Published online Jun 15, 2022. doi: 10.4251/wjgo.v14.i6.1067
Peer-review started: August 19, 2021
First decision: September 12, 2021
Revised: September 22, 2021
Accepted: May 26, 2022
Article in press: May 26, 2022
Published online: June 15, 2022
Processing time: 294 Days and 16.8 Hours
Circular RNAs (circRNAs) have covalently closed loop structures at both ends, exhibiting characteristics dissimilar to those of linear RNAs. Emerging evidence suggests that aberrantly expressed circRNAs play crucial roles in hepatocellular carcinoma (HCC) by affecting the proliferation, apoptosis and invasive capacity of HCC cells. Certain circRNAs may be used as biomarkers to diagnose and predict the prognosis of HCC. Therefore, circRNAs are expected to become novel biomarkers and therapeutic targets for HCC. Herein, we briefly review the characteristics and biological functions of circRNAs, focusing on their roles in HCC to provide new insights for the early diagnosis and targeted therapy of HCC.
Core Tip: Current studies have shown that aberrantly expressed circular RNAs (circRNAs) play crucial roles in hepatocellular carcinoma (HCC) by affecting the proliferation, apoptosis and invasive capacity of HCC cells. Certain circRNAs may be used as potential biomarkers to diagnose and predict the prognosis of HCC. Therefore, circRNAs are expected to become novel biomarkers and therapeutic targets for HCC. Herein, we briefly review the characteristics and biological functions of circRNAs, focusing on their roles in HCC to provide new insights for the early diagnosis and targeted therapy of HCC.
- Citation: Niu ZS, Wang WH. Circular RNAs in hepatocellular carcinoma: Recent advances. World J Gastrointest Oncol 2022; 14(6): 1067-1085
- URL: https://www.wjgnet.com/1948-5204/full/v14/i6/1067.htm
- DOI: https://dx.doi.org/10.4251/wjgo.v14.i6.1067
Early hepatocellular carcinoma (HCC) usually lacks specific symptoms, and most patients have missed the opportunity for effective treatment because they are diagnosed at middle-to-advanced stages. The emergence of novel therapeutic strategies for HCC, such as immunotherapy and molecularly targeted therapies[1], can prolong the survival of HCC patients. Unfortunately, patients with advanced HCC are prone to metastasis and recurrence, and long-term prognosis remains poor[2]. Therefore, identifying new biomarkers for early diagnosis and effective therapeutic targets of HCC is critical.
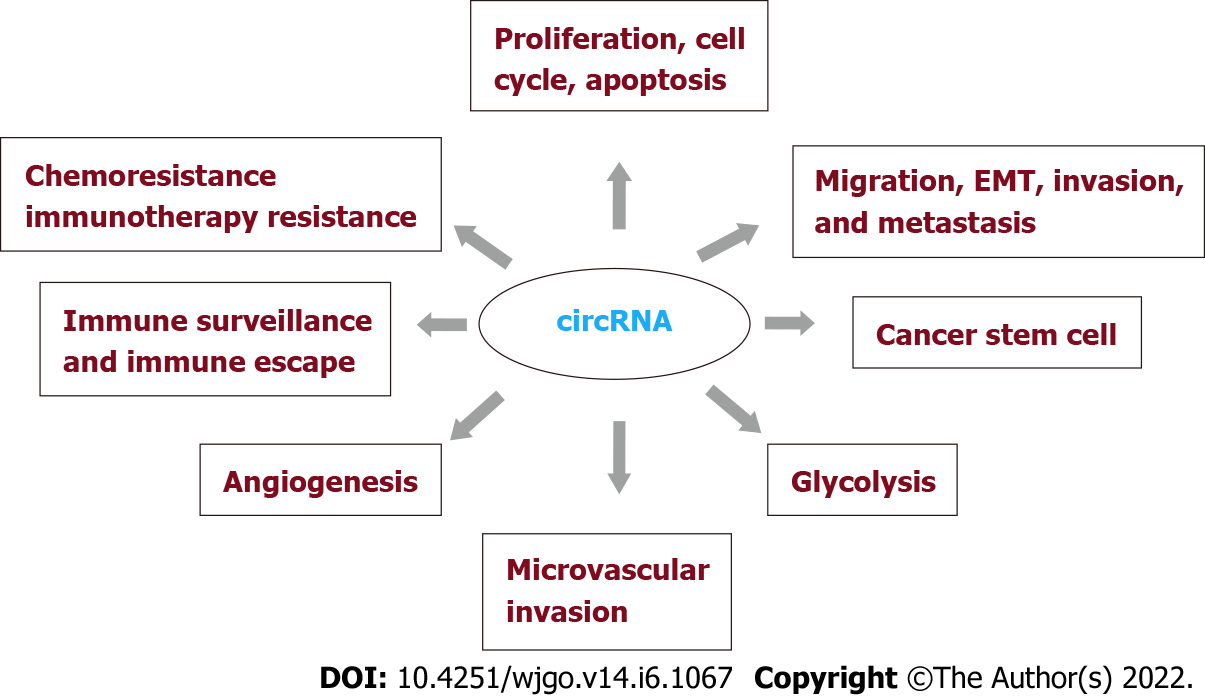
Circular RNAs (circRNAs) are covalently closed loops generated by the back splicing of precursor mRNA (premRNA) molecules, which exist widely in mammalian cells and are characterized by stability, conservative evolution, and cell or tissue specificity. These characteristics endow circRNAs with many biological functions, such as acting as microRNA (miRNA) sponges, regulating the transcription of parental genes, binding RNA binding proteins (RBPs), and encoding proteins and peptides[3]. CircRNAs exert their biological functions mainly at the epigenetic, transcriptional and posttranscriptional levels[4,5]. Dysregulated circRNAs play crucial roles in various diseases, particularly with respect to the occurrence and development of tumors and tumor proliferation, apoptosis and metastasis[6-8]. Currently, aberrantly expressed circRNAs are closely associated with the proliferation, cell cycle, apoptosis, migration, epithelial-mesenchymal transition (EMT), invasion, metastasis, cancer stem cells (CSCs), glycolysis, microvascular invasion (MVI), angiogenesis, immune surveillance, immune escape, chemoresistance, and immunotherapy resistance of HCC. Thus, circRNAs may be promising biomarkers for the diagnosis and prognosis of HCC as well as effective therapeutic targets. Herein, we briefly review the characteristics and biological functions of circRNAs, focusing on their roles in HCC to provide new insights into the early diagnosis and targeted therapy of HCC.
Most circRNAs have the following characteristics: (1) High abundance: The abundance of circRNA expression varies greatly; in some cases, the abundance of circRNAs exceeds 10 times that of their linear RNA counterparts[9]; (2) Stability: The stability of circRNAs is 2.5-5 times higher than that of linear transcripts because the unique covalently closed loop of circRNAs lacks 3’ and 5’ ends, resulting in the absence of ribonuclease binding targets; therefore, circRNAs are not easily degraded[10]; (3) Conservation: CircRNAs are widely present in different species and are evolutionarily conserved. Some studies suggest that most circRNAs in different species are evolutionarily conserved, while a few are not conserved[11]; and (4) Specificity: CircRNAs have tissue and cell specificity, with differential expression in different stages of ontogeny and disease progression[12].
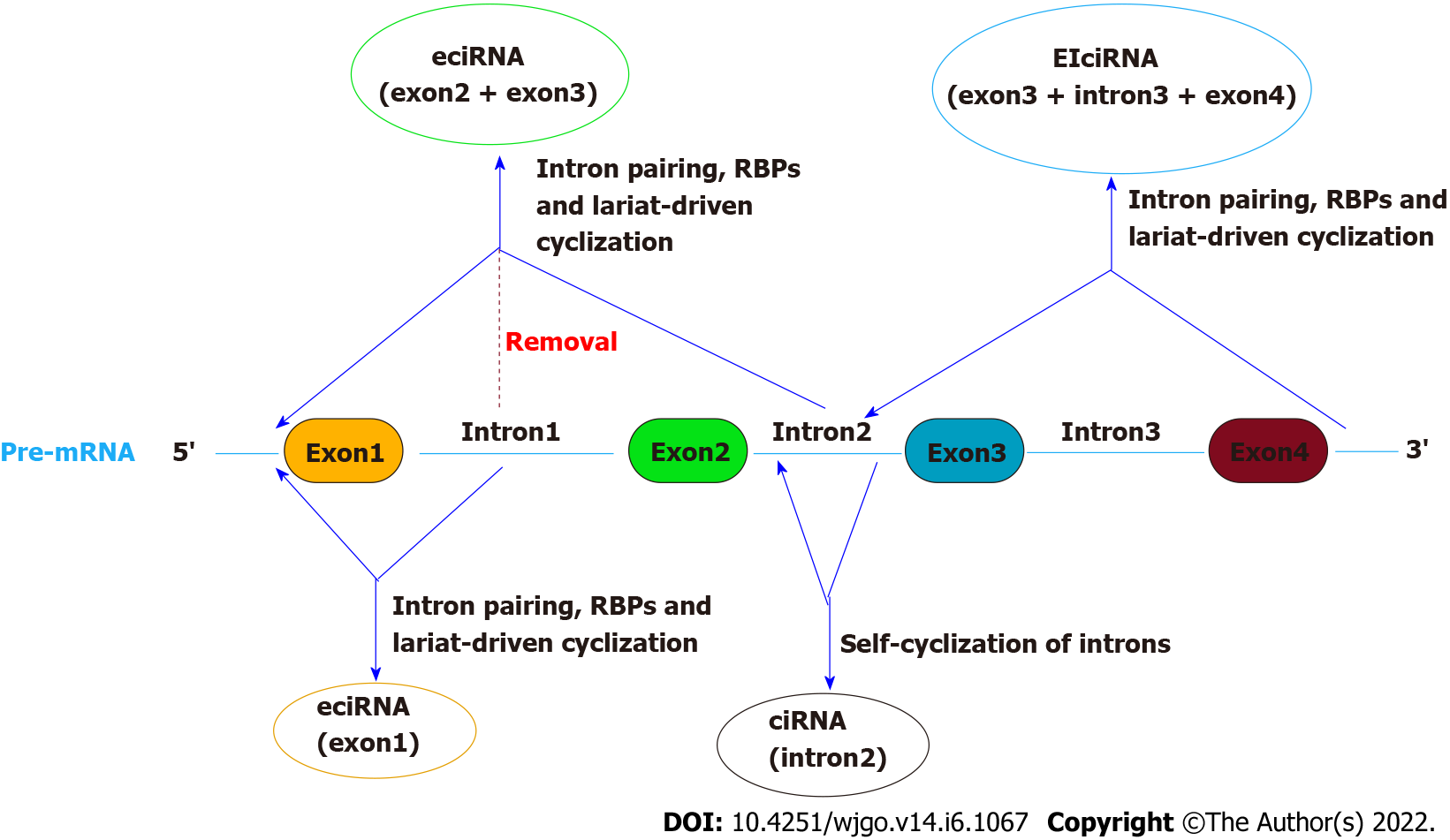
CircRNAs are categorized into four classes based on their origins: Exon circRNAs (ecircRNAs), intron circRNAs (ciRNAs), exon-ciRNAs (EIciRNAs), and intergenic circRNAs[13] (Figure 1). EcircRNAs are predominant and are mainly located in the cytoplasm. CiRNAs and EIciRNAs are located in the nucleus. The generation mechanism of circRNAs is very complex and has not yet been understood. Current studies have shown that the cyclization of circRNAs is principally driven by intron pairing, RBPs or transcription factors and lariat[14].
Intron pairing-driven cyclization or “direct back splicing” is the most common cyclization mode of ecircRNA and EIciRNA, where the special premRNA containing ALU repeats is sheared to form ecircRNA after reverse base complementary pairing[11]. Lariat-driven cyclization or “exon skipping” connects exons at both ends through donor and acceptor sites provided by spliceosomes to form lariat selective splicing to generate ecircRNA[11]. In RBP-driven circulation, RBPs bound to the complementary sequences on both sides of the intron of premRNA interact with each other to form a circular structure and promote the terminal connection at both ends of the head and tail to form ecircRNA[15]. EIciRNAs can be formed if introns are retained between exons during the above three mechanisms[16]. Self-cyclization of introns: When pre-RNA has a 7 nt guanine (G)- and uracil (U)-rich sequence near an exon and an 11 nt cytosine (C)-rich sequence near another exon, the introns escape branching and degradation during the splicing reaction to produce an intron lariat structure and cyclize to form a stable ciRNA[17].
