Published online Dec 15, 2021. doi: 10.4251/wjgo.v13.i12.2129
Peer-review started: July 13, 2021
First decision: August 9, 2021
Revised: August 18, 2021
Accepted: October 25, 2021
Article in press: October 25, 2021
Published online: December 15, 2021
Processing time: 154 Days and 7.1 Hours
BRAFV600E mutated colorectal cancer (CRC) is prone to peritoneal and distant lymph node metastasis and this correlates with a poor prognosis. The BRAFV600E mutation is closely related to the formation of an immunosuppressive microenvironment. However, the correlation between BRAFV600E mutation and changes in local immune microenvironment of CRC is not clear.
To explore the effect and mechanism of BRAFV600E mutant on the immune microenvironment of CRC.
Thirty patients with CRC were included in this study: 20 in a control group and 10 in a treatment group. The density of microvessels and microlymphatic vessels, and M2 subtype macrophages in tumor tissues were detected by immunohistochemistry. Screening and functional analysis of exosomal long noncoding RNAs (lncRNAs) were performed by transcriptomics. The proliferation and migration of human umbilical vein endothelial cells (HUVECs) and human lymphatic endothelial cells (HLECs) were detected by CCK-8 assay and scratch test, respectively. The tube-forming ability of endothelial cells was detected by tube formation assay. The macrophage subtypes were obtained by flow cytometry. The expression of vascular endothelial growth factor (VEGF)-A, basic fibroblast growth factor (bFGF), transforming growth factor (TGF)-β1, VEGF-C, claudin-5, occludin, zonula occludens (ZO)-1, fibroblast activation protein, and α-smooth muscle actin was assessed by western blot analysis. The levels of cytokines interleukin (IL)-6, TGF-β1, and VEGF were assessed by enzyme-linked immunosorbent assay.
BRAFV600E mutation was positively correlated with the increase of preoperative serum carbohydrate antigen 19-9 (P < 0.05), and with poor tumor tissue differentiation in CRC (P < 0.01). Microvascular density and microlymphatic vessel density in BRAFV600E mutant CRC tissues were higher than those in BRAF wild-type CRC (P < 0.05). The number of CD163+ M2 macrophages in BRAFV600E mutant CRC tumor tissue was markedly increased (P < 0.05). Compared with exosomes from CRC cells with BRAF gene silencing, the expression of 13 lncRNAs and 192 mRNAs in the exosomes from BRAFV600E mutant CRC cells was upregulated, and the expression of 22 lncRNAs and 236 mRNAs was downregulated (P < 0.05). The biological functions and signaling pathways predicted by differential lncRNA target genes and differential mRNAs were closely related to angiogenesis, tumor cell proliferation, differentiation, metabolism, and changes in the microenvironment. The proliferation, migration, and tube formation ability of HUVECs and HLECs induced by exosomes in the 1627 cell group (HT29 cells with BRAF gene silencing) was greatly reduced compared with the HT29 cell group (P < 0.05). Compared with the HT29 cell group, the expression levels of VEGF-A, bFGF, TGF-β1, and VEGF-C in the exosomes derived from 1627 cells were reduced. The expression of ZO-1 in HUVECs, and claudin-5, occludin, and ZO-1 in HLECs of the 1627 cell group was higher. Compared with the 1627 cell group, the exosomes of the HT29 cell group promoted the expression of CD163 in macrophages (P < 0.05). IL-6 secretion by macrophages in the HT29 cell group was markedly elevated (P < 0.05), whereas TGF-β1 was decreased (P < 0.05). The levels of IL-6, TGF-β1, and VEGF secreted by fibroblasts in the 1627 cell group decreased, compared with the HT29 cell group (P < 0.05).
BRAFV600E mutant CRC cells can reach the tumor microenvironment by releasing exosomal lncRNAs, and induce the formation of an immunosuppressive microenvironment.
Core Tip: This study revealed that BRAFV600E mutant colorectal cancer (CRC) cells could lead to more angiogenesis and lymphoangiogenesis in the microenvironment by releasing exosomal long noncoding RNAs, inducing the formation of an immunosuppressive microenvironment. Our findings provide a hypothesis for finding new therapeutic strategies for BRAFV600E mutant CRC.
- Citation: Zhi J, Jia XJ, Yan J, Wang HC, Feng B, Xing HY, Jia YT. BRAFV600E mutant colorectal cancer cells mediate local immunosuppressive microenvironment through exosomal long noncoding RNAs. World J Gastrointest Oncol 2021; 13(12): 2129-2148
- URL: https://www.wjgnet.com/1948-5204/full/v13/i12/2129.htm
- DOI: https://dx.doi.org/10.4251/wjgo.v13.i12.2129
Colorectal cancer (CRC) is one of the most common malignant tumors, and ranks third in morbidity and mortality globally[1]. In China, societal and lifestyle changes have tended to increase the morbidity and mortality of CRC[2]. BRAFV600E gene mutation accounts for approximately 10% of patients with metastatic CRC (mCRC), and is a point mutation at nucleotide 1799 of exon 15 (T mutated into A), resulting in a change of the encoded amino acid 600, valine replaced by glutamate (V600E)[3,4]. BRAFV600E mutation can continuously activate the RAS-RAF-MEK-ERK signaling pathway, promote tumor cell proliferation and migration, and induce angiogenesis, thereby reducing tumor cell apoptosis[5-8]. Clinical data reveal that BRAFV600E mutation frequently occurs in elderly women, and the related pathological type is mostly mucinous adenocarcinoma with a high level of tissue differentiation. Around 20% of patients have accompanying microsatellite instability, and most of them develop right colon cancer originating from serrated adenomas[4,9]. Compared with wild-type BRAF, patients with BRAFV600E mutant CRC are prone to peritoneal metastasis and distant lymph node metastasis with a poor prognosis. Generally, the median survival time is < 12 mo. The effective treatment rate reaches only approximately 20%, even with three-drug chemotherapy and targeted combination therapy[10,11]. It is therefore important to explore the mechanism of BRAFV600E mutant CRC with lymph node and peritoneal metastasis and to discover effective therapeutic strategies. This may be applicable to start with its specific immune microenvironment.
