Published online Mar 15, 2022. doi: 10.4251/wjgo.v14.i3.690
Peer-review started: August 10, 2021
First decision: December 4, 2021
Revised: December 25, 2021
Accepted: February 22, 2022
Article in press: February 22, 2022
Published online: March 15, 2022
Processing time: 212 Days and 4.1 Hours
Gastric cancer (GC), a multifactorial disease, is caused by pathogens, such as Helicobacter pylori (H. pylori) and Epstein-Barr virus (EBV), and genetic components.
To investigate microbiomes and host genome instability by cost-effective, low-coverage whole-genome sequencing, as biomarkers for GC subtyping.
Samples from 40 GC patients were collected from Taizhou Hospital, Zhejiang Province, affiliated with Wenzhou Medical University. DNA from the samples was subjected to low-coverage whole-genome sequencing with a median genome coverage of 1.86 × (range: 1.03 × to 3.17 ×) by Illumina × 10, followed by copy number analyses using a customized bioinformatics workflow ultrasensitive chromosomal aneuploidy detector.
Of the 40 GC samples, 20 (50%) were found to be enriched with microbiomes. EBV DNA was detected in 5 GC patients (12.5%). H. pylori DNA was found in 15 (37.5%) patients. The other 20 (50%) patients were found to have relatively higher genomic instability. Copy number amplifications of the oncogenes, ERBB2 and KRAS, were found in 9 (22.5%) and 7 (17.5%) of the GC samples, respectively. EBV enrichment was found to be associated with tumors in the gastric cardia and fundus. H. pylori enrichment was found to be associated with tumors in the pylorus and antrum. Tumors with elevated genomic instability showed no localization and could be observed in any location. Additionally, H. pylori-enriched GC was found to be associated with the Borrmann type II/III and gastritis history. EBV-enriched GC was not associated with gastritis. No statistically significant correlation was observed between genomic instability and gastritis. Furthermore, these three different molecular subtypes showed distinct survival outcomes (P = 0.019). EBV-positive tumors had the best prognosis, whereas patients with high genomic instability (CIN+) showed the worst survival. Patients with H. pylori infection showed intermediate prognosis compared with the other two subtypes.
Thus, using low-coverage whole-genome sequencing, GC can be classified into three categories based on disease etiology; this classification may prove useful for GC diagnosis and precision medicine.
Core Tip: This study investigated the microbiomes and host genome instability via cost-effective low-coverage whole-genome sequencing, to establish the findings for consideration in the development of a biomarker for gastric cancer (GC) subtyping. We believe that our study makes a significant contribution to the literature because it identified three different GC subtypes in the Chinese population, and these were related to different tumorigenesis mechanisms, chronic Epstein-Barr virus infection, Helicobacter pylori infections, and chromosomal instabilities. This discovery may therefore provide guidance for conducting future studies to realize GC treatment and prevention.
- Citation: Ye LP, Mao XL, Zhou XB, Wang Y, Xu SW, He SQ, Qian ZL, Zhang XG, Zhai LJ, Peng JB, Gu BB, Jin XX, Song YQ, Li SW. Cost-effective low-coverage whole-genome sequencing assay for the risk stratification of gastric cancer. World J Gastrointest Oncol 2022; 14(3): 690-702
- URL: https://www.wjgnet.com/1948-5204/full/v14/i3/690.htm
- DOI: https://dx.doi.org/10.4251/wjgo.v14.i3.690
Gastric cancer (GC) is the fourth leading cause of cancer-related deaths worldwide, with an estimated 768793 deaths in 2020, according to the GLOBOCAN of the International Agency for research on Cancer[1]. Most GCs are adenocarcinomas with considerable heterogeneity. According to Lauren’s criteria, GC is classified into three subtypes: intestinal, diffuse, and mixed[2]. World Health Organization (WHO) has divided GC into four subtypes-papillary, tubular, mucinous and poorly cohesive carcinomas[3]. However, these traditional morphology-based classification systems have limited clinical utility due to the molecular heterogeneity of GC. Therefore, it is necessary to develop a robust GC molecular classification to guide clinical practice, determine prognosis, or predict the treatment response.
Infection with microorganisms plays an important role in the development of GC. In contrast to other tumor types, gastric carcinogenesis is closely related to infectious pathogens. Among them, Helicobacter pylori (H. pylori) infection is one of the risk factors for GC, responsible for almost 90% of all noncardia GC[4]. A relationship between H. pylori and GC has been discovered and is characterized as a stepwise inflammatory process that eventually leads to malignancy[5]. In addition to pathogenic bacteria, viral infections significantly contribute to gastric carcinogenesis. Epstein-Barr virus (EBV) is the most characterized gastric oncogenic virus[6]. In a comprehensive molecular analysis of GC conducted by The Cancer Genome Atlas (TCGA), EBV-positive tumors were classified as a distinct subtype with statistical significance (P = 1.5 × 10-18), and ~9% of GC patients were EBV-positive[7]. Although these pathogens infect more than half of the world’s population, fortunately, only a small fraction of those infected develop GC, indicating the complexity that drives gastric tumorigenesis[8].
In addition to microbial infection, alterations in genomic stability also play a key role as drivers of GC. Among these, chromosomal instability (CIN) is one of the most common types of genetic changes, which is usually described as somatic copy number aberrations (SCNAs) accompanied by focal amplification of oncogenes or deletion of tumor suppressor genes[9]. According to information presented in TCGA database, GC can be divided into two distinct subtypes based on the presence or absence of SCNAs[7]. Our previous studies demonstrated that CIN was a valuable prognosis factor in GC using array-based comparative genomic hybridization. Two distinct subtypes of GC were identified, high CIN and low CIN, with distinguished gene expression signatures and different survival outcomes of patients[10,11].
However, these methods are more expensive and sophisticated, limiting their application in clinical practice. Low-coverage whole-genome sequencing (LC-WGS) was first developed as a simple, cost-effective, and reliable technology to identify SCNAs in tumors in 2014[12]. Therefore, the aim of the present study was to develop a robust and cost-effective molecular classification method for GC using LC-WGS to identify candidate drivers of gastric tumorigenesis and to provide a roadmap for gastric risk stratification and targeted therapy trials.
Samples from 40 GC patients were collected and the deadline for the follow-up was May 2021. The study was reviewed and approved by the Institutional Ethics Committee of Taizhou Hospital of Zhejiang Province (Approval No. K20201205), and informed consent was obtained from the patients prior to specimen collection (Table 1).
| EBV+ | H. Pylori+ | ERBB2+ | KRAS+ | P value | |
| Age (yr), mean ± SD | 62.2 ± 6.4 | No significance | |||
| Sex, n | No significance | ||||
| Male | 3 | 13 | 8 | 5 | |
| Female | 2 | 2 | 1 | 2 | |
| Tumor location, n | P = 0.013 (H. Pylori associated with antrum) | ||||
| Cardia/fundus | 5 | 2 | 3 | 1 | |
| Polyrus/antrum | 0 | 13 | 5 | 6 | |
| Other | 0 | 0 | 1 | 0 | |
| Borrmann, n | P = 0.013 (H. Pylori associated with ulcerative) | ||||
| Type I | 1 | 1 | 2 | 2 | |
| Type II | 1 | 7 | 4 | 1 | |
| Type III | 1 | 7 | 2 | 2 | |
| Type IV | 2 | 0 | 1 | 1 | |
| Gastritis, n | No significance | ||||
| Yes | 0 | 4 | 2 | 2 | |
| No | 5 | 11 | 7 | 5 | |
| Vascular invasion, n | No significance | ||||
| Yes | 2 | 9 | 1 | 2 | |
| No | 3 | 6 | 8 | 5 |
DNA was extracted from formalin-fixed paraffin-embedded (FFPE) samples using the Qiagen nucleic acid kits (69504).
