Published online Jan 15, 2022. doi: 10.4251/wjgo.v14.i1.75
Peer-review started: March 17, 2021
First decision: July 27, 2021
Revised: August 11, 2021
Accepted: November 30, 2021
Article in press: November 30, 2021
Published online: January 15, 2022
Processing time: 299 Days and 20.4 Hours
Gastrointestinal (GI) cancers, including colorectal cancer, pancreatic cancer, liver cancer and gastric cancer, are severe social burdens due to high incidence and mortality rates. Bromodomain and extra-terminal (BET) proteins are epigenetic readers consisting of four conserved members (BRD2, BRD3, BRD4 and BRDT). BET family perform pivotal roles in tumorigenesis through transcriptional regulation, thereby emerging as potential therapeutic targets. BET inhibitors, disrupting the interaction between BET proteins and acetylated lysines, have been reported to suppress tumor initiation and progression in most of GI cancers. In this review, we will demonstrate how BET proteins participate in the GI cancers progression and highlight the therapeutic potential of targeting BET proteins for GI cancers treatment.
Core Tip: Bromodomain and extra-terminal (BET) inhibitors, as promising targeted agents, emerge as a new therapeutic avenue for gastrointestinal (GI) cancers. Based on preclinical evidence, BET inhibitors, alone or in combination with other therapies, were effective to suppress the progression of GI cancers.
- Citation: Sun HY, Du ST, Li YY, Deng GT, Zeng FR. Bromodomain and extra-terminal inhibitors emerge as potential therapeutic avenues for gastrointestinal cancers. World J Gastrointest Oncol 2022; 14(1): 75-89
- URL: https://www.wjgnet.com/1948-5204/full/v14/i1/75.htm
- DOI: https://dx.doi.org/10.4251/wjgo.v14.i1.75
Gastrointestinal (GI) cancers, including colorectal cancer (CRC), liver cancer, gastric cancer (GC) and pancreatic cancer, are among the most common malignancies worldwide with high incidence and mortality rates. In the latest global cancer data of 2020, CRC is the second leading cause of cancer death (9.4% of the total cancer deaths), followed by stomach cancer (8.3%), liver cancer (7.7%) and pancreatic cancer (4.6%)[1]. Surgery still remains the only curative treatment for GI cancers[2]. However, most patients are diagnosed as GI cancer at advanced stages or metastases, and thus lose the chance of surgery. Several therapies including chemotherapy[3,4], radiotherapy[5], chemoradiotherapy[6] and immunotherapy[7,8], have been developed for those GI cancers patients who are intolerable to operation. Unfortunately, inevitable toxicity[9], innate or acquired chemo-resistance[10] and low response[11] limit the clinical use of these treatments, highlighting the need for developing new therapeutic strategies.
Bromodomain and extra-terminal (BET) protein inhibitors emerge as a new therapeutic avenue for multiple cancers, including GI cancers. BET inhibitors exert anti-cancer activities by competitively binding to BET proteins and disrupting the interaction between BET proteins and acetylated lysines. Increasing studies have reported that upregulation of BET proteins leads to abnormal transcriptional re
In this review, we will briefly describe the structure and inhibition mechanism of BET proteins and illustrate the role of BET proteins in the initiation and progression of human GI cancers. Then, we will identify whether targeting BET proteins, alone or in combination with other therapies, exhibits potential benefits in GI cancers through preclinical evidence. Finally, we will speculate the outlook of the translation of BET inhibitors into clinic.
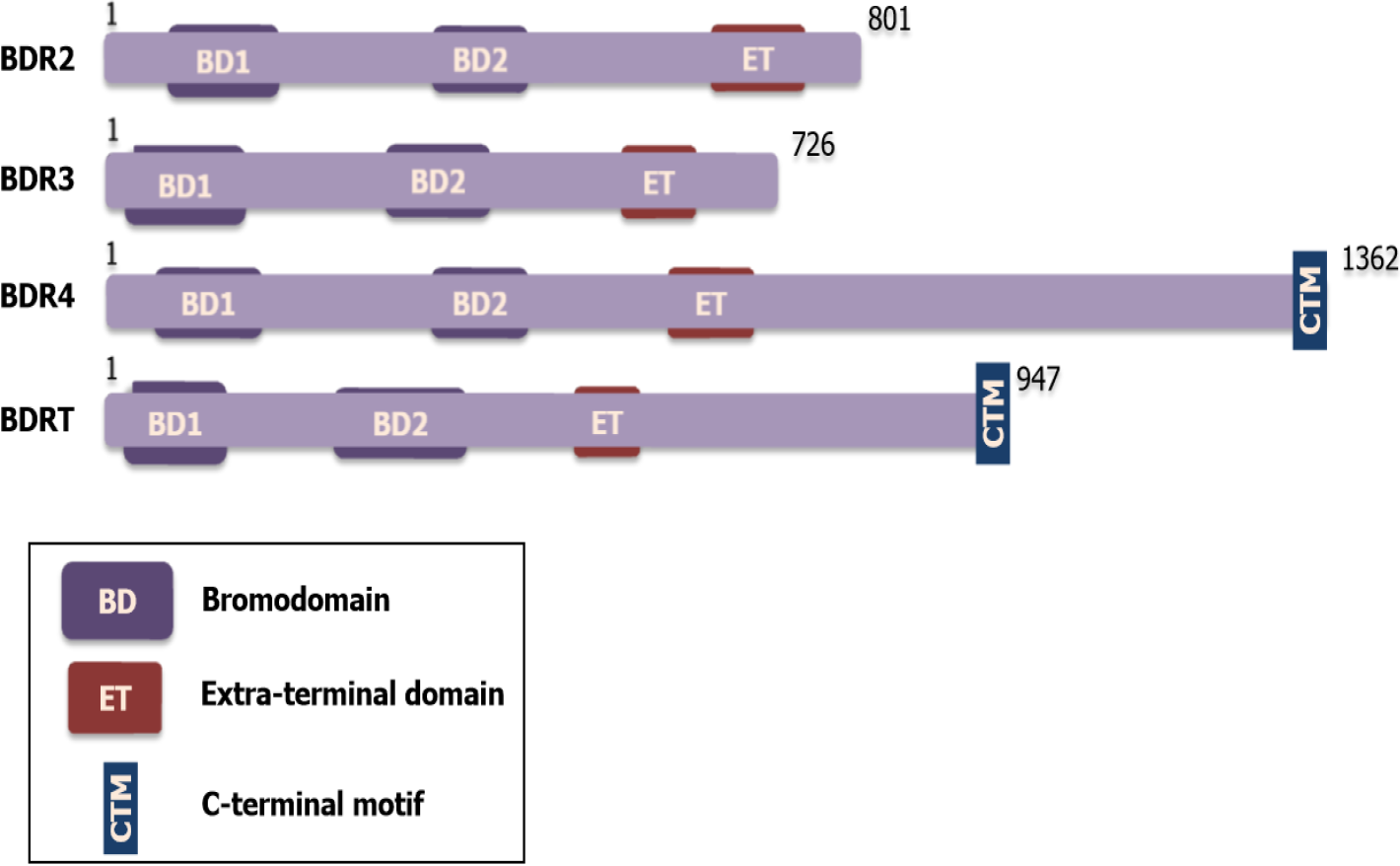
BET family proteins include four subtypes: BRD2 (also known as FSRG1, RING3, RNF3, FSH, or D6S113E), BRD3 (also known as ORFX or RING3L), BRD4 (also known as MCAP or HUNK1) and BRDT (also known as BRD6, CT9, or SPGF21)[13,14]. Each of the BET proteins has a highly conversed structure including two tandem -110 amino acid bromodomains (BD1 and BD2) with direct specificity for acetylated lysines, followed by an extra-terminal (ET) protein-protein interaction domain[15]. Notably, BRD4 and BRDT comprise a C-terminal domain, which functionally recruits transcriptional regulators, like the positive transcription elongation factor b (P-TEFb)[16,17] (Figure 1). The similarity and difference in structure among BET proteins may partly interpret the parallel and differential function in human disease, especially in cancer.
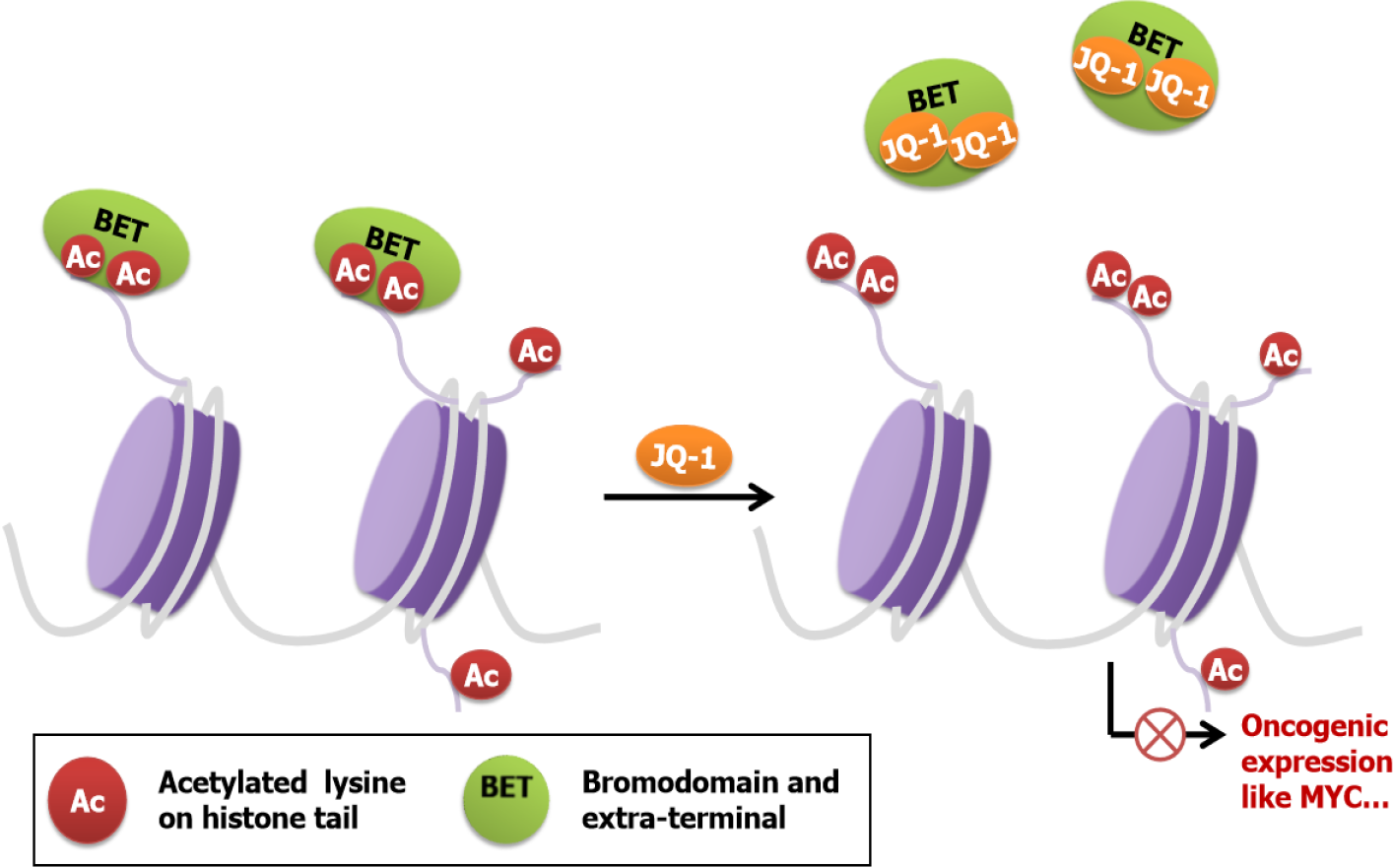
BET proteins have two BDs with the acetylated lysine binding pocket. Compared with acetylated histones, BDs have a higher affinity for small molecules, which provide new possibilities for the development of inhibitors[18]. By occupying the BD pockets, BET inhibitors, such as JQ-1, mimic the binding mode and competitively inhibit binding between acetylated lysines and BDs, resulting in disrupting oncogenic rearrangement and inhibiting the development of some aggressive types of cancer (Figure 2).
