusters in fecal samples from CRC patients, patients with adenomas and healthy subjects, with the CRC patients showing significant enrichment of halophilic archaea and depletion of methanogenic archaea[111].
Several metagenomic analyses of CRC patients have documented an over-representation of Fusobacterium nucleatum (F. nucleatum) in both tissue or stool samples in comparison to healthy controls[112]. Interestingly, in a large cohort of 616 participants, Yachida et al[113] demonstrated that the shift in the GM composition between CRC patients and healthy controls occurred in the very early stages of CRC development. In particular, the relative abundance of F. nucleatum was significantly elevated continuously from intramucosal carcinoma to more advanced stages, while Atopobium parvulum and Actinomyces odontolyticus were significantly increased only in multiple polypoid adenomas and/or intramucosal carcinomas[113]. Recently, in addition to F. nucleatum, several bacteria, such as Bacteroides fragilis, Escherichia coli, Streptococcus bovis, Enterococcus faecalis, Peptostreptococcus anaerobius and Lachnoclostridium spp., have been reported to be enriched in stool or tissue samples of CRC patients compared to healthy ones[114-118].
Moreover, an association between specific bacterial species and antitumor responses have been reported; for instance, a positive correlation between the abundance of Bifidobacterium longum or Ruminococcaceae members and the efficiency of CRC immunotherapy has been documented[119]. Eubacterium limosum, Ruthenibacterium lactatiformans, Fusobacterium ulcerans, Bacteroides uniformis, Paraprevotella xylaniphila and Alistipes senegalensis improved the effectiveness of immune checkpoint inhibitors[120].
Because the GM composition can be modified by probiotic and prebiotic supplementation, which can help maintain intestinal microbial homeostasis and mitigate dysbiosis, many reports have evaluated their effect on colorectal carcinogenesis. Overall, recent systematic reviews suggested that prebiotics may have a protective effect on the progress of CRC, while the administration of certain probiotics in patients with CRC reduced the side effects of chemotherapy, improved the outcomes of surgery, shortened hospital stays and decreased the risk of death[121,122]. However, the findings are still conflicting, and none determined changes in bacterial richness and diversity that are usually reduced in CRC patients. Thus, further studies are needed to better understand the prebiotic and probiotic effects in CRC patients.
Accumulating evidence has suggested that GM modulates the CRC progression, and its metabolites can play a crucial role in this scenario. The rapid development of technologies such as mass spectrometry and nuclear magnetic resonance have documented different profiles of microbial metabolites between CRC patients and healthy subjects. For instance, lower bile acid hydrolase and β-galactosidase abundances and higher levels of leucine, tyrosine, valine, choline, colibactin, gallocin, formyl methionyl leucyl phenylalanine, Bacteroides fragilis toxin and trimethylamine-N-oxide have been associated with CRC development[123-125]. Furthermore, the total amount of short chain fatty acids, the main metabolites produced by the bacterial anerobic fermentation of indigestible polysaccharides that exert various and fundamental functions for the host, was significantly lower in fecal and plasma samples of CRC patients compared to both patients with adenomatous polyps and healthy controls. Therefore, these metabolites could represent novel potential non-invasive diagnostic biomarkers for CRC[126,127]. In addition, recent investigations have reported that the GM plays a critical role in the effectiveness of anti-CRC treatments, including chemotherapy as well as immunosuppressive agents. For instance, it has been reported that the effectiveness of CRC treatment with 5-fluorouracil is enhanced by certain microbial metabolites[128]. The supplementation with probiotics or prebiotics could increase chances of therapeutic success[129].
Since the advent of NGS approaches, many molecular techniques for the diagnosis of CRC from invasive or non-invasive sampling have emerged and have significantly increased the number of known genes and mutations linked to CRC. However, due to the multitude of host and microbial genetic factors and the complexity of the tumor environment, the optimization of a CRC biomarker remains difficult, especially in stool samples, in which the complexity of the lesion environment seems to play a key role[50]. Thus, the development of a biological method to find stable, sensitive and specific markers in non-invasive samples such as feces or plasma remains an arduous challenge to be carried out. Furthermore, despite the great progress in metagenomics methods and bioinformatics tools, WES and WGS are still feasible only in expert centers. Only limited pieces of genomic information are currently clinically relevant for the care of CRC patients, and the list of predictive actionable genomic biomarkers is quite short[86]. Apart from the identification of novel microbial biomarkers, new CRC-associated molecules are under evaluation for CRC screening, such as circular RNA and Piwi-interacting RNA. These advances in the identification of microbial markers and the improvement of non-invasive diagnostic capabilities and their applications in guiding precision cancer therapies are poised to change the diagnosis of CRC and select and monitor the treatments in the near future due to the increasingly adopted precision medicine for the care of CRC patients.
Provenance and peer review: Invited article; Externally peer reviewed.
Peer-review model: Single blind
Specialty type: Oncology
Country/Territory of origin: Italy
Peer-review report’s scientific quality classification
Grade A (Excellent): 0
Grade B (Very good): B
Grade C (Good): C, C
Grade D (Fair): D
Grade E (Poor): 0
P-Reviewer: Bustamante-Lopez LA, Brazil; Galmiche M, France; Wan XH, China; Wen XL, China S-Editor: Fan JR L-Editor: Filipodia P-Editor: Fan JR
| 1. | Siegel RL, Miller KD, Fuchs HE, Jemal A. Cancer statistics, 2022. CA Cancer J Clin. 2022;72:7-33. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 4235] [Cited by in RCA: 11434] [Article Influence: 3811.3] [Reference Citation Analysis (4)] |
| 2. | Xi Y, Xu P. Global colorectal cancer burden in 2020 and projections to 2040. Transl Oncol. 2021;14:101174. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 687] [Cited by in RCA: 1355] [Article Influence: 338.8] [Reference Citation Analysis (5)] |
| 3. | Bénard F, Barkun AN, Martel M, von Renteln D. Systematic review of colorectal cancer screening guidelines for average-risk adults: Summarizing the current global recommendations. World J Gastroenterol. 2018;24:124-138. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in CrossRef: 228] [Cited by in RCA: 204] [Article Influence: 29.1] [Reference Citation Analysis (7)] |
| 4. | Janz T, Lu K, Povlow MR, Urso B. A Review of Colorectal Cancer Detection Modalities, Stool DNA, and Fecal Immunochemistry Testing in Adults Over the Age of 50. Cureus. 2016;8:e931. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 4] [Cited by in RCA: 11] [Article Influence: 1.2] [Reference Citation Analysis (0)] |
