Published online Oct 15, 2022. doi: 10.4251/wjgo.v14.i10.2038
Peer-review started: July 20, 2022
First decision: August 20, 2022
Revised: August 21, 2022
Accepted: September 1, 2022
Article in press: September 1, 2022
Published online: October 15, 2022
Processing time: 86 Days and 1.9 Hours
Gastric cancer (GC) is one of the most prevalent malignant tumors that endangers human health. Early diagnosis is essential for improving the prognosis and survival rate of GC patients. Ring finger protein 180 (RNF180) is involved in the regulation of cell differentiation, proliferation, apoptosis, and tumorigenesis, and aberrant hypermethylation of CpG islands in the promoter is strongly associated with the occurrence and development of GC. Thus, methylated RNF180 can be used as a potential biomarker for GC diagnosis.
To use droplet digital polymerase chain reaction (ddPCR) to quantify the methylation level of the RN180 gene. A reproducible ddPCR assay to detect methylated RNF180 from trace DNA was designed and optimized.
The primer and probe were designed and selected, the conversion time of bisulfite was optimized, the ddPCR system was adjusted by primer concentration, amplification temperature and amplification cycles, and the detection limit of ddPCR was determined.
The best conversion time for blood DNA was 2 h 10 min, and that for plasma DNA was 2 h 10 min and 2 h 30 min. The results of ddPCR were better when the amplification temperature was 56 °C and the number of amplification cycles was 50. Primer concentrations showed little effect on the assay outcome. Therefore, the primer concentration could be adjusted according to the reaction system and DNA input. The assay required at least 0.1 ng of input DNA.
In summary, a ddPCR assay was established to detect methylated RNF180, which is expected to be a new diagnostic biomarker for GC.
Core Tip: Gastric cancer (GC) is one of the most prevalent malignant tumors that endangers human health. Early diagnosis is essential for improving the prognosis and survival rate of GC patients. Ring finger protein 180 (RNF180) is a newly discovered member of the ring finger protein family, and it is an important tumor suppressor gene involved in the construction of the E3 ubiquitin protein ligase in the ubiquitin proteasome system and ubiquitin degradation. Methylated RNF180 can be used as a potential biomarker for GC diagnosis. We aimed to evaluate the droplet digital polymerase chain reaction assay for methylated RNF180 in GC.
- Citation: Guo GH, Xie YB, Jiang T, An Y. Droplet digital polymerase chain reaction assay for methylated ring finger protein 180 in gastric cancer. World J Gastrointest Oncol 2022; 14(10): 2038-2047
- URL: https://www.wjgnet.com/1948-5204/full/v14/i10/2038.htm
- DOI: https://dx.doi.org/10.4251/wjgo.v14.i10.2038
Ring finger protein 180 (RNF180) is a newly discovered member of the ring finger protein family, and it is an important tumor suppressor gene involved in the construction of the E3 ubiquitin protein ligase in the ubiquitin proteasome system and ubiquitin degradation[1]. RNF180 affects many important physiological processes in vivo, including cell growth, differentiation, and tumorigenesis. RNF180 is expressed at low levels in gastric cancer (GC) tissue samples, and it is particularly crucial to the GC pathological stage and overall survival of patients[2]. Hypermethylation in the promoter region is the main mechanism of downregulation or silencing of RNF180 expression in tumors[3]. It is also associated with Helicobacter pylori (H. pylori) infection and GC prognosis[4,5]. Methylation of the RNF180 gene can be used as an independent diagnostic and prognostic biomarker in the clinic[6]. At present, the primary screening methods for GC in the clinic are upper gastrointestinal X-ray examination, gastrointestinal endoscopy, H. pylori antibody detection and plasma pepsinogen detection[7]. Tissue biopsy is the gold standard for pathological diagnosis of GC. The commonly used X-ray examination and endoscopy are not always well suited to the early diagnosis, H. pylori antibody and plasma pepsinogen are not specific enough, and the shortcomings of tissue biopsy are its invasiveness and heterogeneity. The recent development of liquid biopsy technology has been very promising in the early screening of tumors. It is a noninvasive procedure for diagnosis that utilizes circulating free DNA in body fluids and circumvents some of the limitations of conventional tissue biopsy[8,9].
Liquid biopsy has great clinical value for therapy evaluation and prognosis monitoring. Droplet digital polymerase chain reaction (ddPCR) is a novel absolute quantitative technique that divides the reaction system into thousands of units[10]. The fluorescence signal of each reaction unit is detected, and the original concentration is calculated according to a Poisson distribution. Liquid biopsy samples are from blood, urine, and other body fluids, but the tumor DNA content in these specimens is low[11]. Conventional PCR technology cannot meet the requirement of liquid biopsy. Compared with conventional PCR, ddPCR can achieve absolute quantification of trace nucleic acids and is more suitable for clinical liquid biopsy. In this study, a ddPCR assay to detect the methylated RNF180 gene was established and optimized for analyzing plasma and blood samples. It can be used for the screening and early diagnosis of GC and opens new possibilities to use methylated RNF180 as a biomarker of invasive GC.
The samples used in this study were blood samples from patients diagnosed between September 2020 and April 2021. Inclusion criteria: Complete clinicopathological data, clear imaging and pathological diagnosis, and absence of long-term radiotherapy, chemotherapy, and immunotherapy. The exclusion criteria were as follows: Patients with other types of tumors in addition to confirmed GC who had received long-term treatment.