For LC-WGS, libraries were prepared using the Kapa Hyper Prep kit (Roche, CA, United States) with custom adapters (IDT, CA, United States) starting with 50 to 1000 ng of DNA input (median, 471 ng), which was used for low-pass WGS. The 22 Libraries were pooled and sequenced using the 150-base paired-end runs over 1× lane on a HiSeq X10 system (Illumina, CA, United States). Segment copy numbers were derived using a customized workflow ultrasensitive chromosomal aneuploidy detector (UCAD). If the median absolute deviation of the copy ratio (log ratio) between the adjacent bins of the whole-genome was greater than 0.38, indicating poor-quality sequence data, the sample was excluded.
Reads were mapped to the EBV reference genome (gi|82503188). Matches with no more than 1 mismatch were counted as EBV reads. The same approach was applied for H. pylori (gi|261838873). Samples with more than 4 EBV reads were marked as EBV-positive tumor samples. Samples with more than 4 H. pylori reads were marked as H. pylori-positive tumor samples.
Specimens from 40 gastric patients were found to have pathological characteristics of GC. The prepared slides were then sent to pathologists for analysis following the standard protocol. The pathology test results were recorded as tumor type and tumor grade.
The Illumina X10 system was used for DNA extraction and analysis. At least 10 M paired reads were collected for each sample. The reads were mapped to the human reference genome hg19. The genomic coverage was then counted using the software package mpileup[13]. Then, the average coverage for each 200 k bin was calculated, and the Z-score for each bin was normalized using the following formula-1:
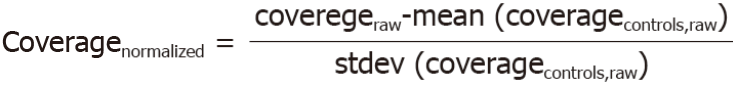
Then, using the circular binary segmentation algorithm from the R package DNACopy[14], significant genomic breakpoints and copy number changes in the genomic segments were found.
We used the R package “DNACopy” to analyze the copy number changes. A P value of < 0.05 was considered to denote a statistically significant binary segmentation. The absolute segment value was used for further analysis. The sensitivity and specificity of UCAD were estimated by receiver operating characteristic curves. For categorical variables, the chi-square test was employed. All statistical analyses were performed using SPSS17.0 (IBM, Foster City, CA, United States).
The associations between the clinicopathological UCAD screening positivity and clinicopathological parameters were analyzed by the proportional trend test[15]. Data were reported as means and standard deviations, medians and interquartile ranges, and hazard ratios or odds ratios with 95%CIs, as appropriate. The missing data were removed from the analyses. All analyses were performed using R software, version 3.4.3 (R Foundation for Statistical Computing, GNU project https://www.r-project.org/). The anonymized data and R code used in the statistical analysis will be made available on request.
In total, 40 FFPE samples were collected. All samples passed QC and were included in this study (Table 1).
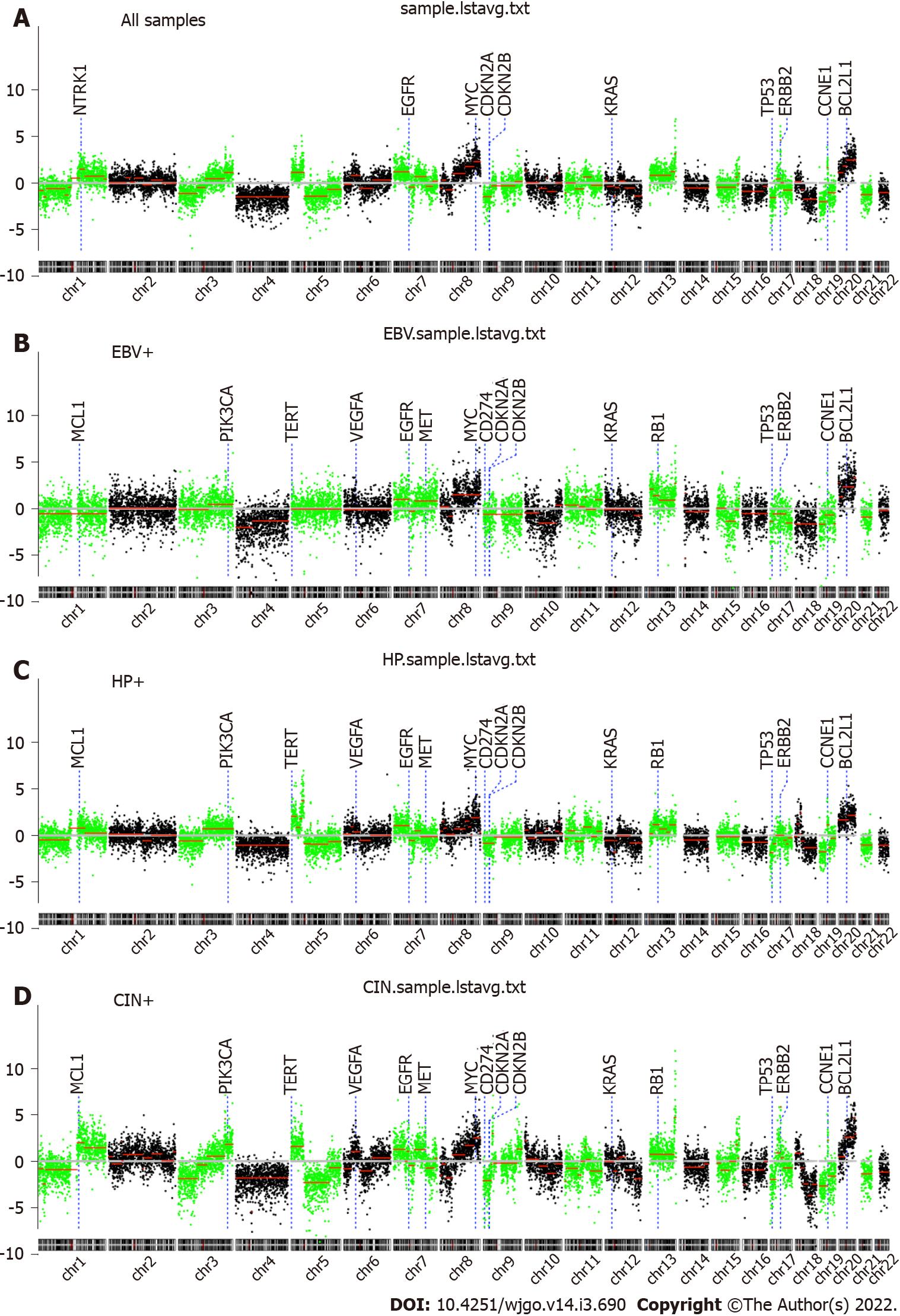
In Figure 1, we summarize the genome-wide copy number variations observed. Interestingly, it was found that chromosomal-arm imbalance was caused by chromosomal breakpoints on the centromeres (Figure 1A for averaged data plot). There were 70 genomic segments with statistically significant copy number changes (details in Table 2). Frequent chromosomal changes in GC include 1p-, 1q+, 3q+, 4-, 5p+, 7p+, 8q+, 9p-, 13+, 17p-, 17q+, and 20q+. The focal events include amplification of 17q12, which contains ERBB2 (chr17:37, 844167-37886679, hg19). Z-scores (formula-1) are listed in Table 2.