The tumor microenvironment (TME) is the local environment facilitating tumor growth and proliferation[12]. In thyroid cancer, the proportion of mast cells in BRAFV600E mutant TME is markedly increased compared with wild-type BRAF, which may be involved in mediating the formation of the immunosuppressive microenvironment, suggesting that BRAFV600E mutation affects the TME[13]. Multiple cells (namely, endothelial cells, fibroblasts, and immune cells) and extracellular components (namely, cytokines, growth factors, hormones, and extracellular matrix) in the TME can induce tumor angiogenesis and lymphangiogenesis and promote chronic inflammation, thereby creating a local immunosuppressive microenvironment, which plays a vital role in tumor occurrence, invasion, and metastasis and drug resistance[14]. Tumor-associated macrophages (TAMs) and cancer-associated fibroblasts (CAFs) are the principal components of the TME. Among them, M2-type macrophages have an immunosuppressive effect. They can promote tumor cell proliferation, infiltration, and metastasis, and their expression level is intimately associated with patient prognosis[15,16]. Meanwhile, CAFs have undergone substantial changes in morphology, proliferation activity, motility, and secretory function, which can facilitate tumor proliferation, invasion, metastasis, and angiogenesis, and their expression level is closely linked to tumor stage and poor prognosis[17]. There is no current research investigating the relationship between BRAFV600E gene mutation and the formation and functional changes of blood vessels, lymphatic vessels, TAMs, and CAFs in the CRC local immune microenvironment.
Exosomes are a type of cystic microvesicle secreted by cells, and the secretion process is active. They are able to carry specific biologically active molecules including lipids, miRNA, and long noncoding RNAs (lncRNAs) into the corresponding target cells and mediate substance transportation and information exchange intercellularly[18]. LncRNAs belong to a class of single-stranded RNA molecules that do not encode proteins. They play a role via transcription, post-transcription, and translation, and participate in tumor occurrence, invasion, and metastasis. Exosomes secreted by tumor cells can modify the TME through lncRNAs, thereby promoting the development of tumors[19,20]. Liang et al[21] noted that the expression of exosome-derived lncRNA RPPH1 in CRC was markedly upregulated. It can prevent ubiquitination by binding to TUBB3 and induce epithelial-mesenchymal transition, and interact with TAMs to promote the polarization of TAMs to M2 subtype, thereby accelerating tumor progression. However, there is still a lack of relevant investigations on whether BRAFV600E mutant CRC cells affect the TME through the release of exosomal lncRNAs.
The present study aimed to investigate the influence of BRAFV600E mutation in CRC on the surrounding immune microenvironment, and to elucidate whether BRAFV600E mutant CRC cell-derived exosomes participate in the formation of an immunosuppressive microenvironment. It is hoped that the results will provide a novel therapeutic strategy for BRAFV600E mutant CRC.
Data of ten BRAFV600E mutant CRC patients who underwent surgical treatment at the Hebei General Hospital from September 2014 to June 2019 were collected. Twenty BRAF wild-type CRC patients were selected as controls. There were 18 male and 12 female patients. The age range was 27-79 years, with an average of 57.57 ± 2.13 years and median of 57 years. The specimens were obtained with informed consent obtained from the patients and under the approval of the Ethics Committee of the Fourth Hospital of Hebei Medical University. The inclusion criteria were: (1) Patients with a pathological diagnosis of CRC, in whom those identified as having BRAFV600E mutation were included in an experimental group, and those identified as having wild-type BRAF were included in a control group; (2) Untreated patients; and (3) Undergoing first surgical treatment for primary CRC. The exclusion criteria were: (1) History of malignant tumors; (2) Current primary tumors in other regions; and (3) Patients with incomplete pathological data.
The human colon cancer cell line HT29 (BRAFV600E mutant) was purchased from the Cell Bank of the Chinese Academy of Sciences. The human colon cancer cell line 1627 was obtained from Shanghai GenePharma Company, which was a HT29 cell strain with the BRAF gene being silenced. Human umbilical vein endothelial cells (HUVECs) and human lymphatic endothelial cells (HLECs) were purchased from ScienCell, San Diego, CA, United States. Human monocytic leukemia cells (THP-1 cells) and human embryonic lung fibroblasts (MRC-5 cells) were obtained from the Cell Bank of the Shanghai Institutes for Biological Sciences, Chinese Academy of Sciences. McCoy’s 5A, RPMI 1640, minimal essential medium, and fetal bovine serum (FBS) were all purchased from Gibco (NY, United States).
