Published online Dec 15, 2022. doi: 10.4251/wjgo.v14.i12.2340
Peer-review started: August 11, 2022
First decision: October 5, 2022
Revised: October 17, 2022
Accepted: November 16, 2022
Article in press: November 16, 2022
Published online: December 15, 2022
Processing time: 123 Days and 2.8 Hours
Esophageal squamous cell carcinoma (ESCC), the predominant type of esophageal cancer, has a 5-year survival rate less than 20%. Although the cause of poor prognosis is the high incidence and mortality of ESCC, the high rate of metastasis after esophageal cancer surgery is the main cause of death after the surgery. Bromodomain-containing protein 4 (BRD4), an epigenetic reader of chromatin-acetylated histones in tumorigenesis and development, plays an essential role in regulating oncogene expression. BRD4 inhibition and BRD4 inhibition-based treatment can potentially suppress ESCC growth. However, the effects and mechanisms of action of BRD4 on ESCC cell migration remain unclear.
To explore the effect of BRD4 on cell migration of ESCC in vitro and its possible molecular mechanism.
Human ESCC cell lines KYSE-450 and KYSE-150 were used. The 3-(4,5-dimethyl-2-thiazolyl)-2,5-diphenyl-2-H-tetrazolium bromide assay was performed to examine cell proliferation, and the transwell migration assay was conducted to test ESCC cell migration. JQ1, a BRD4 inhibitor, was applied to cells, and BRD4 siRNA was transfected into ESCC cells to knockdown endogenous BRD4. GFP-RFP-LC3 adenovirus was infected into ESCC cells to evaluate the effect of JQ1 on autophagy. Western blotting was performed to determine the protein levels of BRD4, E-cadherin, vimentin, AMP-activated protein kinase (AMPK), and p-AMPK.
BRD4 was either downregulated by small interfering RNA or pretreated with JQ1 in ESCC cells, leading to increased tumor migration in ESCC cells in a dose- and time-dependent manner. Inhibition of BRD4 not only significantly suppressed cell proliferation but also strongly increased cell migration by inducing epithelial-mesenchymal transition (EMT). The protein expression of vimentin was increased and E-cadherin decreased in a dose-dependent manner, subsequently promoting autophagy in KYSE-450 and KYSE-150 cells. Pretreatment with JQ1, a BRD4 inhibitor, inhibited BRD4-induced LC3-II activation and upregulated AMPK phosphorylation in a dose-dependent manner. Additionally, an increased number of autophagosomes and autolysosomes were observed in JQ1-treated ESCC cells. The autophagy inhibitor 3-methyladenine (3-MA) reversed the effects of BRD4 knockdown on ESCC cell migration and blocked JQ1-induced cell migration. 3-MA also downregulated the expression of vimentin and upregulation E-cadherin.
BRD4 inhibition enhances cell migration by inducing EMT and autophagy in ESCC cells via the AMPK-modified pathway. Thus, the facilitating role on ESCC cell migration should be considered for BRD4 inhibitor clinical application to ESCC patients.
Core Tip: It has been demonstrated that bromodomain-containing protein 4 (BRD4) as a transcriptional regulator promotes tumor development. Thus, targeting of BRD4 has recently emerged as a promising anti-cancer therapeutic strategy. We present here that BRD4 inhibition suppresses esophageal squamous cell carcinoma (ESCC) cell proliferation, but promotes ESCC cell migration by induction of autophagy, which further facilities epithelial-mesenchymal transition process. Our study implies that the migration-promoting effect should be carefully considered when clinical targeting BRD4 as anti-cancer approach and combination with autophagy inhibitor might be a new therapeutic strategy to avoid the deleterious role of BRD4-targeted strategies.
- Citation: Yang WQ, Liang R, Gao MQ, Liu YZ, Qi B, Zhao BS. Inhibition of bromodomain-containing protein 4 enhances the migration of esophageal squamous cell carcinoma cells by inducing cell autophagy. World J Gastrointest Oncol 2022; 14(12): 2340-2352
- URL: https://www.wjgnet.com/1948-5204/full/v14/i12/2340.htm
- DOI: https://dx.doi.org/10.4251/wjgo.v14.i12.2340
Esophageal squamous cell carcinoma (ESCC), one of the most common and aggressive digestive system cancers, is the main histological type of esophageal carcinoma in East Asian countries such as China[1]. Despite significant advancements in ESCC treatment, the prognosis remains dismal owing to its invasive growth and high frequencies of lymph node metastases[2]. Therefore, understanding the molecular mechanisms of metastasis in ESCC will facilitate the discovery of new therapeutic strategies to promote novel drug development with the goal of improving patient survival.
Bromodomain-containing protein 4 (BRD4) is an epigenetic regulator of the bromodomain and extra-terminal domain (BET) protein family. The structural features of BRD4 are its two bromodomains and one extra-terminal domain[3]. Bromodomain contains a hydrophobic pocket that recognizes acetylated lysine residues and thus acts as the “reader” of lysine acetylation. Therefore, the bromodomain is responsible for transducing the signal carried by acetylated lysine residues. The extra-terminal domain is the focal point for recruiting multiple and varied chromatin or transcriptional regulators[4]. Based on these properties, BRD4 is a pivotal transcriptional and epigenetic regulator. BRD4 has also been demonstrated to play a critical role in cancer development; high expression of BRD4 has been found in several types of cancers, such as colorectal cancer, breast cancer, and lung cancer[5-7]. Several small-molecule inhibitors of BRD4 have been studied, such as JQ1, which has therapeutic uses in combating hematological and solid malignancies[8-10]. In esophageal cancer, inhibition of BRD4 blocked cell proliferation by binding to the promoter region of the chromosome condensation 2 (RCC2) regulator to downregulate RCC2 expression[11]. The finding that enhancer or promoter-associated BRD4 stimulates the expression of oncogenic drivers suggests that BRD4 is a promising target for anti-cancer drug development, but BRD4’s role as a tumor suppressor in tumorigenesis is apparent in lung and breast cancer patient samples[12,13]. Crawford et al[12] also reported that ectopic expression of BRD4 reduced the migration and invasion of the metastatic mouse mammary tumor cell line Mvt-1 without affecting the growth rate, indicating the counteracting oncogenic function of BRD4. Thus, BRD4 probably has diverse functions that are cancer type-dependent. Although the oncogenic function of BRD4 has been well elucidated, given the importance of BRD4-targeted therapy, whether inhibition of BRD4 promotes tumor cell migration has not been clarified.
BRD4 functions as a histone chaperone by interacting with acetylated histones or non-histone proteins and participating in gene expression[14]. In our previous study, we found that trichostatin A (TSA), a histone deacetylase (HDAC) inhibitor, promoted the migration of ESCC cells, and JQ1 attenuated the cell migratory effects induced by TSA and that JQ1 alone promoted ESCC cell migration[15], indicating that BRD4 is involved in hyperacetylation-initiated ESCC cell migration but has an inhibitory effect on ESCC cell migration in the absence of hyperacetylation. Thus, the ability to promote tumor cell migration after inhibition of BRD4 signaling without hyperacetylation represents a crucial research direction regarding the prospect of BRD4 inhibition as an anti-cancer therapeutic approach.
Autophagy is a membrane trafficking process that directs the degradation of cytoplasmic material in lysosomes. Autophagy has been demonstrated to create a tumor-suppressing environment by inhibiting early tumorigenesis through the prevention of chronic tissue damage and regeneration[16]. However, autophagy deteriorates the migration and invasion of tumor cells in vitro while aggravating metastasis in vivo[17]; thus, autophagy elicits a double-sided biological role in tumorigenesis and development. Reports have demonstrated that inhibition of BRD4 induces AMP-activated protein kinase (AMPK)-mTOR-ULK1 modulated autophagy-associated cell death by blocking the BRD4-AMPK interaction in breast cancer cells[18]. Moreover, Jang et al[19] found that JQ1 induced cell autophagy in leukemia stem cells by activating the AMPK/ULK pathway. These reports suggest that inhibition of BRD4 may trigger autophagy via the activation of AMPK signaling.
In this study, we explored the effects of JQ1 on ESCC cell migration and its potential mechanisms of action. We found that JQ1 suppressed ESCC cell proliferation but promoted ESCC cell migration by inducing epithelial-mesenchymal transition (EMT). Knockdown of BRD4 in ESCC cells further confirmed the role of JQ1 in ESCC cell migration. Mechanistically, JQ1 induced autophagy, which is achieved via AMPK activation, which might mediate the promoting role of JQ1 in ESCC cell migration. These data provide new insights into the diverse functions of BRD4 in ESCC cell proliferation and migration, as well as caveats for BRD4-targeted clinical strategies.
