Published online Nov 15, 2023. doi: 10.4251/wjgo.v15.i11.1974
Peer-review started: June 23, 2023
First decision: August 30, 2023
Revised: September 12, 2023
Accepted: October 11, 2023
Article in press: October 11, 2023
Published online: November 15, 2023
Processing time: 145 Days and 7.2 Hours
A series of long non-coding RNAs (lncRNAs) have been reported to play a crucial role in cancer biology. Some previous studies report that lncRNA CDKN2B-AS1 is involved in some human malignancies. However, its role in hepatocellular carcinoma (HCC) has not been fully deciphered.
To decipher the role of CDKN2B-AS1 in the progression of HCC.
CDKN2B-AS1 expression in HCC was detected by quantitative real-time polymerase chain reaction. The malignant phenotypes of Li-7 and SNU-182 cells were detected by the CCK-8 method, EdU method, and flow cytometry, respectively. RNA immunoprecipitation was executed to confirm the interaction between CDKN2B-AS1 and E2F transcription factor 1 (E2F1). Luciferase reporter assay and chromatin immunoprecipitation were performed to verify the binding of E2F1 to the promoter of G protein subunit alpha Z (GNAZ). E2F1 and GNAZ were detected by western blot in HCC cells.
In HCC tissues, CDKN2B-AS1 was upregulated. Depletion of CDKN2B-AS1 inhibited the proliferation of HCC cells, and the depletion of CDKN2B-AS1 also induced cell cycle arrest and apoptosis. CDKN2B-AS1 could interact with E2F1. Depletion of CDKN2B-AS1 inhibited the binding of E2F1 to the GNAZ promoter region. Overexpression of E2F1 reversed the biological effects of depletion of CDKN2B-AS1 on the malignant behaviors of HCC cells.
CDKN2B-AS1 recruits E2F1 to facilitate GNAZ transcription to promote HCC progression.
Core Tip: The high expression of long non-coding RNAs CDKN2B-AS1 in hepatocellular carcinoma (HCC) predicts poor prognosis of the patients, as it facilitates some malignant biological behaviors of HCC cells, including enhanced viability, proliferation, cell cycle progression, and anti-apoptosis ability. This study reveals one mechanism of CDKN2B-AS1 promoting HCC progression, which is the interaction between CDKN2B-AS1 and E2F transcription factor 1 in HCC cells to promote the expression of the oncogene G protein subunit alpha Z.
- Citation: Tao ZG, Yuan YX, Wang GW. Long non-coding RNA CDKN2B-AS1 promotes hepatocellular carcinoma progression via E2F transcription factor 1/G protein subunit alpha Z axis. World J Gastrointest Oncol 2023; 15(11): 1974-1987
- URL: https://www.wjgnet.com/1948-5204/full/v15/i11/1974.htm
- DOI: https://dx.doi.org/10.4251/wjgo.v15.i11.1974
The morbidity and mortality of hepatocellular carcinoma (HCC) patients maintain a high level[1]. Surgical resection can be relatively helpful in treating patients with early-stage HCC[2]. Nevertheless, the early symptoms of HCC are inconspicuous, and most patients are diagnosed only when the tumor has progressed beyond treatment by surgical resection, in situ liver transplantation, etc.[3,4]. In addition, even with early and effective diagnosis and treatment, HCC patients are prone to suffer from recurrence and have adverse prognosis[5].
Long non-coding RNAs (lncRNAs) can exert their biological effects in cell biology by interacting with microRNAs, ncRNAs, and proteins[6,7]. Reportedly, lncRNAs play a key regulatory role in HCC by acting as oncogenes or tumor suppressors[8,9]. Some studies have reported that dysregulation of lncRNA participates in HCC progression. Specifically, lncRNA-PDPK2P is highly expressed in HCC tissues, and it facilitates HCC progression via modulating the PDK1/AKT/caspase 3 axis[10]. Another study reports that lncRNA uc.134 is a tumor-suppressive lncRNA, and it mediates the ubiquitination of LATS1 to repress the expression of oncogene LATS1[11]. High expression of lncRNA CDKN2B-AS1 (CDKN2B-AS1) predicts poor prognosis of HCC patients[12]; additionally, CDKN2B-AS1 can promote the malignancy of HCC cells via modulating miR-424-5p[13]. Nonetheless, the mechanism of CDKN2B-AS1 in modulating HCC progression is not fully explained.
Sustaining cell growth is one of the fundamental hallmarks of carcinogenesis and it results from dysregulation of cell cycle progression. E2F transcription factors are a large family of transcription modulators and are key regulators of the cell cycle; E2F transcription factor 1 (E2F1), a member of the E2F family, is found to interact with SET7/9 to promote HCC progression[14]. G protein subunit alpha Z (GNAZ) is a member of the unique GI/o subfamily encoding GαŽ[10]. GNAZ overexpression promotes the malignancy of HCC cells[11]. In this work, we aim to study the expression characteristics and functions of CDKN2B-AS1 in HCC and the potential mechanisms and confirm that CDKN2B-AS1 can facilitate disease progression by regulating the E2F1/GNAZ axis.
Fifty-five patients diagnosed with HCC at Hangzhou Cancer Hospital who received hepatectomy were included in this study. Surgical tissue specimens were placed in liquid nitrogen for storage. All of the patients were not treated by anti-cancer treatment before surgery. The work was approved by the Ethics Committee of Hangzhou Cancer Hospital.
Li-7, Huh-7, SK-HEP-1, and SNU-387 cell lines (Shanghai Institute of Cell Research), and immortalized liver epithelial cells THLE-2 (ATCC) were cultured in RPMI-1640 medium (Thermo Fisher Scientific) [containing 10% foetal bovine serum (Gibco), 100 U/mL penicillin and 100 mg/mL streptomycin (Invitrogen)]. Culture condition: 5% CO2, 37 °C, saturated humidity.
