Published online Oct 15, 2022. doi: 10.4251/wjgo.v14.i10.1892
Peer-review started: May 19, 2022
First decision: July 13, 2022
Revised: July 15, 2022
Accepted: September 6, 2022
Article in press: September 6, 2022
Published online: October 15, 2022
Processing time: 148 Days and 10.5 Hours
Cancer incidence and mortality are increasing globally, leading to its rising status as a leading cause of death. The Go-Ichi-Ni-San (GINS) complex plays a crucial role in DNA replication and the cell cycle. The GINS complex consists of four subunits encoded by the GINS1, GINS2, GINS3, and GINS4 genes. Recent findings have shown that GINS2 expression is upregulated in many diseases, particularly tumors. For example, increased GINS2 expression has been found in cervical cancer, gastric adenocarcinoma, glioma, non-small cell lung cancer, and pancreatic cancer. It correlates with the clinicopathological characteristics of the tumors. In addition, high GINS2 expression plays a pro-carcinogenic role in tumor development by promoting tumor cell proliferation and migration, inhibiting tumor cell apoptosis, and blocking the cell cycle. This review describes the upregulation of GINS2 expression in most human tumors and the pathway of GINS2 in tumor development. GINS2 may serve as a new marker for tumor diagnosis and a new biological target for therapy.
Core Tip: The Go-Ichi-Ni-San (GINS) complex plays a crucial role in DNA replication and the cell cycle. The GINS complex consists of four subunits encoded by the GINS1, GINS2, GINS3, and GINS4 genes. This review explores the differential expression of GINS2 as a novel target in human cancers. GINS2 is upregulated in most tumors and can influence tumorigenesis and progression through competing endogenous RNA effects and signaling pathways. Therefore, GINS2 may become a new target for the diagnosis and treatment of many cancers.
- Citation: Shan DD, Zheng QX, Chen Z. Go-Ichi-Ni-San 2: A potential biomarker and therapeutic target in human cancers. World J Gastrointest Oncol 2022; 14(10): 1892-1902
- URL: https://www.wjgnet.com/1948-5204/full/v14/i10/1892.htm
- DOI: https://dx.doi.org/10.4251/wjgo.v14.i10.1892
Cancer ranks as the second leading cause of death worldwide, and the burden of cancer is growing, with approximately 9.6 million deaths due to cancer in 2018. Unfortunately, many cancer patients worldwide do not have access to timely, high-quality diagnosis and treatment (World Health Organization, https://www.who.int/cancer/en/). It is therefore crucial to more fully understand how cancer develops and to identify new markers for its diagnosis and new targets for its treatment.
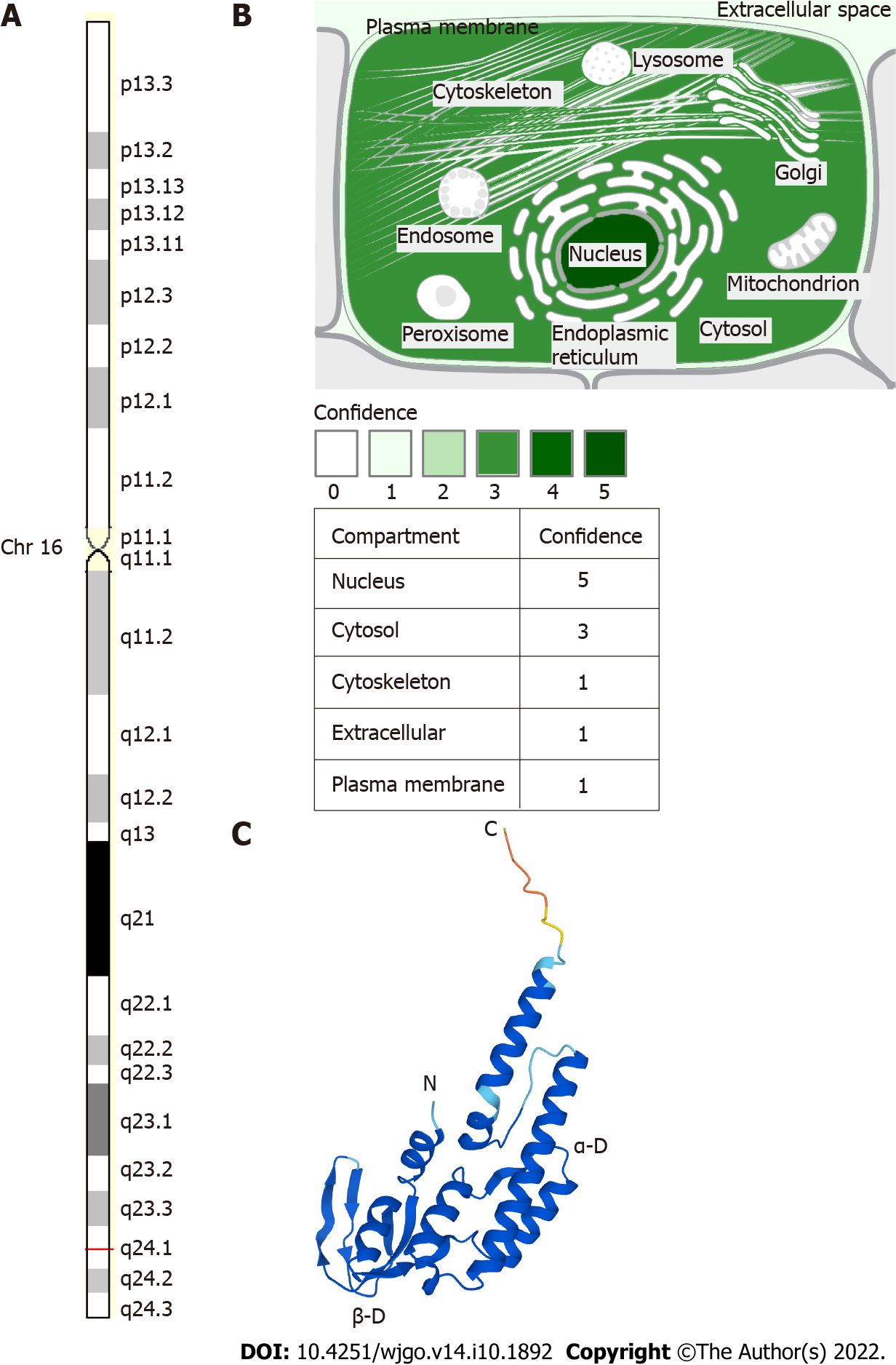
In 2003, Takayama et al[1] described Go-Ichi-Ni-San (GINS) for the first time. The GINS complex is conserved in eukaryotic cells and is essential for DNA replication. When the DNA replication fork is opened, GINS is required to maintain the association between the microchromosome maintenance protein (MCM) and Cdc45 in the large replicator complex[1]. The GINS complex acts as a replicative helicase that unlocks the double-stranded DNA prior to the moving replication fork[2]. The GINS complex consists of four subunits encoded by the GINS1, GINS2, GINS3, and GINS4 genes. GINS2, also known as Psf2, is located in regions 2 and 4 of the long arm of chromosome 16 with a length of 1196 bp[2], as shown in Figure 1. Recent results suggest that GINS2 expression is upregulated in many diseases, especially tumors, and adversely affects prognosis, such as in patients with cervical cancer (CC)[3], breast cancer (BC)[4,5], gastric adenocarcinoma[6], glioma[7], non-small cell lung cancer (NSCLC)[8,9], and pancreatic cancer[10,11].