Bromodomain inhibitor (+) - JQ1 was purchased from APEXBIO (Houston, TX, United States), dissolved in dimethyl sulfoxide (DMSO), and diluted to the desired concentration before use. 3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyl-terazolium bromide (MTT) was purchased from Beyotime Biotechnology (Shanghai, China). 3-methyladenine (3-MA) was purchased from Selleck (Shanghai, China), dissolved in PBS, and diluted to the desired concentration. Anti-E-cadherin (3195), AMPK (5831), p-AMPK (2535), and LC3-II (12741) antibodies were purchased from Cell Signaling Technology (Danvers, MA, United States). Anti-vimentin (WL00742) and anti-GAPDH (WL01114) were purchased from Wanleibio (Shenyang, China).
Two human ESCC cell lines, KYSE-450 (Cobioer Biosciences, Nanjing, China) and KYSE-150 (Cell Bank of the Typical Culture Preservation Committee of the Chinese Academy of Sciences, Shanghai, China), were used. Both cell lines were grown in PRMI-1640 medium (Corning, New York, United States) and supplemented with 10% fetal bovine serum (FBS; Biological Industries, Israel), 100 μg/mL penicillin, and 100 μg/mL streptomycin (Solarbio, Shanghai, China). The cells were cultured at 37 °C in a humidified atmosphere with 50 mL/L CO2. BRD4 small interfering RNA (siBRD4) and control siRNA were purchased from Santa Cruz Biotechnology (sc-43639, Carlsbad, CA, United States). Cells cultured in a 6-well plate at 60% density were transfected with siRNA at a final concentration of 100 nmol/L using Lipofectamine 2000 (Invitrogen, Carlsbad, CA, United States), according to the manufacturer’s protocol. After 24 h, the cells were collected for transwell and western blot assays.
The effect of JQ1 on cellular growth was determined using an MTT assay. Cells (4 × 103 cells/well) were seeded in 96-well plates, incubated for 24 h, and treated with different doses of JQ1 or vehicle. After incubation for 24, 48, and 72 h, cell proliferation was analyzed using an MTT assay. Ten microliters of MTT dye (5 mg/mL) was added to each well and incubated for 4 h. DMSO (100 μL/well) was added to dissolve formazan crystals. Absorbance at 490 nm was measured using a Multisken Spectrum microplate reader (Thermo Fisher Scientific, Carlsbad, CA, United States). Each experiment was repeated thrice.
KYSE-450 and KYSE-150 cells were seeded in six-well plates (KYSE-150 and KYSE-150 2.0 × 105 cells/well). After overnight culture, cells were treated with or without JQ1 for 48 h. Five different fields were selected to observe cell phenotypic changes using a phase-contrast microscope (Nikon).
The cell migration assay was performed using a 6.5 mm transwell chamber with 8 μm micropores (Corning Costar, Manassas, Virginia, United States). Cells at 1 × 105 cells/200 μL per well in serum-free medium were seeded into the transwell chamber with or without application of the drug as follows: JQ1 (APExBIO) or 3-MA (Selleck), both of which were cultured in 24-well plates with 600 μL RPMI-1640 medium supplemented with 10% FBS. After allowing the cells to migrate for 24 h, non-migrated cells on the upper side of the chamber were cleaned with a cotton swab. Migrated cells on the bottom surface of the chamber were fixed with 4% formaldehyde and stained with 0.1% crystal violet. The migrated cells in five different fields of each membrane were captured using a phase-contrast microscope (Nikon), and the migrated cells were counted.
Total protein was extracted from KYSE-150 and KYSE-450 cells using RIPA buffer. A BCA assay (DingGuo, Beijing, China) was used to measure the protein concentration. Next, 30 μg protein from each sample was separated using 10% SDS-PAGE and transferred to a PVDF membrane (Millipore, Billerica, MA, United States). The membrane was blocked using 5% nonfat dry milk for 1 h at room temperature and incubated with primary antibodies overnight at 4 °C (all in a 1:1000 dilution). Next, the membrane was washed thrice by TBST and incubated with HRP-conjugated secondary antibodies (Boster, Wuhan, China) for 1 h. After washing, the PVDF membrane was processed with a BeyoECL chemiluminescence kit (Beyotime Biotechnology, Shanghai, China) and detected using the AmershamTM Imager 600 System (GE Healthcare Bio-Sciences, Pittsburgh, PA, United States).
GFP-RFP-LC3 virus is widely used to detect autophagic flux. The cells were transfected with the GFP-RFP-LC3 expression virus (Hanbio, Wuhan, China) using Lipofectamine 2000, according to the manufacturer’s instructions. After 48 h of transfection, cells were treated with JQ1 for an additional 24 h. GFP-RFP-LC3 fluorescence was observed using a Nikon Eclipse E800 microscope and photographed with a Nikon digital camera DS-U3 (Nikon, Tokyo, Japan). Afterward, autophagosomes (yellow dots) and autolysosomes (red dots) in each cell were counted.
The experimental results are expressed as the mean ± SD. The significance of differences between the two groups was tested by Student’s t test. A value of P < 0.05 was considered statistically significant.
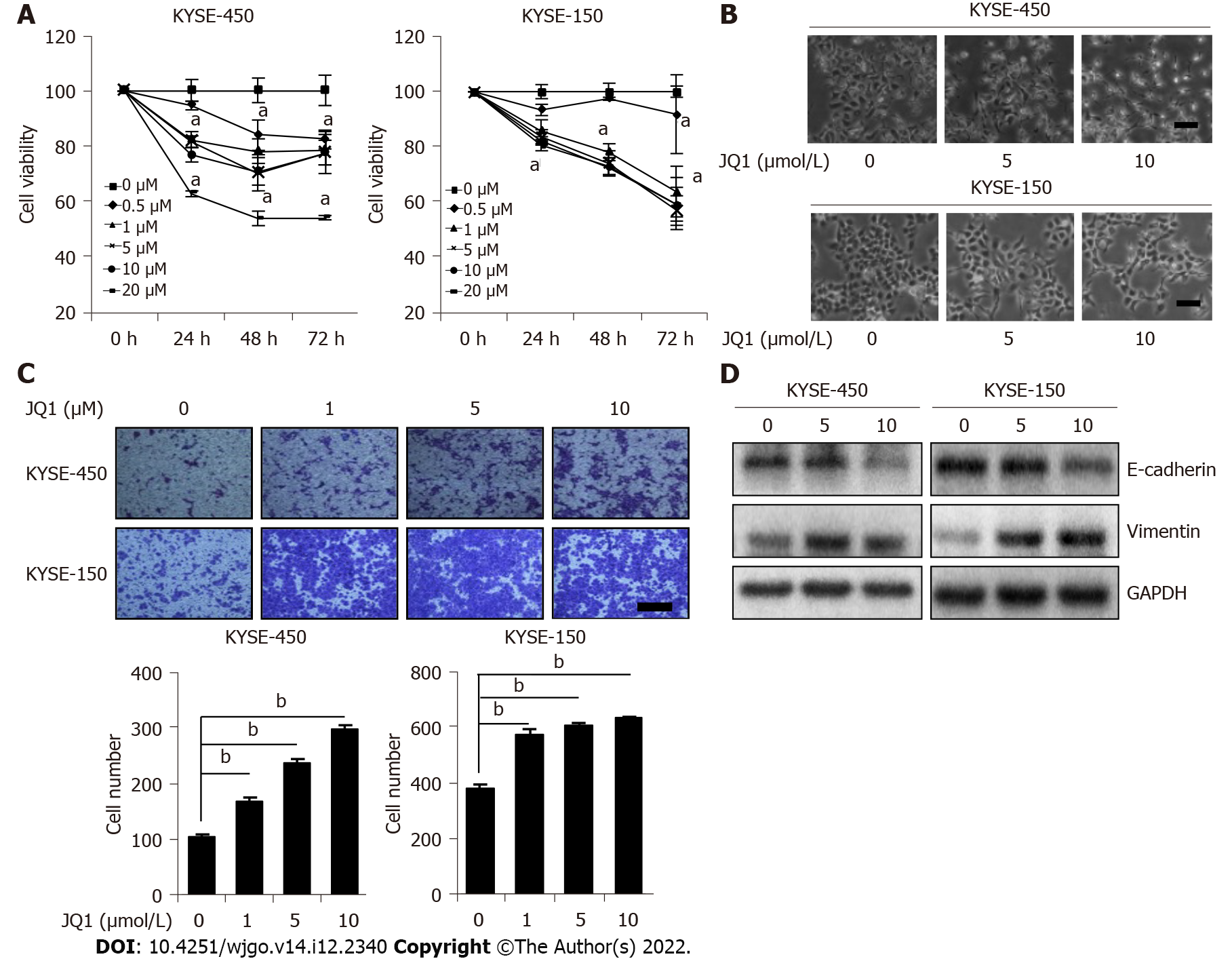
To study the role of BRD4 in ESCC, we first examined its effect on ESCC cell proliferation. A BRD4 inhibitor, JQ1 was applied to KYSE-450 and KYSE-150 cells. The cell viability was measured at various time points. The results showed that JQ1 significantly inhibited KYSE-450 cell proliferation after treatment with all doses (0.5, 1, 5, 10, and 20 μmol/L) at all tested time points (24, 48, and 72 h) (Figure 1A). JQ1 had significant suppressive effects on KYSE-150 cell proliferation at 48 and 72 h after treatment with JQ1 at 1, 5, 10, and 20 μmol/L. When compared with the reaction of KYSE-150 cells to the proliferation-inhibition effects of JQ1, KYSE-450 cells were observed to be more sensitive to JQ1 in a dose-dependent manner. These results indicate an inhibitory effect of JQ1 on the proliferation of esophageal cancer cells. Compared with the control, we also noticed that JQ1-treated KYSE-150 and KYSE-450 cells had stretched and had an elongated spindle-like phenotype, which is a separable feature of the mesenchymal cells (Figure 1B).
