Published online Nov 15, 2023. doi: 10.4251/wjgo.v15.i11.1951
Peer-review started: May 16, 2023
First decision: August 7, 2023
Revised: August 15, 2023
Accepted: September 14, 2023
Article in press: September 14, 2023
Published online: November 15, 2023
Processing time: 182 Days and 20.2 Hours
Tumor recurrence and metastasis lead to a poor prognosis in colorectal cancer (CRC). Necroptosis is closely related to the tumor microenvironment (TME) and affects tumor recurrence and metastasis. We aimed to stratify CRC patients according to necroptosis-related long noncoding RNAs (lncRNAs), which can be used to not only evaluate prognosis and improve precision medicine in clinical practice but also screen potential immunotherapy drugs.
To stratify CRC patients according to necroptosis-related lncRNAs (NRLs), which can be used to not only evaluate prognosis and improve precision medicine in clinical practice but also screen potential immunotherapy drugs.
LncRNA expression profiles were collected from The Cancer Genome Atlas. NRLs were identified by coexpression analysis. Cox regression analysis identified a NRL signature. Then, the value of this signature was comprehensively and multidimensionally evaluated, and its reliability for CRC prognosis prediction was assessed with clinical CRC data and compared with that of six other lncRNA signatures. Gene set enrichment analysis, TME analysis and half-maximal inhibitory concentration (IC50) prediction were also performed according to the risk score (RS) of the signature.
An 8-lncRNA signature significantly associated with overall survival (OS) was constructed, and its reliability was validated with clinical CRC data. Most of the areas under the receiver operating characteristic curves (AUCs) values for 1-, 3- and 5-year OS for this signature were higher than those for the other six lncRNA signatures. OS, disease-specific survival and the progression-free interval were all significantly poorer in the high-risk group. The RS of the signature showed good concordance with the predicted prognosis, with AUCs for 1-, 3- and 5-year OS of 0.79, 0.81 and 0.77, respectively. Additionally, the calibration plots for this signature combined with clinical factors showed that this combination could effectively improve the ability to predict OS. The RS was correlated with tumor stage, lymph node metastasis and distant metastasis. Most of the enriched Kyoto Encyclopedia of Genes and Genomes and Gene Ontology terms were tumor metastasis-related pathways in the high-risk group; these patients showed greater infiltration of immunosuppressive cells, such as cancer-associated fibroblasts, hematopoietic stem cells and M2 macrophages, but less infiltration of infiltrating antitumor effector immune cells, such as cluster of differentiation 8+ T cells and regulatory T cells (Tregs). We explored additional potential immune checkpoint genes and potential immunotherapeutic and chemotherapeutic drugs with relatively low IC50 values.
We identified an NRL signature with strong fidelity that could stably predict prognosis and might be an indicator of the TME of CRC. Furthermore, additional potential immunotherapeutic and chemotherapeutic drugs were explored.
Core Tip: An 8-lncRNA signature significantly associated with overall survival (OS) was constructed, and its fidelity was validated in the OS, disease-specific survival and the progression-free interval, it was also validated with clinical colorectal cancer (CRC) data. The risk score which was correlated with tumor stage, lymph node metastasis and distant metastasis in CRC of the signature showed good concordance with the predicted prognosis. The high-risk group showed greater infiltration of immunosuppressive cells, but fewer infiltrating antitumor effector immune cells. We explored additional potential immune checkpoint genes and immunotherapeutic and chemotherapeutic drugs.
- Citation: Chen ZH, Lin YL, Chen SQ, Yang XY. Identification of necroptosis-related lncRNAs for prognosis prediction and screening of potential drugs in patients with colorectal cancer. World J Gastrointest Oncol 2023; 15(11): 1951-1973
- URL: https://www.wjgnet.com/1948-5204/full/v15/i11/1951.htm
- DOI: https://dx.doi.org/10.4251/wjgo.v15.i11.1951
Despite significant reductions in the incidence and mortality of colorectal cancer (CRC) over several decades, CRC[1] is the third most commonly diagnosed cancer and the second most common malignancy worldwide because of its high rates of tumor recurrence and metastasis, which cause a poor prognosis[2,3]. Systemic treatments, including surgery, chemotherapeutics and targeted therapeutics, have reached a bottleneck for improving patient prognosis. Immunotherapies, such as programmed death-1 (PD-1) and cytotoxic T-lymphocyte antigen 4 inhibitors, have transformed the treatment landscape for CRC patients and become important treatments, especially for advanced CRC patients with microsatellite instability-high (MSI-H) disease, which is highly infiltrated with immune cells and carries a high neoantigen load[4]. Because of the low proportion of CRC patients with MSI-H disease, the achievement of a complete response with immunotherapy is rare in the majority of CRC cases[5].
Necroptosis is an evolutionary form of caspase-independent programmed necrosis that is a vital process in the life cycle of organisms and contributes to the innate immune response by inducing tumor cell death. Necroptosis has emerged as a therapeutic target, as it creates dying cancer cells that can stimulate antitumor immune responses[6], and necroptosis activation can contribute to a clinically favorable immune signature and survival rates, highlighting the novel therapeutic possibility of combining a necroptosis-based therapeutic approach with immune checkpoint inhibitors for more efficient treatment of tumor patients. Necroptosis can enhance antitumor immunity more efficiently than immunogenic apoptosis, implying the high potential of necroptosis as a therapeutic target in cancer[7]. Necroptosis induction significantly improves antitumor immunity by inducing the maturation of dendritic cells and activation of cytotoxic cluster of differentiation (CD) 8+ T cells, which play roles in antitumor immunity[8]. However, many factors, such as genetic instability, contribute to resistance to necroptosis in CRC[9]. Han et al[10] reported that resibufogenin could suppress the growth and metastasis of CRC by inducing necroptosis in vivo. Developing approaches to activate the necroptotic process and subsequently induce antitumor immune cells to exert antitumor effects may become a new direction for tumor immunotherapy.
Long noncoding RNAs (lncRNAs), which are not translated into proteins, can regulate gene expression at multiple levels, such as the transcriptional, chromatin modification and posttranscriptional levels[11]. Furthermore, lncRNAs participate in various biological processes, such as stem cell pluripotency maintenance, cell cycle regulation and cell differentiation. Overexpression of some lncRNAs can inhibit the expression of necroptosis-related proteins and suppress the necroptotic pathway by downregulating membrane receptors, thereby reducing tumor cell necroptosis and antitumor immune cell infiltration[12,13]. In addition, lncRNAs interact with microRNAs (miRNAs) and affect the expression of other miRNA species[14]. Experiments have confirmed that the lncRNA necrosis-related factor targets miR-873 and RIPK1/RIPK3 to regulate cardiomyocyte necroptosis[15]. There are no reports on necroptosis-related lncRNAs (NRLs) widely mentioned as potential immunotherapeutic targets in CRC. Therefore, acquiring more knowledge on NRLs and exploring the internal relationships and mechanisms of action of these lncRNAs could help us understand the roles of necroptosis and lncRNAs in immunotherapy in CRC patients and explore more effective immunotherapeutic methods.
In this study, we aimed to explore the internal relationships and mechanisms of interaction between NRLs and the tumor microenvironment (TME). As lncRNAs in bodily fluids are known to be new cancer biomarkers, we stratified CRC patients according to NRL expression levels, which allowed us to not only evaluate patient prognosis but also improve precision medicine in clinical practice. Additionally, we hoped to identify potential effective immunotherapeutic targets and drugs.
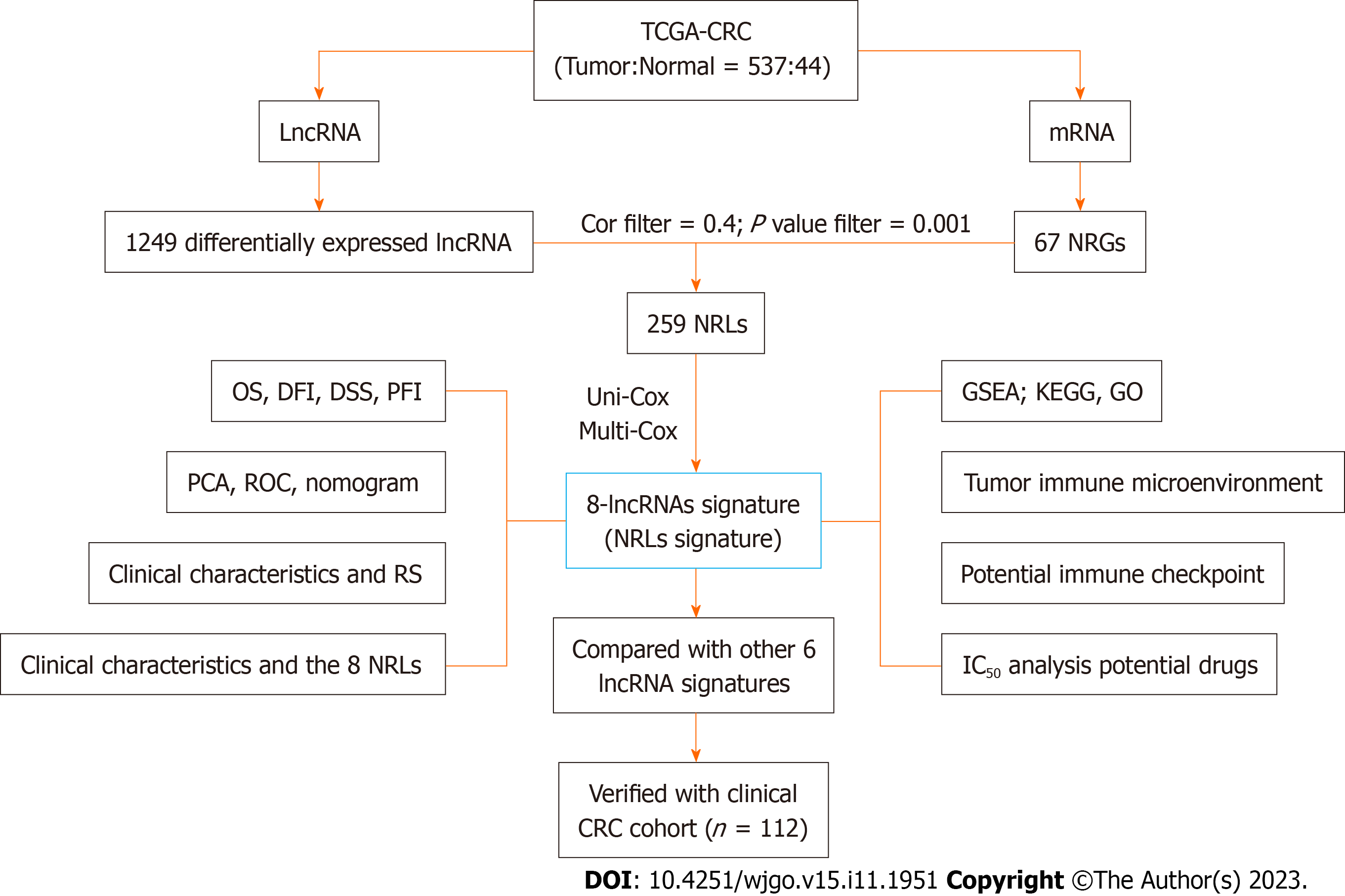
The entire analysis workflow is shown in Figure 1. To obtain RNA-sequencing data [HTSeq counts and HTSeq-fragments per kilobase per million (FPKM)] for colon adenocarcinoma, rectal adenocarcinoma and normal colorectal tissues, matching clinical data were downloaded from The Cancer Genome Atlas (TCGA). The datasets analyzed during the current study are available in the TCGA repository (https://portal.gdc.cancer.gov/). The training cohort was derived from the TCGA-CRC dataset, and the validation cohort was derived from clinical tissue samples from CRC patients (Table 1) who underwent surgery at the First Affiliated Hospital of Fujian Medical University between January 2018 and December 2018. Finally, 121 CRC patients who had complete clinical data and fresh pathological specimens stored in a freezer at -80°C were selected for inclusion in the validation cohort. This study was approved by the Ethics Committee of the First Affiliated Hospital of Fujian Medical University, No. MRCTA, ECFAH of FMU [20190(21)]. The research was approved by the Ethics Committee and includes any relevant details; We also confirmed that all experiments were performed in accordance with relevant guidelines and regulations. The FPKM values of the synthetic matrix were converted into transcripts per million (TPM) values with the data table, dplyr, tidyr, and tibble R packages. As a result, we obtained two synthetic data matrices. The count value matrix was used to identify differentially expressed lncRNAs, while the TPM value matrix was used for the other analyses. We ultimately included 537 patients with relevant clinical information after excluding patients with missing overall survival (OS) values or short OS times (< 30 d).
