Published online Jan 15, 2022. doi: 10.4251/wjgo.v14.i1.253
Peer-review started: June 22, 2021
First decision: July 4, 2021
Revised: July 28, 2021
Accepted: September 16, 2021
Article in press: September 16, 2021
Published online: January 15, 2022
Processing time: 202 Days and 19.4 Hours
Liver cancer is one of the most highly malignant cancers, characterized by easy metastasis and chemoradiotherapy resistance. Emerging evidence indicates that long noncoding RNAs (LncRNAs), including Lnc524369, are highly involved in the initiation, progression, radioresistance, and chemoresistance of hepatocellular carcinoma (HCC). However, the function of Lnc524369 remains unclear.
To explore the function of Lnc524369 in HCC.
To investigate the effect of Lnc524369, tissue from 41 HCC patients were analyzed using CCK8, migration, and invasion assays. Lnc524369 and YWHAZ (also named 14-3-3ζ) mRNA were detected by qPCR, and YWHAZ and RAF1 proteins were detected by western blot in liver cancer cell lines and human HCC tissues. The Cancer Cell Line Encyclopedia (CCLE) databases, STRING database, Human Protein Atlas database, and the TCGA database were used for bioinformatic analysis.
Lnc524369 was significantly upregulated in the nucleus of liver cancer cells and human HCC tissues. Overexpression of Lnc524369 was associated with the proliferation, migration, and invasion of liver cancer cells. YWHAZ and RAF1 proteins and YWHAZ mRNA were overexpressed in liver cancer, which could be attenuated by overexpression of Lnc524369. Lnc524369 and its downstream target YWHAZ and RAF1 proteins were negatively associated with overall survival time.
Lnc524369 might be a promising target of HCC as it can enhance liver cancer progression and decrease the overall survival time of HCC by activating the YWHAZ/RAF1 pathway.
Core Tip: Lnc524369 is expressed at low levels in the cytoplasm but enriched in the nucleus of hepatocellular carcinoma (HCC) cells and might be strongly coexpressed with YWHAZ. Overexpression of Lnc524369 promoted the proliferation, migration, and invasion of liver cancer cells. The Lnc524369-mediated YWHAZ/RAF1 pathway was negatively associated with the overall survival time of HCC patients.
- Citation: Zheng W, Shen GL, Xu KY, Yin QQ, Hui TC, Zhou ZW, Xu CA, Wang SH, Wu WH, Shi LF, Pan HY. Lnc524369 promotes hepatocellular carcinoma progression and predicts poor survival by activating YWHAZ-RAF1 signaling. World J Gastrointest Oncol 2022; 14(1): 253-264
- URL: https://www.wjgnet.com/1948-5204/full/v14/i1/253.htm
- DOI: https://dx.doi.org/10.4251/wjgo.v14.i1.253
Hepatocellular carcinoma (HCC) is the most common pathological type of primary liver cancer[1]. Among all cancers, the incidence and mortality rates of HCC rank sixth and second in the world, respectively[2]. Common risk factors leading to HCC are hepatitis B virus (HBV), hepatitis C virus (HCV), alcoholism, obesity, environmental toxins, and metabolic diseases[3]. At present, approximately 93 million hepatitis B carriers are at least partially responsible for the high incidences of liver fibrosis, cirrhosis, and HCC in China[4]. Due to HCC’s high malignancy and insensitivity to chemoradiotherapy, the potential mechanisms of HCC need to be further clarified.
Long noncoding RNAs (lncRNAs) are a class of RNA transcripts with a length of more than 200 nucleotides that lack protein coding potential[5]. Increasing evidence shows that lncRNAs can regulate many important pathophysiological processes, especially in the occurrence and development of malignant tumors[6]. For example, lncRNAs such as HOTAIR, HULC, and MALAT-1, are closely related to the proliferation, apoptosis, angiogenesis, invasion, metastasis, and prognosis of HCC[6]. Therefore, lncRNAs have therapeutic potential and diagnostic value in HCC. A previous study had suggested that Lnc524369 (ap003469.2) is expressed at low levels in the cytoplasm but enriched in the nucleus of HCC and might be strongly coex
The human liver cancer cell lines Huh7 and HepG2, as well as 41 human HCC samples from Zhejiang Provincial People's Hospital, were used to determine Lnc524369 expression levels. However, due to limited human HCC sample tissue, only 5 of 41 HCC samples (as well as the liver cell lines) were used to examine YWHAZ and RAF1 protein or mRNA levels. The whole study was approved by the Ethics Committee of Zhejiang Provincial People's Hospital.
We analyzed the protein expression of YWHAZ and RAF1 in liver cancer cell lines by using data obtained from the CCLE (http://www.broadinstitute.org/ccle). In addition, we conducted survival analysis of YWHAZ and RAF1 by using the Human Protein Atlas database (http://www.proteinatlas.org/) and the Cancer Genome Atlas (TCGA) database (https://www.cancer.gov/about-nci/organization/ccg/research/structural-genomics/tcga).
RNA was isolated from 1 × 106 Huh7 cells or 15 mg human HCC tissue using a cytoplasmic and nuclear RNA purification kit from Norgen Biotek Corp. (Ontario, Canada). Cells or tissues washed with phosphate-buffered saline (PBS) were lysed with ice-cold lysis solution, and then the lysate was transferred to a microcentrifuge tube and spun at maximum speed for three minutes. The supernatant contained cytoplasmic RNA, while the pellet contained nuclear RNA. Binding solution was then added to the supernatant and pellet separately, and each fraction was mixed and resuspended well. The RNA samples were then bound to separate spin columns using centrifugation. Next, the columns were washed twice with 400 μL of the provided wash solution, and the fractionated RNA was eluted from the columns using 50 μL of the provided elution buffer.
