Published online Feb 15, 2022. doi: 10.4251/wjgo.v14.i2.478
Peer-review started: August 5, 2021
First decision: October 3, 2021
Revised: October 9, 2021
Accepted: January 6, 2022
Article in press: January 6, 2022
Published online: February 15, 2022
Processing time: 189 Days and 13.6 Hours
Stomach adenocarcinoma (STAD) is a leading cause of cancer deaths, but its molecular and prognostic characteristics has never been fully illustrated.
To describe a molecular evaluation of primary STAD and develop new therapies and identify promising prognostic signatures.
We describe a comprehensive molecular evaluation of primary STAD based on comprehensive analysis of energy-metabolism-related gene (EMRG) expression profiles.
On the basis of 86 EMRGs that were significantly associated to patients’ progression-free survival (PFS), we propose a molecular classification dividing gastric cancer into two subtypes: Cluster 1, most of which are young patients and display more immune and stromal cell components in tumor microenvironment and lower tumor priority; and Cluster 2, which show early stages and better PFS. Moreover, we construct a 6-gene signature that can classify the prognostic risk of patients after a three-phase training test and validation process. Compared with patients with low-risk score, patients with high-risk score had shorter overall survival. Furthermore, calibration and DCA analysis plots indicate the excellent predictive performance of the 6-gene signature, and which present higher robustness and clinical usability compared with three previous reported prognostic gene signatures. According to gene set enrichment analysis, gene sets related to the high-risk group were participated in the ECM receptor interaction and hedgehog signaling pathway.
Identification of the EMRG-based molecular subtypes and prognostic gene model provides a roadmap for patient stratification and trials of targeted therapies.
Core Tip: On the basis of 86 energy-metabolism-related gene that were significantly associated to patients’ progression-free survival (PFS), we propose a molecular classification dividing stomach adenocarcinoma into two subtypes: Cluster 1, most of which are young patients and display more immune and stromal cell components in tumor microenvironment and lower tumor priority; and Cluster 2, which show early stages and better PFS. Moreover, by using a three-phase training, test and validation process, we construct a 6-gene signature that can classify the prognostic risk of patients, and which present higher robustness and clinical usability compared with three previous reported prognostic gene signatures.
- Citation: Chang JJ, Wang XY, Zhang W, Tan C, Sheng WQ, Xu MD. Comprehensive molecular characterization and identification of prognostic signature in stomach adenocarcinoma on the basis of energy-metabolism-related genes. World J Gastrointest Oncol 2022; 14(2): 478-497
- URL: https://www.wjgnet.com/1948-5204/full/v14/i2/478.htm
- DOI: https://dx.doi.org/10.4251/wjgo.v14.i2.478
Gastric cancer (GC) is one of the most common malignancies in the digestive system. Within the last decades, the incidence rate of GC has gradually declined in some regions due to effective preventive measures and early diagnosis strategies[1]. However, inoperable GC cases that are diagnosed at an advanced stage still have a poor prognosis[2]. According to the data of GLOBOCAN 2018, GC ranked third in global cancer mortality rate, only behind lung cancer and colorectal cancer in both genders combined[3]. Therefore, there is still urgent need to accurately predict the clinical outcomes of GC patients for the sake of more individualized management.
Reprogrammed metabolic pattern has long been recognized as a hallmark of cancers. Tumor cells can have different manners of nutrient acquisition and consumption compared to normal cells to obtain and maintain malignant features[4]. The most well-known feature of cancer metabolism is the increased glycolysis and lactate production even in an oxygen-rich microenvironment, which is termed as “Warburg effect”[5]. Until now, it was generally believed that glucose is the major source of energy for cancer cells[6]. However, there is a growing awareness that the metabolic phenotype of cancer cells is largely heterogeneous. Some tumor cells primarily utilize glycolysis, while some other tumors have a metabolic property of oxidative phosphorylation (OXPHOS)[7]. Accumulating evidences show that there is a metabolic symbiosis between glycolysis and OXPHOS pathways in tumor cells[8]. For example, the lactate and pyruvate produced by glycolysis can act as the substrates of intermediates in the tricarboxylic acid cycle (TCA) to help generate adenosine triphosphate (ATP) in neighboring cells[9]. Similarly, some other non-glucose nutrients (i.e. free fatty acids, amino acids) may serve as the alternative fuels to fulfill the energy burden of tumor cells[10]. Since the complex metabolic characteristics of tumor cells can greatly influence the clinical fate of malignancies, a deeper understanding of the cancer metabolic fingerprint may be crucial to develop new therapies and identify promising prognostic signatures.
In the present study, we aimed to select key prognostic factors of GC among the 587 energy metabolism genes, and construct a potential metabolism-related model for the survival prediction of GC patients. The model was trained and verified among a total of 339 GC samples from The Cancer Genome Atlas (TCGA) Stomach Adenocarcinoma STAD) dataset and 300 tumor samples from the GSE62254 dataset of the Gene Expression Omnibus (GEO). Moreover, molecular classification of GC based on the expression of energy-metabolism-related genes was also conducted to decipher the underlying role of metabolism in GC.
The TCGA-STAD dataset and GSE62254 dataset were analyzed for signature identification. The “Level 3” RNA sequencing (RNA-seq) data and clinical characteristic information of STAD tumor samples were collected from the TCGA-STAD dataset using the gdc-client tool (https://portal.gdc.cancer.gov/). Gene IDs were converted into official gene symbols according to the Genome Reference Consortium Human Build 38 (GRCh38) assembly. Only genes with average Fragments Per Kilobase per Million (FPKM) value greater than zero in more than 70% samples were included for the analysis. The microarray gene expression profiles and patients’ clinical information of GSE62254 dataset was downloaded from Gene Expression Omnibus database (GEO, https://www.ncbi.nlm.nih.gov/geo/). Probes were mapped to gene symbols according to the corresponding platform file GPL570. The progression-free survival (PFS) period of each STAD patient from the two datasets was calculated, and samples with PFS less than 30 d were excluded from the analysis. A total of 639 STAD subjects were thus analyzed with 339 from TCGA-STAD dataset and 300 from GSE62254 dataset (Table 1).
| Characteristic | Training set (n = 170) | Validation set (n = 169) | P value | Entire TCGA dataset (n = 339) | GSE62254 dataset (n = 300) |
| Age (yr) | 0.701 | ||||
| ≤ 60 | 54 | 58 | 112 | 117 | |
| > 60 | 116 | 111 | 227 | 183 | |
| Progression-free survival | 0.326 | ||||
| Absent | 105 | 114 | 219 | 148 | |
| Present | 65 | 55 | 120 | 152 | |
| Gender | 0.238 | ||||
| Female | 54 | 65 | 119 | 101 | |
| Male | 116 | 104 | 220 | 199 | |
| Grade | 0.278 | ||||
| G1 | 6 | 3 | 9 | - | |
| G2 | 63 | 60 | 123 | - | |
| G3 | 99 | 99 | 198 | - | |
| Gx | 2 | 7 | 9 | - | |
| pT stage | 0.614 | ||||
| T1 | 10 | 7 | 17 | - | |
| T2 | 38 | 35 | 73 | - | |
| T3 | 75 | 83 | 158 | - | |
| T4 | 50 | 41 | 91 | - | |
| pN stage | 0.175 | ||||
| N0 | 52 | 46 | 98 | - | |
| N1 | 54 | 40 | 94 | - | |
| N2 | 30 | 38 | 68 | - | |
| N3/Nx | 33 | 44 | 77 | - | |
| pM stage | 0.707 | ||||
| M0 | 154 | 150 | 304 | - | |
| M1/Mx | 16 | 19 | 35 | - | |
| Tumor Stage | 0.11 | ||||
| Stage I | 24 | 22 | 46 | 30 | |
| Stage II | 57 | 49 | 106 | 96 | |
| Stage III | 58 | 80 | 138 | 95 | |
| Stage IV | 21 | 13 | 34 | 77 | |
| MSI status | 0.793 | ||||
| MSI-H | 21 | 19 | 40 | 68 | |
| MSI-L | 18 | 16 | 34 | ||
| MSS | 65 | 71 | 136 | 186 | |
| EMT | - | - | - | 46 | |
| Lauren classification | 0.083 | ||||
| Diffuse | 18 | 29 | 47 | 142 | |
| Intestinal | 77 | 63 | 140 | 150 | |
| Mixed | 6 | 10 | 16 | 8 | |
| WHO classification | 0.057 | ||||
| Mucinous | 6 | 8 | 14 | - | |
| Papillary | 9 | 7 | 16 | - | |
| Poorly Cohesive | 18 | 29 | 47 | - | |
| Tubular | 59 | 37 | 96 | - | |
| Mixed | 6 | 10 | 16 | - |
The metabolic-related pathways were downloaded from Reactome (https://reactome.org/) and a total of 587 energy metabolism-related genes from 11 pathways were screened out for variate selection (Supplementary Table 1). Among the 587 genes, one gene was not offered in TCGA-STAD dataset, and the FPKM counts of 2 genes were zero. Eventually, two comprehensive matrixes combining the expression levels of the 584 genes and the clinical information of STAD patients from the two independent datasets were generated separately for further analysis.
The non-negative matrix factorization (NMF) approach was applied for clustering analysis based on the gene expression data of TCGA dataset[11]. Firstly, univariate Cox regression analysis was conducted to identify survival-associated genes among the 584 energy metabolism-related genes. Then, the NMF algorithm and 50 runs were performed with the standard “brunet” pattern using the R package NMF[12]. The range of cluster number (k value) was set as 2 to 10, and the minimum number of members per subtype was set to 10. The optimal number of k value was determined by several parameters including the cophenetic correlation coefficient[11], dispersion and silhouette[13] and residual sum of squares (RSS)[14] to ensure a robust clustering.
The enumeration of six tumor-infiltration immune cells (B cell, CD4+ T cell, CD8+ T cell, neutrophil, macrophage, neutrophils and dendritic cell) was estimated using the “Tumor Immune Estimation Resource” (TIMER, https://cistrome.shinyapps.io/timer/) tool[15]. The “Estimation of STromal and Immune cells in MAlignant Tumors using Expression data (ESTIMATE)” algorithm was applied to calculated the ImmuneScore and StromalScore which represent the relative proportion of immune cells and stromal cells in tumor tissues[16]. The ESTIMATEScore is the sum of ImmuneScore and StromalScore and refers to the purity of tumor tissues.
