Published online Jul 15, 2022. doi: 10.4251/wjgo.v14.i7.1239
Peer-review started: November 18, 2021
First decision: January 12, 2022
Revised: January 26, 2022
Accepted: March 26, 2022
Article in press: March 26, 2022
Published online: July 15, 2022
Processing time: 236 Days and 15.7 Hours
Esophageal squamous cell carcinoma (ESCC) is one of the most common malignancies. A total of 45 kinesin superfamily proteins (KIFs) have been identified in humans, among which several family members have demonstrated varied functions in tumor pathobiology via different mechanisms, including regulation of cell cycle progression and metastasis. KIFC3 has microtubule motor activity and is involved in cancer cell invasion and migration, as well as survival. However, the role of KIFC3 in ESCC is still unknown.
To evaluate the role of KIFC3 in ESCC and the underlying mechanisms.
Expression of KIFC3 was evaluated in ESCC tissues and adjacent normal eso
Immunohistochemical staining indicated that KIFC3 was upregulated in ESCC tissues compared with adjacent normal tissues. Kaplan–Meier Plotter revealed that overexpressed KIFC3 was associated with poor prognosis in ESCC patients. Colony formation and EdU assay showed that KIFC3 overexpression promoted cell proliferation, while KIFC3 knockdown inhibited cell proliferation in ESCC cell lines. In addition, cell cycle analysis showed that KIFC3 overexpression promoted cell cycle progression. KIFC3 knockdown suppressed ESCC tumorigenesis in vivo. Transwell assay and western blotting revealed that KIFC3 overexpression promoted cell migration and invasion, as well as epithelial–mesenchymal transition (EMT), while KIFC3 knockdown showed the opposite results. Mechanistically, KIFC3 overexpression promoted β-catenin signaling in KYSE450 cells; however, the role of KIFC3 was abolished by XAV-939, the inhibitor of β-catenin signaling.
KIFC3 was overexpressed in ESCC and was associated with poor prognosis. Furthermore, KIFC3 promoted proliferation, migration and invasion of ESCC via β-catenin signaling and EMT.
Core tip: Esophageal squamous cell carcinoma (ESCC) is one of the most dangerous malignancies affecting human health. However, the mechanism of ESCC is still unclear. We revealed that KIFC3 was upregulated in ESCC. In addition, overexpressed KIFC3 was associated with poor prognosis in ESCC patients. In vitro and in vivo experiments revealed that KIFC3 promoted the proliferation, migration and invasion of ESCC cells by activating epithelial–mesenchymal transition and β-catenin signaling. Our study strongly suggests that KIFC3 may be a potential new therapeutic target for ESCC.
- Citation: Hao WW, Xu F. KIFC3 promotes proliferation, migration and invasion of esophageal squamous cell carcinoma cells by activating EMT and β-catenin signaling. World J Gastrointest Oncol 2022; 14(7): 1239-1251
- URL: https://www.wjgnet.com/1948-5204/full/v14/i7/1239.htm
- DOI: https://dx.doi.org/10.4251/wjgo.v14.i7.1239
Esophageal cancer is one of the most dangerous tumors affecting humans, ranking seventh in incidence and sixth in mortality worldwide[1]. The main histological type of esophageal cancer in western countries is adenocarcinoma, while that in Asian countries is squamous cell carcinoma, where it has high incidence and mortality[2-4]. Although clinicians and researchers have made great efforts to unravel the pathophysiology of esophageal cancer, the mechanism of esophageal squamous cell carcinoma (ESCC) is still unknown[5-7]. Recent research has shown that dysregulation of TP53 and cell cycle regulators are prominent characteristics of ESCC, which may also be detected in precursor lesions, but the molecular progression from dysplasia to invasive ESCC remains unclear[3,8]. Further studies are urgently needed to reveal the underlying molecular mechanisms and discover effective treatment targets to improve ESCC survival.
The kinesin superfamily proteins (KIFs) are a group of proteins that function as microtubule-based motors for transporting cellular cargo and playing crucial roles in chromosomal and spindle movements during mitosis and meiosis[9]. There are 45 KIFs identified in humans; among which, several have demonstrated varied functions in tumor pathobiology via different mechanisms, including regulation of cell cycle progression and metastasis[10-12]. Among these members, KIFC3 has negative end-directed microtubule motor activity and plays roles in Golgi positioning and integration, as well as in apical transport, in epithelial cells[13,14]. Studies of null mice have revealed that KIFC3 is dispensable for normal development and reproduction[15]. Overexpression of KIFC3 may mediate docetaxel resistance in breast cancer cells[16]. KIFC3 expression was positively associated with cell invasion and migration, and overexpressed KIFC3 was associated with shorter overall survival in hepatocellular carcinoma[17]. However, the role of KIFC3 in ESCC has not yet been reported. In view of the importance of cell cycle regulators in the development of ESCC and the involvement of KIFC3 in mitosis, we speculated that this protein might be involved in the occurrence and development of ESCC.
In this study, we aimed to examine KIFC3 expression in ESCC and further explore its role and the relevant mechanisms in ESCC tumor progression, in an attempt to provide promising insights into the mechanism of this disease and possible therapeutic targets.
Primary ESCC tumor tissues and corresponding nontumor tissue specimens were collected from the First Affiliated Hospital of Zhengzhou University. All experiments were carried out in accordance with the Declaration of Helsinki (amended in 2013). All procedures were supervised and approved by the Ethics Committee of the First Affiliated Hospital of Zhengzhou University (No. 2021-KY-0446-001).
The human ESCC cell lines KYSE30, KYSE150, KYSE450, KYSE510 and Eca109 and human normal esophageal epithelial cell line Het-1A were obtained from the Type Culture Collection of the Chinese Academy of Sciences and cultured in RPMI-1640 medium supplemented with fetal bovine serum (FBS) (10%) at 37°C in 5% CO2.
Total proteins were extracted using RIPA buffer supplemented with 1% phenylmethylsulfonyl fluoride and 1% Protease Inhibitor Cocktail. Protein concentration was measured using the BCA kit (Beyotime, China). Proteins were separated using SDS-PAGE and transferred onto a polyvinylidene difluoride membrane. After blocking with 5% fresh milk for 1 h, the membranes were incubated with a specific primary antibody overnight at 4°C, followed by incubation with the corresponding secondary antibody for 1 h. Finally, the bands on the membranes were detected using the ECL detection system (Thermo) and quantified using Quantity One Software version 4.3. Primary antibodies for KIFC3, E-cadherin, N-cadherin, vimentin, proliferating cell nuclear antigen (PCNA), cyclin D1, β-actin, and secondary antibodies were purchased from Proteintech (China), matrix metalloproteinase (MMP)7 and c-Myc were purchased from Abcam (Cambridge, MA, USA), and β-catenin was purchased from Cell Signaling Technology (Danvers, MA, USA).
pLVX-Puro-KIFC3-shRNA- and pLVX-Puro-KIFC3-expressing lentiviruses were designed and provided by Genechem (China). KYSE150 cells were transfected with pLVX-Puro-KIFC3-shRNA- expressing lentiviruses and KYSE450 cells with pLVX-Puro-KIFC3-expressing lentiviruses, while control cells were transfected with empty vectors. After transfection with lentiviruses, 2 μg/mL puromycin dihydrochloride (Beyotime) was added for 6–8 d to establish stable cell lines. Knockdown and overexpression efficiency were validated using western blotting, and the validated cell lines were recorded as KYSE150shNC, KYSE150shKIFC3, KYSE450oeNC and KYSE450oeKIFC3.