The samples were peripheral blood collected on an empty stomach in the morning, and EDTA was used as an anticoagulant. Upon collection, the blood samples were immediately aliquoted into 1.5 mL Eppendorf tubes at 200 μL/tube. Plasma samples were prepared by centrifuging blood at 1500 × g for 10 min. The samples were discarded if hemolysis or lipemia were observed. Plasma was aliquoted into 1000 μL Eppendorf tubes for subsequent experiments. DNA from 200 μL blood samples was extracted according to the instructions of the QIAamp Blood Mini Kit (Qiagen) and eluted in 84 μL Buffer AE. DNA was extracted from 1 mL of plasma sample according to the instructions of the QIAamp MinElute ccfDNA Mini Kit (Qiagen) and eluted in 24 μL ultrafine water. The concentration of double-stranded DNA (dsDNA) was measured by a Qubit dsDNA HS Assay Kit and Qubit 3.0 (Thermo Fisher).
A 20 μL DNA sample was transformed according to the instructions of the EZ DNA Methylation-Gold Kit (Zymo Research), and 22 μL M-Elution Buffer was added to the column matrix to elute DNA. PCR was performed in a thermal cycler with the lid temperature set at 105 °C. Thermal cycling condition was as follows: (1) 98 °C for 10 min; (2) 64 °C for 1 h 30 min, 1 h 50 min, 2 h 10 min, or 2 h 30 min; and (3) Hold at 4°C. The converted DNA was purified according to the cycle-Pure Kit instructions (Omega), and finally, 7 μL elution buffer was added. The single-stranded DNA (ssDNA) Assay Kit and Qubit 3.0 (Thermo Fisher) were used to determine the concentration of ssDNA.
Primers were designed based on the principle that unmethylated cytosine will transform into uracil after bisulfite treatment, while methylated cytosine remains unaltered. Primers and probes were designed so that they could distinguish between methylated and unmethylated sequences. The RNF180 gene sequence was obtained from National Center for Biotechnology Information. Then, the sequence was pasted into the methyl Primer Express software to find the CpG island, and methylation primers and probes were designed according to the transformed sequence. The 5’ end of the probes was modified with the FAM fluorophore, and the 3’ end was modified with BHQ1. Three pairs of primers were designed for ddPCR experiments and tested against 100% methylated control DNA (EpiTect PCR Control DNA Set, Qiagen). After repeated tests, the primers were screened according to their reproducibility, specificity, and detection rate. Then, unmethylated primers and probes were designed for subsequent experiments to calculate methylation rates.
The ddPCR assay was conducted under the same conditions with 100% methylated control DNA at the initial input as shown in Table 1 to determine the lowest limit of detection (LOD). Simultaneously, to find the optimal ddPCR amplification condition, the amplification temperature was set at 56 °C and 59 °C, and the amplification cycles were set at 45 and 50. The ddPCR instructions recommended a final primer concentration of 700-900 nmol and a probe concentration of 250 nmol. Under the optimal cycling conditions, primer concentrations of 700, 750, 800, 850 and 900 nmol were tested to determine the optimal concentration. A negative control (EpiTect PCR Control DNA Set, Qiagen) and no-template control were included in each plate.
| Input DNA (ng) | Conc (copies/μL) | Input DNA (ng) | Conc (copies/μL) |
| 10 | 15.4 | 0.5 | 0.73 |
| 9 | 14 | 0.1 | 0.29 |
| 8 | 12.25 | 0.05 | 0 |
| 7 | 9.35 | 0.01 | 0 |
| 6 | 9.6 | 0.005 | 0 |
| 5 | 7.8 | 0.001 | 0 |
| 4 | 7.56 | 0.0005 | 0 |
| 3 | 4.53 | 0.0001 | 0 |
| 2 | 3.63 | H2O | 0 |
| 1 | 1.4 |
Twenty microliters of quantitative reaction was prepared, mixed with oscillation and briefly centrifuged to remove bubbles. The above reaction mixtures were added to the middle row of the droplet generating cartridge, and 70 μL ddPCR Droplet Reader Oil (BIO-RAD) was added to the bottom row. The reagents were placed smoothly in the droplet generator and the reaction was started. The liquid in the top row of the droplet generating cartridge was transferred to a 96-well plate and then placed into a heat sealer to seal with a film. After sealing, the 96-well plate was placed into a C1000 TouchTM Thermal Cycler (BIO-RAD) for PCR. QuantaSoft was opened after 30 min of preheating of the QX200 Droplet Reader (BIO-RAD). At the end of PCR, the 96-well plate was placed into the Droplet Reader and processed under the sample information setting. When finished, the program was run, and the data were analyzed after droplet reading.