Network construction, hierarchical clustering analysis was performed firstly to remove the outlier samples. As previously described[17], the connection strength between each pair of genes (nodes in the network) was calculated by Pearson correlation analysis. The soft-threshold power β was set to 8 in order to satisfy a scale-free topology with R2 > 0.8. The topology overlapping matrix (TOM) was then constructed from the adjacency matrix to avoid the influence of noise and spurious associations. On the basis of TOM, average linkage clustering using the dynamic tree cut method was subsequently conducted to define co-expression modules. The size of genes in a module should be more than 30. Module eigengenes were further calculated to explore the relationship among modules. Modules with highly correlated eigengenes were merged together and eventually formed a new module network. The cut-off values of module integration parameters were set as height = 0.25, deepSplit = 2, minModuleSize = 30. In order to identify the modules of interest, the correlation between each co-expression module and patients’ clinical features as well as cluster subtypes was further evaluated. Modules with a significant correlation to the energy-metabolism subtypes of STAD patients were defined as key modules for the subsequent selection of hub genes (Spearman correlation coefficient >0.4, P < 0.05). Functional enrichment analysis of genes in the key modules was further conducted using R package clusterProfiler[18].
Since protein-protein interaction (PPI) analysis can help identify hub genes with core functions, PPI among genes in the identified key modules was further explored. The Search Tool for the Retrieval of Interacting Genes (STRING) is a well-known database containing comprehensive PPI information (version 11.0, https://string-db.org/). The PPI network among these genes was thus mapped to the STRING assembly and then visualized by the Cytoscape software. Important nodes in the network were identified by the Cytoscape plugin cytoHubba[19]. The topological analysis method Degree and the centrality analysis methods Closeness and Betweenness were used separately to identify the hub nodes in the PPI network. Among the top 15 hub nodes identified by each method, only genes with consistent high Degree, Closeness, and Betweenness values (larger than the median value) were selected as hub genes.
The factors in the potential prognostic model were selected from the hub genes identified by WGCNA and PPI analyses. Particularly, the 339 STAD samples in the TCGA-STAD dataset were randomly divided into two sets for the training (n = 170) and testing (n = 169) of the model (Table 1). In order to avoid selection bias, 100 times repetition sampling were conducted to the ensure the even distribution of patients’ clinical characteristics between the training and testing sets. The Chi-squared test was performed and two-sided P values > 0.05 for all the parameters were considered to be efficient.
In the training set, univariate Cox regression analysis was firstly performed to identify prognosis-associated genes from the hub genes (P < 0.05). To minimize overfitting, least absolute shrinkage and selection operator (Lasso) regression analysis was further conducted using the R package glmnet for model construction[20,21]. The optimal lambda value was determined through 10-fold cross-validation. The coefficients of the variates included in the constructed model were estimated by the analysis and used to calculate the risk score of each STAD patient. Z-score normalization of risk score was further performed and zero was set as the cut-off value to determine the high-risk and low-risk patients.
The nomogram integrating the identified signature and clinical information was built to improve the predictive capability[22]. The performance of the nomogram was assessed by calibration plot analysis. To assess the superiority of the identified energy metabolism-related prognostic signature, the predictive performance of the present model was further compared with the three other models proposed by previous studies using K-M survival analysis, ROC curve analysis, Harrell's concordance index (C-index), and decision curve analysis (DCA)[23].
Gene set enrichment analysis (GSEA) was performed to identify the functional difference between the high-risk and low-risk STAD patients in the TCGA dataset. Briefly, expression levels of all the protein-coding genes were input for analysis using the GSEA software (version 4.0.3). The classical gene sets of Kyoto Encyclopedia of Genes and Genomes (KEGG) pathways (c2.cp.kegg.v7.0.symbols) were considered to decipher the phenotype. For each analytical pathway, the enrichment score (ES) and the significance of ES were calculated, and the normalized enrichment score (NES) and false discovery rate (FDR) were further calculated to examine functional enrichment results. An FDR cutoff value of 0.05 was considered in this test.
The survival status of the high-risk and low-risk subgroups was compared by Kaplan-Meier (K-M) survival analysis. Time-dependent Receiver operating characteristic (ROC) curve analysis was conducted to assess the prognostic value of the identified model using the R package timeROC[24]. The independence of the prognostic signature in the survival prediction of STAD patients was evaluated using univariate and multivariate Cox regression analyses. The prognostic performance of the signature was similarly evaluated in the TCGA testing and GEO external validation set. The immune status of tumor samples such as immune cells infiltration and tumor purity was compared between different subtypes in the TCGA-STAD dataset using Wilcoxon test. All statistical analyses were using R 3.6.0 (https://mirrors.tuna.tsinghua.edu.cn/CRAN/) with default software parameters. P value < 0.05 was considered significant statistically.
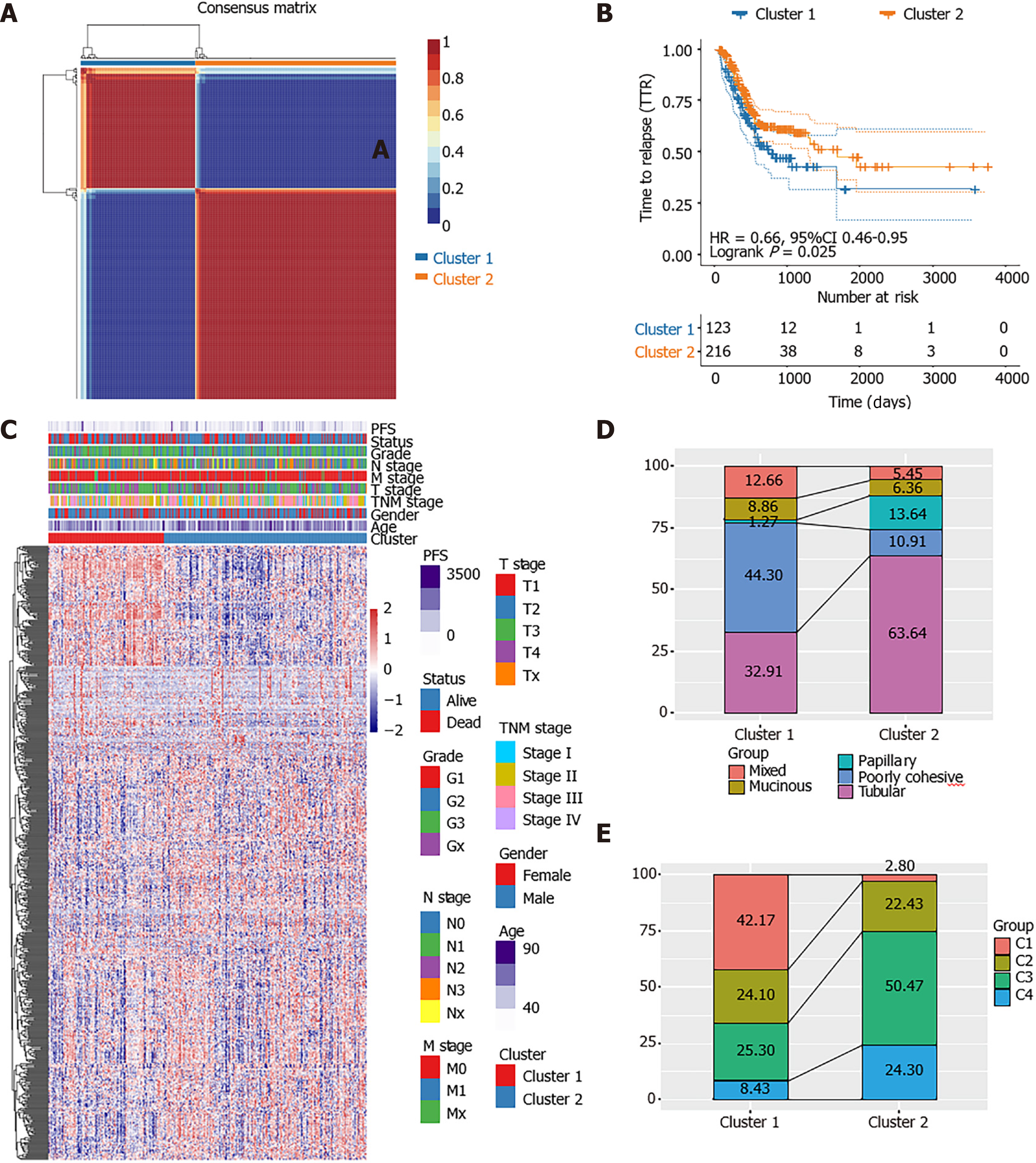
Among the 584 EMRGs, a total of 86 genes were significantly associated with the prognosis of STAD patients according to the results of univariate Cox regression analysis with P < 0.05 (Supplementary Table 2). The NMF analysis based on the expression of the 86 genes eventually identified two distinct subtypes (Cluster1 [n = 123], Cluster2 [n = 216]) among the 336 STAD patients in the TCGA dataset, which might have close association with cancer energy metabolism processes and prognosis in STAD (Figure 1A). Kaplan-Meier survival analysis revealed that the PFS of STAD patients of the two clusters was significantly different (P = 0.025, HR = 0.66, 95%CI 0.46-0.95; Figure 1B). As shown in the heatmap of gene expression across the two clusters (Figure 1C), the great majority of EMRGs presented higher expression levels in Cluster 2 compared with Cluster 1.