We further explored the effect of JQ1 on ESCC cell migration by treating the cells with JQ1 before performing a transwell migration assay. The results revealed that JQ1 at 1, 5, and 10 μmol/L enhanced cell migration more than the control treatment and that this dose-dependent promotion was more apparent in KYSE-450 cells than in KYSE-150 cells (Figure 1C). EMT is an important initiation step during epithelial-derived cancer cell migration, a process that leads to cancer invasion and metastasis[20]. Therefore, we evaluated whether JQ1 promotes cell migration through EMT induction. KYSE-450 and KYSE-150 cells were treated with 5 μmol/L of JQ1 and 10 μmol/L JQ1, respectively. Western blotting was used to examine the EMT marker E-cadherin, which is a marker for epithelia, and vimentin, a mesenchymal marker. We observed that E-cadherin protein levels decreased and vimentin protein levels increased in JQ1-treated cells (Figure 1D). These data suggest that JQ1 facilitates ESCC cell migration by promoting EMT.
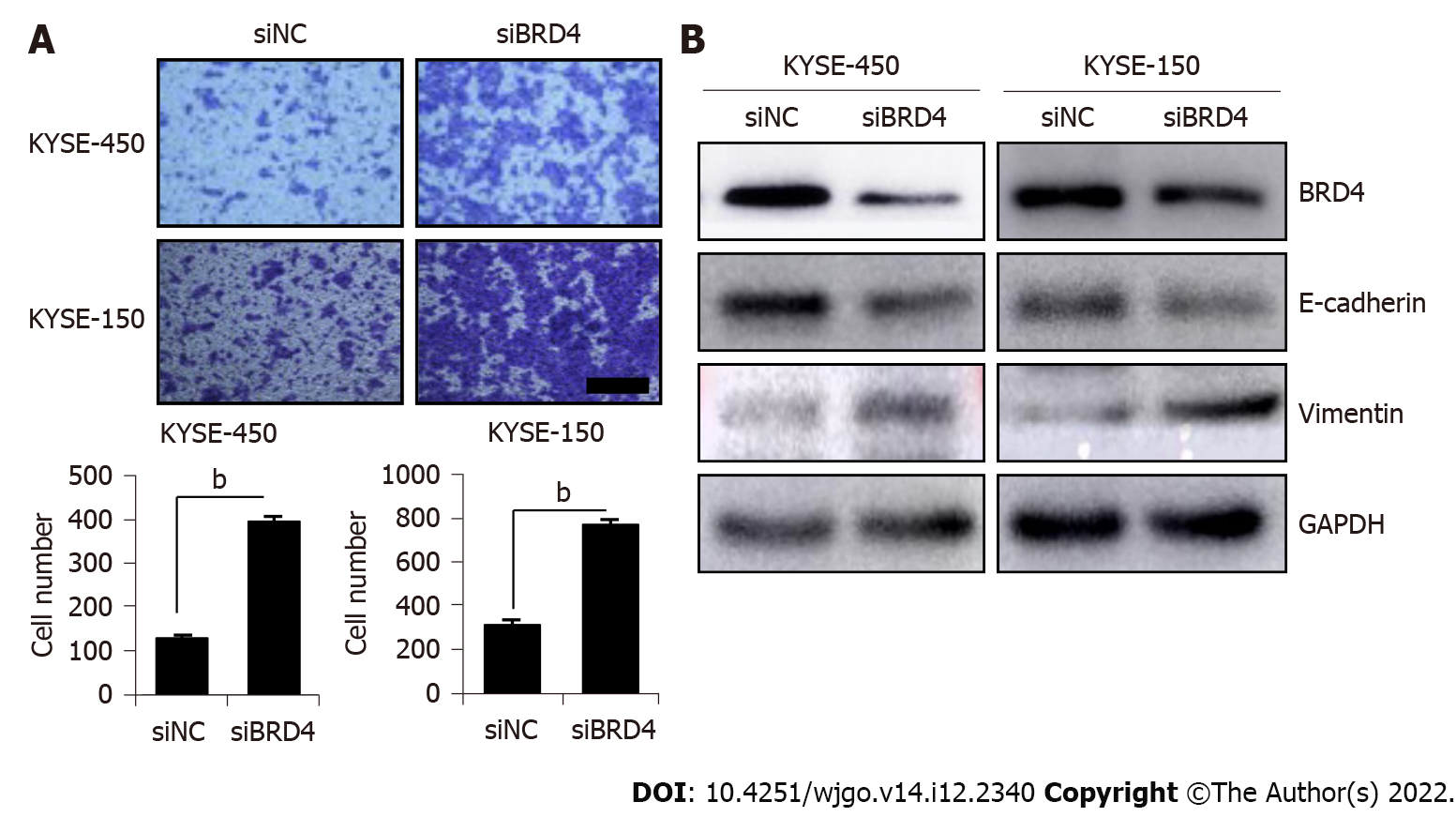
To confirm the role of inhibition BRD4 in ESCC cell migration, siBRD4 was transfected with ESCC cells to knock down endogenous BRD4. Cell migration was examined in siBRD4-transfected KYSE-450 and KYSE-150 cells. As shown in Figure 2A, compared with that in the negative control cells (siNC), a significant increase in the number of migrated cells was observed in siBRD4-transfested KYSE-450 cells and KYSE-150 cells. Western blotting confirmed the knockdown of BRD4 after transfection with siBRD4 in KYSE-450 and KYSE-150 cells. Western blotting also revealed that BRD4 knockdown decreased the level of E-cadherin but increased the level of vimentin in both cell lines (Figure 2B), which is consistent with the results of JQ1 treatment of KYSE-450 and KYSE-150 cells. These findings indicate that inhibition of BRD4 facilitates ESCC cell migration, which is possibly mediated by EMT induction.