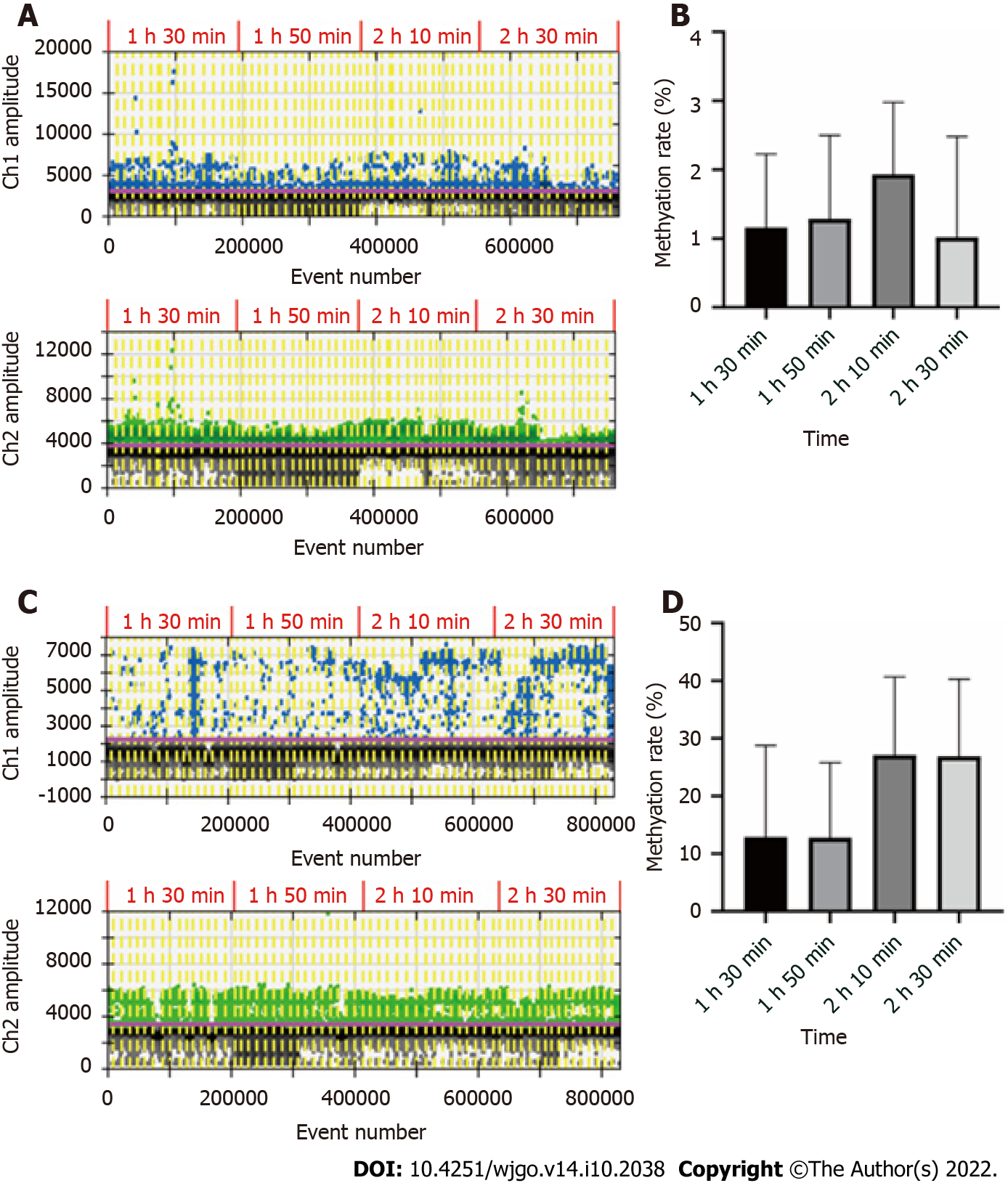
The recommended conversion time was 2 h 30 min according to the instructions of the EZ DNA Methylation-Gold Kit. However, due to the small amount of circulating free DNA in blood and plasma, and the loss after conversion, we decided to optimize the conversion time for our assay. DNA was extracted from the blood and plasma of 16 GC patients for this experiment. After determining the concentration of ssDNA, each DNA sample was divided into 4 conditions. For bisulfite conversion, the initial input amount of blood DNA was 500 ng, the initial input amount of plasma DNA was 10 ng, and the sample volume was adjusted to 20 μL with sterilized deionized water. The conversion time was set at 1 h 30 min, 1 h 50 min, 2 h 10 min or 2 h 30 min, and ddPCR assay was conducted in these 4 conditions. The results are shown in Figure 1. By comparing the methylation rates of the same samples subjected to various conversion times, we concluded that the optimal conversion time for blood DNA was 2 h 10 min, and the optimal time for plasma DNA was 2 h 10 min and 2 h 30 min. To streamline the experimental operation, the conversion time was set as 2 h 10 min in subsequent experiments.