Oncogenic roles of BET proteins family were firstly revealed in the NUT carcinoma. BRD4 and BRD3 are involved in the chromosomal rearrangements of NUT carcinoma by forming BET-NUT fusion protein[19]. The inspirational discovery that BET proteins serve as potential cancer therapeutic targets encourages researchers to look for possible functions of BET proteins in other cancers, including GI cancers. Strikingly, BET proteins (BRD2, BRD4) are overexpressed in GI cancers and have been reported to promote GI cancers progression via multiple mechanisms.
BRD2 was firstly defined as a non-canonical protein kinase[20], which could promote the GI cancers progression by recruiting transcriptional factors and initiating transcriptional regulation. Recent studies demonstrated that BRD2 promoted the progression of CRC, pancreatic ductal adenocarcinoma (PDAC) and GC[21]. Spe
BRD3 was rarely studied in GI cancers. However, recently, some frameshift mu
BRD4 is the most extensively studied BET proteins in GI cancers which is highly expressed in cancer tissues and cell lines, including CRC[27], pancreatic cancer[28], liver cancer[29], and GC[30]. The overexpression of BRD4 promotes GI cancer cell growth, differentiation and metastasis, and correlates with poor outcome of GI cancers patients[31,32]. On one hand, BRD4 could directly bind to the promoter region of oncogenes and induce their overexpression, including c-MYC[33], E2F2[34], caveolin-2[28], PES1[35] and CD276[36]. On the other hand, BRD4 could recognize acetylated lysines on epithelial-to-mesenchymal transition (EMT)-activating transcriptional factors like Twist or Snail, the activation of which facilitated the differentiation and survival of EMT cells and promoted metastatic growth in GI cancers[27,37,38]. Additionally, BRD4 was reported to be recruited to senescence-activated super-enhancers to mediate cellular senescence[39]. The senescent cancer cells induced the secretion of various cytokines and increased CRC cells migration and invasion abilities[40]. In addition to the direct induction of tumorigenesis, BRD4 was also involved in the crosstalk between cancer and cancer-associated fibroblasts. Inhibiting the BRD4 protein changed both transcription and structure of matrisome in PDAC and resulted in better patients’ survival[41]. Moreover, Yasukawa et al[42] also described that BRD4 played an important role in cancer associated fibroblasts in GC[42]. These oncogenic functions suggest that BRD4 is an important molecular target for GI cancers.
Given that BET proteins are important regulators in GI cancer, targeting BET proteins will be a good therapeutic strategy for GI cancers treatment. A series of compounds have been reported as potential therapeutic avenues for GI cancers by targeting BET proteins (Table 1). BET inhibitors share the similar mechanism by displacing BET proteins from chromatin and regulating transcriptional factors. By mediating cell cycle arrest, facilitating apoptosis, and inducing senescence, BET inhibitors functionally inhibit cell proliferation, invasion and migration in most GI cancers including CRC, pancreatic cancer, liver cancer and GC[43]. Mechanically, BET inhibitors exert anti-tumor activity in c-MYC dependent, as well as c-MYC independent manners[44]. BET inhibitors have been widely used in preclinical models, but BET inhibitors alone exhibit limited-single agent activity confronting drug resistance. Combinational therapy with chemotherapy, immunotherapy or other small molecule inhibitors may amplify the clinical outcomes in GI cancers. Herein, we review the application of BET inhibitors in GI cancers.
| GI cancers models | BET inhibitors | Combination with | Targets | Pathway/mechanism | Ref. |
| CRC | JQ-1 | 5-FU | DR5 | Apoptosis | [49] |
| JQ-1 | Bortezomib | MYC, FOXM1 | G2/M arrest | [47] | |
| JQ-1 | - | HGF, MET | Cancer-associated fibroblasts | [98] | |
| Apabetalone | - | APOA1 | Intracellular cholesterol metabolism | [99] | |
| JQ-1 | BEZ235 (PI3K/mTOR inhibitor) | RTKs | Overcome resistance to PI3K/mTOR inhibition | [40] | |
| JQ-1 | Sulforaphane (HDAC3 inhibitor) | ERCC2 | Nucleotide excision repair pathway | [48] | |
| I-BET151, bromosporine | - | BRD4, SNAIL, SLUG | EMT | [100] | |
| SMAD4-defificient CRC | OTX-015 | - | MYC | MYC-p21 axis, G1 cell cycle arrest | [54] |
| Colon cancer | JQ-1 | - | Nkd2, β-catenin, miR-21 | Wnt/β-catenin signaling, apoptosis | [45] |
| Gastric and colon cancer | JQ-1 | Arsenic sulfide | NFATs, c-MYC | Mitochondrial pathway induced cell apoptosis | [51] |
| PDAC | JQ-1 | - | HMGA2 | Block growth of chemoresistant cells | [55] |
| JQ-1 | Olaparib (PARP inhibitor) | BRD2/4, Ku80, RAD51 | DNA damage | [60] | |
| JQ-1 | SAHA (HDAC inhibitor) | p57 | Cell death | [61] | |
| JQ-1 | Gemcitabine | HMGCS2, APOC1 | DNA damage and apoptosis | [62] | |
| CPI203 | - | MYC, GLI, SHH | SHH-GLI signaling pathway, cell cycle progression | [24] | |
| Pancreatic cancer | JQ-1, OTX-015 | Quercetin | BRD4(JQ-1) and hnRNPA1(Quercetin) | Apoptosis | [63] |
| KDM6A null pancreatic cancer | JQ-1 | - | MYC, p63, RUNX3 | Reverse squamous differentiation | [101,102] |
| Liver cancer | JQ-1 | - | BRD4, E2F2 | BRD4-E2F2-cell cycle regulation axis, | [34] |
| JQ-1 | - | PD-L1, PD-L2 | PD-1/PD-L1 signaling | [71] | |
| HCC | JQ-1, I-BET762 | Anti-PD-L1 Ab | BRD4, C/EBPβ, p300 | Suppress M-MDSCs, enhance PD-L1 blockade efficacy | [73] |
| JQ-1 | - | MYC | Impair mitochondrial respiration and glycolysis, induce apoptosis | [66] | |
| Hjp-6-171 | GSK3β inhibitor (CHIR-98014) | β-catenin, NOTUM | WNT pathway | [68] | |
| SF1126 (Pan PI3K/BRD4 Inhibitor) | Sorafenib | BRD4, c-MYC | Ras/Raf/MAPK, PI3K/AKT/mTOR pathways | [90] | |
| JQ-1 | - | PES1 | Cell proliferation, glycolysis | [35] | |
| JQ-1 | Flavopiridol | Mcl-1 | Apoptosis | [67] | |
| JQ-1, OTX-015 | - | SMARCA4 | Down-regulate migration related genes | [65] | |
| CCA2 | JQ-1 | PI3K/mTOR inhibitors | c-Myc, YAP | Overcome resistance to PI3K/mTOR inhibition | [64] |
| Gastric cancer | JQ-1 | - | BRD4, E2F | E2F/miR-106b-5p/p21 axis, cellular senescence | [32] |
| JQ-1 | - | RUNX2 | RUNX2/NID1 signaling, site-specific chromatinremodeling | [75] | |
| JQ1, PNZ5 | - | c-MYC | Apoptosis | [33,74] | |
| iBET-151 | Paclitaxel | RTK | G1 cell cycle arrest | [79] | |
| AZD5153 | - | Sirt5, Mus81 | Sirt5/Mus81/ZEB1 axis, inhibit metastasis | [76] | |
| GAC | JQ-1 | CA3 (YAP inhibitor) | c-MYC | Gal3/RalA/YAP1/c-MYC axis | [78] |
Preclinical data demonstrated that BET inhibitors alone had exhibited efficacy against CRC by inhibiting tumor growth and inducing apoptosis in vivo and vitro[27,45]. However, resistance to BET inhibitors was the major obstacle to CRC treatment. Wang et al[46] raised one possible mechanism that the interaction of STAT3 through BRD4 phosphorylation might result in the resistance of BET inhibitors in CRC. Combining BET inhibitors and other targeted therapies could help to overcome resistance and render CRC more sensitive to BET inhibitors. For example, nuclear factor-kappa B inhibitors[47], PI3K/mTOR inhibitors[40], HDAC3 inhibitor[48] have been reported to sensitize GI cancers to BET inhibitors, and finally achieve synergistical effects.