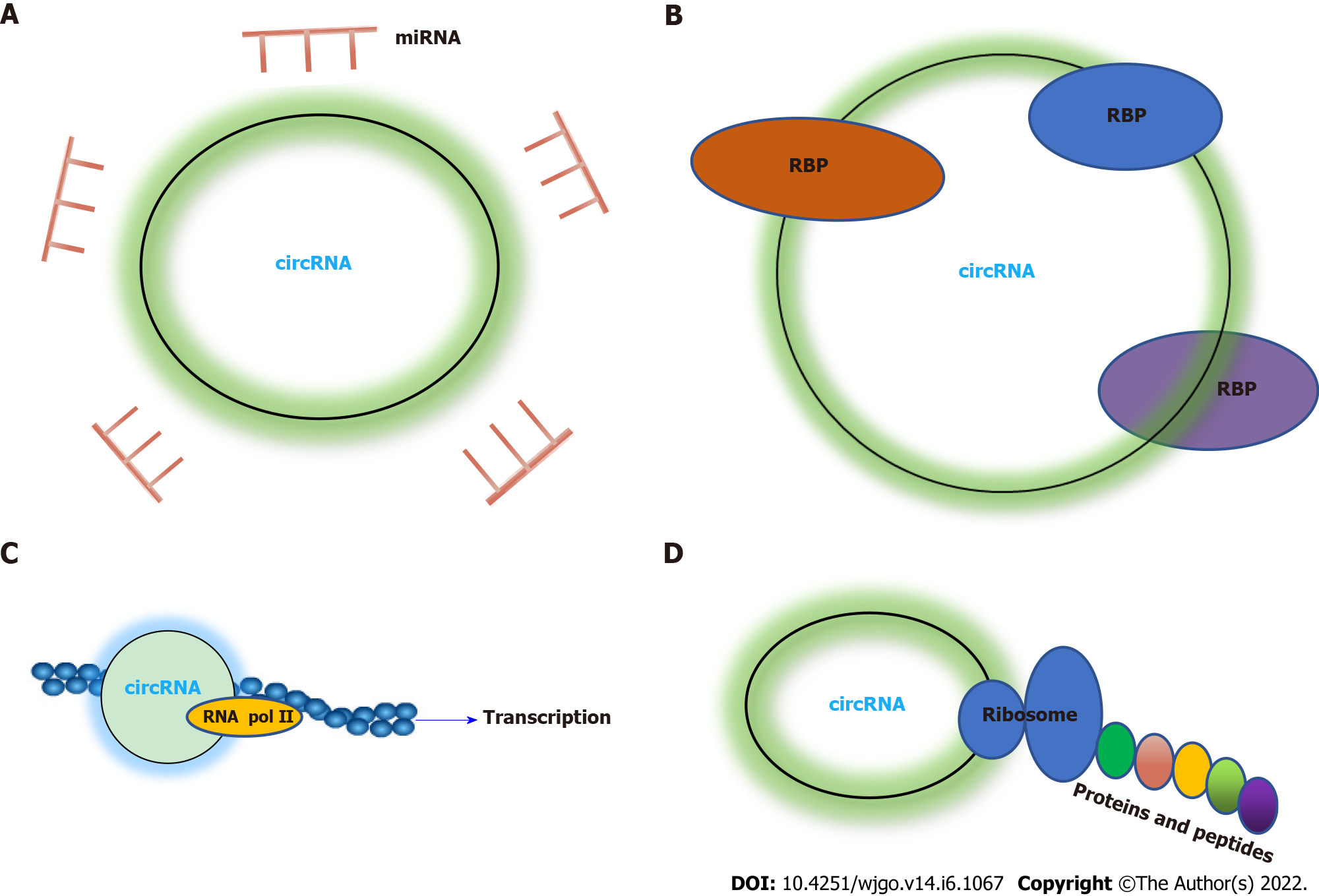
CircRNAs serve in regulatory roles in different biological behaviors through different mechanisms, including acting as sponges of miRNAs, interacting with RBPs, and regulating gene transcription and translation (Figure 2). A recent review analyzed the functions of circRNAs in HCC, of which acting as miRNA sponges accounted for 79.6%[18].
As molecular sponges of miRNAs, circRNAs harbor many miRNA binding sites, which can competitively bind to and restrain the activity of miRNAs[19], thereby regulating the expression of downstream target genes posttranscriptionally. Currently, clinical studies have mainly focused on circRNAs as miRNA molecular sponges[20]. Compared with other types of competing endogenous RNAs, circRNAs have the following advantages. First, circRNAs are not easily degraded by RNA enzymes (RNase or RNA exonucleases)[21,22], which makes the circRNA structure stable and enables the possibility to stably inhibit the performance of miRNA function, with a stronger adsorption capacity for miRNAs than linear mRNAs and long noncoding RNAs. Second, existing studies have shown that the majority of circRNAs are highly expressed and that they can contain substantial miRNA response elements in a single molecule[23-25]; therefore, circRNAs are able to instantly bind or release large amounts of miRNAs to efficiently exert their regulatory roles. For example, cirs-7, also known as CDR1as, is a circRNA containing more than 70 miR-7 binding sites[26], which can bind to miR-7 and act downstream of its mRNA. This molecular axis is widely expressed in various malignancies, including oral squamous cell carcinoma and lung cancer[27,28]. In addition, circRNAs may store and transport miRNAs[29]. For example, CDR1as has both miR-7 and miR-671 binding sites[30], and CDR1as first binds to miR-7 and is transported to subcellular locations, where CDR1as is then degraded by miR-671 to eventually release miR-7[26].
It is worth noting that only circRNAs meeting specific stoichiometric requirements can act as endogenous miRNA sponges, where the abundance of circRNAs as miRNA sponges must match that of miRNAs[31]. Thus, circRNAs as miRNA sponges may not be a universal phenomenon, but one unique to some circRNAs. Only ecircRNAs can act as miRNA sponges, while EIciRNAs and ciRNAs contain few miRNA binding sites that are relatively scattered; thus, EIciRNA and ciRNA may lack the miRNA sponge action ability possessed by ecircRNA[17]. The dysregulation of the circRNA-miRNA-mRNA axis, whether manifesting a promoting or inhibitory role, has been confirmed in many cancers. However, the specific biological mechanism of the circRNA-miRNA-mRNA axis in the occurrence and development of tumors and whether molecular targeted therapy can be improved by intervention in this approach remain to be further studied.
Although most circRNAs are located in the cytoplasm, a fraction exists in the nucleus and participate in regulating RNA transcription. CiRNAs are abundantly expressed in the nucleus and interact with phosphorylated RNA polymerase II to change its transcriptional activity, thereby playing a role in transcriptional regulation[32]. For example, a circRNA (ci-ankrd52), derived from the intron of the ankyrin repeat domain 52 gene, can enhance the expression of its parent gene ankrin52 by interacting with the RNA polymerase II elongation complex[17]. EIciRNAs are intron-preserving circRNAs located near the promoter of their parent genes and bind to RNA polymerase II to improve transcription efficiency by interacting with the 5’ splicing site preserved in introns, thereby promoting the expression of their parent genes[33]. Interestingly, EIciRNAs can act as RBP sponges, like ecircRNAs, and regulate parental gene expression[34]. Additionally, circRNAs can also regulate the expression of parent genes through epigenetic modification. Recently, it has been found that certain circRNAs have N6-methyladenosine (m6A) modifications, and these circRNAs will affect the stability of the parent gene[35].
RBPs are an important class of proteins that participate in posttranscriptional regulation. RBPs interact with circRNAs and play a role in circRNA splicing, replication, folding, stabilization, and localization. The combination of RBPs and circRNAs fulfills roles mainly in the following two ways: (1) RBPs are involved in the action of ceRNA: CircRNAs serve as miRNA “sponges” to modulate mRNA translation, and the potential of these “sponges” is higher than that of their linear counterparts because RBPs participate in the miRNA competition process[36]; and (2) CircRNAs competitively bind to RBPs: CircRNAs play biological roles by binding to RBPs through their specific sequence binding sites[37]. Here we present the most extensively studied RBP, human antigen R (HuR), as an example. HuR, as an RBP, can bind guanylate-rich elements in the 3’ untranslated region (UTR) to prevent mRNA from being degraded and accomplishes the function of stabilizing RNA structure[38,39]. HuR is widely expressed in eukaryotic tissues[40], and circE2F2 binds to HuR and enhances the stability of the mRNA of the HuR target gene E2F2[41]. In contrast, circRHOBTB3 binds to HuR and reduces the stability of the mRNA of HuR target gene PTBP1[42]. In addition, circBACH1 can bind to HuR, facilitate HuR translocation to the cytoplasm and inhibit p27 translation[43].
CircRNAs were previously considered to be noncoding RNAs that cannot be translated into proteins. However, emerging evidence suggests that circRNAs can also be translated into proteins and peptides[44-46]. Some circRNAs initiate protein translation by binding to ribosomes via the internal ribosome entry site (IRES) sequence or after modifying m6A in the 5’UTR[45,47]. In addition, some circRNAs with an open reading frame (ORF) can initiate small proteins or micropeptides[48]. The 40S subunit of eukaryotic ribosomes binds to circRNA and directly initiates in vitro translation[49]. Furthermore, unlike other noncoding RNAs, a few ecircRNAs in the cytoplasm can be translated into functional proteins[11]. Thus, the elements required for circRNA translation are IRES and an m6A sequence or ORF. Although circRNAs have translation ability, the translation efficiency is not high because of the influence of their special ring structure, and the functions of circRNA translation products (proteins and peptides) must be further explored.
Recent studies have confirmed the different critical roles of aberrantly expressed circRNAs in HCC (Figure 3). Here, we summarize the roles of certain circRNAs in HCC (Table 1).
| circRNAs | Dysregulation | Mechanism by competitively binding miRNAs/RBP or m6A modification/mRNA braking | Targets/signaling pathways | Biological functions | Ref. |
| circRNA ZFR | Up-regulated | N/A | MAP2K1 | Promotes HCC proliferation | Cedric et al[50] |
| miR-3619-5p | CTNNB1 Wnt/β-catenin pathway | Promotes HCC proliferation and EMT | Tan et al[99] | ||
| hsa_circ_0091581 | Up-regulated | miR-526b | c-Myc | Promotes HCC proliferation | Wei et al[53] |
| circ_0000517 | Up-regulated | miR-1296-5p | TXNDC5 | Promotes HCC growth and inhibits cell cycle arrest and apoptosis | Zang et al[55] |
| miR-326 | SMAD6 | Promotes HCC invasion and metastasis | He et al[67] | ||
| circSETD3 | Down-regulated | miR-421 | MAPK14 | Inhibits HCC proliferation and induces G1/S arrest | Xu et al[57] |
| N/A | N/A | Predicts MVI of HCC | Wang et al[85] | ||
| Exosomal circ-0051443 | Down-regulated | miR-331-3p | BAK1 | Promotes HCC cell apoptosis and arrests the cell cycle | Chen et al[58] |
| cicrRNA_101368 | Up-regulated | miR-200a | HMGB1/RAGE pathway | Promotes HCC cell migration | Li et al[60] |
| circ-CCND1 | Up-regulated | miR-497-5p | HMGA2 | Promotes HCC proliferation, migration and invasion | Zheng et al[61] |
| hsa_circ_0061395 | Up-regulated | miR-877-5p | PIK3R3 | Promotes HCC proliferation, migration and invasion | Yu et al[62] |
| miR-656-3p | SERBP1 | Li et al[63] | |||
| circRNA-103809 | Up-regulated | miR-377-3p | FGFR1/ERK | Facilitates HCC migration, invasion and EMT | Zhan et al[64] |
| miR-1270 | PLAGL2 | Cao et al[65] | |||
| circ_MMP2 | Up-regulated | miR-136-5p | MMP2 | Promotes HCC metastasis | Liu et al[68] |
| circ-MALAT1 | Up-regulated | mRNA braking | PAX5 | Promotes self-renewal of HCC stem cells | Chen et al[69] |
| miR-6887-3p | JAK2 | ||||
| circZKSCAN1 | Down-regulated | RBP: FMRP | CCAR1, Wnt/β-catenin pathway | Inhibits HCC stem cell activity | Zhu et al[70] |
| circMEG3 | Down-regulated | m6A-METTL3 | HULC and Cbf5 | Inhibits malignant differentiation of human liver CSCs | Jiang et al[71] |
| circ-PRKCI | Up-regulated | miR-1294 and miR-186-5p | FOXK1 | Promotes HCC glycolysis | Chen et al[73] |
| circZFR | Up-regulated | miR-375 | HMGA2 | Promotes HCC glycolysis | Xu et al[75] |
| circMAT2B | Up-regulated | miR-338-3p | PKM2 | Promotes HCC glycolysis | Li et al[77] |
| circ-PRMT5 | Up-regulated | miR-188-5p | HK2 | Enhances HCC glycolysis | Ding et al[79] |
| circ_0001445 | Down-regulated | miR-942-5p | ALX4 | Inhibits HCC metastasis and EMT | Xu et al[81] |
| ciRS-7 | Up-regulated | miR-7 | PIK3CD/p70S6K/mTOR pathway | Predicts hepatic MVI | Xu et al[83] |
| hsa_circ_0068669 | Down-regulated | N/A | N/A | Predicts hepatic MVI | Yao et al[84] |
| circCRIM1 | Up-regulated | miR-378a-3p | SKP2 | Promotes HCC angiogenesis | Ji et al[86] |
| hsa-circ-0046600 | Up-regulated | miR-640 | HIF-1α | Promotes HCC angiogenesis | Zhai et al[87] |
| hsa_circ_0000092 | Up-regulated | miR-338-3p | HN1 | Promotes HCC angiogenesis | Pu et al[89] |
| circGFRA1 | Up-regulated | miR-149 | N/A | Promotes HCC angiogenesis | Yu et al[91] |
| circARSP91 | Down-regulated | N/A | ULBP1 | Enhances HCC innate immune surveillance | Ma et al[93] |
| circTRIM33-12 | Down-regulated | miR-191 | TET1 | Inhibits HCC immune escape | Zhang et al[96] |
| hsa_circ0007456 | Down-regulated | miR-6852-3p | ICAM-1 | Inhibits HCC immune escape | Shi et al[97] |
| hsa_circ_104348 | Up-regulated | miR-187-3p | RTKN2 Wnt/β-catenin pathway | Promotes HCC resistance to sorafenib | Huang et al[100] |
| circβ-catenin | Up-regulated | Translation | Wnt/β-catenin pathway | Facilitates HCC cell growth | Liang et al[101] |
| hsa_circ_0004018 | Down-regulated | miR-626 | DKK3 Wnt/β-catenin pathway | Restrains HCC proliferation and migration | Zhu et al[102] |
| circRNA-ITCH | Down-regulated | miR-7 or miR-214 | c-myc and cyclinD1Wnt/β-catenin | Inhibits HCC proliferation and apoptosis | Yang et al[103] |
| circ-0003418 | Down-regulated | miR-7 and miR-383 | Wnt/β-catenin pathway | Increases HCC sensitivity to cisplatin | Chen et al[104] |
| circ-IGF1R | Up-regulated | N/A | PI3K/AKT pathway | Promotes HCC cell proliferation | Fu et al[106] |
| hsa_circ_0079299 | Down-regulated | N/A | CCNB1PI3K/Akt/mTOR pathway | Inhibits HCC growth | Zheng et al[107] |
| circSOD2 | Up-regulated | miR-502-5p | DNMT3A JAK2/STAT3 pathway | Promotes HCC growth, cell migration and cell cycle progression | Zhao et al[108] |
| circ_0004913 | Down-regulated | miR-184 | HAMP JAK2/STAT3/Akt pathway | Inhibits HCC proliferation, migration, invasion, EMT and glycolysis | Wu et al[109] |
| N/A | N/A | Predicts better prognosis of HCC | Li et al[150] | ||
| circ_0031242 | Up-regulated | miR-924 | POU3F2 | Enhances HCC resistance to cisplatin | Fan et al[112] |
| circARNT2 | Up-regulated | miR-155-5p | PDK1 | Promotes HCC resistance to cisplatin | Li et al[115] |
| circ-G004213 | Down-regulated | miR-513b-5p | PRPF39 | Facilitates HCC sensitivity to cisplatin | Qin et al[117] |
| circUBE2D2 | Up-regulated | miR-889-3p | LDHA | Promotes HCC resistance to sorafenib | Huang et al[121] |
| circFN1 | Up-regulated | miR-1205 | E2F1 | Facilitates HCC resistance to sorafenib | Yang et al[122] |
| circRNA-SORE | Up-regulated | RBP: YBX1 | AKT, Raf1, ERK, c-Myc, and TGF-β1 | Promotes HCC resistance to sorafenib | Xu et al[124] |
| circMEMO1 | Down-regulated | miR-106b-5p | TCF21 | Increases HCC sensitivity to sorafenib | Dong et al[126] |
| circUHRF1 | Up-regulated | miR-449C-5p | TIM-3 | Promotes HCC resistance to PD1 immunotherapy | Zhang et al[131] |
| circMET | Up-regulated | miR-30-5p | Snail/DPP4/CXCL10 axis | Promotes HCC resistance to PD1 immunotherapy | Huang et al[133] |
| Exosomal circ_0070396 | Up-regulated | N/A | N/A | Serves as a biomarker of early diagnosis of HCC | Lyu et al[139] |