Further comparison with World Health Organization classification suggested that Cluster 1 and Cluster 2 were inclined to the poorly cohesive and tubular subtypes, respectively (Figure 1D). Supported by the TCGA project, Adam et al[25] once divided STAD patients into four TCGA subtypes: Epstein–Barr virus positive (C1), micro
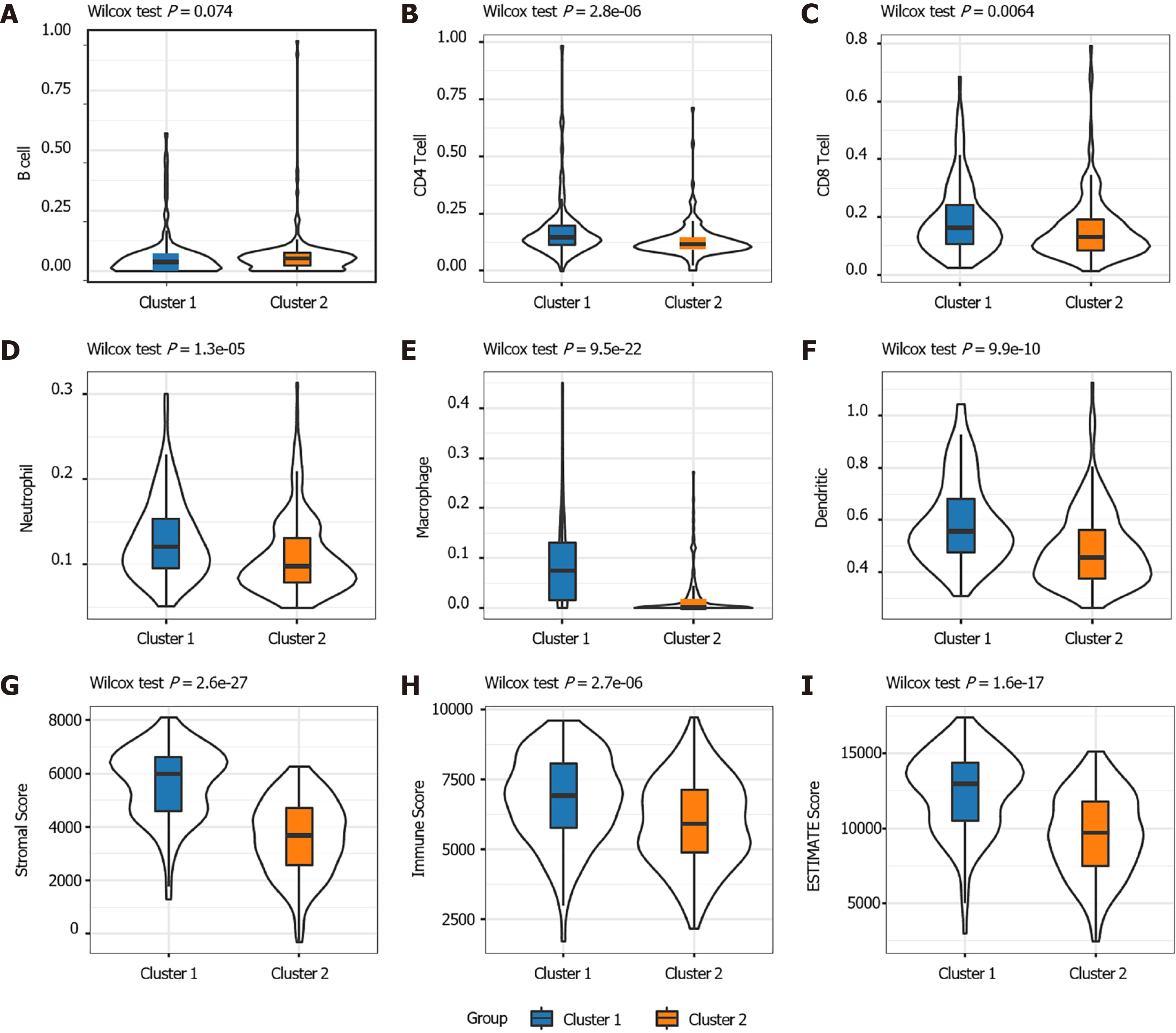
The distribution of the two clusters in STAD patients with different clinical characteristics was further analyzed. It was observed that most of patients with T1 or T2 stage and TNM Stage I were divided into Cluster 2 which had better survival. The Cluster 1 with poor outcomes inversely showed a trend of younger ages (Supplementary Figure 1). The proportions of tumor-infiltration immune cells and the fractions of immune and stromal cell components in tumor microenvironment (TME) were further compared between the two subgroups to explore the association between energy metabolism phenotype and immune status in STAD (Figure 2). The proportions of CD4+ T cells, CD8+ T cells, macrophage, neutrophils and dendritic cell were all significantly higher in Cluster 1 than in Cluster2 (Figure 2A-F). The calculated ImmuneScore, StromalScore, and ESTIMATEScore were also remarkably higher in Cluster 1, which represented more immune and stromal cell components in TME and lower tumor priority for the samples in Cluster 1 (Figure 2G-I). The results further suggested the close association between cancer cell energy metabolism, immune regulation, and clinical outcomes in STAD.
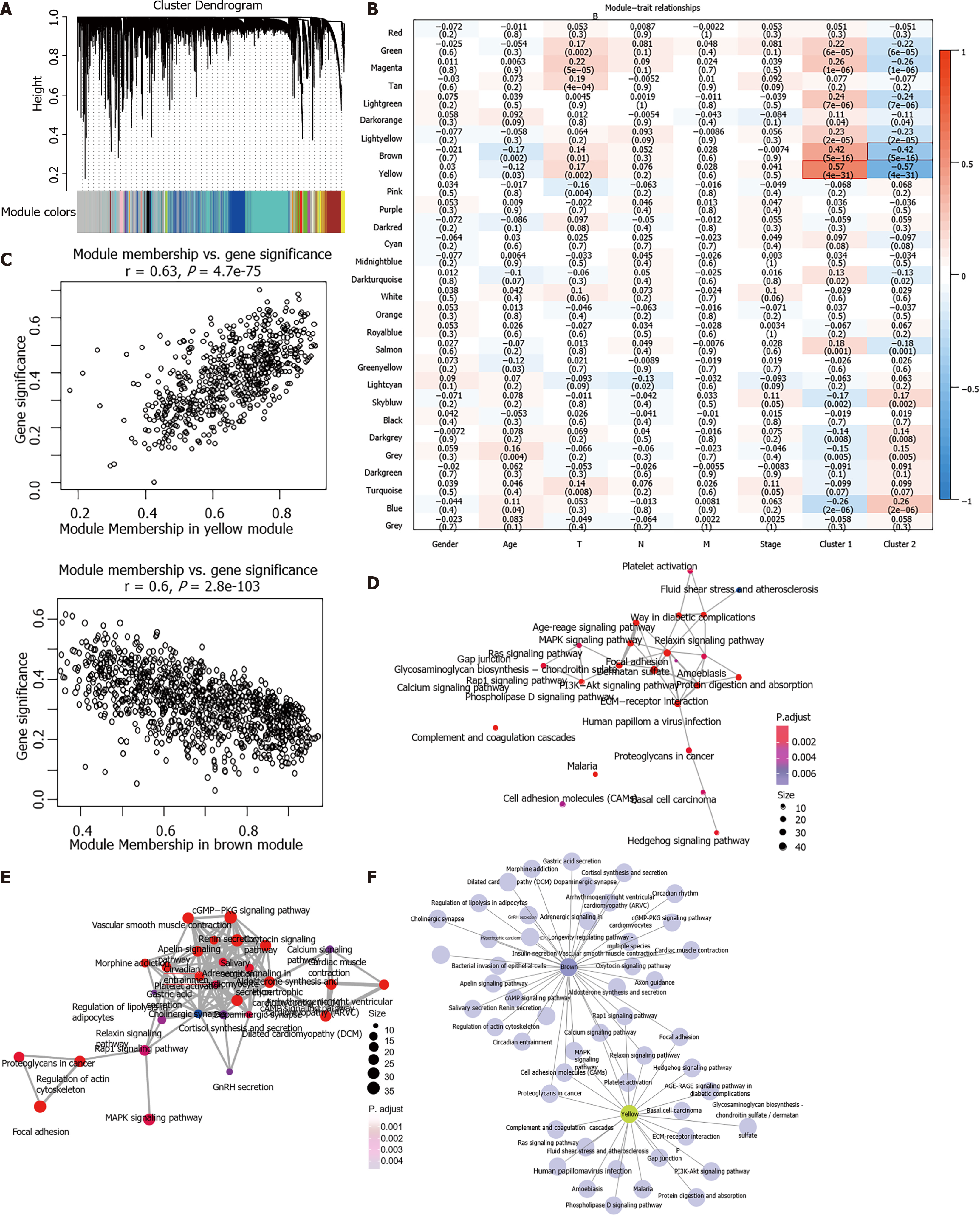
One outlier sample was identified by the hierarchical clustering analysis and removed from WGCNA co-expression analysis (Supplementary Figure 2A-C). Based on the expression of EMRGs in the TCGA dataset, a total of 29 co-expression modules were obtained after module fusion (Figure 3A, grey modules represent gene sets couldn’t be merged). The relationship between the identified modules and clinical characteristics as well as molecular classifications was shown in Figure 3B. It was concluded that Cluster 1 and Cluster 2 were significantly correlated with the yellow and brown module, respectively (r > 0.4, P < 0.05). The correlation between clinical phenotypes and the obtained modules as well as the genes of the modules was listed in Supplementary Table 3. As shown in Figure 3C, members in the yellow module were largely correlated with the Cluster 1 subtype, while members in the brown module were remarkably associated with the Cluster 2 phenotype. Therefore, the two modules having close relationship with energy metabolism-based subtypes of STAD were considered as the key modules, and the genes involved in these key modules were regarded as candidate genes for hub genes identification.
Functional enrichment analysis demonstrated that 23 KEGG pathways (i.e. MAPK signaling pathway, ECM−receptor interaction) were significantly involved in the yellow module (FDR < 0.01; Figure 3D, Supplementary Table 4) and 35 pathways (i.e. cGMP−PKG signaling pathway, Rap1 signaling pathway) significantly involved in the brown module (FDR < 0.01; Figure 3E, Supplementary Table 5). Most of these pathways were classical cancer-related biological processes. Moreover, the crosstalk of pathways was quite limited (Figure 3F), which further demonstrated the functional heterogeneity of the two key modules.