| 5. | Zygulska AL, Pierzchalski P. Novel Diagnostic Biomarkers in Colorectal Cancer. Int J Mol Sci. 2022;23. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 18] [Cited by in RCA: 135] [Article Influence: 45.0] [Reference Citation Analysis (0)] |
| 6. | Onyoh EF, Hsu WF, Chang LC, Lee YC, Wu MS, Chiu HM. The Rise of Colorectal Cancer in Asia: Epidemiology, Screening, and Management. Curr Gastroenterol Rep. 2019;21:36. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 75] [Cited by in RCA: 119] [Article Influence: 19.8] [Reference Citation Analysis (0)] |
| 7. | Fan X, Jin Y, Chen G, Ma X, Zhang L. Gut Microbiota Dysbiosis Drives the Development of Colorectal Cancer. Digestion. 2021;102:508-515. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 42] [Cited by in RCA: 93] [Article Influence: 23.3] [Reference Citation Analysis (0)] |
| 8. | Ng C, Li H, Wu WKK, Wong SH, Yu J. Genomics and metagenomics of colorectal cancer. J Gastrointest Oncol. 2019;10:1164-1170. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 15] [Cited by in RCA: 23] [Article Influence: 3.8] [Reference Citation Analysis (0)] |
| 9. | Ebner DW, Kisiel JB. Stool-Based Tests for Colorectal Cancer Screening: Performance Benchmarks Lead to High Expected Efficacy. Curr Gastroenterol Rep. 2020;22:32. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 16] [Cited by in RCA: 16] [Article Influence: 3.2] [Reference Citation Analysis (0)] |
| 10. | Medical Advisory Secretariat. Fecal occult blood test for colorectal cancer screening: an evidence-based analysis. Ont Health Technol Assess Ser. 2009;9:1-40. [PubMed] |
| 11. | Kościelniak-Merak B, Radosavljević B, Zając A, Tomasik PJ. Faecal Occult Blood Point-of-Care Tests. J Gastrointest Cancer. 2018;49:402-405. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 14] [Cited by in RCA: 15] [Article Influence: 2.5] [Reference Citation Analysis (0)] |
| 12. | Allison JE, Fraser CG, Halloran SP, Young GP. Comparing fecal immunochemical tests: improved standardization is needed. Gastroenterology. 2012;142:422-424. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 42] [Cited by in RCA: 45] [Article Influence: 3.5] [Reference Citation Analysis (0)] |
| 13. | Young GP, Symonds EL, Allison JE, Cole SR, Fraser CG, Halloran SP, Kuipers EJ, Seaman HE. Advances in Fecal Occult Blood Tests: the FIT revolution. Dig Dis Sci. 2015;60:609-622. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 108] [Cited by in RCA: 143] [Article Influence: 14.3] [Reference Citation Analysis (0)] |
| 14. | Allison JE, Fraser CG, Halloran SP, Young GP. Population screening for colorectal cancer means getting FIT: the past, present, and future of colorectal cancer screening using the fecal immunochemical test for hemoglobin (FIT). Gut Liver. 2014;8:117-130. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 108] [Cited by in RCA: 130] [Article Influence: 11.8] [Reference Citation Analysis (0)] |
| 15. | Kortlever TL, van der Vlugt M, Dekker E, Bossuyt PMM. Individualized faecal immunochemical test cut-off based on age and sex in colorectal cancer screening. Prev Med Rep. 2021;23:101447. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 6] [Cited by in RCA: 13] [Article Influence: 3.3] [Reference Citation Analysis (0)] |
| 16. | Imperiale TF, Ransohoff DF, Itzkowitz SH, Levin TR, Lavin P, Lidgard GP, Ahlquist DA, Berger BM. Multitarget stool DNA testing for colorectal-cancer screening. N Engl J Med. 2014;370:1287-1297. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1015] [Cited by in RCA: 1241] [Article Influence: 112.8] [Reference Citation Analysis (1)] |
| 17. | Levin B, Lieberman DA, McFarland B, Andrews KS, Brooks D, Bond J, Dash C, Giardiello FM, Glick S, Johnson D, Johnson CD, Levin TR, Pickhardt PJ, Rex DK, Smith RA, Thorson A, Winawer SJ; American Cancer Society Colorectal Cancer Advisory Group; US Multi-Society Task Force; American College of Radiology Colon Cancer Committee. Screening and surveillance for the early detection of colorectal cancer and adenomatous polyps, 2008: a joint guideline from the American Cancer Society, the US Multi-Society Task Force on Colorectal Cancer, and the American College of Radiology. Gastroenterology. 2008;134:1570-1595. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1423] [Cited by in RCA: 1457] [Article Influence: 85.7] [Reference Citation Analysis (0)] |
| 18. | Niedermaier T, Balavarca Y, Brenner H. Stage-Specific Sensitivity of Fecal Immunochemical Tests for Detecting Colorectal Cancer: Systematic Review and Meta-Analysis. Am J Gastroenterol. 2020;115:56-69. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 34] [Cited by in RCA: 72] [Article Influence: 14.4] [Reference Citation Analysis (0)] |
| 19. | Holme Ø, Løberg M, Kalager M, Bretthauer M, Hernán MA, Aas E, Eide TJ, Skovlund E, Schneede J, Tveit KM, Hoff G. Effect of flexible sigmoidoscopy screening on colorectal cancer incidence and mortality: a randomized clinical trial. JAMA. 2014;312:606-615. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 279] [Cited by in RCA: 311] [Article Influence: 28.3] [Reference Citation Analysis (0)] |
| 20. | Whitlock EP, Lin JS, Liles E, Beil TL, Fu R. Screening for colorectal cancer: a targeted, updated systematic review for the U.S. Preventive Services Task Force. Ann Intern Med. 2008;149:638-658. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 507] [Cited by in RCA: 515] [Article Influence: 30.3] [Reference Citation Analysis (0)] |
| 21. | Jodal HC, Helsingen LM, Anderson JC, Lytvyn L, Vandvik PO, Emilsson L. Colorectal cancer screening with faecal testing, sigmoidoscopy or colonoscopy: a systematic review and network meta-analysis. BMJ Open. 2019;9:e032773. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 110] [Cited by in RCA: 92] [Article Influence: 15.3] [Reference Citation Analysis (0)] |
| 22. | Vogelstein B, Kinzler KW. The multistep nature of cancer. Trends Genet. 1993;9:138-141. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1217] [Cited by in RCA: 1129] [Article Influence: 35.3] [Reference Citation Analysis (0)] |
| 23. | Bettegowda C, Sausen M, Leary RJ, Kinde I, Wang Y, Agrawal N, Bartlett BR, Wang H, Luber B, Alani RM, Antonarakis ES, Azad NS, Bardelli A, Brem H, Cameron JL, Lee CC, Fecher LA, Gallia GL, Gibbs P, Le D, Giuntoli RL, Goggins M, Hogarty MD, Holdhoff M, Hong SM, Jiao Y, Juhl HH, Kim JJ, Siravegna G, Laheru DA, Lauricella C, Lim M, Lipson EJ, Marie SK, Netto GJ, Oliner KS, Olivi A, Olsson L, Riggins GJ, Sartore-Bianchi A, Schmidt K, Shih lM, Oba-Shinjo SM, Siena S, Theodorescu D, Tie J, Harkins TT, Veronese S, Wang TL, Weingart JD, Wolfgang CL, Wood LD, Xing D, Hruban RH, Wu J, Allen PJ, Schmidt CM, Choti MA, Velculescu VE, Kinzler KW, Vogelstein B, Papadopoulos N, Diaz LA Jr. Detection of circulating tumor DNA in early- and late-stage human malignancies. Sci Transl Med. 2014;6:224ra24. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 2770] [Cited by in RCA: 3552] [Article Influence: 322.9] [Reference Citation Analysis (0)] |