| circ_104075 | Up-regulated | miR-582-3p | YAP | Serves as a biomarker of early diagnosis of HCC | Zhang et al[140] |
| has_circ _00224 and hsa_circ _00520 | Up-regulated | N/A | N/A | Serves as biomarkers of early diagnosis of HCC with HCV infection | Matboli et al[141] |
| hsa_circ_0000976 | Up-regulated | N/A | N/A | Serves as biomarkers of early diagnosis of HCC with HBV infection | Yu et al[142] |
| hsa_circ_0007750 | |||||
| hsa_circ_0139897 | |||||
| hsa_circ_0091579 | Up-regulated | N/A | GPC3 | Predicts poorer prognosis of HCC | Zhang et al[144] |
| circ_0000798 | N/A | N/A | Lei et al[145] | ||
| circ_0000267 | Up-regulated | miR-646 | N/A | Predicts poorer prognosis of HCC | Pan et al[146] |
| circASAP1 | miR-326, miR-532-5p | MAPK1 | Hu et al[147] | ||
| circ-ZNF652 | Up-regulated | miR-203/miR-502-5p | Snail-mediated EMT | Predicts poorer prognosis of HCC | Guo et al[148] |
| hsa_circ_0001649 | Down-regulated | N/A | N/A | Predicts better prognosis of HCC | Zhang et al[149] |
| circSETD3 | Down-regulated | miR-421 | MMP1 | Predicts better prognosis of HCC | Xu et al[57] |
| hsa_circ_0036683 | N/A | N/A | Sunagawa et al[151] | ||
| hsa_circ_0005986 | Down-regulated | N/A | N/A | Predicts better prognosis of HCC | Kim et al[152] |
Aberrant cell cycle regulation, uncontrolled cell proliferation and blocked apoptosis are considered the main causes of malignant tumors. Accumulating studies have highlighted the important regulatory roles of circRNAs in HCC proliferation, the cell cycle and apoptosis, among which oncogenic circRNAs accelerate HCC proliferation and suppress cell cycle arrest and apoptosis. For example, circRNA ZFR serves as an oncogene to facilitate the proliferative ability of HCC by upregulating mitogen-activated protein kinase kinase1 (MAP2K1), a promoter of tumor cell proliferation[50,51]. Similarly, c-Myc, a promoter of cell proliferation[52], and hsa_circ_0091581, as an oncogene, facilitates the proliferation of HCC cells by promoting c-Myc expression through sponging miR-526b[53]. Furthermore, TXNDC5, a promoter of tumor cell proliferation and survival[54], and circ_0000517, an oncogene in HCC, promotes tumor growth and inhibits cell cycle arrest and apoptosis by upregulating TXNDC5 through sponging miR-1296–5p[55]. Conversely, the roles of tumor suppressive circRNAs are opposite those of oncogenic circRNAs. For example, MAPK14, a suppressor of cell proliferation in HCC cells[56], and circSETD3, a tumor suppressor of HCC, enhances MAPK14 expression by sponging miR-421 in HCC, thereby inhibiting proliferation and inducing G1/S arrest[57]. Similarly, exosomal circ-0051443, another tumor suppressor of HCC, upregulates the expression of BRI1-associated kinase 1, a regulator of cell death, by sponging miR-331-3p, stimulating apoptosis and impeding the cell cycle[58,59]. The above findings reveal the importance of circRNAs in regulating HCC cell proliferation, the cell cycle and apoptosis.
EMT is an important phenomenon in the occurrence and development of tumors and can promote the migration, infiltration and metastasis of tumor cells. Invasion and metastasis of tumor cells are the main characteristics of malignant tumors and together constitute the primary cause of death in patients with malignant tumors. Elucidating their molecular mechanisms will help to develop effective interventions for cancer. Recently, many circRNAs have been reported to regulate the progression of HCC cells by affecting migration, EMT, invasion and metastasis. For example, circ-101368 promotes high-mobility group (HMG) box 1 protein/advanced glycation end products signaling by sponging miR-200a, facilitating HCC cell migration[60]. Additionally, circ-CCND1 enhances HMGA2 expression by sponging miR-497-5p, thus promoting HCC proliferation, migration and invasion[61]. Similarly, hsa_circ_0061395 upregulates the expression of PIK3R3 and SERBP1 by sponging miR-877-5p and miR-656-3p, respectively, promoting HCC proliferation, invasion and migration[62,63]. Furthermore, circRNA-103809 up-regulates the expression of FGFR1/extracellular signal-regulated kinase and PLAGL2 by sponging miR-377-3p and miR-1270, respectively, and facilitates HCC migration, EMT and invasion[64,65]. Additionally, circ_0000517, another oncogenic circRNA, is related to poor HCC prognosis[66]. Another subsequent study has investigated the possible mechanism of action of circ_0000517 by enhancing the expression of SMAD6 by sponging miR-326 to promote HCC cell invasion and metastasis[67]. Circ_matrix metalloproteinase (MMP) 2 can also promote HCC metastasis, which is the result of enhancing MMP2 expression by sponging miR-136-5p[68]. Thus, circRNAs are critical for regulating HCC migration, EMT, invasion and metastasis.
CSCs are considered the root cause of tumor occurrence, invasion, metastasis, recurrence, and resistance to radiotherapy and chemotherapy because of their self-renewal ability, sustained proliferation potential and therapeutic resistance. CircRNAs and tumor stem cells are closely related to cancer. For example, the high expression of circ-MALAT1 in HCC CSC samples mediated by RBP AU-rich binding factor 1 is closely associated with the regeneration of HCC CSCs. Mechanistically, circ-MALAT1 blocks paired box protein 5 mRNA translation on the ribosome and forms a trimer with the ribosome and mRNA to facilitate self-renewal of CSCs. This blocking mechanism is known as “circRNA braking” and has become another posttranscriptional regulatory mechanism in addition to the function of circRNA subsponges[69]. Additionally, circZKSCAN1 inhibits HCC stem cell activity by mediating the function of fragile X mental retardation protein (FMRP). Regarding the mechanism, circZKSCAN1 competes with FMRP, which serves as RBP, for the target gene cell division cycle and apoptosis regulator 1 (CCAR1), thereby inactivating the Wingless (Wnt) pathway[70]. Similarly, circMEG3 inhibits malignant differentiation of CSCs by restraining highly upregulated in liver cancer and centromere-binding factor 5 in HCC CSCs[71]. The above findings indicate that circRNAs may provide novel treatment strategies for HCC by targeting CSCs.
Aberrant glucose metabolism is the most prominent feature of tumor metabolism. In recent years, numerous studies have shown that circRNAs regulate glucose metabolism, among which oncogenic circRNAs promote glycolysis in HCC cells. For example, Forkhead box K1 (FOXK1) is an inducer of aerobic glycolysis[72], and circ-PRKCI promotes HCC glycolysis by enhancing FOXK1 expression by sponging miR-1294 and miR-186-5p[73]. Similarly, HMGA2 promotes HCC tumor growth and metastasis[74], and circZFR promotes glycolysis in HCC cells by inhibiting miR-375 and increasing HMGA2 expression[75]. Furthermore, PKM2 serves as a mediator of aerobic glycolysis of cancer cells[76], and circMAT2B facilitates HCC glycolysis by strengthening PKM2 expression by acting as a sponge of miR-338-3p[77]. Hexokinase 2 (HK2) is also a regulator of aerobic glycolysis in HCC[78], and circ-PRMT5 promotes HCC glycolysis by sponging miR-188-5p to increase HK2 expression[79]. In contrast, tumor suppressive circRNAs impede HCC glycolysis. For example, aristaless-like homeobox 4 (ALX4) inhibits HCC proliferation and invasion[80], and circ_0001445, a tumor suppressor, enhances ALX4 expression by sponging miR-942-5p, thus inhibiting HCC glycolysis[81]. Collectively, circRNAs have become important regulatory factors in glycolysis in HCC cells, but the specific mechanism of their regulation of metabolism remains to be elucidated. Considering the characteristics of circRNAs in regulating glycolysis in HCC cells, it is possible to interfere with the abnormal expression of downstream genes and some key action sites of specific circRNAs, thereby altering the metabolic pathways of HCC cells and opening up novel therapeutic approaches for HCC.
MVI is a characteristic of HCC and an independent risk factor affecting the prognosis of HCC patients. The exact mechanism by which MVI occurs in HCC has not been fully elucidated. Emerging evidence suggests that circRNAs play important roles in the MVI process of HCC. For example, ciRS-7 (Cdr1as), an oncogene in HCC[82], facilitates HCC MVI by competitively inhibiting miR-7 and interfering with the PI3Kdelta catalytic p110delta/ribosomal protein S6 kinase/mammalian target of rapamycin (mTOR) pathway[83]. Conversely, the downregulation of hsa_circ_0068669, a tumor suppressor, is correlated with HCC MVI[84]. Similarly, low expression of circSETD3, another tumor suppressor, in HCC is associated with the existence of MVI[85]. In summary, circRNAs are associated with the occurrence of MVI in HCC and can be used as indicators for the early detection of MVI and clinical intervention to reduce recurrence and improve the survival rate of patients with HCC.
HCC is a solid tumor rich in blood vessels with obvious vascular hyperplasia and vascular abnor
Abnormal circRNAs may act as tumor antigens in immunocytes to activate antitumor immunity[92]. Natural killer (NK) cells play a pivotal role in tumor immune surveillance. CircARSP91 increases the cytotoxicity of NK cells by elevating UL16-binding protein 1 in HCC, thereby enhancing innate immune surveillance[93].
The immune system monitors and kills tumor cells through specific and nonspecific pathways. When malignant cells appear in the body, the immune system recognizes and eliminates these cells specifically through the immune mechanism to resist the occurrence and development of tumors. However, in some cases, malignant cells can escape the recognition and attack of the immune system through various mechanisms to achieve immune escape in order to survive and proliferate in the body[94]. Current studies have shown that circRNAs play a critical role in tumor immune escape, which is closely associated with drug resistance and tumor recurrence[95]. For example, the low expression of tumor suppressive circTRIM33-12 promotes the immune escape ability of HCC cells by upregulating ten-eleven translocation 1 expression through sponging miR-191[96]. Similarly, hsa_circ0007456, another tumor suppressor, shows low expression in HCC and can promote tumor immune escape by regulating the expression of intercellular adhesion molecule-1 by sponging miR-6852-3p[97]. These findings indicate that circRNAs that regulate immune escape are promising immunotherapeutic targets for HCC.
Various circRNAs mediate the Wnt/beta-catenin (Wnt/β-catenin), phosphoinositide-3-kinase/protein kinase B (PI3K/Akt) or Janus kinase 2/signal transducers and activators of transcription (JAK2/Stat3) pathways by sponging miRNAs to modulate the malignant progression of HCC. In addition to circRNA-miRNA regulation, no study has investigated circRNAs modulating these signaling pathways through direct regulation of processes such as gene transcription and protein translation.
Wnt/β-catenin pathway: Aberrant activation of this pathway is prevalent in HCC occurrence and progression, and this is considered the most frequently activated carcinogenic pathway in HCC[98]. Emerging evidence suggests that circRNAs affect the malignant progression of HCC by mediating the Wnt/β-catenin pathway, among which oncogenic circRNAs can promote HCC progression by triggering the Wnt/β-catenin pathway. For example, circZFR upregulates beta-catenin 1 and activates the Wnt/β-catenin pathway by sponging miR-3619-5p to promote the proliferation and EMT of HCC cells[99]. Similarly, hsa_circ_104348 facilitates HCC proliferation, migration, and invasion by sponging miR-187-3p to elevate rhotekin 2 expression and activate the Wnt/β-catenin pathway[100]. In particular, circβ-catenin, an oncogenic circRNA in HCC, facilitates HCC cell growth by activating the Wnt/β-catenin pathway[101]. Instead, tumor suppressive circRNAs can restrain HCC progression by inhibiting the Wnt/β-catenin pathway. For example, hsa_circ_0004018 enhances Dickkopf-3 expression and inhibits the Wnt/β-catenin pathway by sponging miR-626, thereby restraining HCC proliferation and migration[102]. Similarly, circRNA-ITCH restrains the Wnt/β-catenin pathway and decreases c-myc and cyclin D1 expression by sponging miR-7 or miR-214, thereby inhibiting HCC proliferation and apoptosis[103]. Intriguingly, circ-0003418 plays a tumor suppressor role in HCC and enhances cisplatin sensitivity of HCC cells by restraining the Wnt/β-catenin pathway[104].