THP-1 cells in the logarithmic phase of growth were obtained and placed in a 15-mL centrifuge tube, and centrifuged at 1000 r/min for 5 min. The supernatant was discarded. Following addition of fresh RPMI 1640 culture medium, the cells were resuspended and inoculated in a 12-well plate with 5 × 105 cells per well, supplemented with pharmaceutical manufacturers association (PMA) solution (dissolved in DMSO) (Cayman, MI, United States) at a final concentration of 100 ng/mL, and cultured in a constant temperature incubator for 48 h. The cells were ultimately placed under a biological microscope (Nikon, Tokyo, Japan) and the cell morphology and adhesion were observed.
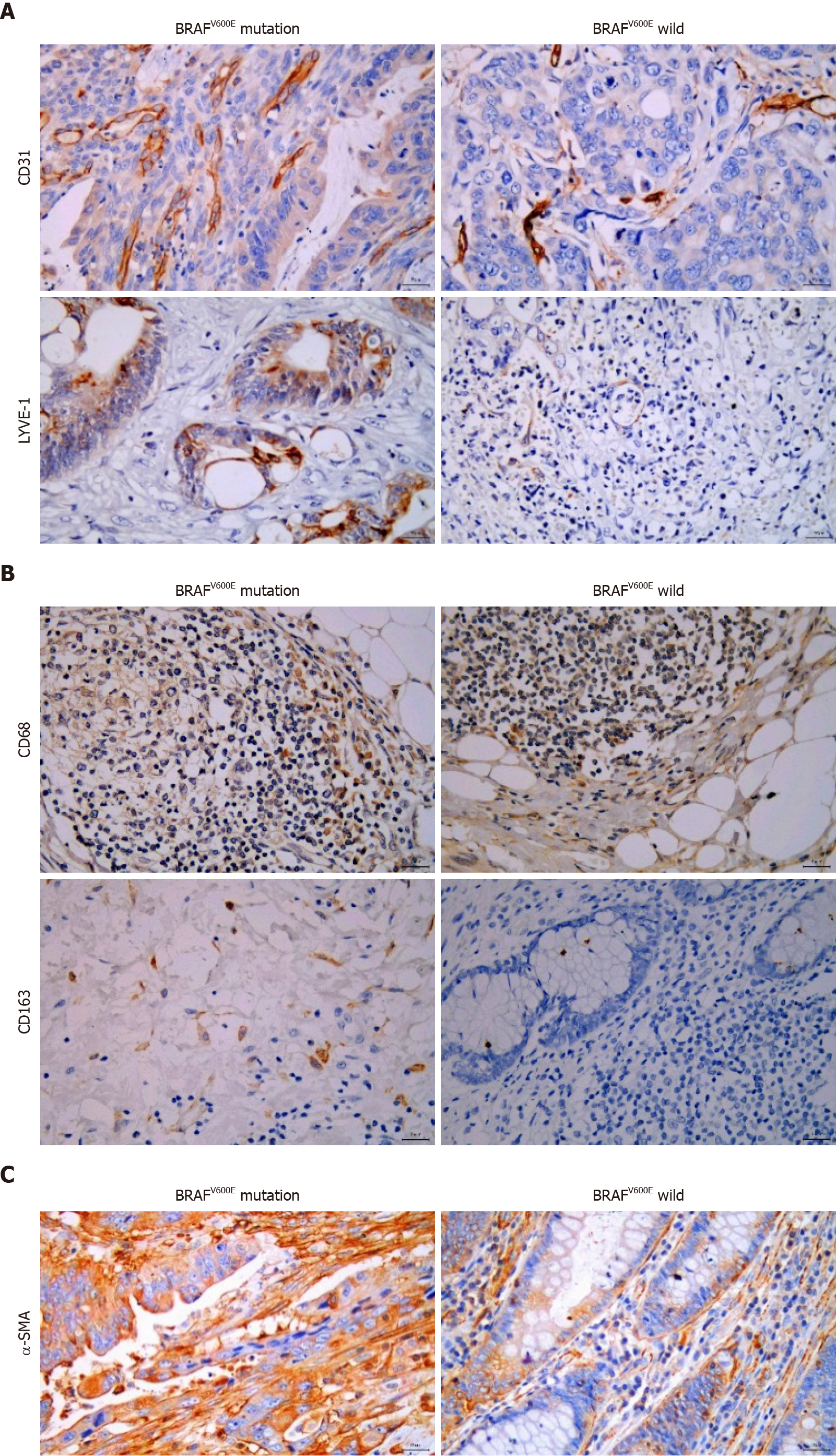
The postoperative tissue samples of CRC patients were collected, embedded in paraffin, and cut into 5-mm sections. After deparaffinization and hydration, antigen retrieval, and incubation in 3% H2O2 for 10 min, the corresponding antibodies were added, including rabbit anti-human CD31 monoclonal antibody (1:2000, Arigo, Hsinchu City, Taiwan), rabbit LYVE-1 polyclonal antibody (1:200, Arigo), mouse anti-human CD68 monoclonal antibody (1:100, BD Biosciences, NJ, United States), rabbit anti-human CD163 monoclonal antibody (1:100, HUABIO, Hangzhou, China), and rabbit anti-human α-smooth muscle actin (SMA) monoclonal antibody (1:100, HUABIO). 3,3-diaminobenzidine (DAB) color development was performed for microscopic observation. A double-blind reading method was adopted by two pathologists. The sections were initially visualized under low magnification (× 100) to determine three fields where cells were most densely distributed. The number of positive cells was counted under high magnification (× 400), and the average number was calculated.