| Characteristics | TCGA-CRC (n = 537) | Clinical-CRC (n = 121) | ||
| All | High risk (n = 269) | Low risk (n = 268) | ||
| Age | ||||
| ≤ 65 | 234 (43.58%) | 114 (42.38%) | 120 (44.78%) | 53 (43.8%) |
| > 65 | 303 (56.42%) | 155 (57.62%) | 148 (55.22%) | 68 (56.2) |
| Sex | ||||
| Male | 287 (53.45%) | 141 (52.42%) | 146 (45.48%) | 61 (50.41%) |
| Female | 250 (46.55%) | 128 (47.58%) | 122 (45.52%) | 60 (49.29%) |
| T Stage | ||||
| T1 | 15 (2.94%) | 8 (2.97%) | 7 (2.61%) | 3 (2.48%) |
| T2 | 93 (17.32%) | 42 (15.61%) | 51 (19.03%) | 17 (14.05%) |
| T3 | 366 (68.16%) | 180 (66.91%) | 186 (69.40%) | 49 (40.5%) |
| T4 | 63 (11.73%) | 39 (14.50%) | 24 (8.96%) | 52 (42.98%) |
| N Stage | ||||
| N0 | 316 (58.85%) | 140 (52.04%) | 176 (65.67%) | 67 (55.37%) |
| N1 | 128 (23.84%) | 67 (24.91%) | 61 (22.76%) | 31 (25.62%) |
| N2 | 92 (17.13%) | 62 (23.05%) | 30 (11.19%) | 23 (19.01%) |
| Unknown | 1 (0.19%) | 0 | 1 (0.38%) | - |
| M Stage | ||||
| M0 | 400 (74.49%) | 190 (70.63%) | 210 (78.36%) | 102 (84.3%) |
| M1 | 75 (13.97%) | 46 (17.1%) | 29 (10.82%) | 19 (15.7%) |
| Unknown | 62 (11.55%) | 33 (12.27%) | 29 (10.82%) | - |
| Stage | ||||
| Stage I | 93 (17.32%) | 41 (15.24%) | 52 (19.40%) | 20 (16.53%) |
| Stage II | 209 (38.92%) | 91 (33.83%) | 118 (44.03%) | 42 (34.71%) |
| Stage III | 149 (27.75%) | 83 (30.86%) | 66 (24.63%) | 40 (33.06%) |
| Stage IV | 76 (14.15%) | 47 (17.47%) | 29 (10.82%) | 19 (15.7%) |
| Unknown | 10 (1.86%) | 7 (2.6%) | 3 (1.12%) | - |
In total, 1249 differentially expressed lncRNAs [log2-fold change (FC) > 1, false discovery rate (FDR) < 0.05, and P < 0.05] were identified after screening the synthetic data matrix with the limma R package and Strawberry Perl (Supple
NRLs were selected as candidate prognostic factors after univariate Cox analysis (uni-Cox) was performed and identified OS-related indicators with a P value < 0.05. Next, multivariate Cox analysis (multi-Cox) was used to construct a signature with the coefficients of the lncRNAs. We calculated the risk score (RS) as follows: RS = coefficient exp lncRNA1 + coefficient exp lncRNA2 + oefficient exp lncRNA3 + ... + coefficient exp lncRNAs. According to the median RS, subgroups including low- and high-risk groups were established[12,15]. We used the chi-square test to analyze the relationships between the model and clinical factors to evaluate the prognostic value of the constructed model.
The differences in OS, disease-free survival, disease-free interval (DFI) and progression-free interval (PFI) between the low- and high-risk groups were analyzed using Kaplan-Meier (K-M) survival analysis and the “survival” and “survminer” R packages. Principal component analysis (PCA) was used to explore the distributions of the different groups. The RS and clinical characteristics were evaluated with uni-Cox and multi-Cox regression analyses to evaluate the independent factors. The 1-, 3-, and 5-year time dependent, clinical characteristic and mixed RS receiver operating characteristic (ROC) curves of the model were plotted with the calculation procedure to evaluate the predicted outcome.
The nomograms for the 1-, 3-, and 5-year OS and correction curves based on the Hosmer–Lemeshow test, which were built with the RS, the clinical model (age, sex, and TNM stage) and the combination model (age, sex, stage and RS) were used to verify the predicted results.
Gene Ontology (GO, go.v7.4.symbols.gmt) and Kyoto Encyclopedia of Genes and Genomes (KEGG, kegg.v7.4.
After preparation for immune analyses, differential cluster analyses of the ESTIMATE score, stromal score, immune score, and tumor purity score were performed to compare the high- and low-risk groups based on the results of the ESTIMATE algorithm (using the “limma” and “vioplot” R packages). We calculated the immune infiltration statuses of CRC patients in the TCGA dataset, including the statuses indicated by analyses with TIMER, CIBERSORT, CBIERSORT-ABS, MCPCOUNTER, XCELL and EPIC, by using TIMER 2.0 (http://timer.cistrome.org/). Then, we compared immune checkpoint activation between the high- and low-risk groups with the ggpubr R package.
The TCGA-CRC gene expression profile and the high- and low-risk group information were used as inputs with the R software package “PRRophetic” to predict half-maximal inhibitory concentration (IC50) values with Genomics of Drug Sensitivity in Cancer (https://www.cancerrxgene.org/)[17].
By reviewing the literature, we identified six different lncRNA-based risk models including the immune-related lncRNA signature (PMID: 30396175)[18], N6-methyladenosine-related lncRNA signature (PMID: 33959151)[19], autophagy-related lncRNA signature (PMID: 34745388)[20], fatty acid metabolism-related lncRNA signature (PMID: 34692467)[21], epithelial-mesenchymal transition-related lncRNA signature (PMID: 33898515)[22] and tumor mutational burden-related lncRNA signature (PMID: 33958876)[23] for comparison with our NRL signature. To make the models comparable, we calculated each RS according to the RS calculation formula provided by the model based on the corresponding gene expression in the same TCGA-CRC patient cohort. Then, we evaluated the 1-, 3-, and 5-year time-dependent ROC curves of the six models according to the RS.
We first used real-time polymerase chain reaction to test the expression of the lncRNAs in the signature and verified the lncRNA expression in 121 clinical CRC patients. The steps of RNA extraction (ER501-01, TransGen Biotech, Beijing, China), reverse transcription (AE341-02, TransGen Biotech, Beijing, China) and amplification were carried out in accordance with the manufacturer’s instructions. The primer sequences for all lncRNAs and the internal reference gene glyceraldehyde-3-phosphate dehydrogenase are shown in Supplementary Table 3. Then, we calculated RSs according to the RS calculation formula for our model. The 1-, 2- and 3-year time dependent, clinical characteristic and mixed RS ROC curves and the K-M survival curve were plotted to validate the predictive ability of the NRL signature.
All statistical analyses were performed using R software (version 4.1.2). A lncRNA and NRG coexpression network was drawn using Cytoscape software (version 3.9.0). The performance of the model was evaluated with K-M curves, time-dependent ROC analysis and Cox regression analysis. A two-tailed P < 0.05 was considered statistically significant.