The cancer cells were seeded in a six-well plate at 1000 cells/well and cultured for 14 d. The colonies were washed once with phosphate-buffered saline (PBS), fixed with 1 mL 4% paraformaldehyde for 15 min, and stained with 1 mL 0.5% crystal violet for 30 min. After washing with deionized distilled water, the images were captured using a digital camera. The number of colonies was then counted.
The cancer cells were seeded in a six-well plate at 2.5 × 105 cells per well. When the cell density was 60%, cells were collected, washed with cold PBS twice, and then fixed in 70% ethanol at 4°C overnight. The cells were washed with cold PBS twice, and then incubated with RNaseA (0.1 mg/mL) and propidium iodide (0.02 mg/mL) at 37°C for 30 min in the dark. The cell cycle was detected using flow cytometry.
A Transwell assay was used to detect cell migration and invasion. For the invasion assay, 100 μL Matrigel (serum-free medium diluted 1:8) was added to the upper chamber of the Transwell chamber (Corning, USA). After shaking well, the gel was incubated at 37°C and solidified for 2–4 h. Cells were seeded at 104 per well in the upper chamber with 100 μL serum-free medium. For the lower chamber, 500 μL medium (20% FBS) was added. After 24 h, the cells that passed through the filter were fixed with 4% paraformaldehyde for 30 min, stained with 0.5% crystal violet for 30 min, and washed with PBS. Finally, the cells were examined under a fluorescence microscope (BX51; Olympus, Japan). The migration assay was similar to the invasion assay, with the difference that no Matrigel was added to the upper chamber.
KYSE150 and KYSE450 cells were seeded onto glass coverslips in 24-well plates overnight. The cells were then washed with PBS and fixed with 4% paraformaldehyde for 15 min at 25°C. Next, the cells were permeabilized for 15 min with 0.3% Triton X-100 in PBS. The cells were then blocked with 5% BSA in PBS for 1 h at 25°C and then incubated overnight at 4°C with the primary antibody against β-catenin (1:100 dilution) overnight; normal goat serum was used as a negative control. The treated cells were incubated with an FITC-labeled or Cy3-labeled goat anti-rabbit IgG (H+L) highly cross-adsorbed secondary antibody (Beyotime) in the dark for 1 h. The nuclei were visualized after staining with 2 μg/mL DAPI (Biosharp, China) for 10 min at 25°C. The glass coverslips were sealed with antifade reagent (Biosharp) and examined under a fluorescence microscope (BX51; Olympus).
All animal research procedures were approved by the Ethics Committee of the First Affiliated Hospital of Zhengzhou University (No. 2021-KY-0446-001). All animal experiments were carried out in accordance with the National Institutes of Health Guide for the Care and Use of Laboratory Animals (NIH Publications No. 8023, revised 1978). Male BALB/c nude mice (age 4 wk) were purchased from Beijing Life River Experimental Animal Technology Co. Ltd. and kept in a temperature-controlled specific pathogen free environment with a regular light/dark cycle and provided with adequate rodent diet and water. Ten mice were randomly divided into KYSE150shNC and KYSE150shKIFC3 groups. There were five mice in each group. KYSE150shNC cells (106) and KYSE150shKIFC3 cells (106) were collected, washed with PBS, suspended in 200 μL PBS, and subcutaneously implanted into the right flank of the dorsal region of nude mice. A Vernier caliper was used to measure tumor sizes every 3 d, and the tumor volume (TV) was calculated using the following formula: TV (mm 3) = 0.5 × d2 × D, where d is the shortest diameter and D is the longest diameter. After nine measurements, the longest diameter of tumor reached 15 mm. All mice were killed and the tumor specimens were collected and weighed.