Cell supernatant was collected and centrifuged at 480 × g for 5 min, followed by 2000 × g for 10 min to remove cell debris. The supernatant was collected and centrifuged at 10000 × g for 30 min to remove macrovesicles. The supernatant was collected again and centrifuged at 100000 × g for 2 h. The supernatant was discarded and the exosomes were resuspended in 200 mL of PBS solution, and stored in a freezer at -80 °C for later use. The morphology of exosomes was identified with a transmission electron microscope (Hitachi, Tokyo, Japan). At room temperature, 10 mL of exosome suspension was dripped on a 2-nm-pore-diameter copper mesh using a pipette and allowed to stand for 2 min. The liquid was absorbed dry using absorbent paper. A quantity of 30 mL 3% tungsten phosphate was dripped onto the copper mesh for negative staining for 5 min, and the liquid was absorbed dry using absorbent paper. After drying, photographs were taken under a transmission electron microscope.
The cell samples were removed from the -80 °C freezer and supplemented with 200 mL of RIPA lysis solution for 20 min to extract the total cell proteins. The BCA protein concentration determination kit was used to quantify the proteins. The proteins were subjected to SDS-PAGE, transferred to polyvinylidene difluoride (PVDF) membranes at 250 mA, and blocked with bovine serum albumin for 2 h. The blocked PVDF membranes were placed directly into the freshly prepared primary antibody working solution at 4 °C overnight. Primary antibodies used included rabbit anti-human CD9 monoclonal antibody (1:1000, Abcam, Cambridge, United Kingdom), rabbit anti-human CD63 monoclonal antibody (1:5000, Abcam), rabbit anti-human BRAF monoclonal antibody (1:1000, Arigo), rabbit anti-zonula occludens (ZO)-1 polyclonal antibody (1:1000, GenTex, Gentex, United States), rabbit anti-claudin-5 polyclonal antibody (1:1000, GenTex), rabbit anti-occludin polyclonal antibody (1:1000, HUABIO), rabbit anti-transforming growth factor (TGF)-1 polyclonal antibody (1:1000, Bioss, Beijing, China), rabbit anti-vascular endothelial growth factor (VEGF)-C polyclonal antibody (1:800, Bioworld, MN, United States), rabbit anti-basic fibroblast growth factor (anti-bFGF) polyclonal antibody (1:600, Bioss), mouse anti-human VEGF-A monoclonal antibody at a concentration of 6 mg/mL (Abcam), rabbit anti-human fibroblast activation protein (FAP) monoclonal antibody (1:1000, Abcam), and rabbit anti-human α-SMA monoclonal antibody (1:1000, Abcam). Following membrane washing on the next day, the corresponding secondary antibody was incubated, and chemiluminescence was detected with ECL substrate. After developing and fixing, the film was scanned using a scanner, and Tanon 1600 software was used for gray scale analysis and quantification.
Cells in the logarithmic phase of growth were selected and seeded in a 96-well plate at 5 × 103 cells/well with a volume of 100 mL per well, and cultured at 37 °C in a 5% CO2 incubator. After 6 h of inoculation, the experimental group was treated with 10 mL exosomes, and the control group with 10 mL PBS solution, and then cultured in the incubator again. Exosomes were cocultured with HUVECs for 24, 48, and 72 h, and cocultured with HLECs for 12, 24 and 36 h; 10 mL of CCK-8 reagent (DOJINDO, Kyushu, Japan) was added to each well and incubated at 37 °C with 5% CO2 for 1 h. Absorbance at 560 nm was measured using a microplate reader (ThermoFisher Scientific, MA, United States).
HUVECs or HLECs were plated in six-well plates. When the cells were evenly spread, they were scratched using a 200-μL pipette tip. PBS solution was used to rinse once for the removal of the suspended cells. To each well, 2 mL of FBS-free culture medium was added. The experimental group was supplemented with 20 μL of exosomes, and the control group with 20 μL of PBS solution. Both groups were cultured at 37 °C in a 5% CO2 incubator. The cells were observed and photographed under a microscope at 0 and 24 h, respectively, to detect the scratch healing. Cell migration rate was calculated as [(scratch area at 0 h - scratch area at 24 h)/scratch area at 0 h] × 100%.
After freezing and thawing the Matrigel matrix (BD Biosciences, United States) containing low levels of growth factors at 4 °C, the homogenate was mixed well with a precooled pipette tip and packaged into precooled Eppendorf tubes. Matrigel matrix was diluted with serum-free medium at a ratio of 1:3, and the 96-well plate was placed on an ice pack. Fifty microliters of diluted Matrigel matrix was added to each well and left to stand at 37 °C for 30 min. Cells were seeded in a 96-well plate at 104 cells/well and supplemented with 100 μL cell suspension in each well. The experimental group was treated with 10 μL of exosomes, and the control group with 10 μL PBS solution, and then cultured at 37 °C in a 5% CO2 incubator. After culturing for 24 h, the cells were visualized and photographed under a low magnification (× 100) microscope and the tube formation ability of endothelial cells was observed.