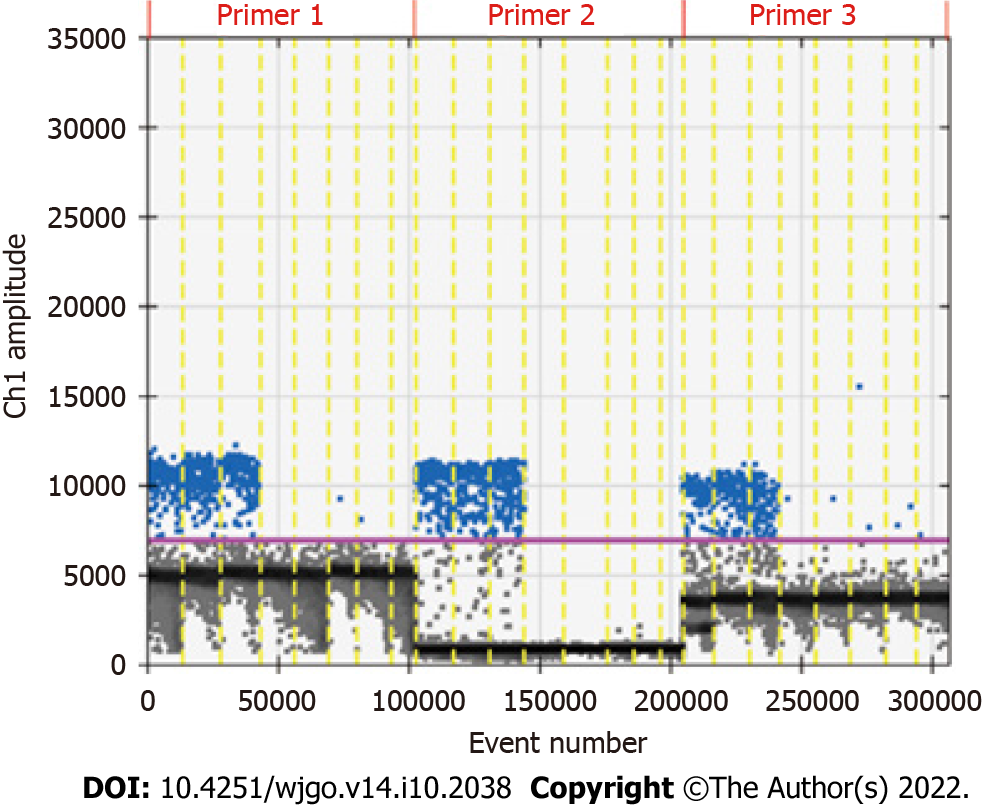
The primers designed for the methylated RNF180 gene was used to conduct ddPCR assays with 100% methylated control DNA, and the data that reached more than 10000 droplets were considered valid data. After repeating experiments, we chose one pair of primers and probes with the best result based on the reproducibility, specificity, and detection rate. The ddPCR results of the tested primers are shown in Figure 2. The second pair of primers with good specificity and reproducibility was selected according to the results shown in the figure. Subsequently, primers and probes for unmethylated RNF180 gene were designed to calculate the methylation rate of the sample.
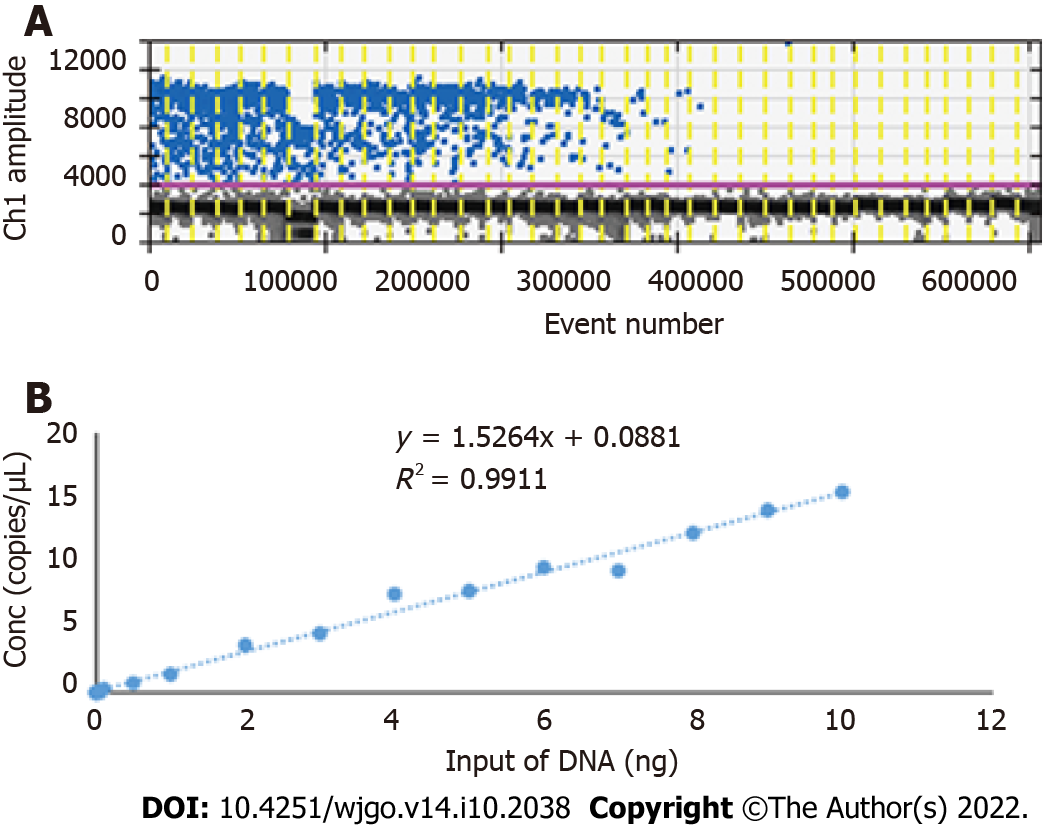
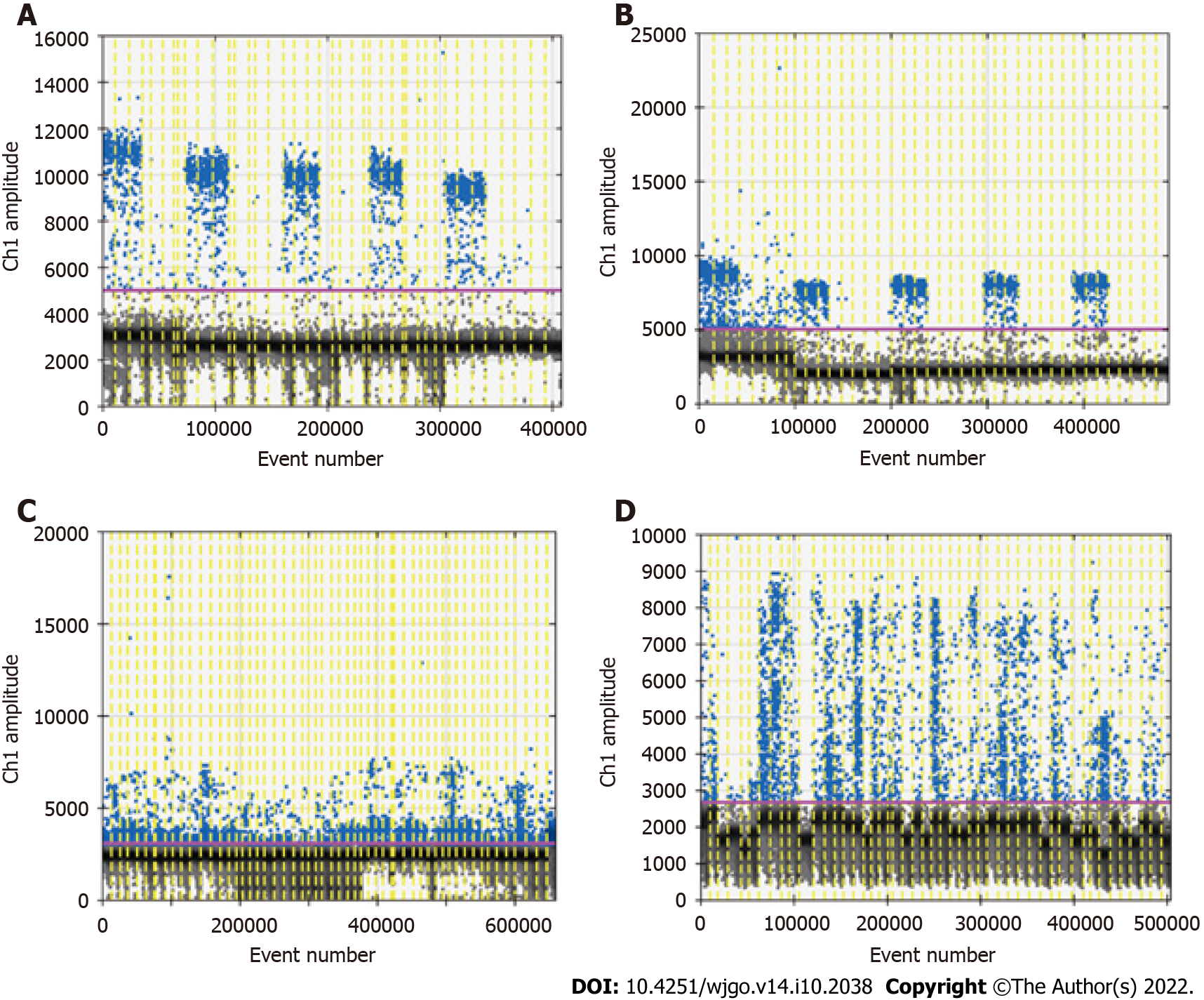
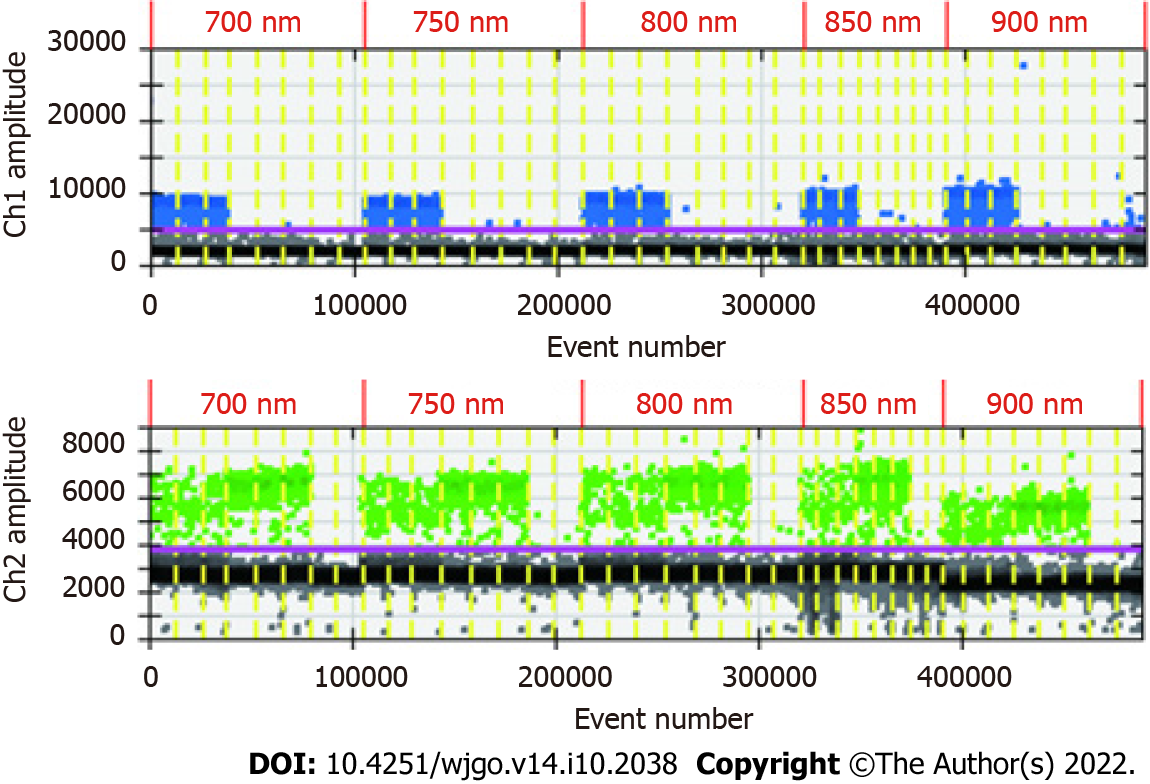
Because of the low concentration of circulating DNA in plasma and inevitable DNA loss after bisulfite conversion, it is conceivable that the plasma trace DNA could not be detected. To address this concern, 100% methylated control DNA with gradient concentration (Table 1) was used to determine the minimum LOD of ddPCR. The results are shown in Figure 3A. The standard curve was drawn based on the above detection data, and the regression equation was y = 1.5264x + 0.0881, R2 = 0.9911, as shown in Figure 3B. We speculated that methylation could not be detected when the input of DNA was less than 0.1 ng. After the bisulfite conversion of blood and plasma DNA, the ssDNA concentrations were all above 0.5 ng/μL. Therefore, ddPCR was reliable for the detection of circulating methylated DNA. To determine the optimal ddPCR amplification condition, the amplification temperatures of 56 °C and 59 °C were compared, and the amplification cycles or 45 and 50 were tested. The result is illustrated in Figure 4. The optimum amplification temperature was 56 °C and the optimum cycle number was 50. Under the reaction conditions of 56 °C and 50 cycles, the primer concentrations of 700 nmol, 750 nmol, 800 nmol, 850 nmol and 900 nmol were compared. As shown in Figure 5, there was little difference among the five primer concentrations; thus, it could be adjusted according to the reaction condition and DNA input amount.