Immunohistochemical staining was performed using an UltraSensitive TM SP kit and DAB kit (Maixin, China). Tumor tissues were embedded in paraffin and cut into 4-μm sections. The deparaffinized sections were incubated with a primary antibody against KIFC3 (1: 200 dilution, Abcam, #ab154419) or Ki-67 (1:100 dilution, Abcam, #ab16667) overnight at 4°C, and normal goat serum was used as a negative control. After washing, the tissue sections were then incubated with a biotinylated anti-rabbit secondary antibody (1:200 dilution, Aspen, #AS-1107) for 1 h at 25°C. The sections were subsequently incubated with horseradish-peroxidase-conjugated streptavidin (Beyotime, #A0303) and developed using 3, 3-diaminobenzidine. An optical microscope (BX51; Olympus) was used to observe the specimens. Two observers, who were blinded to the data of the samples, evaluated, counted and analyzed the positive cells. The proportion of positive tumor cells was scored as follows: 1 (< 10% positive tumor cells), 2 (10%–50% positive tumor cells), 3 (50%–75% positive tumor cells), and 4 (> 75% positive tumor cells). The intensity of staining was graded according to the following criteria: 0 (no staining), 1 (weak staining = light yellow), 2 (moderate staining = yellow brown), and 3 (strong staining = brown). The staining index was calculated as the product of the proportion of positive cells times the staining intensity score (range: 0–12). The median of staining index was used as the cut-off value; the staining index higher than the cut-off value was identified as high expression, while that less than the cut-off value was identified as low expression.
Graphpad Prism 7 was used for all analyses. Data are expressed as the mean ± SD. To analyze the differences between groups, t test and nonparametric tests were used. P < 0.05 was considered statistically significant.
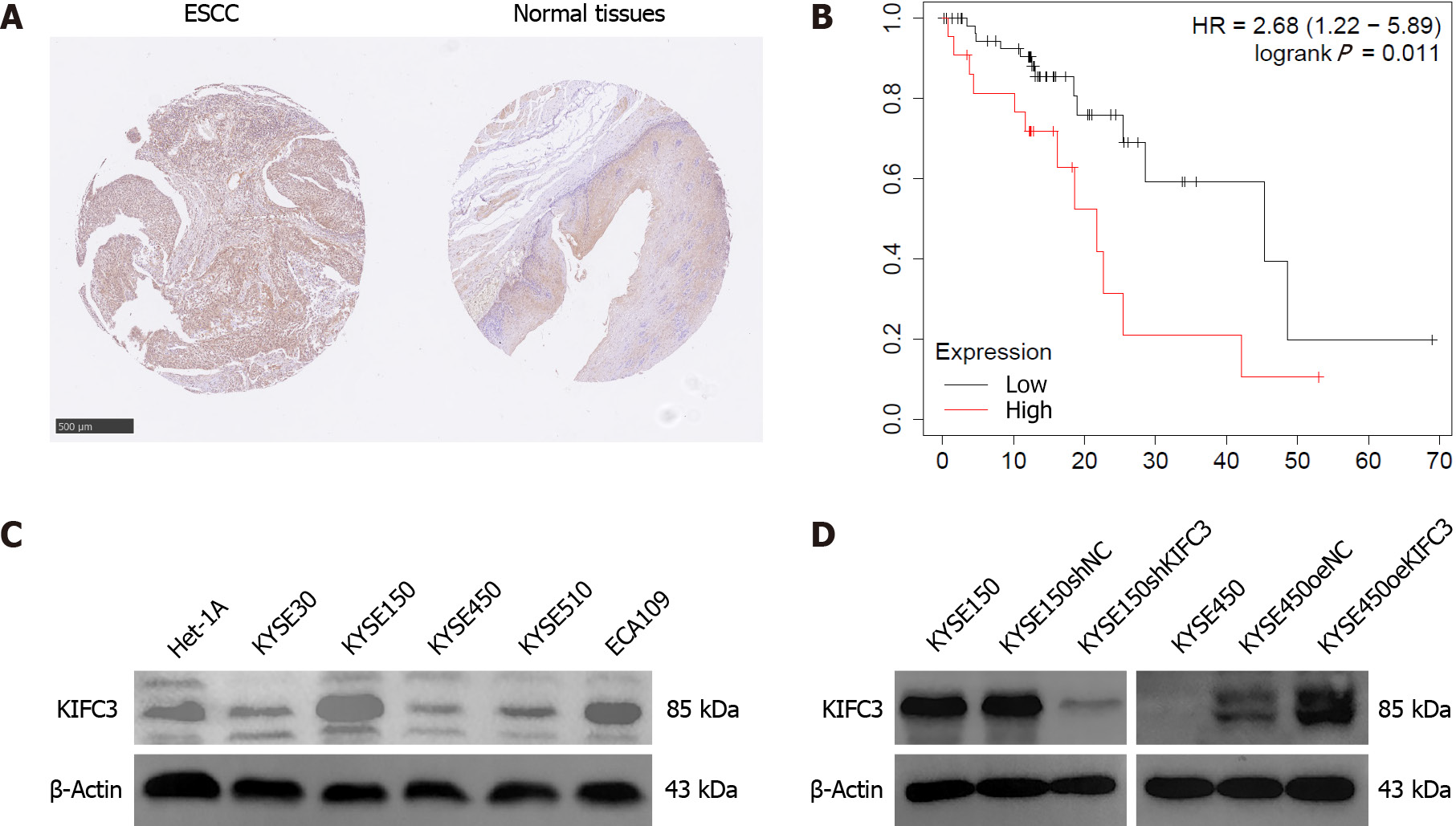
To investigate KIFC3 expression, ESCC tissues and adjacent nontumor tissues were collected from 34 patients (26 male and 8 female, aged 47–72 years) with ESCC. Immunohistochemical assays showed that KIFC3 expression was lower in ESCC tissues than in adjacent nontumor tissues (Figure 1A). Moreover, data from Kaplan–Meier Plotter (https://kmplot.com/analysis) revealed that lower expression of KIFC3 was associated with better overall survival in patients with ESCC (Figure 1B). These results indicate that KIFC3 plays an important role in ESCC development. To explore the role of KIFC3 in ESCC cells, its expression level was evaluated in ESCC cell lines Eca109, KYSE30, KYSE150, KYSE450 and KYSE510, as well as normal esophageal epithelial cell line Het-1A. KYSE450 and KYSE150 cells displayed the lowest and highest expression levels of KIFC3, respectively (Figure 1C). Therefore, KYSE450 cells were used for subsequent overexpression assays, while KYSE150 cells were used for knockdown assays. Western blotting verified that the corresponding cell lines were successfully constructed (Figure 1D).
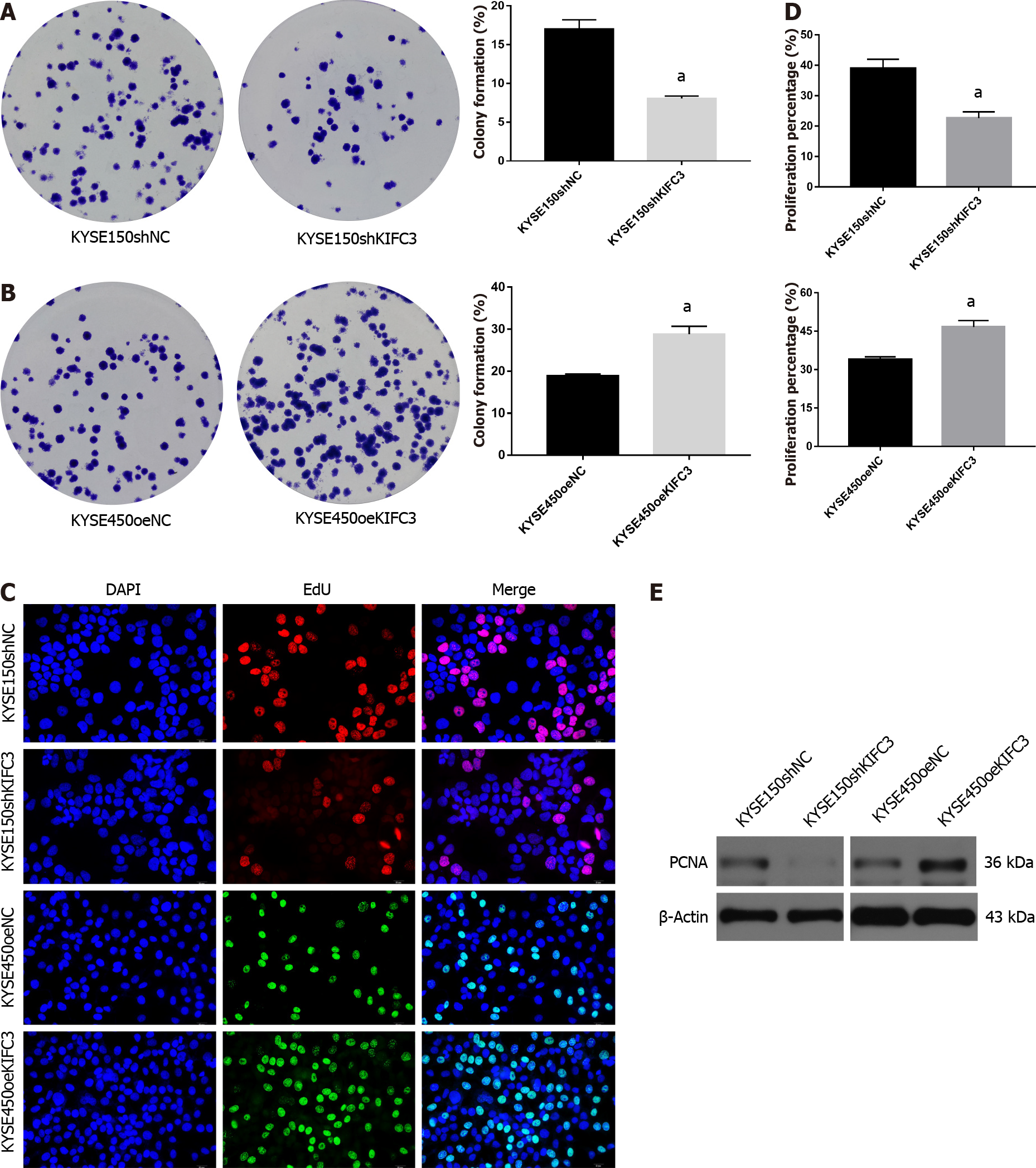
To investigate the effect of KIFC3 in ESCC, colony-formation and EdU assays were performed to observe the proliferation of KYSE150 and KYSE450 cells. The results showed that the colony-formation capacity was decreased after knockdown of KIFC3 in KYSE150 cells, while KIFC3 overexpression promoted colony-formation capacity in KYSE450 cells (Figure 2A and 2B). EdU assay showed that KIFC3 knockdown inhibited the proliferation of KYSE150 cells, while its overexpression promoted proliferation of KYSE450 cells (Figure 2C and 2D). In addition, KIFC3 knockdown inhibited the expression of PCNA, while its overexpression promoted PCNA expression (Figure 2E), which indicated that KIFC3 promoted proliferation in ESCC cells at the molecular level.