In this review, we reviewed associated reports and searched the PubMed database from February 2008 to April 2022 using the keywords “GINS2” and “cancer”. After excluding articles from letters, case reports, reviews, meta-analyses, or conference reports, 55 articles describing the expression of GINS2 in human cancers and its relevance to clinical features, as well as the pathways of GINS2 in tumors, were included for further analysis. We also cited high-quality articles in Reference Citation Analysis (https://www.referencecitationanalysis.com). It is reasonable to assume that GINS2 may become a marker in cancer diagnosis and a biological target for prognosis.
Numerous studies have investigated the expression levels of GINS2 in human tissues. The results show that GINS2 expression is increased in most tumors compared to normal tissues and correlates with various clinicopathological features. It has been demonstrated that GINS2 is expressed at higher levels in tumor tissue than in adjacent normal tissue, such as in CC[3], gastric adenocarcinoma[6], glioma[7], NSCLC[8,9], pancreatic cancer[10,11], and thyroid cancer (TC)[12,13]. Specifically, analysis of potential correlations between GINS2 expression levels and clinicopathological features has indicated that high GINS2 expression levels are closely associated with tumor size[6,10], tumor nodal metastasis (TNM) stage[6,8], pathological grade[7] and vascular permeation[10]. These conclusions imply that GINS2 may act as a tumor promoter. A summary of data obtained from published studies is provided in Table 1.
| Cancer types | Cases | Expression | Clinicopathologic parameters | Ref. |
| Cervical cancer | 155 pairs | Upregulated | Pelvic lymph node metastasis, SCC-Ag, deep stromal invasion, vital status, recurrence | Ouyang et al[3] |
| Gastric adenocarcinoma | 123 pairs | Upregulated | Tumor size, T stage, LN metastasis | Feng et al[6] |
| Glioma | 120 pairs | Upregulated | Uathological grade | Shen et al[7] |
| Glioma | 37 pairs | Upregulated | / | Chi et al[9] |
| Glioma | 63 pairs | Upregulated | TNM stage, clinical stage | Liu et al[8] |
| Pancreatic cancer | 74 pairs | Upregulated | Tumor size, T stage, vascular permeation | Huang et al[10] |
| Pancreatic cancer | 46 pairs | Upregulated | / | Bu et al[11] |
| Ovarian cancer | 30 pairs | Upregulated | / | Zhan et al[14] |
In most tumors, elevated levels of GINS2 expression can increase malignant features such as tumor cell proliferation[3-7], migration[8,13,14], invasion[8,13], epithelial-mesenchymal transition (EMT)[8,10], anti-apoptosis effects[12-16] and cell cycle arrest[7,9,11,12], as shown in Figure 2, which are related to the many mechanisms GINS2 is involved in, as shown in Figure 3 and Table 2.
| Cancer types | Assessed cancer cell lines | Expression | Related genes and pathways | Biological significance | Ref. |
| Bladder cancer | 5637, T24 | Up | miR-515-5p | Proliferation, migration, invasion, EMT | Dai et al[27] |
| Bladder cancer | RT4, T24, J82, 5637 | Up | miR-22-3p | Proliferation, colony formation, anti-apoptosis | Tian et al[26] |
| Breast cancer | MCF10A, T47D, MCF-7, SUM149, SUM159, MDA-MB-231, MDA-MB-468, HS578 | Up | MMP9 | Proliferation, cell cycle, migration, invasion, stem-like feature | Peng et al[4] |
| Breast cancer | HCC-1937, MCF-10A, MDA-MB-231, T-47D, JIMT-1 | Up | / | Proliferation, cell cycle | Rantala et al[5] |
| Cervical cancer | SiHa, HeLa, C33A, Caski, MS751, ME180 | Up | / | Proliferation, migration, invasion | Ouyang et al[3] |
| Colon cancer | HCT116, LS174T, HCT8, SW620 | Up | PTP4A1 | Proliferation, cell cycle, anti-apoptosis | Hu et al[31] |
| Ovarian cancer | SKOV3, CaOV3, OVCAR3 | Up | miR-502-5p | Proliferation, migration, anti-apoptosis | Zhan et al[14] |
| Ovarian cancer | SKOV-3, OVCAR3 | Up | / | Proliferation, anti-apoptosis, cell cycle | Yan et al[15] |
| Gastric adenocarcinoma | KATO-III, MKN45 | Up | / | Proliferation | Feng et al[6] |
| Gliomas | U87, U251 | Up | MCM2, ATM, CHEK2 | Proliferation, cell cycle, anti-apoptosis | Shen et al[7] |
| Leukemia | HL60 | Up | Bax, Bcl2, ATM, CHK2, P53 | Proliferation, cell cycle, anti-apoptosis | Zhang et al[16] |
| Leukemia | K562, NB4 | Up | p38/MAPK | Anti-apoptosis, cell cycle | Gao et al[19] |
| Lung cancer | 95D, A549, NCI-H1299, NCI-H1975 | Up | STAT | Proliferation, growth, colony formation, cell cycle | Sun et al[17] |
| Lung cancer | A549, H460 | Up | p53/GADD45A | Proliferation, anti-apoptosis, cell cycle | Chi et al[9] |
| Lung cancer | H1975, H1299, A549, SPC-A1, H460 | Up | PI3K/Akt, MAPK/ERK | Proliferation, migration, invasion, EMT | Liu et al[8] |
| Pancreatic cancer | PANC-1, BxPC-3 | Up | MAPK/ERK | Proliferation, anti-apoptosis, cell cycle | Zhang et al[18] |
| Pancreatic cancer | Aspc-1, Bxpc-3, PANC-1, Miapaca-2 | Up | MAPK/ERK | EMT | Huang et al[10] |
| Pancreatic cancer | PANC-1, AsPC-1 | Up | / | Proliferation, cell cycle | Bu et al[11] |
| Thyroid cancer | K1, SW579 | Up | CITED2, LOXL2 | Proliferation, anti-apoptosis, cell cycle | Ye et al[12] |
| Thyroid cancer | K1, SW579 | Up | MAPK | Proliferation, migration, invasion, anti-apoptosis | He et al[13] |
BC has high morbidity and mortality rates. However, there is still no cure, and patients diagnosed at a late stage often have a poor survival rate, and therefore it is crucial to better understand the mechanisms of breast cancer development[20]. Matrix metalloproteinases (MMPs) are zinc (Zn2+)-dependent endopeptidases involved in the remodeling of the extracellular matrix during physiopathological processes[21]. MMPs play an important role in development, wound healing, tissue remodeling and angiogenesis, with angiogenesis playing a key role in the growth and development of tumors[22]. MMP9 is one of these MMPs and belongs to the gelatinase family[23]. It degrades gelatine and collagen types IV, V, XI and XVI in tissue remodeling and has a significant impact on tumor invasion and metastasis[24]. Peng et al[4] found that knockdown of GINS2 in breast cancer resulted in a significant reduction in MMP9, and GINS2 may regulate the invasive and stem cell-like properties of breast cancer cells through MMP9. The above findings suggest that the expression of GINS2 may be closely related to the prognosis and survival of BC patients.