PI3K/Akt/mTOR pathway: Aberrant activation of this pathway frequently occurs in HCC and is closely related to HCC growth[105], invasion and metastasis. Current studies support that circRNAs mediate the PI3K/AKT or PI3K/AKT/mTOR pathway to modulate HCC progression. For example, circ-insulin-like growth factor 1 receptor promotes HCC cell proliferation by activating the PI3K/AKT pathway[106]. Additionally, the overexpression of tumor-suppressive hsa_circ_0079299 inhibits HCC growth and retards cell cycle progression partly by mediating the PI3K/Akt/mTOR pathway[107].
JAK2/STAT3 pathway: As a signal transduction pathway stimulated by cytokines, activation of the JAK/STAT pathway is closely related to tumor cell proliferation, apoptosis and differentiation. The JAK2/STAT3 pathway, an important component of the JAK/STAT pathway, is activated in diverse malignant tumors, including HCC. For example, circSOD2 enhances DNA methyltransferase 3A expression and activates the JAK2/STAT3 pathway by sponging miR-502-5p, thereby promoting the growth, migration and cell cycle progression of HCC cells[108]. Additionally, CIRC_0004913 upregulates hepcidin expression and inhibits the JAK2/STAT3/Akt pathway by sponging miR-184 and suppressing HCC proliferation, migration, invasion, EMT and glycolysis[109]. Taken together, the above findings demonstrate that circRNAs modulate the malignant progression of HCC by mediating signaling pathways, such as the Wnt/β-catenin, PI3K/Akt/mTOR and JAK2/Stat3 pathways. These pathway-associated circRNAs may serve as novel therapeutic targets in HCC.
Chemotherapy is a comprehensive treatment for advanced HCC, although the drug resistance of HCC cells considerably limits its efficacy. Multidrug resistance is the principal factor leading to the failure of chemotherapy for HCC, and its mechanism is extremely complex. Therefore, clarifying the mechanisms of drug resistance to improve the drug resistance of patients with HCC is critical. Recent evidence has prioritized the importance of abnormally expressed circRNAs in the chemotherapy resistance of HCC.
Cisplatin resistance: Cisplatin is one of the few most common chemotherapy drugs used to treat HCC. However, thus far, the drug resistance of HCC cells during chemotherapy has been revealed to be the main factor affecting chemotherapy failure[110,111]. Therefore, how to control the occurrence of cisplatin resistance in HCC cells and improve drug sensitivity and therapeutic effects are critical to prolonging the survival of patients with advanced HCC. Current studies have confirmed that circRNAs impact HCC cisplatin resistance. For example, circ_0031242 enhances cisplatin resistance in HCC by sponging miR-924 to enhance the expression of POU class 3 homeobox 2, a promoter of tumor progression and metastasis[112,113]. Additionally, pyruvate dehydrogenase kinase 1 (PDK1), a glycolytic enzyme, is closely associated with chemotherapy resistance[114]. As an oncogene, circARNT2 promotes cisplatin resistance in HCC cells, an activity mechanistically achieved by upregulating PDK1 through sponging miR-155-5p[115]. Analogously, PRPF39 is closely associated with cisplatin sensitivity[116], circ-G004213 promotes HCC cisplatin sensitivity by sponging miR-513b-5p to increase PRPR39 expression[117].
Sorafenib resistance: Sorafenib is an oral multikinase multitarget inhibitor and an important targeted therapy for advanced HCC[118]. However, sorafenib resistance is a common problem in clinical applications, substantially limiting its application[119]. The mechanism leading to sorafenib resistance remains incompletely understood. Therefore, further research on the possible mechanisms of sorafenib resistance and reducing its resistance are crucial for the treatment of HCC. CircRNAs also affect sorafenib resistance in HCC. For example, overexpression of lactate dehydrogenase A (LDHA), an oncogene, facilitates cancer cell invasion and metastasis[120]. CircUBE2D2 promotes sorafenib resistance to HCC, possibly because of the upregulation of LDHA by sponging miR-889-3p[121]. Additionally, circFN1 contributes to sorafenib resistance in HCC cells by elevating the expression of E2F1, a transcription factor associated with cancer chemotherapy resistance, by acting as a miR-1205 sponge[122,123]. Analogously, circRNA-SORE induces sorafenib resistance in HCC by binding to Y-box-binding protein 1, a regulator of EMT in cancer cells[124,125]. In particular, circMEMO1 promotes the sensitivity of HCC to sorafenib by upregulating transcription factor 21 (TCF21) expression by sponging miR-106b-5p[126].
Although the existing evidence partially reveals the critical role of circRNAs in HCC chemotherapy resistance, it suggests that circRNAs associated with chemotherapy resistance offer potential value in predicting and monitoring the efficacy of HCC and even reversing chemotherapy resistance. However, further clinical samples and in vivo experiments are needed to validate the relevant molecular mechanisms involved.
Immunotherapy is currently an effective therapeutic modality for advanced HCC. Immunotherapy enhances antigen presentation, activates the immune response and improves the immunosuppressive status of the tumor microenvironment in different ways, thus improving survival benefits. However, increasing clinical evidence indicates that only 20%-30% of patients treated with programmed death 1 (PD1) and programmed death-ligand 1 are sensitive to immunotherapy, and 70%-80% of patients show an ineffective response because of drug resistance[127]. Therefore, further exploration and under
CircRNAs are characterized by high abundance, stability and conservatism. CircRNAs are not easily degraded by RNA enzymes and stably exist in human tissues, serum, saliva and urine. Additionally, the expression profiles of circRNAs in HCC patients are significantly different from those of normal controls. Thus, abnormally expressed circRNAs may be utilized as biomarkers to diagnose and predict the prognosis of HCC patients[134-136].
There are certain limitations of commonly used clinical diagnostic markers for HCC, such as alpha-fetoprotein (AFP), AFP variants (AFP-L3) and Des-carboxy prothrombin (DCP), and only approximately 1/3 of patients can be diagnosed early[137,138]. The high mortality rate of HCC indicates that exploring new biomarkers for the early diagnosis of HCC is the most reliable strategy to improve the survival rate of HCC patients.
Emerging evidence thus far supports the possibility of utilizing circRNAs as ideal biomarkers to diagnose HCC. For example, exosome CIRC_0070396 has better diagnostic accuracy than AFP with respect to HCC patients[139]. Analogously, the sensitivity (96.0%) and specificity (98.3%) of serum circ_104075 to predict HCC are higher than those of AFP, DCP and AFP-L3, indicating the possibility of employing circ_104075 as an effective serum biomarker for HCC diagnosis[140]. Additionally, compared with AFP, hsa_circ_00224 and hsa_circ_00520 show higher sensitivity and specificity in diagnosing HCC patients with hepatitis C virus infection[141]. Furthermore, the accuracy of plasma hsa_circ_0000976, hsa_circ_0007750, and hsa_circ_0139897 is superior to AFP in diagnosing HCC patients with hepatitis B virus infection[142].
Although the existing evidence supports the feasibility of using specific circRNAs as noninvasive circulating diagnostic biomarkers for the early detection and screening of HCC, further analysis of their sensitivity and specificity and suitable patient populations is warranted. The pathogenesis of HCC is extremely complex and varies among ethnic and regional populations, and circRNAs that can be used as biomarkers in single-center studies may not be applicable to other ethnic and regional populations. Therefore, multicenter trials and large-scale studies are required to verify the performance of serum or plasma circRNAs as biomarkers. Additionally, it is necessary to establish accepted standards, unified detection and analysis methods and to use a rigorous experimental design with the best clinical samples to determine universally representative and practical diagnostic circRNA molecules.
Because of the delay in diagnosis and the high rates of postoperative recurrence and metastasis, the prognosis of HCC patients remains poor[143]. Therefore, exploring more effective HCC markers for prognosis assessment is crucial. Existing evidence has shown the feasibility of circRNAs as biomarkers to predict HCC prognosis. Among these circRNAs, oncogenic circRNAs are associated with worse overall survival (OS) or worse OS and recurrence-free survival (RFS). For example, high expression of hsa_circ_0091579 or circ_0000798 is correlated with shorter OS of HCC patients[144,145]. Similarly, high expression of circ_0000267 or circASAP1 is closely related to poorer OS in HCC patients[146,147]. Additionally, high circ-ZNF652 (hsa_circ_0003258) expression indicates shorter OS and RFS of HCC patients[148]. Conversely, tumor suppressive circRNAs are associated with better OS and RFS or better OS and progression-free survival (PFS). For example, high expression of hsa_circ_0001649 or circ_0004913 signifies longer OS in HCC patients[149,150]. Furthermore, high circSETD3 or hsa_circ_0036683 expression indicates better OS and RFS in HCC patients[57,151]. Moreover, high hsa_circ_0005986 expression implies better OS and PFS in HCC patients[152]. The above findings support the feasibility of the use of circRNAs as biomarkers for predicting HCC prognosis.
In conclusion, circRNAs play important roles in HCC and are expected to be ideal diagnostic biomarkers and therapeutic targets for HCC. However, problems persist that must be solved. First, determining the exact mechanism underlying certain circRNAs in pathogenesis is challenging because of the different nomenclatures of circRNAs, mechanisms of action and tumorigenicities. Second, current studies on circRNAs mainly focus on the function of circRNAs as molecular sponges. We should further explore the biological functions of circRNAs, such as regulating the transcription of parental genes, binding RBPs, and encoding proteins and peptides, in the context of the malignant behavior of HCC. Third, some studies have only investigated circRNA expression in HCC cell lines without detection in clinical samples, and the clinical value of such circRNAs is uncertain. Fourth, most of the studies only knocked down the expression level of circRNAs but did not perform reverse verification by overexpression of circRNAs. Fifth, presently, studies on the pathogenesis of circRNAs in HCC remain in the preliminary stage. The pathogenesis of HCC is complex and heterogeneous, and the disease states of different HCC patients may involve different primary pathogenetic pathways and pathogenic molecules. Exploring the pathogenesis of a certain class of HCC patients with stronger homogeneity at the beginning of the experimental design is crucial to obtain more reproducible conclusions. In summary, we must improve these issues to better clarify the roles and mechanisms of circRNAs in HCC so that circRNAs can become useful diagnostic indicators and therapeutic targets for HCC.
Provenance and peer review: Invited article; Externally peer reviewed.