Real-time PCR was performed to determine Lnc524369 expression in HepG2 and Huh7 cell lines, as well as human HCC and paracancerous tissue. Cells treated with the pcDNA3 control plasmid and the Lnc524369-pcdna3.1 overexpression plasmid, as well as human HCC and paracancerous tissue, were collected for analysis. A cytoplasmic and nuclear RNA purification kit (NGB21000; Norgen Biotek Corp.) was used to extract RNA from cells and tissues. Total RNA was extracted by TRIzol reagent, and cDNA was synthesized according to the instructions of the Quantitect reverse transcription kit (Qiagen, Hilden, German). Using GAPDH as an internal reference, real-time PCR was performed with a Powerup SYBRTM Green master mix kit (Thermo Fisher Scientific, Waltham, MA, United States). The specific primer sequences are shown in Table 1. Each experiment was repeated three times, and three complex holes were set in each sample. The relative expression was calculated by the 2-∆∆Ct method.
| Gene name | Gene accession No. | Primer (5'→3') | Length (bp) |
| Lnc524369 | ENST00000524369.1 | F: CAGCAGAACTGGGTGTTGGA | 90 |
| R: GCGCTGCAGTTTCCTCCTT | |||
| GAPDH | NM_002046.5 | F: CCATGACAACTTTGGTATCGTGGAA | 107 |
| R: GGCCATCACGCCACAGTTTC |
Western blotting was performed to determine YWHAZ and RAF1 expression in the Huh7 cell line, as well as human HCC and paracancerous tissue. Anti-rabbit YWHAZ (1:2000; Proteintech, Rosemont, IL, United States), anti-mouse RAF1 (1:1000; Proteintech), and anti-rabbit GAPDH (internal reference, 1:10000; Abcam, Cambridge, United Kingdom) were added to the cell or tissue samples and incubated overnight at 4 °C. Goat anti-rabbit IgG HRP secondary antibody (1:5000; Thermo Fisher Scientific) was added to the membrane for film exposure. After the film was exposed in a dark room, the optical density of the strips was analyzed by Image-Pro Plus 6.0 software, and the results were analyzed with GAPDH as an internal reference. Each experiment was repeated 3 times, and the results are expressed as the mean ± standard deviation (SD).
The human liver cell lines Huh7 and HepG2 were purchased from the Shanghai Cell Bank of the Chinese Academy of Sciences. They were resuspended in DMEM high glucose (containing 10% FBS and double antibodies). The cells were cultured at 37 °C and 5% CO2 until the confluence was approximately 90%.
According to the sequence of the Lnc524369 gene, the whole gene was synthesized by Shenggong Bioengineering (Shanghai) Co., Ltd. (Shanghai, China) with the addition of BamHI and EcoRI restriction sites the ends of the gene. The synthetic products of the Lnc524369 and pcDNA3.1 plasmids (Invitrogen, Waltham, MA, United States) were digested, purified, and linked with BamHI and EcoRI, respectively, and then transformed into E. coli DH5α competent cells and cultured overnight at 37 °C. The recombinant plasmid Lnc524369-pcDNA3.1 was extracted and sequenced. After identification, an endofree plasma maxi kit (Qiagen) was used to extract the Lnc524369-pcDNA3.1 overexpression plasmid and pcDNA3.1 control plasmid for subsequent transfection experiments.
Huh7 cells were inoculated into 6 well plates at a density of 5 × 105 cells/mL and incubated overnight in a 5% CO2 incubator at 37 °C. When the confluence of the Huh7 cells reached 70%-80%, the Lnc524369-pcDNA3.1 overexpression plasmid and pcDNA3.1 control plasmid were transfected according to the instructions of Lipofectamine 3000 (Thermo Fisher Scientific). After 8 h, the media was replaced with fresh complete culture medium. After 48 h, the total RNA was extracted for subsequent qPCR analysis and the protein was extracted for subsequent western blot detection.
Huh7 cells transfected for 24 h with the pcDNA3.1 control plasmid and the Lnc524369-pcDNA3.1 overexpression plasmid were digested and collected, and the cell density was adjusted. According to the cell density of 2 × 103/100 μL in each well, the cells were recorded as 0 h after adherence. After 0 h, 24 h, 48 h and 72 h, 10 μL CCK8 was added to each well and incubated at 37 °C and 5% CO2 for 2 h. Next, the absorbance value at 450 nm was detected using an enzyme-labeled measurement instrument.
After counting, the density of Huh7 cells was 2 × 105/mL, and 200 μL transwell cell suspension in serum free mediums was added to each transwell chamber (8.0 μm), and 600 μL containing 15% FBS medium was added to the incubator, incubated 48 h in the 5% CO2 incubator at 37 °C. The cells were then fixed with 4% paraformaldehyde for 30 min, and stained with 0.1% crystal violet for 30 min. After washing with PBS, an inverted microscope was used to take pictures and count the cells. Each group was provided with 3 multiple holes. Five visual fields were randomly selected from each well, and the average number of migrating cells was counted.
The 24 well Matrigel invasion chamber was taken out and allowed to recover to room temperature. Then, 500 μL serum-free medium was added and incubated at 37 °C for 2 h. The basement membrane was hydrated and the excess liquid was absorbed for standby. The cells were resuspended in serum-free medium for 24 h, and the cell concentration was adjusted to 2 × 105/mL. Two hundred microliters of cell suspension was added to the upper chamber, and 600 μL of cell culture medium containing 15% FBS was added to the lower chamber. After 48 h incubation in a 5% CO2 incubator at 37 °C, the cells were fixed with 4% paraformaldehyde for 30 min and stained with 0.1% crystal violet for 30 min. After washing with PBS, an inverted microscope was used for counting the cells. Each group included 3 wells, and 5 visual fields were randomly selected under 200 magnification for each well, and the average number of invasive cells was counted.