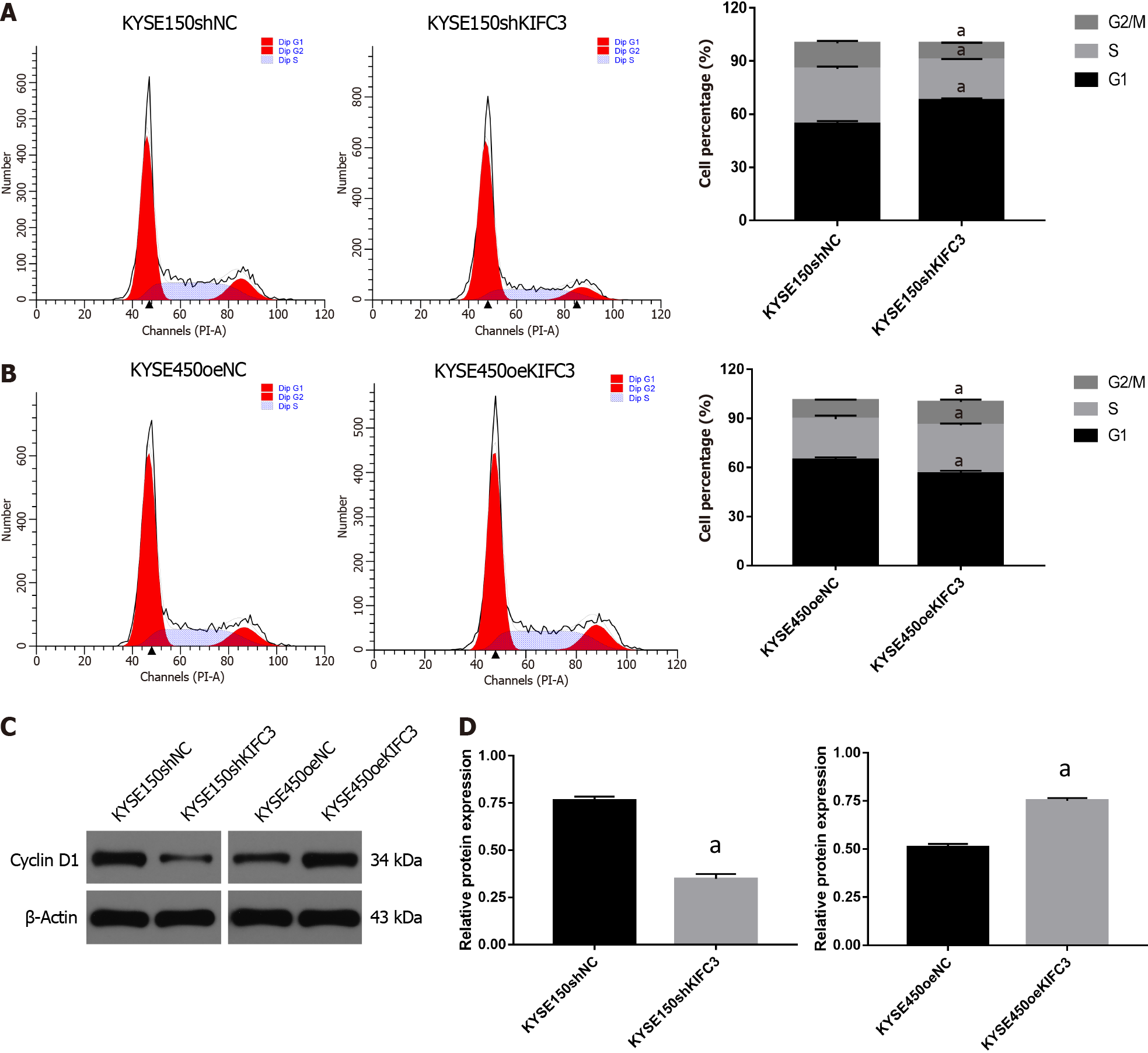
To further investigate the role of KIFC3 in cell cycle progression, cell cycle analysis was conducted. The percentage of G0/1 phase in KYSE150shKIFC3 group was significantly higher, and percentage of S and G2/M phase was lower than that in KYSE150shNC group (Figure 3A), while the percentage of G0/1 phase in KYSE450oeKIFC3 group was significantly lower, and percentage of S and G2/M phase was higher than that in KYSE450oeNC group (Figure 3B). At the molecular level, KIFC3 knockdown caused a decrease while KIFC3 overexpression promoted the expression of cyclin D1, which plays an important role in the transition from G1 to S phase (Figure 3C and 3D). These results indicate that KIFC3 positively regulates cell cycle in ESCC cells.
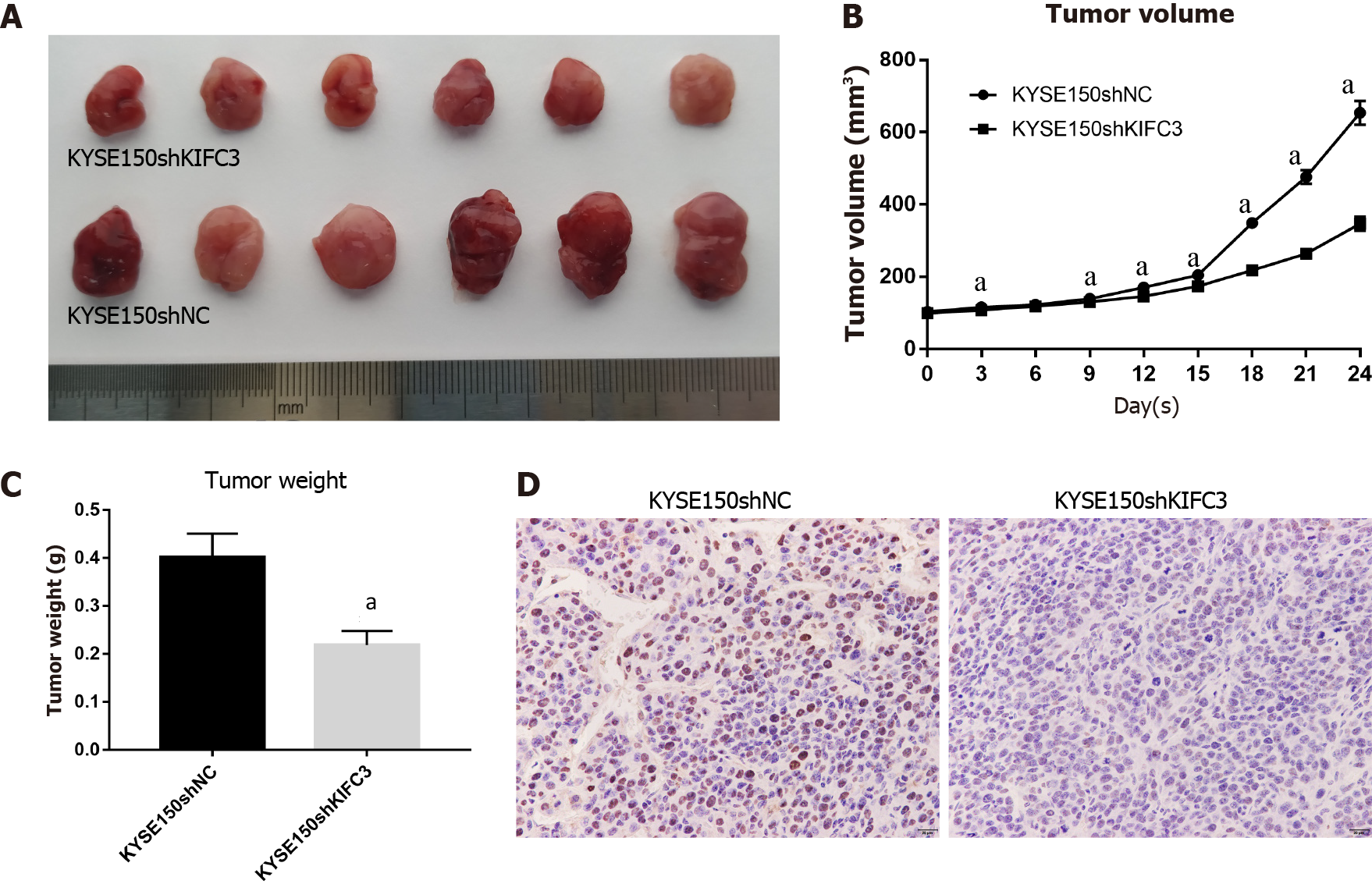
Based on the in vitro data above, we further investigated the role of KIFC3 using xenograft tumors in vivo. All animals were in a fit state during the experiment and all in vivo data were included in the analysis. The transplanted tumors grew rapidly in the KYSE150shNC group but were suppressed in the KYSE150shKIFC3 group (Figure 4A and 4B), and the weight of tumors in the KYSE150shKIFC3 group was significantly lower (Figure 4C). Immunohistochemistry of tumors showed that the expression of Ki-67 was decreased significantly in the KYSE150shKIFC3 group (Figure 4D), proving that KIFC3 knockdown inhibited the proliferation of ESCC in vivo at the molecular level. Taken together, KIFC3 promotes ESCC proliferation in vivo.