Peer-review model: Single blind
Specialty type: Oncology
Country/Territory of origin: China
Peer-review report’s scientific quality classification
Grade A (Excellent): 0
Grade B (Very good): B
Grade C (Good): C
Grade D (Fair): 0
Grade E (Poor): 0
P-Reviewer: Moldogazieva NT, Russia A-Editor: de Melo FF, Brazil S-Editor: Wang JJ L-Editor: A P-Editor: Wang JJ
| 1. | Ghavimi S, Apfel T, Azimi H, Persaud A, Pyrsopoulos NT. Management and Treatment of Hepatocellular Carcinoma with Immunotherapy: A Review of Current and Future Options. J Clin Transl Hepatol. 2020;8:168-176. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 54] [Cited by in RCA: 43] [Article Influence: 8.6] [Reference Citation Analysis (0)] |
| 2. | Toh TB, Lim JJ, Hooi L, Rashid MBMA, Chow EK. Targeting Jak/Stat pathway as a therapeutic strategy against SP/CD44+ tumorigenic cells in Akt/β-catenin-driven hepatocellular carcinoma. J Hepatol. 2020;72:104-118. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 53] [Cited by in RCA: 96] [Article Influence: 19.2] [Reference Citation Analysis (0)] |
| 3. | Peng Z, Fang S, Jiang M, Zhao X, Zhou C, Gong Z. Circular RNAs: Regulatory functions in respiratory tract cancers. Clin Chim Acta. 2020;510:264-271. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 5] [Cited by in RCA: 1] [Article Influence: 0.2] [Reference Citation Analysis (0)] |
| 4. | Wang F, Li X, Li Z, Wang S, Fan J. Functions of Circular RNAs in Regulating Adipogenesis of Mesenchymal Stem Cells. Stem Cells Int. 2020;2020:3763069. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 6] [Cited by in RCA: 5] [Article Influence: 1.0] [Reference Citation Analysis (0)] |
| 5. | Momen-Heravi F, Bala S. Emerging role of non-coding RNA in oral cancer. Cell Signal. 2018;42:134-143. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 90] [Cited by in RCA: 124] [Article Influence: 17.7] [Reference Citation Analysis (0)] |
| 6. | Chen L, Shan G. CircRNA in cancer: Fundamental mechanism and clinical potential. Cancer Lett. 2021;505:49-57. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 41] [Cited by in RCA: 312] [Article Influence: 78.0] [Reference Citation Analysis (0)] |
| 7. | Chen Q, Chen Z, Cao S, Guo B, Chen Y, Feng Z, Wang J, Guo G, Chen X, Huang X. Role of CircRNAs_100395 in Proliferation and Metastases of Liver Cancer. Med Sci Monit. 2019;25:6181-6192. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 16] [Cited by in RCA: 23] [Article Influence: 3.8] [Reference Citation Analysis (0)] |
| 8. | Zhang X, Zhang Q, Zhang K, Wang F, Qiao X, Cui J. Circ SMARCA5 Inhibited Tumor Metastasis by Interacting with SND1 and Downregulating the YWHAB Gene in Cervical Cancer. Cell Transplant. 2021;30:963689720983786. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 16] [Cited by in RCA: 21] [Article Influence: 5.3] [Reference Citation Analysis (0)] |
| 9. | Xiao MS, Ai Y, Wilusz JE. Biogenesis and Functions of Circular RNAs Come into Focus. Trends Cell Biol. 2020;30:226-240. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 137] [Cited by in RCA: 240] [Article Influence: 48.0] [Reference Citation Analysis (0)] |
| 10. | Enuka Y, Lauriola M, Feldman ME, Sas-Chen A, Ulitsky I, Yarden Y. Circular RNAs are long-lived and display only minimal early alterations in response to a growth factor. Nucleic Acids Res. 2016;44:1370-1383. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 318] [Cited by in RCA: 527] [Article Influence: 52.7] [Reference Citation Analysis (0)] |
| 11. | Jeck WR, Sorrentino JA, Wang K, Slevin MK, Burd CE, Liu J, Marzluff WF, Sharpless NE. Circular RNAs are abundant, conserved, and associated with ALU repeats. RNA. 2013;19:141-157. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 3481] [Cited by in RCA: 3419] [Article Influence: 284.9] [Reference Citation Analysis (0)] |
| 12. | Hallajzadeh J, Amirani E, Mirzaei H, Shafabakhsh R, Mirhashemi SM, Sharifi M, Yousefi B, Mansournia MA, Asemi Z. Circular RNAs: new genetic tools in melanoma. Biomark Med. 2020;14:563-571. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 10] [Cited by in RCA: 16] [Article Influence: 3.2] [Reference Citation Analysis (0)] |
| 13. | Zhao X, Cai Y, Xu J. Circular RNAs: Biogenesis, Mechanism, and Function in Human Cancers. Int J Mol Sci. 2019;20. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 118] [Cited by in RCA: 177] [Article Influence: 29.5] [Reference Citation Analysis (0)] |
| 14. | Li P, Zhu K, Mo Y, Deng X, Jiang X, Shi L, Guo C, Zhang W, Zeng Z, Li G, Xiong W, Zhang S, Gong Z. Research Progress of circRNAs in Head and Neck Cancers. Front Oncol. 2021;11:616202. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 5] [Cited by in RCA: 8] [Article Influence: 2.0] [Reference Citation Analysis (0)] |
| 15. | Lasda E, Parker R. Circular RNAs Co-Precipitate with Extracellular Vesicles: A Possible Mechanism for circRNA Clearance. PLoS One. 2016;11:e0148407. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 243] [Cited by in RCA: 321] [Article Influence: 35.7] [Reference Citation Analysis (0)] |
| 16. | Li Z, Huang C, Bao C, Chen L, Lin M, Wang X, Zhong G, Yu B, Hu W, Dai L, Zhu P, Chang Z, Wu Q, Zhao Y, Jia Y, Xu P, Liu H, Shan G. Exon-intron circular RNAs regulate transcription in the nucleus. Nat Struct Mol Biol. 2015;22:256-264. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1619] [Cited by in RCA: 2221] [Article Influence: 222.1] [Reference Citation Analysis (0)] |
| 17. | Zhang Y, Zhang XO, Chen T, Xiang JF, Yin QF, Xing YH, Zhu S, Yang L, Chen LL. Circular intronic long noncoding RNAs. Mol Cell. 2013;51:792-806. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1810] [Cited by in RCA: 1753] [Article Influence: 146.1] [Reference Citation Analysis (0)] |
| 18. | Cheng D, Wang J, Dong Z, Li X. Cancer-related circular RNA: diverse biological functions. Cancer Cell Int. 2021;21:11. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 23] [Cited by in RCA: 54] [Article Influence: 13.5] [Reference Citation Analysis (0)] |
| 19. | Kristensen LS, Andersen MS, Stagsted LVW, Ebbesen KK, Hansen TB, Kjems J. The biogenesis, biology and characterization of circular RNAs. Nat Rev Genet. 2019;20:675-691. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 3273] [Cited by in RCA: 3149] [Article Influence: 524.8] [Reference Citation Analysis (0)] |
| 20. | Sun Z, Chen C, Su Y, Wang W, Yang S, Zhou Q, Wang G, Li Z, Song J, Zhang Z, Yuan W, Liu J. Regulatory mechanisms and clinical perspectives of circRNA in digestive system neoplasms. J Cancer. 2019;10:2885-2891. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 20] [Cited by in RCA: 31] [Article Influence: 5.2] [Reference Citation Analysis (0)] |
| 21. | Xia L, Song M, Sun M, Wang F, Yang C. Circular RNAs as Biomarkers for Cancer. Adv Exp Med Biol. 2018;1087:171-187. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 19] [Cited by in RCA: 25] [Article Influence: 3.6] [Reference Citation Analysis (0)] |
| 22. | Wei X, Shi Y, Dai Z, Wang P, Meng X, Yin B. Underlying metastasis mechanism and clinical application of exosomal circular RNA in tumors (Review). Int J Oncol. 2021;58:289-297. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 6] [Cited by in RCA: 23] [Article Influence: 5.8] [Reference Citation Analysis (0)] |
| 23. | Harper KL, Mcdonnell E, Whitehouse A. CircRNAs: From anonymity to novel regulators of gene expression in cancer (Review). Int J Oncol. 2019;55:1183-1193. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 6] [Cited by in RCA: 8] [Article Influence: 1.3] [Reference Citation Analysis (0)] |
| 24. | Solé C, Mentxaka G, Lawrie CH. The Use of circRNAs as Biomarkers of Cancer. Methods Mol Biol. 2021;2348:307-341. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 7] [Cited by in RCA: 18] [Article Influence: 4.5] [Reference Citation Analysis (0)] |
| 25. | Shen L, Zhang Y, Zhou W, Peng Z, Hong X. Circular RNA expression in ovarian endometriosis. Epigenomics. 2018;10:559-572. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 19] [Cited by in RCA: 25] [Article Influence: 3.6] [Reference Citation Analysis (0)] |
| 26. | Memczak S, Jens M, Elefsinioti A, Torti F, Krueger J, Rybak A, Maier L, Mackowiak SD, Gregersen LH, Munschauer M, Loewer A, Ziebold U, Landthaler M, Kocks C, le Noble F, Rajewsky N. Circular RNAs are a large class of animal RNAs with regulatory potency. Nature. 2013;495:333-338. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 6253] [Cited by in RCA: 6024] [Article Influence: 502.0] [Reference Citation Analysis (0)] |
| 27. | Dou Z, Gao L, Ren W, Zhang H, Wang X, Li S, Zheng J, Kong X, Chi P, Zhi K. CiRS-7 functions as a ceRNA of RAF-1/PIK3CD to promote metastatic progression of oral squamous cell carcinoma via MAPK/AKT signaling pathways. Exp Cell Res. 2020;396:112290. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 13] [Cited by in RCA: 26] [Article Influence: 5.2] [Reference Citation Analysis (0)] |
| 28. | Su C, Han Y, Zhang H, Li Y, Yi L, Wang X, Zhou S, Yu D, Song X, Xiao N, Cao X, Liu Z. CiRS-7 targeting miR-7 modulates the progression of non-small cell lung cancer in a manner dependent on NF-κB signalling. J Cell Mol Med. 2018;22:3097-3107. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 71] [Cited by in RCA: 107] [Article Influence: 15.3] [Reference Citation Analysis (0)] |
| 29. | Misir S, Hepokur C, Aliyazicioglu Y, Enguita FJ. Circular RNAs serve as miRNA sponges in breast cancer. Breast Cancer. 2020;27:1048-1057. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 14] [Cited by in RCA: 30] [Article Influence: 6.0] [Reference Citation Analysis (0)] |
| 30. | Piwecka M, Glažar P, Hernandez-Miranda LR, Memczak S, Wolf SA, Rybak-Wolf A, Filipchyk A, Klironomos F, Cerda Jara CA, Fenske P, Trimbuch T, Zywitza V, Plass M, Schreyer L, Ayoub S, Kocks C, Kühn R, Rosenmund C, Birchmeier C, Rajewsky N. Loss of a mammalian circular RNA locus causes miRNA deregulation and affects brain function. Science. 2017;357. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 457] [Cited by in RCA: 332] [Article Influence: 23.7] [Reference Citation Analysis (0)] |
| 31. | Aufiero S, Reckman YJ, Pinto YM, Creemers EE. Circular RNAs open a new chapter in cardiovascular biology. Nat Rev Cardiol. 2019;16:503-514. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 163] [Cited by in RCA: 267] [Article Influence: 44.5] [Reference Citation Analysis (0)] |
| 32. | Sarkar D, Diermeier SD. Circular RNAs: Potential Applications as Therapeutic Targets and Biomarkers in Breast Cancer. Noncoding RNA. 2021;7. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 13] [Cited by in RCA: 16] [Article Influence: 4.0] [Reference Citation Analysis (0)] |
| 33. | Hsiao KY, Sun HS, Tsai SJ. Circular RNA - New member of noncoding RNA with novel functions. Exp Biol Med (Maywood). 2017;242:1136-1141. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 252] [Cited by in RCA: 330] [Article Influence: 41.3] [Reference Citation Analysis (0)] |
| 34. | Du WW, Yang W, Liu E, Yang Z, Dhaliwal P, Yang BB. Foxo3 circular RNA retards cell cycle progression via forming ternary complexes with p21 and CDK2. Nucleic Acids Res. 2016;44:2846-2858. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 955] [Cited by in RCA: 1303] [Article Influence: 144.8] [Reference Citation Analysis (0)] |
| 35. | Zhou C, Molinie B, Daneshvar K, Pondick JV, Wang J, Van Wittenberghe N, Xing Y, Giallourakis CC, Mullen AC. Genome-Wide Maps of m6A circRNAs Identify Widespread and Cell-Type-Specific Methylation Patterns that Are Distinct from mRNAs. Cell Rep. 2017;20:2262-2276. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 282] [Cited by in RCA: 343] [Article Influence: 42.9] [Reference Citation Analysis (0)] |
| 36. | Dell’Orco M, Oliver RJ, Perrone-Bizzozero N. HuD Binds to and Regulates Circular RNAs Derived From Neuronal Development- and Synaptic Plasticity-Associated Genes. Front Genet. 2020;11:790. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 12] [Cited by in RCA: 31] [Article Influence: 6.2] [Reference Citation Analysis (0)] |
| 37. | Wang Z, Lei X, Wu FX. Identifying Cancer-Specific circRNA-RBP Binding Sites Based on Deep Learning. Molecules. 2019;24. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 44] [Cited by in RCA: 54] [Article Influence: 9.0] [Reference Citation Analysis (0)] |
| 38. | García-Mauriño SM, Rivero-Rodríguez F, Velázquez-Cruz A, Hernández-Vellisca M, Díaz-Quintana A, De la Rosa MA, Díaz-Moreno I. RNA Binding Protein Regulation and Cross-Talk in the Control of AU-rich mRNA Fate. Front Mol Biosci. 2017;4:71. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 133] [Cited by in RCA: 130] [Article Influence: 16.3] [Reference Citation Analysis (0)] |
| 39. | Grammatikakis I, Abdelmohsen K, Gorospe M. Posttranslational control of HuR function. Wiley Interdiscip Rev RNA. 2017;8. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 130] [Cited by in RCA: 189] [Article Influence: 21.0] [Reference Citation Analysis (0)] |