SPSS 19.0 software (IBM Corp., Armonk, NY, United States) was used for statistical analysis. The results were expressed as the mean ± SD. Independent sample t tests or paired sample t tests were used for mean comparisons between the two groups, and one-way ANOVA was used for multigroup mean comparisons. The Spearman method was used for correlation analysis. Kaplan–Meier method was used for survival analysis. When P < 0.05 (*), the difference was significant; when P < 0.01 (**), the difference was very significant; and when P < 0.001 (***), the difference was extremely significant.
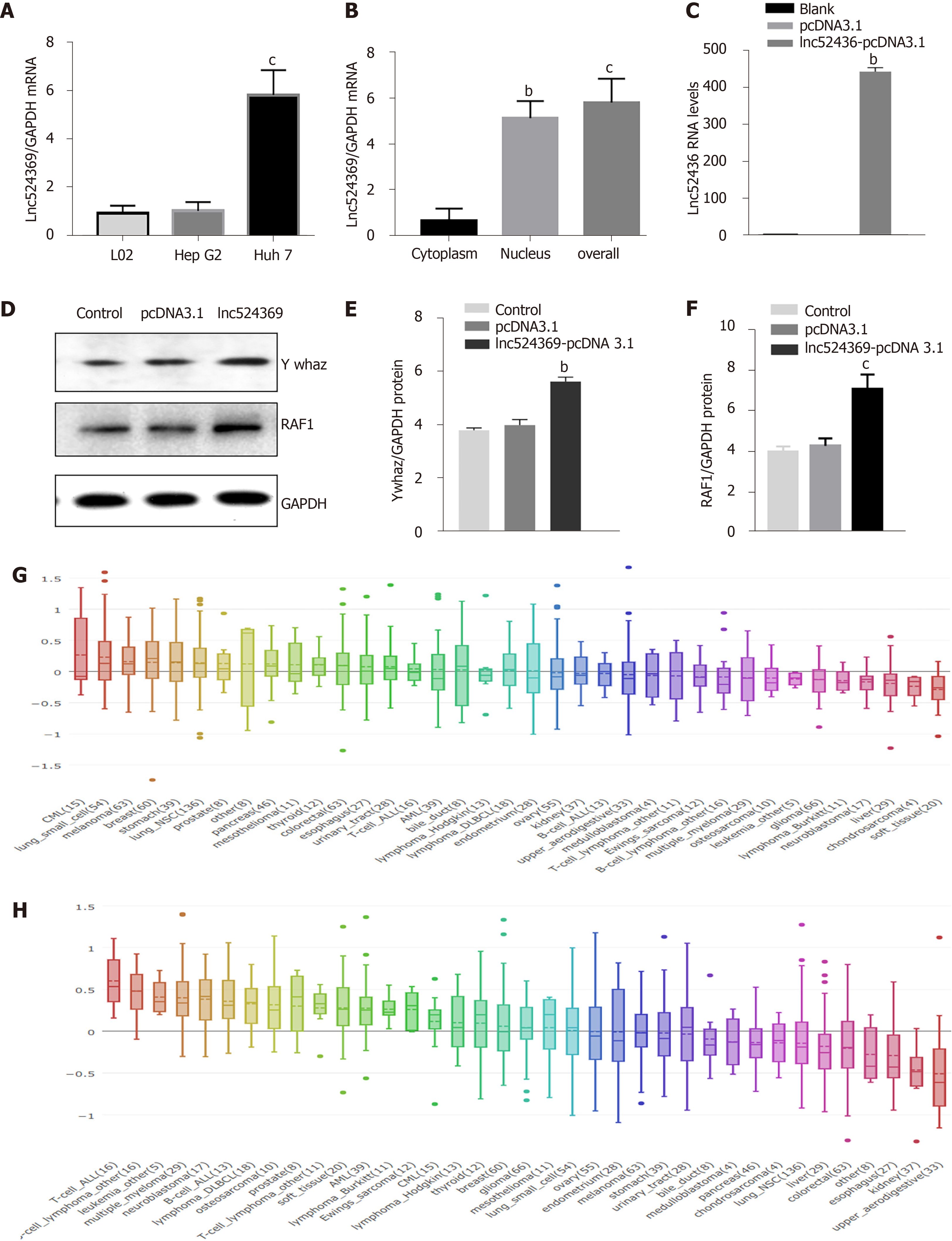
The Lnc524369 level was relatively higher in Huh7 cells than in HepG2 cells and L02 cells (Figure 1A) and enriched in the nucleus of Huh7 cells (Figure 1B). Considering the weaker invasion and lower Lnc524369 level of HepG2, Huh7 cells were used to investigate overexpression of Lnc524369.
Real-time PCR results showed that compared with the blank and pcDNA3.1 transfection groups, lnc52436-pcDNA3.1 was upregulated 443-fold in Huh7 cells, as shown in Figure 1C (P < 0.01). Compared with the blank and pcDNA3.1 transfection groups, the YWHAZ protein and mRNA levels, as well as the RAF1 protein levels in the Lnc52436-pcDNA3.1 transfection group were significantly increased, as shown in Figures 1D-F and 2B (P < 0.01).