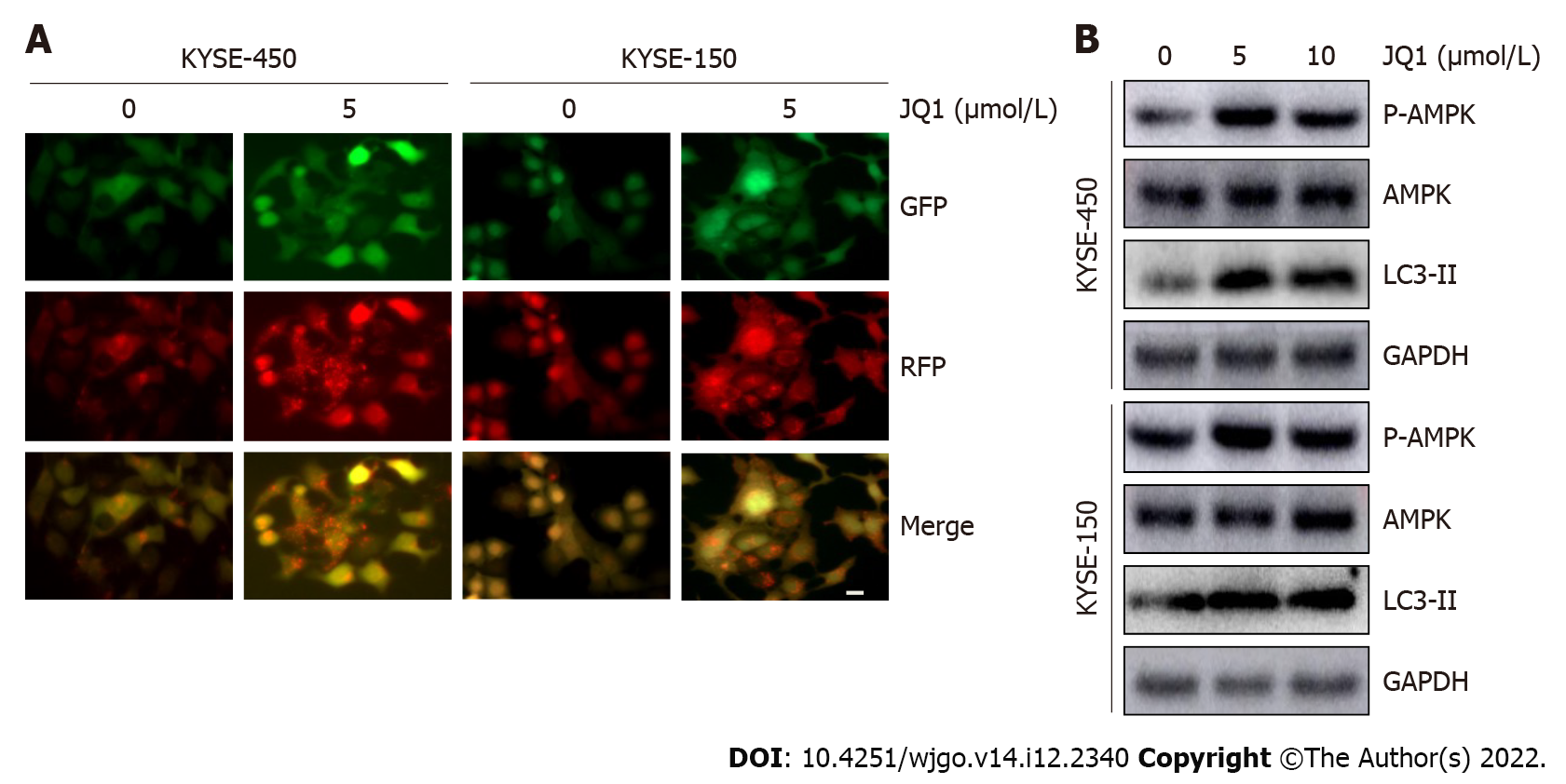
Increasing evidence has revealed that autophagy can promote the metastasis of cancer cells[21,22]. In a study on acute myeloid leukemia, JQ1 induced cell autophagy with little effect on cell apoptosis[19]; therefore, we speculated that autophagy might participate in JQ1-induced ESCC cell migration. To explore the role of JQ1 in autophagy, the GFP-RFP-LC3 double fluorescent autophagy indicator system was used to mark and track changes in LC3 and autophagy flow. KYSE-450 and KYSE-150 cells were infected with GFP-RFP-LC3 virus, as shown in Figure 3A. The number of red dots (indicating autolysosomes) and yellow dots (indicating autophagosomes) were significantly increased after treatment with 5 μmol/L of JQ1. Western blots in Figure 3B showed that the level of LC3-II, an autophagy marker, was significantly increased after treatment with JQ1 at doses of 5 and 10 μmol/L (Figure 3B). AMPK is known to regulate many cellular processes, including autophagy; therefore, we examined AMPK phosphorylation. The addition of JQ1 caused an increase in the level of phosphorylated AMPK (Figure 3B) in KYSE-450 and KYSE-150 cells. These results collectively suggest that JQ1 induces autophagy, which may be triggered by AMPK activation in ESCC cells.
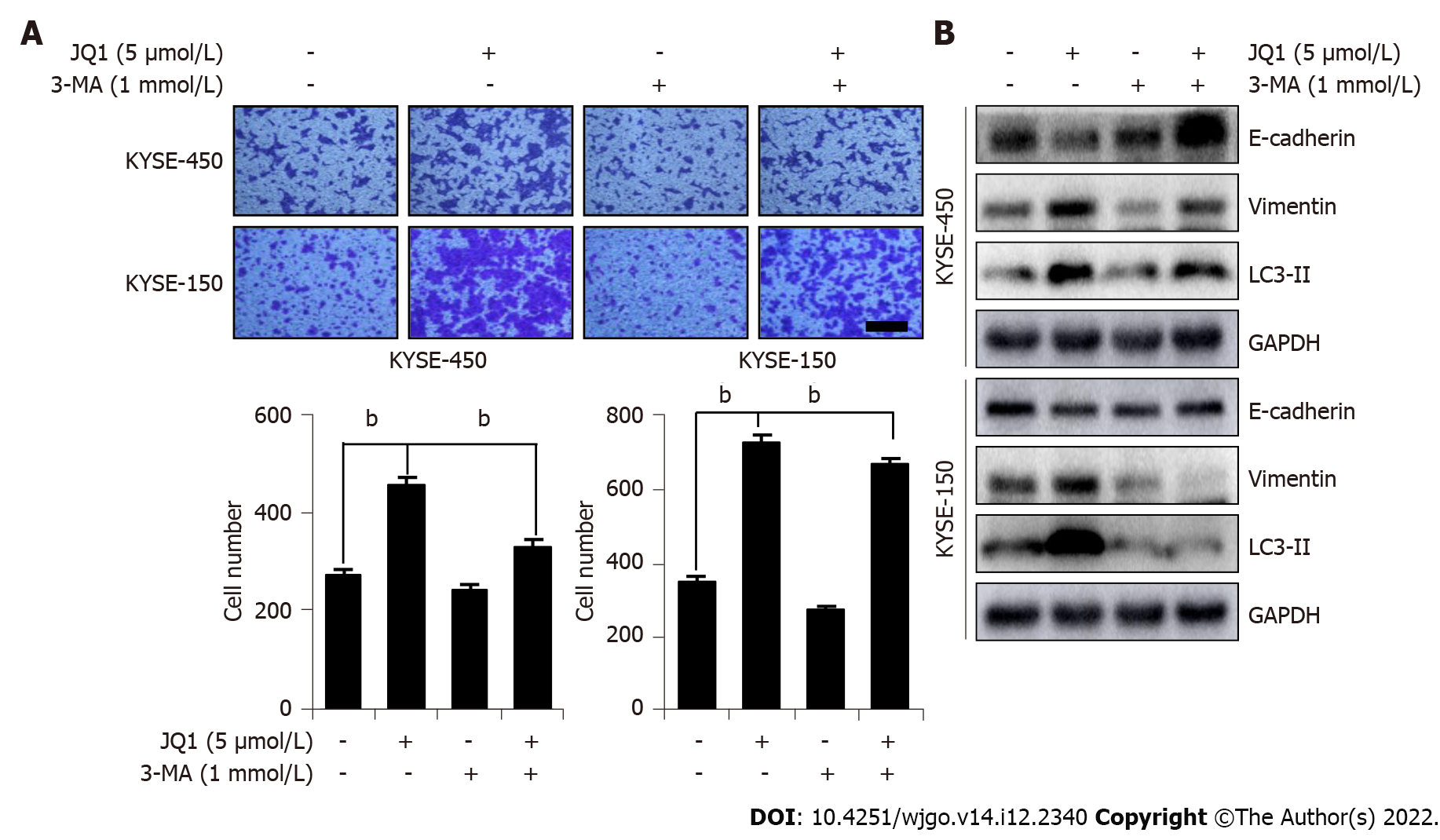
To examine whether JQ1-induced cell migration was due to autophagy activation, we used 3-MA, a classic autophagy inhibitor, to arrest autophagy at an early stage. 3-MA works by inhibiting the class III phosphoinositide 3-kinase (PI3K) complex, thus exhibiting an inhibitory role in the formation and development of autophagosomes. As shown in Figure 4, compared with that in cells treated with only JQ1, a significant decrease in the number of migrated cells was observed in cells treated with both JQ1 and 3-MA. This finding indicates that JQ1 facilitates ESCC migration via the induction of autophagy. Next, we explored whether the JQ1-promoted EMT process is related to the induction of autophagy. As shown in Figure 4B, 3-MA treatment noticeably blocked the JQ1-induced upregulation of LC3-II, indicating that 3-MA prevents JQ1-induced autophagy. Moreover, it was revealed that combination treatment with 3-MA and JQ1 increased the level of E-cadherin while lowering the level of vimentin when compared with that of JQ1 alone treatment, which showed activation of the EMT process. These findings suggest that JQ1-induced ESCC migration may be related to the autophagy-activated EMT process in ESCC cells.