Bladder cancer has a high incidence of cancer of the urinary system, and 150000 people die of bladder cancer each year[25]. Targets for the effective diagnosis and treatment of bladder cancer are vital. Tian et al[26] found that GINS2 mRNA was a downstream target of miR-22-3p in bladder cancer cells and that knockdown of GINS2 suppressed the phenotype of tumor cells. Similar results were found in bladder cancer cells by Dai et al[27].
The incidence and mortality rate of colon cancer remain high and pose a substantial global burden[28]. Exploring new targets for colon cancer is particularly critical. In cells, protein tyrosine phosphatases (PTPs) have a vital role in regulating tyrosine phosphorylation levels and numerous physiological processes[29]. PTP4A1 belongs to the tripentenyl PTP subclass (PTP4A1/2/3)[30]. Hu et al[31] found that GINS2 interacted with PTP4A1 to regulate the proliferation and apoptosis of colon cancer cells. This finding indicates that GINS2 may be a potential new molecular target for colon cancer.
In 2018, the worldwide incidence of ovarian cancer (OC) was 6.6 per 100000[32]. Zhan et al[14] found that miR-502-5p can inhibit GINS2 expression through the activity of a competing endogenous RNA, which inhibits OC progression by suppressing OC cell growth and promoting apoptosis. In summary, GINS2 can be used as a downstream molecule to influence OC development, and GINS2 may be a new OC target.
Gliomas are the most commonly occurring primary malignancies in the brain, with significantly higher recurrence and mortality rates[33]. In addition, the prognosis of patients is poor, methods to significantly improve patient survival are lacking, and research into the mechanisms of glioma is urgently needed. Minichromosome maintenance complex component 2 (MCM2) belongs to the minichromosome maintenance protein complex and consists of 6 highly conserved proteins (MCM2-7)[34]. Ataxia telangiectasia mutated (ATM) is an important upstream signaling molecule that controls the cell cycle and phosphorylates and activates CHEK2 during DNA replication or upon stimulation by other substances, halting the cell cycle[35-37]. Shen et al[7] used laser confocal microscopy to reveal the relationship between MCM2 and ATM in glioma cells. Additionally, it was reported that inhibition of GINS2 expression reduced cell proliferation and tumorigenicity and that GINS2 could block the cell cycle by regulating MCM2, ATM, CHEK2 and other downstream molecules[7]. GINS2 could be a prognostic indicator and potential therapeutic target for glioma.
Leukaemia is a blood cancer that originates in the bone marrow and is one of the leading causes of death from tumors in humans. In 2016, there were 467000 new cases of leukaemia and 310000 deaths from leukaemia worldwide. Early detection of effective treatment options for leukaemia can help reduce mortality[38]. Mitogen-activated protein kinase (MAPK) is a serine/threonine-protein kinase found in eukaryotic cells that can be activated by various internal and external stimuli. Upon activation, MAPK transmits extracellular signals to the nucleus and affects cellular functions by modulating the activity of transcription factors to alter the expression of related genes[39]. The p38/MAPK signaling pathway is a member of the MAPK superfamily. Gao et al[19] reported that GINS2 knockdown caused cell cycle arrest in chronic granulocytic leukaemia cells and acute promyelocytic leukaemia cells at the G2 phase through activation of p38/MAPK.
ATM-Chk2 and ATM-Chk1 are key signaling pathways that mediate the DNA damage response, and activation of these pathways is critical for the coordination of checkpoint and DNA repair processes. The DNA damage response is crucial to both cancer progression and treatment. p53 oncogene mutations are a way to evade the oncogenic barrier during tumor progression[40]. The findings of Zhang et al[16] suggest that the ATM, Chk2 and p53 genes may play a role in the pathogenic signaling pathway of human acute promyelocytic leukaemia when the GINS2 gene is downregulated. The above studies suggest that GINS2 contributes to the diagnosis and treatment of leukaemia.
Lung cancer is the leading cause of cancer deaths, with NSCLC accounting for approximately 85% of all lung cancers[41]. Patients with NSCLC are often at an advanced stage at the time of detection. A better understanding of the development and evolution of NSCLC is needed to improve this situation. GADD45A is a protein whose expression is regulated over the entire cell cycle, with levels of this protein at their highest in the G1 phase and significantly reduced in the S phase. p53 is a member of the GADD45 (growth arrest and DNA damage induction) family and is responsible for maintaining genomic stability. Wild-type p53 protein arrests cell proliferation, inhibits cell division at the G1 checkpoint and contributes to the repair of damaged DNA. p53 mutations predispose cells to cellular malignancy and tumor formation during the S-phase of damaged DNA. GADD45A-mediated G2-M arrest was found to be dependent on wild-type p53, which controls cell proliferation/apoptosis by regulating cell cycle phases[42]. The results of Chi et al[9] showed that GINS2 expression was increased in NSCLC tissues and cell lines and could promote cell proliferation and inhibit apoptosis via the p53/GADD45A pathway.