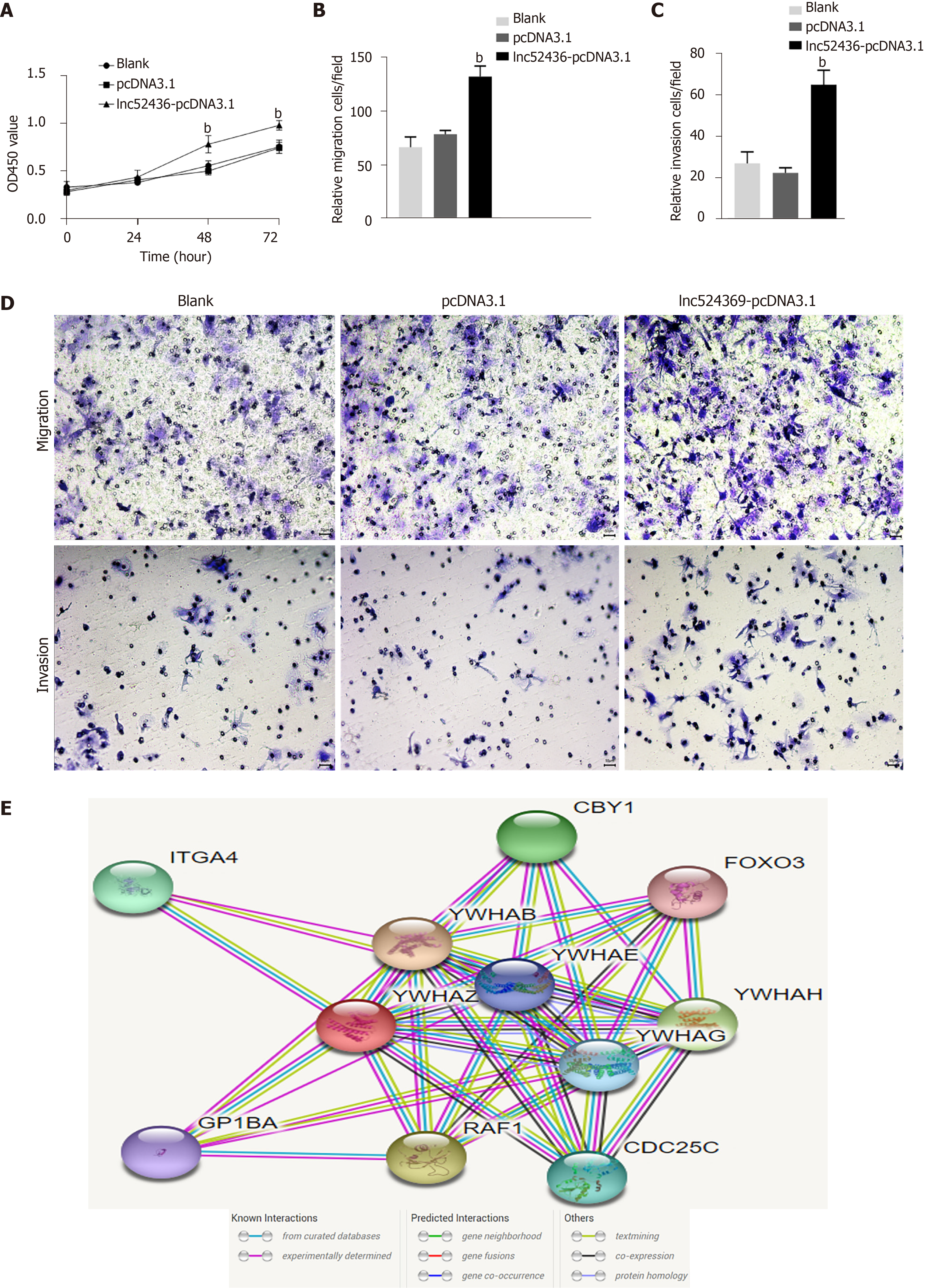
The CCK-8 assay was used to detect the viability of Huh7 cells after overexpression of Lnc524369. Compared with the blank and pcDNA3.1 groups, the viability of Huh7 cells increased significantly from 48 h (Figure 3A, P < 0.01). This result indicated that the proliferation of liver cancer cells was enhanced by overexpression of Lnc524369. Transwell assay results showed that compared with the blank and pcDNA3.1 transfection groups, the number of migrated Lnc52436-pcDNA3.1 cells was significantly increased, as shown in Figure 3B and 3D (P < 0.01). This result indicated that the migration of liver cancer cells could be enhanced by overexpression of Lnc524369. Transwell assay results showed that compared with the blank and pcDNA3.1 transfection groups, the number of cells passing through the basement membrane in the Lnc52436-pcDNA3.1 transfection group was significantly increased (Figure 3C and 3D) (P < 0.01). This result indicated that the invasion of liver cancer cells could be enhanced by overexpression of Lnc524369.
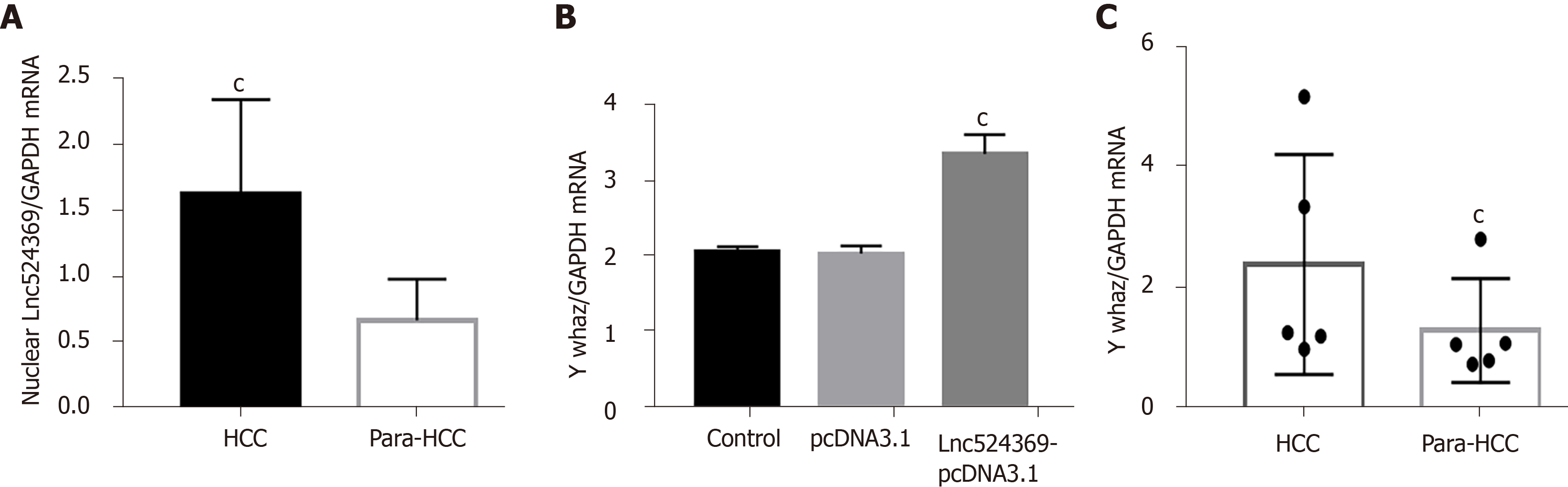
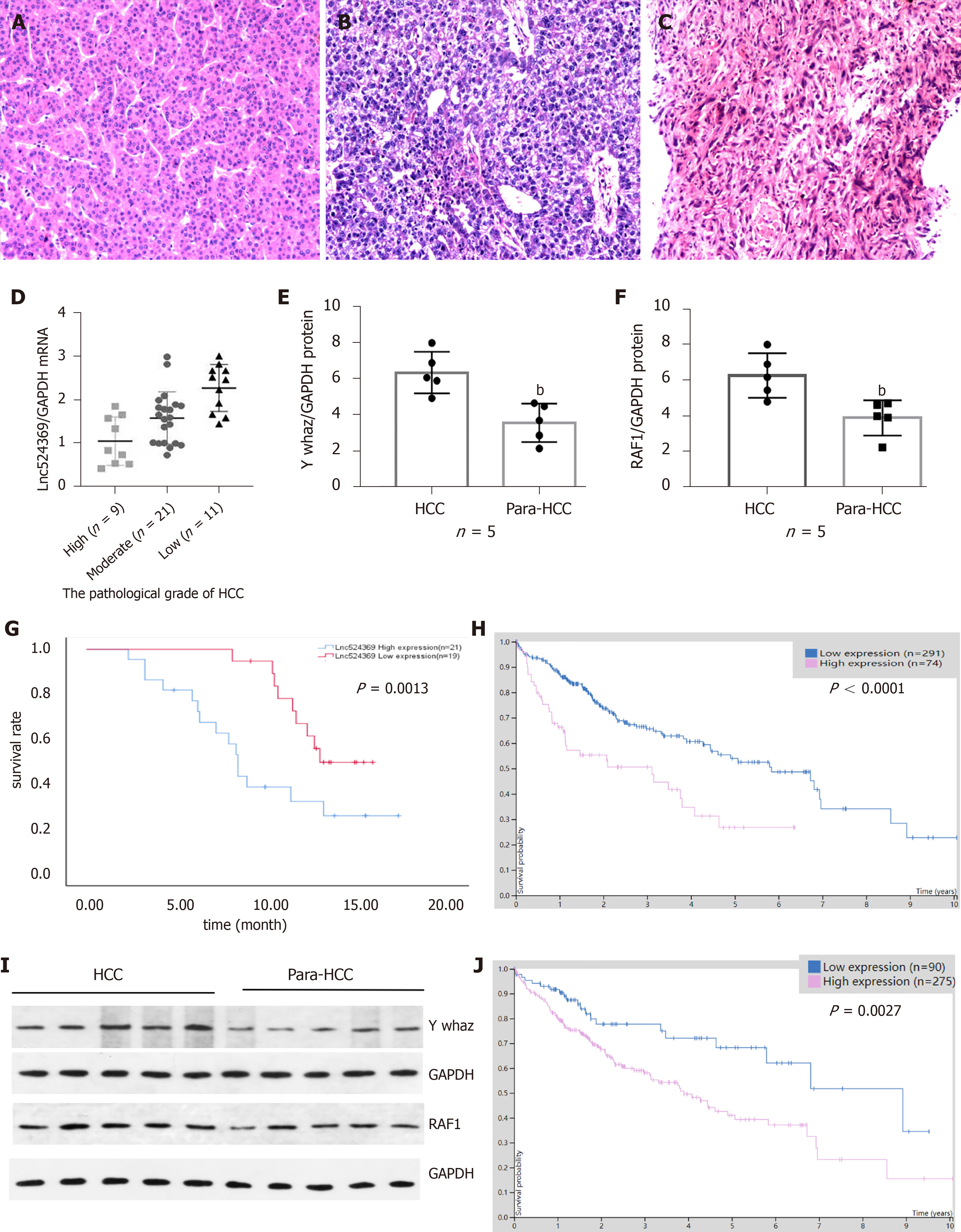
Forty-one confirmed HCC patients were included in this study according to the HCC guidelines[12], and their baseline characteristics are shown in Table 2. Human HCC and para-HCC tissues were acquired by surgical resection, which was further confirmed by two histopathological doctors. The relative nuclear Lnc524369 