| 24. | Li H, Zhang J, Tong JHM, Chan AWH, Yu J, Kang W, To KF. Targeting the Oncogenic p53 Mutants in Colorectal Cancer and Other Solid Tumors. Int J Mol Sci. 2019;20. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 41] [Cited by in RCA: 66] [Article Influence: 11.0] [Reference Citation Analysis (0)] |
| 25. | Macedo MP, Andrade Lde B, Coudry R, Crespo R, Gomes M, Lisboa BC, Aguiar S Jr, Soares FA, Carraro DM, Cunha IW. Multiple mutations in the Kras gene in colorectal cancer: review of the literature with two case reports. Int J Colorectal Dis. 2011;26:1241-1248. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 28] [Cited by in RCA: 30] [Article Influence: 2.1] [Reference Citation Analysis (0)] |
| 26. | Mei ZB, Duan CY, Li CB, Cui L, Ogino S. Prognostic role of tumor PIK3CA mutation in colorectal cancer: a systematic review and meta-analysis. Ann Oncol. 2016;27:1836-1848. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 112] [Cited by in RCA: 113] [Article Influence: 12.6] [Reference Citation Analysis (0)] |
| 27. | Fiore D, Piscopo C, Proto MC, Vasaturo M, Dal Piaz F, Fusco BM, Pagano C, Laezza C, Bifulco M, Gazzerro P. N6-Isopentenyladenosine Inhibits Colorectal Cancer and Improves Sensitivity to 5-Fluorouracil-Targeting FBXW7 Tumor Suppressor. Cancers (Basel). 2019;11. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 9] [Cited by in RCA: 11] [Article Influence: 1.8] [Reference Citation Analysis (0)] |
| 28. | Yu J, Wu WK, Li X, He J, Li XX, Ng SS, Yu C, Gao Z, Yang J, Li M, Wang Q, Liang Q, Pan Y, Tong JH, To KF, Wong N, Zhang N, Chen J, Lu Y, Lai PB, Chan FK, Li Y, Kung HF, Yang H, Wang J, Sung JJ. Novel recurrently mutated genes and a prognostic mutation signature in colorectal cancer. Gut. 2015;64:636-645. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 122] [Cited by in RCA: 148] [Article Influence: 14.8] [Reference Citation Analysis (0)] |
| 29. | Carethers JM. DNA testing and molecular screening for colon cancer. Clin Gastroenterol Hepatol. 2014;12:377-381. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 24] [Cited by in RCA: 28] [Article Influence: 2.5] [Reference Citation Analysis (0)] |
| 30. | Carethers JM, Jung BH. Genetics and Genetic Biomarkers in Sporadic Colorectal Cancer. Gastroenterology. 2015;149:1177-1190.e3. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 254] [Cited by in RCA: 340] [Article Influence: 34.0] [Reference Citation Analysis (0)] |
| 31. | Stefanski CD, Prosperi JR. Wnt-Independent and Wnt-Dependent Effects of APC Loss on the Chemotherapeutic Response. Int J Mol Sci. 2020;21. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 15] [Cited by in RCA: 16] [Article Influence: 3.2] [Reference Citation Analysis (0)] |
| 32. | Mei Z, Shao YW, Lin P, Cai X, Wang B, Ding Y, Ma X, Wu X, Xia Y, Zhu D, Shu Y, Fu Z, Gu Y. SMAD4 and NF1 mutations as potential biomarkers for poor prognosis to cetuximab-based therapy in Chinese metastatic colorectal cancer patients. BMC Cancer. 2018;18:479. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 27] [Cited by in RCA: 34] [Article Influence: 4.9] [Reference Citation Analysis (0)] |
| 33. | Armaghany T, Wilson JD, Chu Q, Mills G. Genetic alterations in colorectal cancer. Gastrointest Cancer Res. 2012;5:19-27. [PubMed] |
| 34. | Tan C, Du X. KRAS mutation testing in metastatic colorectal cancer. World J Gastroenterol. 2012;18:5171-5180. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in RCA: 103] [Reference Citation Analysis (0)] |
| 35. | Gausachs M, Borras E, Chang K, Gonzalez S, Azuara D, Delgado Amador A, Lopez-Doriga A, San Lucas FA, Sanjuan X, Paules MJ, Taggart MW, Davies GE, Ehli EA, Fowler J, Moreno V, Pineda M, You YN, Lynch PM, Lazaro C, Navin NE, Scheet PA, Hawk ET, Capella G, Vilar E. Mutational Heterogeneity in APC and KRAS Arises at the Crypt Level and Leads to Polyclonality in Early Colorectal Tumorigenesis. Clin Cancer Res. 2017;23:5936-5947. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 22] [Cited by in RCA: 22] [Article Influence: 2.8] [Reference Citation Analysis (0)] |
| 36. | Ye P, Cai P, Xie J, Wei Y. The diagnostic accuracy of digital PCR, ARMS and NGS for detecting KRAS mutation in cell-free DNA of patients with colorectal cancer: A protocol for systematic review and meta-analysis. Medicine (Baltimore). 2020;99:e20708. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 2] [Cited by in RCA: 1] [Article Influence: 0.2] [Reference Citation Analysis (0)] |
| 37. | Mauri G, Vitiello PP, Sogari A, Crisafulli G, Sartore-Bianchi A, Marsoni S, Siena S, Bardelli A. Liquid biopsies to monitor and direct cancer treatment in colorectal cancer. Br J Cancer. 2022;127:394-407. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 49] [Cited by in RCA: 68] [Article Influence: 22.7] [Reference Citation Analysis (0)] |
| 38. | Güttlein L, Luca MR, Esteso F, Fresno C, Mariani J, Otero Pizarro M, Brest E, Starapoli S, Kreimberg K, Teves P, Mendoza Bertelli A, R Girotti M, Salanova R, O'Connor JM. Liquid biopsy for KRAS, NRAS and BRAF mutation testing in advanced colorectal cancer patients: the Argentinean experience. Future Oncol. 2022;18:3277-3287. [RCA] [PubMed] [DOI] [Full Text] [Cited by in RCA: 6] [Reference Citation Analysis (0)] |
| 39. | Johnson DH, Kisiel JB, Burger KN, Mahoney DW, Devens ME, Ahlquist DA, Sweetser S. Multitarget stool DNA test: clinical performance and impact on yield and quality of colonoscopy for colorectal cancer screening. Gastrointest Endosc. 2017;85:657-665.e1. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 37] [Cited by in RCA: 42] [Article Influence: 5.3] [Reference Citation Analysis (0)] |
| 40. | Zou H, Harrington J, Rego RL, Ahlquist DA. A novel method to capture methylated human DNA from stool: implications for colorectal cancer screening. Clin Chem. 2007;53:1646-1651. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 33] [Cited by in RCA: 33] [Article Influence: 1.8] [Reference Citation Analysis (0)] |
| 41. | Knudsen AB, Rutter CM, Peterse EFP, Lietz AP, Seguin CL, Meester RGS, Perdue LA, Lin JS, Siegel RL, Doria-Rose VP, Feuer EJ, Zauber AG, Kuntz KM, Lansdorp-Vogelaar I. Colorectal Cancer Screening: An Updated Modeling Study for the US Preventive Services Task Force. JAMA. 2021;325:1998-2011. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 145] [Cited by in RCA: 213] [Article Influence: 53.3] [Reference Citation Analysis (0)] |
| 42. | Volk RJ, Leal VB, Jacobs LE, Wolf AMD, Brooks DD, Wender RC, Smith RA. From guideline to practice: New shared decision-making tools for colorectal cancer screening from the American Cancer Society. CA Cancer J Clin. 2018;68:246-249. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 24] [Cited by in RCA: 31] [Article Influence: 4.4] [Reference Citation Analysis (0)] |
| 43. | Heigh RI, Yab TC, Taylor WR, Hussain FT, Smyrk TC, Mahoney DW, Domanico MJ, Berger BM, Lidgard GP, Ahlquist DA. Detection of colorectal serrated polyps by stool DNA testing: comparison with fecal immunochemical testing for occult blood (FIT). PLoS One. 2014;9:e85659. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 53] [Cited by in RCA: 62] [Article Influence: 5.6] [Reference Citation Analysis (0)] |