Studies have shown that noncanonical nuclear factor-kappaB (NF-κB) transcription factors regulate several normal cellular and tissue processes, such as inflammatory responses, immunity, cell growth, and apoptosis[43,44]. NF-κB is an important “transcription factor”, and aberrant activation of NF-κB signaling has been implicated in the pathogenesis of many diseases, especially tumors[45-47]. Tumor necrosis factor-inducible protein 3 (TNFAIP3) encodes TNFAIP3 (also known as A20) and is a critical negative regulator of NF-κB signaling[48]. Family members of transcription signal transducers and activators (STATs) have been identified as key proteins involved in cytokine signaling and interferon-related antiviral activity[49-51]. Their signaling activities are involved in many normal physiological cellular processes, including proliferation, differentiation, apoptosis, and angiogenesis. However, aberrant STAT regulation can lead to various pathological events, such as malignant cell transformation and metastasis[52]. Sun et al[17] found that after GINS2 gene knockout, the expression of STAT1 and STAT2 proteins increased, which inhibited tumor migration and proliferation. The protein expression of TNFAIP3 increased, suggesting that TNFAIP3 participates in the activity of GINS2 and could be involved in the spread and migration of NSCLC.
Both the PI3K/Akt and MEK/extracellular signal-regulated kinase (ERK) pathways have been reported to be associated with EMT in tumors[53,54]. Liu et al[8] also found that GINS2 could enhance the proliferation, migration, invasion and EMT of NSCLC cells in vivo and in vitro and further demonstrated that GINS2 could regulate the PI3K/Akt and MEK/ERK signaling pathways. In conclusion, GINS2 may be a therapeutic target for NSCLC.
Due to the adverse survival prognosis of pancreatic cancer, the number of deaths is almost as high as the number of patients, and morbidity and mortality rates have remained stable or increased slightly in many countries[55]. It is therefore of interest to identify new targets for the diagnosis and treatment of pancreatic cancer. ERKs belong to the MAPK family and function in signaling cascades that transmit extracellular signals to cells. MAPK cascades are key signaling elements that regulate key processes such as cell proliferation, differentiation, and stress responses[56-58]. The ERK cascade is a tightly controlled cascade responsible for fundamental cellular processes. Excessive activation of proteins and kinases in the ERK pathway has been shown to contribute to a variety of diseases, including cancer, inflammation, developmental disorders, and neurological disorders[59,60]. Huang et al[10] found that overexpression of GINS2 in pancreatic cancer could stimulate EMT in vitro. In MiaPaCa-2 and PANC-1 cells with high GINS2 expression, GINS2 colocalized and coprecipitated with ERK, suggesting that GINS2 interacts closely with the MAPK/ERK pathway. Zhang et al[18] used small interfering RNA to reduce GINS2 expression and explored its mechanism of action in pancreatic cancer. Their results showed that GINS2 interference inhibited pancreatic cancer cell viability through the MAPK/ERK pathway, induced cell cycle arrest and promoted apoptosis in pancreatic cancer cell lines. The above findings suggest that GINS2 may play a negative role in pancreatic cancer and has a guiding role in treating pancreatic cancer.
Since the 1980s, the incidence of TC has increased rapidly in most parts of the world. However, the aetiology of TC is not well understood, and the study of its development is particularly critical in its prevention and treatment[55]. Cbp/p300-interacting transcription factor 2 (CITED2) has a Glu/asp-rich carboxy-terminal domain and is a non-DNA-binding transcriptional coregulator. CITED2 can directly bind to host transcription factors and coactivators, interacting with them to activate gene transcription and affect their function[61]. Several studies have demonstrated that interference with CITED2 can induce apoptosis[62]. Lysine oxidase class 2 (LOXL2) is a member of the lysine oxidase (LOX) family, and some researchers have found that overexpression of LOXL2 activates cell growth in BC. In addition, LOXL2 can directly bind to substrate-like 1 of myristoylation alanine-rich kinase (MARCKSL1), activate the FAK/Akt/mTOR signaling pathway, and inhibit MARCKSL1-induced apoptosis[63]. Ye et al[12] found that GINS2 plays a role in TC cell proliferation and apoptosis by regulating the expression of CITED2 and LOXL2 in TC cells. He et al[13] reported that GINS2 plays a vital role in the survival, migration and invasion of TC cells and regulates the MAPK signaling pathway. GINS2 may be a potential biomarker for TC diagnosis or prognosis and a drug target for treatment.
Most studies have shown that GINS2 expression is upregulated in tumor tissues such as CC, gastric adenocarcinoma, glioma, pancreatic cancer and OC compared to adjacent normal tissues, while GINS2 expression levels correlate with various clinicopathological parameters such as tumor size and TNM stage. These findings suggest that GINS2 can promote tumor progression by regulating tumor cell proliferation, apoptosis, migration, the cell cycle and EMT. In addition, at the cellular level, GINS2 affects the function of several pro- or oncogenic molecules through several signaling pathways, leading to poor patient prognosis. These results imply that GINS2 may be a new target in the diagnosis and treatment of certain tumors.
Currently, there are few publications on interfering with GINS2 in tumor therapy, and no corresponding inhibitors have been reported. In contrast, GINS2 expression is increased in the vast majority of tumors compared to normal tissues, which may make it possible to interfere with GINS2 expression and inhibit GINS2 protein function as an effective way to control tumor development. In future research, potent agents can be explored through molecular docking based on the GINS2 structure, for example.
In conclusion, a better understanding of the role of GINS2 in clinicopathological features and mechanisms of tumor development may help improve diagnostic and therapeutic outcomes. Further studies on GINS2 and its regulatory mechanisms may help improve prevention and treatment based on patient biological and pathological characteristics.
The authors gratefully acknowledge all the people that have made this study.
Provenance and peer review: Unsolicited article; Externally peer reviewed.