RNAiso Plus kit (Takara) and PrimeScriptTM RT Reagent Kit (Takara) were applied for RNA extraction and cDNA synthesis. Quantitative real-time polymerase chain reaction (qRT-PCR) was performed with SYBR Premix Ex TaqTM II Kit (Takara). For the sequences of primers, please check Table 1.
| Name | Primer sequences |
| CDKN2B-AS1 | Forward: 5’-AGGAGGCTGAATGTCAGTTTT-3’ |
| Reverse: 5’-AGCGGTTTAGTTTAATTTCGCTT-3’ | |
| E2F1 | Forward: 5’-GGACCUGGAAACUGACCAUTT-3’ |
| Reverse: 5’-GACCACCUGAUGAAUAUCUTT-3’ | |
| GNAZ | Forward: 5’-GGTCTACATCCAACGTCAGTTC-3’ |
| Reverse: 5’-TCTGTCACTGCGTCAAACAC-3’ | |
| GAPDH | Forward: 5’-TCTTCACCACCATGGAGAAG-3’ |
| Reverse: 5’-GCAGAGATGATGACCCTTTTG-3’ | |
| U6 | Forward: 5’-GCTTCGGCAGCACATATACTAAAAT-3’ |
| Reverse: 5’-CGCTTCACGAATTTGCGTGTCAT-3’ |
Two CDKN2B-AS1 small interfering RNAs (siRNAs), E2F1 overexpression plasmid (oe-E2F1), and controls [siRNA control (si-NC), plasmid (NC)] were created by GenePharma Co., Ltd. When the Li-7 and SNU-182 cells grew to a density of 50%-60%, cells were transfected using Lipofectamine® 2000 (Invitrogen). Briefly, the cells were cultured with serum-free medium for 24 h. Next, the transfection reagent was mixed with serum-free medium and vectors/oligonucleotides, and incubated for 8 h. Next, the cells were mixed with the transfection reagent containing vectors/oligonucleotides, and incubated for 12 h. Subsequently, the cells were cultured with a complete medium with serum, and 24 h later, the cells were collected, and qPCR was applied for validation of the transfection efficacy.
Li-7 and SNU-182 cells were inoculated in 96-well plates (5 × 103 cells/well). MTT reagent (5 mg/mL; Sigma) was added (20 μL per well) after the cells were cultured overnight and then incubated for 4 h at 37 °C. Then, dimethyl sulfoxide (DMSO; Sigma) was added (150 μL per well) and the cells were shaken at low speed for 15 min to fully dissolve the crystals. Next, the absorbance [optical density (OD)] value finally was measured at 450 nm light wavelength with a microplate reader. Then the growth curve of the cells was plotted according to the OD value.
The transfected Li-7 and SNU-182 cells were taken and inoculated on 24-well plates at 1.5 × 105 cells per well. EdU solution (RiboBio, Guangzhou, China) (50 μM) was added, and then the cells were incubator (2h, at 37 °C). Li-7 and SNU-182 cells were fixed in phosphate-buffered saline (PBS) containing 4% paraformaldehyde (Sigma) for 30 min. After washing with PBS, the cells were stained with 1 × Apollo staining solution and DAPI (Solarbio) (protected from light, room temperature, 20 min). After PBS washing, the EdU-positive cells were observed under fluorescence microscopy. At least three visual fields were captured, and the cells were counted. The percentage of proliferating cells (%) = EdU positive cells/DAPI positive cells × 100 %.
To assess the cell cycle, 1 × 107 Li-7 and SNU-182 cells were fixed in ethanol and placed at -20 °C for 24 h. After washing twice with PBS, Li-7, and SNU-182 cells were resuspended in 100 μL RNase A (BD Bioscience) and incubated (at 37 °C for 30 min). Then Li-7 and SNU-182 cells were stained with 400 μL of propidium iodide (PI) (50 μg/mL; BD Biosciences) (5 min at 4 °C in the dark). Finally, a FACScan flow cytometer (BD Bioscience) was utilized to sort the cells.
For apoptosis analysis, 1 × 106 Li-7 and SNU-182 single cell suspensions (in 1 mL PBS) were centrifuged (400 × g, 5 min, 4 °C). Li-7 and SNU-182 cells were then resuspended and marked with 10 μL of Annexin V-fluorescein isothiocyanate and 10 μL of PI (BD Biosciences) (protected from light, 30 min, 4 °C). Finally, apoptosis was detected by flow cytometry within 1 h.
The PARIS™ kit (Ambion, Austin, TX) was utilized in this assay. Briefly, 5 × 106 Li-7 and SNU-182 cells were re-suspended in 0.5 mL resuspension buffer and next homogenization was performed. After centrifugation (400 × g, 15 min), the supernatant was collected (cytoplasmic fraction). The pellet was then dissolved in a mixture of PBS (0.3 mL), nuclear isolation buffer (0.3 mL), and RNase-free H2O (0.3 mL) (on ice, 20 min), and then centrifugation was performed (nuclear fraction). Next, qRT-PCR was performed.
Li-7 and SNU-182 cells (5 × 106 cells) were fixed with 0.3% formaldehyde. Then Li-7 and SNU-182 cells were resuspended in RIPA buffer (Sigma) and incubated on ice with shaking for 30 min. Cell lysates were incubated with 5 μg of anti-E2F1 antibody (ab288369, Abcam) or anti-immunoglobulin (Ig)G antibody conjugated to magnetic beads (ab150077, Abcam) (4 °C, 8 h). The beads were washed with RIPA buffer, followed by elution, reverse cross-linking, and subsequent RNA extraction. Finally, qPCR was conducted[15].
The bicinchoninic acid protein assay kit (Beyotime) was used to detect the protein concentration of the samples. Protein samples were mixed with loading buffer and separated by sodium-dodecyl sulfate gel electrophoresis, and the proteins were transferred onto polyvinylidene fluoride membranes (Millipore) by electrotransfer. After blocking, the membranes were incubated with primary anti-E2F1 antibody (ab288369, 1:1000), anti-GNAZ antibody (ab154846, 1:1000), and anti-GAPDH antibody (ab9485, 1:1000) (4 °C, 8 h), then incubated with secondary antibody (ab205718, 1:5000) (room temperature, 1 h). All of the antibodies were obtained from Abcam. An enhanced chemiluminescence kit (Promega) was finally used to develop the blot on the membrane under an imaging device.
The PROMO database was utilized to show the potential E2F1 binding motif in the GNAZ promoter. The wild type and mutated recombinant vectors containing the predicted sequence were co-transfected into Li-7 and SNU-182 cells with the E2F1 overexpression plasmid or its negative control. At 48 h after transfection, luciferase activity was assessed with the kit (Promega). Firefly luciferase activity was used to show the binding intensity between E2F1 and GNAZ promoter, with Renilla luciferase activity as the internal reference.
An EZ-CHIPTM Chromatin Immunoprecipitation Kit (Millipore) was used for this assay. Transfected Li-7 and SNU-182 cells (1 × 107 cells) were then taken and cultured (24 h, 37 °C). Cells were fixed with 10% formaldehyde for 10 min and then terminated with glycine. Cells were then collected and centrifuged to obtain cell precipitates. Subsequently, cells were lysed by adding cell lysis solution (Sigma) and centrifuged to obtain cell nuclear precipitates. Then sonication was performed and the sonically sheared cell nuclear lysates were incubated with control IgG antibody or E2F1 antibody, respectively, at 4 °C overnight. Afterward, the DNA bound to the E2F1 transcription factor was centrifuged and eluted using a fresh elution buffer, and the DNA was then extracted. qRT-PCR was conducted with the purified DNA.