expression level was significantly higher in HCC tissues than in para-HCC tissues (P < 0.001) (Figure 2A). There was a positive correlation between Lnc524369 and the pathological grade of HCC (correlation coefficient RS: 0.604, P < 0.001) (Figure 4D). The survival rate of patients with high expression of Lnc524369 was significantly lower than that of patients with low expression (P = 0.013) (Figure 4G).
| Higher expression (n = 22) | Lower expression (n = 19) | t/χ2 | P | ||
| Age | 57.68 ± 10.77 | 54.21 ± 11.44 | 1.000 | 0.688 | |
| Male | 13 (59.1%) | 12 (63.16%) | 0.004 | 0.952 | |
| Pathological grading | Low | 9 (40.91%) | 2 (10.53%) | ||
| Moderate | 11 (50.00%) | 10 (52.63%) | - | 0.009 | |
| High | 2 (9.09%) | 7 (36.84%) | |||
YWHAZ mRNA and protein levels were determined in our five included HCC samples by real-time PCR and western blot. Our results showed that the YWHAZ mRNA level and protein were significantly higher in HCC tissues than in para-HCC tissues (P < 0.05) (Figures 2C, 4E and 4I); RAF1 protein was also significantly higher in HCC tissues than in para-HCC tissues (P < 0.05) (Figure 4F and 4I). In addition, the YWHAZ mRNA data of 235 live and 130 deceased HCC patients (264 male; 119 female) in the TCGA database were included for survival analysis. The survival probability of patients with high YWHAZ and RAF1 mRNA expression was significantly decreased compared with those patients with low YWHAZ expression (P < 0.001) (Figure 4H and 4J).