According to the statistical report on global cancer incidence and mortality in 2020 issued by the International Agency for Research on Cancer, GC has been listed as the fifth most common malignant tumor in the world, with 1089103 new cases compared with the previous year. As GC is usually diagnosed at an advanced stage, it has a high mortality rate. GC accounted for 7.7% of total cancer deaths globally in 2020, making it the fourth most common cause of cancer-related deaths[12]. RNF180 is a tumor suppressor gene involved in many important physiological processes in vivo, such as cell growth, differentiation, and tumorigenesis[13]. When RNF180 was abnormally expressed, a variety of physical, chemical, and biological factors were affected, resulting in decreased gene expression or corresponding protein dysfunction. It has been confirmed that the inactivation of RNF180 is associated with apoptosis, tumor invasion and metastasis[14,15]. Downregulation of RNF180 gene expression was related to hypermethylation in the promoter region, and the methylated RNF180 gene played an important role in the occurrence, development, prognosis of GC and infection of H. pylori[16,17]. DNA methylation is the most common chemical modification of nucleic acids. Mechanistically, methyltransferases catalyze methylation by selectively adding the methyl group to cytosine of CG dinucleotide and turning it into 5-methyl cytosine. DNA methylation occurs mainly in DNA fragments with high CG content and between 300 and 3000 bp in length (CpG islands)[18]. Currently, many studies have confirmed that DNA methylation can cause alterations in chromatin structure and DNA stability, thereby controlling gene expression and participating in the regulation of many biological processes, such as aging, nervous system development, occurrence and development of cancer, tumor heterogeneity, and drug resistance[19,20].