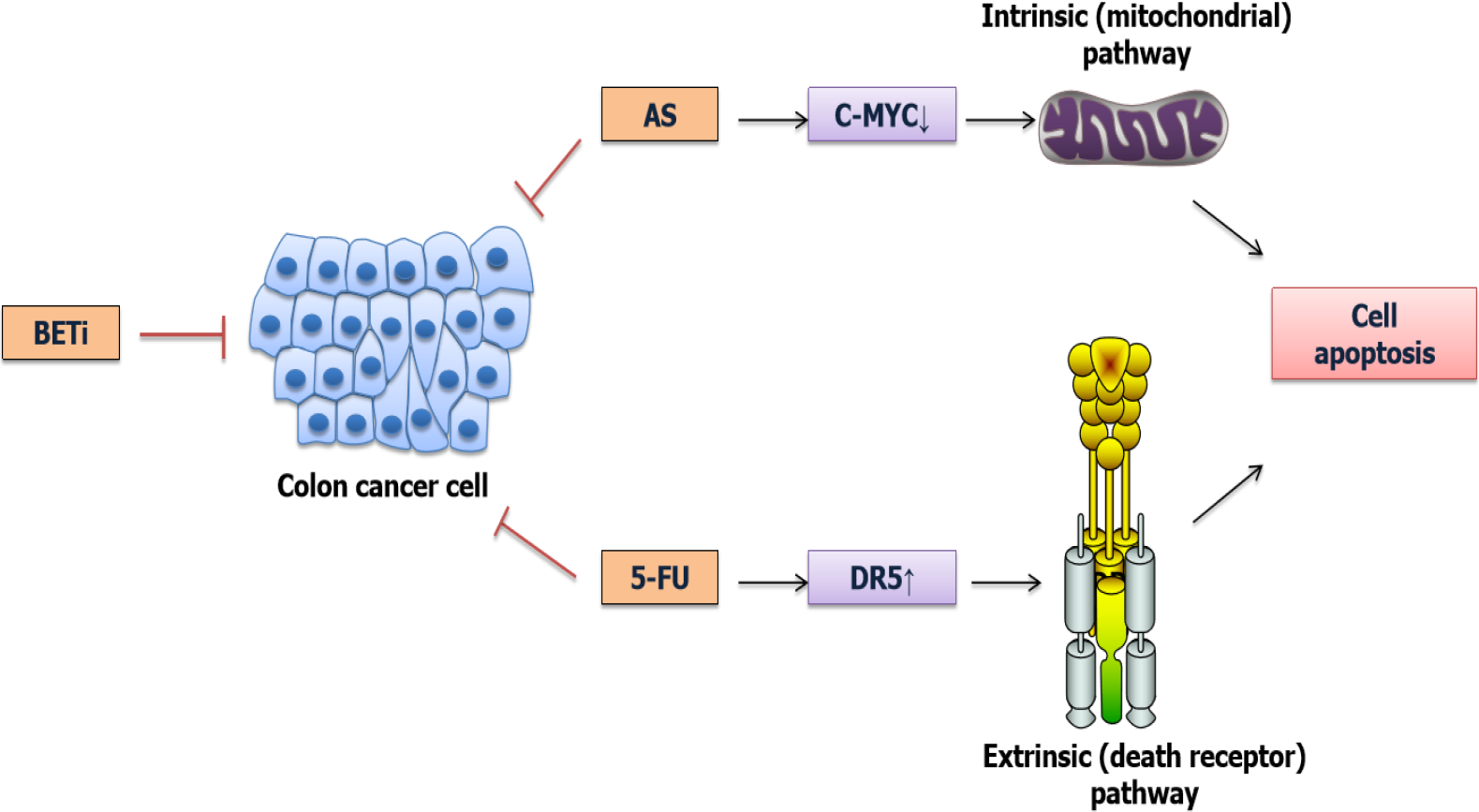
Moreover, BET inhibition could be used in combination with chemotherapy to enhance chemotherapy effect via increasing the apoptosis induction[49]. For example, BET inhibitors could increase the sensitivity of CRC cells to 5-fluorouracil[50] and Arsenic sulfide[51,52] (Figure 3). More importantly, this combination therapy could decrease the side effect of chemotherapeutic drugs[53]. Moreover, BET inhibitors conferred a synthetic lethality with loss of SMAD4 in CRC cells by restoring the loss of c-MYC repression[54], suggesting that BET inhibitors were essential for the treatment of SMAD4-deficient CRC.
BET inhibitors not only effectively inhibited PDAC cell growth in three-dimensional collagen partly by repressing c-MYC expression, but also conducted its efficacy in a MYC-independent way by repressing the expression of FOSL1[55]. However, clinical studies suggested that BET inhibitors monotherapies were not effective revenues for PDAC treatment[56]. Drug resistance assumed the major responsibility for treatment failure. The main mechanism of resistance was associated with either up-regulating or stabilizing c-MYC expression. Loss of FBP1[57] , aberrant expression of ADAR1[58], high levels of GLI[24] and overexpression of PES1[59] could explain the up-regulation of c-MYC in pancreatic cancer.
To improve the efficacy of BET inhibitor on PDAC, several studies evaluated the efficiency of BET inhibitors in combination with other agents. Encouragingly, BET inhibitors could synergize with other target therapy in preclinical PDAC models. For example, BET inhibitor attenuated the DNA repair through decreasing Ku80 and RAD51 proteins, and sensitized the PDAC to PARP inhibitors[60]. Another team also illustrated that BET inhibitors synergizing with HDAC inhibitors enhanced the efficacy of inducing cell death via de-repressing p57[61]. In addition to being combined with target therapies, BET augmented the efficiency of chemotherapeutic drugs like Gemcitabine by increasing DNA damage and apoptosis[62]. Besides, BET inhibitors combined with Quercetin suppress hnRNPA1 leading to better therapeutic effect compared with monotherapy[63].
BET inhibitors exhibit anti-tumorigenic effects on both hepatocellular carcinoma (HCC) and cholangiocarcinoma (CCA), but in different manners. JQ-1 inhibited CCA growth in a MYC-dependent way[64], while JQ-1 played its anti-tumor role in HCC by suppressing E2F2-cell cycle regulation circuit[34] or the expression of SMARCA4[65]. Notably, Yin et al[66] stated that JQ-1 exerted more cytotoxicity on MYC-positive HCC cells than sorafenib (first-line drug for advanced HCC) by inducing more apoptosis. This team further demonstrated that EGFR signaling contributed to the JQ1 resistance by stabilizing MYC. Zhang et al[67] arrived at a different resistance mechanism that upregulation of Mcl-1 was a major contributor to the resistance to BET inhibitor in HCC cells. They further found that BET inhibitors, in combination with other drugs capable of down-regulating Mcl-1 had a synergic effect in human HCC. Liu et al[68] reported another resistance mechanism and the reactivation of WNT pathway in liver cancer cells could increase the sensitivity of HCC to BET inhibitor[68].
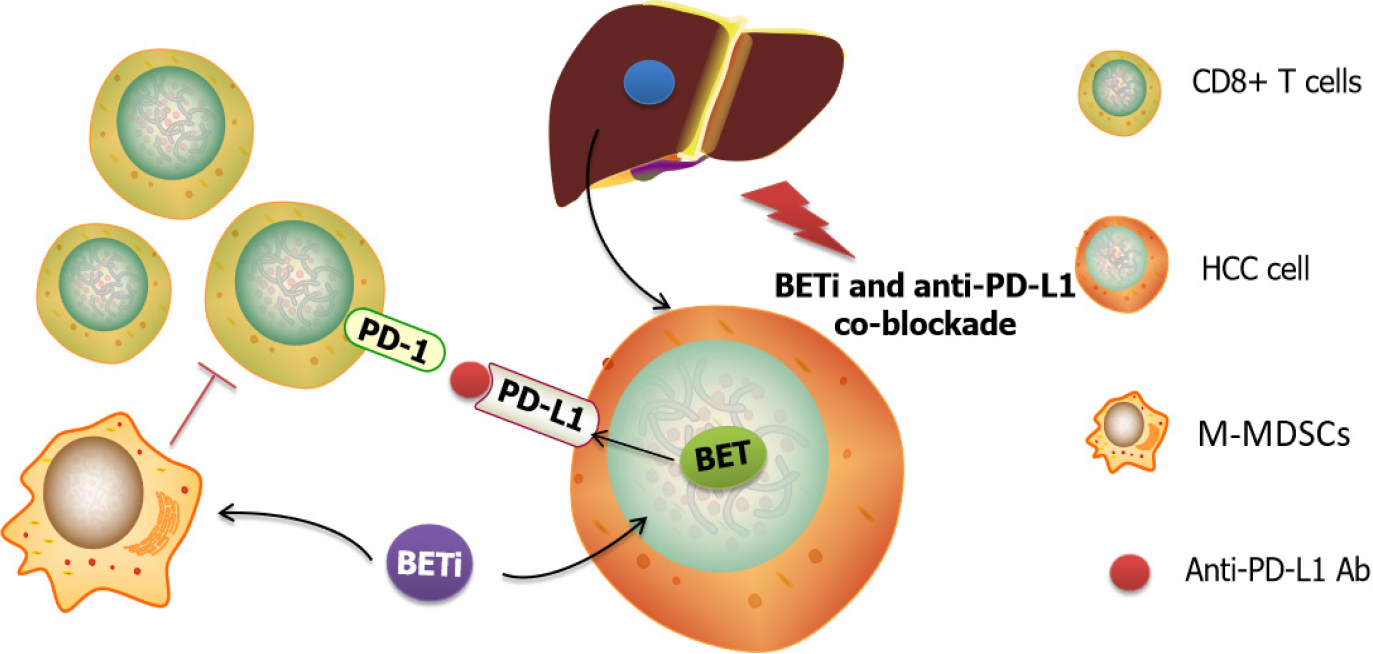
BET inhibitor were also reported to impact the immunotherapy efficacy in HCC (Figure 4). Several studies had shown that BET inhibition could enhance anti-tumor immunity via modulating programmed cell death-ligand 1 (PD-L1) expression[69,70]. Liu et al[71] demonstrated that JQ-1 could decrease the total mRNA and protein levels of PD-L1 in liver cancer cell lines. However, Liu et al[72] reported that JQ1 upregulated the expression of PD-L1 on the plasma membrane in vivo and in vitro, but did not change the total levels of PD-L1 mRNA and protein. Another study conducted by Cheng and his colleague[73] reported that I-BET762, exerted a synergistic effect with anti-PD-L1 in the HCC model leading to augment tumor infiltrating lymphocytes. Altogether, the mechanism by which BET inhibitors modulate immunotherapy is different, but the phenotypic enhancement of immunotherapy by BET inhibitors is assured.