| 44. | Zhai RL, Xu F, Zhang P, Zhang WL, Wang H, Wang JL, Cai KL, Long YP, Lu XM, Tao KX, Wang GB. The Diagnostic Performance of Stool DNA Testing for Colorectal Cancer: A Systematic Review and Meta-Analysis. Medicine (Baltimore). 2016;95:e2129. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 16] [Cited by in RCA: 15] [Article Influence: 1.7] [Reference Citation Analysis (0)] |
| 45. | Tagore KS, Lawson MJ, Yucaitis JA, Gage R, Orr T, Shuber AP, Ross ME. Sensitivity and specificity of a stool DNA multitarget assay panel for the detection of advanced colorectal neoplasia. Clin Colorectal Cancer. 2003;3:47-53. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 104] [Cited by in RCA: 106] [Article Influence: 4.8] [Reference Citation Analysis (0)] |
| 46. | Olson JE, Kirsch EJ, Edwards V DK, Kirt CR, Kneedler B, Laffin JJ, Weaver AL, St Sauver JL, Yost KJ, Finney Rutten LJ. Colorectal cancer outcomes after screening with the multi-target stool DNA assay: protocol for a large-scale, prospective cohort study (the Voyage study). BMJ Open Gastroenterol. 2020;7:e000353. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 4] [Cited by in RCA: 7] [Article Influence: 1.4] [Reference Citation Analysis (0)] |
| 47. | Weiser E, Parks PD, Swartz RK, Thomme JV, Lavin PT, Limburg P, Berger BM. Cross-sectional adherence with the multi-target stool DNA test for colorectal cancer screening: Real-world data from a large cohort of older adults. J Med Screen. 2021;28:18-24. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 16] [Cited by in RCA: 40] [Article Influence: 8.0] [Reference Citation Analysis (0)] |
| 48. | Ma ZY, Chan CSY, Lau KS, Ng L, Cheng YY, Leung WK. Application of droplet digital polymerase chain reaction of plasma methylated septin 9 on detection and early monitoring of colorectal cancer. Sci Rep. 2021;11:23446. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 1] [Cited by in RCA: 15] [Article Influence: 3.8] [Reference Citation Analysis (0)] |
| 49. | Hindson BJ, Ness KD, Masquelier DA, Belgrader P, Heredia NJ, Makarewicz AJ, Bright IJ, Lucero MY, Hiddessen AL, Legler TC, Kitano TK, Hodel MR, Petersen JF, Wyatt PW, Steenblock ER, Shah PH, Bousse LJ, Troup CB, Mellen JC, Wittmann DK, Erndt NG, Cauley TH, Koehler RT, So AP, Dube S, Rose KA, Montesclaros L, Wang S, Stumbo DP, Hodges SP, Romine S, Milanovich FP, White HE, Regan JF, Karlin-Neumann GA, Hindson CM, Saxonov S, Colston BW. High-throughput droplet digital PCR system for absolute quantitation of DNA copy number. Anal Chem. 2011;83:8604-8610. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 2056] [Cited by in RCA: 1967] [Article Influence: 140.5] [Reference Citation Analysis (0)] |
| 50. | Reinert T, Schøler LV, Thomsen R, Tobiasen H, Vang S, Nordentoft I, Lamy P, Kannerup AS, Mortensen FV, Stribolt K, Hamilton-Dutoit S, Nielsen HJ, Laurberg S, Pallisgaard N, Pedersen JS, Ørntoft TF, Andersen CL. Analysis of circulating tumour DNA to monitor disease burden following colorectal cancer surgery. Gut. 2016;65:625-634. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 277] [Cited by in RCA: 342] [Article Influence: 38.0] [Reference Citation Analysis (0)] |
| 51. | Taly V, Pekin D, Benhaim L, Kotsopoulos SK, Le Corre D, Li X, Atochin I, Link DR, Griffiths AD, Pallier K, Blons H, Bouché O, Landi B, Hutchison JB, Laurent-Puig P. Multiplex picodroplet digital PCR to detect KRAS mutations in circulating DNA from the plasma of colorectal cancer patients. Clin Chem. 2013;59:1722-1731. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 378] [Cited by in RCA: 380] [Article Influence: 31.7] [Reference Citation Analysis (0)] |
| 52. | Vivancos A, Aranda E, Benavides M, Élez E, Gómez-España MA, Toledano M, Alvarez M, Parrado MRC, García-Barberán V, Diaz-Rubio E. Comparison of the Clinical Sensitivity of the Idylla Platform and the OncoBEAM RAS CRC Assay for KRAS Mutation Detection in Liquid Biopsy Samples. Sci Rep. 2019;9:8976. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 25] [Cited by in RCA: 37] [Article Influence: 6.2] [Reference Citation Analysis (0)] |
| 53. | Holm M, Andersson E, Osterlund E, Ovissi A, Soveri LM, Anttonen AK, Kytölä S, Aittomäki K, Osterlund P, Ristimäki A. Detection of KRAS mutations in liquid biopsies from metastatic colorectal cancer patients using droplet digital PCR, Idylla, and next generation sequencing. PLoS One. 2020;15:e0239819. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 40] [Cited by in RCA: 55] [Article Influence: 11.0] [Reference Citation Analysis (0)] |
| 54. | Yu F, Makrigiorgos A, Leong KW, Makrigiorgos GM. Sensitive detection of microsatellite instability in tissues and liquid biopsies: Recent developments and updates. Comput Struct Biotechnol J. 2021;19:4931-4940. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 1] [Cited by in RCA: 19] [Article Influence: 4.8] [Reference Citation Analysis (0)] |
| 55. | Herring E, Kanaoka S, Tremblay É, Beaulieu JF. Droplet digital PCR for quantification of ITGA6 in a stool mRNA assay for the detection of colorectal cancers. World J Gastroenterol. 2017;23:2891-2898. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in CrossRef: 15] [Cited by in RCA: 18] [Article Influence: 2.3] [Reference Citation Analysis (0)] |
| 56. | Vega-Benedetti AF, Loi E, Moi L, Orrù S, Ziranu P, Pretta A, Lai E, Puzzoni M, Ciccone L, Casadei-Gardini A, Cabras F, Fortunato F, Restivo A, Zorcolo L, Scartozzi M, Zavattari P. Colorectal Cancer Early Detection in Stool Samples Tracing CpG Islands Methylation Alterations Affecting Gene Expression. Int J Mol Sci. 2020;21. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 9] [Cited by in RCA: 27] [Article Influence: 5.4] [Reference Citation Analysis (0)] |
| 57. | Liu Y, Wu H, Zhou Q, Song Q, Rui J, Zou B, Zhou G. Digital quantification of gene methylation in stool DNA by emulsion-PCR coupled with hydrogel immobilized bead-array. Biosens Bioelectron. 2017;92:596-601. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 10] [Cited by in RCA: 14] [Article Influence: 1.6] [Reference Citation Analysis (0)] |
| 58. | Garrigou S, Perkins G, Garlan F, Normand C, Didelot A, Le Corre D, Peyvandi S, Mulot C, Niarra R, Aucouturier P, Chatellier G, Nizard P, Perez-Toralla K, Zonta E, Charpy C, Pujals A, Barau C, Bouché O, Emile JF, Pezet D, Bibeau F, Hutchison JB, Link DR, Zaanan A, Laurent-Puig P, Sobhani I, Taly V. A Study of Hypermethylated Circulating Tumor DNA as a Universal Colorectal Cancer Biomarker. Clin Chem. 2016;62:1129-1139. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 89] [Cited by in RCA: 106] [Article Influence: 11.8] [Reference Citation Analysis (0)] |
| 59. | Rowlands V, Rutkowski AJ, Meuser E, Carr TH, Harrington EA, Barrett JC. Optimisation of robust singleplex and multiplex droplet digital PCR assays for high confidence mutation detection in circulating tumour DNA. Sci Rep. 2019;9:12620. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 48] [Cited by in RCA: 81] [Article Influence: 13.5] [Reference Citation Analysis (0)] |
| 60. | Huang H, Springborn S, Haug K, Bartow K, Samra H, Menon S, Mackinnon AC. Evaluation, Validation, and Implementation of the Idylla System as Rapid Molecular Testing for Precision Medicine. J Mol Diagn. 2019;21:862-872. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 19] [Cited by in RCA: 26] [Article Influence: 4.3] [Reference Citation Analysis (0)] |