Data of all experiments (performed in triplicate) were expressed as mean ± SD, and analyzed by SPSS 20.0 (IBM), with independent samples t-test or one-way ANOVA. P < 0.05 signified statistical significance.
First of all, we investigate the expression characteristics and clinical significance of CDKN2B-AS1 in HCC. StarBase database suggested that CDKN2B-AS1 expression was higher in HCC samples than (vs paracancerous samples) (Figure 1A). Subsequent qPCR assays indicated that, in HCC samples, CDKN2B-AS1 was remarkably overexpressed (vs paracancerous tissues) (Figure 1B). Consistently, the expression level of CDKN2B-AS1 was promoted in cancer cell lines (vs normal hepatic epithelial cell line THLE-2) (Figure 1C). In addition, by analysis of the data in the GEPIA database, it was known that high CDKN2B-AS1 expression predicted a shorter survival time of patients (Figure 1D).
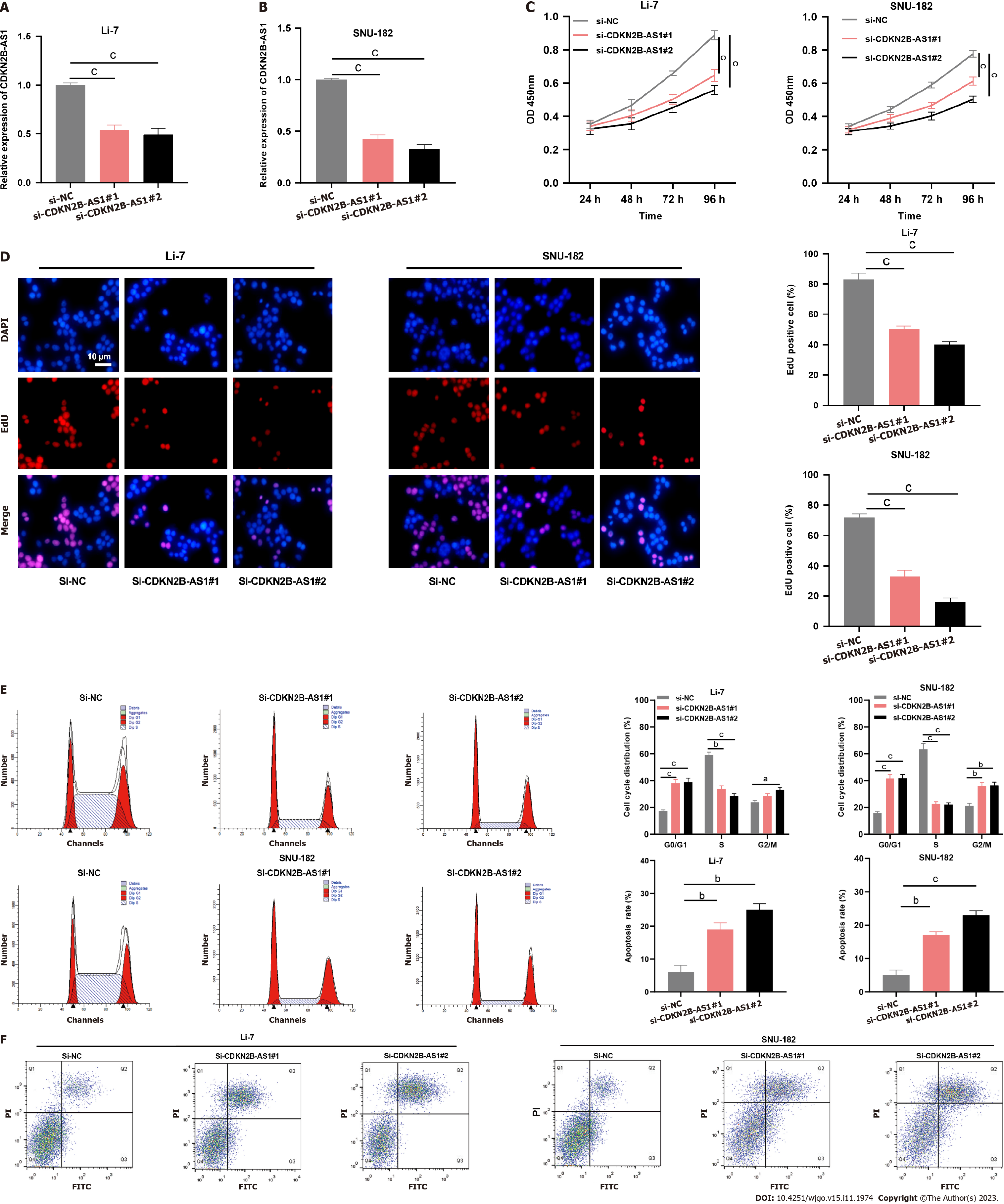
Next, we explored the biological function of CDKN2B-AS1 in modulating the malignant biological behaviors of HCC cells. Since CDKN2B-AS1 expression was most aberrantly expressed in Li-7 and SNU-182 cells, they were chosen for further investigation. As shown, transfection of CDKN2B-AS1 siRNA significantly reduced CDKN2B-AS1 expression in Li-7 and SNU-182 cells (Figures 2A and B). Functional assays showed depletion of CDKN2B-AS1 significantly suppressed Li-7 and SNU-182 cell viability (Figures 2C and D). Furthermore, by flow cytometry analysis, we found that the knockdown of CDKN2B-AS1 resulted in Li-7 and SNU-182 cells being blocked in the G0/G1 phase and promoted apoptosis (Figures 2E and F). In conclusion, these data unveiled that CDKN2B-AS1 enhanced the malignancy of HCC cells.