All the clinicopathologic characteristics of the CRC patients included in this study are shown in Table 2. The expression of 67 NRGs (Supplementary Figure 1A) and 14142 lncRNAs was identified from the TCGA cohort. The 1249 differentially expressed lncRNAs (log2-FC > 1, FDR < 0.05, and P < 0.05) (Supplementary Figure 1B) included 1075 upregulated and 174 downregulated lncRNAs (Supplementary Figure 1C). Finally, the 259 NRLs with a Pearson correlation coefficient > 0.4 and P < 0.001 were selected. The network figure and corresponding data connecting the NRGs and NRLs are shown in Supplementary Figure 1D and Supplementary Table 1.
| Variables | Univariate analysis | Multivariate analysis | |||
| HR (95%CI) | P value | Coefficient | HR (95%CI) | P value | |
| AP001469.3 | 1.671 (1.045-2.673) | 0.032 | 0.396 | 1.485 (0.888-2.484) | 0.132 |
| AC007128.1 | 1.599 (1.016-2.516) | 0.043 | 0.500 | 1.648 (0.976-2.784) | 0.062 |
| LINC02381 | 1.619 (1.1.1-2.381) | 0.014 | 0.390 | 1.477 (0.934-2.337) | 0.095 |
| AC099850.3 | 0.747 (0.559-0.998) | 0.048 | -0.292 | 0.747 (0.536-1.041) | 0.085 |
| AC010973.2 | 2.285 (1.431-3.649) | 0.001 | 0.776 | 2.172 (1.3-3.629) | 0.003 |
| MIR4435-2HG | 1.766 (1.103-2.827) | 0.018 | 0.747 | 2.110 (1.192-3.736) | 0.010 |
| AC245100.7 | 2.084 (1.397-3.110) | 0.000 | 0.471 | 1.602 (1.046-2.455) | 0.030 |
| AL137782.1 | 0.384 (0.177-0.835) | 0.016 | -1.127 | 0.324 (0.153-0.688) | 0.003 |
There were 29 NRLs significantly correlated with OS according to uni-Cox analysis (Supplementary Figure 2A). Among them, 26 lncRNAs had hazard ratios (HRs) greater than 1, and the P values were all less than 0.05 (Supple
The TCGA-CRC patients were divided into low- and high-risk (n = 268 vs. 269) groups with an optimal cutoff value of 0.9558 for the RS, and the clinicopathological characteristics are shown in Table 2. As the RS increased, the patient mortality risk increased gradually, while the survival risk decreased gradually (Figure 2A and B). In addition, the high-risk group had not only a higher mortality rate but also a higher N grade, M grade and stage, with greater lymph node and distant metastasis rates (Figure 2C).
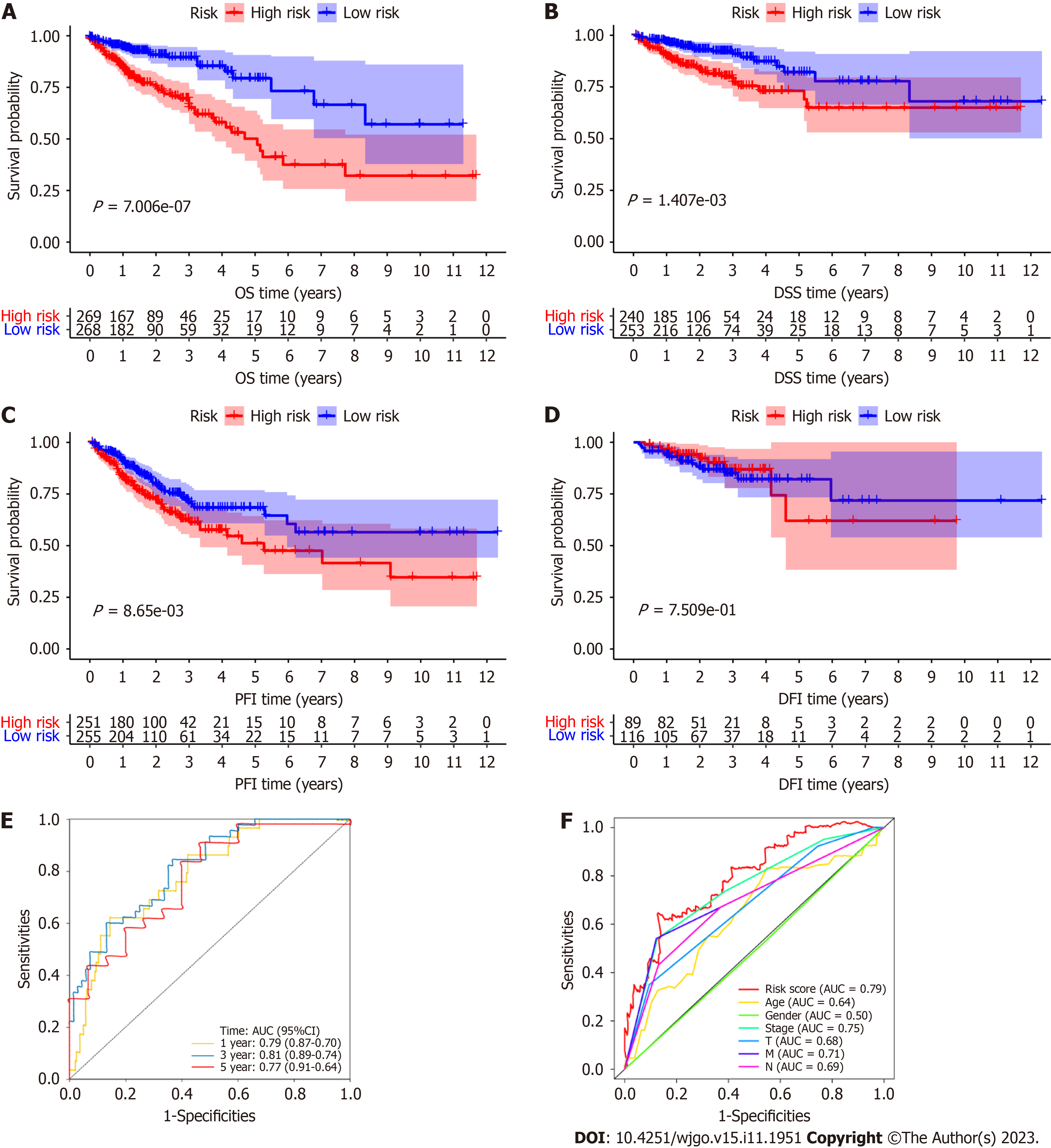
The OS of the high-risk group was significantly poorer than that of the low-risk group (P < 0.0001; Figure 3A), and the disease-specific survival (DSS) and PFI were significantly poorer than those of the low-risk group (P < 0.01; Figure 3B and C). However, the DFI was not significantly different between the high- and low-risk groups (P > 0.05; Figure 3D). All of the K-M analysis results showed that the CRC patients in the high-risk group had a poorer prognosis and shorter OS, DSS, and PFI. The areas under the receiver operating characteristic curves (AUCs) of the RS for 1-, 3- and 5-year OS were 0.79, 0.81 and 0.77, respectively (Figure 3E), and the AUC for 1-year OS was higher than the AUC of any other clinicopathological characteristic, such as age, sex, tumor invasion depth (T stage), lymph node metastasis (N stage), distant metastasis (M stage) or overall TNM stage (Figure 3F). All of these results indicate that the model is moderately sensitive and specific for predicting the prognosis of TCGA-CRC patients.
PCA was used to compare the efficiencies of different gene set groups (all genes, differentially expressed lncRNAs, NRGs, all NRLs and our signature lncRNAs) in separating CRC patients in the TCGA database according to OS. We found that our NRL signature could best distribute CRC patients into high- and low-risk groups (Supple
To further explore the prognostic value of the NRL signature for CRC patients stratified by clinicopathological characteristics, we divided patients into different groups according to age, sex, T stage, N stage, M stage or total stage, and the results showed that the RS of CRC patients was positively associated with N stage, M stage and total stage but not significantly correlated with age, sex or T stage in the different stratified analyses (Supplementary Figure 4).
The HR for the RS and 95% confidence interval (95%CI) calculated by uni-Cox regression analysis were 1.309 and 1.222-1.403 (P < 0.001), respectively (Figure 4A), and those calculated by multi-Cox regression analysis were 1.214 and 1.123-1.312 (P < 0.001), respectively. In addition, age (1.030, 1.009-1.080; P = 0.005) and T stage (1.750, 1.082-2.831; P = 0.023) were the other two independent prognostic parameters (Figure 4B) identified by multi-Cox regression analysis. Then, we built a nomogram for predicting the 1-, 3-, and 5-year OS rates of CRC patients according to these three independent prognostic factors (RS, age, and T stage) (Figure 4C). Calibration plots showed that the performance of the nomogram was best in predicting 1-, 3- and 5-year OS (Figure 4D) and that the total RS of the nomogram could clearly divide the high- and low-risk groups; the high-risk group had a poorer prognosis by K-M analysis (Figure 4E). The AUCs of the total RSs for 1-, 3- and 5-year OS were 0.76, 0.84 and 0.86, respectively (Figure 4F).
In conclusion, our predictive model may increase the predictive sensitivity and specificity of traditional clinical models and provide some information that may be helpful in the clinical prognostic assessment and best management of CRC patients.