| Chrom | Loc.start | Loc.end | Seg.mean | LogP | Key genes |
| Chr04 | 0 | 190800000 | -1.63 | -100.00 | FHIT |
| Chr05 | 45800000 | 131000000 | -1.49 | -100.00 | |
| Chr13 | 19000000 | 110000000 | 0.87 | -88.38 | |
| Chr18 | 30800000 | 77800000 | -1.86 | -86.67 | |
| Chr03 | 0 | 66200000 | -1.23 | -80.98 | |
| Chr07 | 0 | 55600000 | 1.28 | -75.03 | EGFR |
| Chr20 | 30200000 | 62800000 | 2.54 | -74.06 | |
| Chr16 | 0 | 76200000 | -0.93 | -67.73 | |
| Chr08 | 90600000 | 122000000 | 1.84 | -59.87 | |
| Chr05 | 0 | 45600000 | 1.29 | -56.55 | TERT |
| Chr01 | 170000000 | 240000000 | 0.78 | -55.86 | |
| Chr21 | 9400000 | 47800000 | -1.33 | -54.61 | |
| Chr09 | 0 | 29800000 | -1.56 | -50.52 | CDKN2A |
| Chr08 | 47800000 | 90400000 | 1.02 | -49.62 | |
| Chr08 | 122200000 | 146000000 | 2.42 | -49.24 | MYC |
| Chr01 | 29600000 | 98400000 | -0.69 | -47.14 | |
| Chr19 | 0 | 28800000 | -2.03 | -43.15 | |
| Chr10 | 50800000 | 111400000 | -0.77 | -41.27 | |
| Chr03 | 165800000 | 197800000 | 1.19 | -40.36 | |
| Chr22 | 16000000 | 51000000 | -1.11 | -38.57 | |
| Chr14 | 19000000 | 107000000 | -0.57 | -37.54 | |
| Chr12 | 107600000 | 133600000 | -1.44 | -36.53 | |
| Chr15 | 20000000 | 88200000 | -0.60 | -34.61 | |
| Chr01 | 151200000 | 169800000 | 1.47 | -32.21 | |
| Chr17 | 0 | 21800000 | -1.75 | -31.78 | TP53 |
| Chr03 | 96200000 | 165600000 | 0.48 | -30.10 | |
| Chr19 | 32200000 | 58800000 | -1.15 | -29.94 | |
| Chr07 | 76400000 | 117800000 | 0.66 | -28.48 | |
| Chr05 | 131200000 | 180600000 | -0.70 | -28.10 | |
| Chr17 | 49200000 | 80800000 | -0.85 | -26.77 | |
| Chr06 | 26000000 | 57000000 | 0.80 | -25.75 | |
| Chr12 | 73400000 | 107400000 | -0.64 | -23.98 | |
| Chr06 | 57200000 | 104200000 | -0.63 | -22.49 | |
| Chr20 | 0 | 17000000 | 1.09 | -22.19 | |
| Chr20 | 17200000 | 30000000 | 1.73 | -18.89 | |
| Chr01 | 98600000 | 109600000 | -1.31 | -18.38 | |
| Chr11 | 67000000 | 96000000 | 0.58 | -17.22 | |
| Chr08 | 20200000 | 47600000 | -0.82 | -16.59 | |
| Chr03 | 66400000 | 96000000 | -0.54 | -16.41 | |
| Chr01 | 19800000 | 29400000 | -1.60 | -15.48 | |
| Chr02 | 92200000 | 122200000 | 0.51 | -14.89 | |
| Chr11 | 37000000 | 66800000 | -0.69 | -14.39 | |
| Chr02 | 153200000 | 193600000 | 0.37 | -14.32 | |
| Chr09 | 30000000 | 115400000 | -0.36 | -13.72 | |
| Chr01 | 0 | 19600000 | -0.77 | -12.44 | |
| Chr10 | 200000 | 5600000 | 1.32 | -10.20 | |
| Chr02 | 62400000 | 76000000 | 0.58 | -9.30 | |
| Chr12 | 7400000 | 31200000 | -0.40 | -9.25 | |
| Chr13 | 110200000 | 113800000 | 3.69 | -8.57 | |
| Chr06 | 104400000 | 170800000 | 0.24 | -8.31 | |
| hr18 | 23000000 | 30600000 | -0.85 | -7.70 | |
| Chr12 | 31400000 | 38200000 | -1.65 | -7.02 | |
| Chr15 | 88400000 | 95200000 | 0.80 | -6.84 | |