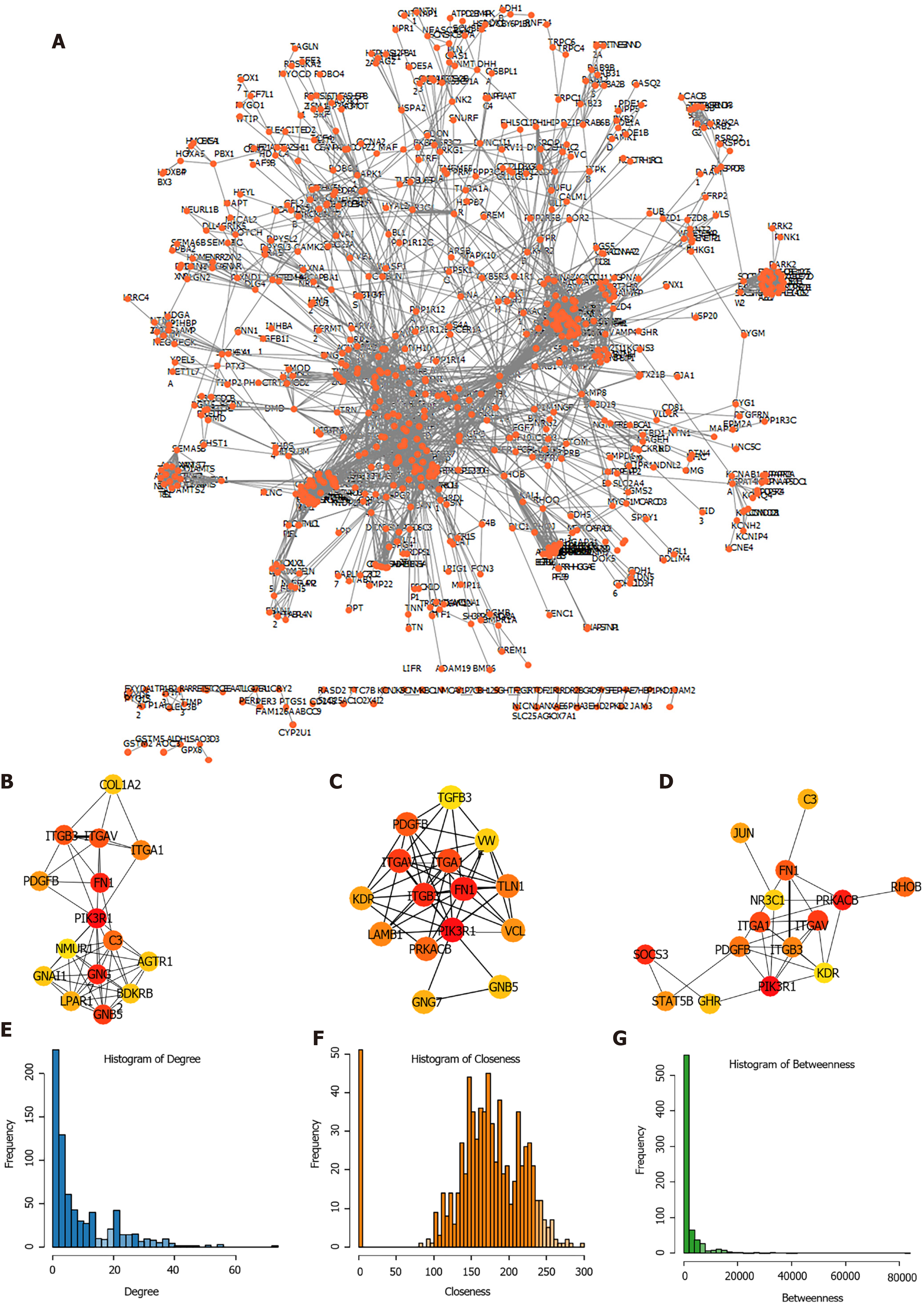
Subsequently, the expression of the candidate genes in key modules was mapped to STRING database to construct PPI network. A total of 3585 PPI pairs with a score higher than 0.9 were matched among the 1713 co-expression genes (Figure 4A, Supplementary Table 5). The top hub genes identified by the Degree (Figure 4B), Closeness (Figure 4C), and Betweenness (Figure 4D) methods were largely consistent (Supplementary Table 6). The topological properties of the PPI network were also evaluated and the distributions of degree, closeness, and betweenness were shown in Figure 4E-G. A total of 220 genes that satisfied high degree, closeness, and betwe
The clinical information of STAD patients in the training (n = 170), testing (n = 169) and external validation (n = 300) sets used for model construction and evaluation was listed in Table 1. In the training set, after the selection of univariate Cox regression and Lasso regression analysis (Supplementary Figure 2D and E), six genes (DYNC1I1, GPER1, MFAP2, ARRB1, C3 and GLI1) out of the 220 hub genes were included in the prognostic model (Table 2). And a gene-based prognostic model was established to evaluate the survival risk of each patient as follows: Risk score = 0.38585 × expDYNC1I1 + 0.10411 × expGPER1 + 0.04476 × expMFAP2 - 0.70386 × expARRB1 + 0.09187 × expC3 + 0.21797 × expGLI1.
| Symbol | Coefficient | Hazard ratio | Z-score | P value | Low 95%CI | High 95%CI |
| DYNC1I1 | 0.38585 | 1.4709 | 2.307 | 0.021 | 1.060 | 2.041 |
| GPER1 | 0.10411 | 1.1097 | 0.648 | 0.517 | 0.810 | 1.520 |
| MFAP2 | 0.04476 | 1.0458 | 0.399 | 0.690 | 0.839 | 1.303 |
| ARRB1 | -0.70386 | 0.4947 | -3.499 | ≤ 0.001 | 0.334 | 0.734 |
| C3 | 0.09187 | 1.0962 | 1.081 | 0.280 | 0.928 | 1.295 |
| GLI1 | 0.21797 | 1.2435 | 1.245 | 0.213 | 0.882 | 1.752 |
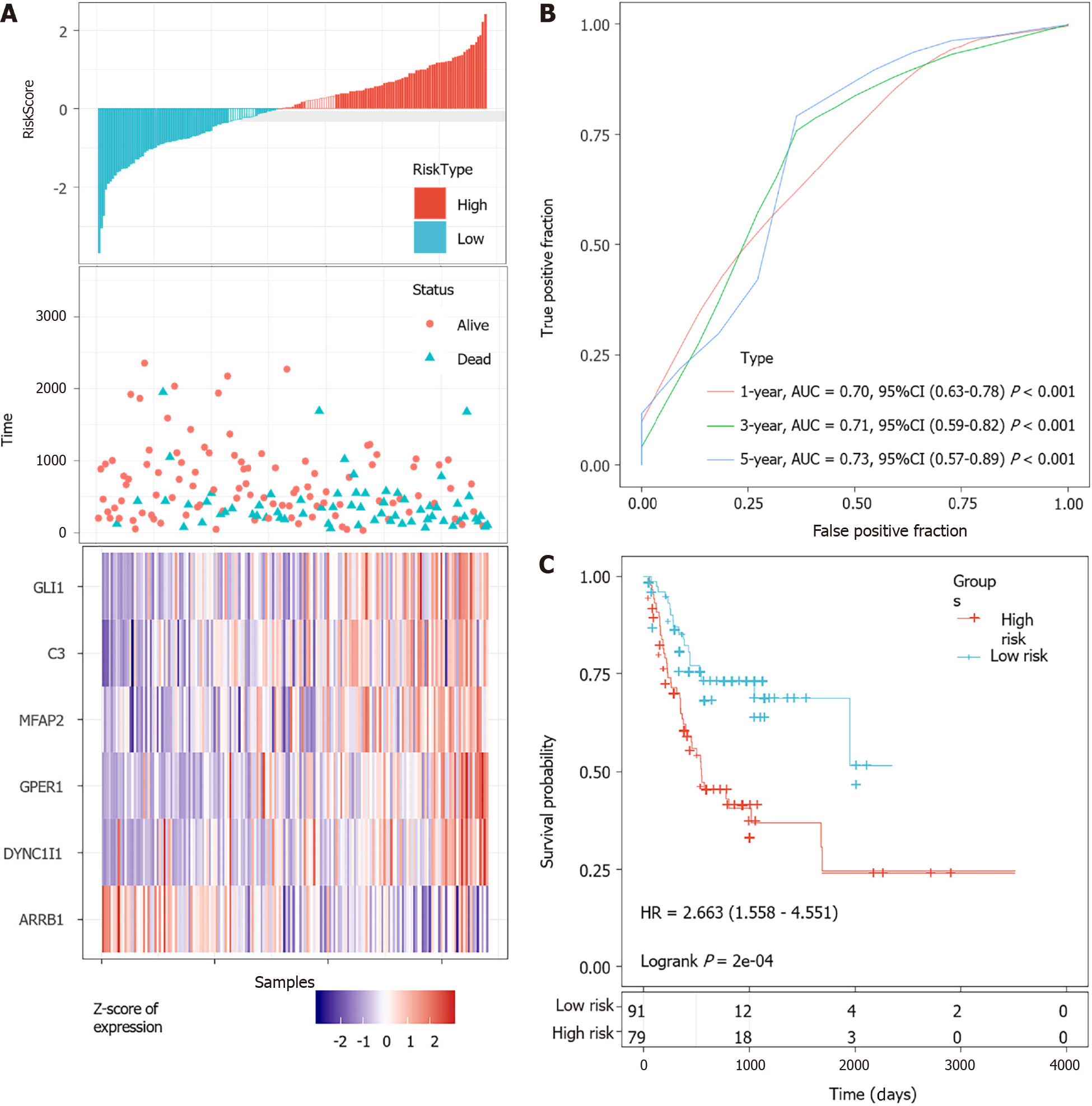
According to the cut-off value of normalized risk score (Z-score = 0), STAD patients were divided into high- and low-risk groups. The distribution of risk scores in the training set was shown in Figure 5A, which showed that expression levels of DYNC1I1, GPER1, MFAP2, C3, and GLI1 were positively correlated with risk scores, while ARRB1 levels was negatively correlated with risk scores. It was concluded that higher ARRB1 expression was associated with a worse prognosis and was a favorable prognostic, while the other 5 genes were identified as unfavorable prognostic factors for STAD patients. The AUCs of 1-year, 3-year and 5-year ROC curves for the 6-gene signature to predict STAD survival were 0.70, 0.71 and 0.73, respectively (Figure 5B). Kaplan-Meier survival analysis confirmed that the high-risk group had significantly worse PFS than the low-risk group (Figure 5C).