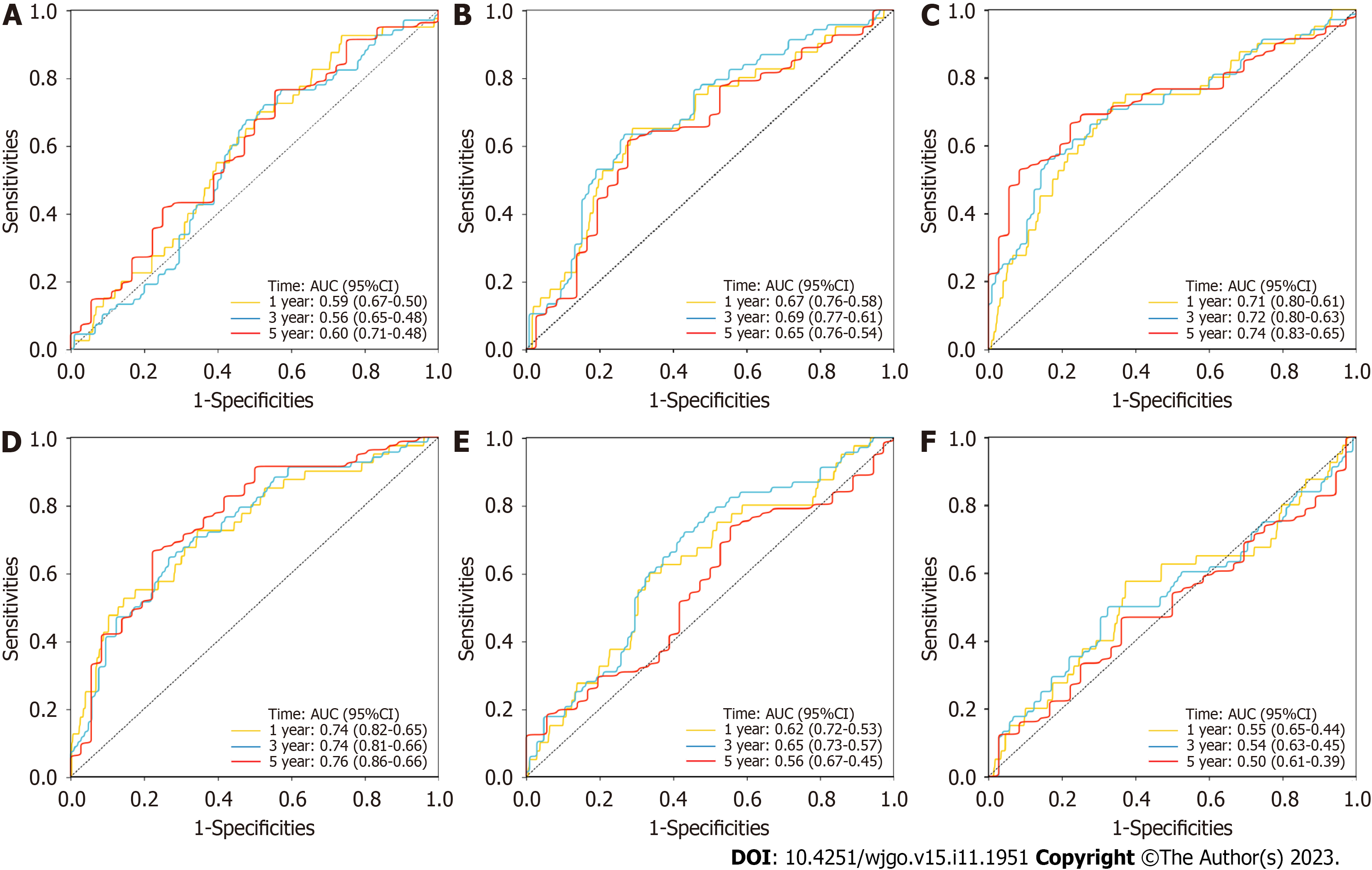
Many lncRNA-related models that have been derived from the TCGA database for predicting the prognosis of CRC patients, such as the immune-related lncRNA signature, N6-methyladenosine-related lncRNA signature and autophagy-related lncRNA signature (Figure 5). Most of these AUCs for 1-, 3- and 5-year OS were lower than those for the NRL signature, which had better prognostic efficacy.
The AUCs for 1-, 2- and 3-year OS were 0.74, 0.68 and 0.66, respectively, for the CRC patients with clinical data, which were suggested that this model could be used to predict the prognosis of CRC patients (Figure 6A). The AUC for the clinical data RS for 1-year OS was higher than the AUC for any other clinicopathological characteristic, such as age, sex, T stage, N stage, M stage, or total stage (Figure 6B). OS was significantly poorer in the high-risk group (n = 61) than in the low-risk group (n = 60), which was defined by the median RS as the cutoff (P < 0.01, Figure 6C).
In the clinical CRC cohort, the clinical value of the signature including the RS and multiple clinical characteristics was examined by uni-Cox and multi-Cox regression analysis. The HR for the RS and 95%CI calculated by uni-Cox regression analysis were 54.17 and 5.99−490.03 (P < 0.001), respectively (Supplementary Figure 5A), and those calculated by multi-Cox regression analysis were 39.43 and 54.71−330.12 (P < 0.001), respectively. In addition, age (1.032, 1.006−1.059; P = 0.014) was the other independent prognostic parameter (Supplementary Figure 5B) identified by multi-Cox regression analysis. AC099850.3 and AL137782.1 were protective factors significantly associated with better OS in CRC patients, and the other lncRNAs were also shown to be risk factors for significantly poorer OS with the best expression cutoff point in the K-M method (Supplementary Figure 5C-J). In summary, most of these factors have prognostic value in clinical CRC cohort patients.
Then, the expression levels of the 8 lncRNAs were stratified by clinicopathological characteristics, and we found that the expression of AC007128.1, MIR4435-2HG and AC245100.7 was related to N stage, M stage and total stage (all P < 0.05). The expression of LINC02381 was related to N stage, and the expression of AP001469.3 was related to M stage (all P < 0.05), but none of the 8 lncRNAs were related to age, sex or T stage in the TCGA-CRC cohort (Figure 6D-G). In the clinical CRC patient cohort, the RSs were significantly related to T stage, N stage, M stage and total stage. The expression of AC007128.1, AP001469.3 and MIR4435-2HG was related to N stage, M stage and total stage, and MIR4435-2HG was also related to T stage (all P < 0.05). The expression of AC245100.7 was related to M stage (P < 0.05), but the other lncRNAs were not related to any of the clinicopathological characteristics (Figure 6H-K, Supplementary Figure 6).
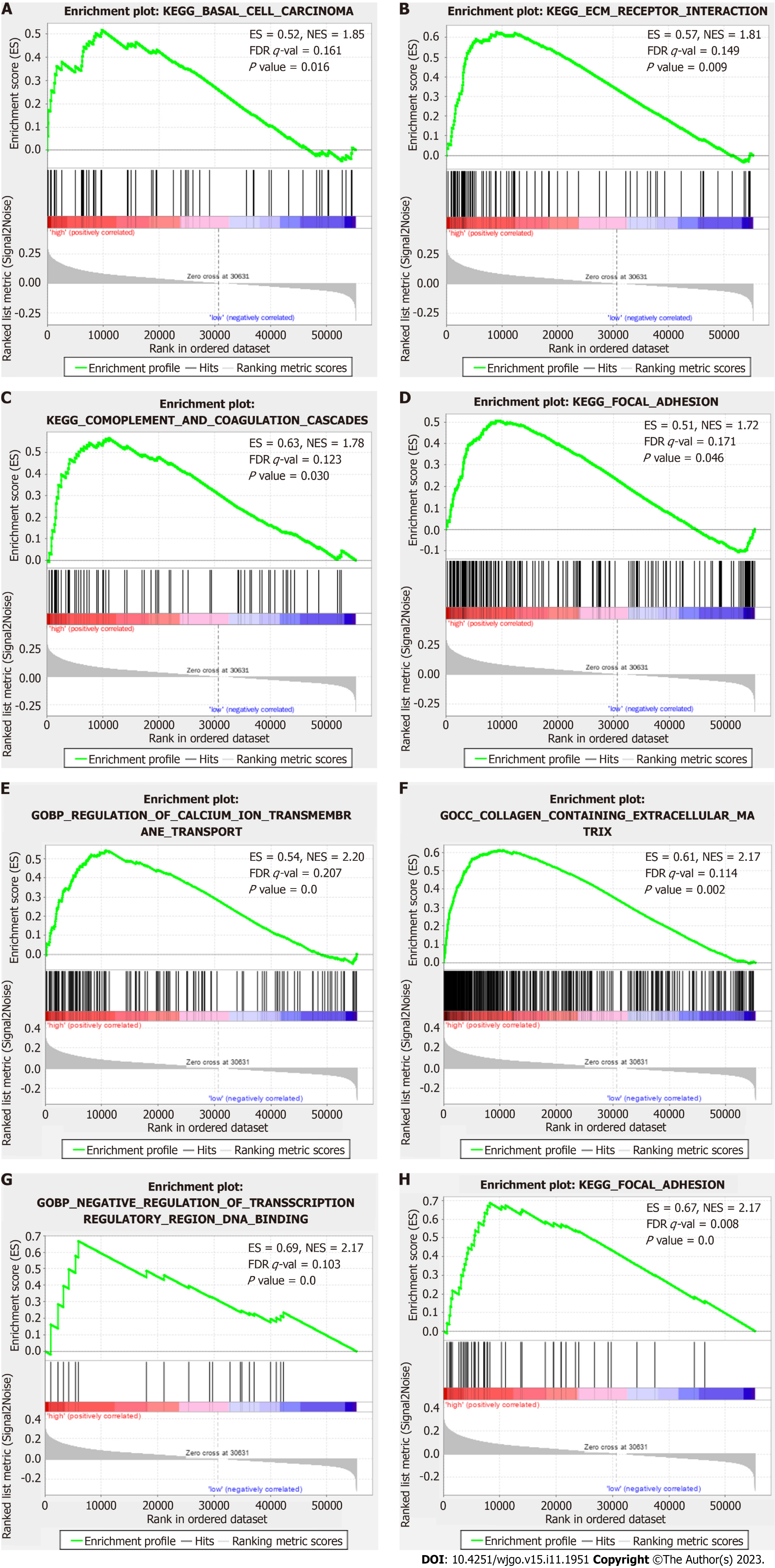
GSEA results showed that the top ten enriched KEGG terms in the high-risk patient group were in cancer- and tumor metastasis-related pathways, such as “basal cell carcinoma”, “complement and coagulation cascades”, “extracellular matrix (ECM) receptor interaction” and “focal adhesion” (Figure 7A-D). The top ten enriched GO terms in the high-risk patient group were tumor metastasis- and ion channel-related pathways, such as “regulation of calcium ion trans
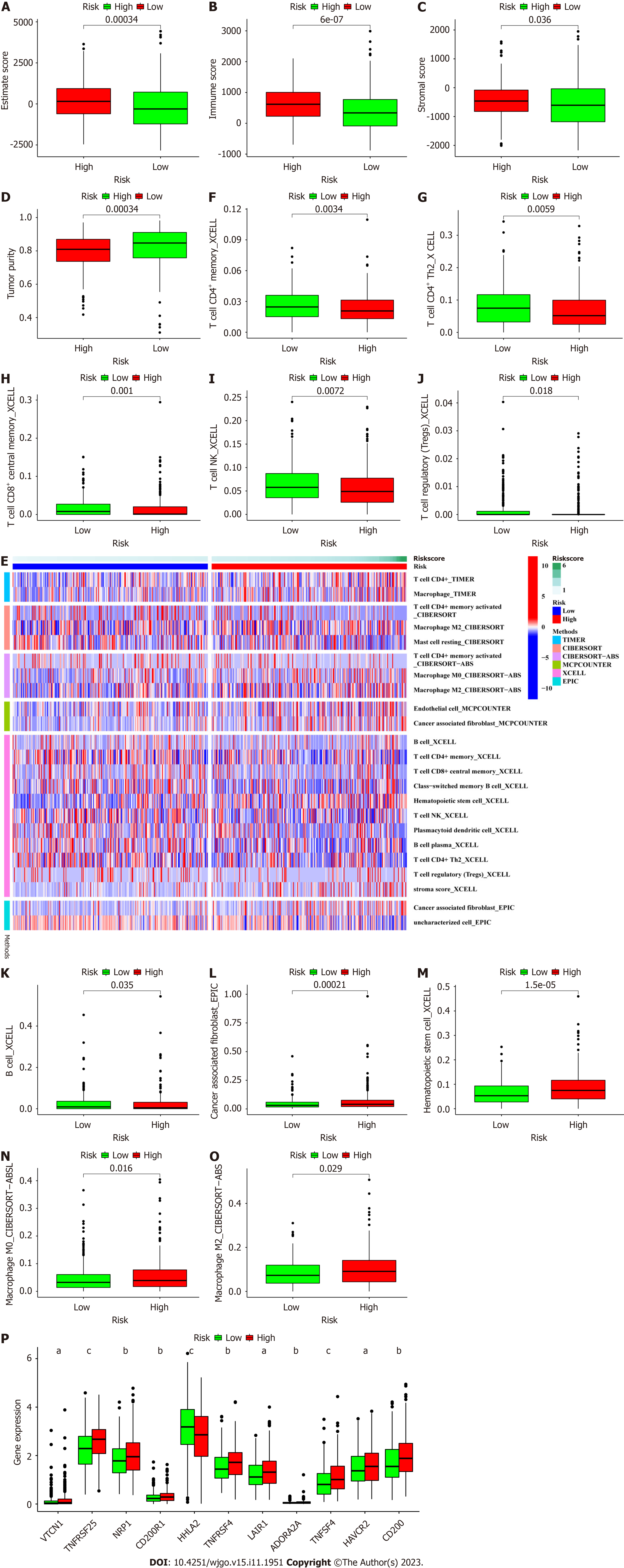