| Chr16 | 76400000 | 90000000 | -0.45 | -6.61 | |
| Chr12 | 52200000 | 58200000 | 0.72 | -6.00 | |
| Chr19 | 29000000 | 32000000 | 1.50 | -6.00 | |
| Chr08 | 0 | 6200000 | -0.65 | -5.82 | |
| Chr10 | 123000000 | 126800000 | 1.09 | -5.72 | |
| Chr11 | 96200000 | 134800000 | -0.26 | -5.51 | |
| Chr17 | 37600000 | 39600000 | 4.54 | -5.50 | |
| Chr02 | 122400000 | 153000000 | -0.28 | -5.38 | |
| Chr07 | 55800000 | 76200000 | -0.63 | -5.35 | |
| Chr15 | 99200000 | 102200000 | 1.24 | -5.04 | |
| Chr11 | 0 | 4000000 | -1.16 | -4.92 | |
| Chr12 | 38400000 | 52000000 | -0.46 | -4.85 | |
| Chr01 | 109800000 | 119800000 | -0.44 | -4.03 | |
| Chr01 | 120000000 | 151000000 | 0.52 | -4.01 | |
| Chr07 | 118000000 | 158800000 | -0.21 | -3.77 | |
| Chr17 | 22000000 | 37400000 | 0.44 | -3.48 | |
| Chr08 | 6400000 | 20000000 | 0.27 | -3.01 |
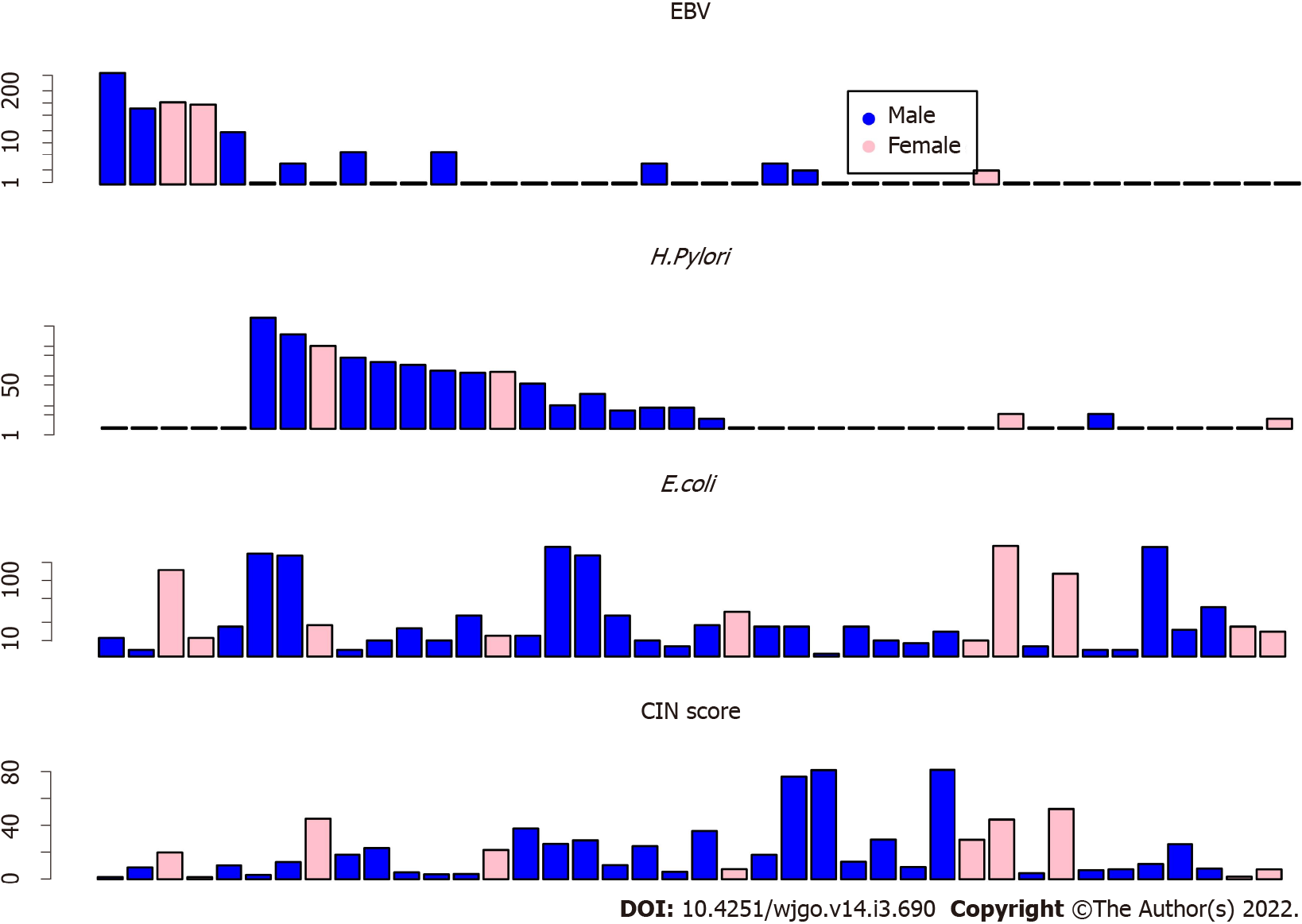
CIN scores were summarized by the formula, CIN = sum (Lchr × Zchr), where Lchr is the length of the chromosome segment and Zchr is the Z score of the segment. By using the CIN score cutoff value of 20, 19 (47.5%) patients had low CIN scores. The other 21 (52.5%) patients had elevated CIN scores (Figure 2).
Forty GC tissues were analyzed, and twenty (50% of the total) GC samples were found to be enriched with microbiomes. As shown in Figure 2, the samples of patients with abundant EBV DNA showed less abundance of H. pylori DNA (Figure 2, top), which may suggest that EBV and H. pylori are different drivers of GC tumorigenesis. As a control, the random distribution of Escherichia coli DNA may suggest less contribution of this microbiome to GC tumorigenesis. Furthermore, patients with low EBV and H. pylori DNA showed high CIN scores (Figure 2 bottom).
EBV DNA was detected in 5 GC patients (12.5%). H. pylori DNA was found in 15 (37.5%) patients. The other 20 (50%) patients were found to have relatively higher genomic instability. The copy number amplifications of the oncogenes, ERBB2 and KRAS, were found in 9 (22.5%) and 7 (17.5%) GC samples, respectively.
EBV-positive GC showed a relatively stable genome (Figure 1B). The patients with positive H. pylori statuses showed an unstable genome (Figure 1C), where chr5p amplifications are frequently located in TERT (5p15.33). The other patients with H. pylori- and EBV-negative GC were also characterized by an unstable genome, where ERBB2 amplifications were significantly enriched (Figure 1D).
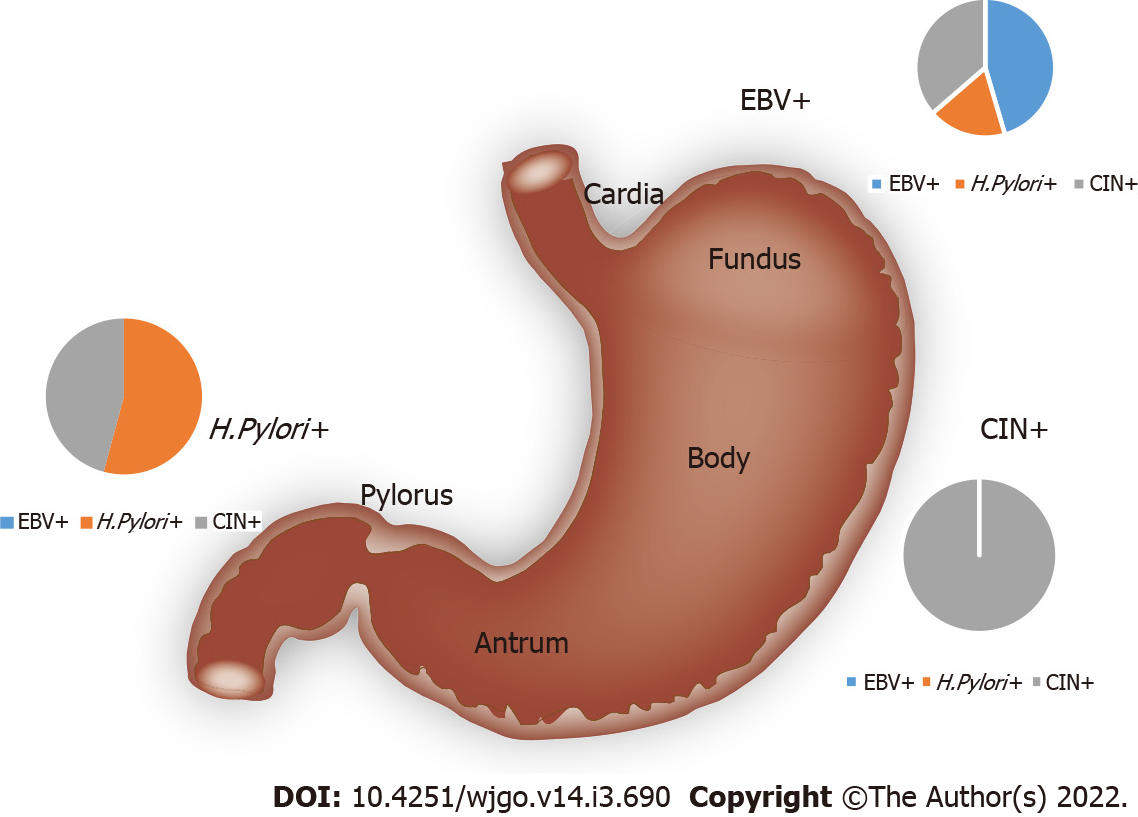
As shown in Table 1, EBV enrichment was found to be associated with tumors in the gastric cardia and fundus (P = 0.013). H. pylori enrichment was found to be associated with tumors in the pylorus and antrum. Tumors with elevated genomic instability could be found in any location (Figure 3).