Upregulation or translocation of the BET family frequently occurs in different tumor types, including hematological malignancies and solid tumors[23]. Targeting BET family members is a promising therapeutic strategy in anti-cancer medicine development. For instance, BRD4, a member of the BET family, has been reported as a novel therapeutic candidate target for incurable subtypes of human squamous carcinoma, such as respiratory mucosa cancer[24]. JQ1, a BET bromodomain inhibitor, exhibits anti-cancer activity by competitively displacing BRD4 to bind nuclear chromatin[25], repressing the transcription of BRD4-controlled downstream genes, such as c-Myc[26]. JQ1 has been demonstrated to suppress multi-organ cancer cell proliferation, and multi-organ cancer cell migration and invasion[27-29]. However, Nagarajan et al[30] reported that BRD4 was required for epithelium-specific gene expression and cellular phenotype expression in mammary epithelial cells, and knockdown of BRD4 or application of JQ1 promoted epithelial transformation and migration of mammary cells. Thus, concerns associated with the induction of unwanted cell characteristics should be cautiously considered in the clinical use of BET domain inhibitors. In this study, our data revealed that JQ1 suppressed the growth of ESCC cells (Figure 1A), which is consistent with reports on JQ1’s anti-tumor effect involving anti-ESCC cell proliferation. Our data also revealed that JQ1 and BRD4 knockdown promoted ESCC cell migration. Thus, improving the understanding of the mechanism underlying the promoting role of JQ1 on cell migration is urgently necessary to design strategies to improve its efficiency and overcome its role in promoting cell migration.
BRD4 is not only recruited to histones by acetylated lysine but also interacts directly with transcription factors that determine cell-specific functions and fates[31-33]. The two tandem bromodomain domains of BRD4 recognize acetylated lysine residues in nucleosomal histones and recruit transcriptional proteins to chromatin. The literature has suggested that the first bromodomain of BRD4 may specifically bind to acetylated histones, and the second bromodomain may bind to acetylated lysine residues in cell-specific transcription factors[31,34]. We previously reported that JQ1 or knockdown of BRD4 noticeably counteracted the promoting effect of TSA (an HDAC inhibitor that causes histone acetylation) on ESCC cell migration and that this counteractive mechanism might be involved in the recruitment of BRD4 to TSA-induced acetylated histones[15]. Shi et al[31] reported that by overexpressing twist and BRD4 in HEK293 cells, TSA increases the interaction between twist and BRD4 via twist acetylation promotion. By binding to acetylated twist, BRD4 is recruited to twist target gene promoters/enhancers to direct gene transcription (i.e., directing the transcription of WNT5A in basal-like breast cancer cells), ultimately resulting in BRD4-regulated cell migration and invasion processes. In summary, the function of BRD4 is regulated by post-translational modifications and interactor switches that reshape the genomic landscape, leading to the reorganization of transcriptional programs at specific genetic loci[35]. This reorganization results in differing downstream gene diversification and effects on the migratory behavior of tumor cells. Based on these results, our hypothesis was that the function of BRD4 is dependent on the level of acetylated histones or transcription factors/cofactors. In our study, either JQ1 or knockdown of BRD4 significantly promoted the migration of ESCC cells (Figures 1B and 2A), effectively showing that ESCC cell migration was opposite to that of other cells. This finding indicates a protective role of BRD4 in ESCC cell migration. Our data suggest that inhibition of BRD4 not only represses ESCC cell proliferation but also activates cancer progressing genes that affect cell migration to disrupt the therapeutic anti-cancer effects of BRD4 inhibition in ESCC. Combination treatment strategies that selectively overcome JQ1-induced cell migration could potentially provide maximum anti-cancer therapeutic benefits.
We explored the possible mechanism by which JQ1 promotes ESCC cell migration and provided new insights that support the clinical application of JQ1. A study reported that ubenimex, a classical anti-cancer drug, inhibited glioma cell autophagy to enhance JQ1 sensitivity, which induces cell death by upregulation of hexamethylene bisacetamide-inducible protein 1. The most notable finding in that study was that cell migration and autophagy did not respond to JQ1-only treatment, in contrast with the increased inhibition of cell migration and increased autophagy in cells treated with ubenimex-adjuvant JQ1[36].
Autophagy participates in various intracellular processes; therefore, disordered autophagy is involved in the progression of many diseases and some processes, such as cancer, lysosomal storage diseases, neurodegenerative diseases, aging, development and immune function[37-39]. In addition, the regulation of autophagy is unique and selective. In some growth situations, BRD4 acts as a transcriptional suppressor by working with the methyltransferase EHMT2 to negatively regulate autophagy. Moreover, inhibition of BRD4 resulted in increased autophagy. When nutrient deprivation occurs, AMPK signaling cascades, rather than BRD4, and binds to chromatin, which promotes autophagy gene activation and cell survival[40,41]. Thus, BRD4 can negatively regulate cellular autophagy, and the unique role of BRD4 in the selective regulation of autophagy may facilitate future therapeutic strategies for treating various diseases. In addition, activation of AMPK, a major metabolic energy sensor, triggers activation of downstream autophagy[42,43]. In this study, we found that AMPK may mediate JQ1-induced autophagy in ESCC cells and detect increased phosphorylation of AMPK. Compared with those in the control, the levels of LC3-II and autophagosome/autolysosome increased in JQ1-treated cells because of the activation of AMPK signaling (Figure 3A and B). Utilizing the autophagy inhibitor 3-MA reduced the effect of JQ1-induced upregulation of LC3-II. Further research is necessary to explore the molecular mechanisms of autophagy in relation to JQ1-induced cell migration in esophageal squamous cell carcinoma.
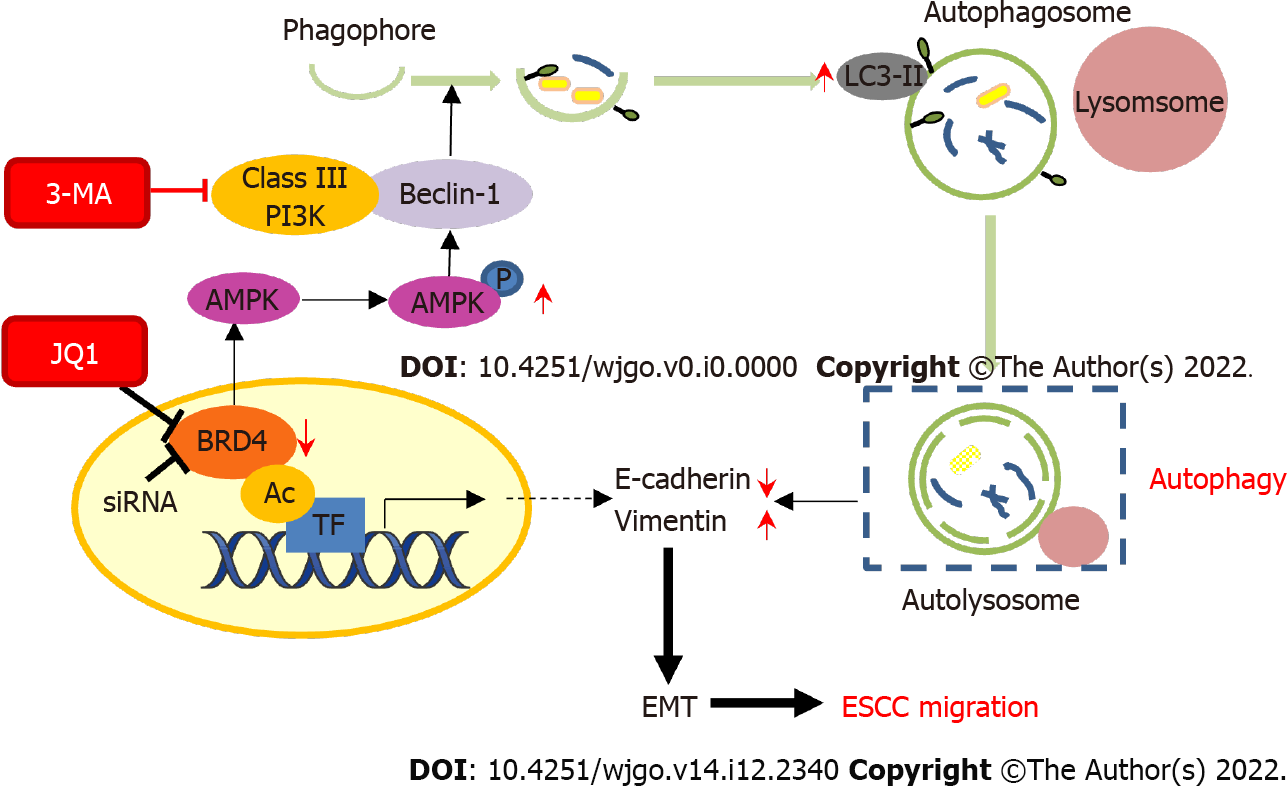
Autophagy has been shown to play an important role in cancer metastasis[44]. In HepG2 cells, fluid shear stress induces cell migration and invasion by activating autophagy[45]. The induction of autophagy leads to loss of the metastatic phenotype by promoting autophagy-mediated degradation of Snail and Twist in breast cancer cells[46]. The literature has also reported that sirtuin-1 induced EMT by promoting autophagy degradation of E-cadherin in melanoma cells[47]. In addition, in a study on hepatocellular carcinoma, plant homeodomain finger protein 8, an EMT activator, promoted metastasis via FIP200-dependent autophagic degradation of E-cadherin[48]. In this study, we demonstrated that JQ1-induced autophagy might promote EMT. Inhibition of autophagy blocks JQ1-induced cell migration and EMT. The transwell assay showed that 3-MA treatment suppressed JQ1-induced cell migration in KYSE-450 and KYSE-150 cell lines (Figure 4A). EMT drives migratory properties that cause adherent cells to adopt a mesenchymal phenotype and enhance cell fate plasticity[49]. E-cadherin is an epithelial marker, and β-catenin and vimentin are mesenchymal markers involved in EMT. In this study, we showed that inhibition of BRD4, either by siBRD4 transfection or by JQ1 treatment, not only facilitated ESCC cell migration by promoting the EMT process (Figure 1C, 2A and B) but also induced the upregulation of E-cadherin and downregulation of vimentin. Compared with those in JQ1-only treated cells, the protein level of E-cadherin was increased and vimentin was reduced in ESCC cells treated with JQ1 combined with 3-MA (Figure 4B). This study showed that cell autophagy is involved in BRD4 regulated cell migration in ESCC. Therefore, the downregulation of BRD4 or JQ1 treatment augments cell migration by promoting autophagy in ESCC cells. Our data indicate that inhibition of autophagy is a potential therapeutic strategy for JQ1-treated ESCC (Figure 5).