| 61. | Uguen A, Troncone G. A review on the Idylla platform: towards the assessment of actionable genomic alterations in one day. J Clin Pathol. 2018;71:757-762. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 28] [Cited by in RCA: 37] [Article Influence: 5.3] [Reference Citation Analysis (0)] |
| 62. | Van Haele M, Vander Borght S, Ceulemans A, Wieërs M, Metsu S, Sagaert X, Weynand B. Rapid clinical mutational testing of KRAS, BRAF and EGFR: a prospective comparative analysis of the Idylla technique with high-throughput next-generation sequencing. J Clin Pathol. 2020;73:35-41. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 21] [Cited by in RCA: 29] [Article Influence: 4.8] [Reference Citation Analysis (0)] |
| 63. | Janku F, Huang HJ, Claes B, Falchook GS, Fu S, Hong D, Ramzanali NM, Nitti G, Cabrilo G, Tsimberidou AM, Naing A, Piha-Paul SA, Wheler JJ, Karp DD, Holley VR, Zinner RG, Subbiah V, Luthra R, Kopetz S, Overman MJ, Kee BK, Patel S, Devogelaere B, Sablon E, Maertens G, Mills GB, Kurzrock R, Meric-Bernstam F. BRAF Mutation Testing in Cell-Free DNA from the Plasma of Patients with Advanced Cancers Using a Rapid, Automated Molecular Diagnostics System. Mol Cancer Ther. 2016;15:1397-1404. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 59] [Cited by in RCA: 70] [Article Influence: 7.8] [Reference Citation Analysis (0)] |
| 64. | Zwaenepoel K, Holmgaard Duelund J, De Winne K, Maes V, Weyn C, Lambin S, Dendooven R, Broeckx G, Steiniche T, Pauwels P. Clinical Performance of the Idylla MSI Test for a Rapid Assessment of the DNA Microsatellite Status in Human Colorectal Cancer. J Mol Diagn. 2020;22:386-395. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 18] [Cited by in RCA: 29] [Article Influence: 4.8] [Reference Citation Analysis (0)] |
| 65. | ICGC/TCGA Pan-Cancer Analysis of Whole Genomes Consortium. Pan-cancer analysis of whole genomes. Nature. 2020;578:82-93. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 2285] [Cited by in RCA: 1834] [Article Influence: 366.8] [Reference Citation Analysis (0)] |
| 66. | Li X, Xu J, Li L, Mu X, Wang Y, Li X. Evaluation of a Fully Automated Idylla Test System for Microsatellite Instability in Colorectal Cancer. Clin Colorectal Cancer. 2019;18:e316-e323. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 12] [Cited by in RCA: 16] [Article Influence: 2.7] [Reference Citation Analysis (0)] |
| 67. | Zanke BW, Greenwood CM, Rangrej J, Kustra R, Tenesa A, Farrington SM, Prendergast J, Olschwang S, Chiang T, Crowdy E, Ferretti V, Laflamme P, Sundararajan S, Roumy S, Olivier JF, Robidoux F, Sladek R, Montpetit A, Campbell P, Bezieau S, O'Shea AM, Zogopoulos G, Cotterchio M, Newcomb P, McLaughlin J, Younghusband B, Green R, Green J, Porteous ME, Campbell H, Blanche H, Sahbatou M, Tubacher E, Bonaiti-Pellié C, Buecher B, Riboli E, Kury S, Chanock SJ, Potter J, Thomas G, Gallinger S, Hudson TJ, Dunlop MG. Genome-wide association scan identifies a colorectal cancer susceptibility locus on chromosome 8q24. Nat Genet. 2007;39:989-994. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 584] [Cited by in RCA: 559] [Article Influence: 31.1] [Reference Citation Analysis (0)] |
| 68. | Broderick P, Carvajal-Carmona L, Pittman AM, Webb E, Howarth K, Rowan A, Lubbe S, Spain S, Sullivan K, Fielding S, Jaeger E, Vijayakrishnan J, Kemp Z, Gorman M, Chandler I, Papaemmanuil E, Penegar S, Wood W, Sellick G, Qureshi M, Teixeira A, Domingo E, Barclay E, Martin L, Sieber O; CORGI Consortium, Kerr D, Gray R, Peto J, Cazier JB, Tomlinson I, Houlston RS. A genome-wide association study shows that common alleles of SMAD7 influence colorectal cancer risk. Nat Genet. 2007;39:1315-1317. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 406] [Cited by in RCA: 395] [Article Influence: 21.9] [Reference Citation Analysis (0)] |
| 69. | Tomlinson IP, Webb E, Carvajal-Carmona L, Broderick P, Howarth K, Pittman AM, Spain S, Lubbe S, Walther A, Sullivan K, Jaeger E, Fielding S, Rowan A, Vijayakrishnan J, Domingo E, Chandler I, Kemp Z, Qureshi M, Farrington SM, Tenesa A, Prendergast JG, Barnetson RA, Penegar S, Barclay E, Wood W, Martin L, Gorman M, Thomas H, Peto J, Bishop DT, Gray R, Maher ER, Lucassen A, Kerr D, Evans DG; CORGI Consortium, Schafmayer C, Buch S, Völzke H, Hampe J, Schreiber S, John U, Koessler T, Pharoah P, van Wezel T, Morreau H, Wijnen JT, Hopper JL, Southey MC, Giles GG, Severi G, Castellví-Bel S, Ruiz-Ponte C, Carracedo A, Castells A; EPICOLON Consortium, Försti A, Hemminki K, Vodicka P, Naccarati A, Lipton L, Ho JW, Cheng KK, Sham PC, Luk J, Agúndez JA, Ladero JM, de la Hoya M, Caldés T, Niittymäki I, Tuupanen S, Karhu A, Aaltonen L, Cazier JB, Campbell H, Dunlop MG, Houlston RS. A genome-wide association study identifies colorectal cancer susceptibility loci on chromosomes 10p14 and 8q23.3. Nat Genet. 2008;40:623-630. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 440] [Cited by in RCA: 444] [Article Influence: 26.1] [Reference Citation Analysis (0)] |
| 70. | Del Vecchio F, Mastroiaco V, Di Marco A, Compagnoni C, Capece D, Zazzeroni F, Capalbo C, Alesse E, Tessitore A. Next-generation sequencing: recent applications to the analysis of colorectal cancer. J Transl Med. 2017;15:246. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 69] [Cited by in RCA: 72] [Article Influence: 9.0] [Reference Citation Analysis (0)] |
| 71. | Cancer Genome Atlas Research Network, Weinstein JN, Collisson EA, Mills GB, Shaw KR, Ozenberger BA, Ellrott K, Shmulevich I, Sander C, Stuart JM. The Cancer Genome Atlas Pan-Cancer analysis project. Nat Genet. 2013;45:1113-1120. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 4788] [Cited by in RCA: 5550] [Article Influence: 462.5] [Reference Citation Analysis (0)] |
| 72. | Goodwin S, McPherson JD, McCombie WR. Coming of age: ten years of next-generation sequencing technologies. Nat Rev Genet. 2016;17:333-351. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 2404] [Cited by in RCA: 2435] [Article Influence: 304.4] [Reference Citation Analysis (0)] |
| 73. | Mamlouk S, Childs LH, Aust D, Heim D, Melching F, Oliveira C, Wolf T, Durek P, Schumacher D, Bläker H, von Winterfeld M, Gastl B, Möhr K, Menne A, Zeugner S, Redmer T, Lenze D, Tierling S, Möbs M, Weichert W, Folprecht G, Blanc E, Beule D, Schäfer R, Morkel M, Klauschen F, Leser U, Sers C. DNA copy number changes define spatial patterns of heterogeneity in colorectal cancer. Nat Commun. 2017;8:14093. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 81] [Cited by in RCA: 78] [Article Influence: 9.8] [Reference Citation Analysis (0)] |
| 74. | Xie YH, Chen YX, Fang JY. Comprehensive review of targeted therapy for colorectal cancer. Signal Transduct Target Ther. 2020;5:22. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 905] [Cited by in RCA: 1032] [Article Influence: 206.4] [Reference Citation Analysis (0)] |
| 75. | Zheng K, Wan H, Zhang J, Shan G, Chai N, Li D, Fang N, Liu L, Du R, Wu Q, Li X, Zhang C. A novel NGS-based microsatellite instability (MSI) status classifier with 9 loci for colorectal cancer patients. J Transl Med. 2020;18:215. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 8] [Cited by in RCA: 27] [Article Influence: 5.4] [Reference Citation Analysis (0)] |