Additionally, H. pylori-enriched GC was found to be associated with Borrmann type II/III and gastritis history (P = 0.013). The EBV-enriched GC was not associated with gastritis. There was no statistically significant correlation between genomic instability and gastritis.
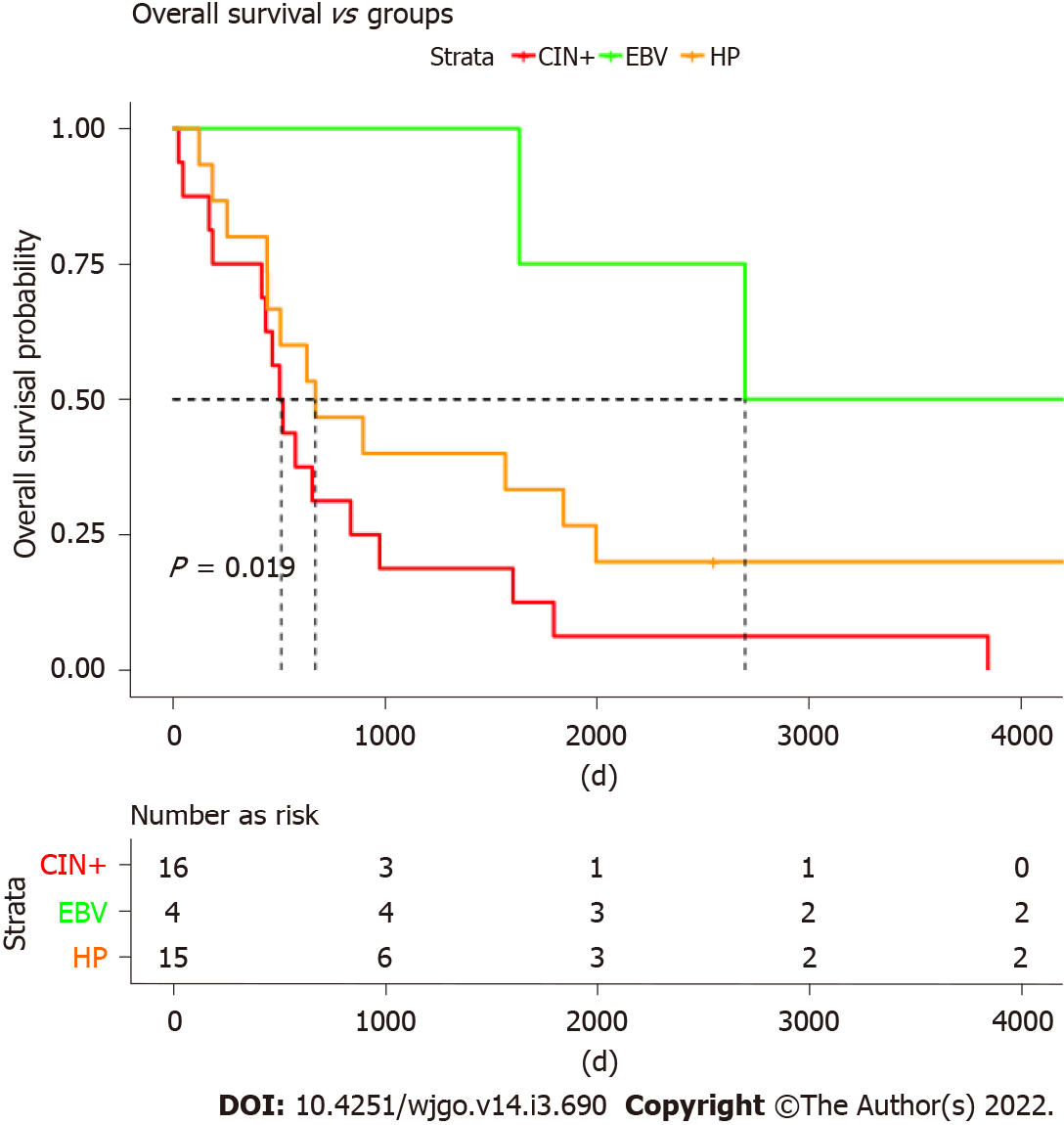
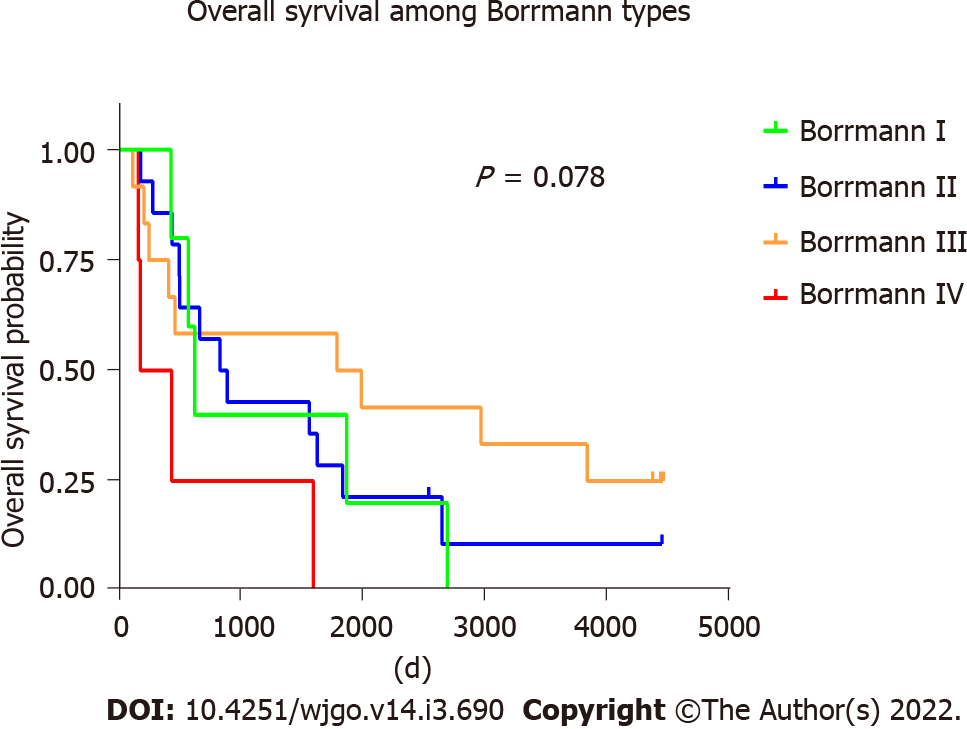
Furthermore, we analyzed the overall survival (OS) of different molecular groups. As shown in Figure 4, patients with different molecular subtypes show distinct prognoses (long rank test, P = 0.019). EBV-positive tumors showed the best OS, with a median OS of ~2800 d. CIN+ patients were found to have the worst survival, with a median OS less than 500 d. Patients with H. pylori infections also showed worse survival than those infected with EBV but tended to be better than CIN+ patients. Moreover, the OS was investigated among four Borrmann types of GC, and no significant difference was found (log-rank test, P = 0.078) (Figure 5).