Current methods of GC screening and diagnosis in clinical practice all have certain shortcomings, such as invasiveness, sampling deviation, low accuracy and specificity, and long detection time. Liquid biopsy is a noninvasive procedure that uses circulating free DNA, circulating tumor cells or other compounds in patients’ body fluids for diagnosis. Currently, circulating tumor cells, exosomes and circulating nucleic acids are the main materials used in liquid biopsy of cancer. These circulating biomarkers are excreted from the tumor site into the blood, urine, saliva, or cerebrospinal fluid and can provide better insights into the evolution of tumor dynamics during treatment and disease progression. The most widely reported method has been detecting circulating free DNA in blood or plasma[21,22]. The detection of methylated DNA in GC patient plasma by liquid biopsy requires a highly sensitive and accurate method. DdPCR technology has the advantages of high accuracy, high sensitivity, and absolute quantitation and can be used to detect trace DNA. The ddPCR system emulsifies the aqueous PCR mixture into thermally stable oil droplets, while the real-time PCR of nucleic acid quantitation requires a standard curve obtained by diluting samples with known concentrations, which will be affected by laboratory and daily errors. A standard curve was not necessary for ddPCR, which can achieve absolute quantification by counting fluorescence-positive droplets and total droplets according to a Poisson distribution[23].
In this study, we developed a reliable ddPCR assay for the detection of methylated RNF180 in trace DNA. The results indicated that ddPCR could detect the methylated sites of trace circulating free DNA in plasma and achieve absolute quantification. When the DNA input was more than 0.1 ng, ddPCR could analyze the methylated sites of plasma DNA more accurately. This study proved that methylated RNF180 in blood or plasma can be further studied as a potential diagnostic biomarker of GC, and ddPCR assays of methylated DNA have promising applications in tumor screening and diagnosis. At present, this study only established a ddPCR detection method for the methylated RNF180 gene, but according to the experimental results, other methylated sites can also be analyzed by ddPCR technology. The subsequent detection of other GC methylation biomarkers in plasma can be explored.
In summary, a ddPCR assay was established to detect methylated RNF180, which is expected to be a new diagnostic biomarker for GC.