THP-1 cells were induced with PMA for 48 h and the culture medium was discarded following three cycles of washing with PBS. Trypsin (0.5 mL) was supplied to each well for digestion, and 1.5 mL of culture medium containing 10% FBS was utilized to terminate the digestion. The cells were then collected and centrifuged at 1000 r/min for 5 min. The supernatant was discarded, and the cells were resuspended in PBS and centrifuged again to obtain the precipitate. Mouse anti-human CD68 monoclonal antibody (5 μL; BD Biosciences) and rabbit anti-human CD163 monoclonal antibody (5 μL; BD Biosciences) were added and incubated at 4 °C in the dark for 30 min. Following two cycles of washing with PBS, the cells were centrifuged at 1000 r/min for 5 min, resuspended in 300 μL PBS, and loaded on a flow cytometer (Beckman Coulter, CA, United States) for determination.
After cells were cultured for 48 h, the medium was collected and centrifuged at 1000 r/min for 5 min. An enzyme-linked immunosorbent assay (ELISA) kit (MULTI SCIENCES, Hangzhou, China) was used to detect the expression of interleukin (IL)-6, TGF-β1, and VEGF in the culture medium. IL-6 antibody, TGF-β1 antibody, VEGF antibody, and horseradish peroxidase-labeled streptavidin were all diluted 1:100.
AllPrep RNA/LncRNA kit (Qiagen, Hilden, Germany) was used to extract total RNA from exosomes. The purity of RNA was determined using QubitRNA kit (Thermo Fischer Scientific). The RNA concentration was accurately quantified utilizing Qubit. The integrity of RNA was assessed using Agilent 2100.
Exosome samples of HT29 and 1627 cells were initially obtained and total exosomal RNA was extracted. Ovation Solo RNA kit (NugEN) was used for library construction of exosomal lncRNAs. The constructed library was sequenced through the Illumina Hiseq 4000 platform where the double-terminal 250-300 nt transcripts were generated. The raw data in Fastq format were processed using perl to obtain sequencing data. By logging in to the TopHat2 system, the sequenced data transcripts were aligned and analyzed in light of the reference genome. Cufflinks software was used to compare the analyzed data for transcript splicing and the transcriptome was obtained. Quantitative analysis was performed on the transcript. Finally, the transcript data set was retrieved using the RefSeq database as an mRNA data set. Cuffmerge software was used to merge the obtained transcripts after splicing, delete the transcripts with unclear mid-strand direction, and obtain a mRNA transcript data set. LncRNAs were ultimately screened from the combined transcript set.
The target genes of lncRNAs were predicted through cis-acting and trans-acting, including annotating the function of its target gene mRNA. The GO seq software and KOBAS software were used to enrich differential lncRNA target genes and differential mRNAs in the three processes of biological process, cell component, and molecular function. The biological functions involved in differential lncRNA target genes and differential mRNAs were analyzed. KOBAS software was used to enrich the differential lncRNA target genes and differential mRNAs with KEGG pathways. The signal transduction pathways involved in differential lncRNA target genes and differential mRNA were also analyzed.
SPSS 21.0 software was used for statistical analyses. Enumerative data are expressed as percentages and analyzed using the χ2 test or Fisher’s exact test. The enumerative data are expressed as the mean ± SD. The two groups of enumerative data were tested by two independent samples t-tests. One-way analysis of variance was applied for multiple groups of samples. P < 0.05 was considered statistically significant.
We collected and analyzed the clinical data of 10 BRAFV600E mutant and 20 BRAF wild-type CRC patients. In CRC, BRAFV600E mutation was positively correlated with the increase of preoperative serum carbohydrate antigen (CA)19-9 (P < 0.05), and it was correlated with poor tumor tissue differentiation (P < 0.01). However, no correlation was revealed with gender, age, location, mucous tissue, T stage, TNM stage, lymph node metastasis, nerve invasion, vascular tumor thrombus, preoperative carcinoembryonic antigen (CEA) level, or preoperative platelet count of the patients (Table 1).