Peer-review model: Single blind
Specialty type: Oncology
Country/Territory of origin: China
Peer-review report’s scientific quality classification
Grade A (Excellent): 0
Grade B (Very good): B
Grade C (Good): C, C
Grade D (Fair): 0
Grade E (Poor): 0
P-Reviewer: Fazilat-Panah D, Iran; Luo Y, China; Tousidonis M, Spain S-Editor: Wang JJ L-Editor: A P-Editor: Wang JJ
| 1. | Takayama Y, Kamimura Y, Okawa M, Muramatsu S, Sugino A, Araki H. GINS, a novel multiprotein complex required for chromosomal DNA replication in budding yeast. Genes Dev. 2003;17:1153-1165. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 268] [Cited by in RCA: 289] [Article Influence: 13.1] [Reference Citation Analysis (0)] |
| 2. | Gambus A, Jones RC, Sanchez-Diaz A, Kanemaki M, van Deursen F, Edmondson RD, Labib K. GINS maintains association of Cdc45 with MCM in replisome progression complexes at eukaryotic DNA replication forks. Nat Cell Biol. 2006;8:358-366. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 591] [Cited by in RCA: 608] [Article Influence: 32.0] [Reference Citation Analysis (0)] |
| 3. | Ouyang F, Liu J, Xia M, Lin C, Wu X, Ye L, Song L, Li J, Wang J, Guo P, He M. GINS2 is a novel prognostic biomarker and promotes tumor progression in early-stage cervical cancer. Oncol Rep. 2017;37:2652-2662. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 33] [Cited by in RCA: 52] [Article Influence: 6.5] [Reference Citation Analysis (0)] |
| 4. | Peng L, Song Z, Chen D, Linghu R, Wang Y, Zhang X, Kou X, Yang J, Jiao S. GINS2 regulates matrix metallopeptidase 9 expression and cancer stem cell property in human triple negative Breast cancer. Biomed Pharmacother. 2016;84:1568-1574. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 21] [Cited by in RCA: 32] [Article Influence: 3.6] [Reference Citation Analysis (0)] |
| 5. | Rantala JK, Edgren H, Lehtinen L, Wolf M, Kleivi K, Vollan HK, Aaltola AR, Laasola P, Kilpinen S, Saviranta P, Iljin K, Kallioniemi O. Integrative functional genomics analysis of sustained polyploidy phenotypes in breast cancer cells identifies an oncogenic profile for GINS2. Neoplasia. 2010;12:877-888. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 36] [Cited by in RCA: 42] [Article Influence: 3.0] [Reference Citation Analysis (0)] |
| 6. | Feng H, Zeng J, Gao L, Zhou Z, Wang L. GINS Complex Subunit 2 Facilitates Gastric Adenocarcinoma Proliferation and Indicates Poor Prognosis. Tohoku J Exp Med. 2021;255:111-121. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1] [Cited by in RCA: 2] [Article Influence: 0.5] [Reference Citation Analysis (0)] |
| 7. | Shen YL, Li HZ, Hu YW, Zheng L, Wang Q. Loss of GINS2 inhibits cell proliferation and tumorigenesis in human gliomas. CNS Neurosci Ther. 2019;25:273-287. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 11] [Cited by in RCA: 23] [Article Influence: 3.3] [Reference Citation Analysis (0)] |
| 8. | Liu X, Sun L, Zhang S, Li W. GINS2 facilitates epithelial-to-mesenchymal transition in non-small-cell lung cancer through modulating PI3K/Akt and MEK/ERK signaling. J Cell Physiol. 2020;235:7747-7756. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 19] [Cited by in RCA: 37] [Article Influence: 6.2] [Reference Citation Analysis (0)] |
| 9. | Chi F, Wang Z, Li Y, Chang N. Knockdown of GINS2 inhibits proliferation and promotes apoptosis through the p53/GADD45A pathway in non-small-cell lung cancer. Biosci Rep. 2020;40. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 11] [Cited by in RCA: 23] [Article Influence: 5.8] [Reference Citation Analysis (0)] |
| 10. | Huang L, Chen S, Fan H, Ji D, Chen C, Sheng W. GINS2 promotes EMT in pancreatic cancer via specifically stimulating ERK/MAPK signaling. Cancer Gene Ther. 2021;28:839-849. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 25] [Cited by in RCA: 28] [Article Influence: 7.0] [Reference Citation Analysis (0)] |
| 11. | Bu F, Zhu X, Yi X, Luo C, Lin K, Zhu J, Hu C, Liu Z, Zhao J, Huang C, Zhang W, Huang J. Expression Profile of GINS Complex Predicts the Prognosis of Pancreatic Cancer Patients. Onco Targets Ther. 2020;13:11433-11444. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 22] [Cited by in RCA: 23] [Article Influence: 4.6] [Reference Citation Analysis (0)] |
| 12. | Ye Y, Song YN, He SF, Zhuang JH, Wang GY, Xia W. GINS2 promotes cell proliferation and inhibits cell apoptosis in thyroid cancer by regulating CITED2 and LOXL2. Cancer Gene Ther. 2019;26:103-113. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 15] [Cited by in RCA: 29] [Article Influence: 4.1] [Reference Citation Analysis (0)] |
| 13. | He S, Zhang M, Ye Y, Song Y, Ma X, Wang G, Zhuang J, Xia W, Zhao B. GINS2 affects cell proliferation, apoptosis, migration and invasion in thyroid cancer via regulating MAPK signaling pathway. Mol Med Rep. 2021;23. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 1] [Cited by in RCA: 6] [Article Influence: 1.5] [Reference Citation Analysis (0)] |
| 14. | Zhan L, Yang J, Liu Y, Cheng Y, Liu H. MicroRNA miR-502-5p inhibits ovarian cancer genesis by downregulation of GINS complex subunit 2. Bioengineered. 2021;12:3336-3347. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 3] [Cited by in RCA: 13] [Article Influence: 3.3] [Reference Citation Analysis (0)] |
| 15. | Yan T, Liang W, Jiang E, Ye A, Wu Q, Xi M. GINS2 regulates cell proliferation and apoptosis in human epithelial ovarian cancer. Oncol Lett. 2018;16:2591-2598. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 5] [Cited by in RCA: 13] [Article Influence: 1.9] [Reference Citation Analysis (0)] |
| 16. | Zhang X, Zhong L, Liu BZ, Gao YJ, Gao YM, Hu XX. Effect of GINS2 on proliferation and apoptosis in leukemic cell line. Int J Med Sci. 2013;10:1795-1804. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 27] [Cited by in RCA: 32] [Article Influence: 2.7] [Reference Citation Analysis (0)] |