The differences in immune cell infiltration and the TME between the different RS groups were analyzed. The ESTIMATE analysis results showed that the high-risk group had a higher ESTIMATE (microenvironment) score, immune score and stromal score but a lower tumor purity than the low-risk group (all P < 0.001), which signified different TMEs in the two groups (Figure 8A-D). The heatmap (Figure 8E) for immune cell infiltration showed that more immune cells were associated with the high-risk group on different platforms (Supplementary Table 6); the immune cell populations included CD4+ memory T cells, CD4+ Th2 cells, CD8+ central memory T cells, Natural killer T (NKT) cells, regulatory T cells (Tregs) and B cells identified by XCELL (Figure 8F-K); CD4+ T cells and M2 macrophages identified by TIMER; activated CD4+ memory T cells identified by CIBERSORT; and M2 macrophages identified by CIBERSORT−ABS (all P < 0.05) (Supplementary Figure 7). In addition, we found significantly reduced numbers of antitumor immune cells, such as CD4+ Th2 cells, CD8+ central memory T cells, NKT cells, Tregs and B cells (Figure 8F-K)[25], and increased numbers of protumor immune cells, such as cancer-associated fibroblasts (CAFs), hematopoietic stem cells, M0 macrophages and M2 macrophages[26], in the TME of the high-risk group, which suggested a poor prognosis and might be a direction for immunotherapy in CRC (Figure 8L-O)[27].
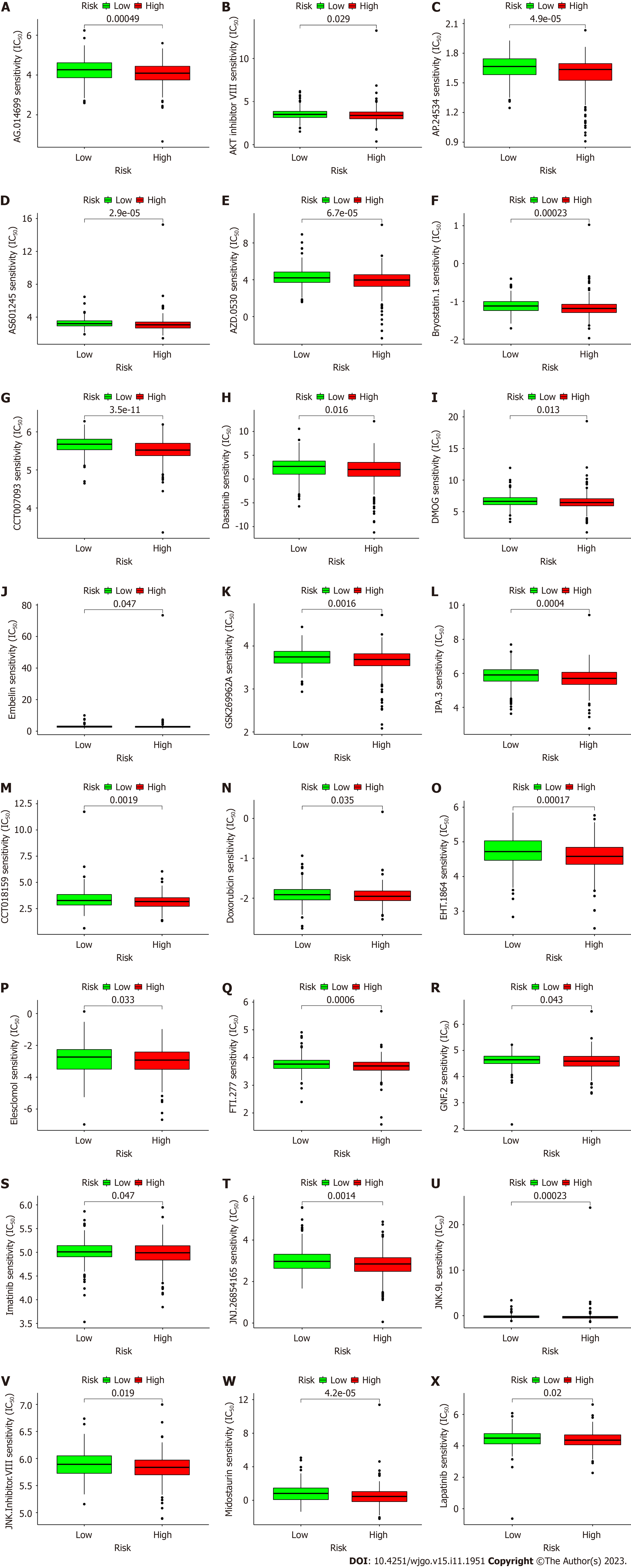
Most immune checkpoint molecules, such as VTCN1[28], TNFRSF25 and NRP1, also showed greater activation in the high-risk group (Figure 8P), which implied that we could choose appropriate checkpoint inhibitors for CRC patients. Moreover, we found that 16 immunotherapeutic anticancer drugs and 22 chemotherapeutic or targeted drugs showed lower IC50 values in the high-risk group when applied for CRC therapy (Figure 9; Supplementary Figures 7 and 8). These observations will assist in the discovery of potential drugs for effective antitumor immunotherapy and chemotherapy to improve the therapeutic efficacy and prognosis of patients in high-risk groups.
In recent years, many lncRNAs have been found to play an important regulatory role in the occurrence and progression of CRC[11] which has become a research hotspot. However, the relationship between them and the internal detailed mechanism of action are not completely clear. Many r studies have been carried out not only in the in-depth exploration of the detailed mechanism of action of a single lncRNA but also on the interaction mechanism between lncRNAs. More
As a modulated form of necrosis, necroptosis is an evolutionary form of caspase-independent programmed necrosis that is a vital process in the life cycle of organisms. It can induce swelling of organelles, rupture of cell membranes, decom
Therefore, some NRL signatures have been used to evaluate the prognosis of patients with lung carcinoma[32], esophageal carcinoma[33], stomach carcinoma[34], and so on. However, the prognostic value of NRLs in CRC has not been documented. Here, we identified a new model including an 8-NRL signature that was associated with CRC OS. The fidelity of this signature was tested with clinical datasets and compared with that of other signatures. The AUCs for OS demonstrated that the NRL signature was more reliable than 6 other signatures. This NRL signature showed a perfect clustering capability and could be treated as a potential tool for CRC molecular signatures. Although the signature showed potential as a prognostic biomarker for CRC, other independent large clinical datasets need to be evaluated to assess the fidelity of this signature as a prognostic biomarker for CRC. Most of these lncRNAs may play crucial roles in necroptosis in CRC. MIR4435-2HG likely has adverse effects on prognosis and could be a potential therapeutic target in CRC[35,36]. MIR4435-2HG has been associated with AXL, which reprograms the immunological microenvironment when PD-1 inhibitors are administered as an immunotherapy[37]. AC007128.1 plays an important role and is associated with many NRGs that contribute to immunotherapy, such as MAPK8[38] and BRAF[39], but has not been reported in CRC. AC007128.1 expression is upregulated and associated with a poor prognosis in esophageal squamous cell carcinoma; it may promote epithelial-mesenchymal transition by increasing the activation of the MAPK/ERK and MAPK/p38 signaling pathways in its cells[40]. However, the specific mechanisms of most lncRNAs are unknown; for example, AP001469.3 and AC245100.7 have not been reported in CRC or other cancers and need to be explored through more in-depth basic experiments.