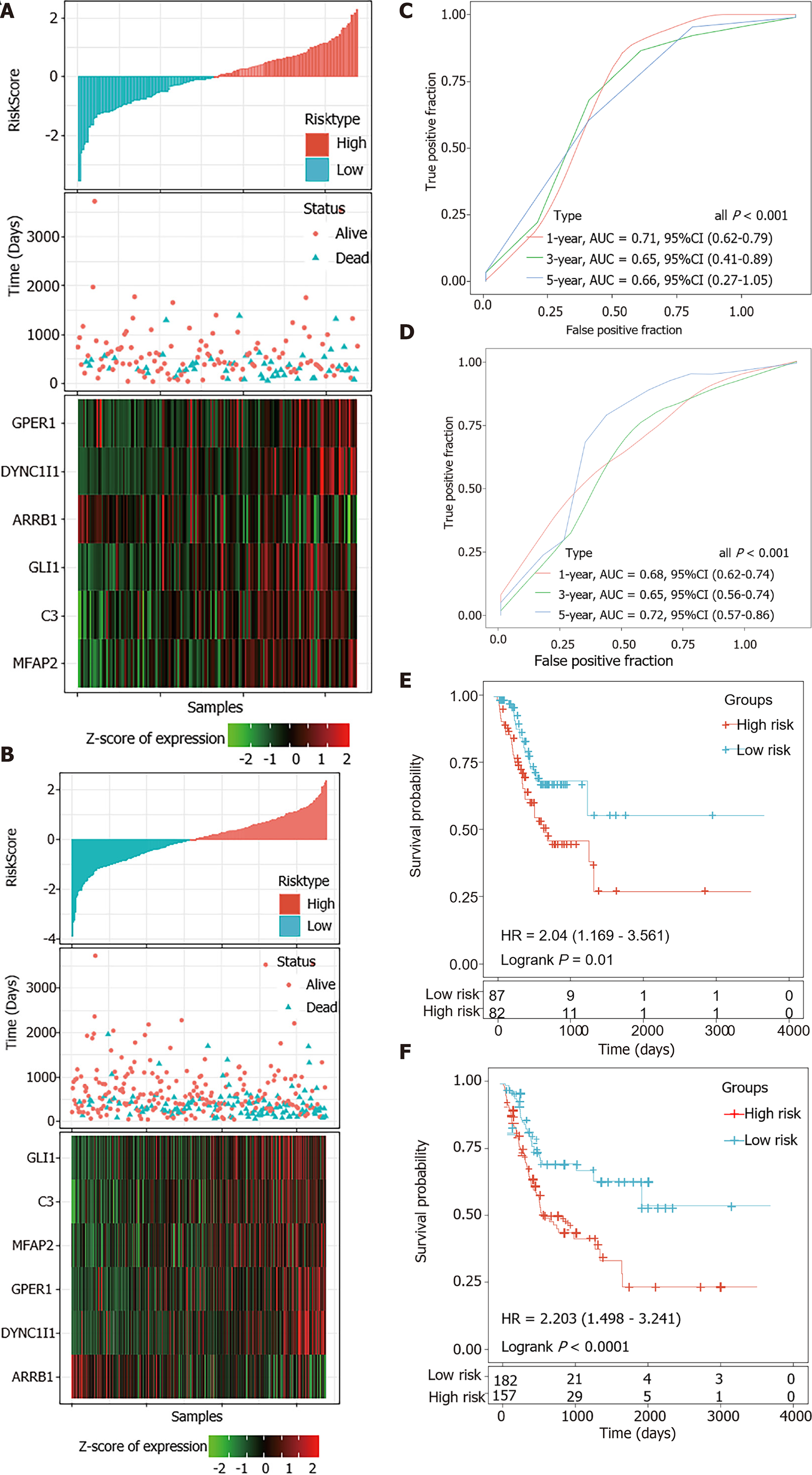
The risk scores of STAD patients in the testing and internal validation sets were further calculated using the same coefficients. Patients were sub-grouped using the same cutoff value as the training set. The corresponding ROC curve and Kaplan-Meier survival curves for the TCGA testing set and the entire TCGA dataset showed that the AUCs of the signature remained high and the high-risk groups had consistently shorter PFS periods than the low-risk groups (Figure 6).
A total of 300 STAD samples in GSE62254 were analyzed for the external validation of the signature. In this dataset, ARRB1 was a consistent protective factor while the other five genes were still risk factors for STAD survival (Figure 7A). The robustness of the signature was further verified (Figure 7B and C).
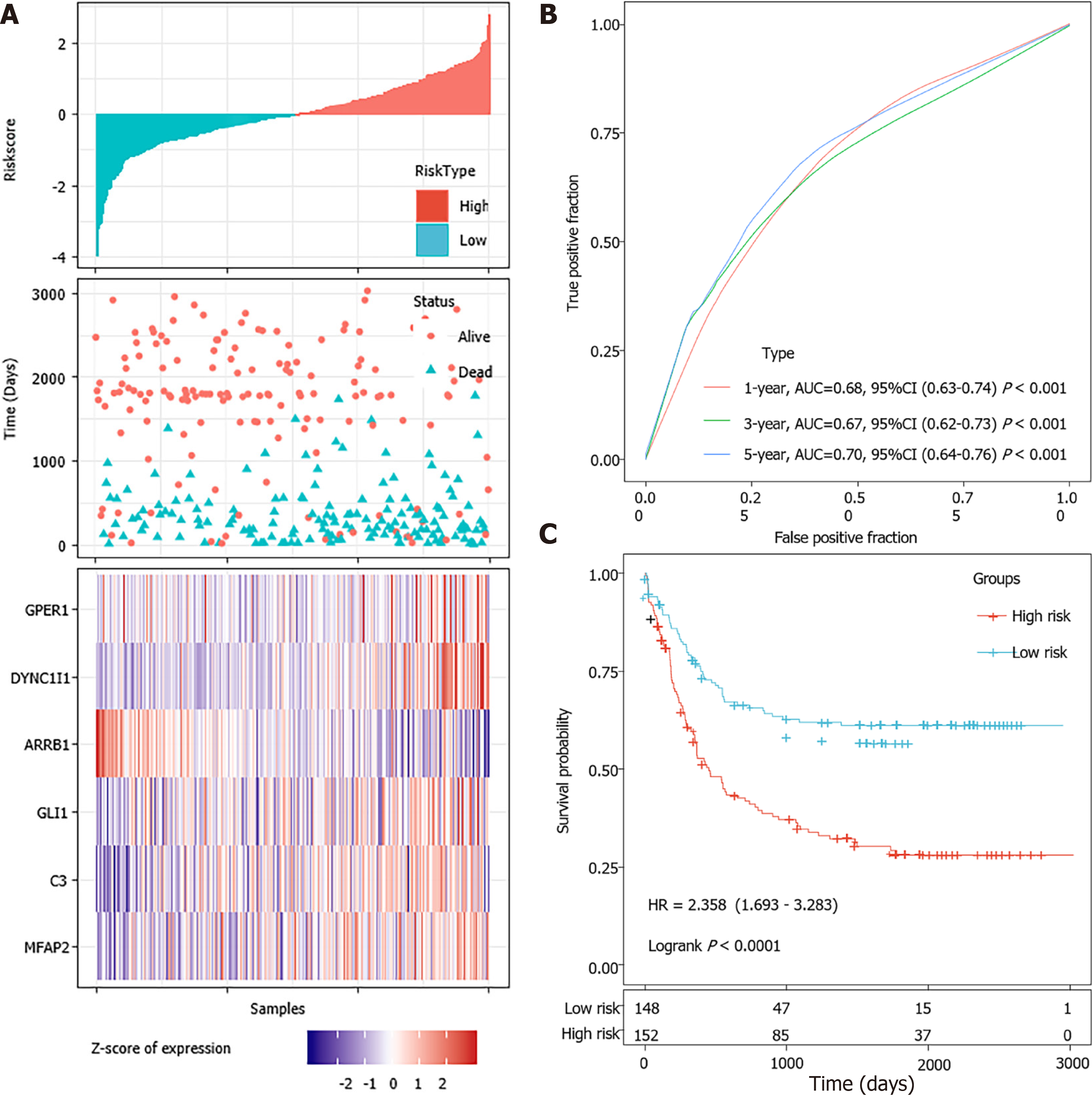
The predictive performance of the prognostic model was evaluated among the 339 STAD patients with varied clinical features in the TCGA dataset. The results of subgroup survival analyses revealed that the 6-gene signature could effectively discriminate high-risk and low-risk patients among the elder, both sexes, all stage, Lauren intestinal type and microsatellite instability-high (MSI-H) subgroups, which expanded its potential application (Supplementary Figure 3A). Univariate and multivariate Cox regression analyses were further performed to evaluate the clinical independence of the identified signature. It was proved that the calculated risk score could independently predict the PFS of STAD patients without the interference of other clinical parameters in the TCGA dataset (Supplementary Figure 3B).