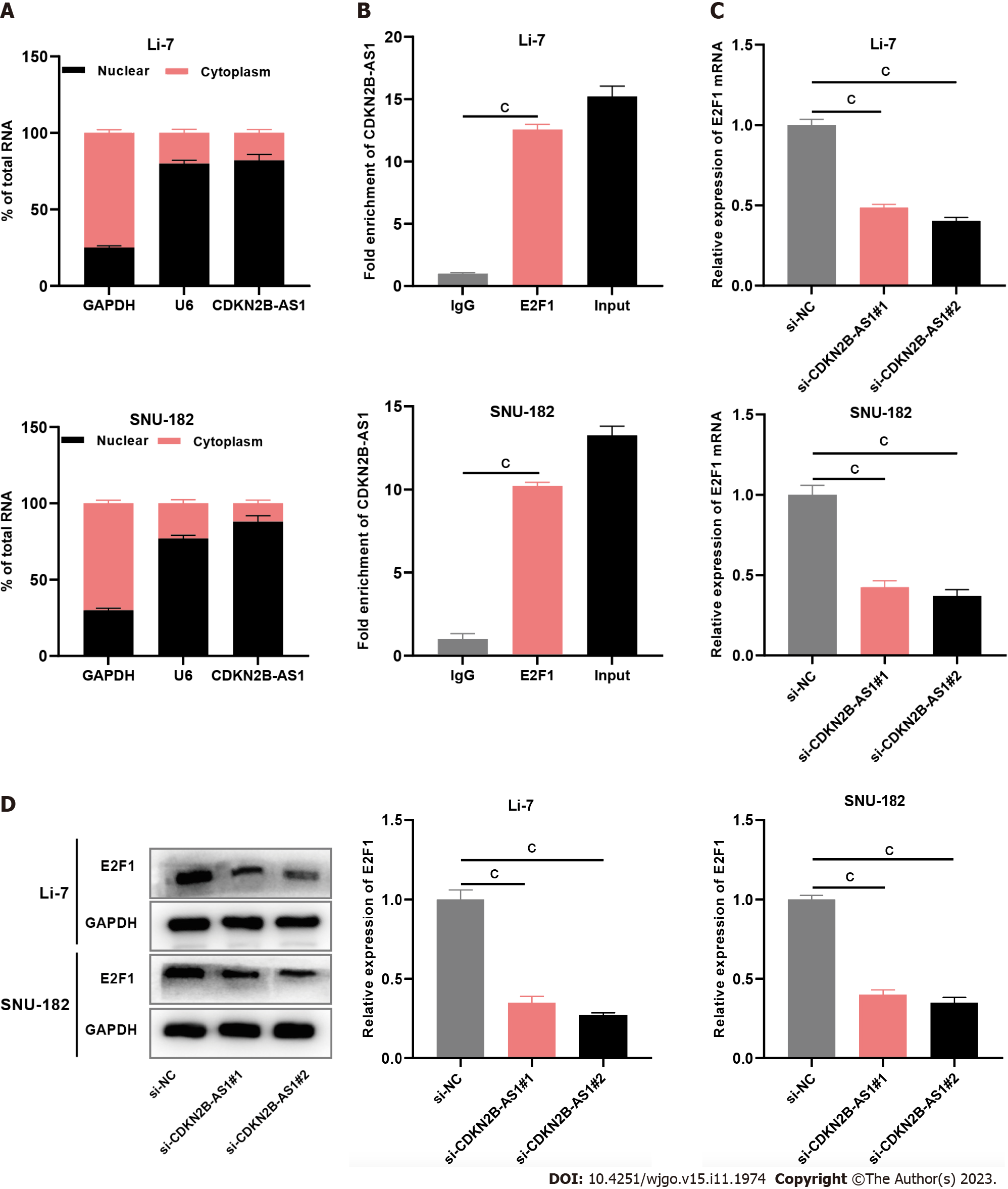
Subsequently, we tried to explore the downstream mechanism of CDKN2B-AS1. The results of nucleoplasmic separation experiments showed that most CDKN2B-AS1 transcripts were present in the nuclei of Li-7 and SNU-182 cells (Figure 3A), this suggested that CDKN2B-AS1 probably exerts its biological function at the transcriptional level. Through the RIP experiment, we found that compared with the IgG group, CDKN2B-AS1 was remarkably enriched by anti-E2F1 antibody (Figure 3B). Subsequently, the qRT-PCR analysis demonstrated that transfection with CDKN2B-AS1 siRNA significantly reduced E2F1 mRNA expression in the cells (Figure 3C). Immunoblotting showed that depletion of CDKN2B-AS1 significantly inhibited the expression level of E2F1 protein in Li-7 and SNU-182 cells (Figure 3D).
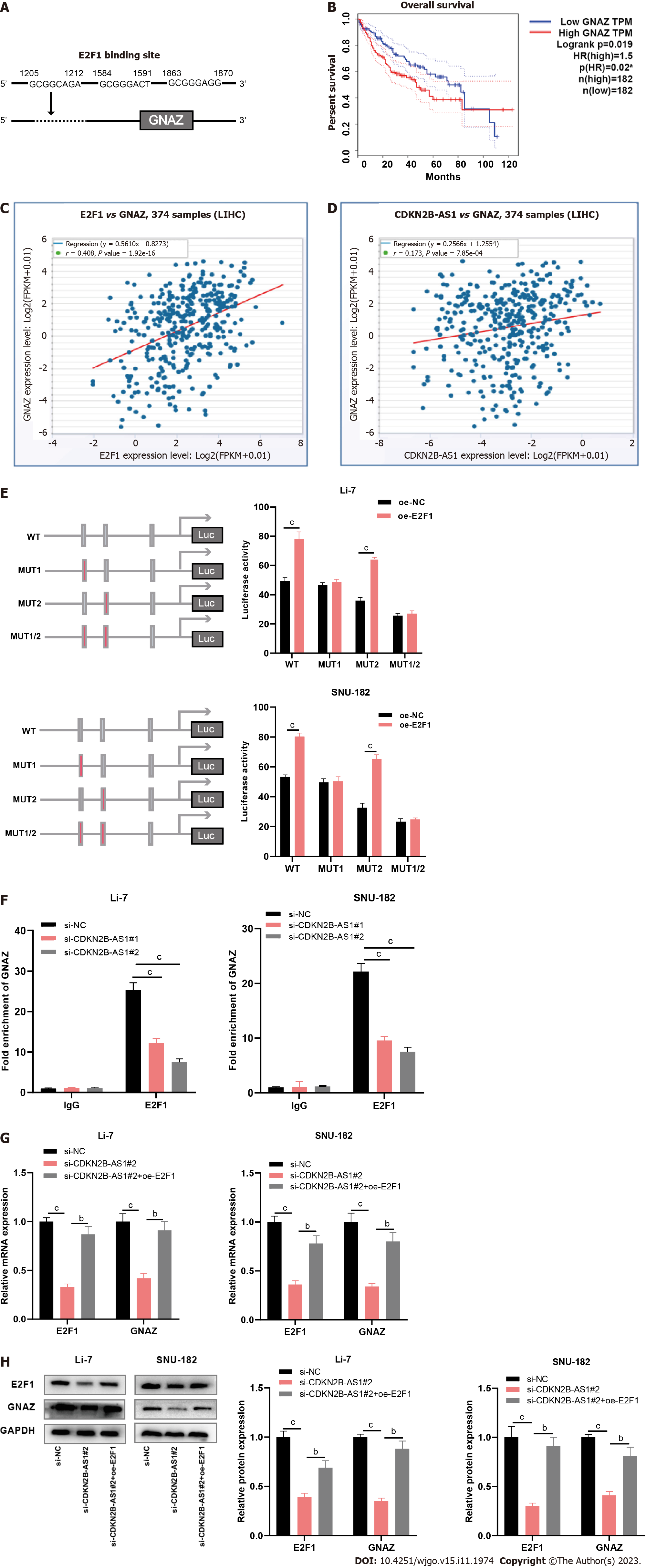
Finally, we tried to screen out the downstream effects modulated by CDKN2B-AS1/E2F1. PROMO database implied that E2F1 could bind to the GNAZ promoter sequence (Figure 4A). StarBase database showed that high GNAZ expression hinted at shorter overall survival in HCC patients (Figure 4B); besides, GNAZ expression level was positively correlated with either E2F1 expression or CDKN2B-AS1 expression in HCC tissue samples (Figures 4C and D). Moreover, the binding sequence was mutated and a luciferase reporter assay was conducted. The results unveiled that site 3 was a specific site for the binding of E2F1 protein to the GNAZ promoter (Figure 4E). Chromatin immunoprecipitation analysis demonstrated that the depletion of CDKN2B-AS1 decreased the binding between E2F1 and GNAZ (Figure 4F). As expected, CDKN2B-AS1 knockdown significantly inhibited E2F1 and GNAZ expression in Li-7 and SNU-182 cells, which was reversed by E2F1 overexpression (Figures 4G and H).