Recently, lncRNAs have emerged as critical molecules in multiple biological processes involved in virus infection, metabolic diseases, vascular diseases, stem cell biology, fibrosis, and cancer[13-16]. Currently, the roles of lncRNAs have been widely reported in the progression of HCC and liver cancer stem cells[17]. A large number of lncRNAs localize in the nucleus and are highly involved in several cellular components, biological processes, and molecular functions, such as chromatin organization, structural scaffolds of nuclear domains, and transcriptional and posttranscriptional gene expression[18]. Previous fractionation-then-sequencing data from human HCC tissues have shown that Lnc524369 is enriched in the nucleus of liver cancer but not the cytoplasm[7]. In our study, we also confirmed that Lnc524369 expression was enriched in the nucleus of Huh 7 cells.
Furthermore, we illustrated that the expression of nuclear Lnc524369 was significantly increased in human HCC tissues compared to HCC adjacent tissues (P < 0.01). Additionally, we found that the overexpression of Lnc524369 could clearly promote the proliferation, migration, and invasion of liver cancer cells and simultaneously enhance YWHAZ and RAF1 expression. This result suggested that Lnc524369 might positively correlate with the expression of YWHAZ and RAF1 in the development of HCC. Lnc524369 was significantly correlated with a poor survival rate of HCC patients.
YWHAZ has been shown to be commonly upregulated in multiple cancers, especially HCC, as it can promote tumorigenesis, metastasis, and chemoresistance in the progression of cancer[19-21]. Our bioinformatic analysis showed that RAF1 was strongly coexpressed with YWHAZ (Figure 3E). RAF1 is also highly involved in the development of HCC[22]. Again, our analysis of the CCLE databases revealed that YWHAZ and RAF1 were frequently expressed in liver cancer cell lines. Our study indicated that YWHAZ transcriptional and posttranscriptional levels were both overexpressed in HCC tissues compared to HCC adjacent tissues (P < 0.01). In addition, we found that YWHAZ and RAF1 mRNA levels were negatively related to overall survival time in the TCGA database. Previous studies have suggested that multiple noncoding RNAs, such as miR-451a, miR-22, and the long noncoding RNA MIR4435-2HG, are involved in HCC proliferation, invasion, and metastasis by targeting YWHAZ[8,9,11]. These results indicate that the overexpression of YWHAZ plays a critical role in the initiation and progression of HCC; therefore, the YWHAZ/RAF1 protein might have significant clinical potential for targeted therapy and early diagnosis.
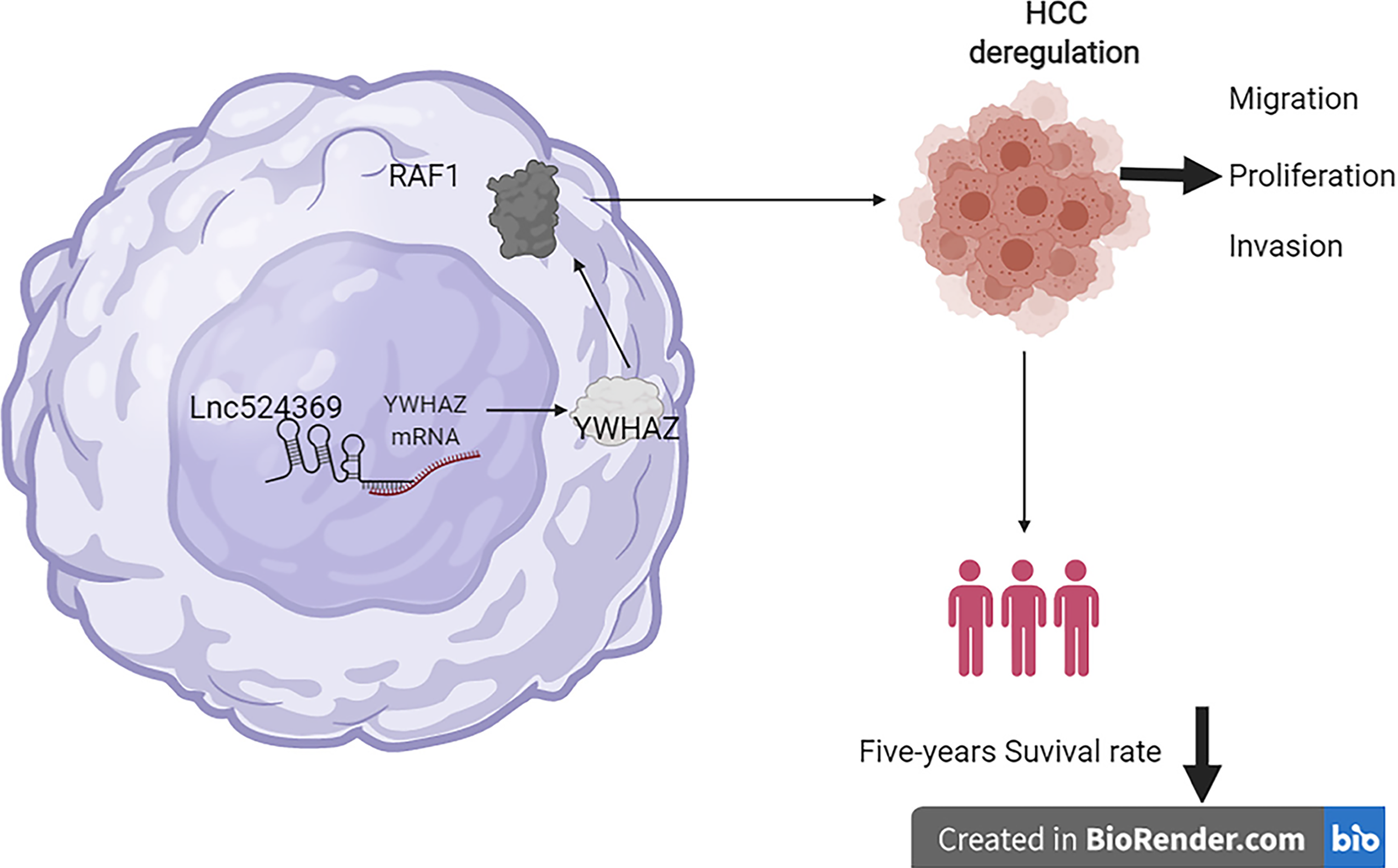
There are some limitations in this study that will be improved in further studies. First, the expression of Lnc524369, YWHAZ, and RAF1 should be further knocked down for malignant function confirmation; second, the effect of Lnc524369 and YWHAZ in rodent models should be further studied; third, the included number of HCC patients for this study should be further enlarged. In summary, Lnc524369 and its downstream targets YWHAZ and RAF1 play a crucial role in the development of HCC and are negatively associated with the overall survival times of HCC patients, which provides new insight into the early diagnosis and targeted treatment of HCC (Figure 5).