| 76. | Lawler M, Alsina D, Adams RA, Anderson AS, Brown G, Fearnhead NS, Fenwick SW, Halloran SP, Hochhauser D, Hull MA, Koelzer VH, McNair AGK, Monahan KJ, Näthke I, Norton C, Novelli MR, Steele RJC, Thomas AL, Wilde LM, Wilson RH, Tomlinson I; Bowel Cancer UK Critical Research Gaps in Colorectal Cancer Initiative. Critical research gaps and recommendations to inform research prioritisation for more effective prevention and improved outcomes in colorectal cancer. Gut. 2018;67:179-193. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 71] [Cited by in RCA: 69] [Article Influence: 9.9] [Reference Citation Analysis (1)] |
| 77. | Mathai RA, Vidya RVS, Reddy BS, Thomas L, Udupa K, Kolesar J, Rao M. Potential Utility of Liquid Biopsy as a Diagnostic and Prognostic Tool for the Assessment of Solid Tumors: Implications in the Precision Oncology. J Clin Med. 2019;8. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 67] [Cited by in RCA: 96] [Article Influence: 16.0] [Reference Citation Analysis (0)] |
| 78. | Myint NNM, Verma AM, Fernandez-Garcia D, Sarmah P, Tarpey PS, Al-Aqbi SS, Cai H, Trigg R, West K, Howells LM, Thomas A, Brown K, Guttery DS, Singh B, Pringle HJ, McDermott U, Shaw JA, Rufini A. Circulating tumor DNA in patients with colorectal adenomas: assessment of detectability and genetic heterogeneity. Cell Death Dis. 2018;9:894. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 32] [Cited by in RCA: 39] [Article Influence: 5.6] [Reference Citation Analysis (0)] |
| 79. | Stahler A, Stintzing S, von Einem JC, Westphalen Benedikt CB, Heinrich K, Krämer N, Michl M, Modest DP, Fischer von Weikersthal L, Decker T, Kiani A, Heintges T, Kahl C, Kullmann F, Scheithauer W, Moehler M, Kaiser F, Kirchner T, Jung A, Heinemann V. Corrigendum to 'Single-nucleotide variants, tumour mutational burden and microsatellite instability in patients with metastatic colorectal cancer: Next-generation sequencing results of the FIRE-3 trial'. [European Journal of Cancer 137 (2020) 250-259]. Eur J Cancer. 2022;169:223-225. [RCA] [PubMed] [DOI] [Full Text] [Reference Citation Analysis (0)] |
| 80. | Leary RJ, Kinde I, Diehl F, Schmidt K, Clouser C, Duncan C, Antipova A, Lee C, McKernan K, De La Vega FM, Kinzler KW, Vogelstein B, Diaz LA Jr, Velculescu VE. Development of personalized tumor biomarkers using massively parallel sequencing. Sci Transl Med. 2010;2:20ra14. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 393] [Cited by in RCA: 392] [Article Influence: 26.1] [Reference Citation Analysis (0)] |
| 81. | Gould GM, Grauman PV, Theilmann MR, Spurka L, Wang IE, Melroy LM, Chin RG, Hite DH, Chu CS, Maguire JR, Hogan GJ, Muzzey D. Detecting clinically actionable variants in the 3' exons of PMS2 via a reflex workflow based on equivalent hybrid capture of the gene and its pseudogene. BMC Med Genet. 2018;19:176. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 7] [Cited by in RCA: 9] [Article Influence: 1.3] [Reference Citation Analysis (0)] |
| 82. | Corti G, Bartolini A, Crisafulli G, Novara L, Rospo G, Montone M, Negrino C, Mussolin B, Buscarino M, Isella C, Barault L, Siravegna G, Siena S, Marsoni S, Di Nicolantonio F, Medico E, Bardelli A. A Genomic Analysis Workflow for Colorectal Cancer Precision Oncology. Clin Colorectal Cancer. 2019;18:91-101.e3. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 18] [Cited by in RCA: 30] [Article Influence: 5.0] [Reference Citation Analysis (0)] |
| 83. | Battaglin F, Naseem M, Lenz HJ, Salem ME. Microsatellite instability in colorectal cancer: overview of its clinical significance and novel perspectives. Clin Adv Hematol Oncol. 2018;16:735-745. [PubMed] |
| 84. | Xiao J, Li W, Huang Y, Huang M, Li S, Zhai X, Zhao J, Gao C, Xie W, Qin H, Cai S, Bai Y, Lan P, Zou Y. A next-generation sequencing-based strategy combining microsatellite instability and tumor mutation burden for comprehensive molecular diagnosis of advanced colorectal cancer. BMC Cancer. 2021;21:282. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 17] [Cited by in RCA: 58] [Article Influence: 14.5] [Reference Citation Analysis (3)] |
| 85. | Gurjao C, Zhong R, Haruki K, Li YY, Spurr LF, Lee-Six H, Reardon B, Ugai T, Zhang X, Cherniack AD, Song M, Van Allen EM, Meyerhardt JA, Nowak JA, Giovannucci EL, Fuchs CS, Wu K, Ogino S, Giannakis M. Discovery and Features of an Alkylating Signature in Colorectal Cancer. Cancer Discov. 2021;11:2446-2455. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 39] [Cited by in RCA: 48] [Article Influence: 12.0] [Reference Citation Analysis (0)] |
| 86. | Chang YS, Lee CC, Ke TW, Chang CM, Chao DS, Huang HY, Chang JG. Molecular characterization of colorectal cancer using whole-exome sequencing in a Taiwanese population. Cancer Med. 2019;8:3738-3747. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 6] [Cited by in RCA: 9] [Article Influence: 1.5] [Reference Citation Analysis (0)] |
| 87. | Diederichs S, Bartsch L, Berkmann JC, Fröse K, Heitmann J, Hoppe C, Iggena D, Jazmati D, Karschnia P, Linsenmeier M, Maulhardt T, Möhrmann L, Morstein J, Paffenholz SV, Röpenack P, Rückert T, Sandig L, Schell M, Steinmann A, Voss G, Wasmuth J, Weinberger ME, Wullenkord R. The dark matter of the cancer genome: aberrations in regulatory elements, untranslated regions, splice sites, non-coding RNA and synonymous mutations. EMBO Mol Med. 2016;8:442-457. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 158] [Cited by in RCA: 200] [Article Influence: 22.2] [Reference Citation Analysis (0)] |
| 88. | Ishaque N, Abba ML, Hauser C, Patil N, Paramasivam N, Huebschmann D, Leupold JH, Balasubramanian GP, Kleinheinz K, Toprak UH, Hutter B, Benner A, Shavinskaya A, Zhou C, Gu Z, Kerssemakers J, Marx A, Moniuszko M, Kozlowski M, Reszec J, Niklinski J, Eils J, Schlesner M, Eils R, Brors B, Allgayer H. Whole genome sequencing puts forward hypotheses on metastasis evolution and therapy in colorectal cancer. Nat Commun. 2018;9:4782. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 67] [Cited by in RCA: 102] [Article Influence: 14.6] [Reference Citation Analysis (0)] |
| 89. | Dashti H, Dehzangi I, Bayati M, Breen J, Beheshti A, Lovell N, Rabiee HR, Alinejad-Rokny H. Integrative analysis of mutated genes and mutational processes reveals novel mutational biomarkers in colorectal cancer. BMC Bioinformatics. 2022;23:138. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 4] [Cited by in RCA: 11] [Article Influence: 3.7] [Reference Citation Analysis (0)] |
| 90. | Yang L. A Practical Guide for Structural Variation Detection in the Human Genome. Curr Protoc Hum Genet. 2020;107:e103. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 8] [Cited by in RCA: 31] [Article Influence: 6.2] [Reference Citation Analysis (0)] |
| 91. | Watson CM, Crinnion LA, Simmonds J, Camm N, Adlard J, Bonthron DT. Long-read nanopore sequencing enables accurate confirmation of a recurrent PMS2 insertion-deletion variant located in a region of complex genomic architecture. Cancer Genet. 2021;256-257:122-126. [RCA] [PubMed] [DOI] [Full Text] [Cited by in RCA: 4] [Reference Citation Analysis (0)] |