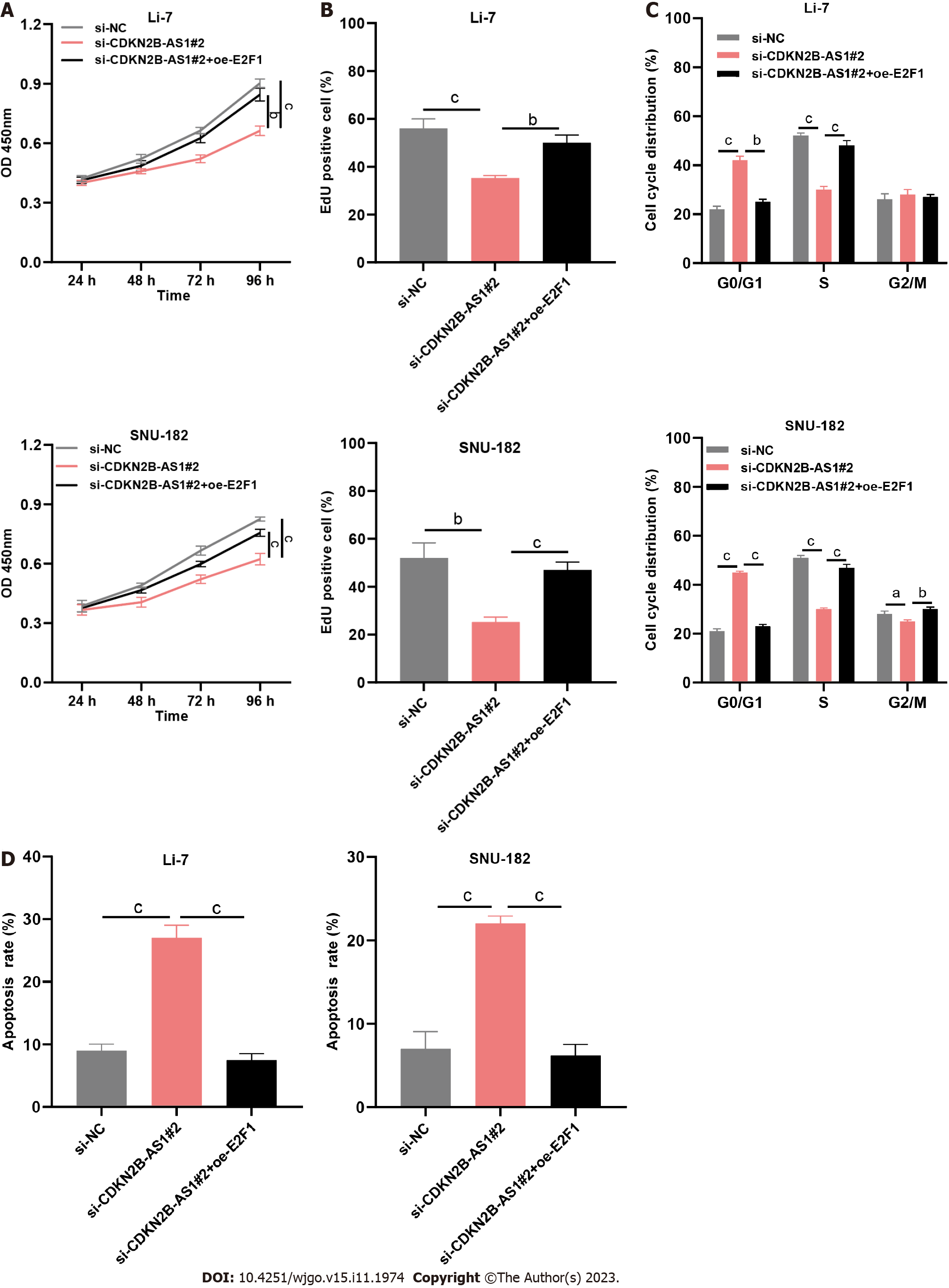
To decipher the role of the CDKN2B-AS1/E2F1/GNAZ axis, we co-transfected si-NC, CDKN2B-AS1 siRNA, and CDKN2B-AS1 siRNA + E2F1 overexpression plasmid into Li-7 and SNU-182 cells. As shown, depletion of CDKN2B-AS1 markedly inhibited the growth of Li-7 and SNU-182 cells, while E2F1 overexpression reversed this effect (Figures 5A and B). Similarly, E2F1 overexpression counteracted the suppressive and promotive effects of CDKN2B-AS1 depletion on cell cycle progression and apoptosis of Li-7 and SNU-182 cells (Figures 5C and D).