Methylated ring finger protein 180 (RNF180) can be used as a potential biomarker for gastric cancer (GC) diagnosis.
Standard and sensitive of methylation detection methods for in plasma are urgently needed in clinical practice.
We aimed to use droplet digital polymerase chain reaction (ddPCR) to quantify the methylation level of the RNF180 gene. A reproducible ddPCR assay to detect methylated RNF180 from trace DNA was designed and optimized.
The primer and probe were designed and selected, the conversion time of bisulfite was optimized, the ddPCR system was adjusted by primer concentration, amplification temperature and amplification cycles, and the detection limit of ddPCR was determined.
The best conversion time for blood DNA was 2 h 10 min, and that for plasma DNA was 2 h 10 min and 2 h 30 min. The results of ddPCR were better when the amplification temperature was 56 °C and the number of amplification cycles was 50. Primer concentrations showed little effect on the assay outcome. Therefore, the primer concentration could be adjusted according to the reaction system and DNA input. The assay required at least 0.1 ng of input DNA.
In summary, a ddPCR assay was established to detect methylated RNF180, which is expected to be a new diagnostic biomarker for GC.
The standard procedure of ddPCR in clinical practice should be performed and evaluated.
Provenance and peer review: Unsolicited article; Externally peer reviewed.
Peer-review model: Single blind
Specialty type: Oncology
Country/Territory of origin: China
Peer-review report’s scientific quality classification
Grade A (Excellent): 0
Grade B (Very good): B, B, B
Grade C (Good): 0
Grade D (Fair): 0
Grade E (Poor): 0
P-Reviewer: Albillos A, Spain; Valery PC, Australia; Wagner-Skacel J, Austria S-Editor: Wang JJ L-Editor: A P-Editor: Wang JJ
| 1. | Ogawa M, Mizugishi K, Ishiguro A, Koyabu Y, Imai Y, Takahashi R, Mikoshiba K, Aruga J. Rines/RNF180, a novel RING finger gene-encoded product, is a membrane-bound ubiquitin ligase. Genes Cells. 2008;13:397-409. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 23] [Cited by in RCA: 37] [Article Influence: 2.2] [Reference Citation Analysis (0)] |
| 2. | Sun W, Ma G, Zhang L, Wang P, Zhang N, Wu Z, Dong Y, Cai F, Chen L, Liu H, Liang H, Deng J. DNMT3A-mediated silence in ADAMTS9 expression is restored by RNF180 to inhibit viability and motility in gastric cancer cells. Cell Death Dis. 2021;12:428. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 24] [Cited by in RCA: 23] [Article Influence: 5.8] [Reference Citation Analysis (0)] |
| 3. | Wu Z, Liu H, Sun W, Du Y, He W, Guo S, Chen L, Zhao Z, Wang P, Liang H, Deng J. RNF180 mediates STAT3 activity by regulating the expression of RhoC via the proteasomal pathway in gastric cancer cells. Cell Death Dis. 2020;11:881. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 26] [Cited by in RCA: 25] [Article Influence: 5.0] [Reference Citation Analysis (0)] |
| 4. | Han F, Sun LP, Liu S, Xu Q, Liang QY, Zhang Z, Cao HC, Yu J, Fan DM, Nie YZ, Wu KC, Yuan Y. Promoter methylation of RNF180 is associated with H.pylori infection and serves as a marker for gastric cancer and atrophic gastritis. Oncotarget. 2016;7:24800-24809. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 9] [Cited by in RCA: 17] [Article Influence: 2.1] [Reference Citation Analysis (0)] |
| 5. | Deng J, Liang H, Ying G, Zhang R, Wang B, Yu J, Fan D, Hao X. Methylation of CpG sites in RNF180 DNA promoter prediction poor survival of gastric cancer. Oncotarget. 2014;5:3173-3183. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 21] [Cited by in RCA: 23] [Article Influence: 2.1] [Reference Citation Analysis (0)] |
| 6. | Zhang X, Zhang X, Sun B, Lu H, Wang D, Yuan X, Huang Z. Detection of aberrant promoter methylation of RNF180, DAPK1 and SFRP2 in plasma DNA of patients with gastric cancer. Oncol Lett. 2014;8:1745-1750. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 18] [Cited by in RCA: 27] [Article Influence: 2.5] [Reference Citation Analysis (0)] |
| 7. | Hamashima C; Systematic Review Group and Guideline Development Group for Gastric Cancer Screening Guidelines. Update version of the Japanese Guidelines for Gastric Cancer Screening. Jpn J Clin Oncol. 2018;48:673-683. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 281] [Cited by in RCA: 260] [Article Influence: 37.1] [Reference Citation Analysis (0)] |
| 8. | Chivu-Economescu M, Necula L, Matei L, Dragu D, Bleotu C, Diaconu CC. Clinical Applications of Liquid Biopsy in Gastric Cancer. Front Med (Lausanne). 2021;8:749250. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 9] [Cited by in RCA: 8] [Article Influence: 2.0] [Reference Citation Analysis (1)] |
| 9. | Cheung AH, Chow C, To KF. Latest development of liquid biopsy. J Thorac Dis. 2018;10:S1645-S1651. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 40] [Cited by in RCA: 60] [Article Influence: 8.6] [Reference Citation Analysis (0)] |