| 40. | Levidou G, Kotta-Loizou I, Tasoulas J, Papadopoulos T, Theocharis S. Clinical Significance and Biological Role of HuR in Head and Neck Carcinomas. Dis Markers. 2018;2018:4020937. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 12] [Cited by in RCA: 15] [Article Influence: 2.1] [Reference Citation Analysis (0)] |
| 41. | Zhang M, Xu Y, Zhang Y, Li B, Lou G. Circular RNA circE2F2 promotes malignant progression of ovarian cancer cells by upregulating the expression of E2F2 protein via binding to HuR protein. Cell Signal. 2021;84:110014. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 9] [Cited by in RCA: 24] [Article Influence: 6.0] [Reference Citation Analysis (0)] |
| 42. | Chen J, Wu Y, Luo X, Jin D, Zhou W, Ju Z, Wang D, Meng Q, Wang H, Fu X, Xu J, Song Z. Circular RNA circRHOBTB3 represses metastasis by regulating the HuR-mediated mRNA stability of PTBP1 in colorectal cancer. Theranostics. 2021;11:7507-7526. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 27] [Cited by in RCA: 132] [Article Influence: 33.0] [Reference Citation Analysis (0)] |
| 43. | Liu B, Yang G, Wang X, Liu J, Lu Z, Wang Q, Xu B, Liu Z, Li J. CircBACH1 (hsa_circ_0061395) promotes hepatocellular carcinoma growth by regulating p27 repression via HuR. J Cell Physiol. 2020;235:6929-6941. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 32] [Cited by in RCA: 63] [Article Influence: 12.6] [Reference Citation Analysis (0)] |
| 44. | Wesselhoeft RA, Kowalski PS, Anderson DG. Engineering circular RNA for potent and stable translation in eukaryotic cells. Nat Commun. 2018;9:2629. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 220] [Cited by in RCA: 513] [Article Influence: 73.3] [Reference Citation Analysis (0)] |
| 45. | Diallo LH, Tatin F, David F, Godet AC, Zamora A, Prats AC, Garmy-Susini B, Lacazette E. How are circRNAs translated by non-canonical initiation mechanisms? Biochimie. 2019;164:45-52. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 33] [Cited by in RCA: 60] [Article Influence: 10.0] [Reference Citation Analysis (0)] |
| 46. | Prats AC, David F, Diallo LH, Roussel E, Tatin F, Garmy-Susini B, Lacazette E. Circular RNA, the Key for Translation. Int J Mol Sci. 2020;21. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 81] [Cited by in RCA: 90] [Article Influence: 18.0] [Reference Citation Analysis (0)] |
| 47. | Yang Y, Fan X, Mao M, Song X, Wu P, Zhang Y, Jin Y, Yang Y, Chen LL, Wang Y, Wong CC, Xiao X, Wang Z. Extensive translation of circular RNAs driven by N6-methyladenosine. Cell Res. 2017;27:626-641. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 960] [Cited by in RCA: 1403] [Article Influence: 175.4] [Reference Citation Analysis (0)] |
| 48. | Chekulaeva M, Rajewsky N. Roles of Long Noncoding RNAs and Circular RNAs in Translation. Cold Spring Harb Perspect Biol. 2019;11. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 26] [Cited by in RCA: 36] [Article Influence: 6.0] [Reference Citation Analysis (0)] |
| 49. | Shi Y, Jia X, Xu J. The new function of circRNA: translation. Clin Transl Oncol. 2020;22:2162-2169. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 53] [Cited by in RCA: 133] [Article Influence: 26.6] [Reference Citation Analysis (0)] |
| 50. | Cedric BC, Souraka TDM, Feng YL, Kisembo P, Tu JC. CircRNA ZFR stimulates the proliferation of hepatocellular carcinoma through upregulating MAP2K1. Eur Rev Med Pharmacol Sci. 2020;24:9924-9931. [RCA] [PubMed] [DOI] [Full Text] [Cited by in RCA: 6] [Reference Citation Analysis (0)] |
| 51. | Shneider BL, Cortes-Santiago N, Schady DA, Krishnamoorthy S, Thevananther S, Rajapakshe K, Perera D, Huang S, Coarfa C. Constitutive activation of mitogen-activated protein kinase kinase (MEK1) in ileal enterocytes leads to dysplasia and a predisposition to cancer. Am J Physiol Gastrointest Liver Physiol. 2021;320:G366-G379. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1] [Cited by in RCA: 1] [Article Influence: 0.3] [Reference Citation Analysis (0)] |
| 52. | Mastronikolis N, Ragos V, Kyrodimos E, Chrysovergis A, Papanikolaou V, Mastronikolis S, Stamatelopoulos A, Tsiambas E. Mechanisms of C-myc oncogenic activity in head and neck squamous cell carcinoma. J BUON. 2019;24:2242-2244. [PubMed] |
| 53. | Wei X, Zheng W, Tian P, He Y, Liu H, Peng M, Li X, Liu X. Oncogenic hsa_circ_0091581 promotes the malignancy of HCC cell through blocking miR-526b from degrading c-MYC mRNA. Cell Cycle. 2020;19:817-824. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 23] [Cited by in RCA: 41] [Article Influence: 8.2] [Reference Citation Analysis (0)] |
| 54. | Chawsheen HA, Ying Q, Jiang H, Wei Q. A critical role of the thioredoxin domain containing protein 5 (TXNDC5) in redox homeostasis and cancer development. Genes Dis. 2018;5:312-322. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 21] [Cited by in RCA: 44] [Article Influence: 6.3] [Reference Citation Analysis (0)] |
| 55. | Zang H, Li Y, Zhang X, Huang G. Circ_0000517 Contributes to Hepatocellular Carcinoma Progression by Upregulating TXNDC5 via Sponging miR-1296-5p. Cancer Manag Res. 2020;12:3457-3468. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 16] [Cited by in RCA: 35] [Article Influence: 7.0] [Reference Citation Analysis (0)] |
| 56. | Fang Z, Wu L, Dai H, Hu P, Wang B, Han Q, Xu Y, Lv S, Zhu Y, Gan M, Zhou W, Zhang W. The role of vesicular overexpressed in cancer pro-survival protein 1 in hepatocellular carcinoma proliferation. Cancer Biomark. 2020;28:9-20. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 5] [Cited by in RCA: 6] [Article Influence: 1.5] [Reference Citation Analysis (0)] |
| 57. | Xu L, Feng X, Hao X, Wang P, Zhang Y, Zheng X, Li L, Ren S, Zhang M, Xu M. CircSETD3 (Hsa_circ_0000567) acts as a sponge for microRNA-421 inhibiting hepatocellular carcinoma growth. J Exp Clin Cancer Res. 2019;38:98. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 143] [Cited by in RCA: 153] [Article Influence: 25.5] [Reference Citation Analysis (0)] |
| 58. | Chen W, Quan Y, Fan S, Wang H, Liang J, Huang L, Chen L, Liu Q, He P, Ye Y. Exosome-transmitted circular RNA hsa_circ_0051443 suppresses hepatocellular carcinoma progression. Cancer Lett. 2020;475:119-128. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 121] [Cited by in RCA: 213] [Article Influence: 42.6] [Reference Citation Analysis (0)] |
| 59. | Kim SY, Shang Y, Joo SH, Kim SK, Nam KH. Overexpression of BAK1 causes salicylic acid accumulation and deregulation of cell death control genes. Biochem Biophys Res Commun. 2017;484:781-786. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 11] [Cited by in RCA: 11] [Article Influence: 1.4] [Reference Citation Analysis (0)] |
| 60. | Li S, Gu H, Huang Y, Peng Q, Zhou R, Yi P, Chen R, Huang Z, Hu X, Tang D. Circular RNA 101368/miR-200a axis modulates the migration of hepatocellular carcinoma through HMGB1/RAGE signaling. Cell Cycle. 2018;17:2349-2359. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 40] [Cited by in RCA: 54] [Article Influence: 7.7] [Reference Citation Analysis (0)] |
| 61. | Zheng S, Hou J, Chang Y, Zhao D, Yang H, Yang J. CircRNA Circ-CCND1 Aggravates Hepatocellular Carcinoma Tumorigenesis by Regulating the miR-497-5p/HMGA2 Axis. Mol Biotechnol. 2022;64:178-186. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 2] [Cited by in RCA: 9] [Article Influence: 2.3] [Reference Citation Analysis (0)] |
| 62. | Yu Y, Bian L, Liu R, Wang Y, Xiao X. Circular RNA hsa_circ_0061395 accelerates hepatocellular carcinoma progression via regulation of the miR-877-5p/PIK3R3 axis. Cancer Cell Int. 2021;21:10. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 18] [Cited by in RCA: 36] [Article Influence: 9.0] [Reference Citation Analysis (0)] |
| 63. | Li G, Du P, He J, Li Y. CircRNA circBACH1 (hsa_circ_0061395) serves as a miR-656-3p sponge to facilitate hepatocellular carcinoma progression through increasing SERBP1 expression. Biochem Biophys Res Commun. 2021;556:1-8. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 4] [Cited by in RCA: 13] [Article Influence: 3.3] [Reference Citation Analysis (0)] |
| 64. | Zhan W, Liao X, Chen Z, Li L, Tian T, Yu L, Wang W, Hu Q. Circular RNA hsa_circRNA_103809 promoted hepatocellular carcinoma development by regulating miR-377-3p/FGFR1/ERK axis. J Cell Physiol. 2020;235:1733-1745. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 53] [Cited by in RCA: 71] [Article Influence: 11.8] [Reference Citation Analysis (0)] |
| 65. | Cao Y, Tao Q, Kao X, Zhu X. Hsa-circRNA-103809 Promotes Hepatocellular Carcinoma Development via MicroRNA-1270/PLAG1 Like Zinc Finger 2 Axis. Dig Dis Sci. 2021;66:1524-1532. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 11] [Cited by in RCA: 20] [Article Influence: 5.0] [Reference Citation Analysis (0)] |
| 66. | Wang X, Wang X, Li W, Zhang Q, Chen J, Chen T. Up-Regulation of hsa_circ_0000517 Predicts Adverse Prognosis of Hepatocellular Carcinoma. Front Oncol. 2019;9:1105. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 37] [Cited by in RCA: 35] [Article Influence: 5.8] [Reference Citation Analysis (0)] |
| 67. | He S, Guo Z, Kang Q, Wang X, Han X. Circular RNA hsa_circ_0000517 modulates hepatocellular carcinoma advancement via the miR-326/SMAD6 axis. Cancer Cell Int. 2020;20:360. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 21] [Cited by in RCA: 22] [Article Influence: 4.4] [Reference Citation Analysis (0)] |
| 68. | Liu D, Kang H, Gao M, Jin L, Zhang F, Chen D, Li M, Xiao L. Exosome-transmitted circ_MMP2 promotes hepatocellular carcinoma metastasis by upregulating MMP2. Mol Oncol. 2020;14:1365-1380. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 44] [Cited by in RCA: 100] [Article Influence: 20.0] [Reference Citation Analysis (0)] |
| 69. | Chen L, Kong R, Wu C, Wang S, Liu Z, Liu S, Li S, Chen T, Mao C. Circ-MALAT1 Functions as Both an mRNA Translation Brake and a microRNA Sponge to Promote Self-Renewal of Hepatocellular Cancer Stem Cells. Adv Sci (Weinh). 2020;7:1900949. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 75] [Cited by in RCA: 78] [Article Influence: 15.6] [Reference Citation Analysis (0)] |
| 70. | Zhu YJ, Zheng B, Luo GJ, Ma XK, Lu XY, Lin XM, Yang S, Zhao Q, Wu T, Li ZX, Liu XL, Wu R, Liu JF, Ge Y, Yang L, Wang HY, Chen L. Circular RNAs negatively regulate cancer stem cells by physically binding FMRP against CCAR1 complex in hepatocellular carcinoma. Theranostics. 2019;9:3526-3540. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 189] [Cited by in RCA: 201] [Article Influence: 33.5] [Reference Citation Analysis (0)] |
| 71. | Jiang X, Xing L, Chen Y, Qin R, Song S, Lu Y, Xie S, Wang L, Pu H, Gui X, Li T, Xu J, Li J, Jia S, Lu D. CircMEG3 inhibits telomerase activity by reducing Cbf5 in human liver cancer stem cells. Mol Ther Nucleic Acids. 2021;23:310-323. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 19] [Cited by in RCA: 36] [Article Influence: 7.2] [Reference Citation Analysis (0)] |
| 72. | Sukonina V, Ma H, Zhang W, Bartesaghi S, Subhash S, Heglind M, Foyn H, Betz MJ, Nilsson D, Lidell ME, Naumann J, Haufs-Brusberg S, Palmgren H, Mondal T, Beg M, Jedrychowski MP, Taskén K, Pfeifer A, Peng XR, Kanduri C, Enerbäck S. FOXK1 and FOXK2 regulate aerobic glycolysis. Nature. 2019;566:279-283. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 60] [Cited by in RCA: 132] [Article Influence: 22.0] [Reference Citation Analysis (0)] |
| 73. | Chen W, Li Y, Zhong J, Wen G. circ-PRKCI targets miR-1294 and miR-186-5p by downregulating FOXK1 expression to suppress glycolysis in hepatocellular carcinoma. Mol Med Rep. 2021;23. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 8] [Cited by in RCA: 22] [Article Influence: 5.5] [Reference Citation Analysis (0)] |
| 74. | Cui H, Song R, Wu J, Wang W, Chen X, Yin J. MicroRNA-337 regulates the PI3K/AKT and Wnt/β-catenin signaling pathways to inhibit hepatocellular carcinoma progression by targeting high-mobility group AT-hook 2. Am J Cancer Res. 2018;8:405-421. [PubMed] |
| 75. | Xu R, Yin S, Zheng M, Pei X, Ji X. Circular RNA circZFR Promotes Hepatocellular Carcinoma Progression by Regulating miR-375/HMGA2 Axis. Dig Dis Sci. 2021;66:4361-4373. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 9] [Cited by in RCA: 19] [Article Influence: 4.8] [Reference Citation Analysis (0)] |
| 76. | Fu J, Xiong Z, Huang C, Li J, Yang W, Han Y, Paiboonrungruan C, Major MB, Chen KN, Kang X, Chen X. Hyperactivity of the transcription factor Nrf2 causes metabolic reprogramming in mouse esophagus. J Biol Chem. 2019;294:327-340. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 64] [Cited by in RCA: 69] [Article Influence: 11.5] [Reference Citation Analysis (0)] |
| 77. | Li Q, Pan X, Zhu D, Deng Z, Jiang R, Wang X. Circular RNA MAT2B Promotes Glycolysis and Malignancy of Hepatocellular Carcinoma Through the miR-338-3p/PKM2 Axis Under Hypoxic Stress. Hepatology. 2019;70:1298-1316. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 237] [Cited by in RCA: 220] [Article Influence: 36.7] [Reference Citation Analysis (0)] |