JQ-1 exerts an anti-cancer effect on GC as well. Interestingly, JQ-1 has race specificity on GC that Asians rendered more resistance to BET inhibitors than Brazilians[74]. Recently, Zhou et al[75] noted that JQ-1 suppressed proliferation, migration and invasion of GC cells via targeting RUNX2/NID1 axis, while BET inhibitor AZD5153 inhibited GC metastasis by regulating Mus81 at both RNA and protein levels[76]. Kim et al[77] revealed new BRD4 inhibitor that showed efficiency in I-BET762 resistant GC cell lines[77]. Additionally, through blocking the expression of c-MYC and YAP1, JQ-1 reduced gastric adenocarcinoma cell growth induced by Gal-3, and the anti-cancer activity could be improved in combination with YAP inhibitors[78]. Other combina
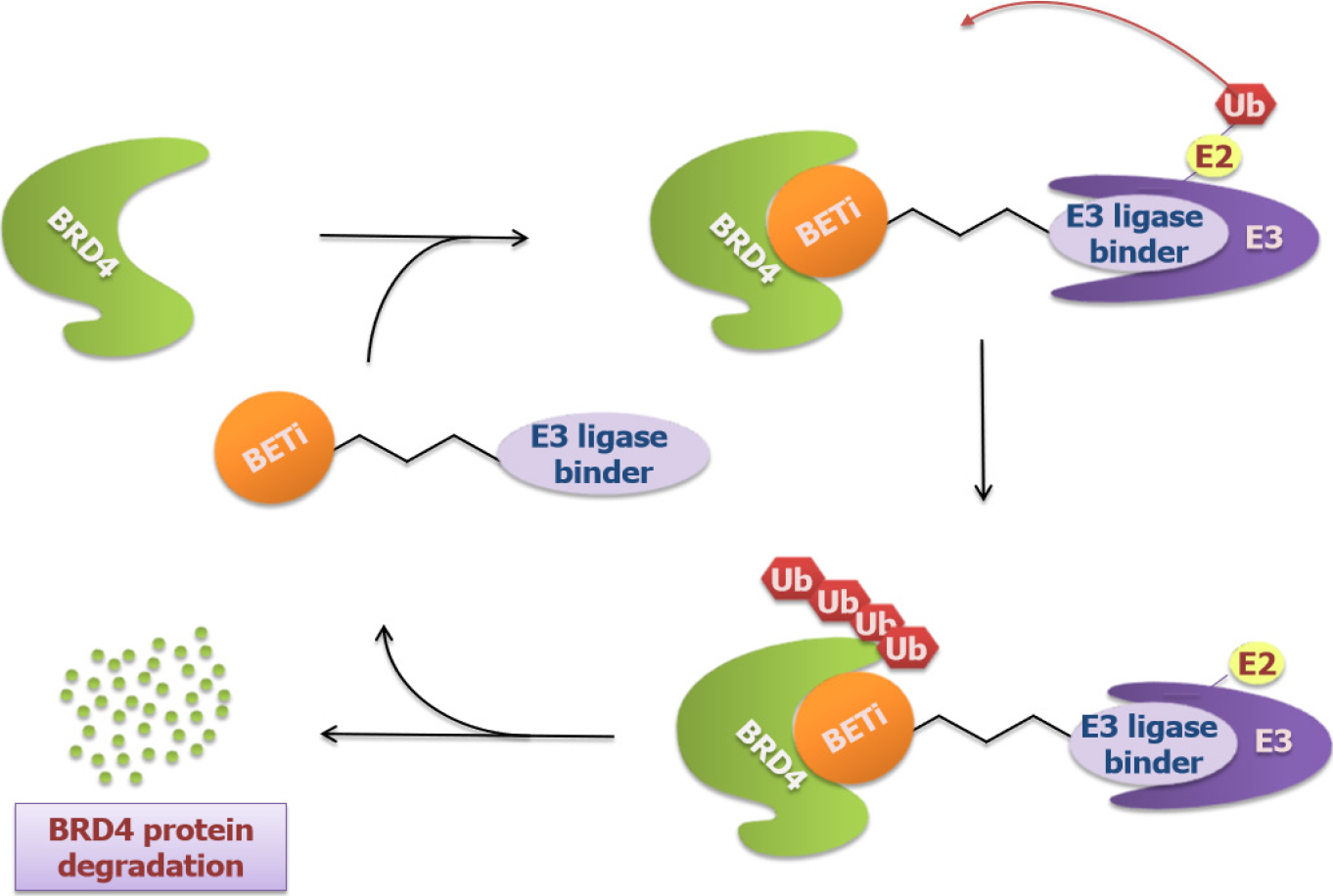
Though exhibiting promising outcomes in GI cancers, BET inhibitors showed therapeutic limitations due to their reversibility, often followed by re-accumulating BET proteins and removing inhibition of c-MYC[19]. This motivated new BET targeting molecules using Proteolysis Targeting Chimeras (PROTACs) technology to be invented like ARV-825 and A1874. These molecules, also called BRD4-degrading PROTACs, are heterobifunctional compounds that contain two binders with one recruiting an E3 ubiquitin ligase cereblon (CRBN) and the other targeting BRD4 proteins based on BET inhibitors. Data has shown that these molecules induce effective and selective degradation of BRD4[80] (Figure 5). The approach to target BRD4 degradation instead of inhibition resulted in more potent suppression of c-MYC as well as c-MYC-dependent genes and led to a longer-lasting effect in GI cancers. For example, Lu et al[81] stated that ARV-825 was superior to OTX-015 and JQ-1 in the suppression of c-MYC expression in CCA and thus exerted more inhibition on CCA cell proliferation and apoptosis. Minko[82] reported a similar anticancer activity of ARV-825 in pancreatic cancer and this activity exhibited in both 2D cell culture and 3D multicellular tumor spheroid models. Additionally, Qin et al[83] showed that A1874 down-regulated c-MYC, Bcl-2, and cyclin D1 in colon cancer cells and had an anti-colon cancer activity by inhibiting cell proliferation, invasion and migration. Strikingly, A1874 presented to be much more effective than other BET inhibitors including JQ1 and I-BET151. However, after long-term exposure to BRD4-degrading PROTACs, resistance exists[84]. Downregulating the expression of CRBN is a common mechanism of resistance. In terms of this issue, Otto et al[85] proposed an alternative avenue to prevent the development of resistance, which might be the use of several PROTACs to recruit different E3 Ligases.
BET inhibitors, including I-BET762 (NCT01587703), INCB057643 (NCT02711137), INCB054329 (NCT02431260), AZD5153 (NCT03205176) and OTX-015(NCT02698176) have entered Clinical Trial for diverse cancers[86], but the majority of them remain in the Phase I/II. Here, we are concentrating on the trials of BET inhibitors alone or in combination with other inhibitors in GI cancers (Table 2).
| Drug | Combination with | Condition | Status | Clinical phase | Trial ID |
| INCB054329 | - | Solid Tumors and Hematologic Malignancy (CRPC, BC, HGSC, CRC, Ewing sarcoma, Pancreatic adenocarcinoma, AML, MDS, MF, MM) | Terminated due to PK variability | Phase I/II | NCT02431260 |
| INCB057643 | Gemcitabine; Paclitaxel; Rucaparib; Abiraterone; Ruxolitinib; Azacitidine | Solid Tumors (CRPC, BC, HGSC, CRC, Glioblastoma multiforme, Ewing sarcoma, Pancreatic adenocarcinoma, AML, MDS) | Terminated due to safety issues | Phase I/II | NCT02711137 |
| AZD5153 | Olaparib | Malignant Solid Tumors, Lymphoma, Ovarian Cancer, Breast Cancer, Pancreatic Cancer, Prostate Cancer | Active, not recruiting | Phase I | NCT03205176 |
| I-BET762 (Molibresib, GSK525762) | Entinostat | Solid tumors (Advanced Malignant Solid Neoplasm, Refractory Malignant Solid Neoplasm, Refractory Pancreatic Carcinoma, Stage II/IIA/IIB/III/IV Pancreatic cancer AJCC v8, Unresectable Pancreatic Carcinoma) or Lymphomas | Withdrawn (Other-Protocol moved to Disapprove) | Phase I | NCT03925428 |
| SF1126 | - | Advanced Hepatocellular Carcinoma | Active, not recruiting | Phase I | NCT03059147 |
I-BET762 (Molibresib) is a pan-BET inhibitor that remarkably inhibits the PDAC cell proliferation by down-regulating c-MYC and reducing protein levels of ERK1/2. Remarkably, the anti-tumor effect can be enhanced combined with gemcitabine[87]. NCT03925428 is a phase I clinical trial that tests the side effects and best dose of I-BET 762 combined with entinostat in solid tumors or lymphomas advanced or refractory, including PDAC. However, the study was withdrawn because other protocol moved to disapprove.
INCB054329 and INCB057643 are two small-molecule BET inhibitors which exhibit anti-cancer activity by reducing the expression level of c-MYC[88,89]. Phase I/II dose-escalation, safety and tolerability studies of INCB054329 and INCB057643 were conducted in subjects with advanced malignancies including GI cancers. INCB054329 was terminated due to an unfavorable clinical Pharmacokinetic (PK) profile (NCT02431260). INCB057643 compared with INCB054329 has a longer half-life and a shorter PK variability. However, patients received INCB057643 resulted in treatment discontinuance or dose interruption or dose reduction due to TRAEs and the study ultimately terminated in 2020 (NCT02711137).
AZD5153 is a novel BRD4 inhibitor, effecting Mus81 down-regulation and sup
Dual PI3K/BRD4 Inhibitor SF1126 blocks both the Ras/Raf/MAPK and PI3K/ AKT/mTOR pathways and disrupts c-MYC expression as well[90]. And a Phase I clinical trial of SF1126 has completed in humans with well toleration and efficacy in solid tumor including CRC[91]. Recently, SF1126 is being tested in combination with Nivolumab in patients with advanced HCC and this study is expected to be completed by October 2022 (NCT03059147).
With high bioavailability and biosafety, SF1126 has completed a Phase I clinical study and steps into a Phase II study in advanced HCC. And AZD5153 shows an optimistic preclinical result in GC treatment. All these evidences demonstrate that BET inhibitors constitute a promising field of clinical research in GI cancers. Continued progresses are required especially in exploring rational combinations to open new possibilities for BET inhibitors as anti-GI cancers agents.
BET inhibitors have emerged as a new possible strategy for the treatment of GI cancers in recent years. However, either nondurable cytotoxic effects, such as thrombocytopenia and GI disorders[92] or drug resistance make BET inhibitors fail to be administrated as single agents by far. To achieve better selectivity and reduce unwanted toxicities, BET inhibitors continue to be updated, increasing their potential in cancer treatment.
The first-generation pan-BET inhibitors have been identified to suppress GI cancer in preclinical results, however, the inevitable side effects limit their clinical applications. Hence, drug discovery efforts concentrate on selectively inhibiting BET proteins[93]. Selective BD inhibitors achieved almost equally efficiency in cancer to the pan-BET inhibitors[94] and showed less toxicity[95]. A set of selective BD inhibitors help to understand the role of BD in cancers and further focusing on specific BD perturbations may provide more efficiency and tolerability in GI cancers treatment.
Another approach to acquire selective inhibition is to target each BET family members. Since BRD4 is the predominant BET protein that mediates the development of GI cancers, selective BRD4 inhibition may have a better outlook. New BRD4 degraders ARV-825 and A1874 that have already shown their antitumor efficiency in preclinical results support further clinical development of BET inhibitors in GI cancers.
Other strategy to improve the efficacy and pharmacokinetic property of BET inhibitors is via modulating their structure. After modification, these major clinical stage BET inhibitors acquire better tumor killing capacity with minimal IC50 in multiple solid tumors[96]. The optimistic preclinical result makes it possible to treat GI cancer with single agents.
Additionally, synergistic inhibition provides an optimistic prospect for increasing the efficacy of BET inhibitors. The preclinical and clinical results verify high potential in combinational therapy. The resistance to BET inhibitors will be overcome if combined with drugs targeting the pathways that cause resistance[47]. Besides, the dosage will be decreased dramatically if combined with drugs rendering GI cancers more sensitive to BET inhibitors[97]. Without a doubt, BET inhibitors emerge as a promising avenue for the GI cancers treatment.
Provenance and peer review: Invited article; Externally peer reviewed.