| 92. | Pradhan B, Cajuso T, Katainen R, Sulo P, Tanskanen T, Kilpivaara O, Pitkänen E, Aaltonen LA, Kauppi L, Palin K. Detection of subclonal L1 transductions in colorectal cancer by long-distance inverse-PCR and Nanopore sequencing. Sci Rep. 2017;7:14521. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 18] [Cited by in RCA: 20] [Article Influence: 2.5] [Reference Citation Analysis (0)] |
| 93. | Galvan A, Ioannidis JP, Dragani TA. Beyond genome-wide association studies: genetic heterogeneity and individual predisposition to cancer. Trends Genet. 2010;26:132-141. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 114] [Cited by in RCA: 115] [Article Influence: 7.7] [Reference Citation Analysis (0)] |
| 94. | Cheng Y, Ling Z, Li L. The Intestinal Microbiota and Colorectal Cancer. Front Immunol. 2020;11:615056. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 363] [Cited by in RCA: 329] [Article Influence: 65.8] [Reference Citation Analysis (0)] |
| 95. | Azevedo MM, Pina-Vaz C, Baltazar F. Microbes and Cancer: Friends or Faux? Int J Mol Sci. 2020;21. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 13] [Cited by in RCA: 58] [Article Influence: 11.6] [Reference Citation Analysis (0)] |
| 96. | Bokulich NA, Chung J, Battaglia T, Henderson N, Jay M, Li H, D Lieber A, Wu F, Perez-Perez GI, Chen Y, Schweizer W, Zheng X, Contreras M, Dominguez-Bello MG, Blaser MJ. Antibiotics, birth mode, and diet shape microbiome maturation during early life. Sci Transl Med. 2016;8:343ra82. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 742] [Cited by in RCA: 989] [Article Influence: 123.6] [Reference Citation Analysis (0)] |
| 97. | Panek M, Čipčić Paljetak H, Barešić A, Perić M, Matijašić M, Lojkić I, Vranešić Bender D, Krznarić Ž, Verbanac D. Methodology challenges in studying human gut microbiota - effects of collection, storage, DNA extraction and next generation sequencing technologies. Sci Rep. 2018;8:5143. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 94] [Cited by in RCA: 128] [Article Influence: 18.3] [Reference Citation Analysis (0)] |
| 98. | Hill CJ, Brown JR, Lynch DB, Jeffery IB, Ryan CA, Ross RP, Stanton C, O'Toole PW. Effect of room temperature transport vials on DNA quality and phylogenetic composition of faecal microbiota of elderly adults and infants. Microbiome. 2016;4:19. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 49] [Cited by in RCA: 54] [Article Influence: 6.0] [Reference Citation Analysis (0)] |
| 99. | Kennedy NA, Walker AW, Berry SH, Duncan SH, Farquarson FM, Louis P, Thomson JM; UK IBD Genetics Consortium, Satsangi J, Flint HJ, Parkhill J, Lees CW, Hold GL. The impact of different DNA extraction kits and laboratories upon the assessment of human gut microbiota composition by 16S rRNA gene sequencing. PLoS One. 2014;9:e88982. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 204] [Cited by in RCA: 217] [Article Influence: 19.7] [Reference Citation Analysis (0)] |
| 100. | Walker AW, Martin JC, Scott P, Parkhill J, Flint HJ, Scott KP. 16S rRNA gene-based profiling of the human infant gut microbiota is strongly influenced by sample processing and PCR primer choice. Microbiome. 2015;3:26. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 179] [Cited by in RCA: 191] [Article Influence: 19.1] [Reference Citation Analysis (0)] |
| 101. | McBurney MI, Davis C, Fraser CM, Schneeman BO, Huttenhower C, Verbeke K, Walter J, Latulippe ME. Establishing What Constitutes a Healthy Human Gut Microbiome: State of the Science, Regulatory Considerations, and Future Directions. J Nutr. 2019;149:1882-1895. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 165] [Cited by in RCA: 172] [Article Influence: 28.7] [Reference Citation Analysis (0)] |
| 102. | Hess JF, Kohl TA, Kotrová M, Rönsch K, Paprotka T, Mohr V, Hutzenlaub T, Brüggemann M, Zengerle R, Niemann S, Paust N. Library preparation for next generation sequencing: A review of automation strategies. Biotechnol Adv. 2020;41:107537. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 34] [Cited by in RCA: 104] [Article Influence: 20.8] [Reference Citation Analysis (0)] |
| 103. | Durazzi F, Sala C, Castellani G, Manfreda G, Remondini D, De Cesare A. Comparison between 16S rRNA and shotgun sequencing data for the taxonomic characterization of the gut microbiota. Sci Rep. 2021;11:3030. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 204] [Cited by in RCA: 290] [Article Influence: 72.5] [Reference Citation Analysis (0)] |
| 104. | Niccolai E, Russo E, Baldi S, Ricci F, Nannini G, Pedone M, Stingo FC, Taddei A, Ringressi MN, Bechi P, Mengoni A, Fani R, Bacci G, Fagorzi C, Chiellini C, Prisco D, Ramazzotti M, Amedei A. Significant and Conflicting Correlation of IL-9 With Prevotella and Bacteroides in Human Colorectal Cancer. Front Immunol. 2020;11:573158. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 44] [Cited by in RCA: 43] [Article Influence: 10.8] [Reference Citation Analysis (0)] |
| 105. | Russo E, Bacci G, Chiellini C, Fagorzi C, Niccolai E, Taddei A, Ricci F, Ringressi MN, Borrelli R, Melli F, Miloeva M, Bechi P, Mengoni A, Fani R, Amedei A. Preliminary Comparison of Oral and Intestinal Human Microbiota in Patients with Colorectal Cancer: A Pilot Study. Front Microbiol. 2017;8:2699. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 87] [Cited by in RCA: 81] [Article Influence: 11.6] [Reference Citation Analysis (0)] |
| 106. | Sobhani I, Amiot A, Le Baleur Y, Levy M, Auriault ML, Van Nhieu JT, Delchier JC. Microbial dysbiosis and colon carcinogenesis: could colon cancer be considered a bacteria-related disease? Therap Adv Gastroenterol. 2013;6:215-229. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 97] [Cited by in RCA: 91] [Article Influence: 7.6] [Reference Citation Analysis (0)] |
| 107. | Tjalsma H, Boleij A, Marchesi JR, Dutilh BE. A bacterial driver-passenger model for colorectal cancer: beyond the usual suspects. Nat Rev Microbiol. 2012;10:575-582. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 484] [Cited by in RCA: 667] [Article Influence: 51.3] [Reference Citation Analysis (0)] |
| 108. | Wang Y, Zhang C, Hou S, Wu X, Liu J, Wan X. Analyses of Potential Driver and Passenger Bacteria in Human Colorectal Cancer. Cancer Manag Res. 2020;12:11553-11561. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 33] [Cited by in RCA: 29] [Article Influence: 5.8] [Reference Citation Analysis (0)] |
| 109. | Luan C, Xie L, Yang X, Miao H, Lv N, Zhang R, Xiao X, Hu Y, Liu Y, Wu N, Zhu Y, Zhu B. Dysbiosis of fungal microbiota in the intestinal mucosa of patients with colorectal adenomas. Sci Rep. 2015;5:7980. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 133] [Cited by in RCA: 131] [Article Influence: 13.1] [Reference Citation Analysis (0)] |
| 110. | Coker OO, Nakatsu G, Dai RZ, Wu WKK, Wong SH, Ng SC, Chan FKL, Sung JJY, Yu J. Enteric fungal microbiota dysbiosis and ecological alterations in colorectal cancer. Gut. 2019;68:654-662. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 207] [Cited by in RCA: 340] [Article Influence: 56.7] [Reference Citation Analysis (0)] |