| 78. | Liu Z, Ning F, Cai Y, Sheng H, Zheng R, Yin X, Lu Z, Su L, Chen X, Zeng C, Wang H, Liu L. The EGFR-P38 MAPK axis up-regulates PD-L1 through miR-675-5p and down-regulates HLA-ABC via hexokinase-2 in hepatocellular carcinoma cells. Cancer Commun (Lond). 2021;41:62-78. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 27] [Cited by in RCA: 34] [Article Influence: 8.5] [Reference Citation Analysis (0)] |
| 79. | Ding Z, Guo L, Deng Z, Li P. Circ-PRMT5 enhances the proliferation, migration and glycolysis of hepatoma cells by targeting miR-188-5p/HK2 axis. Ann Hepatol. 2020;19:269-279. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 25] [Cited by in RCA: 41] [Article Influence: 10.3] [Reference Citation Analysis (0)] |
| 80. | Zhao J, Chen HQ, Yang HF, Li Y, Chen DJ, Huang YJ, He LX, Zheng CF, Wang LQ, Wang J, Zhang N, Cao J, Liu JY, Shu WQ, Liu WB. Epigenetic silencing of ALX4 regulates microcystin-LR induced hepatocellular carcinoma through the P53 pathway. Sci Total Environ. 2019;683:317-330. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 16] [Cited by in RCA: 22] [Article Influence: 3.7] [Reference Citation Analysis (0)] |
| 81. | Xu Q, Zhou L, Yang G, Meng F, Wan Y, Wang L, Zhang L. Overexpression of circ_0001445 decelerates hepatocellular carcinoma progression by regulating miR-942-5p/ALX4 axis. Biotechnol Lett. 2020;42:2735-2747. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 8] [Cited by in RCA: 14] [Article Influence: 2.8] [Reference Citation Analysis (0)] |
| 82. | Chen J, Yang J, Fei X, Wang X, Wang K. CircRNA ciRS-7: a Novel Oncogene in Multiple Cancers. Int J Biol Sci. 2021;17:379-389. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 17] [Cited by in RCA: 56] [Article Influence: 14.0] [Reference Citation Analysis (0)] |
| 83. | Xu L, Zhang M, Zheng X, Yi P, Lan C, Xu M. The circular RNA ciRS-7 (Cdr1as) acts as a risk factor of hepatic microvascular invasion in hepatocellular carcinoma. J Cancer Res Clin Oncol. 2017;143:17-27. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 233] [Cited by in RCA: 259] [Article Influence: 32.4] [Reference Citation Analysis (0)] |
| 84. | Yao T, Chen Q, Shao Z, Song Z, Fu L, Xiao B. Circular RNA 0068669 as a new biomarker for hepatocellular carcinoma metastasis. J Clin Lab Anal. 2018;32:e22572. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 27] [Cited by in RCA: 34] [Article Influence: 4.9] [Reference Citation Analysis (0)] |
| 85. | Wang P, Xu LL, Zheng XB, Hu YT, Zhang JF, Ren SS, Hao XY, Li L, Zhang M, Xu MQ. Correlation between the expressions of circular RNAs in peripheral venous blood and clinicopathological features in hepatocellular carcinoma. Ann Transl Med. 2020;8:338. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 9] [Cited by in RCA: 13] [Article Influence: 2.6] [Reference Citation Analysis (0)] |
| 86. | Ji Y, Yang S, Yan X, Zhu L, Yang W, Yang X, Yu F, Shi L, Zhu X, Lu Y, Zhang C, Lu H, Zhang F. CircCRIM1 Promotes Hepatocellular Carcinoma Proliferation and Angiogenesis by Sponging miR-378a-3p and Regulating SKP2 Expression. Front Cell Dev Biol. 2021;9:796686. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 5] [Cited by in RCA: 21] [Article Influence: 5.3] [Reference Citation Analysis (0)] |
| 87. | Zhai Z, Fu Q, Liu C, Zhang X, Jia P, Xia P, Liu P, Liao S, Qin T, Zhang H. Emerging Roles Of hsa-circ-0046600 Targeting The miR-640/HIF-1α Signalling Pathway In The Progression Of HCC. Onco Targets Ther. 2019;12:9291-9302. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 20] [Cited by in RCA: 33] [Article Influence: 5.5] [Reference Citation Analysis (0)] |
| 88. | Farhadi P, Yarani R, Kiani S, Mansouri K. Perfluorocarbon as an adjuvant for tumor anti-angiogenic therapy: Relevance to hypoxia and HIF-1. Med Hypotheses. 2021;146:110357. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 2] [Cited by in RCA: 6] [Article Influence: 1.2] [Reference Citation Analysis (0)] |
| 89. | Pu J, Wang J, Li W, Lu Y, Wu X, Long X, Luo C, Wei H. hsa_circ_0000092 promotes hepatocellular carcinoma progression through up-regulating HN1 expression by binding to microRNA-338-3p. J Cell Mol Med. 2020;. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 37] [Cited by in RCA: 43] [Article Influence: 43.0] [Reference Citation Analysis (0)] |
| 90. | Liu Z, Yang D, Li Y, Jiao Y, Lv G. HN1 as a diagnostic and prognostic biomarker for liver cancer. Biosci Rep. 2020;40. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 9] [Cited by in RCA: 8] [Article Influence: 1.6] [Reference Citation Analysis (0)] |
| 91. | Yu YX, Ge TW, Zhang P. Circular RNA circGFRA1 promotes angiogenesis, cell proliferation and migration of hepatocellular carcinoma by combining with miR-149. Eur Rev Med Pharmacol Sci. 2020;24:11058-11064. [RCA] [PubMed] [DOI] [Full Text] [Cited by in RCA: 9] [Reference Citation Analysis (0)] |
| 92. | Xu Z, Li P, Fan L, Wu M. The Potential Role of circRNA in Tumor Immunity Regulation and Immunotherapy. Front Immunol. 2018;9:9. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 58] [Cited by in RCA: 106] [Article Influence: 15.1] [Reference Citation Analysis (0)] |
| 93. | Ma Y, Zhang C, Zhang B, Yu H, Yu Q. circRNA of AR-suppressed PABPC1 91 bp enhances the cytotoxicity of natural killer cells against hepatocellular carcinoma via upregulating UL16 binding protein 1. Oncol Lett. 2019;17:388-397. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 17] [Cited by in RCA: 42] [Article Influence: 6.0] [Reference Citation Analysis (0)] |
| 94. | Schreiber RD, Old LJ, Smyth MJ. Cancer immunoediting: integrating immunity's roles in cancer suppression and promotion. Science. 2011;331:1565-1570. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 4947] [Cited by in RCA: 4541] [Article Influence: 324.4] [Reference Citation Analysis (0)] |
| 95. | Bourquin C, Pommier A, Hotz C. Harnessing the immune system to fight cancer with Toll-like receptor and RIG-I-like receptor agonists. Pharmacol Res. 2020;154:104192. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 36] [Cited by in RCA: 47] [Article Influence: 9.4] [Reference Citation Analysis (0)] |
| 96. | Zhang PF, Wei CY, Huang XY, Peng R, Yang X, Lu JC, Zhang C, Gao C, Cai JB, Gao PT, Gao DM, Shi GM, Ke AW, Fan J. Circular RNA circTRIM33-12 acts as the sponge of MicroRNA-191 to suppress hepatocellular carcinoma progression. Mol Cancer. 2019;18:105. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 170] [Cited by in RCA: 173] [Article Influence: 28.8] [Reference Citation Analysis (0)] |
| 97. | Shi M, Li ZY, Zhang LM, Wu XY, Xiang SH, Wang YG, Zhang YQ. Hsa_circ_0007456 regulates the natural killer cell-mediated cytotoxicity toward hepatocellular carcinoma via the miR-6852-3p/ICAM-1 axis. Cell Death Dis. 2021;12:94. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 45] [Cited by in RCA: 50] [Article Influence: 12.5] [Reference Citation Analysis (0)] |
| 98. | Asaoka Y, Tanaka A. Clinical implications of WNT/β-catenin signaling for hepatocellular carcinoma. Glob Health Med. 2020;2:269-272. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 4] [Cited by in RCA: 4] [Article Influence: 0.8] [Reference Citation Analysis (0)] |
| 99. | Tan A, Li Q, Chen L. CircZFR promotes hepatocellular carcinoma progression through regulating miR-3619-5p/CTNNB1 axis and activating Wnt/β-catenin pathway. Arch Biochem Biophys. 2019;661:196-202. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 56] [Cited by in RCA: 84] [Article Influence: 12.0] [Reference Citation Analysis (0)] |
| 100. | Huang G, Liang M, Liu H, Huang J, Li P, Wang C, Zhang Y, Lin Y, Jiang X. CircRNA hsa_circRNA_104348 promotes hepatocellular carcinoma progression through modulating miR-187-3p/RTKN2 axis and activating Wnt/β-catenin pathway. Cell Death Dis. 2020;11:1065. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 54] [Cited by in RCA: 242] [Article Influence: 48.4] [Reference Citation Analysis (0)] |
| 101. | Liang WC, Wong CW, Liang PP, Shi M, Cao Y, Rao ST, Tsui SK, Waye MM, Zhang Q, Fu WM, Zhang JF. Translation of the circular RNA circβ-catenin promotes liver cancer cell growth through activation of the Wnt pathway. Genome Biol. 2019;20:84. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 228] [Cited by in RCA: 364] [Article Influence: 60.7] [Reference Citation Analysis (0)] |
| 102. | Zhu P, Liang H, Huang X, Zeng Q, Liu Y, Lv J, Ming L. Circular RNA Hsa_circ_0004018 Inhibits Wnt/β-Catenin Signaling Pathway by Targeting microRNA-626/DKK3 in Hepatocellular Carcinoma. Onco Targets Ther. 2020;13:9351-9364. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 30] [Cited by in RCA: 29] [Article Influence: 5.8] [Reference Citation Analysis (0)] |
| 103. | Yang B, Zhao J, Huo T, Zhang M, Wu X. Effects of CircRNA-ITCH on proliferation and apoptosis of hepatocellular carcinoma cells through inhibiting Wnt/β-catenin signaling pathway. J BUON. 2020;25:1368-1374. [PubMed] |
| 104. | Chen H, Liu S, Li M, Huang P, Li X. circ_0003418 Inhibits Tumorigenesis And Cisplatin Chemoresistance Through Wnt/β-Catenin Pathway In Hepatocellular Carcinoma. Onco Targets Ther. 2019;12:9539-9549. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 35] [Cited by in RCA: 63] [Article Influence: 10.5] [Reference Citation Analysis (0)] |
| 105. | Sun EJ, Wankell M, Palamuthusingam P, McFarlane C, Hebbard L. Targeting the PI3K/Akt/mTOR Pathway in Hepatocellular Carcinoma. Biomedicines. 2021;9. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 13] [Cited by in RCA: 159] [Article Influence: 39.8] [Reference Citation Analysis (0)] |
| 106. | Fu HW, Lin X, Zhu YX, Lan X, Kuang Y, Wang YZ, Ke ZG, Yuan T, Chen P. Circ-IGF1R has pro-proliferative and anti-apoptotic effects in HCC by activating the PI3K/AKT pathway. Gene. 2019;716:144031. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 31] [Cited by in RCA: 51] [Article Influence: 8.5] [Reference Citation Analysis (0)] |
| 107. | Zheng H, Chen T, Li C, Xu C, Ding C, Chen J, Ju S, Zhang Z, Liang Z, Cui Z, Zhao J. A circular RNA hsa_circ_0079929 inhibits tumor growth in hepatocellular carcinoma. Cancer Manag Res. 2019;11:443-454. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 29] [Cited by in RCA: 41] [Article Influence: 6.8] [Reference Citation Analysis (0)] |
| 108. | Zhao Z, Song J, Tang B, Fang S, Zhang D, Zheng L, Wu F, Gao Y, Chen C, Hu X, Weng Q, Yang Y, Tu J, Ji J. CircSOD2 induced epigenetic alteration drives hepatocellular carcinoma progression through activating JAK2/STAT3 signaling pathway. J Exp Clin Cancer Res. 2020;39:259. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 19] [Cited by in RCA: 67] [Article Influence: 13.4] [Reference Citation Analysis (0)] |
| 109. | Wu M, Sun T, Xing L. Circ_0004913 Inhibits Cell Growth, Metastasis, and Glycolysis by Absorbing miR-184 to Regulate HAMP in Hepatocellular Carcinoma. Cancer Biother Radiopharm. 2020;. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 16] [Cited by in RCA: 25] [Article Influence: 12.5] [Reference Citation Analysis (0)] |
| 110. | Zeng T, Luo L, Huang Y, Ye X, Lin J. Upregulation of miR-138 Increases Sensitivity to Cisplatin in Hepatocellular Carcinoma by Regulating EZH2. Biomed Res Int. 2021;2021:6665918. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 3] [Cited by in RCA: 9] [Article Influence: 2.3] [Reference Citation Analysis (0)] |
| 111. | Zhao G, Zhang A, Sun S, Ding Y. Long non-coding RNA LINC00173 enhances cisplatin resistance in hepatocellular carcinoma via the microRNA-641/RAB14 axis. Oncol Lett. 2021;21:371. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 5] [Cited by in RCA: 13] [Article Influence: 3.3] [Reference Citation Analysis (0)] |
| 112. | Fan W, Chen L, Wu X, Zhang T. Circ_0031242 Silencing Mitigates the Progression and Drug Resistance in DDP-Resistant Hepatoma Cells by the miR-924/POU3F2 Axis. Cancer Manag Res. 2021;13:743-755. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 7] [Cited by in RCA: 16] [Article Influence: 4.0] [Reference Citation Analysis (0)] |
| 113. | Zhao G, Wei Z, Guo Y. MicroRNA-107 is a novel tumor suppressor targeting POU3F2 in melanoma. Biol Res. 2020;53:11. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 12] [Cited by in RCA: 19] [Article Influence: 3.8] [Reference Citation Analysis (0)] |
| 114. | Qian Y, Wu X, Wang H, Hou G, Han X, Song W. MicroRNA-4290 suppresses PDK1-mediated glycolysis to enhance the sensitivity of gastric cancer cell to cisplatin. Braz J Med Biol Res. 2020;53:e9330. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 15] [Cited by in RCA: 25] [Article Influence: 5.0] [Reference Citation Analysis (0)] |
| 115. | Li Y, Zhang Y, Zhang S, Huang D, Li B, Liang G, Wu Y, Jiang Q, Li L, Lin C, Wei Z, Meng L. circRNA circARNT2 Suppressed the Sensitivity of Hepatocellular Carcinoma Cells to Cisplatin by Targeting the miR-155-5p/PDK1 Axis. Mol Ther Nucleic Acids. 2021;23:244-254. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 21] [Cited by in RCA: 42] [Article Influence: 8.4] [Reference Citation Analysis (0)] |