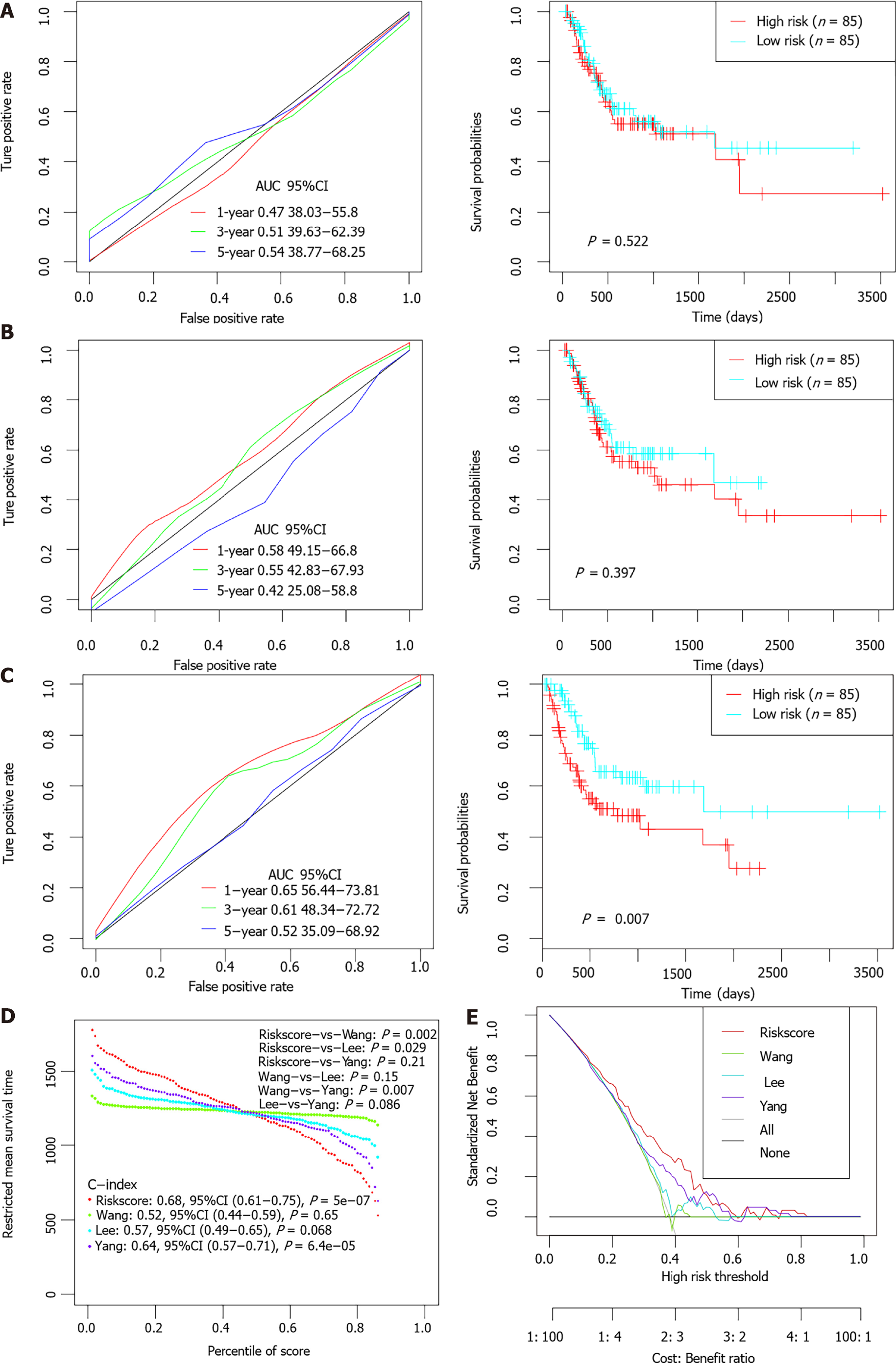
Previous studies had identified several prognostic models for survival prediction of STAD patients. The predictive performance of the present 6-gene signature was further compared with three previous models (a 5-gene signature proposed by Wang et al[26], a 6-gene signature proposed by Lee et al[27], and a 10 immune-related gene signature proposed by Yang et al[28]. For normalization, gene expression levels in each model were uniformly extracted from the original matrix of the TCGA-STAD dataset. The risk score of each STAD patient was calculated accordingly based on the corresponding coefficients provided by each model. Patients were divided into high-and low-risk groups separately according to the median value of normalized risk score for each signature. The comparative plots of Kaplan-Meier survival curve and ROC curves were shown in Figure 8A-C. Restricted mean survival time (RMST) was applied to calculated and compared the C-index of all signatures. The AUCs of the present 6-EMRG models were relatively higher and more stable than the other signatures, and the C-index was the highest among the four models (Figure 8D). DCA curves further demonstrated that the 6-gene signature had better clinical utility than the other models in the survival prediction of STAD patients (Figure 8E).
ssGSEA was performed to determine the potential related pathways according to patients’ prognostic risk in the entire TCGA dataset, and pathways with FDR < 0.05 were derived. By dividing samples into high-risk group and low-risk group based on whether the Riskscore is greater than 0, and analyzed the enriched pathway in both groups by using GSEA, we found that 10 pathways were significantly enriched in the high-risk group, such as ECM receptor interaction, hedgehog signaling pathway and etc.; while only citrate cycle TCA cycle was significantly enriched in the low-risk group (P < 0.05; Supplementary Figure 4). Thus, the 6-gene signature may be involved in the development and progression of STAD by participating in these pathways.
Cumulative evidence has revealed that metabolic reprogramming in cancer has extensive ties with oncogenesis and immune disorder[29,30]. In GC, previous studies suggested that the metabolic alteration in GC was typically characterized by increased glycolysis and repressed aerobic respiration for glucose metabolism, elevated consumption of some amino acids (especially glutamine) for amino acid metabolism, and upregulated fatty acid β-oxidation and oxidative degradation for lipid metabolism and others[31-33]. Moreover, there is a complex interplay among these reprogrammed metabolic pathways which forms the unique metabolic contexture in GC[34].
The detection of aberrant metabolomics also contributes to the identification of novel biomarkers for GC diagnosis or prognostic prediction, and the discovery of potential targets for GC treatment. For example, there are significant differences in metabolic profiles not only between GC patients and normal controls but also among different pathological GC subtypes, and the metabolic alterations have helped identify several promising biomarkers such as 3-hydroxypropionic acid and pyruvic acid in serum, phenylalanine in gastric juice, and alanine in urine[35-37]. Chen et al[38] once discovered proline and serine metabolites that could significantly discriminate metastatic animal models with GC from the non-metastatic samples. GC patients with higher levels of proline, p-cresol and 4-hydroxybenzoic acid in urine might have a worse prognosis according to a population-based study[39]. Taken together, the distinct features of energy metabolism in GC are worth investigating and may indicate novel biomarkers related to metabolism. However, the accurate detection of metabolites in biological samples is still hampered by some technical defects such as lack of optimized study methods, limited coverage in metabolomics fingerprints and interference caused by unwanted sources[40]. Moreover, the abundance of some metabolites can be quite low even less then the detection limit[41]. Gene expression profiling, with the advantage of being convenient and precise, can give a whole picture of tumor properties based on quantitative data[42]. By analyzing the expression levels of energy metabolism-related genes in GC tumor tissue, the metabolic characteristics of GC can be comprehensively interpreted from another dimension.
In the present study, a total of 587 energy metabolism-related genes were selected from the Reactome database. These genes are main participants in the key pathways of carbohydrate, protein, and lipid metabolism. Based on the expression data of the TCGA-STAD dataset, GC patients were divided into two metabolic subtypes using the NMF algorithm. Significant difference was observed in patients’ clinical characteristics and survival state between the two subtypes. This phenomenon further demonstrated the important role of energy metabolism in the development and long-term survival of GC. In addition, previous evidence has proved that metabolic interventions have crucial function in the modulation of cancer immunology[43]. In this study, when the tumor-infiltration immune cells and non-tumor components in TME were compared between the two groups, it could be observed that the proportion of almost all the immune cells and the fraction of immune components were significantly different between the two subtypes with varied metabolic features, which strongly indicated the close relationship between tumor metabolism and immunology in GC. Combined with findings from previous research, the results of this study confirmed the significance of identifying potential prognostic biomarkers from metabolism-related genes.
In order to select the hub genes that may significantly modulate cancer metabolism in GC, WGCNA co-expression analysis was firstly conducted and the PPI network was constructed. A total of 20 genes that strongly correlated with the two metabolic subtypes and had the most connections within the PPI network were screened out and considered as candidates for the construction of the prognostic model. Using the Lasso regression analysis, a six-gene (DYNC1I1, GPER1, MFAP2, ARRB1, C3 and GLI1) signature was identified after the verification of the training, testing, and external validation sets which included a total of 639 GC patients from the TCGA-STAD and GSE62254 dataset. The model interpreted the information of gene expression into risk score for the accurate estimation of prognosis in GC. The results of survival analyses and time-dependent ROC analyses in each set revealed that the signature had stable performance in discriminating high-risk and low-risk GC patients. Notably, the 5-year AUCs for the signature in the whole TCGA-STAD dataset and GSE62254 dataset were 0.72 and 0.70, respectively. Furthermore, subgroup analysis confirmed that the signature performed well in risk prediction among GC patients with different clinical and pathological features. When clinicopathologic parameters were taken into consideration, the constructed risk-score system could still independently predict the prognosis of GC patients. A nomogram integrating the calculated risk score and clinical information was ultimately constructed for the accurate prediction of survival probability of GC patients. The nomogram showed confident clinical utility and outperformed the individual predictor in GC.
Among the six-energy metabolism-related genes, DYNC1I1, GPER1, MFAP2, C3 and GLI1 were risk factors while ARRB1 was a protective factor for clinical outcomes in GC. The prognostic value of the five risk genes have been sporadically reported in previous studies, while the protective value of ARRB1 in GC was rarely identified[44-49]. Functional enrichment analysis revealed that this metabolism-related signature was significantly involved in some classical cancer-related pathways. The interaction between the six genes and tumor metabolism and progression in GC deserves further investigation.
Several previous studies have also identified specific prognostic models for the risk prediction of GC. For example, Lv et al[50] proposed a seven-gene signature which contained TGFB1, EGF, MKI67, ILF3, INCENP, TNPO2 and CHAF1A. Jiang et al[51] identified a biomarker consisting of 16 immune-related genes such as HSPA1A, HSPA1B, HSPA5 et al. Yang et al[28] discovered another immune-related signature containing 10 genes such as NRP1 and TNFRSF18 that was totally different from that of Jiang et al[51]. The prognostic performance of the present model was further compared with that of the three previous models. Among the four different signatures, this six-gene biomarker had the highest C-index and AUC values. It could be concluded that this energy metabolism-related signature outperforms some previous biomarkers in the survival prediction of GC patients, and has great potential to be used for clinical application in the future.