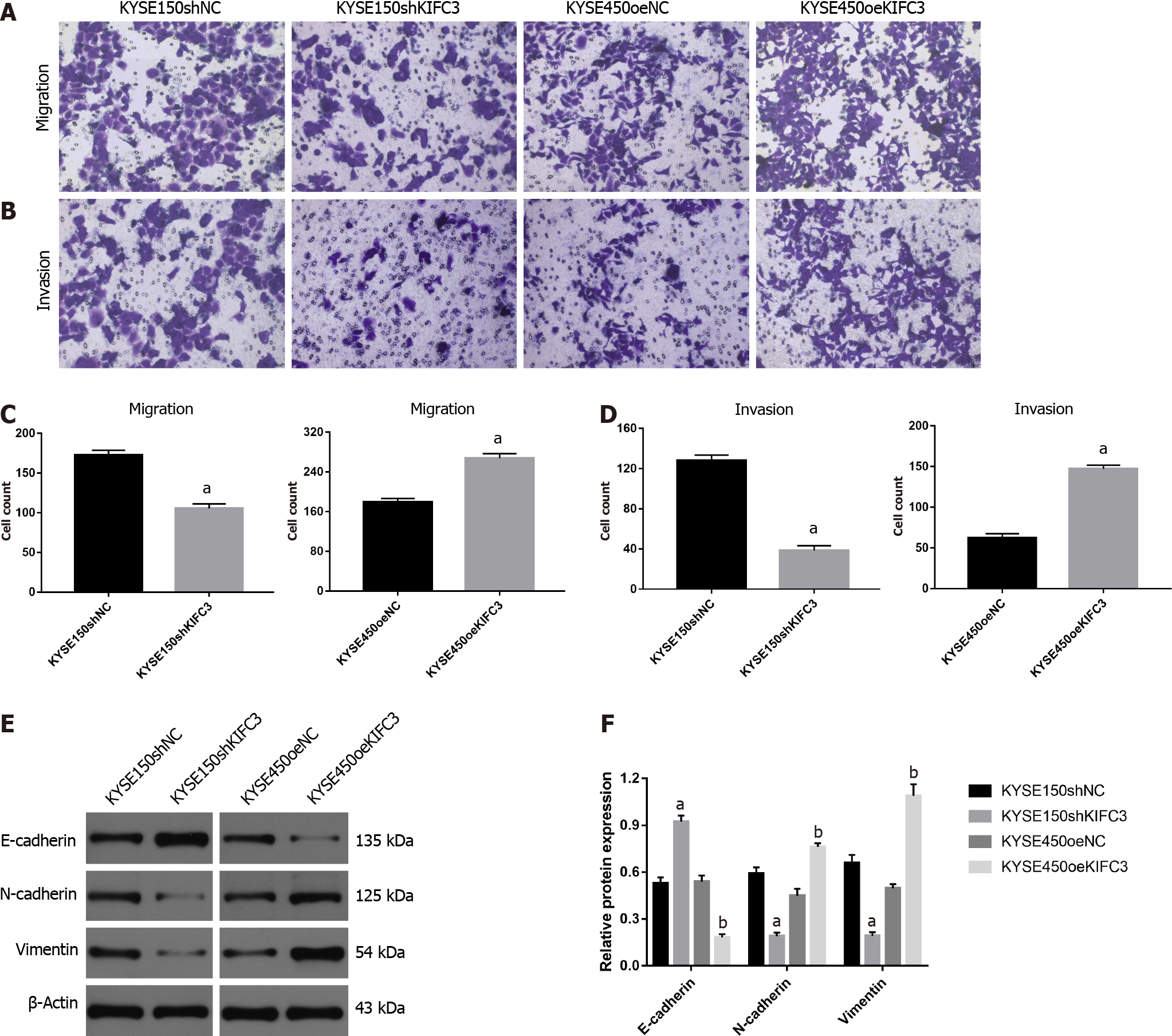
KIFC3 promotes migration and invasion via epithelial–mesenchymal transition in ESCC cells
To detect whether KIFC3 exerts effects on the migration and invasion of ESCC cells, Transwell migration and invasion assays were used. Accelerated cell migration was observed in the KYSE450oeKIFC3 group compared with the KYSE450oeNC group, while it was less active in the KYSE150shKIFC3 group than in the KYSE150shNC group (Figure 5A and 5C). Transwell invasion showed similar results (Figure 5B and 5D). To further explore the molecular mechanism of KIFC3-promoted migration and invasion, we detected the expression of E-cadherin, N-cadherin and vimentin, which are key molecules of epithelial–mesenchymal transition (EMT). The results indicated that N-cadherin and vimentin, which are associated with the mesenchyme phenotype and indicate a higher possibility of tumor migration and invasion, were decreased, while E-cadherin, which is associated with the epithelial phenotype, increased after KIFC3 knockdown. In KIFC3-overexpressing cells, the expression of N-cadherin, vimentin and E-cadherin showed the opposite results (Figure 5E and 5F). These results indicate that KIFC3 promotes migration and invasion through EMT in ESCC cells.