| 116. | Stark AL, Delaney SM, Wheeler HE, Im HK, Dolan ME. Functional consequences of PRPF39 on distant genes and cisplatin sensitivity. Hum Mol Genet. 2012;21:4348-4355. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 6] [Cited by in RCA: 8] [Article Influence: 0.6] [Reference Citation Analysis (0)] |
| 117. | Qin L, Zhan Z, Wei C, Li X, Zhang T, Li J. HsacircRNAG004213 promotes cisplatin sensitivity by regulating miR513b5p/PRPF39 in liver cancer. Mol Med Rep. 2021;23. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 12] [Cited by in RCA: 32] [Article Influence: 8.0] [Reference Citation Analysis (0)] |
| 118. | Demir T, Lee SS, Kaseb AO. Systemic therapy of liver cancer. Adv Cancer Res. 2021;149:257-294. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 19] [Cited by in RCA: 62] [Article Influence: 15.5] [Reference Citation Analysis (0)] |
| 119. | Yu J, Wang N, Gong Z, Liu L, Yang S, Chen GG, Lai PBS. Cytochrome P450 1A2 overcomes nuclear factor kappa B-mediated sorafenib resistance in hepatocellular carcinoma. Oncogene. 2021;40:492-507. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 10] [Cited by in RCA: 25] [Article Influence: 5.0] [Reference Citation Analysis (0)] |
| 120. | Feng Y, Xiong Y, Qiao T, Li X, Jia L, Han Y. Lactate dehydrogenase A: A key player in carcinogenesis and potential target in cancer therapy. Cancer Med. 2018;7:6124-6136. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 453] [Cited by in RCA: 438] [Article Influence: 62.6] [Reference Citation Analysis (0)] |
| 121. | Huang H, Peng J, Yi S, Ding C, Ji W, Huang Q, Zeng S. Circular RNA circUBE2D2 functions as an oncogenic factor in hepatocellular carcinoma sorafenib resistance and glycolysis. Am J Transl Res. 2021;13:6076-6086. [PubMed] |
| 122. | Yang C, Dong Z, Hong H, Dai B, Song F, Geng L, Lu J, Yang J, Sui C, Xu M. circFN1 Mediates Sorafenib Resistance of Hepatocellular Carcinoma Cells by Sponging miR-1205 and Regulating E2F1 Expression. Mol Ther Nucleic Acids. 2020;22:421-433. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 19] [Cited by in RCA: 40] [Article Influence: 8.0] [Reference Citation Analysis (0)] |
| 123. | Fang Z, Lin M, Li C, Liu H, Gong C. A comprehensive review of the roles of E2F1 in colon cancer. Am J Cancer Res. 2020;10:757-768. [PubMed] |
| 124. | Xu J, Ji L, Liang Y, Wan Z, Zheng W, Song X, Gorshkov K, Sun Q, Lin H, Zheng X, Chen J, Jin RA, Liang X, Cai X. CircRNA-SORE mediates sorafenib resistance in hepatocellular carcinoma by stabilizing YBX1. Signal Transduct Target Ther. 2020;5:298. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 96] [Cited by in RCA: 308] [Article Influence: 61.6] [Reference Citation Analysis (0)] |
| 125. | Kwon E, Todorova K, Wang J, Horos R, Lee KK, Neel VA, Negri GL, Sorensen PH, Lee SW, Hentze MW, Mandinova A. The RNA-binding protein YBX1 regulates epidermal progenitors at a posttranscriptional level. Nat Commun. 2018;9:1734. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 36] [Cited by in RCA: 67] [Article Influence: 9.6] [Reference Citation Analysis (0)] |
| 126. | Dong ZR, Ke AW, Li T, Cai JB, Yang YF, Zhou W, Shi GM, Fan J. CircMEMO1 modulates the promoter methylation and expression of TCF21 to regulate hepatocellular carcinoma progression and sorafenib treatment sensitivity. Mol Cancer. 2021;20:75. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 46] [Cited by in RCA: 48] [Article Influence: 12.0] [Reference Citation Analysis (0)] |
| 127. | Salmaninejad A, Valilou SF, Shabgah AG, Aslani S, Alimardani M, Pasdar A, Sahebkar A. PD-1/PD-L1 pathway: Basic biology and role in cancer immunotherapy. J Cell Physiol. 2019;234:16824-16837. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 172] [Cited by in RCA: 307] [Article Influence: 51.2] [Reference Citation Analysis (0)] |
| 128. | Zhao L, Yu G, Han Q, Cui C, Zhang B. TIM-3: An emerging target in the liver diseases. Scand J Immunol. 2020;91:e12825. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 9] [Cited by in RCA: 25] [Article Influence: 5.0] [Reference Citation Analysis (0)] |
| 129. | Tan S, Xu Y, Wang Z, Wang T, Du X, Song X, Guo X, Peng J, Zhang J, Liang Y, Lu J, Gao C, Wu Z, Li C, Li N, Gao L, Liang X, Ma C. Tim-3 Hampers Tumor Surveillance of Liver-Resident and Conventional NK Cells by Disrupting PI3K Signaling. Cancer Res. 2020;80:1130-1142. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 40] [Cited by in RCA: 103] [Article Influence: 17.2] [Reference Citation Analysis (0)] |
| 130. | Sanchez-Correa B, Lopez-Sejas N, Duran E, Labella F, Alonso C, Solana R, Tarazona R. Modulation of NK cells with checkpoint inhibitors in the context of cancer immunotherapy. Cancer Immunol Immunother. 2019;68:861-870. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 40] [Cited by in RCA: 42] [Article Influence: 7.0] [Reference Citation Analysis (0)] |
| 131. | Zhang PF, Gao C, Huang XY, Lu JC, Guo XJ, Shi GM, Cai JB, Ke AW. Cancer cell-derived exosomal circUHRF1 induces natural killer cell exhaustion and may cause resistance to anti-PD1 therapy in hepatocellular carcinoma. Mol Cancer. 2020;19:110. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 391] [Cited by in RCA: 402] [Article Influence: 80.4] [Reference Citation Analysis (0)] |
| 132. | Li Y, Zhai Y, Song Q, Zhang H, Cao P, Ping J, Liu X, Guo B, Liu G, Song J, Zhang Y, Yang A, Yan H, Yang L, Cui Y, Ma Y, Xing J, Shen X, Liu T, An J, Bei JX, Jia W, Kang L, Liu L, Yuan D, Hu Z, Shen H, Lu L, Wang X, Li H, He F, Zhou G. Genome-Wide Association Study Identifies a New Locus at 7q21.13 Associated with Hepatitis B Virus-Related Hepatocellular Carcinoma. Clin Cancer Res. 2018;24:906-915. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 24] [Cited by in RCA: 44] [Article Influence: 5.5] [Reference Citation Analysis (0)] |
| 133. | Huang XY, Zhang PF, Wei CY, Peng R, Lu JC, Gao C, Cai JB, Yang X, Fan J, Ke AW, Zhou J, Shi GM. Circular RNA circMET drives immunosuppression and anti-PD1 therapy resistance in hepatocellular carcinoma via the miR-30-5p/snail/DPP4 axis. Mol Cancer. 2020;19:92. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 163] [Cited by in RCA: 164] [Article Influence: 32.8] [Reference Citation Analysis (0)] |
| 134. | Seimiya T, Otsuka M, Iwata T, Shibata C, Tanaka E, Suzuki T, Koike K. Emerging Roles of Exosomal Circular RNAs in Cancer. Front Cell Dev Biol. 2020;8:568366. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 66] [Cited by in RCA: 64] [Article Influence: 12.8] [Reference Citation Analysis (0)] |
| 135. | Moldogazieva NT, Zavadskiy SP, Terentiev AA. Genomic Landscape of Liquid Biopsy for Hepatocellular Carcinoma Personalized Medicine. Cancer Genomics Proteomics. 2021;18:369-383. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 6] [Cited by in RCA: 23] [Article Influence: 5.8] [Reference Citation Analysis (0)] |
| 136. | Sukowati CHC, Cabral LKD, Tiribelli C, Pascut D. Circulating Long and Circular Noncoding RNA as Non-Invasive Diagnostic Tools of Hepatocellular Carcinoma. Biomedicines. 2021;9. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 27] [Cited by in RCA: 29] [Article Influence: 7.3] [Reference Citation Analysis (0)] |
| 137. | Debes JD, Romagnoli PA, Prieto J, Arrese M, Mattos AZ, Boonstra A; On Behalf Of The Escalon Consortium. Serum Biomarkers for the Prediction of Hepatocellular Carcinoma. Cancers (Basel). 2021;13. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 47] [Cited by in RCA: 45] [Article Influence: 11.3] [Reference Citation Analysis (0)] |
| 138. | Ozgor D, Otan E. HCC and Tumor Biomarkers: Does One Size Fits All? J Gastrointest Cancer. 2020;51:1122-1126. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 4] [Cited by in RCA: 5] [Article Influence: 1.3] [Reference Citation Analysis (0)] |
| 139. | Lyu L, Yang W, Yao J, Wang H, Zhu J, Jin A, Liu T, Wang B, Zhou J, Fan J, Yang X, Guo W. The diagnostic value of plasma exosomal hsa_circ_0070396 for hepatocellular carcinoma. Biomark Med. 2021;15:359-371. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 38] [Cited by in RCA: 39] [Article Influence: 9.8] [Reference Citation Analysis (0)] |
| 140. | Zhang X, Xu Y, Qian Z, Zheng W, Wu Q, Chen Y, Zhu G, Liu Y, Bian Z, Xu W, Zhang Y, Sun F, Pan Q, Wang J, Du L, Yu Y. circRNA_104075 stimulates YAP-dependent tumorigenesis through the regulation of HNF4a and may serve as a diagnostic marker in hepatocellular carcinoma. Cell Death Dis. 2018;9:1091. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 159] [Cited by in RCA: 185] [Article Influence: 26.4] [Reference Citation Analysis (0)] |
| 141. | Matboli M, Shafei AE, Ali MA, Ashry AM, Kamal KM, Agag MA, Reda I, Tash EF, Ali M. circRNAs (hsa_circ_00156, hsa_circ _000224, and hsa_circ _000520) are novel potential biomarkers in hepatocellular carcinoma. J Cell Biochem. 2018;. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 23] [Cited by in RCA: 28] [Article Influence: 4.7] [Reference Citation Analysis (0)] |
| 142. | Yu J, Ding WB, Wang MC, Guo XG, Xu J, Xu QG, Yang Y, Sun SH, Liu JF, Qin LX, Liu H, Yang F, Zhou WP. Plasma circular RNA panel to diagnose hepatitis B virus-related hepatocellular carcinoma: A large-scale, multicenter study. Int J Cancer. 2020;146:1754-1763. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 49] [Cited by in RCA: 83] [Article Influence: 13.8] [Reference Citation Analysis (0)] |
| 143. | Parizadeh SM, Jafarzadeh-Esfehani R, Ghandehari M, Goldani F, Parizadeh SMR, Hassanian SM, Ghayour-Mobarhan M, Ferns GA, Avan A. MicroRNAs as Potential Diagnostic and Prognostic Biomarkers in Hepatocellular Carcinoma. Curr Drug Targets. 2019;20:1129-1140. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 17] [Cited by in RCA: 21] [Article Influence: 4.2] [Reference Citation Analysis (0)] |
| 144. | Zhang C, Zhang C, Lin J, Wang H. Circular RNA Hsa_Circ_0091579 Serves as a Diagnostic and Prognostic Marker for Hepatocellular Carcinoma. Cell Physiol Biochem. 2018;51:290-300. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 31] [Cited by in RCA: 34] [Article Influence: 4.9] [Reference Citation Analysis (0)] |
| 145. | Lei B, Zhou J, Xuan X, Tian Z, Zhang M, Gao W, Lin Y, Ni B, Pang H, Fan W. Circular RNA expression profiles of peripheral blood mononuclear cells in hepatocellular carcinoma patients by sequence analysis. Cancer Med. 2019;8:1423-1433. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 27] [Cited by in RCA: 37] [Article Influence: 6.2] [Reference Citation Analysis (0)] |
| 146. | Pan H, Tang L, Jiang H, Li X, Wang R, Gao J, Li Q. Enhanced expression of circ_0000267 in hepatocellular carcinoma indicates poor prognosis and facilitates cell progression by sponging miR-646. J Cell Biochem. 2019;. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 21] [Cited by in RCA: 32] [Article Influence: 5.3] [Reference Citation Analysis (0)] |
| 147. | Hu ZQ, Zhou SL, Li J, Zhou ZJ, Wang PC, Xin HY, Mao L, Luo CB, Yu SY, Huang XW, Cao Y, Fan J, Zhou J. Circular RNA Sequencing Identifies CircASAP1 as a Key Regulator in Hepatocellular Carcinoma Metastasis. Hepatology. 2020;72:906-922. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 112] [Cited by in RCA: 174] [Article Influence: 34.8] [Reference Citation Analysis (0)] |
| 148. | Guo J, Duan H, Li Y, Yang L, Yuan L. A novel circular RNA circ-ZNF652 promotes hepatocellular carcinoma metastasis through inducing snail-mediated epithelial-mesenchymal transition by sponging miR-203/miR-502-5p. Biochem Biophys Res Commun. 2019;513:812-819. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 38] [Cited by in RCA: 41] [Article Influence: 6.8] [Reference Citation Analysis (0)] |
| 149. | Zhang X, Qiu S, Luo P, Zhou H, Jing W, Liang C, Tu J. Down-regulation of hsa_circ_0001649 in hepatocellular carcinoma predicts a poor prognosis. Cancer Biomark. 2018;22:135-142. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 30] [Cited by in RCA: 44] [Article Influence: 6.3] [Reference Citation Analysis (0)] |
| 150. | Li X, Yang J, Yang X, Cao T. Dysregulated circ_0004913, circ_0008160, circ_0000517, and their potential as biomarkers for disease monitoring and prognosis in hepatocellular carcinoma. J Clin Lab Anal. 2021;35:e23785. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 4] [Cited by in RCA: 7] [Article Influence: 1.8] [Reference Citation Analysis (0)] |
| 151. | Sunagawa Y, Yamada S, Sonohara F, Kurimoto K, Tanaka N, Suzuki Y, Inokawa Y, Takami H, Hayashi M, Kanda M, Tanaka C, Nakayama G, Koike M, Kodera Y. Genome-wide identification and characterization of circular RNA in resected hepatocellular carcinoma and background liver tissue. Sci Rep. 2021;11:6016. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 1] [Cited by in RCA: 10] [Article Influence: 2.5] [Reference Citation Analysis (0)] |
| 152. | Kim G, Han JR, Park SY, Tak WY, Kweon YO, Lee YR, Han YS, Park JG, Kang MK, Lee HW, Lee WK, Kim D, Jang SY, Hur K. Circular noncoding RNA hsa_circ_0005986 as a prognostic biomarker for hepatocellular carcinoma. Sci Rep. 2021;11:14930. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 2] [Cited by in RCA: 5] [Article Influence: 1.3] [Reference Citation Analysis (0)] |