| 17. | Sun D, Zong Y, Cheng J, Li Z, Xing L, Yu J. GINS2 attenuates the development of lung cancer by inhibiting the STAT signaling pathway. J Cancer. 2021;12:99-110. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 5] [Cited by in RCA: 14] [Article Influence: 3.5] [Reference Citation Analysis (0)] |
| 18. | Zhang M, He S, Ma X, Ye Y, Wang G, Zhuang J, Song Y, Xia W. GINS2 affects cell viability, cell apoptosis, and cell cycle progression of pancreatic cancer cells via MAPK/ERK pathway. J Cancer. 2020;11:4662-4670. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 6] [Cited by in RCA: 16] [Article Influence: 3.2] [Reference Citation Analysis (0)] |
| 19. | Gao Y, Wang S, Liu B, Zhong L. Roles of GINS2 in K562 human chronic myelogenous leukemia and NB4 acute promyelocytic leukemia cells. Int J Mol Med. 2013;31:1402-1410. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 6] [Cited by in RCA: 8] [Article Influence: 0.7] [Reference Citation Analysis (0)] |
| 20. | Salod Z, Singh Y. A five-year (2015 to 2019) analysis of studies focused on breast cancer prediction using machine learning: A systematic review and bibliometric analysis. J Public Health Res. 2020;9:1792. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 11] [Cited by in RCA: 8] [Article Influence: 1.6] [Reference Citation Analysis (0)] |
| 21. | Vu HT, Hoang TX, Kim JY. All-Trans Retinoic Acid Enhances Matrix Metalloproteinase 2 Expression and Secretion in Human Myeloid Leukemia THP-1 Cells. Biomed Res Int. 2018;2018:5971080. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 9] [Cited by in RCA: 17] [Article Influence: 2.4] [Reference Citation Analysis (0)] |
| 22. | Gialeli C, Theocharis AD, Karamanos NK. Roles of matrix metalloproteinases in cancer progression and their pharmacological targeting. FEBS J. 2011;278:16-27. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1046] [Cited by in RCA: 1208] [Article Influence: 80.5] [Reference Citation Analysis (0)] |
| 23. | Bronisz E, Kurkowska-Jastrzębska I. Matrix Metalloproteinase 9 in Epilepsy: The Role of Neuroinflammation in Seizure Development. Mediators Inflamm. 2016;2016:7369020. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 33] [Cited by in RCA: 60] [Article Influence: 6.7] [Reference Citation Analysis (0)] |
| 24. | Curran S, Dundas SR, Buxton J, Leeman MF, Ramsay R, Murray GI. Matrix metalloproteinase/tissue inhibitors of matrix metalloproteinase phenotype identifies poor prognosis colorectal cancers. Clin Cancer Res. 2004;10:8229-8234. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 117] [Cited by in RCA: 123] [Article Influence: 6.2] [Reference Citation Analysis (0)] |
| 25. | Sanli O, Dobruch J, Knowles MA, Burger M, Alemozaffar M, Nielsen ME, Lotan Y. Bladder cancer. Nat Rev Dis Primers. 2017;3:17022. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 420] [Cited by in RCA: 586] [Article Influence: 73.3] [Reference Citation Analysis (0)] |
| 26. | Tian Y, Guan Y, Su Y, Yang T, Yu H. TRPM2-AS Promotes Bladder Cancer by Targeting miR-22-3p and Regulating GINS2 mRNA Expression. Onco Targets Ther. 2021;14:1219-1237. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 3] [Cited by in RCA: 12] [Article Influence: 3.0] [Reference Citation Analysis (0)] |
| 27. | Dai G, Huang C, Yang J, Jin L, Fu K, Yuan F, Zhu J, Xue B. LncRNA SNHG3 promotes bladder cancer proliferation and metastasis through miR-515-5p/GINS2 axis. J Cell Mol Med. 2020;24:9231-9243. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 24] [Cited by in RCA: 55] [Article Influence: 11.0] [Reference Citation Analysis (0)] |
| 28. | Siegel RL, Miller KD, Jemal A. Cancer statistics, 2019. CA Cancer J Clin. 2019;69:7-34. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 13300] [Cited by in RCA: 15470] [Article Influence: 2578.3] [Reference Citation Analysis (2)] |
| 29. | Alonso A, Sasin J, Bottini N, Friedberg I, Osterman A, Godzik A, Hunter T, Dixon J, Mustelin T. Protein tyrosine phosphatases in the human genome. Cell. 2004;117:699-711. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1394] [Cited by in RCA: 1465] [Article Influence: 69.8] [Reference Citation Analysis (0)] |
| 30. | Sacchetti C, Bai Y, Stanford SM, Di Benedetto P, Cipriani P, Santelli E, Piera-Velazquez S, Chernitskiy V, Kiosses WB, Ceponis A, Kaestner KH, Boin F, Jimenez SA, Giacomelli R, Zhang ZY, Bottini N. PTP4A1 promotes TGFβ signaling and fibrosis in systemic sclerosis. Nat Commun. 2017;8:1060. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 28] [Cited by in RCA: 43] [Article Influence: 5.4] [Reference Citation Analysis (0)] |
| 31. | Hu H, Ye L, Liu Z. GINS2 regulates the proliferation and apoptosis of colon cancer cells through PTP4A1. Mol Med Rep. 2022;25. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 1] [Cited by in RCA: 2] [Article Influence: 0.7] [Reference Citation Analysis (0)] |
| 32. | Ferlay J, Colombet M, Soerjomataram I, Mathers C, Parkin DM, Piñeros M, Znaor A, Bray F. Estimating the global cancer incidence and mortality in 2018: GLOBOCAN sources and methods. Int J Cancer. 2019;144:1941-1953. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 3585] [Cited by in RCA: 4898] [Article Influence: 699.7] [Reference Citation Analysis (1)] |
| 33. | Wiedmann MKH, Brunborg C, Di Ieva A, Lindemann K, Johannesen TB, Vatten L, Helseth E, Zwart JA. The impact of body mass index and height on the risk for glioblastoma and other glioma subgroups: a large prospective cohort study. Neuro Oncol. 2017;19:976-985. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 15] [Cited by in RCA: 28] [Article Influence: 4.0] [Reference Citation Analysis (0)] |
| 34. | Zheng G, Kanchwala M, Xing C, Yu H. MCM2-7-dependent cohesin loading during S phase promotes sister-chromatid cohesion. Elife. 2018;7. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 55] [Cited by in RCA: 54] [Article Influence: 7.7] [Reference Citation Analysis (0)] |