However, there are still some limitations of this study. For example, the analysis was based on just retrospective data and needs to be verified in a prospective cohort containing samples from multi-centers before clinical application. Deeper mechanism research was also needed to elucidate the exact functions of the identified signature in GC.
In summary, by analyzing the expression levels of energy metabolism-related genes in GC tumor tissues, two different clusters with varied clinical characteristics, clinical outcomes, and immune status were identified in the TCGA-STAD dataset. A prognostic signature containing six metabolism-related genes and a novel nomogram was identified for the accurate risk prediction of GC patients.
Energy metabolism has always been a hallmark of cancer cells and the complex metabolic characteristics of tumor cells can greatly influence the clinical fate of malignancies.
A deep understanding of the cancer metabolic fingerprint may be crucial to the development of new therapies and in identifying promising prognostic signatures.
To select key prognostic factors of gastric cancer (GC) among the 587 energy metabolism genes, and construct a potential metabolism-related model for the survival prediction of GC patients.
We trained and verified the energy metabolism-related gene signature among a total of 339 GC samples from The Cancer Genome Atlas (TCGA) Stomach Adenocarcinoma STAD) dataset and 300 tumor samples from the GSE62254 dataset of the Gene Expression Omnibus.
We successfully created a prognostic model based on energy-metabolism-related gene expression profiles in primary stomach adenocarcinoma based on an analysis of the TCGA-STAD and GSE62254 datasets. We were able to divide and identify different subtypes for prognosis and develop a risk score based on 6 gene signatures to potentially stratify the prognosis of individuals which was validated in a second cohort.
In summary, by analyzing the expression levels of energy metabolism-related genes in GC tumor tissues, two different clusters with varied clinical characteristics, clinical outcomes, and immune status were identified in the TCGA-STAD dataset. A prognostic signature containing six metabolism-related genes and a novel nomogram was identified for the accurate risk prediction of GC patients.
This study demonstrates the possibility of the risk score calculated with combination of gene expression in energy metabolism-related prognostic models.
The authors would like to thank all researchers who contributed to the The Cancer Genome Atlas and Gene Expression Omnibus data sets included. An earlier version of it has been presented as preprint in Research square according to the following link: https://www.researchsquare.com/article/rs-138125/v1.
Provenance and peer review: Unsolicited article; Externally peer reviewed.
Peer-review model: Single blind
Specialty type: Gastroenterology and hepatology
Country/Territory of origin: China
Peer-review report’s scientific quality classification
Grade A (Excellent): 0
Grade B (Very good): B
Grade C (Good): 0
Grade D (Fair): 0
Grade E (Poor): 0
P-Reviewer: Tanabe S S-Editor: Wang LL L-Editor: Filipodia P-Editor: Yu HG
| 1. | Sitarz R, Skierucha M, Mielko J, Offerhaus GJA, Maciejewski R, Polkowski WP. Gastric cancer: epidemiology, prevention, classification, and treatment. Cancer Manag Res. 2018;10:239-248. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 743] [Cited by in RCA: 725] [Article Influence: 103.6] [Reference Citation Analysis (0)] |
| 2. | Rawla P, Barsouk A. Epidemiology of gastric cancer: global trends, risk factors and prevention. Prz Gastroenterol. 2019;14:26-38. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 297] [Cited by in RCA: 724] [Article Influence: 103.4] [Reference Citation Analysis (1)] |
| 3. | Bray F, Ferlay J, Soerjomataram I, Siegel RL, Torre LA, Jemal A. Global cancer statistics 2018: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J Clin. 2018;68:394-424. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 53206] [Cited by in RCA: 55777] [Article Influence: 7968.1] [Reference Citation Analysis (132)] |
| 4. | DeBerardinis RJ, Chandel NS. Fundamentals of cancer metabolism. Sci Adv. 2016;2:e1600200. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 1598] [Cited by in RCA: 2054] [Article Influence: 228.2] [Reference Citation Analysis (0)] |
| 5. | Liberti MV, Locasale JW. The Warburg Effect: How Does it Benefit Cancer Cells? Trends Biochem Sci. 2016;41:211-218. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1910] [Cited by in RCA: 3250] [Article Influence: 361.1] [Reference Citation Analysis (1)] |
| 6. | Tekade RK, Sun X. The Warburg effect and glucose-derived cancer theranostics. Drug Discov Today. 2017;22:1637-1653. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 83] [Cited by in RCA: 109] [Article Influence: 13.6] [Reference Citation Analysis (0)] |
| 7. | Nayak AP, Kapur A, Barroilhet L, Patankar MS. Oxidative Phosphorylation: A Target for Novel Therapeutic Strategies Against Ovarian Cancer. Cancers (Basel). 2018;10. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 57] [Cited by in RCA: 98] [Article Influence: 2.2] [Reference Citation Analysis (0)] |
| 8. | Lee M, Yoon JH. Metabolic interplay between glycolysis and mitochondrial oxidation: The reverse Warburg effect and its therapeutic implication. World J Biol Chem. 2015;6:148-161. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in CrossRef: 110] [Cited by in RCA: 117] [Article Influence: 11.7] [Reference Citation Analysis (4)] |
| 9. | Griguer CE, Oliva CR, Gillespie GY. Glucose metabolism heterogeneity in human and mouse malignant glioma cell lines. J Neurooncol. 2005;74:123-133. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 128] [Cited by in RCA: 142] [Article Influence: 7.1] [Reference Citation Analysis (0)] |
| 10. | Keenan MM, Chi JT. Alternative fuels for cancer cells. Cancer J. 2015;21:49-55. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 74] [Cited by in RCA: 99] [Article Influence: 9.9] [Reference Citation Analysis (0)] |
| 11. | Brunet JP, Tamayo P, Golub TR, Mesirov JP. Metagenes and molecular pattern discovery using matrix factorization. Proc Natl Acad Sci U S A. 2004;101:4164-4169. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1255] [Cited by in RCA: 1138] [Article Influence: 54.2] [Reference Citation Analysis (0)] |
| 12. | Gaujoux R, Seoighe C. A flexible R package for nonnegative matrix factorization. BMC Bioinformatics. 2010;11:367. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 747] [Cited by in RCA: 967] [Article Influence: 64.5] [Reference Citation Analysis (0)] |
| 13. | Lovmar L, Ahlford A, Jonsson M, Syvänen AC. Silhouette scores for assessment of SNP genotype clusters. BMC Genomics. 2005;6:35. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 73] [Cited by in RCA: 81] [Article Influence: 4.1] [Reference Citation Analysis (0)] |
| 14. | Stephanie Glen. Sum of Squares: Residual Sum, Total Sum, Explained Sum, Within From StatisticsHowTo.com: Elementary Statistics for the rest of us!. Available from: https://www.statisticshowto.com/residual-sum-squares/. |
| 15. | Tang Z, Li C, Kang B, Gao G, Zhang Z. GEPIA: a web server for cancer and normal gene expression profiling and interactive analyses. Nucleic Acids Res. 2017;45:W98-W102. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 5550] [Cited by in RCA: 7064] [Article Influence: 883.0] [Reference Citation Analysis (0)] |
| 16. | Yoshihara K, Shahmoradgoli M, Martínez E, Vegesna R, Kim H, Torres-Garcia W, Treviño V, Shen H, Laird PW, Levine DA, Carter SL, Getz G, Stemke-Hale K, Mills GB, Verhaak RG. Inferring tumour purity and stromal and immune cell admixture from expression data. Nat Commun. 2013;4:2612. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 3056] [Cited by in RCA: 6426] [Article Influence: 584.2] [Reference Citation Analysis (0)] |
| 17. | Langfelder P, Horvath S. WGCNA: an R package for weighted correlation network analysis. BMC Bioinformatics. 2008;9:559. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 10254] [Cited by in RCA: 16367] [Article Influence: 962.8] [Reference Citation Analysis (0)] |
| 18. | Yu G, Wang LG, Han Y, He QY. clusterProfiler: an R package for comparing biological themes among gene clusters. OMICS. 2012;16:284-287. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 11591] [Cited by in RCA: 22107] [Article Influence: 1700.5] [Reference Citation Analysis (0)] |
| 19. | Chin CH, Chen SH, Wu HH, Ho CW, Ko MT, Lin CY. cytoHubba: identifying hub objects and sub-networks from complex interactome. BMC Syst Biol. 2014;8 Suppl 4:S11. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 1658] [Cited by in RCA: 3738] [Article Influence: 339.8] [Reference Citation Analysis (0)] |
| 20. | Friedman J, Hastie T, Tibshirani R. Regularization Paths for Generalized Linear Models via Coordinate Descent. J Stat Softw. 2010;33:1-22. [PubMed] |
| 21. | Candia J, Tsang JS. eNetXplorer: an R package for the quantitative exploration of elastic net families for generalized linear models. BMC Bioinformatics. 2019;20:189. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 18] [Cited by in RCA: 31] [Article Influence: 5.2] [Reference Citation Analysis (0)] |
| 22. | Lubsen J, Pool J, van der Does E. A practical device for the application of a diagnostic or prognostic function. Methods Inf Med. 1978;17:127-129. [PubMed] |
| 23. | Kerr KF, Brown MD, Zhu K, Janes H. Assessing the Clinical Impact of Risk Prediction Models With Decision Curves: Guidance for Correct Interpretation and Appropriate Use. J Clin Oncol. 2016;34:2534-2540. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 224] [Cited by in RCA: 456] [Article Influence: 50.7] [Reference Citation Analysis (0)] |