Peer-review model: Single blind
Specialty type: Oncology
Country/Territory of origin: China
Peer-review report’s scientific quality classification
Grade A (Excellent): 0
Grade B (Very good): B
Grade C (Good): 0
Grade D (Fair): 0
Grade E (Poor): 0
P-Reviewer: Biondi A S-Editor: Fan JR L-Editor: A P-Editor: Fan JR
| 1. | Sung H, Ferlay J, Siegel RL, Laversanne M, Soerjomataram I, Jemal A, Bray F. Global Cancer Statistics 2020: GLOBOCAN Estimates of Incidence and Mortality Worldwide for 36 Cancers in 185 Countries. CA Cancer J Clin. 2021;71:209-249. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 75126] [Cited by in RCA: 64542] [Article Influence: 16135.5] [Reference Citation Analysis (176)] |
| 2. | Stoica AF, Chang CH, Pauklin S. Molecular Therapeutics of Pancreatic Ductal Adenocarcinoma: Targeted Pathways and the Role of Cancer Stem Cells. Trends Pharmacol Sci. 2020;41:977-993. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 18] [Cited by in RCA: 23] [Article Influence: 4.6] [Reference Citation Analysis (0)] |
| 3. | Moertel CG. Chemotherapy of gastrointestinal cancer. N Engl J Med. 1978;299:1049-1052. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 158] [Cited by in RCA: 142] [Article Influence: 3.0] [Reference Citation Analysis (0)] |
| 4. | Chen X, Zeh HJ, Kang R, Kroemer G, Tang D. Cell death in pancreatic cancer: from pathogenesis to therapy. Nat Rev Gastroenterol Hepatol. 2021;18:804-823. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 72] [Cited by in RCA: 220] [Article Influence: 55.0] [Reference Citation Analysis (0)] |
| 5. | Debenham BJ, Hu KS, Harrison LB. Present status and future directions of intraoperative radiotherapy. Lancet Oncol. 2013;14:e457-e464. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 31] [Cited by in RCA: 28] [Article Influence: 2.3] [Reference Citation Analysis (0)] |
| 6. | Joshi SS, Badgwell BD. Current treatment and recent progress in gastric cancer. CA Cancer J Clin. 2021;71:264-279. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 382] [Cited by in RCA: 1082] [Article Influence: 270.5] [Reference Citation Analysis (0)] |
| 7. | Long J, Lin J, Wang A, Wu L, Zheng Y, Yang X, Wan X, Xu H, Chen S, Zhao H. PD-1/PD-L blockade in gastrointestinal cancers: lessons learned and the road toward precision immunotherapy. J Hematol Oncol. 2017;10:146. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 64] [Cited by in RCA: 83] [Article Influence: 10.4] [Reference Citation Analysis (0)] |
| 8. | Golan T, Hammel P, Reni M, Van Cutsem E, Macarulla T, Hall MJ, Park JO, Hochhauser D, Arnold D, Oh DY, Reinacher-Schick A, Tortora G, Algül H, O'Reilly EM, McGuinness D, Cui KY, Schlienger K, Locker GY, Kindler HL. Maintenance Olaparib for Germline BRCA-Mutated Metastatic Pancreatic Cancer. N Engl J Med. 2019;381:317-327. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1137] [Cited by in RCA: 1626] [Article Influence: 271.0] [Reference Citation Analysis (0)] |
| 9. | Dunn C, Hong W, Gibbs P, Ackland S, Sjoquist K, Tebbutt NC, Price T, Burge M. Personalizing First-Line Systemic Therapy in Metastatic Colorectal Cancer: Is There a Role for Initial Low-Intensity Therapy in 2021 and Beyond? Clin Colorectal Cancer. 2021;20:245-255. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1] [Cited by in RCA: 1] [Article Influence: 0.3] [Reference Citation Analysis (0)] |
| 10. | Zhang N, Ng AS, Cai S, Li Q, Yang L, Kerr D. Novel therapeutic strategies: targeting epithelial-mesenchymal transition in colorectal cancer. Lancet Oncol. 2021;22:e358-e368. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 69] [Cited by in RCA: 201] [Article Influence: 50.3] [Reference Citation Analysis (0)] |
| 11. | Nussbaum YI, Manjunath Y, Suvilesh KN, Warren WC, Shyu CR, Kaifi JT, Ciorba MA, Mitchem JB. Current and Prospective Methods for Assessing Anti-Tumor Immunity in Colorectal Cancer. Int J Mol Sci. 2021;22. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 6] [Cited by in RCA: 4] [Article Influence: 1.0] [Reference Citation Analysis (0)] |
| 12. | Doroshow DB, Eder JP, LoRusso PM. BET inhibitors: a novel epigenetic approach. Ann Oncol. 2017;28:1776-1787. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 196] [Cited by in RCA: 277] [Article Influence: 39.6] [Reference Citation Analysis (0)] |
| 13. | Florence B, Faller DV. You bet-cha: a novel family of transcriptional regulators. Front Biosci. 2001;6:D1008-D1018. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 47] [Cited by in RCA: 104] [Article Influence: 4.3] [Reference Citation Analysis (0)] |
| 14. | Wang N, Wu R, Tang D, Kang R. The BET family in immunity and disease. Signal Transduct Target Ther. 2021;6:23. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 54] [Cited by in RCA: 181] [Article Influence: 45.3] [Reference Citation Analysis (0)] |
| 15. | Dhalluin C, Carlson JE, Zeng L, He C, Aggarwal AK, Zhou MM. Structure and ligand of a histone acetyltransferase bromodomain. Nature. 1999;399:491-496. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1221] [Cited by in RCA: 1275] [Article Influence: 49.0] [Reference Citation Analysis (0)] |
| 16. | Itzen F, Greifenberg AK, Bösken CA, Geyer M. Brd4 activates P-TEFb for RNA polymerase II CTD phosphorylation. Nucleic Acids Res. 2014;42:7577-7590. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 148] [Cited by in RCA: 185] [Article Influence: 16.8] [Reference Citation Analysis (0)] |
| 17. | Wu SY, Chiang CM. The double bromodomain-containing chromatin adaptor Brd4 and transcriptional regulation. J Biol Chem. 2007;282:13141-13145. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 459] [Cited by in RCA: 537] [Article Influence: 29.8] [Reference Citation Analysis (0)] |
| 18. | Filippakopoulos P, Knapp S. Targeting bromodomains: epigenetic readers of lysine acetylation. Nat Rev Drug Discov. 2014;13:337-356. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 920] [Cited by in RCA: 1016] [Article Influence: 92.4] [Reference Citation Analysis (0)] |
| 19. | Stathis A, Bertoni F. BET Proteins as Targets for Anticancer Treatment. Cancer Discov. 2018;8:24-36. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 239] [Cited by in RCA: 360] [Article Influence: 45.0] [Reference Citation Analysis (0)] |
| 20. | Belkina AC, Denis GV. BET domain co-regulators in obesity, inflammation and cancer. Nat Rev Cancer. 2012;12:465-477. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 564] [Cited by in RCA: 582] [Article Influence: 44.8] [Reference Citation Analysis (0)] |
| 21. | Chen Z, Li Z, Soutto M, Wang W, Piazuelo MB, Zhu S, Guo Y, Maturana MJ, Corvalan AH, Chen X, Xu Z, El-Rifai WM. Integrated Analysis of Mouse and Human Gastric Neoplasms Identifies Conserved microRNA Networks in Gastric Carcinogenesis. Gastroenterology. 2019;156:1127-1139.e8. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 27] [Cited by in RCA: 36] [Article Influence: 6.0] [Reference Citation Analysis (0)] |
| 22. | Zhu Z, Song J, Guo Y, Huang Z, Chen X, Dang X, Huang Y, Wang Y, Ou W, Yang Y, Yu W, Liu CY, Cui L. LAMB3 promotes tumour progression through the AKT-FOXO3/4 axis and is transcriptionally regulated by the BRD2/acetylated ELK4 complex in colorectal cancer. Oncogene. 2020;39:4666-4680. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 37] [Cited by in RCA: 48] [Article Influence: 9.6] [Reference Citation Analysis (0)] |
| 23. | Sherman MH, Yu RT, Tseng TW, Sousa CM, Liu S, Truitt ML, He N, Ding N, Liddle C, Atkins AR, Leblanc M, Collisson EA, Asara JM, Kimmelman AC, Downes M, Evans RM. Stromal cues regulate the pancreatic cancer epigenome and metabolome. Proc Natl Acad Sci U S A. 2017;114:1129-1134. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 96] [Cited by in RCA: 114] [Article Influence: 14.3] [Reference Citation Analysis (0)] |
| 24. | Huang Y, Nahar S, Nakagawa A, Fernandez-Barrena MG, Mertz JA, Bryant BM, Adams CE, Mino-Kenudson M, Von Alt KN, Chang K, Conery AR, Hatton C, Sims RJ 3rd, Fernandez-Zapico ME, Wang X, Lillemoe KD, Fernández-Del Castillo C, Warshaw AL, Thayer SP, Liss AS. Regulation of GLI Underlies a Role for BET Bromodomains in Pancreatic Cancer Growth and the Tumor Microenvironment. Clin Cancer Res. 2016;22:4259-4270. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 31] [Cited by in RCA: 39] [Article Influence: 4.3] [Reference Citation Analysis (0)] |
| 25. | Cho J, Chang YH, Heo YJ, Kim S, Kim NK, Park JO, Kang WK, Lee J, Kim KM. Four distinct immune microenvironment subtypes in gastric adenocarcinoma with special reference to microsatellite instability. ESMO Open. 2018;3:e000326. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 37] [Cited by in RCA: 61] [Article Influence: 8.7] [Reference Citation Analysis (0)] |
| 26. | Tan M, Brusgaard K, Gerdes AM, Mortensen MB, Detlefsen S, Schaffalitzky de Muckadell OB, Joergensen MT. Whole genome sequencing identifies rare germline variants enriched in cancer related genes in first degree relatives of familial pancreatic cancer patients. Clin Genet. 2021;100:551-562. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 3] [Cited by in RCA: 4] [Article Influence: 1.0] [Reference Citation Analysis (0)] |
| 27. | Hu Y, Zhou J, Ye F, Xiong H, Peng L, Zheng Z, Xu F, Cui M, Wei C, Wang X, Wang Z, Zhu H, Lee P, Zhou M, Jiang B, Zhang DY. BRD4 inhibitor inhibits colorectal cancer growth and metastasis. Int J Mol Sci. 2015;16:1928-1948. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 55] [Cited by in RCA: 76] [Article Influence: 7.6] [Reference Citation Analysis (0)] |
| 28. | Jiao F, Han T, Yuan C, Liang Y, Cui J, Zhuo M, Wang L. Caveolin-2 is regulated by BRD4 and contributes to cell growth in pancreatic cancer. Cancer Cell Int. 2020;20:55. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 10] [Cited by in RCA: 18] [Article Influence: 3.6] [Reference Citation Analysis (0)] |