After reviewing the literature, we determined that this study is the first to show that JQ1 or inhibition of BRD4 augments the migration of ESCC cells by inducing autophagy, which promotes the EMT process. To determine whether BRD4 effects are specific for ESCC cells, we observed the effect of JQ1 on human glioma cell line U251 cell migration, and found that JQ1 obviously repressed U251 cell migration (data not shown). On the basis of our observations, we speculate that the promoting effect on cell migration induced by BRD4 inhibition is not ESCC cell specific. In the future work, we will explore whether this role occurs on other types of cancer cell. Collectively, our work implies that the migration-promoting effect should be carefully considered when applying JQ1 or targeting BRD4 as an anti-cancer approach. JQ1, in combination with an autophagy inhibitor, might be a new therapeutic strategy to overcome the effects of JQ1 on cancer cell migration.
Our research shows that the downregulation of BRD4 by JQ1 treatment or knockdown of BRD4 promotes the upregulation of AMPK phosphorylation, which might induce autophagy by activating Beclin-1 (a mammalian homolog of yeast Atg6) and class III PI3K complex signaling. Once these signaling pathways are activated, LC3-II level increases, which generates autophagosomes and autolysosomes. Autophagy leads to EMT molecular changes that promote events related to the migration behavior of ESCC cells.
Bromodomain-containing protein 4 (BRD4) as a transcriptional regulator promotes tumor development. Thus, targeting of BRD4 has recently emerged as a promising anti-cancer therapeutic strategy. Although it has been reported that BRD4 inhibition repressed esophageal squamous cell carcinoma (ESCC) cell proliferation, the role of BRD4 inhibition in ESCC cell migration remains unclear.
To explore the role of targeting BRD4 on ESCC cell migration for developing BRD4 inhibitor combination therapies when clinical application of BRD4 inhibitor as anti-cancer therapy.
To explore the effect of BRD4 inhibition on ESCC cell migration and the potential mechanism.
Human ESCC cell lines KYSE-450 and KYSE-150 were cultured. 3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyl-terazolium bromide assay was performed to examine cell proliferation and transwell assay was conducted to cell migration. JQ1 was used to inhibit BRD4 function and siBRD4 was transfected into ESCC cells to knockdown endogenous BRD4. GFP-RFP-LC3 adenovirus was infected into ESCC cells to evaluate the effect of JQ1 on autophagy. Western blot was performed to determine the protein levels of BRD4, E-cadherin, vimentin, AMP-activated protein kinase (AMPK), and p-AMPK.
JQ1 inhibited ESCC cell proliferation, but JQ1 or knockdown of BRD4 promoted ESCC cell migration as well as epithelial-mesenchymal transition (EMT). Application of JQ1 increased autophagosomes and autolysosomes in ESCC cells and enhanced level of LC3-II and AMPK phosphorylation in a dose-dependent manner. The autophagy inhibitor 3-MA blocked JQ1-induced cell migration and EMT.
Inhibition of BRD4 promotes ESCC cell migration and EMT mediated by activation of autophagy.
The migration-promoting effect should be carefully considered when applying JQ1 or targeting BRD4 as an anti-cancer approach. JQ1, in combination with an autophagy inhibitor, might be a new therapeutic strategy to overcome the effects of JQ1 on cancer cell migration.
We thank Chen-Fang Qi who works at Cedars-Sinai Medical Center, 8700 Beverly Blvd, Los Angeles, CA 90048, United States, for providing language editorial assistance.
Provenance and peer review: Unsolicited article; Externally peer reviewed.
Peer-review model: Single blind
Specialty type: Gastroenterology and hepatology
Country/Territory of origin: China
Peer-review report’s scientific quality classification
Grade A (Excellent): 0
Grade B (Very good): B, B
Grade C (Good): C
Grade D (Fair): 0
Grade E (Poor): 0
P-Reviewer: Anandan H, India; Gassler N, Germany S-Editor: Zhang H L-Editor: A P-Editor: Zhang H
| 1. | Lin Y, Totsuka Y, He Y, Kikuchi S, Qiao Y, Ueda J, Wei W, Inoue M, Tanaka H. Epidemiology of esophageal cancer in Japan and China. J Epidemiol. 2013;23:233-242. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 304] [Cited by in RCA: 429] [Article Influence: 35.8] [Reference Citation Analysis (0)] |
| 2. | Huang JX, Chen WC, Lin M, Zhang YL, Li FY, Song ZX, Xiao W, Chen P, Qian RY, Salminen E, Yu H. Clinicopathological significance of cyclooxygenase-2 and cell cycle-regulatory proteins expression in patients with esophageal squamous cell carcinoma. Dis Esophagus. 2012;25:121-129. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 10] [Cited by in RCA: 11] [Article Influence: 0.8] [Reference Citation Analysis (0)] |
| 3. | Zhang W, Prakash C, Sum C, Gong Y, Li Y, Kwok JJ, Thiessen N, Pettersson S, Jones SJ, Knapp S, Yang H, Chin KC. Bromodomain-containing protein 4 (BRD4) regulates RNA polymerase II serine 2 phosphorylation in human CD4+ T cells. J Biol Chem. 2012;287:43137-43155. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 132] [Cited by in RCA: 148] [Article Influence: 11.4] [Reference Citation Analysis (0)] |
| 4. | Belkina AC, Denis GV. BET domain co-regulators in obesity, inflammation and cancer. Nat Rev Cancer. 2012;12:465-477. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 564] [Cited by in RCA: 582] [Article Influence: 44.8] [Reference Citation Analysis (0)] |
| 5. | Hu Y, Zhou J, Ye F, Xiong H, Peng L, Zheng Z, Xu F, Cui M, Wei C, Wang X, Wang Z, Zhu H, Lee P, Zhou M, Jiang B, Zhang DY. BRD4 inhibitor inhibits colorectal cancer growth and metastasis. Int J Mol Sci. 2015;16:1928-1948. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 55] [Cited by in RCA: 76] [Article Influence: 7.6] [Reference Citation Analysis (0)] |
| 6. | Marcotte R, Sayad A, Brown KR, Sanchez-Garcia F, Reimand J, Haider M, Virtanen C, Bradner JE, Bader GD, Mills GB, Pe'er D, Moffat J, Neel BG. Functional Genomic Landscape of Human Breast Cancer Drivers, Vulnerabilities, and Resistance. Cell. 2016;164:293-309. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 250] [Cited by in RCA: 337] [Article Influence: 37.4] [Reference Citation Analysis (0)] |
| 7. | Liao YF, Wu YB, Long X, Zhu SQ, Jin C, Xu JJ, Ding JY. High level of BRD4 promotes non-small cell lung cancer progression. Oncotarget. 2016;7:9491-9500. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 33] [Cited by in RCA: 48] [Article Influence: 6.0] [Reference Citation Analysis (0)] |
| 8. | Herrmann H, Blatt K, Shi J, Gleixner KV, Cerny-Reiterer S, Müllauer L, Vakoc CR, Sperr WR, Horny HP, Bradner JE, Zuber J, Valent P. Small-molecule inhibition of BRD4 as a new potent approach to eliminate leukemic stem- and progenitor cells in acute myeloid leukemia AML. Oncotarget. 2012;3:1588-1599. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 118] [Cited by in RCA: 129] [Article Influence: 10.8] [Reference Citation Analysis (0)] |
| 9. | Shimamura T, Chen Z, Soucheray M, Carretero J, Kikuchi E, Tchaicha JH, Gao Y, Cheng KA, Cohoon TJ, Qi J, Akbay E, Kimmelman AC, Kung AL, Bradner JE, Wong KK. Efficacy of BET bromodomain inhibition in Kras-mutant non-small cell lung cancer. Clin Cancer Res. 2013;19:6183-6192. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 144] [Cited by in RCA: 172] [Article Influence: 14.3] [Reference Citation Analysis (0)] |