Because Lnc524369 can enhance liver cancer progression and decrease the overall survival time of HCC by activating the YWHAZ/RAF1 pathway, might be a promising target of HCC.
Long noncoding RNAs, including Lnc524369, have the potential to regulate unknown cellular and molecular mechanisms in the initiation, progression, diagnosis, and prognosis of hepatocellular carcinoma (HCC). However, a critical gap in our knowledge is understanding how nucleus-enriched Lnc524369 promotes liver cancer growth.
To discover specific targets for the diagnosis and treatment of HCC.
To investigate the underlying mechanisms of Lnc524369 in HCC.
The expression of Lnc524369, YWHAZ, and RAF1 was determined by qPCR or western blot. CCK-8, migration, and invasion assays were used to investigate Lnc524369 function. Forty-one HCC patients, the Cancer Cell Line Encyclopedia databases, STRING database, Human Protein Atlas database and the TCGA database were used for survival analysis.
Lnc524369 was significantly upregulated in the nucleus of HCC, which promoted the proliferation, migration, and invasion of liver cancer cells by upregulating YWHAZ and RAF1 expression. Lnc524369 and its downstream target YWHAZ/RAF1 protein could predict the poor overall survival time of HCC patients.
The Lnc524369-mediated YWHAZ/RAF1 pathway is highly involved in the progr
In the future, we will reveal the critical role of Lnc524369, which might enhance the early diagnosis of HCC and facilitate the further development of targeted therapy.
Provenance and peer review: Unsolicited article; Externally peer reviewed.
Peer-review model: Single blind
Specialty type: Oncology
Country/Territory of origin: China
Peer-review report’s scientific quality classification
Grade A (Excellent): 0
Grade B (Very good): 0
Grade C (Good): C
Grade D (Fair): 0
Grade E (Poor): 0
P-Reviewer: Youness RA S-Editor: Wang JJ L-Editor: Filipodia P-Editor: Yu HG
| 1. | Massarweh NN, El-Serag HB. Epidemiology of Hepatocellular Carcinoma and Intrahepatic Cholangiocarcinoma. Cancer Control. 2017;24:1073274817729245. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 350] [Cited by in RCA: 434] [Article Influence: 54.3] [Reference Citation Analysis (1)] |
| 2. | Bray F, Ferlay J, Soerjomataram I, Siegel RL, Torre LA, Jemal A. Global cancer statistics 2018: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J Clin. 2018;68:394-424. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 53206] [Cited by in RCA: 55806] [Article Influence: 7972.3] [Reference Citation Analysis (132)] |
| 3. | Fujiwara N, Friedman SL, Goossens N, Hoshida Y. Risk factors and prevention of hepatocellular carcinoma in the era of precision medicine. J Hepatol. 2018;68:526-549. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 423] [Cited by in RCA: 523] [Article Influence: 74.7] [Reference Citation Analysis (0)] |
| 4. | Trépo C, Chan HL, Lok A. Hepatitis B virus infection. Lancet. 2014;384:2053-2063. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1004] [Cited by in RCA: 1157] [Article Influence: 105.2] [Reference Citation Analysis (0)] |
| 5. | Quinn JJ, Chang HY. Unique features of long non-coding RNA biogenesis and function. Nat Rev Genet. 2016;17:47-62. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 2868] [Cited by in RCA: 2718] [Article Influence: 302.0] [Reference Citation Analysis (0)] |
| 6. | Cai Z, Xu K, Li Y, Lv Y, Bao J, Qiao L. Long noncoding RNA in liver cancer stem cells. Discov Med. 2017;24:87-93. [PubMed] |
| 7. | Chow EY, Zhang J, Qin H, Chan TF. Characterization of Hepatocellular Carcinoma Cell Lines Using a Fractionation-Then-Sequencing Approach Reveals Nuclear-Enriched HCC-Associated lncRNAs. Front Genet. 2019;10:1081. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 11] [Cited by in RCA: 11] [Article Influence: 1.8] [Reference Citation Analysis (0)] |
| 8. | Wei GY, Hu M, Zhao L, Guo WS. MiR-451a suppresses cell proliferation, metastasis and EMT via targeting YWHAZ in hepatocellular carcinoma. Eur Rev Med Pharmacol Sci. 2019;23:5158-5167. [RCA] [PubMed] [DOI] [Full Text] [Cited by in RCA: 18] [Reference Citation Analysis (0)] |
| 9. | Chen M, Hu W, Xiong CL, Qu Z, Yin CQ, Wang YH, Luo CL, Guan Q, Yuan CH, Wang FB. miR-22 targets YWHAZ to inhibit metastasis of hepatocellular carcinoma and its down-regulation predicts a poor survival. Oncotarget. 2016;7:80751-80764. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 30] [Cited by in RCA: 43] [Article Influence: 6.1] [Reference Citation Analysis (0)] |