GC has a high incidence and fatality rate in China. It shows high histopathological and molecular heterogeneity[16]. The disease involves multiple genes with different genetic events occurring at different stages. The heredity of GC, individual differences, and the complexity of molecular mechanisms necessitates its characterization by gene groups or cluster[7]. Molecular subtyping of GC involves the screening of the genes or protein markers related to tumorigenesis, diagnosis, and prognosis. The currently studied genes and protein markers include oncogenes, tumor suppressor genes, intercellular adhesion molecules, growth factors, and certain hormone receptors[17]. Although most of them show poor sensitivity, specificity, or reliability, a few have been recognized as effective biomarkers. Gastric adenocarcinoma, the most common type of GC, shows a remarkable heterogeneity among different patients with a high mortality due to the tumor’s innate aggressiveness. Despite recent advances in diagnosis and treatment, the 5-year OS remains poor. Moreover, gastrointestinal stroma tumor is a rare but highly curable cancer and has a satisfactory prognosis with a 5-year OS ranging from 60%-85%[18,19]. Notably, multimodal complications associated with radical gastrectomy during perioperative period should be addressed. Anastomotic fistula, one of the main surgical complications that raises the risk of local recurrence and worsens the overall prognosis, has been reported to be positively correlated with the neutrophil/lymphocyte ratio[20]. The traditional morphology-based subtyping systems include the Lauren classification (intestinal, diffuse, and mixed) and WHO-based classification (papillary, tubular, mucinous, and poorly cohesive). For the prediction of lymph node metastasis risk, a modified WHO classification can be used to distinguish GCs into the differentiated and undifferentiated types[21]. However, the dysregulation of oncogenes and suppressors owing to multiple genetic and epigenetic alterations has been shown in several studies to be a significant driver of tumorigenesis[22]. The current morphology-based clinical classification of GC can neither convey the molecular heterogeneity of GC nor can it guide clinical practice in predicting the prognosis or treatment response of patients with advanced GC. Although subclassification by molecular testing may add complexity to the classification, it is essential to identify specific GC subtypes based on the molecular and genetic features for the precise and selective targeting of anticancer therapies[23].
A recent publication by TCGA project proposed a molecular classification of GC, which divided it into four subtypes[7]: (1) EBV-positive type, characterized by frequent PIK3CA mutations; DNA hypermethylation; and JAK2, CD274, and PDCD1LG2 amplification; (2) Microsatellite unstable type, which has a high mutation rate, including the activation of gene mutations that encode oncogene signaling pathway proteins; (3) The genome stable type, which mostly occurs in the diffuse histology and is caused by RHOA mutation or THO family GTPase activation protein gene fusion phenomenon; and (4) CIN type, which has an aneuploid chromosome and the receptor tyrosine kinase, which is amplified in situ.
In the present study, we identified three GC subtypes through WGS: (1) EBV-positive GC; (2) H. pylori-positive GC; and (3) CIN type GC. Exclusivity was observed among the three subtypes, indicating different modes of tumorigenesis among the subtypes. The three subtypes showed a different genetic pattern. The CIN group was enriched in ERBB2amplification, and the H. pylori group was enriched in H. pylori DNzA and 5p (TERT) copy number gains. The distinct genetic patterns may suggest a different treatment approach for each GC subtype, which may require further research. In addition, patients with different molecular subtypes showed distinct prognoses by long rank test (P = 0.019), in which CIN+ patients were found to have the worst survival with a median OS less than 500 d. However, no significant difference of OS was found among the Borrmann types (P = 0.078), a classic GC classification widely used currently. It may indicate that the molecular subtypes in our study have advantages in guiding the prognosis of patients with GC. Nonetheless, owing to the limited sample sizes in this study, additional clinical evidence is needed to support this argument.
Further analyses showed that H. pylori positive tumors were associated with gastritis history, which may suggest chronic infections. H. pylori colonization causes chronic inflammation as well as a significant increase in the possibility of developing GC[24]. Currently, a persistent H. pylori infection is the strongest risk factor for the development of GC. Once H. pylori colonizes the gastric epithelium, it may persist for the host lifetime, which may increase the risk of developing GC[25]. Since H. pylori inhabits the gastric epithelium of half of the population and has been linked to 38% of the cases of gastric adenocarcinoma included in this study, it is critical to further understand the interrelationship between the host and microbial factors to reduce the risk of gastric adenocarcinoma, which necessitates further studies in this direction.
Approximately, 40% of GCs are characterized by high CIN. Among them, ERBB2 amplifications were frequently found in this study. HER2, also known as ERBB2, belongs to the ERBB family of proteins, including EGFR (or HER1), HER3, and HER4. Trastuzumab is a humanized monoclonal antibody that binds to HER2 specifically and inhibits its homodimerization and phosphorylation, which results in the inhibition of the proliferation of HER2-overexpressing tumor cells.
In the present study, approximately 60% of the GC cases were linked to microbiomes, including chronic EBV and H. pylori infections. Animal studies have also shown that H. pylori eradication treatment at the early stage has considerable potential to reduce the incidence of H. pylori-associated GC. Early clinical evidence has shown that H. pylori eradication may help prevent the progression of gastric precancerous lesions in some cases. Additionally, H. pylori eradication might be the most efficient method for preventing GC. The current clinical data in humans support the idea that the removal of H. pylori leads to a reduced risk of developing GC. It is even more useful in patients without intestinal metaplasia or atrophic gastritis[26]. However, the mechanism through which H. pylori induces tumorigenesis requires further investigation.
EBV can be found in the vast majority of the general population (at least 90%). However, typically, EBV causes a silent infection in the patient and does not lead to clinically positive symptoms[27]. In some adolescents and young individuals, EBV infection usually leads to infectious mononucleosis with fever, fatigue, headache, lymphadenopathy, sore throat, hepatomegaly, and rash. EBV can also cause B-cell lymphomas, Hodgkin’s disease, GC, and nasopharyngeal carcinoma[28]. Hence, targeting EBV may be an approach to prevent EBV-related GC. However, the latent EBV load in healthy individuals becomes generally stable over time, maintaining a “set point”[29]. Currently, there exists no efficient treatment regimen for the complete clearance of EBV infection.