Recent studies have revealed an important role for necroptosis in metastasis and implied the potential of targeting necroptosis with antimetastatic and advanced-stage cancer therapies[41]. Additionally, the GSEA results showed that most of the enriched KEGG and GO terms in the high-risk patient group were related to tumor metastasis-related pathways, implying the important role of necroptosis in CRC metastasis, which has received little attention and research to date[10,42]. In this study, the RS was correlated with tumor stage, as well as the N stage and M stage, which suggested that the poor prognosis of the high-risk group might be closely related to the high risks of lymph node metastasis and distant metastasis and that this model would also be useful for detecting lymphatic metastasis, distant metastasis and total stage in CRC.
The definite relationships between immune cell infiltration or function and metastasis in the TME indicate that we should pay much attention[43]. Necroptosis was found to be involved in antitumor immunity, possibly by altering TME functions during cancer immunotherapy[44]. Consequently, we also systematically investigated the correlation between the TME, immune cell infiltration, immune checkpoints and the NRLs. We look forward to using these results in the future to predict the prognostic risk of CRC and explore potential new targets and approaches for clinical treatment. The high-risk group had a high immune score, ESTIMATE score and stomal score, which are traditionally considered to indicate a “hot tumor” and stronger responses to immunotherapy[45]. However, the infiltrated immune cells and their functions are the most important factors in immunotherapy. The high-risk group was infiltrated with more immunosuppressive cells, such as CAFs[46], hematopoietic stem cells[47] and M2 macrophages[48,49], and fewer antitumor effector immune cells, such as CD8+ T cells[50] and Tregs[51], which may be the reason for the increased metastasis and poor prognosis in the high-risk group. However, selecting effective treatment methods and improving therapeutic efficacy in these high-risk patients remain difficult issues. Traditional immune checkpoint therapy has achieved only poor efficacy in CRC, and it is necessary to identify other potential therapeutic targets or develop combination therapies including immune checkpoint therapy as future directions for exploration. Our results found that increased potential immune checkpoint gene expression was closely related to the high-risk group, and more potential immunotherapeutic and chemotherapeutic drugs had lower IC50 values in the high-risk group than in the low-risk group[52-55].
However, there are some limitations to our study. First, although the method of identifying NRLs by coexpression analysis is the most common, it is not accurate and comprehensive enough. Second, our results are mainly based on the TCGA dataset. While we only have a small set of clinical samples for external validation, we expect other large-scale case series data to be used to validate and evaluate the suitability of signatures. Third, we only found that MIR4435-2HG has poor prognosis effects in CRC, but other lncRNAs may have similar effects and may become potential therapeutic targets for CRC. Further research and exploration of the detailed molecular mechanisms of these key lncRNAs in CRC are needed through in vitro and in vivo experiments.
Overall, we identified an NRL signature for CRC prognosis prediction and subtyping using lncRNA expression profiles. This signature will not only greatly improve the ability to predict CRC prognosis but also allow exploration of the reasons and mechanism underlying the increased TME infiltration of more immunosuppressive cells and decreased TME infiltration of antitumor effector immune cells in high-risk group patients, which leads to distant metastases. This signature found additional potential immune checkpoint genes and immunotherapeutic and chemotherapeutic drugs, and future in-depth research will identify more potential biomarkers and treatment targets for CRC.
Overall, we identified an NRL signature for CRC prognosis prediction and subtyping using lncRNA expression profiles. This signature will not only greatly improve the ability to predict CRC prognosis but also allow exploration of the reasons and mechanism underlying the increased TME infiltration of more immunosuppressive cells and decreased TME infiltration of antitumor effector immune cells in high-risk group patients, which leads to distant metastases. This signature found additional potential immune checkpoint genes and immunotherapeutic and chemotherapeutic drugs, and future in-depth research will identify more potential biomarkers and treatment targets for CRC.
Tumor recurrence and metastasis lead to a poor prognosis in colorectal cancer (CRC). Systemic treatments, including surgery, chemotherapeutics and targeted therapeutics, have reached a bottleneck for improving patient prognosis. Necroptosis induction significantly improves antitumor immunity by inducing the maturation of dendritic cells and activation of cytotoxic cluster of differentiation 8+ T cells, which play roles in antitumor immunity.
We aimed to explore the internal relationships and mechanisms of interaction between necroptosis-related long noncoding RNAs (lncRNAs) (NRLs) and the tumor microenvironment. As lncRNAs in bodily fluids are known to be new cancer biomarkers, we stratified CRC patients according to NRL expression levels, which allowed us to not only evaluate patient prognosis but also improve precision medicine in clinical practice. Additionally, we hoped to identify potential effective immunotherapeutic targets and drugs.
To identify a NRL signature and to comprehensively and multidimensionally evaluated the value of this signature, and its reliability for CRC prognosis prediction was assessed with clinical CRC data and compared with that of six other lncRNA signatures.
Identification of NRLs and development and preliminary validation of the risk signature with univariate Cox analysis and multivariate Cox analysis. Then, the value of this signature was comprehensively and multidimensionally evaluated, and its reliability for CRC prognosis prediction was assessed with clinical CRC data and compared with that of six other lncRNA signatures. Gene Set Enrichment Analysis, tumor microenvironment (TME) analysis and half-maximal inhibitory concentration (IC50) prediction were also performed according to the risk score (RS) of the signature.
An 8-lncRNA signature significantly associated with overall survival (OS) was constructed, and its reliability was validated with clinical CRC data. Most of the areas under the receiver operating characteristic curves values for this signature were higher than those for the other six lncRNA signatures. OS, disease-specific survival and progression-free interval were all significantly poorer in the high-risk group. The RS of the signature showed good concordance with the predicted prognosis. Additionally, the calibration plots for this signature combined with clinical factors showed that this combination could effectively improve the ability to predict OS. The RS was correlated with tumor stage, lymph node metastasis and distant metastasis. Most of the enriched Kyoto Encyclopedia of Genes and Genomes and Gene Ontology terms were tumor metastasis-related pathways in the high-risk group; these patients showed greater infiltration of immunosuppressive cells, but less infiltration of infiltrating antitumor effector immune cells. We explored additional potential immune checkpoint genes and potential immunotherapeutic and chemotherapeutic drugs with relatively low IC50 values.
We identified an NRL signature for CRC prognosis prediction and subtyping using lncRNA expression profiles. This signature will not only greatly improve the ability to predict CRC prognosis but also allow exploration of the reasons and mechanism underlying the increased TME infiltration of more immunosuppressive cells and decreased TME infiltration of antitumor effector immune cells in high-risk group patients, which leads to distant metastases. This signature found additional potential immune checkpoint genes and immunotherapeutic and chemotherapeutic drugs, and future in-depth research will identify more potential biomarkers and treatment targets for CRC.
We identified an NRL signature with strong fidelity that could stably predict prognosis and might be an indicator of the TME of CRC. Furthermore, additional potential immunotherapeutic and chemotherapeutic drugs were explored.
We thank the reviewers for their constructive comments. We thank the Cancer Research Center and the Central Laboratory of the First Affiliated Hospital of Fujian Medical University for providing the research and experimental platform.
Provenance and peer review: Unsolicited article; Externally peer reviewed.