| 10. | Asangani IA, Dommeti VL, Wang X, Malik R, Cieslik M, Yang R, Escara-Wilke J, Wilder-Romans K, Dhanireddy S, Engelke C, Iyer MK, Jing X, Wu YM, Cao X, Qin ZS, Wang S, Feng FY, Chinnaiyan AM. Therapeutic targeting of BET bromodomain proteins in castration-resistant prostate cancer. Nature. 2014;510:278-282. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 645] [Cited by in RCA: 795] [Article Influence: 72.3] [Reference Citation Analysis (0)] |
| 11. | Wu Q, Liu F, Ge M, Laster KV, Wei L, Du R, Jiang M, Zhang J, Zhi Y, Jin G, Zhao S, Kim DJ, Dong Z, Liu K. BRD4 drives esophageal squamous cell carcinoma growth by promoting RCC2 expression. Oncogene. 2022;41:347-360. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 5] [Cited by in RCA: 24] [Article Influence: 8.0] [Reference Citation Analysis (0)] |
| 12. | Crawford NP, Alsarraj J, Lukes L, Walker RC, Officewala JS, Yang HH, Lee MP, Ozato K, Hunter KW. Bromodomain 4 activation predicts breast cancer survival. Proc Natl Acad Sci U S A. 2008;105:6380-6385. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 138] [Cited by in RCA: 159] [Article Influence: 9.4] [Reference Citation Analysis (0)] |
| 13. | Fernandez P, Scaffidi P, Markert E, Lee JH, Rane S, Misteli T. Transformation resistance in a premature aging disorder identifies a tumor-protective function of BRD4. Cell Rep. 2014;9:248-260. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 52] [Cited by in RCA: 48] [Article Influence: 4.4] [Reference Citation Analysis (0)] |
| 14. | Kanno T, Kanno Y, LeRoy G, Campos E, Sun HW, Brooks SR, Vahedi G, Heightman TD, Garcia BA, Reinberg D, Siebenlist U, O'Shea JJ, Ozato K. BRD4 assists elongation of both coding and enhancer RNAs by interacting with acetylated histones. Nat Struct Mol Biol. 2014;21:1047-1057. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 229] [Cited by in RCA: 251] [Article Influence: 22.8] [Reference Citation Analysis (0)] |
| 15. | Liu D, Liu Y, Qi B, Gu C, Huo S, Zhao B. Trichostatin A promotes esophageal squamous cell carcinoma cell migration and EMT through BRD4/ERK1/2-dependent pathway. Cancer Med. 2021;10:5235-5245. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 4] [Cited by in RCA: 13] [Article Influence: 3.3] [Reference Citation Analysis (0)] |
| 16. | White E. The role for autophagy in cancer. J Clin Invest. 2015;125:42-46. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 802] [Cited by in RCA: 1033] [Article Influence: 103.3] [Reference Citation Analysis (0)] |
| 17. | Sharifi MN, Mowers EE, Drake LE, Collier C, Chen H, Zamora M, Mui S, Macleod KF. Autophagy Promotes Focal Adhesion Disassembly and Cell Motility of Metastatic Tumor Cells through the Direct Interaction of Paxillin with LC3. Cell Rep. 2016;15:1660-1672. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 186] [Cited by in RCA: 254] [Article Influence: 28.2] [Reference Citation Analysis (0)] |
| 18. | Ouyang L, Zhang L, Liu J, Fu L, Yao D, Zhao Y, Zhang S, Wang G, He G, Liu B. Discovery of a Small-Molecule Bromodomain-Containing Protein 4 (BRD4) Inhibitor That Induces AMP-Activated Protein Kinase-Modulated Autophagy-Associated Cell Death in Breast Cancer. J Med Chem. 2017;60:9990-10012. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 76] [Cited by in RCA: 75] [Article Influence: 9.4] [Reference Citation Analysis (0)] |
| 19. | Jang JE, Eom JI, Jeung HK, Cheong JW, Lee JY, Kim JS, Min YH. AMPK-ULK1-Mediated Autophagy Confers Resistance to BET Inhibitor JQ1 in Acute Myeloid Leukemia Stem Cells. Clin Cancer Res. 2017;23:2781-2794. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 76] [Cited by in RCA: 101] [Article Influence: 11.2] [Reference Citation Analysis (0)] |
| 20. | Behrens J, Mareel MM, Van Roy FM, Birchmeier W. Dissecting tumor cell invasion: epithelial cells acquire invasive properties after the loss of uvomorulin-mediated cell-cell adhesion. J Cell Biol. 1989;108:2435-2447. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 654] [Cited by in RCA: 695] [Article Influence: 19.3] [Reference Citation Analysis (0)] |
| 21. | Liu H, Ma Y, He HW, Zhao WL, Shao RG. SPHK1 (sphingosine kinase 1) induces epithelial-mesenchymal transition by promoting the autophagy-linked lysosomal degradation of CDH1/E-cadherin in hepatoma cells. Autophagy. 2017;13:900-913. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 69] [Cited by in RCA: 101] [Article Influence: 14.4] [Reference Citation Analysis (0)] |
| 22. | Li J, Yang B, Zhou Q, Wu Y, Shang D, Guo Y, Song Z, Zheng Q, Xiong J. Autophagy promotes hepatocellular carcinoma cell invasion through activation of epithelial-mesenchymal transition. Carcinogenesis. 2013;34:1343-1351. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 226] [Cited by in RCA: 239] [Article Influence: 19.9] [Reference Citation Analysis (0)] |
| 23. | Spriano F, Stathis A, Bertoni F. Targeting BET bromodomain proteins in cancer: The example of lymphomas. Pharmacol Ther. 2020;215:107631. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 26] [Cited by in RCA: 51] [Article Influence: 10.2] [Reference Citation Analysis (0)] |
| 24. | French CA, Miyoshi I, Aster JC, Kubonishi I, Kroll TG, Dal Cin P, Vargas SO, Perez-Atayde AR, Fletcher JA. BRD4 bromodomain gene rearrangement in aggressive carcinoma with translocation t(15;19). Am J Pathol. 2001;159:1987-1992. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 159] [Cited by in RCA: 159] [Article Influence: 6.6] [Reference Citation Analysis (0)] |
| 25. | Filippakopoulos P, Qi J, Picaud S, Shen Y, Smith WB, Fedorov O, Morse EM, Keates T, Hickman TT, Felletar I, Philpott M, Munro S, McKeown MR, Wang Y, Christie AL, West N, Cameron MJ, Schwartz B, Heightman TD, La Thangue N, French CA, Wiest O, Kung AL, Knapp S, Bradner JE. Selective inhibition of BET bromodomains. Nature. 2010;468:1067-1073. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 3566] [Cited by in RCA: 3310] [Article Influence: 220.7] [Reference Citation Analysis (0)] |
| 26. | Delmore JE, Issa GC, Lemieux ME, Rahl PB, Shi J, Jacobs HM, Kastritis E, Gilpatrick T, Paranal RM, Qi J, Chesi M, Schinzel AC, McKeown MR, Heffernan TP, Vakoc CR, Bergsagel PL, Ghobrial IM, Richardson PG, Young RA, Hahn WC, Anderson KC, Kung AL, Bradner JE, Mitsiades CS. BET bromodomain inhibition as a therapeutic strategy to target c-Myc. Cell. 2011;146:904-917. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 2297] [Cited by in RCA: 2331] [Article Influence: 166.5] [Reference Citation Analysis (0)] |
| 27. | Zhou S, Zhang S, Wang L, Huang S, Yuan Y, Yang J, Wang H, Li X, Wang P, Zhou L, Xu Y, Gao H, Zhang Y, Lv Y, Zou X. BET protein inhibitor JQ1 downregulates chromatin accessibility and suppresses metastasis of gastric cancer via inactivating RUNX2/NID1 signaling. Oncogenesis. 2020;9:33. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 22] [Cited by in RCA: 66] [Article Influence: 13.2] [Reference Citation Analysis (0)] |
| 28. | Wen N, Guo B, Zheng H, Xu L, Liang H, Wang Q, Wang D, Chen X, Zhang S, Li Y, Zhang L. Bromodomain inhibitor jq1 induces cell cycle arrest and apoptosis of glioma stem cells through the VEGF/PI3K/AKT signaling pathway. Int J Oncol. 2019;55:879-895. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 19] [Cited by in RCA: 48] [Article Influence: 8.0] [Reference Citation Analysis (0)] |