| 24. | Blanche P, Dartigues JF, Jacqmin-Gadda H. Estimating and comparing time-dependent areas under receiver operating characteristic curves for censored event times with competing risks. Stat Med. 2013;32:5381-5397. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 564] [Cited by in RCA: 1132] [Article Influence: 94.3] [Reference Citation Analysis (0)] |
| 25. | Cancer Genome Atlas Research Network. Comprehensive molecular characterization of gastric adenocarcinoma. Nature. 2014;513:202-209. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 5015] [Cited by in RCA: 4845] [Article Influence: 440.5] [Reference Citation Analysis (2)] |
| 26. | Wang Z, Yan Z, Zhang B, Rao Z, Zhang Y, Liu J, Yu L, Zhao Y, Yang B, Wu T, Gao J. Identification of a 5-gene signature for clinical and prognostic prediction in gastric cancer patients upon microarray data. Med Oncol. 2013;30:678. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 20] [Cited by in RCA: 22] [Article Influence: 1.8] [Reference Citation Analysis (0)] |
| 27. | Cho JY, Lim JY, Cheong JH, Park YY, Yoon SL, Kim SM, Kim SB, Kim H, Hong SW, Park YN, Noh SH, Park ES, Chu IS, Hong WK, Ajani JA, Lee JS. Gene expression signature-based prognostic risk score in gastric cancer. Clin Cancer Res. 2011;17:1850-1857. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 257] [Cited by in RCA: 280] [Article Influence: 20.0] [Reference Citation Analysis (0)] |
| 28. | Yang W, Lai Z, Li Y, Mu J, Yang M, Xie J, Xu J. Immune signature profiling identified prognostic factors for gastric cancer. Chin J Cancer Res. 2019;31:463-470. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 31] [Cited by in RCA: 40] [Article Influence: 6.7] [Reference Citation Analysis (0)] |
| 29. | Andrejeva G, Rathmell JC. Similarities and Distinctions of Cancer and Immune Metabolism in Inflammation and Tumors. Cell Metab. 2017;26:49-70. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 253] [Cited by in RCA: 271] [Article Influence: 33.9] [Reference Citation Analysis (0)] |
| 30. | Hirschey MD, DeBerardinis RJ, Diehl AME, Drew JE, Frezza C, Green MF, Jones LW, Ko YH, Le A, Lea MA, Locasale JW, Longo VD, Lyssiotis CA, McDonnell E, Mehrmohamadi M, Michelotti G, Muralidhar V, Murphy MP, Pedersen PL, Poore B, Raffaghello L, Rathmell JC, Sivanand S, Vander Heiden MG, Wellen KE; Target Validation Team. Dysregulated metabolism contributes to oncogenesis. Semin Cancer Biol. 2015;35 Suppl:S129-S150. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 228] [Cited by in RCA: 223] [Article Influence: 22.3] [Reference Citation Analysis (0)] |
| 31. | Yuan LW, Yamashita H, Seto Y. Glucose metabolism in gastric cancer: The cutting-edge. World J Gastroenterol. 2016;22:2046-2059. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in CrossRef: 64] [Cited by in RCA: 89] [Article Influence: 9.9] [Reference Citation Analysis (0)] |
| 32. | Cluntun AA, Lukey MJ, Cerione RA, Locasale JW. Glutamine Metabolism in Cancer: Understanding the Heterogeneity. Trends Cancer. 2017;3:169-180. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 411] [Cited by in RCA: 546] [Article Influence: 68.3] [Reference Citation Analysis (0)] |
| 33. | Agustsson T, Rydén M, Hoffstedt J, van Harmelen V, Dicker A, Laurencikiene J, Isaksson B, Permert J, Arner P. Mechanism of increased lipolysis in cancer cachexia. Cancer Res. 2007;67:5531-5537. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 203] [Cited by in RCA: 219] [Article Influence: 12.2] [Reference Citation Analysis (0)] |
| 34. | Xiao S, Zhou L. Gastric cancer: Metabolic and metabolomics perspectives (Review). Int J Oncol. 2017;51:5-17. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 69] [Cited by in RCA: 116] [Article Influence: 14.5] [Reference Citation Analysis (0)] |
| 35. | Ikeda A, Nishiumi S, Shinohara M, Yoshie T, Hatano N, Okuno T, Bamba T, Fukusaki E, Takenawa T, Azuma T, Yoshida M. Serum metabolomics as a novel diagnostic approach for gastrointestinal cancer. Biomed Chromatogr. 2012;26:548-558. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 100] [Cited by in RCA: 118] [Article Influence: 8.4] [Reference Citation Analysis (0)] |
| 36. | Deng K, Lin S, Zhou L, Li Y, Chen M, Wang Y. High levels of aromatic amino acids in gastric juice during the early stages of gastric cancer progression. PLoS One. 2012;7:e49434. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 30] [Cited by in RCA: 37] [Article Influence: 2.8] [Reference Citation Analysis (0)] |
| 37. | Chan AW, Mercier P, Schiller D, Bailey R, Robbins S, Eurich DT, Sawyer MB, Broadhurst D. (1)H-NMR urinary metabolomic profiling for diagnosis of gastric cancer. Br J Cancer. 2016;114:59-62. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 68] [Cited by in RCA: 84] [Article Influence: 8.4] [Reference Citation Analysis (0)] |
| 38. | Chen JL, Tang HQ, Hu JD, Fan J, Hong J, Gu JZ. Metabolomics of gastric cancer metastasis detected by gas chromatography and mass spectrometry. World J Gastroenterol. 2010;16:5874-5880. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in CrossRef: 48] [Cited by in RCA: 53] [Article Influence: 3.5] [Reference Citation Analysis (0)] |
| 39. | Chen Y, Zhang J, Guo L, Liu L, Wen J, Xu L, Yan M, Li Z, Zhang X, Nan P, Jiang J, Ji J, Cai W, Zhuang H, Wang Y, Zhu Z, Yu Y. A characteristic biosignature for discrimination of gastric cancer from healthy population by high throughput GC-MS analysis. Oncotarget. 2016;7:87496-87510. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 33] [Cited by in RCA: 38] [Article Influence: 5.4] [Reference Citation Analysis (0)] |
| 40. | Scalbert A, Brennan L, Fiehn O, Hankemeier T, Kristal BS, van Ommen B, Pujos-Guillot E, Verheij E, Wishart D, Wopereis S. Mass-spectrometry-based metabolomics: limitations and recommendations for future progress with particular focus on nutrition research. Metabolomics. 2009;5:435-458. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 417] [Cited by in RCA: 384] [Article Influence: 24.0] [Reference Citation Analysis (0)] |
| 41. | Kang YP, Ward NP, DeNicola GM. Recent advances in cancer metabolism: a technological perspective. Exp Mol Med. 2018;50:1-16. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 38] [Cited by in RCA: 41] [Article Influence: 5.9] [Reference Citation Analysis (0)] |
| 42. | Nevins JR, Potti A. Mining gene expression profiles: expression signatures as cancer phenotypes. Nat Rev Genet. 2007;8:601-609. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 137] [Cited by in RCA: 139] [Article Influence: 7.7] [Reference Citation Analysis (0)] |
| 43. | O'Sullivan D, Sanin DE, Pearce EJ, Pearce EL. Metabolic interventions in the immune response to cancer. Nat Rev Immunol. 2019;19:324-335. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 135] [Cited by in RCA: 209] [Article Influence: 34.8] [Reference Citation Analysis (0)] |
| 44. | Gong LB, Wen T, Li Z, Xin X, Che XF, Wang J, Liu YP, Qu XJ. DYNC1I1 Promotes the Proliferation and Migration of Gastric Cancer by Up-Regulating IL-6 Expression. Front Oncol. 2019;9:491. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 14] [Cited by in RCA: 27] [Article Influence: 4.5] [Reference Citation Analysis (0)] |
| 45. | Tian S, Zhan N, Li R, Dong W. Downregulation of G Protein-Coupled Estrogen Receptor (GPER) is Associated with Reduced Prognosis in Patients with Gastric Cancer. Med Sci Monit. 2019;25:3115-3126. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 7] [Cited by in RCA: 21] [Article Influence: 3.5] [Reference Citation Analysis (0)] |
| 46. | Yao LW, Wu LL, Zhang LH, Zhou W, Wu L, He K, Ren JC, Deng YC, Yang DM, Wang J, Mu GG, Xu M, Zhou J, Xiang GA, Ding QS, Yang YN, Yu HG. MFAP2 is overexpressed in gastric cancer and promotes motility via the MFAP2/integrin α5β1/FAK/ERK pathway. Oncogenesis. 2020;9:17. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 40] [Cited by in RCA: 40] [Article Influence: 8.0] [Reference Citation Analysis (0)] |
| 47. | Wang JK, Wang WJ, Cai HY, Du BB, Mai P, Zhang LJ, Ma W, Hu YG, Feng SF, Miao GY. MFAP2 promotes epithelial-mesenchymal transition in gastric cancer cells by activating TGF-β/SMAD2/3 signaling pathway. Onco Targets Ther. 2018;11:4001-4017. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 27] [Cited by in RCA: 41] [Article Influence: 5.9] [Reference Citation Analysis (0)] |
| 48. | Ye J, Ren Y, Chen J, Song W, Chen C, Cai S, Tan M, Yuan Y, He Y. Prognostic Significance of Preoperative and Postoperative Complement C3 Depletion in Gastric Cancer: A Three-Year Survival Investigation. Biomed Res Int. 2017;2017:2161840. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 7] [Cited by in RCA: 8] [Article Influence: 1.0] [Reference Citation Analysis (0)] |
| 49. | Shao X, Kuai X, Pang Z, Zhang L, Wu L, Xu L, Zhou C. Correlation of Gli1 and HER2 expression in gastric cancer: Identification of novel target. Sci Rep. 2018;8:397. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 13] [Cited by in RCA: 18] [Article Influence: 2.6] [Reference Citation Analysis (0)] |
| 50. | Lv X, Zhao Y, Zhang L, Zhou S, Zhang B, Zhang Q, Jiang L, Li X, Wu H, Zhao L, Wei M, He M. Development of a novel gene signature in patients without Helicobacter pylori infection gastric cancer. J Cell Biochem. 2020;121:1842-1854. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 3] [Cited by in RCA: 7] [Article Influence: 1.2] [Reference Citation Analysis (0)] |
| 51. | Jiang B, Sun Q, Tong Y, Wang Y, Ma H, Xia X, Zhou Y, Zhang X, Gao F, Shu P. An immune-related gene signature predicts prognosis of gastric cancer. Medicine (Baltimore). 2019;98:e16273. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 21] [Cited by in RCA: 24] [Article Influence: 4.0] [Reference Citation Analysis (0)] |