| 111. | Coker OO, Wu WKK, Wong SH, Sung JJY, Yu J. Altered Gut Archaea Composition and Interaction With Bacteria Are Associated With Colorectal Cancer. Gastroenterology. 2020;159:1459-1470.e5. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 46] [Cited by in RCA: 99] [Article Influence: 19.8] [Reference Citation Analysis (0)] |
| 112. | Janati AI, Karp I, Laprise C, Sabri H, Emami E. Detection of Fusobaterium nucleatum in feces and colorectal mucosa as a risk factor for colorectal cancer: a systematic review and meta-analysis. Syst Rev. 2020;9:276. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 16] [Cited by in RCA: 23] [Article Influence: 4.6] [Reference Citation Analysis (0)] |
| 113. | Yachida S, Mizutani S, Shiroma H, Shiba S, Nakajima T, Sakamoto T, Watanabe H, Masuda K, Nishimoto Y, Kubo M, Hosoda F, Rokutan H, Matsumoto M, Takamaru H, Yamada M, Matsuda T, Iwasaki M, Yamaji T, Yachida T, Soga T, Kurokawa K, Toyoda A, Ogura Y, Hayashi T, Hatakeyama M, Nakagama H, Saito Y, Fukuda S, Shibata T, Yamada T. Metagenomic and metabolomic analyses reveal distinct stage-specific phenotypes of the gut microbiota in colorectal cancer. Nat Med. 2019;25:968-976. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 404] [Cited by in RCA: 879] [Article Influence: 146.5] [Reference Citation Analysis (0)] |
| 114. | Choi S, Chung J, Cho ML, Park D, Choi SS. Analysis of changes in microbiome compositions related to the prognosis of colorectal cancer patients based on tissue-derived 16S rRNA sequences. J Transl Med. 2021;19:485. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in RCA: 22] [Reference Citation Analysis (0)] |
| 115. | de Almeida CV, Taddei A, Amedei A. The controversial role of Enterococcus faecalis in colorectal cancer. Therap Adv Gastroenterol. 2018;11:1756284818783606. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 117] [Cited by in RCA: 106] [Article Influence: 15.1] [Reference Citation Analysis (0)] |
| 116. | Liang JQ, Li T, Nakatsu G, Chen YX, Yau TO, Chu E, Wong S, Szeto CH, Ng SC, Chan FKL, Fang JY, Sung JJY, Yu J. A novel faecal Lachnoclostridium marker for the non-invasive diagnosis of colorectal adenoma and cancer. Gut. 2020;69:1248-1257. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 230] [Cited by in RCA: 213] [Article Influence: 42.6] [Reference Citation Analysis (0)] |
| 117. | Long X, Wong CC, Tong L, Chu ESH, Ho Szeto C, Go MYY, Coker OO, Chan AWH, Chan FKL, Sung JJY, Yu J. Peptostreptococcus anaerobius promotes colorectal carcinogenesis and modulates tumour immunity. Nat Microbiol. 2019;4:2319-2330. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 124] [Cited by in RCA: 327] [Article Influence: 54.5] [Reference Citation Analysis (0)] |
| 118. | Deng Q, Wang C, Yu K, Wang Y, Yang Q, Zhang J, Xu X. Streptococcus bovis Contributes to the Development of Colorectal Cancer via Recruiting CD11b⁺TLR-4⁺ Cells. Med Sci Monit. 2020;26:e921886. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 7] [Cited by in RCA: 25] [Article Influence: 5.0] [Reference Citation Analysis (0)] |
| 119. | Xia C, Cai Y, Ren S, Xia C. Role of microbes in colorectal cancer therapy: Cross-talk between the microbiome and tumor microenvironment. Front Pharmacol. 2022;13:1051330. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 4] [Cited by in RCA: 8] [Article Influence: 2.7] [Reference Citation Analysis (0)] |
| 120. | Koulouridi A, Messaritakis I, Gouvas N, Tsiaoussis J, Souglakos J. Immunotherapy in Solid Tumors and Gut Microbiota: The Correlation-A Special Reference to Colorectal Cancer. Cancers (Basel). 2020;13. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 7] [Cited by in RCA: 17] [Article Influence: 3.4] [Reference Citation Analysis (0)] |
| 121. | Dikeocha IJ, Al-Kabsi AM, Eid EEM, Hussin S, Alshawsh MA. Probiotics supplementation in patients with colorectal cancer: a systematic review of randomized controlled trials. Nutr Rev. 2021;80:22-49. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 9] [Cited by in RCA: 35] [Article Influence: 8.8] [Reference Citation Analysis (0)] |
| 122. | Kim CE, Yoon LS, Michels KB, Tranfield W, Jacobs JP, May FP. The Impact of Prebiotic, Probiotic, and Synbiotic Supplements and Yogurt Consumption on the Risk of Colorectal Neoplasia among Adults: A Systematic Review. Nutrients. 2022;14. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 13] [Cited by in RCA: 16] [Article Influence: 5.3] [Reference Citation Analysis (0)] |
| 123. | Nannini G, Meoni G, Tenori L, Ringressi MN, Taddei A, Niccolai E, Baldi S, Russo E, Luchinat C, Amedei A. Fecal metabolomic profiles: A comparative study of patients with colorectal cancer vs adenomatous polyps. World J Gastroenterol. 2021;27:6430-6441. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in CrossRef: 12] [Cited by in RCA: 13] [Article Influence: 3.3] [Reference Citation Analysis (1)] |
| 124. | Chung L, Thiele Orberg E, Geis AL, Chan JL, Fu K, DeStefano Shields CE, Dejea CM, Fathi P, Chen J, Finard BB, Tam AJ, McAllister F, Fan H, Wu X, Ganguly S, Lebid A, Metz P, Van Meerbeke SW, Huso DL, Wick EC, Pardoll DM, Wan F, Wu S, Sears CL, Housseau F. Bacteroides fragilis Toxin Coordinates a Pro-carcinogenic Inflammatory Cascade via Targeting of Colonic Epithelial Cells. Cell Host Microbe. 2018;23:203-214.e5. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 345] [Cited by in RCA: 377] [Article Influence: 53.9] [Reference Citation Analysis (0)] |
| 125. | Li J, Zhang AH, Wu FF, Wang XJ. Alterations in the Gut Microbiota and Their Metabolites in Colorectal Cancer: Recent Progress and Future Prospects. Front Oncol. 2022;12:841552. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 30] [Cited by in RCA: 49] [Article Influence: 16.3] [Reference Citation Analysis (0)] |
| 126. | Niccolai E, Baldi S, Ricci F, Russo E, Nannini G, Menicatti M, Poli G, Taddei A, Bartolucci G, Calabrò AS, Stingo FC, Amedei A. Evaluation and comparison of short chain fatty acids composition in gut diseases. World J Gastroenterol. 2019;25:5543-5558. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in CrossRef: 110] [Cited by in RCA: 99] [Article Influence: 16.5] [Reference Citation Analysis (0)] |
| 127. | Baldi S, Menicatti M, Nannini G, Niccolai E, Russo E, Ricci F, Pallecchi M, Romano F, Pedone M, Poli G, Renzi D, Taddei A, Calabrò AS, Stingo FC, Bartolucci G, Amedei A. Free Fatty Acids Signature in Human Intestinal Disorders: Significant Association between Butyric Acid and Celiac Disease. Nutrients. 2021;13. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 45] [Cited by in RCA: 36] [Article Influence: 9.0] [Reference Citation Analysis (0)] |
| 128. | Yuan L, Zhang S, Li H, Yang F, Mushtaq N, Ullah S, Shi Y, An C, Xu J. The influence of gut microbiota dysbiosis to the efficacy of 5-Fluorouracil treatment on colorectal cancer. Biomed Pharmacother. 2018;108:184-193. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 81] [Cited by in RCA: 152] [Article Influence: 21.7] [Reference Citation Analysis (0)] |
| 129. | De Almeida CV, de Camargo MR, Russo E, Amedei A. Role of diet and gut microbiota on colorectal cancer immunomodulation. World J Gastroenterol. 2019;25:151-162. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in CrossRef: 95] [Cited by in RCA: 95] [Article Influence: 15.8] [Reference Citation Analysis (5)] |