| 29. | Wang L, Wu X, Huang P, Lv Z, Qi Y, Wei X, Yang P, Zhang F. JQ1, a small molecule inhibitor of BRD4, suppresses cell growth and invasion in oral squamous cell carcinoma. Oncol Rep. 2016;36:1989-1996. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 28] [Cited by in RCA: 40] [Article Influence: 4.4] [Reference Citation Analysis (0)] |
| 30. | Nagarajan S, Bedi U, Budida A, Hamdan FH, Mishra VK, Najafova Z, Xie W, Alawi M, Indenbirken D, Knapp S, Chiang CM, Grundhoff A, Kari V, Scheel CH, Wegwitz F, Johnsen SA. BRD4 promotes p63 and GRHL3 expression downstream of FOXO in mammary epithelial cells. Nucleic Acids Res. 2017;45:3130-3145. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 7] [Cited by in RCA: 18] [Article Influence: 2.3] [Reference Citation Analysis (0)] |
| 31. | Shi J, Wang Y, Zeng L, Wu Y, Deng J, Zhang Q, Lin Y, Li J, Kang T, Tao M, Rusinova E, Zhang G, Wang C, Zhu H, Yao J, Zeng YX, Evers BM, Zhou MM, Zhou BP. Disrupting the interaction of BRD4 with diacetylated Twist suppresses tumorigenesis in basal-like breast cancer. Cancer Cell. 2014;25:210-225. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 331] [Cited by in RCA: 388] [Article Influence: 35.3] [Reference Citation Analysis (0)] |
| 32. | Wu SY, Lee AY, Lai HT, Zhang H, Chiang CM. Phospho switch triggers Brd4 chromatin binding and activator recruitment for gene-specific targeting. Mol Cell. 2013;49:843-857. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 223] [Cited by in RCA: 282] [Article Influence: 23.5] [Reference Citation Analysis (0)] |
| 33. | Roe JS, Mercan F, Rivera K, Pappin DJ, Vakoc CR. BET Bromodomain Inhibition Suppresses the Function of Hematopoietic Transcription Factors in Acute Myeloid Leukemia. Mol Cell. 2015;58:1028-1039. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 275] [Cited by in RCA: 279] [Article Influence: 27.9] [Reference Citation Analysis (0)] |
| 34. | Shi J, Vakoc CR. The mechanisms behind the therapeutic activity of BET bromodomain inhibition. Mol Cell. 2014;54:728-736. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 691] [Cited by in RCA: 711] [Article Influence: 64.6] [Reference Citation Analysis (0)] |
| 35. | Chiang CM. Brd4 engagement from chromatin targeting to transcriptional regulation: selective contact with acetylated histone H3 and H4. F1000 Biol Rep. 2009;1:98. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 94] [Cited by in RCA: 105] [Article Influence: 6.6] [Reference Citation Analysis (0)] |
| 36. | Han L, Zhao Q, Liang X, Wang X, Zhang Z, Ma Z, Zhao M, Wang A, Liu S. Ubenimex enhances Brd4 inhibition by suppressing HEXIM1 autophagic degradation and suppressing the Akt pathway in glioma cells. Oncotarget. 2017;8:45643-45655. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 6] [Cited by in RCA: 7] [Article Influence: 1.0] [Reference Citation Analysis (0)] |
| 37. | Mizushima N, Levine B, Cuervo AM, Klionsky DJ. Autophagy fights disease through cellular self-digestion. Nature. 2008;451:1069-1075. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 5421] [Cited by in RCA: 5292] [Article Influence: 311.3] [Reference Citation Analysis (0)] |
| 38. | Martinez-Vicente M, Cuervo AM. Autophagy and neurodegeneration: when the cleaning crew goes on strike. Lancet Neurol. 2007;6:352-361. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 336] [Cited by in RCA: 348] [Article Influence: 19.3] [Reference Citation Analysis (0)] |
| 39. | Kimmelman AC. The dynamic nature of autophagy in cancer. Genes Dev. 2011;25:1999-2010. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 426] [Cited by in RCA: 475] [Article Influence: 33.9] [Reference Citation Analysis (0)] |
| 40. | Wen X, Klionsky DJ. BRD4 is a newly characterized transcriptional regulator that represses autophagy and lysosomal function. Autophagy. 2017;13:1801-1803. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 17] [Cited by in RCA: 28] [Article Influence: 3.5] [Reference Citation Analysis (0)] |
| 41. | Sakamaki JI, Wilkinson S, Hahn M, Tasdemir N, O'Prey J, Clark W, Hedley A, Nixon C, Long JS, New M, Van Acker T, Tooze SA, Lowe SW, Dikic I, Ryan KM. Bromodomain Protein BRD4 Is a Transcriptional Repressor of Autophagy and Lysosomal Function. Mol Cell. 2017;66:517-532.e9. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 190] [Cited by in RCA: 193] [Article Influence: 24.1] [Reference Citation Analysis (0)] |
| 42. | Pineda-Ramírez N, Alquisiras-Burgos I, Ortiz-Plata A, Ruiz-Tachiquín ME, Espinoza-Rojo M, Aguilera P. Resveratrol Activates Neuronal Autophagy Through AMPK in the Ischemic Brain. Mol Neurobiol. 2020;57:1055-1069. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 38] [Cited by in RCA: 83] [Article Influence: 13.8] [Reference Citation Analysis (0)] |
| 43. | Wu Q, Li J, Li Z, Sun S, Zhu S, Wang L, Wu J, Yuan J, Zhang Y, Wang C. Exosomes from the tumour-adipocyte interplay stimulate beige/brown differentiation and reprogram metabolism in stromal adipocytes to promote tumour progression. J Exp Clin Cancer Res. 2019;38:223. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 90] [Cited by in RCA: 116] [Article Influence: 19.3] [Reference Citation Analysis (0)] |
| 44. | Choi AM, Ryter SW, Levine B. Autophagy in human health and disease. N Engl J Med. 2013;368:651-662. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1792] [Cited by in RCA: 1913] [Article Influence: 159.4] [Reference Citation Analysis (0)] |
| 45. | Yan Z, Su G, Gao W, He J, Shen Y, Zeng Y, Liu X. Fluid shear stress induces cell migration and invasion via activating autophagy in HepG2 cells. Cell Adh Migr. 2019;13:152-163. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 15] [Cited by in RCA: 29] [Article Influence: 4.8] [Reference Citation Analysis (0)] |
| 46. | Lv Q, Wang W, Xue J, Hua F, Mu R, Lin H, Yan J, Lv X, Chen X, Hu ZW. DEDD interacts with PI3KC3 to activate autophagy and attenuate epithelial-mesenchymal transition in human breast cancer. Cancer Res. 2012;72:3238-3250. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 117] [Cited by in RCA: 144] [Article Influence: 11.1] [Reference Citation Analysis (0)] |
| 47. | Sun T, Jiao L, Wang Y, Yu Y, Ming L. SIRT1 induces epithelial-mesenchymal transition by promoting autophagic degradation of E-cadherin in melanoma cells. Cell Death Dis. 2018;9:136. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 52] [Cited by in RCA: 77] [Article Influence: 11.0] [Reference Citation Analysis (0)] |
| 48. | Zhou W, Gong L, Wu Q, Xing C, Wei B, Chen T, Zhou Y, Yin S, Jiang B, Xie H, Zhou L, Zheng S. PHF8 upregulation contributes to autophagic degradation of E-cadherin, epithelial-mesenchymal transition and metastasis in hepatocellular carcinoma. J Exp Clin Cancer Res. 2018;37:215. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 16] [Cited by in RCA: 40] [Article Influence: 5.7] [Reference Citation Analysis (0)] |
| 49. | Aiello NM, Kang Y. Context-dependent EMT programs in cancer metastasis. J Exp Med. 2019;216:1016-1026. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 327] [Cited by in RCA: 422] [Article Influence: 70.3] [Reference Citation Analysis (0)] |