| 10. | Pinheiro LB, Coleman VA, Hindson CM, Herrmann J, Hindson BJ, Bhat S, Emslie KR. Evaluation of a droplet digital polymerase chain reaction format for DNA copy number quantification. Anal Chem. 2012;84:1003-1011. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 679] [Cited by in RCA: 780] [Article Influence: 55.7] [Reference Citation Analysis (0)] |
| 11. | Diefenbach RJ, Lee JH, Rizos H. Methylated circulating tumor DNA as a biomarker in cutaneous melanoma. Melanoma Manag. 2020;7:MMT46. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 5] [Cited by in RCA: 7] [Article Influence: 1.4] [Reference Citation Analysis (0)] |
| 12. | Sung H, Ferlay J, Siegel RL, Laversanne M, Soerjomataram I, Jemal A, Bray F. Global Cancer Statistics 2020: GLOBOCAN Estimates of Incidence and Mortality Worldwide for 36 Cancers in 185 Countries. CA Cancer J Clin. 2021;71:209-249. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 75126] [Cited by in RCA: 64133] [Article Influence: 16033.3] [Reference Citation Analysis (174)] |
| 13. | Han F, Liu S, Jing J, Li H, Yuan Y, Sun LP. Identification of High-Frequency Methylation Sites in RNF180 Promoter Region Affecting Expression and Their Relationship with Prognosis of Gastric Cancer. Cancer Manag Res. 2020;12:3389-3399. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 1] [Cited by in RCA: 4] [Article Influence: 0.8] [Reference Citation Analysis (0)] |
| 14. | Cheung KF, Lam CN, Wu K, Ng EK, Chong WW, Cheng AS, To KF, Fan D, Sung JJ, Yu J. Characterization of the gene structure, functional significance, and clinical application of RNF180, a novel gene in gastric cancer. Cancer. 2012;118:947-959. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 45] [Cited by in RCA: 51] [Article Influence: 3.6] [Reference Citation Analysis (0)] |
| 15. | Wang H, Lu Y, Wang M, Wu Y, Wang X, Li Y. Roles of E3 ubiquitin ligases in gastric cancer carcinogenesis and their effects on cisplatin resistance. J Mol Med (Berl). 2021;99:193-212. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 3] [Cited by in RCA: 19] [Article Influence: 4.8] [Reference Citation Analysis (0)] |
| 16. | Deng J, Liang H, Zhang R, Hou Y, Liu Y, Ying G, Pan Y, Hao X. Clinical and experimental role of ring finger protein 180 on lymph node metastasis and survival in gastric cancer. Br J Surg. 2016;103:407-416. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 25] [Cited by in RCA: 23] [Article Influence: 2.6] [Reference Citation Analysis (0)] |
| 17. | Xie XM, Deng JY, Hou YC, Cui JL, Wu WP, Ying GG, Dong QP, Hao XS, Liang H. Evaluating the clinical feasibility: The direct bisulfite genomic sequencing for examination of methylated status of E3 ubiquitin ligase RNF180 DNA promoter to predict the survival of gastric cancer. Cancer Biomark. 2015;15:259-265. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 12] [Cited by in RCA: 14] [Article Influence: 1.4] [Reference Citation Analysis (0)] |
| 18. | Lissa D, Robles AI. Methylation analyses in liquid biopsy. Transl Lung Cancer Res. 2016;5:492-504. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 40] [Cited by in RCA: 56] [Article Influence: 6.2] [Reference Citation Analysis (0)] |
| 19. | Liu C, Jiao C, Wang K, Yuan N. DNA Methylation and Psychiatric Disorders. Prog Mol Biol Transl Sci. 2018;157:175-232. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 61] [Cited by in RCA: 46] [Article Influence: 6.6] [Reference Citation Analysis (0)] |
| 20. | Shinjo K, Hara K, Nagae G, Umeda T, Katsushima K, Suzuki M, Murofushi Y, Umezu Y, Takeuchi I, Takahashi S, Okuno Y, Matsuo K, Ito H, Tajima S, Aburatani H, Yamao K, Kondo Y. A novel sensitive detection method for DNA methylation in circulating free DNA of pancreatic cancer. PLoS One. 2020;15:e0233782. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 12] [Cited by in RCA: 28] [Article Influence: 5.6] [Reference Citation Analysis (0)] |
| 21. | Petit J, Carroll G, Gould T, Pockney P, Dun M, Scott RJ. Cell-Free DNA as a Diagnostic Blood-Based Biomarker for Colorectal Cancer: A Systematic Review. J Surg Res. 2019;236:184-197. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 31] [Cited by in RCA: 52] [Article Influence: 8.7] [Reference Citation Analysis (0)] |
| 22. | Lehmann-Werman R, Neiman D, Zemmour H, Moss J, Magenheim J, Vaknin-Dembinsky A, Rubertsson S, Nellgård B, Blennow K, Zetterberg H, Spalding K, Haller MJ, Wasserfall CH, Schatz DA, Greenbaum CJ, Dorrell C, Grompe M, Zick A, Hubert A, Maoz M, Fendrich V, Bartsch DK, Golan T, Ben Sasson SA, Zamir G, Razin A, Cedar H, Shapiro AM, Glaser B, Shemer R, Dor Y. Identification of tissue-specific cell death using methylation patterns of circulating DNA. Proc Natl Acad Sci U S A. 2016;113:E1826-E1834. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 381] [Cited by in RCA: 452] [Article Influence: 50.2] [Reference Citation Analysis (0)] |
| 23. | Kuypers J, Jerome KR. Applications of Digital PCR for Clinical Microbiology. J Clin Microbiol. 2017;55:1621-1628. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 107] [Cited by in RCA: 168] [Article Influence: 21.0] [Reference Citation Analysis (0)] |