LncRNAs are one of the hot spots in cancer research nowadays[16]. Although lncRNAs lack protein-coding capabilities, many studies in the last decade have shown that they are important in regulating gene expression and protein function: In the nucleus, lncRNA participates in histone modification, mediate DNA methylation, affect chromatin remodeling, and regulate the variable splicing of mRNA; in the cytoplasm, lncRNA acts as an functional element to regulate the translation of mRNA, or decoys miRNA as molecular sponge, or directly interacts with protein to affect its biological activity and subcellular location[17,18]. In HCC, many lncRNAs have been identified and their associations with HCC have been reported[15,19,20]. For example, lncRNA HAND2-AS1 promotes self-renewal of HCC cells through BMP signaling[21]. LncRNA-SNHG7 expression level is linked with the pathological and prognostic characteristics of patients with HCC[22]. In addition, CDKN2B-AS1 has been validated to play carcinogenic roles in some malignancies including lung carcinoma[23], ovarian carcinoma[24], laryngeal squamous cell carcinoma[25], cervical carcinoma[26] and HCC[13,27]. Here, our data suggested that, in HCC, the expression level of CDKN2B-AS1 was increased, and it hinted at an adverse prognosis. In vitro assays confirmed that depletion of CDKN2B-AS1 inhibited the malignancy of HCC cells. These demonstrations are consistent with previous reports, which show CDKN2B-AS1 as an oncogene in HCC[27].
E2F1, a part of the E2F family of transcription factors, is recognized as an important promoter for cell entry into the S phase, and a non-negligible modulator in physiological and pathological processes[28]. E2F1 is aberrantly expressed in a variety of human malignancies and acts as a pro-oncogene[29]. For example, E2F1 promotes bladder carcinoma cell proliferation by binding to the promoter of RAD54L[30]. In HCC, E2F1 is also aberrantly expressed, and it promotes the malignancy of HCC cells by regulating DDX11 transcription and activating PI3K/AKT/mTOR signaling[31]. Some recent studies report that lncRNAs synergistically work with E2F1 to modulate gene transcription. For example, lncRNA NR-104098, interacted with E2F1, suppresses EZH2 transcription, suppressing acute myeloid leukemia progression[32]. Here, we found that CDKN2B-AS1 interacted with E2F1. Further studies confirmed that GNAZ was a transcriptional target of E2F1. Importantly, the knockdown of CDKN2B-AS1 attenuated the recruitment of E2F1 to GNAZ. Furthermore, rescue experiments confirmed that E2F1 mediated the carcinogenic effect of CDKN2B-AS1.
G protein-coupled receptors (GPCRs) are crucial in the transduction of a variety of signaling and are considered attractive targets for the treatment of many diseases[33]. GNAZ is one of the notable members of the GPCR family[11]. High GNAZ expression leads to a surge in leukocyte and lymphocyte counts in patients with laparoscopic lymphoma, thereby reducing overall patient survival[34]. Notably, GNAZ expression level is reported to be associated with unsatisfactory prognosis of HCC patients, and GNAZ overexpression leads to enhanced aggressiveness of the cells[11]. In this study, our data showed that CDKN2B-AS1 promoted GNAZ transcription by enhancing the binding of E2F1 in the GNAZ promoter, facilitating HCC cell proliferation and inhibiting apoptosis.
Nevertheless, there are some shortcomings in the present work. Firstly, in vivo experiments are required to further validate the tumor-promoting properties of CDKN2B-AS1 in HCC. In addition, knockout rather than knockdown of the CDKN2B-AS1 in HCC cell lines or induced pluripotent stem cell derived hepatocytes should be performed to validate the functions of this gene as a more authentic and objective evidence. Also, it would be interesting to explore whether CDKN2B-AS1 depletion will affect other malignant biological behaviors of HCC such as chemosensitivity and radiosensitivity. Furthermore, the other downstream targets of the CDKN2B-AS1/E2F1 axis should be explored, and this will further clarify the mechanism by which CDKN2B-AS1 modulates the phenotypes of HCC. Last but not least, more tissue samples from HCC patients should be collected to analyze the clinical significance of CDKN2B-AS1 and to evaluate the potential of CDKN2B-AS1 as a biomarker.
CDKN2B-AS1 expression was upregulated in HCC and hinted at poor prognosis in the patients. We confirmed that CDKN2B-AS1 promoted GNAZ transcription through the recruitment of E2F1, thereby promoting the malignancy of HCC cells.
Long non-coding RNAs (lncRNAs) have been implicated in cancer biology, with lncRNA CDKN2B-AS1 being reported to associate with several human cancers, though its role in hepatocellular carcinoma (HCC) remains unclear.
The motivation behind this research is to better understand the mechanisms underlying the development and progression of HCC, and to explore whether CDKN2B-AS1 could serve as a potential therapeutic target for HCC.
This study aims to investigate the role of the lncRNA CDKN2B-AS1 in HCC progression.
This study investigated the role of CDKN2B-AS1 in HCC progression by measuring its expression in HCC using quantitative real-time polymerase chain reaction. Effects on proliferation, cell cycle, and apoptosis of Li-7 and SNU-182 cells were then assessed using the CCK-8 assay, the EdU assay, and flow cytometry. RNA immunoprecipitation was performed to verify the interaction between CDKN2B-AS1 and E2F transcription factor 1 (E2F1). The binding of E2F1 to the promoter of G protein subunit alpha Z (GNAZ) was confirmed using luciferase reporter assay and Chromatin immunoprecipitation. And western blot was utilized to confirm the expression of E2F1 and GNAZ in HCC cells.
Upregulation of CDKN2B-AS1 was identified in HCC tissues. Inhibited proliferation, induced cell cycle arrest as well as apoptosis were detected in HCC cells with silenced CDKN2B-AS1. In addition, CDKN2B-AS1 was found to interact with E2F1, and its depletion significantly inhibited the binding of E2F1 to the GNAZ promoter region. It has also been found that these effects caused by CDKN2B-AS1 knockdown, can be reversed by E2F1 overexpression.
In conclusion, the promotion of HCC progression is facilitated by CDKN2B-AS1 recruiting E2F1 to enhance GNAZ transcription.
This research suggests that CDKN2B-AS1 may serve as a potential therapeutic target for HCC. Further research could investigate the effectiveness of CDKN2B-AS1 inhibition as a treatment for HCC. Additionally, this study provides a better understanding of the mechanisms underlying HCC progression and could inform the development of new diagnostic and treatment approaches for this disease.
Provenance and peer review: Unsolicited article; Externally peer reviewed.