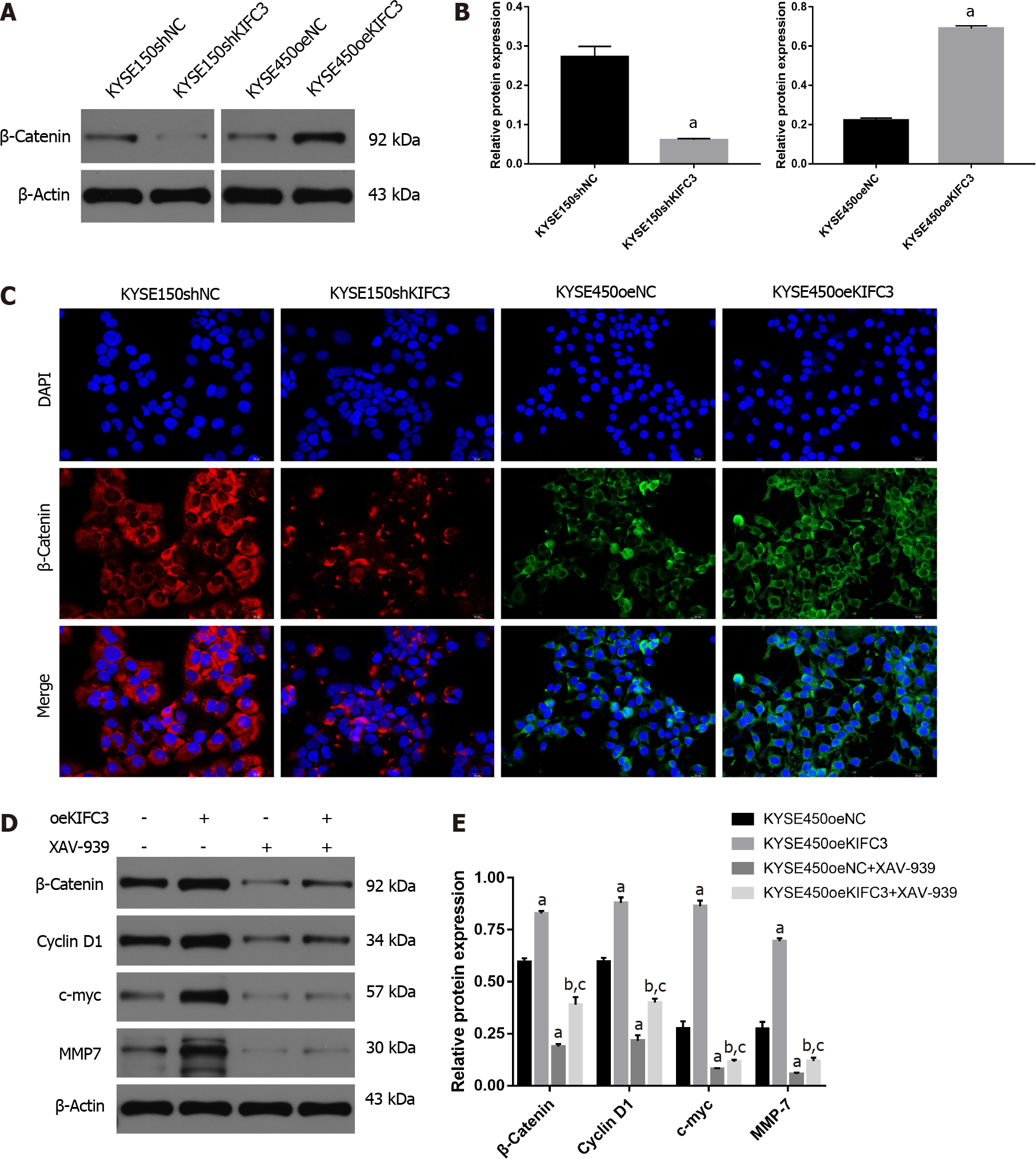
Previous studies have demonstrated that β-catenin plays an essential role in the proliferation, migration, and invasion of ESCC cells. In our study, western blotting showed that β-catenin protein expression in KYSE150shKIFC3 group was decreased significantly compared with that in the KYSE150shNC group (Figure 6A and 6B), while expression of β-catenin was increased significantly in the KYSE450oeKIFC3 group compared to that in the KYSE450oeNC group (Figure 6A and 6B). Immunofluorescence showed the same results; β-catenin expression was decreased after KIFC3 knockdown and increased after KIFC3 overexpression (Figure 6C). These results suggest that KIFC3 promotes β-catenin expression in ESCC cells.
To explore the role of β-catenin signaling in KIFC3-promoted proliferation, migration and invasion, we used XAV-939, an inhibitor of β-catenin. KYSE450oeNC and KYSE450oeKIFC3 cells were treated with XAV-939, and the expression of proteins downstream of β-catenin, which play an important role in proliferation, migration, and invasion was evaluated. Although the expression of c-myc, cyclin D1 and MMP7 was increased after KIFC3 overexpression (P < 0.05), when β-catenin was inhibited by XAV-939, the levels of c-myc, cyclin D1 and MMP7 were decreased even when KIFC3 was overexpressed (P < 0.05) (Figure 6D and 6E). These results suggest that KIFC3 promotes the progression of ESCC via β-catenin signaling.
ESCC is one of the most malignant tumors that impose a significant medical burden on the health system. Although several gene mutations that could increase the susceptibility to ESCC have been identified[8], the exact mechanism that induces tumor progression is still unclear. KIFC3 has been reported to be overexpressed in a number of cancers and may be involved in cell cycle and cell proliferation, which suggests that KIFC3 is an oncogene, thus arousing our interest in its possible role in ESCC progression.
In the present study, we found that KIFC3 was upregulated in ESCC tissues compared to adjacent nontumor tissues. Data from the Kaplan–Meier Plotter website indicated that high levels of KIFC3 expression were correlated with poor ESCC prognosis. These findings indicate that KIFC3 may be involved in the development of ESCC, and that high expression of KIFC3 may be a prognostic factor in ESCC. Functional assays showed that knockdown of KIFC3 suppressed proliferation in KYSE150 cells, and the cell cycle was arrested compared with that in KYSE150shNC cells. Migration and invasion were inhibited in KYSE150shKIFC3 cells. Compared with KYSE450oeNC cells, cell proliferation, migration, and invasion were activated in KYSE450oeKIFC3 cells, further demonstrating that KIFC3 promoted the progression of ESCC cells. Furthermore, KIFC3 knockdown inhibited ESCC proliferation in vivo. Given the above results, we may consider KIFC3 as a tumor marker associated with the progression and prognosis of ESCC.
EMT plays a potential role in the promotion of tumor invasiveness, metastasis and resistance to apoptotic stimuli, and it is marked by downregulation of epithelial biomarkers, such as E-cadherin, and the upregulation of mesenchymal biomarkers, such as N-cadherin and vimentin. Cells undergoing EMT can acquire greater mobility and become prometastatic[18-20]. EMT is known to play an important role in the metastasis of ESCC[21-23]. In addition, members of the KIF family are involved in the regulation of EMT and thus affect the migration and invasion of tumors[24-26]. In our study, KIFC3 knockdown suppressed the migration and invasion of KYSE150 cells, accompanied by the upregulation of E-cadherin and downregulation of N-cadherin and vimentin, while KIFC3 overexpression showed the opposite results in KYSE450 cells. These results indicate that KIFC3 promotes the migration and invasion of ESCC cells by inhibiting EMT.
β-Catenin signaling has been shown to be associated with the regulation of processes such as proliferation and invasion, which are involved in the occurrence and progression of cancers[27,28]. Moreover, the deactivation of β-catenin has been a potential treatment target in ESCC[29-31]. Thus, regulation of β-catenin may play an important role in the development of ESCC. It has been reported that in HEK293 cells depleted of centrosomes, normal accumulation of β-catenin in response to Wnt signaling is attenuated[32]. KIFC3 regulates centrosome cohesion in a microtubule-dependent manner[14]. Collectively, the relationship between KIFC3 and β-catenin is worth exploring. In the present study, KIFC3 overexpression promoted the expression of β-catenin and downstream molecules of β-catenin signaling, such as cyclin D1, c-myc and MMP7. However, after treatment with XAV-939, an inhibitor of β-catenin, KIFC3-induced upregulation of cyclin D1, c-Myc, and MMP7 was blocked, which means that β-catenin plays a vital role in KIFC3-induced tumor progression. Although further investigation is required to clarify the exact mechanism underlying the effect of KIFC3 on β-catenin, our research demonstrated that KIFC3 promotes tumor progression via β-catenin signaling in ESCC.