| Variable | Cases (%) | BRAFV600E mutation | P value | |
| Yes (%) | No (%) | |||
| Gender | ||||
| Male | 18 (60) | 6 (60) | 12 (60) | 0.326 |
| Female | 12 (40) | 4 (40) | 8 (40) | |
| Age (yr) | ||||
| ≥ 65 | 10 (33.3) | 4 (40) | 6 (30) | 0.440 |
| < 65 | 20 (66.7) | 6 (60) | 14 (70) | |
| Location | ||||
| Right colon | 9 (30) | 2 (20) | 7 (35) | 0.345 |
| Left colon | 9 (30) | 5 (50) | 4 (20) | |
| Rectum | 12 (40) | 3 (30) | 9 (45) | |
| Mucous tissue | ||||
| Positive | 6 (20) | 1 (10) | 5 (25) | 0.326 |
| Negative | 24 (80) | 9 (90) | 15 (75) | |
| Differentiated degree | ||||
| High/moderate differentiation | 20 (66.7) | 3 (30) | 17 (85) | 0.005a |
| Poor differentiation | 10 (33.3) | 7 (70) | 3 (15) | |
| T stage | ||||
| T2 | 4 (13.3) | 0 | 4 (20) | 0.211 |
| T3 | 3 (10) | 2 (20) | 1 (5) | |
| T4 | 23 (76.7) | 8 (80) | 15 (75) | |
| TNM stage | ||||
| I + II | 13 (43.3) | 2 (20) | 11 (55) | 0.074 |
| III + IV | 17 (56.7) | 8 (80) | 9 (45) | |
| Lymph node | ||||
| Positive | 17 (56.7) | 8 (80) | 9 (45) | 0.074 |
| Negative | 13 (43.3) | 2 (20) | 11 (55) | |
| Nerve invasion | ||||
| Positive | 7 (23.3) | 4 (40) | 3 (15) | 0.143 |
| Negative | 23 (76.7) | 6 (60) | 17 (85) | |
| Vessel carcinoma embolus | ||||
| Positive | 3 (10) | 1 (10) | 2 (10) | 0.652 |
| Negative | 27 (90) | 9 (90) | 18 (90) | |
| CA19-9 (U/mL) | ||||
| High | 12 (40) | 7 (70) | 5 (25) | 0.024a |
| Normal | 18 (60) | 3 (30) | 15 (75) | |
| CEA (ng/mL) | ||||
| High | 11 (36.7) | 3 (30) | 8 (40) | 0.452 |
| Normal | 19 (63.3) | 7 (70) | 12 (60) | |
| PLT count (× 109/L) | ||||
| High | 12 (40) | 5 (50) | 7 (35) | 0.344 |
| Normal | 18 (60) | 5 (50) | 13 (65) | |
To explore the influence of BRAFV600E mutation on the TME, we identified the formation of tumor blood vessels and lymph vessels in BRAFV600E mutation and BRAF wild-type CRC tissues. Microvascular density (MVD) and microlymphatic vessel density (MLVD) in BRAFV600E mutant CRC tissues were higher than those in BRAF wild-type CRC (P < 0.05) (Figure 1A and Table 2). The number of CD163+ M2 macrophages in BRAFV600E mutant CRC tumor tissue was markedly increased (P < 0.05) (Figure 1B and Table 3), whereas the number of CD68+ M1 macrophages was not significantly different (Figure 1B and Table 3). Additionally, the density of fibroblasts exhibited no significant difference (Figure 1C and Table 4). These results suggest that BRAFV600E mutation promotes the formation of microvessels and microlymphatic vessels in tumor tissues, increases the infiltration of M2 macrophages, and induces an immunosuppressive microenvironment.
| BRAFV600E mutation | MVD | MLVD | ||
| mean ± SD | P value | mean ± SD | P value | |
| + | 38.53 ± 17.11 | 0.030 | 9.67 ± 5.63 | 0.0001 |