Peer-review model: Single blind
Specialty type: Gastroenterology and hepatology
Country/Territory of origin: China
Peer-review report’s scientific quality classification
Grade A (Excellent): A
Grade B (Very good): B, B
Grade C (Good): 0
Grade D (Fair): 0
Grade E (Poor): 0
P-Reviewer: Demir FB, Turkey; Sahin TT, Turkey; Sanal MG, India S-Editor: Wang JJ L-Editor: A P-Editor: Yuan YY
| 1. | McGlynn KA, Petrick JL, El-Serag HB. Epidemiology of Hepatocellular Carcinoma. Hepatology. 2021;73 Suppl 1:4-13. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 820] [Cited by in RCA: 1321] [Article Influence: 330.3] [Reference Citation Analysis (1)] |
| 2. | Morse MA, Sun W, Kim R, He AR, Abada PB, Mynderse M, Finn RS. The Role of Angiogenesis in Hepatocellular Carcinoma. Clin Cancer Res. 2019;25:912-920. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 188] [Cited by in RCA: 386] [Article Influence: 55.1] [Reference Citation Analysis (0)] |
| 3. | Yi PS, Li Y, Yan S, Wu B, Lan C, Li JS. Surgery combined with post-operative trancatheter arterial chemoembolization improves survival of intermediate hepatocellular carcinoma. Scand J Gastroenterol. 2019;54:240-245. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 6] [Cited by in RCA: 2] [Article Influence: 0.3] [Reference Citation Analysis (0)] |
| 4. | Greten TF, Lai CW, Li G, Staveley-O'Carroll KF. Targeted and Immune-Based Therapies for Hepatocellular Carcinoma. Gastroenterology. 2019;156:510-524. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 168] [Cited by in RCA: 199] [Article Influence: 33.2] [Reference Citation Analysis (0)] |
| 5. | Mullath A, Krishna M. Hepatocellular carcinoma - time to take the ticket. World J Gastrointest Surg. 2019;11:287-295. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in CrossRef: 3] [Cited by in RCA: 3] [Article Influence: 0.5] [Reference Citation Analysis (0)] |
| 6. | Liao K, Xu J, Yang W, You X, Zhong Q, Wang X. The research progress of LncRNA involved in the regulation of inflammatory diseases. Mol Immunol. 2018;101:182-188. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 57] [Cited by in RCA: 78] [Article Influence: 11.1] [Reference Citation Analysis (0)] |
| 7. | Wang JY, Yang Y, Ma Y, Wang F, Xue A, Zhu J, Yang H, Chen Q, Chen M, Ye L, Wu H, Zhang Q. Potential regulatory role of lncRNA-miRNA-mRNA axis in osteosarcoma. Biomed Pharmacother. 2020;121:109627. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 90] [Cited by in RCA: 160] [Article Influence: 32.0] [Reference Citation Analysis (0)] |
| 8. | Esposito R, Bosch N, Lanzós A, Polidori T, Pulido-Quetglas C, Johnson R. Hacking the Cancer Genome: Profiling Therapeutically Actionable Long Non-coding RNAs Using CRISPR-Cas9 Screening. Cancer Cell. 2019;35:545-557. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 124] [Cited by in RCA: 160] [Article Influence: 26.7] [Reference Citation Analysis (0)] |
| 9. | Li Y, Jiang T, Zhou W, Li J, Li X, Wang Q, Jin X, Yin J, Chen L, Zhang Y, Xu J. Pan-cancer characterization of immune-related lncRNAs identifies potential oncogenic biomarkers. Nat Commun. 2020;11:1000. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 159] [Cited by in RCA: 295] [Article Influence: 59.0] [Reference Citation Analysis (0)] |
| 10. | Vancura P, Abdelhadi S, Csicsely E, Baba K, Tosini G, Iuvone PM, Spessert R. Gnaz couples the circadian and dopaminergic system to G protein-mediated signaling in mouse photoreceptors. PLoS One. 2017;12:e0187411. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 9] [Cited by in RCA: 9] [Article Influence: 1.1] [Reference Citation Analysis (0)] |
| 11. | Tian F, Cai D. Overexpressed GNAZ predicts poor outcome and promotes G0/G1 cell cycle progression in hepatocellular carcinoma. Gene. 2022;807:145964. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1] [Cited by in RCA: 1] [Article Influence: 0.3] [Reference Citation Analysis (0)] |
| 12. | Sui J, Miao Y, Han J, Nan H, Shen B, Zhang X, Zhang Y, Wu Y, Wu W, Liu T, Xu S, Yang S, Yin L, Pu Y, Liang G. Systematic analyses of a novel lncRNA-associated signature as the prognostic biomarker for Hepatocellular Carcinoma. Cancer Med. 2018;7:3240-3256. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 20] [Cited by in RCA: 31] [Article Influence: 4.4] [Reference Citation Analysis (0)] |
| 13. | Shen X, Li Y, He F, Kong J. LncRNA CDKN2B-AS1 Promotes Cell Viability, Migration, and Invasion of Hepatocellular Carcinoma via Sponging miR-424-5p. Cancer Manag Res. 2020;12:6807-6819. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 9] [Cited by in RCA: 11] [Article Influence: 2.2] [Reference Citation Analysis (0)] |
| 14. | Gu Y, Wang X, Liu H, Li G, Yu W, Ma Q. SET7/9 promotes hepatocellular carcinoma progression through regulation of E2F1. Oncol Rep. 2018;40:1863-1874. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 5] [Cited by in RCA: 24] [Article Influence: 3.4] [Reference Citation Analysis (0)] |
| 15. | Lv E, Sheng J, Yu C, Rao D, Huang W. LncRNA influence sequential steps of hepatocellular carcinoma metastasis. Biomed Pharmacother. 2021;136:111224. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 8] [Cited by in RCA: 26] [Article Influence: 6.5] [Reference Citation Analysis (0)] |
| 16. | Charles Richard JL, Eichhorn PJA. Platforms for Investigating LncRNA Functions. SLAS Technol. 2018;23:493-506. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 124] [Cited by in RCA: 133] [Article Influence: 19.0] [Reference Citation Analysis (0)] |
| 17. | Ma Y, Zhang J, Wen L, Lin A. Membrane-lipid associated lncRNA: A new regulator in cancer signaling. Cancer Lett. 2018;419:27-29. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 76] [Cited by in RCA: 120] [Article Influence: 17.1] [Reference Citation Analysis (0)] |