KIFC3 is upregulated in ESCC, and KIFC3 overexpression is associated with poor prognosis in ESCC patients. KIFC3 promotes the proliferation, migration, and invasion of ESCC cells by activating EMT and β-catenin signaling. Overall, our study provides a comprehensive understanding of KIFC3 in ESCC and its underlying mechanisms, which strongly suggests that KIFC3 may be a potential new therapeutic target for ESCC treatment.
Esophageal squamous cell carcinoma (ESCC) is one of the most common malignancies. The mechanism of ESCC is still unclear.
Kinesin family member (KIF)C3 has microtubule motor activity and may be involved in mitotic progression. KIFC3 was also shown to be involved in cell invasion and migration, as well as in survival, in hepatocellular carcinoma.
To elucidate the role of KIFC3 in ESCC and the underlying mechanisms.
The expression of KIFC3 was evaluated in ESCC tissues and normal tissues. In addition, KIFC3 knockdown and KIFC3-overexpressing cell lines were constructed and then colony formation, EdU assays, cell cycle analysis, Transwell assays, and western blotting were performed to explore the underlying mechanisms of action. A xenograft tumor model in nude mice was used to verify the role of KIFC3 in tumorigenesis.
We showed that KIFC3 was upregulated in ESCC tissues and was associated with poor prognosis. KIFC3 promoted cell proliferation, mitosis progression, migration and invasion. In addition, KIFC3 knockdown suppressed ESCC tumorigenesis in an in vivo model. Mechanistically, we validated the involvement of KIFC3 β-catenin signaling and epithelial–mesenchymal transition (EMT) in ESCC progression.
We found that KIFC3 was overexpressed in ESCC, and the expression of this protein was associated with prognosis in ESCC patients. Furthermore, KIFC3 promoted proliferation, migration, and invasion of ESCC via β-catenin signaling and EMT.
Although the detailed mechanism underlying the effect of KIFC3 in promoting ESCC progression should be studied more carefully in our next research, we believe our research strongly suggests that KIFC3 may be a potential new therapeutic target for ESCC treatment.
Provenance and peer review: Unsolicited article; Externally peer reviewed.
Peer-review model: Single blind
Specialty type: Oncology
Country/Territory of origin: China
Peer-review report’s scientific quality classification
Grade A (Excellent): 0
Grade B (Very good): B, B
Grade C (Good): 0
Grade D (Fair): 0
Grade E (Poor): 0
P-Reviewer: Exbrayat JM, France; Oguma J, Japan S-Editor: Wu YXJ L-Editor: Kerr C P-Editor: Yuan YY
| 1. | Siegel RL, Miller KD, Fuchs HE, Jemal A. Cancer Statistics, 2021. CA Cancer J Clin. 2021;71:7-33. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 8287] [Cited by in RCA: 11936] [Article Influence: 2984.0] [Reference Citation Analysis (4)] |
| 2. | Fan J, Liu Z, Mao X, Tong X, Zhang T, Suo C, Chen X. Global trends in the incidence and mortality of esophageal cancer from 1990 to 2017. Cancer Med. 2020;9:6875-6887. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 31] [Cited by in RCA: 83] [Article Influence: 16.6] [Reference Citation Analysis (0)] |
| 3. | Smyth EC, Lagergren J, Fitzgerald RC, Lordick F, Shah MA, Lagergren P, Cunningham D. Oesophageal cancer. Nat Rev Dis Primers. 2017;3:17048. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 587] [Cited by in RCA: 737] [Article Influence: 92.1] [Reference Citation Analysis (2)] |
| 4. | Thrumurthy SG, Chaudry MA, Thrumurthy SSD, Mughal M. Oesophageal cancer: risks, prevention, and diagnosis. BMJ. 2019;366:l4373. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 53] [Cited by in RCA: 62] [Article Influence: 10.3] [Reference Citation Analysis (0)] |
| 5. | Cao W, Lee H, Wu W, Zaman A, McCorkle S, Yan M, Chen J, Xing Q, Sinnott-Armstrong N, Xu H, Sailani MR, Tang W, Cui Y, Liu J, Guan H, Lv P, Sun X, Sun L, Han P, Lou Y, Chang J, Wang J, Gao Y, Guo J, Schenk G, Shain AH, Biddle FG, Collisson E, Snyder M, Bivona TG. Multi-faceted epigenetic dysregulation of gene expression promotes esophageal squamous cell carcinoma. Nat Commun. 2020;11:3675. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 64] [Cited by in RCA: 78] [Article Influence: 15.6] [Reference Citation Analysis (0)] |
| 6. | Triantafyllou T, Wijnhoven BPL. Current status of esophageal cancer treatment. Chin J Cancer Res. 2020;32:271-286. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 6] [Cited by in RCA: 14] [Article Influence: 2.8] [Reference Citation Analysis (0)] |
| 7. | Yang YM, Hong P, Xu WW, He QY, Li B. Advances in targeted therapy for esophageal cancer. Signal Transduct Target Ther. 2020;5:229. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 209] [Cited by in RCA: 314] [Article Influence: 62.8] [Reference Citation Analysis (0)] |
| 8. | Liu X, Zhang M, Ying S, Zhang C, Lin R, Zheng J, Zhang G, Tian D, Guo Y, Du C, Chen Y, Chen S, Su X, Ji J, Deng W, Li X, Qiu S, Yan R, Xu Z, Wang Y, Cui J, Zhuang S, Yu H, Zheng Q, Marom M, Sheng S, Hu S, Li R, Su M. Genetic Alterations in Esophageal Tissues From Squamous Dysplasia to Carcinoma. Gastroenterology. 2017;153:166-177. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 94] [Cited by in RCA: 134] [Article Influence: 16.8] [Reference Citation Analysis (0)] |
| 9. | Hirokawa N, Noda Y, Okada Y. Kinesin and dynein superfamily proteins in organelle transport and cell division. Curr Opin Cell Biol. 1998;10:60-73. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 251] [Cited by in RCA: 253] [Article Influence: 9.4] [Reference Citation Analysis (0)] |
| 10. | Qureshi Z, Ahmad M, Yang WX, Tan FQ. Kinesin 12 (KIF15) contributes to the development and tumorigenicity of prostate cancer. Biochem Biophys Res Commun. 2021;576:7-14. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 3] [Cited by in RCA: 9] [Article Influence: 2.3] [Reference Citation Analysis (0)] |
| 11. | Wu YP, Ke ZB, Zheng WC, Chen YH, Zhu JM, Lin F, Li XD, Chen SH, Cai H, Zheng QS, Wei Y, Xue XY, Xu N. Kinesin family member 18B regulates the proliferation and invasion of human prostate cancer cells. Cell Death Dis. 2021;12:302. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 15] [Cited by in RCA: 18] [Article Influence: 4.5] [Reference Citation Analysis (0)] |
| 12. | Zou JX, Duan Z, Wang J, Sokolov A, Xu J, Chen CZ, Li JJ, Chen HW. Kinesin family deregulation coordinated by bromodomain protein ANCCA and histone methyltransferase MLL for breast cancer cell growth, survival, and tamoxifen resistance. Mol Cancer Res. 2014;12:539-549. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 102] [Cited by in RCA: 135] [Article Influence: 12.3] [Reference Citation Analysis (0)] |
| 13. | Xu Y, Takeda S, Nakata T, Noda Y, Tanaka Y, Hirokawa N. Role of KIFC3 motor protein in Golgi positioning and integration. J Cell Biol. 2002;158:293-303. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 65] [Cited by in RCA: 62] [Article Influence: 2.7] [Reference Citation Analysis (0)] |