| - | 27.54 ± 9.54 | 3.81 ± 2.06 | ||
| BRAFV600E mutation | CD68+ macrophage | CD163+ M2 macrophage | ||
| mean ± SD | P value | mean ± SD | P value | |
| + | 18.43 ± 13.53 | 0.664 | 8.33 ± 5.93 | 0.040 |
| - | 20.96 ± 15.34 | 3.67 ± 3.02 | ||
| BRAFV600E mutation | CAFs | P value | |
| Low expression (cases) | High expression (cases) | ||
| + | 2 | 8 | 1 |
| - | 3 | 17 | |
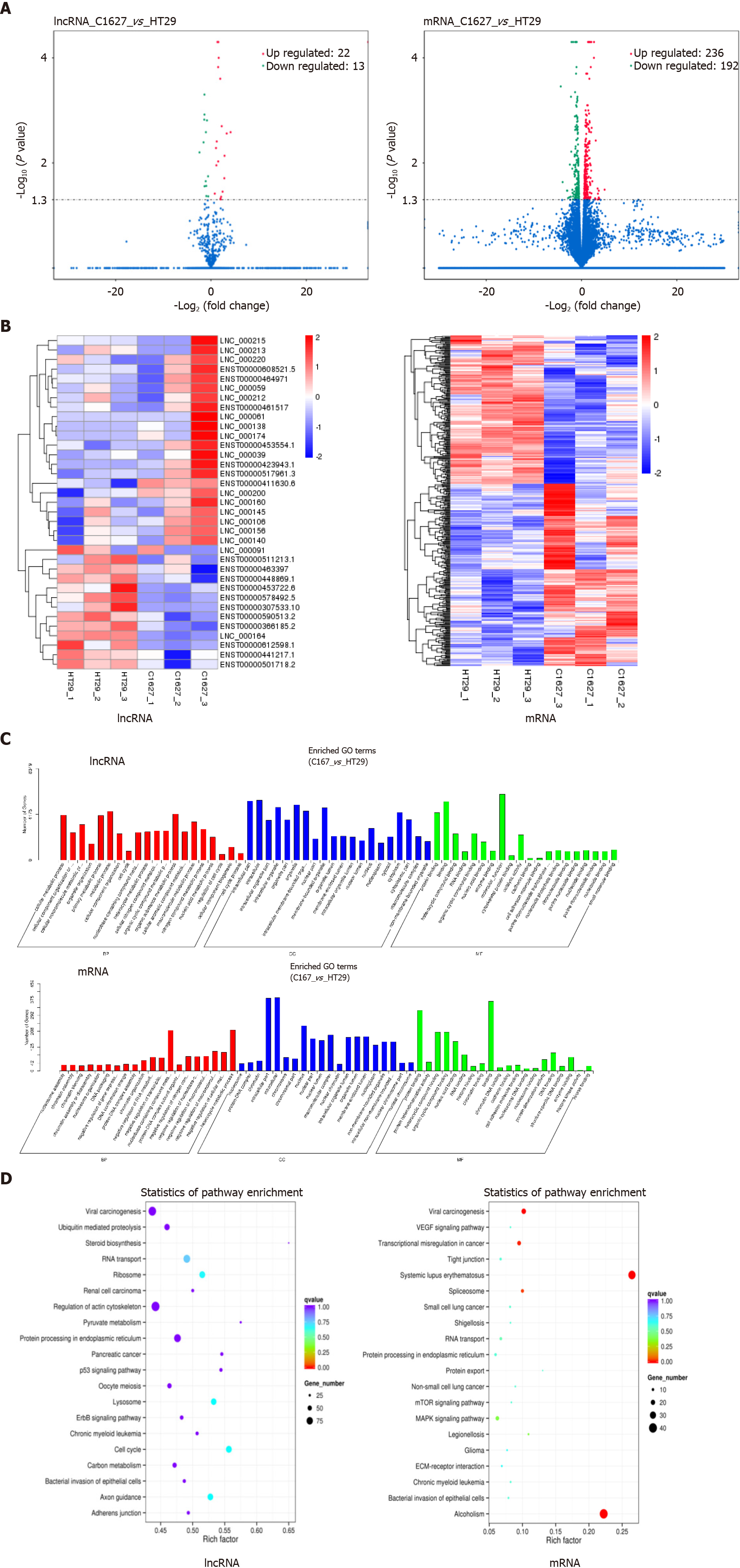
We performed transcriptomics analysis on exosomes derived from BRAFV600E mutant CRC cells and those with BRAF gene silencing. Compared with the exosomes from cells with BRAF gene silencing, the expression of 13 lncRNAs and 192 mRNAs in the exosomes of BRAFV600E mutant CRC cells was upregulated, and the expression of 22 lncRNAs and 236 mRNAs was downregulated (P < 0.05) (Figure 2A). Using cluster analysis charts, the distribution of differentially expressed lncRNAs and mRNAs in exosomes of BRAFV600E mutant CRC cells and those with BRAF gene silencing was further exhibited (Figure 2B).
To illustrate the biological functions of differential lncRNAs and mRNAs and the signaling pathways involved, we conducted GO enrichment analysis and KEGG signaling pathway analysis. GO enrichment analysis included three aspects of biological process, cell component, and molecular function. Differential lncRNA target genes and mRNAs presented similar biological functions, and they are mainly involved in nucleic acid metabolism, macromolecular metabolism, nitride metabolism, and RNA metabolism. Cellular components include mostly the formation of nuclei, organelles, and nucleosomes. Molecular functions involve regulation of cell adhesion, cytoskeletal remodeling, gene expression regulation, and protein binding (Figure 2C).
KEGG results indicated that the target genes of differential lncRNAs were mainly involved in the p53 pathway, ErbB pathway, steroid synthesis pathway, actin cytoskeleton regulation pathway, pyruvate metabolism pathway, cell cycle regulation pathway, and the pathway of protein processing in the endoplasmic reticulum (Figure 2D). Differential mRNAs are mainly involved in the VEGF pathway, mammalian target of rapamycin pathway, mitogen-activated protein kinase pathway, and the pathway of protein processing in the endoplasmic reticulum (Figure 2D). The biological functions and signaling pathways predicted by differential lncRNA target genes and differential mRNAs coincided with each other. Furthermore, it was closely related to angiogenesis, tumor cell proliferation, differentiation, metabolism, and changes in the microenvironment, suggesting that BRAFV600E mutant CRC cells could reach the TME by releasing exosomal lncRNAs, and induce formation of an immunosuppressive microenvironment through mRNAs.