| 18. | Rajagopal T, Talluri S, Akshaya RL, Dunna NR. HOTAIR LncRNA: A novel oncogenic propellant in human cancer. Clin Chim Acta. 2020;503:1-18. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 52] [Cited by in RCA: 121] [Article Influence: 24.2] [Reference Citation Analysis (0)] |
| 19. | Yang Y, Chen Q, Piao HY, Wang B, Zhu GQ, Chen EB, Xiao K, Zhou ZJ, Shi GM, Shi YH, Wu WZ, Fan J, Zhou J, Dai Z. HNRNPAB-regulated lncRNA-ELF209 inhibits the malignancy of hepatocellular carcinoma. Int J Cancer. 2020;146:169-180. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 29] [Cited by in RCA: 21] [Article Influence: 4.2] [Reference Citation Analysis (0)] |
| 20. | Qi W, Li Z, Xia L, Dai J, Zhang Q, Wu C, Xu S. LncRNA GABPB1-AS1 and GABPB1 regulate oxidative stress during erastin-induced ferroptosis in HepG2 hepatocellular carcinoma cells. Sci Rep. 2019;9:16185. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 148] [Cited by in RCA: 174] [Article Influence: 29.0] [Reference Citation Analysis (0)] |
| 21. | Wang Y, Zhu P, Luo J, Wang J, Liu Z, Wu W, Du Y, Ye B, Wang D, He L, Ren W, Sun X, Chen R, Tian Y, Fan Z. LncRNA HAND2-AS1 promotes liver cancer stem cell self-renewal via BMP signaling. EMBO J. 2019;38:e101110. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 71] [Cited by in RCA: 118] [Article Influence: 19.7] [Reference Citation Analysis (0)] |
| 22. | Shen A, Ma J, Hu X, Cui X. High expression of lncRNA-SNHG7 is associated with poor prognosis in hepatocellular carcinoma. Oncol Lett. 2020;19:3959-3963. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 6] [Cited by in RCA: 8] [Article Influence: 1.6] [Reference Citation Analysis (0)] |
| 23. | Wang G, Xu G, Wang W. Long Noncoding RNA CDKN2B-AS1 Facilitates Lung Cancer Development Through Regulating miR-378b/NR2C2. Onco Targets Ther. 2020;13:10641-10649. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 10] [Cited by in RCA: 18] [Article Influence: 3.6] [Reference Citation Analysis (0)] |
| 24. | Xu C, Zhai J, Fu Y. LncRNA CDKN2B-AS1 promotes the progression of ovarian cancer by miR-143-3p/SMAD3 axis and predicts a poor prognosis. Neoplasma. 2020;67:782-793. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 10] [Cited by in RCA: 10] [Article Influence: 2.0] [Reference Citation Analysis (0)] |
| 25. | Cui X, Yu T, Shang J, Xiao D, Wang X. Long Non-Coding RNA CDKN2B-AS1 Facilitates Laryngeal Squamous Cell Cancer Through Regulating miR-497/CDK6 Pathway. Onco Targets Ther. 2019;12:8853-8862. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 17] [Cited by in RCA: 16] [Article Influence: 2.7] [Reference Citation Analysis (0)] |
| 26. | Zhang Y, Zhao L, Yang S, Cen Y, Zhu T, Wang L, Xia L, Liu Y, Zou J, Xu J, Li Y, Cheng X, Lu W, Wang X, Xie X. CircCDKN2B-AS1 interacts with IMP3 to stabilize hexokinase 2 mRNA and facilitate cervical squamous cell carcinoma aerobic glycolysis progression. J Exp Clin Cancer Res. 2020;39:281. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 21] [Cited by in RCA: 51] [Article Influence: 10.2] [Reference Citation Analysis (0)] |
| 27. | Huang Y, Xiang B, Liu Y, Wang Y, Kan H. LncRNA CDKN2B-AS1 promotes tumor growth and metastasis of human hepatocellular carcinoma by targeting let-7c-5p/NAP1L1 axis. Cancer Lett. 2018;437:56-66. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 98] [Cited by in RCA: 133] [Article Influence: 19.0] [Reference Citation Analysis (0)] |
| 28. | Denechaud PD, Fajas L, Giralt A. E2F1, a Novel Regulator of Metabolism. Front Endocrinol (Lausanne). 2017;8:311. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 106] [Cited by in RCA: 148] [Article Influence: 18.5] [Reference Citation Analysis (0)] |
| 29. | Shen C, Li J, Chang S, Che G. Advancement of E2F1 in Common Tumors. Zhongguo Fei Ai Za Zhi. 2020;23:921-926. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in RCA: 5] [Reference Citation Analysis (0)] |
| 30. | Mun JY, Baek SW, Park WY, Kim WT, Kim SK, Roh YG, Jeong MS, Yang GE, Lee JH, Chung JW, Choi YH, Chu IS, Leem SH. E2F1 Promotes Progression of Bladder Cancer by Modulating RAD54L Involved in Homologous Recombination Repair. Int J Mol Sci. 2020;21. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 12] [Cited by in RCA: 24] [Article Influence: 4.8] [Reference Citation Analysis (0)] |
| 31. | Yu Y, Zhao D, Li K, Cai Y, Xu P, Li R, Li J, Chen X, Chen P, Cui G. E2F1 mediated DDX11 transcriptional activation promotes hepatocellular carcinoma progression through PI3K/AKT/mTOR pathway. Cell Death Dis. 2020;11:273. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 37] [Cited by in RCA: 52] [Article Influence: 10.4] [Reference Citation Analysis (0)] |
| 32. | Feng Y, Hu S, Li L, Zhang S, Liu J, Xu X, Zhang M, Du T, Du Y, Peng X, Chen F. LncRNA NR-104098 Inhibits AML Proliferation and Induces Differentiation Through Repressing EZH2 Transcription by Interacting With E2F1. Front Cell Dev Biol. 2020;8:142. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 11] [Cited by in RCA: 13] [Article Influence: 2.6] [Reference Citation Analysis (0)] |
| 33. | Wein MN, Foretz M, Fisher DE, Xavier RJ, Kronenberg HM. Salt-Inducible Kinases: Physiology, Regulation by cAMP, and Therapeutic Potential. Trends Endocrinol Metab. 2018;29:723-735. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 94] [Cited by in RCA: 96] [Article Influence: 13.7] [Reference Citation Analysis (0)] |
| 34. | Mundt F, Merrien M, Nygren L, Sutton LA, Christensson B, Wahlin BE, Rosenquist R, Sander B, Wasik AM. Expression of GNAZ, encoding the Gα(z) protein, predicts survival in mantle cell lymphoma. Br J Haematol. 2019;185:708-712. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 2] [Cited by in RCA: 3] [Article Influence: 0.5] [Reference Citation Analysis (0)] |