| 14. | Hata S, Pastor Peidro A, Panic M, Liu P, Atorino E, Funaya C, Jäkle U, Pereira G, Schiebel E. The balance between KIFC3 and EG5 tetrameric kinesins controls the onset of mitotic spindle assembly. Nat Cell Biol. 2019;21:1138-1151. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 25] [Cited by in RCA: 42] [Article Influence: 7.0] [Reference Citation Analysis (0)] |
| 15. | Yang Z, Xia Ch, Roberts EA, Bush K, Nigam SK, Goldstein LS. Molecular cloning and functional analysis of mouse C-terminal kinesin motor KifC3. Mol Cell Biol. 2001;21:765-770. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 21] [Cited by in RCA: 23] [Article Influence: 1.0] [Reference Citation Analysis (0)] |
| 16. | De S, Cipriano R, Jackson MW, Stark GR. Overexpression of kinesins mediates docetaxel resistance in breast cancer cells. Cancer Res. 2009;69:8035-8042. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 88] [Cited by in RCA: 109] [Article Influence: 6.8] [Reference Citation Analysis (0)] |
| 17. | Chen J, Li S, Zhou S, Cao S, Lou Y, Shen H, Yin J, Li G. Kinesin superfamily protein expression and its association with progression and prognosis in hepatocellular carcinoma. J Cancer Res Ther. 2017;13:651-659. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 51] [Cited by in RCA: 70] [Article Influence: 10.0] [Reference Citation Analysis (0)] |
| 18. | Lamouille S, Xu J, Derynck R. Molecular mechanisms of epithelial-mesenchymal transition. Nat Rev Mol Cell Biol. 2014;15:178-196. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 4715] [Cited by in RCA: 6233] [Article Influence: 566.6] [Reference Citation Analysis (0)] |
| 19. | Mittal V. Epithelial Mesenchymal Transition in Tumor Metastasis. Annu Rev Pathol. 2018;13:395-412. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 478] [Cited by in RCA: 1009] [Article Influence: 168.2] [Reference Citation Analysis (0)] |
| 20. | Navas T, Kinders RJ, Lawrence SM, Ferry-Galow KV, Borgel S, Hollingshead MG, Srivastava AK, Alcoser SY, Makhlouf HR, Chuaqui R, Wilsker DF, Konaté MM, Miller SB, Voth AR, Chen L, Vilimas T, Subramanian J, Rubinstein L, Kummar S, Chen AP, Bottaro DP, Doroshow JH, Parchment RE. Clinical Evolution of Epithelial-Mesenchymal Transition in Human Carcinomas. Cancer Res. 2020;80:304-318. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 68] [Cited by in RCA: 70] [Article Influence: 14.0] [Reference Citation Analysis (0)] |
| 21. | Cai R, Wang P, Zhao X, Lu X, Deng R, Wang X, Su Z, Hong C, Lin J. LTBP1 promotes esophageal squamous cell carcinoma progression through epithelial-mesenchymal transition and cancer-associated fibroblasts transformation. J Transl Med. 2020;18:139. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 10] [Cited by in RCA: 25] [Article Influence: 5.0] [Reference Citation Analysis (0)] |
| 22. | Tang Q, Lento A, Suzuki K, Efe G, Karakasheva T, Long A, Giroux V, Islam M, Wileyto EP, Klein-Szanto AJ, Nakagawa H, Bass A, Rustgi AK. Rab11-FIP1 mediates epithelial-mesenchymal transition and invasion in esophageal cancer. EMBO Rep. 2021;22:e48351. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 14] [Cited by in RCA: 19] [Article Influence: 4.8] [Reference Citation Analysis (0)] |
| 23. | Zhang J, Luo A, Huang F, Gong T, Liu Z. SERPINE2 promotes esophageal squamous cell carcinoma metastasis by activating BMP4. Cancer Lett. 2020;469:390-398. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 22] [Cited by in RCA: 43] [Article Influence: 7.2] [Reference Citation Analysis (0)] |
| 24. | Gao Y, Zheng H, Li L, Zhou C, Chen X, Zhou X, Cao Y. KIF3C Promotes Proliferation, Migration, and Invasion of Glioma Cells by Activating the PI3K/AKT Pathway and Inducing EMT. Biomed Res Int. 2020;2020:6349312. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 7] [Cited by in RCA: 21] [Article Influence: 4.2] [Reference Citation Analysis (0)] |
| 25. | Han J, Wang F, Lan Y, Wang J, Nie C, Liang Y, Song R, Zheng T, Pan S, Pei T, Xie C, Yang G, Liu X, Zhu M, Wang Y, Liu Y, Meng F, Cui Y, Zhang B, Meng X, Zhang J, Liu L. KIFC1 regulated by miR-532-3p promotes epithelial-to-mesenchymal transition and metastasis of hepatocellular carcinoma via gankyrin/AKT signaling. Oncogene. 2019;38:406-420. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 74] [Cited by in RCA: 75] [Article Influence: 12.5] [Reference Citation Analysis (0)] |
| 26. | Li J, Diao H, Guan X, Tian X. Kinesin Family Member C1 (KIFC1) Regulated by Centrosome Protein E (CENPE) Promotes Proliferation, Migration, and Epithelial-Mesenchymal Transition of Ovarian Cancer. Med Sci Monit. 2020;26:e927869. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 2] [Cited by in RCA: 12] [Article Influence: 2.4] [Reference Citation Analysis (0)] |
| 27. | Liu C, Wang L, Liu X, Tan Y, Tao L, Xiao Y, Deng P, Wang H, Deng Q, Lin Y, Jie H, Zhang H, Zhang J, Peng Y, Zhou Z, Sun Q, Cen X, Zhao Y. Cytoplasmic SHMT2 drives the progression and metastasis of colorectal cancer by inhibiting β-catenin degradation. Theranostics. 2021;11:2966-2986. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 20] [Cited by in RCA: 44] [Article Influence: 11.0] [Reference Citation Analysis (0)] |
| 28. | Zhang Y, Wang X. Targeting the Wnt/β-catenin signaling pathway in cancer. J Hematol Oncol. 2020;13:165. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 212] [Cited by in RCA: 915] [Article Influence: 183.0] [Reference Citation Analysis (0)] |
| 29. | Wang T, Wang J, Ren W, Liu ZL, Cheng YF, Zhang XM. Combination treatment with artemisinin and oxaliplatin inhibits tumorigenesis in esophageal cancer EC109 cell through Wnt/β-catenin signaling pathway. Thorac Cancer. 2020;11:2316-2324. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 4] [Cited by in RCA: 12] [Article Influence: 2.4] [Reference Citation Analysis (0)] |
| 30. | Wei B, Wang Y, Wang J, Cai X, Xu L, Wu J, Liu W, Gu Y, Guo W, Xu Q. Apatinib suppresses tumor progression and enhances cisplatin sensitivity in esophageal cancer via the Akt/β-catenin pathway. Cancer Cell Int. 2020;20:198. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 10] [Cited by in RCA: 18] [Article Influence: 3.6] [Reference Citation Analysis (0)] |
| 31. | Zhang L, Zhou S, Guo E, Chen X, Yang J, Li X. DCLK1 inhibition attenuates tumorigenesis and improves chemosensitivity in esophageal squamous cell carcinoma by inhibiting β-catenin/c-Myc signaling. Pflugers Arch. 2020;472:1041-1049. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 6] [Cited by in RCA: 4] [Article Influence: 0.8] [Reference Citation Analysis (0)] |
| 32. | Vora SM, Fassler JS, Phillips BT. Centrosomes are required for proper β-catenin processing and Wnt response. Mol Biol Cell. 2020;31:1951-1961. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 8] [Cited by in RCA: 7] [Article Influence: 1.4] [Reference Citation Analysis (0)] |