Peer-review model: Single blind
Specialty type: Oncology
Country/Territory of origin: China
Peer-review report’s scientific quality classification
Grade A (Excellent): 0
Grade B (Very good): B
Grade C (Good): C, C, C, C
Grade D (Fair): 0
Grade E (Poor): 0
P-Reviewer: Emran TB, Bangladesh; Gupta R, India; Sheykhhasan M, Iran; Tanabe S, Japan S-Editor: Lin C L-Editor: A P-Editor: Xu ZH
| 1. | Siegel RL, Miller KD, Fuchs HE, Jemal A. Cancer Statistics, 2021. CA Cancer J Clin. 2021;71:7-33. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 8287] [Cited by in RCA: 11901] [Article Influence: 2975.3] [Reference Citation Analysis (4)] |
| 2. | Sung H, Ferlay J, Siegel RL, Laversanne M, Soerjomataram I, Jemal A, Bray F. Global Cancer Statistics 2020: GLOBOCAN Estimates of Incidence and Mortality Worldwide for 36 Cancers in 185 Countries. CA Cancer J Clin. 2021;71:209-249. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 75126] [Cited by in RCA: 64142] [Article Influence: 16035.5] [Reference Citation Analysis (174)] |
| 3. | Agarwal P, Le DT, Boland PM. Immunotherapy in colorectal cancer. Adv Cancer Res. 2021;151:137-196. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 6] [Cited by in RCA: 20] [Article Influence: 5.0] [Reference Citation Analysis (0)] |
| 4. | Feng M, Zhao Z, Yang M, Ji J, Zhu D. T-cell-based immunotherapy in colorectal cancer. Cancer Lett. 2021;498:201-209. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 12] [Cited by in RCA: 55] [Article Influence: 11.0] [Reference Citation Analysis (0)] |
| 5. | Um W, Ko H, You DG, Lim S, Kwak G, Shim MK, Yang S, Lee J, Song Y, Kim K, Park JH. Necroptosis-Inducible Polymeric Nanobubbles for Enhanced Cancer Sonoimmunotherapy. Adv Mater. 2020;32:e1907953. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 67] [Cited by in RCA: 89] [Article Influence: 17.8] [Reference Citation Analysis (0)] |
| 6. | Zhang DW, Shao J, Lin J, Zhang N, Lu BJ, Lin SC, Dong MQ, Han J. RIP3, an energy metabolism regulator that switches TNF-induced cell death from apoptosis to necrosis. Science. 2009;325:332-336. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1352] [Cited by in RCA: 1644] [Article Influence: 102.8] [Reference Citation Analysis (0)] |
| 7. | Aaes TL, Kaczmarek A, Delvaeye T, De Craene B, De Koker S, Heyndrickx L, Delrue I, Taminau J, Wiernicki B, De Groote P, Garg AD, Leybaert L, Grooten J, Bertrand MJ, Agostinis P, Berx G, Declercq W, Vandenabeele P, Krysko DV. Vaccination with Necroptotic Cancer Cells Induces Efficient Anti-tumor Immunity. Cell Rep. 2016;15:274-287. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 221] [Cited by in RCA: 347] [Article Influence: 38.6] [Reference Citation Analysis (0)] |
| 8. | Lomphithak T, Akara-Amornthum P, Murakami K, Hashimoto M, Usubuchi H, Iwabuchi E, Unno M, Cai Z, Sasano H, Jitkaew S. Tumor necroptosis is correlated with a favorable immune cell signature and programmed death-ligand 1 expression in cholangiocarcinoma. Sci Rep. 2021;11:11743. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 23] [Cited by in RCA: 32] [Article Influence: 8.0] [Reference Citation Analysis (0)] |
| 9. | Seneviratne D, Ma J, Tan X, Kwon YK, Muhammad E, Melhem M, DeFrances MC, Zarnegar R. Genomic instability causes HGF gene activation in colon cancer cells, promoting their resistance to necroptosis. Gastroenterology. 2015;148:181-191.e17. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 29] [Cited by in RCA: 40] [Article Influence: 4.0] [Reference Citation Analysis (0)] |
| 10. | Han Q, Ma Y, Wang H, Dai Y, Chen C, Liu Y, Jing L, Sun X. Resibufogenin suppresses colorectal cancer growth and metastasis through RIP3-mediated necroptosis. J Transl Med. 2018;16:201. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 50] [Cited by in RCA: 99] [Article Influence: 14.1] [Reference Citation Analysis (0)] |
| 11. | Chen S, Shen X. Long noncoding RNAs: functions and mechanisms in colon cancer. Mol Cancer. 2020;19:167. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 67] [Cited by in RCA: 208] [Article Influence: 41.6] [Reference Citation Analysis (0)] |
| 12. | Jiang N, Zhang X, Gu X, Li X, Shang L. Progress in understanding the role of lncRNA in programmed cell death. Cell Death Discov. 2021;7:30. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 52] [Cited by in RCA: 129] [Article Influence: 32.3] [Reference Citation Analysis (0)] |
| 13. | Sheykhhasan M, Tanzadehpanah H, Ahmadieh Yazdi A, Mahaki H, Seyedebrahimi R, Akbari M, Manoochehri H, Kalhor N, Dama P. FLVCR1-AS1 and FBXL19-AS1: Two Putative lncRNA Candidates in Multiple Human Cancers. Noncoding RNA. 2022;9. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 12] [Cited by in RCA: 5] [Article Influence: 1.7] [Reference Citation Analysis (0)] |
| 14. | Akhbari MH, Zafari Z, Sheykhhasan M. Competing Endogenous RNAs (ceRNAs) in Colorectal Cancer: A Review. Expert Rev Mol Med. 2022;24:e27. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 23] [Cited by in RCA: 15] [Article Influence: 5.0] [Reference Citation Analysis (0)] |
| 15. | Wang K, Liu F, Liu CY, An T, Zhang J, Zhou LY, Wang M, Dong YH, Li N, Gao JN, Zhao YF, Li PF. The long noncoding RNA NRF regulates programmed necrosis and myocardial injury during ischemia and reperfusion by targeting miR-873. Cell Death Differ. 2016;23:1394-1405. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 139] [Cited by in RCA: 180] [Article Influence: 20.0] [Reference Citation Analysis (0)] |
| 16. | Zhao Z, Liu H, Zhou X, Fang D, Ou X, Ye J, Peng J, Xu J. Necroptosis-Related lncRNAs: Predicting Prognosis and the Distinction between the Cold and Hot Tumors in Gastric Cancer. J Oncol. 2021;2021:6718443. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 65] [Cited by in RCA: 141] [Article Influence: 35.3] [Reference Citation Analysis (0)] |
| 17. | Geeleher P, Cox NJ, Huang RS. Clinical drug response can be predicted using baseline gene expression levels and in vitro drug sensitivity in cell lines. Genome Biol. 2014;15:R47. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 380] [Cited by in RCA: 625] [Article Influence: 56.8] [Reference Citation Analysis (0)] |
| 18. | Qin F, Xu H, Wei G, Ji Y, Yu J, Hu C, Yuan C, Ma Y, Qian J, Li L, Huo J. A Prognostic Model Based on the Immune-Related lncRNAs in Colorectal Cancer. Front Genet. 2021;12:658736. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 9] [Cited by in RCA: 16] [Article Influence: 4.0] [Reference Citation Analysis (0)] |
| 19. | Zhao Z, Yang YB, Li XY, Li XG, Chu XD, Lin ZB, Zhang YR, Guo YG, Ding H, Pan YL, Wang L, Pan JH. Comprehensive Analysis of N6-Methyladenosine-Related lncRNA Signature for Predicting Prognosis and Immune Cell Infiltration in Patients with Colorectal Cancer. Dis Markers. 2021;2021:8686307. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 7] [Cited by in RCA: 3] [Article Influence: 0.8] [Reference Citation Analysis (0)] |
| 20. | Xu G, Yang M, Wang Q, Zhao L, Zhu S, Zhu L, Xu T, Cao R, Li C, Liu Q, Xiong W, Su Y, Dong J. A Novel Prognostic Prediction Model for Colorectal Cancer Based on Nine Autophagy-Related Long Noncoding RNAs. Front Oncol. 2021;11:613949. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 4] [Cited by in RCA: 9] [Article Influence: 2.3] [Reference Citation Analysis (0)] |
| 21. | Peng Y, Xu C, Wen J, Zhang Y, Wang M, Liu X, Zhao K, Wang Z, Liu Y, Zhang T. Fatty Acid Metabolism-Related lncRNAs Are Potential Biomarkers for Predicting the Overall Survival of Patients With Colorectal Cancer. Front Oncol. 2021;11:704038. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 4] [Cited by in RCA: 18] [Article Influence: 4.5] [Reference Citation Analysis (0)] |
| 22. | Liu C, Hu C, Li J, Jiang L, Zhao C. Identification of Epithelial-Mesenchymal Transition-Related lncRNAs that Associated With the Prognosis and Immune Microenvironment in Colorectal Cancer. Front Mol Biosci. 2021;8:633951. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 3] [Cited by in RCA: 4] [Article Influence: 1.0] [Reference Citation Analysis (0)] |
| 23. | Ding C, Shan Z, Li M, Xia Y, Jin Z. Exploration of the Associations of lncRNA Expression Patterns with Tumor Mutation Burden and Prognosis in Colon Cancer. Onco Targets Ther. 2021;14:2893-2909. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 4] [Cited by in RCA: 12] [Article Influence: 3.0] [Reference Citation Analysis (0)] |
| 24. | Bennett IM, Farfano HM, Bogani F, Primak A, Liddell PA, Otero L, Sereno L, Silber JJ, Moore AL, Moore TA, Gust D. Active transport of Ca2+ by an artificial photosynthetic membrane. Nature. 2002;420:398-401. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 141] [Cited by in RCA: 129] [Article Influence: 5.6] [Reference Citation Analysis (0)] |
| 25. | Wang C, Lu Y, Chen L, Gao T, Yang Q, Zhu C, Chen Y. Th9 cells are subjected to PD-1/PD-L1-mediated inhibition and are capable of promoting CD8 T cell expansion through IL-9R in colorectal cancer. Int Immunopharmacol. 2020;78:106019. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 18] [Cited by in RCA: 39] [Article Influence: 6.5] [Reference Citation Analysis (0)] |
| 26. | Choi YW, Kim YH, Oh SY, Suh KW, Kim YS, Lee GY, Yoon JE, Park SS, Lee YK, Park YJ, Kim HS, Park SH, Kim JH, Park TJ. Senescent Tumor Cells Build a Cytokine Shield in Colorectal Cancer. Adv Sci (Weinh). 2021;8:2002497. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 29] [Cited by in RCA: 51] [Article Influence: 12.8] [Reference Citation Analysis (0)] |
| 27. | Jary M, Liu WW, Yan D, Bai I, Muranyi A, Colle E, Brocheriou I, Turpin A, Radosevic-Robin N, Bourgoin P, Penault-Llorca F, Cohen R, Vernerey D, André T, Borg C, Shanmugam K, Svrcek M. Immune microenvironment in patients with mismatch-repair-proficient oligometastatic colorectal cancer exposed to chemotherapy: the randomized MIROX GERCOR cohort study. Mol Oncol. 2022;16:2260-2273. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in RCA: 11] [Reference Citation Analysis (0)] |
| 28. | Ding S, Lv X, Liu Z, Zhan S, Xu Y, Zhang X, Liu C, Cao L. Overexpression of B7-H4 is associated with infiltrating immune cells and poor prognosis in metastatic colorectal cancer. Int Immunopharmacol. 2021;90:107144. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 12] [Cited by in RCA: 18] [Article Influence: 4.5] [Reference Citation Analysis (0)] |