| 10. | Zhao JF, Zhao Q, Hu H, Liao JZ, Lin JS, Xia C, Chang Y, Liu J, Guo AY, He XX. The ASH1-miR-375-YWHAZ Signaling Axis Regulates Tumor Properties in Hepatocellular Carcinoma. Mol Ther Nucleic Acids. 2018;11:538-553. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 31] [Cited by in RCA: 43] [Article Influence: 6.1] [Reference Citation Analysis (0)] |
| 11. | Shen X, Ding Y, Lu F, Yuan H, Luan W. Long noncoding RNA MIR4435-2HG promotes hepatocellular carcinoma proliferation and metastasis through the miR-22-3p/YWHAZ axis. Am J Transl Res. 2020;12:6381-6394. [PubMed] |
| 12. | Zhou J, Sun HC, Wang Z, Cong WM, Wang JH, Zeng MS, Yang JM, Bie P, Liu LX, Wen TF, Han GH, Wang MQ, Liu RB, Lu LG, Ren ZG, Chen MS, Zeng ZC, Liang P, Liang CH, Chen M, Yan FH, Wang WP, Ji Y, Cheng WW, Dai CL, Jia WD, Li YM, Li YX, Liang J, Liu TS, Lv GY, Mao YL, Ren WX, Shi HC, Wang WT, Wang XY, Xing BC, Xu JM, Yang JY, Yang YF, Ye SL, Yin ZY, Zhang BH, Zhang SJ, Zhou WP, Zhu JY, Liu R, Shi YH, Xiao YS, Dai Z, Teng GJ, Cai JQ, Wang WL, Dong JH, Li Q, Shen F, Qin SK, Fan J. Guidelines for Diagnosis and Treatment of Primary Liver Cancer in China (2017 Edition). Liver Cancer. 2018;7:235-260. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 328] [Cited by in RCA: 444] [Article Influence: 63.4] [Reference Citation Analysis (0)] |
| 13. | Liu Y, Feng J, Sun M, Yang G, Yuan H, Wang Y, Bu Y, Zhao M, Zhang S, Zhang X. Long non-coding RNA HULC activates HBV by modulating HBx/STAT3/miR-539/APOBEC3B signaling in HBV-related hepatocellular carcinoma. Cancer Lett. 2019;454:158-170. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 48] [Cited by in RCA: 64] [Article Influence: 10.7] [Reference Citation Analysis (0)] |
| 14. | Li Y, Xu K, Chen S, Cao Y, Zhan H. Roles of Identified Long Noncoding RNA in Diabetic Nephropathy. J Diabetes Res. 2019;2019:5383010. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 25] [Cited by in RCA: 37] [Article Influence: 6.2] [Reference Citation Analysis (0)] |
| 15. | Chen S, Sun X, Wu S, Jiang J, Zhu C, Xu K. Role of identified noncoding RNA in erectile dysfunction. Andrologia. 2020;52:e13596. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 5] [Cited by in RCA: 6] [Article Influence: 1.2] [Reference Citation Analysis (0)] |
| 16. | Prabhakar B, Lee S, Bochanis A, He W, Manautou JE, Rasmussen TP. lnc-RHL, a novel long non-coding RNA required for the differentiation of hepatocytes from human bipotent progenitor cells. Cell Prolif. 2021;54:e12978. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 4] [Cited by in RCA: 5] [Article Influence: 1.3] [Reference Citation Analysis (0)] |
| 17. | Lv H, Lv G, Han Q, Yang W, Wang H. Noncoding RNAs in liver cancer stem cells: The big impact of little things. Cancer Lett. 2018;418:51-63. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 30] [Cited by in RCA: 39] [Article Influence: 5.6] [Reference Citation Analysis (0)] |
| 18. | Sun Q, Hao Q, Prasanth KV. Nuclear Long Noncoding RNAs: Key Regulators of Gene Expression. Trends Genet. 2018;34:142-157. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 427] [Cited by in RCA: 435] [Article Influence: 62.1] [Reference Citation Analysis (0)] |
| 19. | Liu Y, Guo J, Shen K, Wang R, Chen C, Liao Z, Zhou J. Paclitaxel Suppresses Hepatocellular Carcinoma Tumorigenesis Through Regulating Circ-BIRC6/miR-877-5p/YWHAZ Axis. Onco Targets Ther. 2020;13:9377-9388. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 13] [Cited by in RCA: 25] [Article Influence: 5.0] [Reference Citation Analysis (0)] |
| 20. | Zhao J, Fu X, Chen H, Min L, Sun J, Yin J, Guo J, Li H, Tang Z, Ruan Y, Wang X, Sun Y, Huang L. G3BP1 interacts with YWHAZ to regulate chemoresistance and predict adjuvant chemotherapy benefit in gastric cancer. Br J Cancer. 2021;124:425-436. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 14] [Cited by in RCA: 39] [Article Influence: 7.8] [Reference Citation Analysis (0)] |
| 21. | Hui C, Tian L, He X. Circular RNA circNHSL1 Contributes to Gastric Cancer Progression Through the miR-149-5p/YWHAZ Axis. Cancer Manag Res. 2020;12:7117-7130. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 15] [Cited by in RCA: 24] [Article Influence: 4.8] [Reference Citation Analysis (0)] |
| 22. | Ghousein A, Mosca N, Cartier F, Charpentier J, Dupuy JW, Raymond AA, Bioulac-Sage P, Grosset CF. miR-4510 blocks hepatocellular carcinoma development through RAF1 targeting and RAS/RAF/MEK/ERK signalling inactivation. Liver Int. 2020;40:240-251. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 17] [Cited by in RCA: 26] [Article Influence: 5.2] [Reference Citation Analysis (0)] |