| 35. | Li Y, Li L, Wu Z, Wang L, Wu Y, Li D, Ma U, Shao J, Yu H, Wang D. Silencing of ATM expression by siRNA technique contributes to glioma stem cell radiosensitivity in vitro and in vivo. Oncol Rep. 2017;38:325-335. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 15] [Cited by in RCA: 15] [Article Influence: 1.9] [Reference Citation Analysis (0)] |
| 36. | Han B, Cai J, Gao W, Meng X, Gao F, Wu P, Duan C, Wang R, Dinislam M, Lin L, Kang C, Jiang C. Loss of ATRX suppresses ATM dependent DNA damage repair by modulating H3K9me3 to enhance temozolomide sensitivity in glioma. Cancer Lett. 2018;419:280-290. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 32] [Cited by in RCA: 54] [Article Influence: 7.7] [Reference Citation Analysis (0)] |
| 37. | Blake SM, Stricker SH, Halavach H, Poetsch AR, Cresswell G, Kelly G, Kanu N, Marino S, Luscombe NM, Pollard SM, Behrens A. Inactivation of the ATMIN/ATM pathway protects against glioblastoma formation. Elife. 2016;5. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 15] [Cited by in RCA: 17] [Article Influence: 1.9] [Reference Citation Analysis (0)] |
| 38. | Global Burden of Disease Cancer Collaboration, Fitzmaurice C, Akinyemiju TF, Al Lami FH, Alam T, Alizadeh-Navaei R, Allen C, Alsharif U, Alvis-Guzman N, Amini E, Anderson BO, Aremu O, Artaman A, Asgedom SW, Assadi R, Atey TM, Avila-Burgos L, Awasthi A, Ba Saleem HO, Barac A, Bennett JR, Bensenor IM, Bhakta N, Brenner H, Cahuana-Hurtado L, Castañeda-Orjuela CA, Catalá-López F, Choi JJ, Christopher DJ, Chung SC, Curado MP, Dandona L, Dandona R, das Neves J, Dey S, Dharmaratne SD, Doku DT, Driscoll TR, Dubey M, Ebrahimi H, Edessa D, El-Khatib Z, Endries AY, Fischer F, Force LM, Foreman KJ, Gebrehiwot SW, Gopalani SV, Grosso G, Gupta R, Gyawali B, Hamadeh RR, Hamidi S, Harvey J, Hassen HY, Hay RJ, Hay SI, Heibati B, Hiluf MK, Horita N, Hosgood HD, Ilesanmi OS, Innos K, Islami F, Jakovljevic MB, Johnson SC, Jonas JB, Kasaeian A, Kassa TD, Khader YS, Khan EA, Khan G, Khang YH, Khosravi MH, Khubchandani J, Kopec JA, Kumar GA, Kutz M, Lad DP, Lafranconi A, Lan Q, Legesse Y, Leigh J, Linn S, Lunevicius R, Majeed A, Malekzadeh R, Malta DC, Mantovani LG, McMahon BJ, Meier T, Melaku YA, Melku M, Memiah P, Mendoza W, Meretoja TJ, Mezgebe HB, Miller TR, Mohammed S, Mokdad AH, Moosazadeh M, Moraga P, Mousavi SM, Nangia V, Nguyen CT, Nong VM, Ogbo FA, Olagunju AT, Pa M, Park EK, Patel T, Pereira DM, Pishgar F, Postma MJ, Pourmalek F, Qorbani M, Rafay A, Rawaf S, Rawaf DL, Roshandel G, Safiri S, Salimzadeh H, Sanabria JR, Santric Milicevic MM, Sartorius B, Satpathy M, Sepanlou SG, Shackelford KA, Shaikh MA, Sharif-Alhoseini M, She J, Shin MJ, Shiue I, Shrime MG, Sinke AH, Sisay M, Sligar A, Sufiyan MB, Sykes BL, Tabarés-Seisdedos R, Tessema GA, Topor-Madry R, Tran TT, Tran BX, Ukwaja KN, Vlassov VV, Vollset SE, Weiderpass E, Williams HC, Yimer NB, Yonemoto N, Younis MZ, Murray CJL, Naghavi M. Global, Regional, and National Cancer Incidence, Mortality, Years of Life Lost, Years Lived With Disability, and Disability-Adjusted Life-Years for 29 Cancer Groups, 1990 to 2016: A Systematic Analysis for the Global Burden of Disease Study. JAMA Oncol. 2018;4:1553-1568. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 1105] [Cited by in RCA: 1167] [Article Influence: 166.7] [Reference Citation Analysis (0)] |
| 39. | Mariani E, Pulsatelli L, Facchini A. Signaling pathways in cartilage repair. Int J Mol Sci. 2014;15:8667-8698. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 97] [Cited by in RCA: 113] [Article Influence: 10.3] [Reference Citation Analysis (0)] |
| 40. | Smith J, Tho LM, Xu N, Gillespie DA. The ATM-Chk2 and ATR-Chk1 pathways in DNA damage signaling and cancer. Adv Cancer Res. 2010;108:73-112. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 772] [Cited by in RCA: 932] [Article Influence: 66.6] [Reference Citation Analysis (0)] |
| 41. | Herbst RS, Heymach JV, Lippman SM. Lung cancer. N Engl J Med. 2008;359:1367-1380. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1876] [Cited by in RCA: 1999] [Article Influence: 117.6] [Reference Citation Analysis (0)] |
| 42. | Jin S, Mazzacurati L, Zhu X, Tong T, Song Y, Shujuan S, Petrik KL, Rajasekaran B, Wu M, Zhan Q. Gadd45a contributes to p53 stabilization in response to DNA damage. Oncogene. 2003;22:8536-8540. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 68] [Cited by in RCA: 74] [Article Influence: 3.4] [Reference Citation Analysis (0)] |
| 43. | Hoffmann A, Baltimore D. Circuitry of nuclear factor kappaB signaling. Immunol Rev. 2006;210:171-186. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 690] [Cited by in RCA: 739] [Article Influence: 41.1] [Reference Citation Analysis (0)] |
| 44. | Gilmore TD, Kalaitzidis D, Liang MC, Starczynowski DT. The c-Rel transcription factor and B-cell proliferation: a deal with the devil. Oncogene. 2004;23:2275-2286. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 105] [Cited by in RCA: 111] [Article Influence: 5.3] [Reference Citation Analysis (0)] |
| 45. | Qiao L, Zhang H, Yu J, Francisco R, Dent P, Ebert MP, Röcken C, Farrell G. Constitutive activation of NF-kappaB in human hepatocellular carcinoma: evidence of a cytoprotective role. Hum Gene Ther. 2006;17:280-290. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 55] [Cited by in RCA: 59] [Article Influence: 3.1] [Reference Citation Analysis (0)] |
| 46. | Mann AP, Verma A, Sethi G, Manavathi B, Wang H, Fok JY, Kunnumakkara AB, Kumar R, Aggarwal BB, Mehta K. Overexpression of tissue transglutaminase leads to constitutive activation of nuclear factor-kappaB in cancer cells: delineation of a novel pathway. Cancer Res. 2006;66:8788-8795. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 154] [Cited by in RCA: 162] [Article Influence: 9.0] [Reference Citation Analysis (0)] |