| 29. | Niu X, Wang W, Liang T, Li S, Yang C, Xu X, Li L, Liu S. CPI-203 improves the efficacy of anti-PD-1 therapy by inhibiting the induced PD-L1 overexpression in liver cancer. Cancer Sci. 2021;. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 1] [Cited by in RCA: 1] [Article Influence: 0.3] [Reference Citation Analysis (0)] |
| 30. | Zhu Y, Yang W, Ji G, Lin N, Wu W, Xiong P, Zheng C, Yan L, Wan P, Wang Y. Bromodomain protein 4 is a novel predictor of survival for gastric carcinoma. Oncotarget. 2017;8:31092-31100. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 5] [Cited by in RCA: 6] [Article Influence: 0.8] [Reference Citation Analysis (0)] |
| 31. | Zhang P, Dong Z, Cai J, Zhang C, Shen Z, Ke A, Gao D, Fan J, Shi G. BRD4 promotes tumor growth and epithelial-mesenchymal transition in hepatocellular carcinoma. Int J Immunopathol Pharmacol. 2015;28:36-44. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 46] [Cited by in RCA: 45] [Article Influence: 4.5] [Reference Citation Analysis (0)] |
| 32. | Dong X, Hu X, Chen J, Hu D, Chen LF. BRD4 regulates cellular senescence in gastric cancer cells via E2F/miR-106b/p21 axis. Cell Death Dis. 2018;9:203. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 53] [Cited by in RCA: 60] [Article Influence: 8.6] [Reference Citation Analysis (0)] |
| 33. | Ba M, Long H, Yan Z, Wang S, Wu Y, Tu Y, Gong Y, Cui S. BRD4 promotes gastric cancer progression through the transcriptional and epigenetic regulation of c-MYC. J Cell Biochem. 2018;119:973-982. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 36] [Cited by in RCA: 59] [Article Influence: 7.4] [Reference Citation Analysis (0)] |
| 34. | Hong SH, Eun JW, Choi SK, Shen Q, Choi WS, Han JW, Nam SW, You JS. Epigenetic reader BRD4 inhibition as a therapeutic strategy to suppress E2F2-cell cycle regulation circuit in liver cancer. Oncotarget. 2016;7:32628-32640. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 33] [Cited by in RCA: 40] [Article Influence: 5.7] [Reference Citation Analysis (0)] |
| 35. | Fan P, Wang B, Meng Z, Zhao J, Jin X. PES1 is transcriptionally regulated by BRD4 and promotes cell proliferation and glycolysis in hepatocellular carcinoma. Int J Biochem Cell Biol. 2018;104:1-8. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 17] [Cited by in RCA: 31] [Article Influence: 4.4] [Reference Citation Analysis (0)] |
| 36. | Zhao J, Meng Z, Xie C, Yang C, Liu Z, Wu S, Wang B, Fan P, Jin X, Wu H. B7-H3 is regulated by BRD4 and promotes TLR4 expression in pancreatic ductal adenocarcinoma. Int J Biochem Cell Biol. 2019;108:84-91. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 15] [Cited by in RCA: 25] [Article Influence: 4.2] [Reference Citation Analysis (0)] |
| 37. | Wang LT, Wang SN, Chiou SS, Liu KY, Chai CY, Chiang CM, Huang SK, Yokoyama KK, Hsu SH. TIP60-dependent acetylation of the SPZ1-TWIST complex promotes epithelial-mesenchymal transition and metastasis in liver cancer. Oncogene. 2019;38:518-532. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 24] [Cited by in RCA: 29] [Article Influence: 4.8] [Reference Citation Analysis (0)] |
| 38. | Qin ZY, Wang T, Su S, Shen LT, Zhu GX, Liu Q, Zhang L, Liu KW, Zhang Y, Zhou ZH, Zhang XN, Wen LZ, Yao YL, Sun WJ, Guo Y, Liu KJ, Liu L, Wang XW, Wei YL, Wang J, Xiao HL, Liu P, Bian XW, Chen DF, Wang B. BRD4 Promotes Gastric Cancer Progression and Metastasis through Acetylation-Dependent Stabilization of Snail. Cancer Res. 2019;79:4869-4881. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 26] [Cited by in RCA: 84] [Article Influence: 14.0] [Reference Citation Analysis (0)] |
| 39. | Tasdemir N, Banito A, Roe JS, Alonso-Curbelo D, Camiolo M, Tschaharganeh DF, Huang CH, Aksoy O, Bolden JE, Chen CC, Fennell M, Thapar V, Chicas A, Vakoc CR, Lowe SW. BRD4 Connects Enhancer Remodeling to Senescence Immune Surveillance. Cancer Discov. 2016;6:612-629. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 197] [Cited by in RCA: 299] [Article Influence: 33.2] [Reference Citation Analysis (0)] |
| 40. | Lee HS, Lee S, Cho KH. Cotargeting BET proteins overcomes resistance arising from PI3K/mTOR blockade-induced protumorigenic senescence in colorectal cancer. Int J Cancer. 2020;147:2824-2837. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 2] [Cited by in RCA: 8] [Article Influence: 1.6] [Reference Citation Analysis (0)] |
| 41. | Honselmann KC, Finetti P, Birnbaum DJ, Monsalve CS, Wellner UF, Begg SKS, Nakagawa A, Hank T, Li A, Goldsworthy MA, Sharma H, Bertucci F, Birnbaum D, Tai E, Ligorio M, Ting DT, Schilling O, Biniossek ML, Bronsert P, Ferrone CR, Keck T, Mino-Kenudson M, Lillemoe KD, Warshaw AL, Fernández-Del Castillo C, Liss AS. Neoplastic-Stromal Cell Cross-talk Regulates Matrisome Expression in Pancreatic Cancer. Mol Cancer Res. 2020;18:1889-1902. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 9] [Cited by in RCA: 12] [Article Influence: 2.4] [Reference Citation Analysis (0)] |
| 42. | Yasukawa Y, Hattori N, Iida N, Takeshima H, Maeda M, Kiyono T, Sekine S, Seto Y, Ushijima T. SAA1 is upregulated in gastric cancer-associated fibroblasts possibly by its enhancer activation. Carcinogenesis. 2021;42:180-189. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 7] [Cited by in RCA: 20] [Article Influence: 5.0] [Reference Citation Analysis (0)] |
| 43. | Filippakopoulos P, Qi J, Picaud S, Shen Y, Smith WB, Fedorov O, Morse EM, Keates T, Hickman TT, Felletar I, Philpott M, Munro S, McKeown MR, Wang Y, Christie AL, West N, Cameron MJ, Schwartz B, Heightman TD, La Thangue N, French CA, Wiest O, Kung AL, Knapp S, Bradner JE. Selective inhibition of BET bromodomains. Nature. 2010;468:1067-1073. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 3566] [Cited by in RCA: 3310] [Article Influence: 220.7] [Reference Citation Analysis (0)] |
| 44. | Delmore JE, Issa GC, Lemieux ME, Rahl PB, Shi J, Jacobs HM, Kastritis E, Gilpatrick T, Paranal RM, Qi J, Chesi M, Schinzel AC, McKeown MR, Heffernan TP, Vakoc CR, Bergsagel PL, Ghobrial IM, Richardson PG, Young RA, Hahn WC, Anderson KC, Kung AL, Bradner JE, Mitsiades CS. BET bromodomain inhibition as a therapeutic strategy to target c-Myc. Cell. 2011;146:904-917. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 2297] [Cited by in RCA: 2330] [Article Influence: 166.4] [Reference Citation Analysis (0)] |
| 45. | Zhang Y, Tian S, Xiong J, Zhou Y, Song H, Liu C. JQ-1 Inhibits Colon Cancer Proliferation via Suppressing Wnt/β-Catenin Signaling and miR-21. Chem Res Toxicol. 2018;31:302-307. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 15] [Cited by in RCA: 22] [Article Influence: 3.1] [Reference Citation Analysis (0)] |
| 46. | Wang W, Tang YA, Xiao Q, Lee WC, Cheng B, Niu Z, Oguz G, Feng M, Lee PL, Li B, Yang ZH, Chen YF, Lan P, Wu XJ, Yu Q. Stromal induction of BRD4 phosphorylation Results in Chromatin Remodeling and BET inhibitor Resistance in Colorectal Cancer. Nat Commun. 2021;12:4441. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 11] [Cited by in RCA: 68] [Article Influence: 17.0] [Reference Citation Analysis (0)] |
| 47. | Wu T, Wang G, Chen W, Zhu Z, Liu Y, Huang Z, Huang Y, Du P, Yang Y, Liu CY, Cui L. Co-inhibition of BET proteins and NF-κB as a potential therapy for colorectal cancer through synergistic inhibiting MYC and FOXM1 expressions. Cell Death Dis. 2018;9:315. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 23] [Cited by in RCA: 36] [Article Influence: 5.1] [Reference Citation Analysis (0)] |
| 48. | Kapoor S, Gustafson T, Zhang M, Chen YS, Li J, Nguyen N, Perez JET, Dashwood WM, Rajendran P, Dashwood RH. Deacetylase Plus Bromodomain Inhibition Downregulates ERCC2 and Suppresses the Growth of Metastatic Colon Cancer Cells. Cancers (Basel). 2021;13. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 7] [Cited by in RCA: 13] [Article Influence: 3.3] [Reference Citation Analysis (0)] |
| 49. | Tan X, Tong J, Wang YJ, Fletcher R, Schoen RE, Yu J, Shen L, Zhang L. BET Inhibitors Potentiate Chemotherapy and Killing of SPOP-Mutant Colon Cancer Cells via Induction of DR5. Cancer Res. 2019;79:1191-1203. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 26] [Cited by in RCA: 47] [Article Influence: 7.8] [Reference Citation Analysis (0)] |
| 50. | Cheng X, Huang Z, Long D, Jin W. BET inhibitor bromosporine enhances 5-FU effect in colorectal cancer cells. Biochem Biophys Res Commun. 2020;521:840-845. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 6] [Cited by in RCA: 10] [Article Influence: 1.7] [Reference Citation Analysis (0)] |
| 51. | Tan Z, Zhang X, Kang T, Zhang L, Chen S. Arsenic sulfide amplifies JQ1 toxicity via mitochondrial pathway in gastric and colon cancer cells. Drug Des Devel Ther. 2018;12:3913-3927. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 10] [Cited by in RCA: 9] [Article Influence: 1.3] [Reference Citation Analysis (0)] |
| 52. | Zhang L, Tong Y, Zhang X, Pan M, Chen S. Arsenic sulfide combined with JQ1, chemotherapy agents, or celecoxib inhibit gastric and colon cancer cell growth. Drug Des Devel Ther. 2015;9:5851-5862. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 5] [Cited by in RCA: 14] [Article Influence: 1.4] [Reference Citation Analysis (0)] |
| 53. | Lei L, Xie X, He L, Chen K, Lv Z, Zhou B, Li Y, Hu W, Zhou Z. The bromodomain and extra-terminal domain inhibitor JQ1 synergistically sensitizes human colorectal cancer cells to topoisomerase I inhibitors through repression of Mre11-mediated DNA repair pathway. Invest New Drugs. 2021;39:362-376. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 5] [Cited by in RCA: 6] [Article Influence: 1.2] [Reference Citation Analysis (0)] |
| 54. | Shi C, Yang EJ, Liu Y, Mou PK, Ren G, Shim JS. Bromodomain and extra-terminal motif (BET) inhibition is synthetic lethal with loss of SMAD4 in colorectal cancer cells via restoring the loss of MYC repression. Oncogene. 2021;40:937-950. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 9] [Cited by in RCA: 15] [Article Influence: 3.0] [Reference Citation Analysis (0)] |