| 29. | Lin Y, Pan X, Chen Z, Lin S, Shen Z, Chen S. Prognostic value and immune infiltration of novel signatures in colon cancer microenvironment. Cancer Cell Int. 2021;21:679. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 1] [Cited by in RCA: 6] [Article Influence: 1.5] [Reference Citation Analysis (0)] |
| 30. | Nestarenkaite A, Fadhil W, Rasmusson A, Susanti S, Hadjimichael E, Laurinaviciene A, Ilyas M, Laurinavicius A. Immuno-Interface Score to Predict Outcome in Colorectal Cancer Independent of Microsatellite Instability Status. Cancers (Basel). 2020;12. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 2] [Cited by in RCA: 16] [Article Influence: 3.2] [Reference Citation Analysis (0)] |
| 31. | Snyder AG, Oberst A. The Antisocial Network: Cross Talk Between Cell Death Programs in Host Defense. Annu Rev Immunol. 2021;39:77-101. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 44] [Cited by in RCA: 82] [Article Influence: 20.5] [Reference Citation Analysis (0)] |
| 32. | Lu Y, Luo X, Wang Q, Chen J, Zhang X, Li Y, Chen Y, Li X, Han S. A Novel Necroptosis-Related lncRNA Signature Predicts the Prognosis of Lung Adenocarcinoma. Front Genet. 2022;13:862741. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 14] [Cited by in RCA: 42] [Article Influence: 14.0] [Reference Citation Analysis (0)] |
| 33. | Luo Z, Ding E, Yu L, Wang W, Guo Q, Li X, Wang Y, Li T, Zhang Y, Zhang X. Identification of hub necroptosis-related lncRNAs for prognosis prediction of esophageal carcinoma. Aging (Albany NY). 2023;15:4794-4819. [RCA] [PubMed] [DOI] [Full Text] [Cited by in RCA: 5] [Reference Citation Analysis (0)] |
| 34. | Wang N, Liu D. Identification and Validation a Necroptosisrelated Prognostic Signature and Associated Regulatory Axis in Stomach Adenocarcinoma. Onco Targets Ther. 2021;14:5373-5383. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 36] [Cited by in RCA: 58] [Article Influence: 14.5] [Reference Citation Analysis (0)] |
| 35. | Lam JH, Hong M, Koo SL, Chua CWL, Lim KL, Wee F, Wan WK, Leow WQ, Yeo JG, Tan IBH, Yeong J, Lim TKH, Lim TS. CD30(+)OX40(+) Treg is associated with improved overall survival in colorectal cancer. Cancer Immunol Immunother. 2021;70:2353-2365. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 4] [Cited by in RCA: 21] [Article Influence: 5.3] [Reference Citation Analysis (0)] |
| 36. | Hardman C, Ho S, Shimizu A, Luu-Nguyen Q, Sloane JL, Soliman MSA, Marsden MD, Zack JA, Wender PA. Synthesis and evaluation of designed PKC modulators for enhanced cancer immunotherapy. Nat Commun. 2020;11:1879. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 16] [Cited by in RCA: 33] [Article Influence: 6.6] [Reference Citation Analysis (0)] |
| 37. | Chen MH, Wang YH, Sun BJ, Yu LM, Chen QQ, Han XX, Liu YH. HIF-1α activator DMOG inhibits alveolar bone resorption in murine periodontitis by regulating macrophage polarization. Int Immunopharmacol. 2021;99:107901. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 6] [Cited by in RCA: 47] [Article Influence: 11.8] [Reference Citation Analysis (0)] |
| 38. | Wang H, Zhang H, Wang Y, Yang L, Wang D. Embelin can protect mice from thioacetamide-induced acute liver injury. Biomed Pharmacother. 2019;118:109360. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 6] [Cited by in RCA: 24] [Article Influence: 4.0] [Reference Citation Analysis (0)] |
| 39. | Mousset CM, Hobo W, Ji Y, Fredrix H, De Giorgi V, Allison RD, Kester MGD, Falkenburg JHF, Schaap NPM, Jansen JH, Gattinoni L, Dolstra H, van der Waart AB. Ex vivo AKT-inhibition facilitates generation of polyfunctional stem cell memory-like CD8(+) T cells for adoptive immunotherapy. Oncoimmunology. 2018;7:e1488565. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 27] [Cited by in RCA: 42] [Article Influence: 6.0] [Reference Citation Analysis (0)] |
| 40. | Chen J, Song Y, Li M, Zhang Y, Lin T, Sun J, Wang D, Liu Y, Guo J, Yu W. Comprehensive analysis of ceRNA networks reveals prognostic lncRNAs related to immune infiltration in colorectal cancer. BMC Cancer. 2021;21:255. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 36] [Cited by in RCA: 35] [Article Influence: 8.8] [Reference Citation Analysis (0)] |
| 41. | Bolik J, Krause F, Stevanovic M, Gandraß M, Thomsen I, Schacht SS, Rieser E, Müller M, Schumacher N, Fritsch J, Wichert R, Galun E, Bergmann J, Röder C, Schafmayer C, Egberts JH, Becker-Pauly C, Saftig P, Lucius R, Schneider-Brachert W, Barikbin R, Adam D, Voss M, Hitzl W, Krüger A, Strilic B, Sagi I, Walczak H, Rose-John S, Schmidt-Arras D. Inhibition of ADAM17 impairs endothelial cell necroptosis and blocks metastasis. J Exp Med. 2022;219. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 6] [Cited by in RCA: 44] [Article Influence: 14.7] [Reference Citation Analysis (0)] |
| 42. | Yan J, Wan P, Choksi S, Liu ZG. Necroptosis and tumor progression. Trends Cancer. 2022;8:21-27. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 58] [Cited by in RCA: 268] [Article Influence: 89.3] [Reference Citation Analysis (0)] |
| 43. | Strilic B, Yang L, Albarrán-Juárez J, Wachsmuth L, Han K, Müller UC, Pasparakis M, Offermanns S. Tumour-cell-induced endothelial cell necroptosis via death receptor 6 promotes metastasis. Nature. 2016;536:215-218. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 276] [Cited by in RCA: 382] [Article Influence: 42.4] [Reference Citation Analysis (0)] |
| 44. | Sprooten J, De Wijngaert P, Vanmeerbeerk I, Martin S, Vangheluwe P, Schlenner S, Krysko DV, Parys JB, Bultynck G, Vandenabeele P, Garg AD. Necroptosis in Immuno-Oncology and Cancer Immunotherapy. Cells. 2020;9. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 106] [Cited by in RCA: 126] [Article Influence: 25.2] [Reference Citation Analysis (0)] |
| 45. | Galon J, Bruni D. Approaches to treat immune hot, altered and cold tumours with combination immunotherapies. Nat Rev Drug Discov. 2019;18:197-218. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1151] [Cited by in RCA: 2203] [Article Influence: 367.2] [Reference Citation Analysis (0)] |
| 46. | Zhang C, Wang XY, Zhang P, He TC, Han JH, Zhang R, Lin J, Fan J, Lu L, Zhu WW, Jia HL, Zhang JB, Chen JH. Cancer-derived exosomal HSPC111 promotes colorectal cancer liver metastasis by reprogramming lipid metabolism in cancer-associated fibroblasts. Cell Death Dis. 2022;13:57. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 7] [Cited by in RCA: 147] [Article Influence: 49.0] [Reference Citation Analysis (0)] |
| 47. | Zhang C, Zhou C, Wu XJ, Yang M, Yang ZH, Xiong HZ, Zhou CP, Lu YX, Li Y, Li XN. Human CD133-positive hematopoietic progenitor cells initiate growth and metastasis of colorectal cancer cells. Carcinogenesis. 2014;35:2771-2777. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 16] [Cited by in RCA: 20] [Article Influence: 1.8] [Reference Citation Analysis (0)] |
| 48. | Yang C, Dou R, Wei C, Liu K, Shi D, Zhang C, Liu Q, Wang S, Xiong B. Tumor-derived exosomal microRNA-106b-5p activates EMT-cancer cell and M2-subtype TAM interaction to facilitate CRC metastasis. Mol Ther. 2021;29:2088-2107. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 23] [Cited by in RCA: 149] [Article Influence: 37.3] [Reference Citation Analysis (0)] |
| 49. | Wu X, Lan X, Hu W, Zhang W, Lai X, Xu S, Li J, Qiu W, Wang W, Xiao J, Wang F, Ding Y, Liang L. CMTM6 expression in M2 macrophages is a potential predictor of PD-1/PD-L1 inhibitor response in colorectal cancer. Cancer Immunol Immunother. 2021;70:3235-3248. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 8] [Cited by in RCA: 21] [Article Influence: 5.3] [Reference Citation Analysis (0)] |
| 50. | Lalos A, Tülek A, Tosti N, Mechera R, Wilhelm A, Soysal S, Daester S, Kancherla V, Weixler B, Spagnoli GC, Eppenberger-Castori S, Terracciano L, Piscuoglio S, von Flüe M, Posabella A, Droeser RA. Prognostic significance of CD8+ T-cells density in stage III colorectal cancer depends on SDF-1 expression. Sci Rep. 2021;11:775. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 22] [Cited by in RCA: 22] [Article Influence: 5.5] [Reference Citation Analysis (0)] |
| 51. | Dong X, Yang Z, Yang H, Li D, Qiu X. Long Non-coding RNA MIR4435-2HG Promotes Colorectal Cancer Proliferation and Metastasis Through miR-206/YAP1 Axis. Front Oncol. 2020;10:160. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 40] [Cited by in RCA: 68] [Article Influence: 13.6] [Reference Citation Analysis (0)] |
| 52. | Aguilera TA, Rafat M, Castellini L, Shehade H, Kariolis MS, Hui AB, Stehr H, von Eyben R, Jiang D, Ellies LG, Koong AC, Diehn M, Rankin EB, Graves EE, Giaccia AJ. Reprogramming the immunological microenvironment through radiation and targeting Axl. Nat Commun. 2016;7:13898. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 106] [Cited by in RCA: 149] [Article Influence: 16.6] [Reference Citation Analysis (0)] |
| 53. | Noman MZ, Berchem G, Janji B. Targeting autophagy blocks melanoma growth by bringing natural killer cells to the tumor battlefield. Autophagy. 2018;14:730-732. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 23] [Cited by in RCA: 32] [Article Influence: 4.6] [Reference Citation Analysis (0)] |
| 54. | Guisier F, Dubos-Arvis C, Viñas F, Doubre H, Ricordel C, Ropert S, Janicot H, Bernardi M, Fournel P, Lamy R, Pérol M, Dauba J, Gonzales G, Falchero L, Decroisette C, Assouline P, Chouaid C, Bylicki O. Efficacy and Safety of Anti-PD-1 Immunotherapy in Patients With Advanced NSCLC With BRAF, HER2, or MET Mutations or RET Translocation: GFPC 01-2018. J Thorac Oncol. 2020;15:628-636. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 89] [Cited by in RCA: 217] [Article Influence: 43.4] [Reference Citation Analysis (0)] |
| 55. | Zhang S, Li J, Gao H, Tong Y, Li P, Wang Y, Du L, Wang C. lncRNA Profiles Enable Prognosis Prediction and Subtyping for Esophageal Squamous Cell Carcinoma. Front Cell Dev Biol. 2021;9:656554. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 5] [Cited by in RCA: 5] [Article Influence: 1.3] [Reference Citation Analysis (0)] |