In the present study, we subtyped GC into four groups using a cost-effective LC-WGS assay. The subtypes showed exclusive genetic features similar to each other, which may suggest different carcinogenesis processes and clinical outcomes of GC. We further analyzed the prognosis of the groups, and CIN+ groups showed poor prognosis. Moreover, EBV-positive patients showed better prognosis than that of negative patients, with median OS around 2800 d. The different clinical outcomes may help clinicians with differential treatment decisions; for example, adjuvant treatment might be recommended for CIN+ patients due to poor prognosis expectations.
The cost of this UCAD assay of LC-WGS is estimated to be ~$ 50 per patient. With the rapid reductions in next-generation sequencing costs, the UCAD assay is expected to become much more cost-effective in the near future. Conventionally, multiple assays, including H. Pylori, EBV, and copy number variation assays (most of these assays use the FISH technique, such as HER2 FISH) are performed separately. This leads to a high cost burden for GC patients. Secondly, due to the utility of the WGS technique, the UCAD assay captures not only human DNA but also microbiome DNA, which makes it a more informative technique for GC subtyping than other methods. Collectively, the new technique may help guide GC precision therapy in a cost-effective manner.
This study has a few limitations. The most important limitation of the present study is the limited number of patients recruited. Although statistically significant findings were reported, such as OS for each subtype, the conclusions should be confirmed by increasing the patient numbers. In addition, we only studied the copy number variations and microbiomes (EBV, H. Pylori) as GC subtyping biomarkers. Other molecular changes, including methylation and oncogene single nucleotide variations, were not included in this study. In future studies, the potential molecular subtyping markers in addition to the CNV and microbiome markers must be investigated.
In the present study, we identified three different GC subtypes associated with different tumorigenesis mechanisms in the Chinese population-chronic EBV infection, H. pylori infection, and CIN. Additionally, there were significant differences in the survival outcomes of patients among the three molecular subtypes. Therefore, these findings may beinstructive for future research on the treatment and prevention of GC.
These findings from our research may be instructive for future research on the treatment and prevention of gastric cancer (GC).
Thus, using low-coverage whole-genome sequencing, GC can be classified into three categories based on disease etiology; this classification may prove useful for GC diagnosis and precision medicine.
Epstein-Barr virus (EBV) enrichment was found to be associated with tumors in the gastric cardia and fundus. Helicobacter pylori (H. pylori) enrichment was found to be associated with tumors in the pylorus and antrum.
DNA from the 40 GC patients were subjected to low-coverage whole-genome sequencing by Illumina × 10, followed by copy number analyses using a customized bioinformatics workflow ultrasensitive chromosomal aneuploidy detector. EBV-positive tumors had the best prognosis, whereas patients with higher genomic instability showed the worst survival.
To investigate biomarkers for GC sub-typing by cost-effective, low-coverage whole-genome sequencing.
To search for new biomarkers of GC subtypes.
GC, a multifactorial disease, is caused by pathogens like H. pylori or EBV and by genetic components.
We wish to acknowledge Professor. Tao-Hsin Tung and Dr. Mei-Xian Zhang, Evidence-based Medicine Center, Taizhou Hospital of Zhejiang Province Affiliated to Wenzhou Medical University, Linhai 317000, Zhejiang Province, China.
Provenance and peer review: Invited article; Externally peer reviewed.
Peer-review model: Single blind
Specialty type: Gastroenterology and hepatology
Country/Territory of origin: China
Peer-review report’s scientific quality classification
Grade A (Excellent): 0
Grade B (Very good): B
Grade C (Good): 0
Grade D (Fair): 0
Grade E (Poor): 0
P-Reviewer: Socea B S-Editor: Zhang H L-Editor: A P-Editor: Zhang H
| 1. | Sung H, Ferlay J, Siegel RL, Laversanne M, Soerjomataram I, Jemal A, Bray F. Global Cancer Statistics 2020: GLOBOCAN Estimates of Incidence and Mortality Worldwide for 36 Cancers in 185 Countries. CA Cancer J Clin. 2021;71:209-249. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 75126] [Cited by in RCA: 64461] [Article Influence: 16115.3] [Reference Citation Analysis (176)] |
| 2. | Lauren P. The two histological main types of gastric carcinoma: diffuse and so-called intestinal-type carcinoma. An attempt at a histo-clinical classification. Acta Pathol Microbiol Scand. 1965;64:31-49. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 4011] [Cited by in RCA: 4321] [Article Influence: 149.0] [Reference Citation Analysis (0)] |
| 3. | Assarzadegan N, Montgomery E. What is New in the 2019 World Health Organization (WHO) Classification of Tumors of the Digestive System: Review of Selected Updates on Neuroendocrine Neoplasms, Appendiceal Tumors, and Molecular Testing. Arch Pathol Lab Med. 2021;145:664-677. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 38] [Cited by in RCA: 110] [Article Influence: 27.5] [Reference Citation Analysis (0)] |
| 4. | Plummer M, Franceschi S, Vignat J, Forman D, de Martel C. Global burden of gastric cancer attributable to Helicobacter pylori. Int J Cancer. 2015;136:487-490. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 643] [Cited by in RCA: 695] [Article Influence: 69.5] [Reference Citation Analysis (0)] |
| 5. | Moss SF. The Clinical Evidence Linking Helicobacter pylori to Gastric Cancer. Cell Mol Gastroenterol Hepatol. 2017;3:183-191. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 149] [Cited by in RCA: 205] [Article Influence: 22.8] [Reference Citation Analysis (0)] |
| 6. | de Martel C, Ferlay J, Franceschi S, Vignat J, Bray F, Forman D, Plummer M. Global burden of cancers attributable to infections in 2008: a review and synthetic analysis. Lancet Oncol. 2012;13:607-615. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1583] [Cited by in RCA: 1716] [Article Influence: 132.0] [Reference Citation Analysis (1)] |
| 7. | Cancer Genome Atlas Research Network. Comprehensive molecular characterization of gastric adenocarcinoma. Nature. 2014;513:202-209. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 5015] [Cited by in RCA: 4845] [Article Influence: 440.5] [Reference Citation Analysis (2)] |
| 8. | Selgrad M, Malfertheiner P, Fini L, Goel A, Boland CR, Ricciardiello L. The role of viral and bacterial pathogens in gastrointestinal cancer. J Cell Physiol. 2008;216:378-388. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 38] [Cited by in RCA: 38] [Article Influence: 2.2] [Reference Citation Analysis (0)] |