| 55. | Sahai V, Kumar K, Knab LM, Chow CR, Raza SS, Bentrem DJ, Ebine K, Munshi HG. BET bromodomain inhibitors block growth of pancreatic cancer cells in three-dimensional collagen. Mol Cancer Ther. 2014;13:1907-1917. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 72] [Cited by in RCA: 82] [Article Influence: 7.5] [Reference Citation Analysis (0)] |
| 56. | Hessmann E, Johnsen SA, Siveke JT, Ellenrieder V. Epigenetic treatment of pancreatic cancer: is there a therapeutic perspective on the horizon? Gut. 2017;66:168-179. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 79] [Cited by in RCA: 98] [Article Influence: 12.3] [Reference Citation Analysis (0)] |
| 57. | Wang B, Fan P, Zhao J, Wu H, Jin X. FBP1 Loss contributes to BET inhibitors resistance by undermining c-Myc expression in pancreatic ductal adenocarcinoma. J Exp Clin Cancer Res. 2018;37:224. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 23] [Cited by in RCA: 36] [Article Influence: 5.1] [Reference Citation Analysis (0)] |
| 58. | Sun Y, Fan J, Wang B, Meng Z, Ren D, Zhao J, Liu Z, Li D, Jin X, Wu H. The aberrant expression of ADAR1 promotes resistance to BET inhibitors in pancreatic cancer by stabilizing c-Myc. Am J Cancer Res. 2020;10:148-163. [PubMed] |
| 59. | Jin X, Fang R, Fan P, Zeng L, Zhang B, Lu X, Liu T. PES1 promotes BET inhibitors resistance and cells proliferation through increasing c-Myc expression in pancreatic cancer. J Exp Clin Cancer Res. 2019;38:463. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 16] [Cited by in RCA: 29] [Article Influence: 4.8] [Reference Citation Analysis (0)] |
| 60. | Miller AL, Fehling SC, Garcia PL, Gamblin TL, Council LN, van Waardenburg RCAM, Yang ES, Bradner JE, Yoon KJ. The BET inhibitor JQ1 attenuates double-strand break repair and sensitizes models of pancreatic ductal adenocarcinoma to PARP inhibitors. EBioMedicine. 2019;44:419-430. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 71] [Cited by in RCA: 84] [Article Influence: 14.0] [Reference Citation Analysis (0)] |
| 61. | Mazur PK, Herner A, Mello SS, Wirth M, Hausmann S, Sánchez-Rivera FJ, Lofgren SM, Kuschma T, Hahn SA, Vangala D, Trajkovic-Arsic M, Gupta A, Heid I, Noël PB, Braren R, Erkan M, Kleeff J, Sipos B, Sayles LC, Heikenwalder M, Heßmann E, Ellenrieder V, Esposito I, Jacks T, Bradner JE, Khatri P, Sweet-Cordero EA, Attardi LD, Schmid RM, Schneider G, Sage J, Siveke JT. Combined inhibition of BET family proteins and histone deacetylases as a potential epigenetics-based therapy for pancreatic ductal adenocarcinoma. Nat Med. 2015;21:1163-1171. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 327] [Cited by in RCA: 327] [Article Influence: 32.7] [Reference Citation Analysis (0)] |
| 62. | Miller AL, Garcia PL, Fehling SC, Gamblin TL, Vance RB, Council LN, Chen D, Yang ES, van Waardenburg RCAM, Yoon KJ. The BET Inhibitor JQ1 Augments the Antitumor Efficacy of Gemcitabine in Preclinical Models of Pancreatic Cancer. Cancers (Basel). 2021;13. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 8] [Cited by in RCA: 18] [Article Influence: 4.5] [Reference Citation Analysis (0)] |
| 63. | Pham TND, Stempel S, Shields MA, Spaulding C, Kumar K, Bentrem DJ, Matsangou M, Munshi HG. Quercetin Enhances the Anti-Tumor Effects of BET Inhibitors by Suppressing hnRNPA1. Int J Mol Sci. 2019;20. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 27] [Cited by in RCA: 44] [Article Influence: 7.3] [Reference Citation Analysis (0)] |
| 64. | Miao X, Liu C, Jiang Y, Wang Y, Kong D, Wu Z, Wang X, Tian R, Yu X, Zhu X, Gong W. BET protein inhibition evidently enhances sensitivity to PI3K/mTOR dual inhibition in intrahepatic cholangiocarcinoma. Cell Death Dis. 2021;12:1020. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 15] [Cited by in RCA: 14] [Article Influence: 3.5] [Reference Citation Analysis (0)] |
| 65. | Choi HI, An GY, Baek M, Yoo E, Chai JC, Lee YS, Jung KH, Chai YG. BET inhibitor suppresses migration of human hepatocellular carcinoma by inhibiting SMARCA4. Sci Rep. 2021;11:11799. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 9] [Cited by in RCA: 19] [Article Influence: 4.8] [Reference Citation Analysis (0)] |
| 66. | Yin Y, Sun M, Zhan X, Wu C, Geng P, Sun X, Wu Y, Zhang S, Qin J, Zhuang Z, Liu Y. EGFR signaling confers resistance to BET inhibition in hepatocellular carcinoma through stabilizing oncogenic MYC. J Exp Clin Cancer Res. 2019;38:83. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 20] [Cited by in RCA: 28] [Article Influence: 4.7] [Reference Citation Analysis (0)] |
| 67. | Zhang HP, Li GQ, Zhang Y, Guo WZ, Zhang JK, Li J, Lv JF, Zhang SJ. Upregulation of Mcl-1 inhibits JQ1-triggered anticancer activity in hepatocellular carcinoma cells. Biochem Biophys Res Commun. 2018;495:2456-2461. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 16] [Cited by in RCA: 15] [Article Influence: 2.1] [Reference Citation Analysis (0)] |
| 68. | Liu Y, Xue M, Cao D, Qin L, Wang Y, Miao Z, Wang P, Hu X, Shen J, Xiong B. Multi-omics characterization of WNT pathway reactivation to ameliorate BET inhibitor resistance in liver cancer cells. Genomics. 2021;113:1057-1069. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 3] [Cited by in RCA: 4] [Article Influence: 1.0] [Reference Citation Analysis (0)] |
| 69. | Zhu H, Bengsch F, Svoronos N, Rutkowski MR, Bitler BG, Allegrezza MJ, Yokoyama Y, Kossenkov AV, Bradner JE, Conejo-Garcia JR, Zhang R. BET Bromodomain Inhibition Promotes Anti-tumor Immunity by Suppressing PD-L1 Expression. Cell Rep. 2016;16:2829-2837. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 236] [Cited by in RCA: 330] [Article Influence: 41.3] [Reference Citation Analysis (0)] |
| 70. | Hogg SJ, Vervoort SJ, Deswal S, Ott CJ, Li J, Cluse LA, Beavis PA, Darcy PK, Martin BP, Spencer A, Traunbauer AK, Sadovnik I, Bauer K, Valent P, Bradner JE, Zuber J, Shortt J, Johnstone RW. BET-Bromodomain Inhibitors Engage the Host Immune System and Regulate Expression of the Immune Checkpoint Ligand PD-L1. Cell Rep. 2017;18:2162-2174. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 178] [Cited by in RCA: 237] [Article Influence: 29.6] [Reference Citation Analysis (0)] |
| 71. | Liu K, Zhou Z, Gao H, Yang F, Qian Y, Jin H, Guo Y, Liu Y, Li H, Zhang C, Guo J, Wan Y, Chen R. JQ1, a BET-bromodomain inhibitor, inhibits human cancer growth and suppresses PD-L1 expression. Cell Biol Int. 2019;43:642-650. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 15] [Cited by in RCA: 25] [Article Influence: 4.2] [Reference Citation Analysis (0)] |
| 72. | Liu C, Miao X, Wang Y, Wen L, Cheng X, Kong D, Zhao P, Song D, Wang X, Ding X, Xia H, Wang W, Sun Q, Gong W. Bromo- and extraterminal domain protein inhibition improves immunotherapy efficacy in hepatocellular carcinoma. Cancer Sci. 2020;111:3503-3515. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 4] [Cited by in RCA: 17] [Article Influence: 3.4] [Reference Citation Analysis (0)] |
| 73. | Liu M, Zhou J, Liu X, Feng Y, Yang W, Wu F, Cheung OK, Sun H, Zeng X, Tang W, Mok MTS, Wong J, Yeung PC, Lai PBS, Chen Z, Jin H, Chen J, Chan SL, Chan AWH, To KF, Sung JJY, Chen M, Cheng AS. Targeting monocyte-intrinsic enhancer reprogramming improves immunotherapy efficacy in hepatocellular carcinoma. Gut. 2020;69:365-379. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 80] [Cited by in RCA: 134] [Article Influence: 26.8] [Reference Citation Analysis (0)] |
| 74. | Montenegro RC, Clark PG, Howarth A, Wan X, Ceroni A, Siejka P, Nunez-Alonso GA, Monteiro O, Rogers C, Gamble V, Burbano R, Brennan PE, Tallant C, Ebner D, Fedorov O, O'Neill E, Knapp S, Dixon D, Müller S. BET inhibition as a new strategy for the treatment of gastric cancer. Oncotarget. 2016;7:43997-44012. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 41] [Cited by in RCA: 42] [Article Influence: 4.7] [Reference Citation Analysis (0)] |
| 75. | Zhou S, Zhang S, Wang L, Huang S, Yuan Y, Yang J, Wang H, Li X, Wang P, Zhou L, Xu Y, Gao H, Zhang Y, Lv Y, Zou X. BET protein inhibitor JQ1 downregulates chromatin accessibility and suppresses metastasis of gastric cancer via inactivating RUNX2/NID1 signaling. Oncogenesis. 2020;9:33. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 22] [Cited by in RCA: 66] [Article Influence: 13.2] [Reference Citation Analysis (0)] |
| 76. | Yin Y, Liu W, Shen Q, Zhang P, Wang L, Tao R, Li H, Ma X, Zeng X, Cheong JH, Song S, Ajani JA, Mills GB, Tao K, Peng G. The DNA Endonuclease Mus81 Regulates ZEB1 Expression and Serves as a Target of BET4 Inhibitors in Gastric Cancer. Mol Cancer Ther. 2019;18:1439-1450. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 5] [Cited by in RCA: 12] [Article Influence: 2.0] [Reference Citation Analysis (0)] |
| 77. | Kim YH, Kim M, Kim JE, Yoo M, Lee HK, Lee CO, Jung KY, Kim Y, Choi SU, Park CH. Novel brd4 inhibitors with a unique scaffold exhibit antitumor effects. Oncol Lett. 2021;21:473. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 1] [Cited by in RCA: 1] [Article Influence: 0.3] [Reference Citation Analysis (0)] |
| 78. | Ajani JA, Estrella JS, Chen Q, Correa AM, Ma L, Scott AW, Jin J, Liu B, Xie M, Sudo K, Shiozaki H, Badgwell B, Weston B, Lee JH, Bhutani MS, Onodera H, Suzuki K, Suzuki A, Ding S, Hofstetter WL, Johnson RL, Bresalier RS, Song S. Galectin-3 expression is prognostic in diffuse type gastric adenocarcinoma, confers aggressive phenotype, and can be targeted by YAP1/BET inhibitors. Br J Cancer. 2018;118:52-61. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 15] [Cited by in RCA: 21] [Article Influence: 3.0] [Reference Citation Analysis (0)] |