| 47. | Prasad S, Ravindran J, Aggarwal BB. NF-kappaB and cancer: how intimate is this relationship. Mol Cell Biochem. 2010;336:25-37. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 293] [Cited by in RCA: 315] [Article Influence: 21.0] [Reference Citation Analysis (0)] |
| 48. | Yu MP, Xu XS, Zhou Q, Deuitch N, Lu MP. Haploinsufficiency of A20 (HA20): updates on the genetics, phenotype, pathogenesis and treatment. World J Pediatr. 2020;16:575-584. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 29] [Cited by in RCA: 51] [Article Influence: 10.2] [Reference Citation Analysis (0)] |
| 49. | Wegenka UM, Lütticken C, Buschmann J, Yuan J, Lottspeich F, Müller-Esterl W, Schindler C, Roeb E, Heinrich PC, Horn F. The interleukin-6-activated acute-phase response factor is antigenically and functionally related to members of the signal transducer and activator of transcription (STAT) family. Mol Cell Biol. 1994;14:3186-3196. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 9] [Cited by in RCA: 53] [Article Influence: 1.7] [Reference Citation Analysis (0)] |
| 50. | Sadowski HB, Shuai K, Darnell JE Jr, Gilman MZ. A common nuclear signal transduction pathway activated by growth factor and cytokine receptors. Science. 1993;261:1739-1744. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 505] [Cited by in RCA: 546] [Article Influence: 17.1] [Reference Citation Analysis (0)] |
| 51. | Darnell JE Jr, Kerr IM, Stark GR. Jak-STAT pathways and transcriptional activation in response to IFNs and other extracellular signaling proteins. Science. 1994;264:1415-1421. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 4322] [Cited by in RCA: 4587] [Article Influence: 148.0] [Reference Citation Analysis (0)] |
| 52. | Bowman T, Garcia R, Turkson J, Jove R. STATs in oncogenesis. Oncogene. 2000;19:2474-2488. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1376] [Cited by in RCA: 1417] [Article Influence: 56.7] [Reference Citation Analysis (0)] |
| 53. | Pan H, Jiang T, Cheng N, Wang Q, Ren S, Li X, Zhao C, Zhang L, Cai W, Zhou C. Long non-coding RNA BC087858 induces non-T790M mutation acquired resistance to EGFR-TKIs by activating PI3K/AKT and MEK/ERK pathways and EMT in non-small-cell lung cancer. Oncotarget. 2016;7:49948-49960. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 70] [Cited by in RCA: 94] [Article Influence: 13.4] [Reference Citation Analysis (0)] |
| 54. | Ha GH, Park JS, Breuer EK. TACC3 promotes epithelial-mesenchymal transition (EMT) through the activation of PI3K/Akt and ERK signaling pathways. Cancer Lett. 2013;332:63-73. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 84] [Cited by in RCA: 98] [Article Influence: 8.2] [Reference Citation Analysis (0)] |
| 55. | Sung H, Ferlay J, Siegel RL, Laversanne M, Soerjomataram I, Jemal A, Bray F. Global Cancer Statistics 2020: GLOBOCAN Estimates of Incidence and Mortality Worldwide for 36 Cancers in 185 Countries. CA Cancer J Clin. 2021;71:209-249. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 75126] [Cited by in RCA: 64628] [Article Influence: 16157.0] [Reference Citation Analysis (176)] |
| 56. | Sabio G, Davis RJ. TNF and MAP kinase signalling pathways. Semin Immunol. 2014;26:237-245. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 378] [Cited by in RCA: 533] [Article Influence: 48.5] [Reference Citation Analysis (0)] |
| 57. | Plotnikov A, Zehorai E, Procaccia S, Seger R. The MAPK cascades: signaling components, nuclear roles and mechanisms of nuclear translocation. Biochim Biophys Acta. 2011;1813:1619-1633. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 539] [Cited by in RCA: 681] [Article Influence: 45.4] [Reference Citation Analysis (0)] |
| 58. | Keshet Y, Seger R. The MAP kinase signaling cascades: a system of hundreds of components regulates a diverse array of physiological functions. Methods Mol Biol. 2010;661:3-38. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 366] [Cited by in RCA: 443] [Article Influence: 29.5] [Reference Citation Analysis (0)] |
| 59. | Kim JY, Lee SG, Chung JY, Kim YJ, Park JE, Koh H, Han MS, Park YC, Yoo YH, Kim JM. Ellipticine induces apoptosis in human endometrial cancer cells: the potential involvement of reactive oxygen species and mitogen-activated protein kinases. Toxicology. 2011;289:91-102. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 37] [Cited by in RCA: 44] [Article Influence: 3.1] [Reference Citation Analysis (0)] |
| 60. | Gupta J, Nebreda AR. Roles of p38α mitogen-activated protein kinase in mouse models of inflammatory diseases and cancer. FEBS J. 2015;282:1841-1857. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 91] [Cited by in RCA: 120] [Article Influence: 12.0] [Reference Citation Analysis (0)] |
| 61. | Chou YT, Yang YC. Post-transcriptional control of Cited2 by transforming growth factor beta. Regulation via Smads and Cited2 coding region. J Biol Chem. 2006;281:18451-18462. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 30] [Cited by in RCA: 33] [Article Influence: 1.7] [Reference Citation Analysis (0)] |
| 62. | Minemura H, Takagi K, Sato A, Takahashi H, Miki Y, Shibahara Y, Watanabe M, Ishida T, Sasano H, Suzuki T. CITED2 in breast carcinoma as a potent prognostic predictor associated with proliferation, migration and chemoresistance. Cancer Sci. 2016;107:1898-1908. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 10] [Cited by in RCA: 17] [Article Influence: 1.9] [Reference Citation Analysis (0)] |
| 63. | Kim BR, Dong SM, Seo SH, Lee JH, Lee JM, Lee SH, Rho SB. Lysyl oxidase-like 2 (LOXL2) controls tumor-associated cell proliferation through the interaction with MARCKSL1. Cell Signal. 2014;26:1765-1773. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 21] [Cited by in RCA: 28] [Article Influence: 2.5] [Reference Citation Analysis (0)] |