| 9. | Ottini L, Falchetti M, Lupi R, Rizzolo P, Agnese V, Colucci G, Bazan V, Russo A. Patterns of genomic instability in gastric cancer: clinical implications and perspectives. Ann Oncol. 2006;17 Suppl 7:vii97-vi102. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 105] [Cited by in RCA: 119] [Article Influence: 7.0] [Reference Citation Analysis (0)] |
| 10. | Wang X, Liu Y, Shao D, Qian Z, Dong Z, Sun Y, Xing X, Cheng X, Du H, Hu Y, Li Y, Li L, Dong B, Li Z, Wu A, Wu X, Bu Z, Zong X, Zhu G, Ji Q, Wen XZ, Zhang LH, Ji JF. Recurrent amplification of MYC and TNFRSF11B in 8q24 is associated with poor survival in patients with gastric cancer. Gastric Cancer. 2016;19:116-127. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 31] [Cited by in RCA: 41] [Article Influence: 4.6] [Reference Citation Analysis (0)] |
| 11. | Qian Z, Zhu G, Tang L, Wang M, Zhang L, Fu J, Huang C, Fan S, Sun Y, Lv J, Dong H, Gao B, Su X, Yu D, Zang J, Zhang X, Ji J, Ji Q. Whole genome gene copy number profiling of gastric cancer identifies PAK1 and KRAS gene amplification as therapy targets. Genes Chromosomes Cancer. 2014;53:883-894. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 37] [Cited by in RCA: 57] [Article Influence: 5.2] [Reference Citation Analysis (0)] |
| 12. | Scheinin I, Sie D, Bengtsson H, van de Wiel MA, Olshen AB, van Thuijl HF, van Essen HF, Eijk PP, Rustenburg F, Meijer GA, Reijneveld JC, Wesseling P, Pinkel D, Albertson DG, Ylstra B. DNA copy number analysis of fresh and formalin-fixed specimens by shallow whole-genome sequencing with identification and exclusion of problematic regions in the genome assembly. Genome Res. 2014;24:2022-2032. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 267] [Cited by in RCA: 359] [Article Influence: 32.6] [Reference Citation Analysis (0)] |
| 13. | Camargo MC, Murphy G, Koriyama C, Pfeiffer RM, Kim WH, Herrera-Goepfert R, Corvalan AH, Carrascal E, Abdirad A, Anwar M, Hao Z, Kattoor J, Yoshiwara-Wakabayashi E, Eizuru Y, Rabkin CS, Akiba S. Determinants of Epstein-Barr virus-positive gastric cancer: an international pooled analysis. Br J Cancer. 2011;105:38-43. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 128] [Cited by in RCA: 136] [Article Influence: 9.7] [Reference Citation Analysis (0)] |
| 14. | Yan M, Schwaederle M, Arguello D, Millis SZ, Gatalica Z, Kurzrock R. HER2 expression status in diverse cancers: review of results from 37,992 patients. Cancer Metastasis Rev. 2015;34:157-164. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 268] [Cited by in RCA: 343] [Article Influence: 34.3] [Reference Citation Analysis (0)] |
| 15. | Wilson EB. Probable Inference, the Law of Succession, and Statistical Inference. J Am Stat Assoc. 1927;22:209-212. |
| 16. | GBD 2017 Stomach Cancer Collaborators. The global, regional, and national burden of stomach cancer in 195 countries, 1990-2017: a systematic analysis for the Global Burden of Disease study 2017. Lancet Gastroenterol Hepatol. 2020;5:42-54. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 425] [Cited by in RCA: 435] [Article Influence: 87.0] [Reference Citation Analysis (0)] |
| 17. | Zhou C, Zhong X, Song Y, Shi J, Wu Z, Guo Z, Sun J, Wang Z. Prognostic Biomarkers for Gastric Cancer: An Umbrella Review of the Evidence. Front Oncol. 2019;9:1321. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 5] [Cited by in RCA: 11] [Article Influence: 1.8] [Reference Citation Analysis (0)] |
| 18. | Ceausu M, Socea B, Ciobotaru VP, Constantin VD, Enache S, Enache V, Bancu A, Socea LI, Șerban D, Predescu D, Smarandache CG, Ceausu Z. A multidisciplinary approach in the diagnostic challenge of GIST. Exp Ther Med. 2021;22:1063. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 2] [Cited by in RCA: 2] [Article Influence: 0.5] [Reference Citation Analysis (0)] |
| 19. | Call J, Walentas CD, Eickhoff JC, Scherzer N. Survival of gastrointestinal stromal tumor patients in the imatinib era: life raft group observational registry. BMC Cancer. 2012;12:90. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 33] [Cited by in RCA: 40] [Article Influence: 3.1] [Reference Citation Analysis (0)] |
| 20. | Radulescu D, Baleanu VD, Padureanu V, Radulescu PM, Bordu S, Patrascu S, Socea B, Bacalbasa N, Surlin MV, Georgescu I, Georgescu EF. Neutrophil/Lymphocyte Ratio as Predictor of Anastomotic Leak after Gastric Cancer Surgery. Diagnostics (Basel). 2020;10. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 27] [Cited by in RCA: 26] [Article Influence: 5.2] [Reference Citation Analysis (0)] |
| 21. | Wang Q, Liu G, Hu C. Molecular Classification of Gastric Adenocarcinoma. Gastroenterology Res. 2019;12:275-282. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 34] [Cited by in RCA: 59] [Article Influence: 9.8] [Reference Citation Analysis (0)] |
| 22. | Cheng Y, He C, Wang M, Ma X, Mo F, Yang S, Han J, Wei X. Targeting epigenetic regulators for cancer therapy: mechanisms and advances in clinical trials. Signal Transduct Target Ther. 2019;4:62. [PubMed] |
| 23. | Locke WJ, Guanzon D, Ma C, Liew YJ, Duesing KR, Fung KYC, Ross JP. DNA Methylation Cancer Biomarkers: Translation to the Clinic. Front Genet. 2019;10:1150. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 159] [Cited by in RCA: 300] [Article Influence: 50.0] [Reference Citation Analysis (0)] |
| 24. | Salama NR, Hartung ML, Müller A. Life in the human stomach: persistence strategies of the bacterial pathogen Helicobacter pylori. Nat Rev Microbiol. 2013;11:385-399. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 405] [Cited by in RCA: 479] [Article Influence: 39.9] [Reference Citation Analysis (2)] |
| 25. | Wroblewski LE, Peek RM Jr, Wilson KT. Helicobacter pylori and gastric cancer: factors that modulate disease risk. Clin Microbiol Rev. 2010;23:713-739. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 817] [Cited by in RCA: 1012] [Article Influence: 67.5] [Reference Citation Analysis (1)] |
| 26. | Cheung TK, Wong BC. Treatment of Helicobacter pylori and prevention of gastric cancer. J Dig Dis. 2008;9:8-13. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 20] [Cited by in RCA: 22] [Article Influence: 1.3] [Reference Citation Analysis (0)] |
| 27. | Cohen JI. Optimal treatment for chronic active Epstein-Barr virus disease. Pediatr Transplant. 2009;13:393-396. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 48] [Cited by in RCA: 36] [Article Influence: 2.3] [Reference Citation Analysis (0)] |
| 28. | Kimura H, Cohen JI. Chronic Active Epstein-Barr Virus Disease. Front Immunol. 2017;8:1867. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 87] [Cited by in RCA: 123] [Article Influence: 15.4] [Reference Citation Analysis (0)] |
| 29. | Hatton OL, Harris-Arnold A, Schaffert S, Krams SM, Martinez OM. The interplay between Epstein-Barr virus and B lymphocytes: implications for infection, immunity, and disease. Immunol Res. 2014;58:268-276. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 97] [Cited by in RCA: 139] [Article Influence: 13.9] [Reference Citation Analysis (0)] |