| 79. | Kang SK, Bae HJ, Kwon WS, Che J, Kim TS, Chung HC, Rha SY. Transcriptome analysis of iBET-151, a BET inhibitor alone and in combination with paclitaxel in gastric cancer cells. Genomics Inform. 2020;18:e37. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 1] [Cited by in RCA: 1] [Article Influence: 0.3] [Reference Citation Analysis (0)] |
| 80. | Zengerle M, Chan KH, Ciulli A. Selective Small Molecule Induced Degradation of the BET Bromodomain Protein BRD4. ACS Chem Biol. 2015;10:1770-1777. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 531] [Cited by in RCA: 758] [Article Influence: 75.8] [Reference Citation Analysis (0)] |
| 81. | Lu Q, Ding X, Huang T, Zhang S, Li Y, Xu L, Chen G, Ying Y, Wang Y, Feng Z, Wang L, Zou X. BRD4 degrader ARV-825 produces long-lasting loss of BRD4 protein and exhibits potent efficacy against cholangiocarcinoma cells. Am J Transl Res. 2019;11:5728-5739. [PubMed] |
| 82. | Minko T. Nanoformulation of BRD4-Degrading PROTAC: Improving Druggability To Target the 'Undruggable' MYC in Pancreatic Cancer. Trends Pharmacol Sci. 2020;41:684-686. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 11] [Cited by in RCA: 31] [Article Influence: 6.2] [Reference Citation Analysis (0)] |
| 83. | Qin AC, Jin H, Song Y, Gao Y, Chen YF, Zhou LN, Wang SS, Lu XS. The therapeutic effect of the BRD4-degrading PROTAC A1874 in human colon cancer cells. Cell Death Dis. 2020;11:805. [RCA] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 9] [Cited by in RCA: 41] [Article Influence: 8.2] [Reference Citation Analysis (0)] |
| 84. | Shirasaki R, Matthews GM, Gandolfi S, de Matos Simoes R, Buckley DL, Raja Vora J, Sievers QL, Brüggenthies JB, Dashevsky O, Poarch H, Tang H, Bariteau MA, Sheffer M, Hu Y, Downey-Kopyscinski SL, Hengeveld PJ, Glassner BJ, Dhimolea E, Ott CJ, Zhang T, Kwiatkowski NP, Laubach JP, Schlossman RL, Richardson PG, Culhane AC, Groen RWJ, Fischer ES, Vazquez F, Tsherniak A, Hahn WC, Levy J, Auclair D, Licht JD, Keats JJ, Boise LH, Ebert BL, Bradner JE, Gray NS, Mitsiades CS. Functional Genomics Identify Distinct and Overlapping Genes Mediating Resistance to Different Classes of Heterobifunctional Degraders of Oncoproteins. Cell Rep. 2021;34:108532. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 25] [Cited by in RCA: 74] [Article Influence: 18.5] [Reference Citation Analysis (0)] |
| 85. | Otto C, Schmidt S, Kastner C, Denk S, Kettler J, Müller N, Germer CT, Wolf E, Gallant P, Wiegering A. Targeting bromodomain-containing protein 4 (BRD4) inhibits MYC expression in colorectal cancer cells. Neoplasia. 2019;21:1110-1120. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 25] [Cited by in RCA: 48] [Article Influence: 8.0] [Reference Citation Analysis (0)] |
| 86. | Alqahtani A, Choucair K, Ashraf M, Hammouda DM, Alloghbi A, Khan T, Senzer N, Nemunaitis J. Bromodomain and extra-terminal motif inhibitors: a review of preclinical and clinical advances in cancer therapy. Future Sci OA. 2019;5:FSO372. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 193] [Cited by in RCA: 196] [Article Influence: 32.7] [Reference Citation Analysis (0)] |
| 87. | Xie F, Huang M, Lin X, Liu C, Liu Z, Meng F, Wang C, Huang Q. The BET inhibitor I-BET762 inhibits pancreatic ductal adenocarcinoma cell proliferation and enhances the therapeutic effect of gemcitabine. Sci Rep. 2018;8:8102. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 27] [Cited by in RCA: 31] [Article Influence: 4.4] [Reference Citation Analysis (0)] |
| 88. | Falchook G, Rosen S, LoRusso P, Watts J, Gupta S, Coombs CC, Talpaz M, Kurzrock R, Mita M, Cassaday R, Harb W, Peguero J, Smith DC, Piha-Paul SA, Szmulewitz R, Noel MS, Yeleswaram S, Liu P, Switzky J, Zhou G, Zheng F, Mehta A. Development of 2 Bromodomain and Extraterminal Inhibitors With Distinct Pharmacokinetic and Pharmacodynamic Profiles for the Treatment of Advanced Malignancies. Clin Cancer Res. 2020;26:1247-1257. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 33] [Cited by in RCA: 64] [Article Influence: 10.7] [Reference Citation Analysis (0)] |
| 89. | Leal AS, Liu P, Krieger-Burke T, Ruggeri B, Liby KT. The Bromodomain Inhibitor, INCB057643, Targets Both Cancer Cells and the Tumor Microenvironment in Two Preclinical Models of Pancreatic Cancer. Cancers (Basel). 2020;13. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 6] [Cited by in RCA: 15] [Article Influence: 3.0] [Reference Citation Analysis (0)] |
| 90. | Singh AR, Joshi S, Burgoyne AM, Sicklick JK, Ikeda S, Kono Y, Garlich JR, Morales GA, Durden DL. Single Agent and Synergistic Activity of the "First-in-Class" Dual PI3K/BRD4 Inhibitor SF1126 with Sorafenib in Hepatocellular Carcinoma. Mol Cancer Ther. 2016;15:2553-2562. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 36] [Cited by in RCA: 51] [Article Influence: 5.7] [Reference Citation Analysis (0)] |
| 91. | Mahadevan D, Chiorean EG, Harris WB, Von Hoff DD, Stejskal-Barnett A, Qi W, Anthony SP, Younger AE, Rensvold DM, Cordova F, Shelton CF, Becker MD, Garlich JR, Durden DL, Ramanathan RK. Phase I pharmacokinetic and pharmacodynamic study of the pan-PI3K/mTORC vascular targeted pro-drug SF1126 in patients with advanced solid tumours and B-cell malignancies. Eur J Cancer. 2012;48:3319-3327. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 88] [Cited by in RCA: 105] [Article Influence: 8.1] [Reference Citation Analysis (0)] |
| 92. | Halder TG, Soldi R, Sharma S. Bromodomain and extraterminal domain protein bromodomain inhibitor based cancer therapeutics. Curr Opin Oncol. 2021;33:526-531. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 7] [Cited by in RCA: 9] [Article Influence: 2.3] [Reference Citation Analysis (0)] |
| 93. | Petretich M, Demont EH, Grandi P. Domain-selective targeting of BET proteins in cancer and immunological diseases. Curr Opin Chem Biol. 2020;57:184-193. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 23] [Cited by in RCA: 43] [Article Influence: 8.6] [Reference Citation Analysis (0)] |
| 94. | Gilan O, Rioja I, Knezevic K, Bell MJ, Yeung MM, Harker NR, Lam EYN, Chung CW, Bamborough P, Petretich M, Urh M, Atkinson SJ, Bassil AK, Roberts EJ, Vassiliadis D, Burr ML, Preston AGS, Wellaway C, Werner T, Gray JR, Michon AM, Gobbetti T, Kumar V, Soden PE, Haynes A, Vappiani J, Tough DF, Taylor S, Dawson SJ, Bantscheff M, Lindon M, Drewes G, Demont EH, Daniels DL, Grandi P, Prinjha RK, Dawson MA. Selective targeting of BD1 and BD2 of the BET proteins in cancer and immunoinflammation. Science. 2020;368:387-394. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 255] [Cited by in RCA: 291] [Article Influence: 58.2] [Reference Citation Analysis (0)] |
| 95. | Faivre EJ, McDaniel KF, Albert DH, Mantena SR, Plotnik JP, Wilcox D, Zhang L, Bui MH, Sheppard GS, Wang L, Sehgal V, Lin X, Huang X, Lu X, Uziel T, Hessler P, Lam LT, Bellin RJ, Mehta G, Fidanze S, Pratt JK, Liu D, Hasvold LA, Sun C, Panchal SC, Nicolette JJ, Fossey SL, Park CH, Longenecker K, Bigelow L, Torrent M, Rosenberg SH, Kati WM, Shen Y. Selective inhibition of the BD2 bromodomain of BET proteins in prostate cancer. Nature. 2020;578:306-310. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 176] [Cited by in RCA: 274] [Article Influence: 54.8] [Reference Citation Analysis (0)] |
| 96. | Yin M, Guo Y, Hu R, Cai WL, Li Y, Pei S, Sun H, Peng C, Li J, Ye R, Yang Q, Wang N, Tao Y, Chen X, Yan Q. Potent BRD4 inhibitor suppresses cancer cell-macrophage interaction. Nat Commun. 2020;11:1833. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 46] [Cited by in RCA: 114] [Article Influence: 22.8] [Reference Citation Analysis (0)] |
| 97. | Bechter O, Schöffski P. Make your best BET: The emerging role of BET inhibitor treatment in malignant tumors. Pharmacol Ther. 2020;208:107479. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 48] [Cited by in RCA: 80] [Article Influence: 16.0] [Reference Citation Analysis (0)] |
| 98. | Wen D, Wang Y, Zhu Z, Huang Z, Cui L, Wu T, Liu CY. Bromodomain and Extraterminal (BET) protein inhibition suppresses tumor progression and inhibits HGF-MET signaling through targeting cancer-associated fibroblasts in colorectal cancer. Biochim Biophys Acta Mol Basis Dis. 2020;1866:165923. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 2] [Cited by in RCA: 7] [Article Influence: 1.4] [Reference Citation Analysis (0)] |
| 99. | Aguirre-Portolés C, Feliu J, Reglero G, Ramírez de Molina A. ABCA1 overexpression worsens colorectal cancer prognosis by facilitating tumour growth and caveolin-1-dependent invasiveness, and these effects can be ameliorated using the BET inhibitor apabetalone. Mol Oncol. 2018;12:1735-1752. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 41] [Cited by in RCA: 88] [Article Influence: 12.6] [Reference Citation Analysis (0)] |
| 100. | Kato Y, Kondo S, Itakura T, Tokunaga M, Hatayama S, Katayama K, Sugimoto Y. SNAIL- and SLUG-induced side population phenotype of HCT116 human colorectal cancer cells and its regulation by BET inhibitors. Biochem Biophys Res Commun. 2020;521:152-157. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 5] [Cited by in RCA: 6] [Article Influence: 1.0] [Reference Citation Analysis (0)] |
| 101. | Andricovich J, Perkail S, Kai Y, Casasanta N, Peng W, Tzatsos A. Loss of KDM6A Activates Super-Enhancers to Induce Gender-Specific Squamous-like Pancreatic Cancer and Confers Sensitivity to BET Inhibitors. Cancer Cell. 2018;33:512-526.e8. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 173] [Cited by in RCA: 236] [Article Influence: 33.7] [Reference Citation Analysis (0)] |
| 102. | Garcia PL, Miller AL, Gamblin TL, Council LN, Christein JD, Arnoletti JP, Heslin MJ, Reddy S, Richardson JH, Cui X, van Waardenburg RCAM, Bradner JE, Yang ES, Yoon KJ. JQ1 Induces DNA Damage and Apoptosis, and Inhibits Tumor Growth in a Patient-Derived Xenograft Model of Cholangiocarcinoma. Mol Cancer Ther. 2018;17:107-118. [RCA] [DOI] [Full Text] [Cited by in Crossref: 21] [Cited by in RCA: 30] [Article Influence: 3.8] [Reference Citation Analysis (0)] |