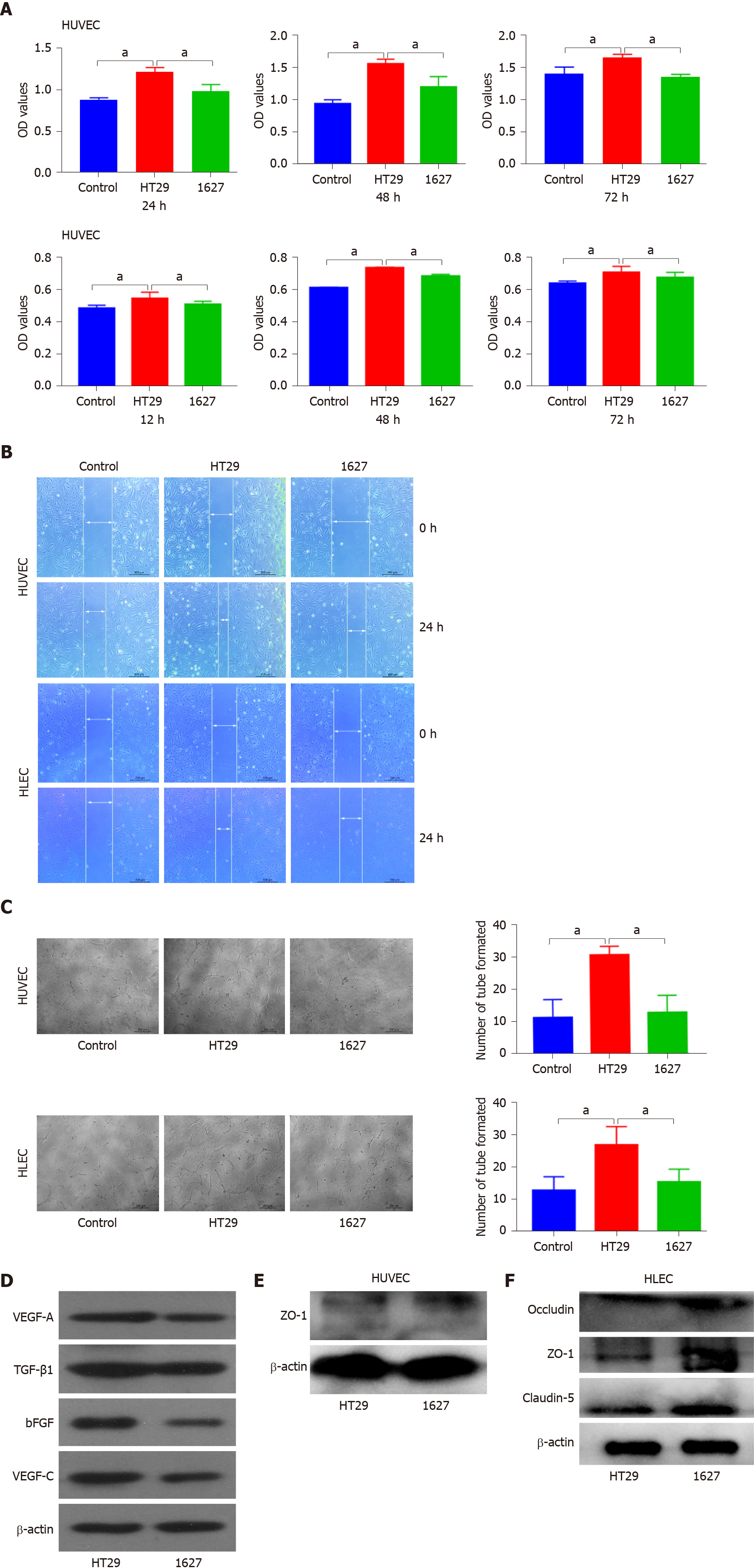
Exosomes derived from HT29 and 1627 cells were cocultured with HUVECs or HLECs to detect cell proliferation. OD values of HUVECs in the HT29 group and 1627 group at 24 h were 1.215 ± 0.032 and 0.986 ± 0.046, respectively; 1.563 ± 0.035 and 1.200 ± 0.163 at 48 h, respectively; and 1.661 ± 0.031 and 1.369 ± 0.020 at 72 h, respectively. At 24, 48 and 72 h, HUVEC proliferation induced by exosomes in the 1627 cell group was reduced compared with the HT29 cell group (P < 0.05), but there was no significant difference in HUVEC proliferation between the 1627 cell group and control group (Figure 2A). The OD values of HLECs in the control group, HT29 cell group, and 1627 cell group at 12 h were 0.477 ± 0.006, 0.526 ± 0.007, and 0.500 ± 0.004, respectively; 0.622 ± 0.003, 0.728 ± 0.010, and 0.680 ± 0.010 at 24 h, respectively; and 0.644 ± 0.006, 0.725 ± 0.009, and 0.682 ± 0.014 at 36 h, respectively. At 12, 24, and 36 h, the proliferation of HLECs induced by exosomes in the 1627 cell group was greatly reduced compared with the HT29 cell group (P < 0.05) and the proliferation of HLECs in the 1627 cell group was greater than that of the control group (P < 0.05) (Figure 3A).
Light microscopy indicated that the average migration rate of HUVECs in the HT29 cell group was 82.863% ± 3.095% and that of the 1627 cell group was 45.067% ± 2.895% at 24 h after scratch. The average migration rate of HLECs in the HT29 cell group was 42.393% ± 0.247%, and that of the 1627 cell group was 23.327% ± 1.434%. Compared with the 1627 cell group, the migration of HUVECs and HLECs induced by exosomes in the HT29 cell group was substantially elevated (P < 0.01) (Figure 3B).
After 24 h of incubation, the average number of tube formations by HUVECs in the HT29 cell group and 1627 cell group was 30.625 ± 0.925 and 12.750 ± 1.887, respectively, at low magnification. The average number of tube formations by HLECs in the HT29 cell group and 1627 cell group was 26.750 ± 2.016 and 15.375 ± 1.413, respectively, at low magnification. Compared with the 1627 cell group, the tube formation ability of HUVECs and HLECs induced by exosomes in the HT29 cell group was markedly higher than that in the 1627 cell group (P < 0.01) (Figure 3C).
Western blot analysis indicated that compared with the HT29 cell group, the expression of VEGF-A, bFGF, TGF-β1, and VEGF-C proteins in the exosomes derived from 1627 cells was reduced (Figure 3D). However, the expression of ZO-1 in HUVECs and that of claudin-5, occludin, and ZO-1 in HLECs in the 1627 cell group were higher than those in the HT29 cell group (Figure 3E and F). Neither the 1627 nor the HT29 group failed to display occludin and claudin-5 protein. This suggests that exosomes derived from BRAFV600E mutant CRC cells promote angiogenesis and lymphoangiogenesis.
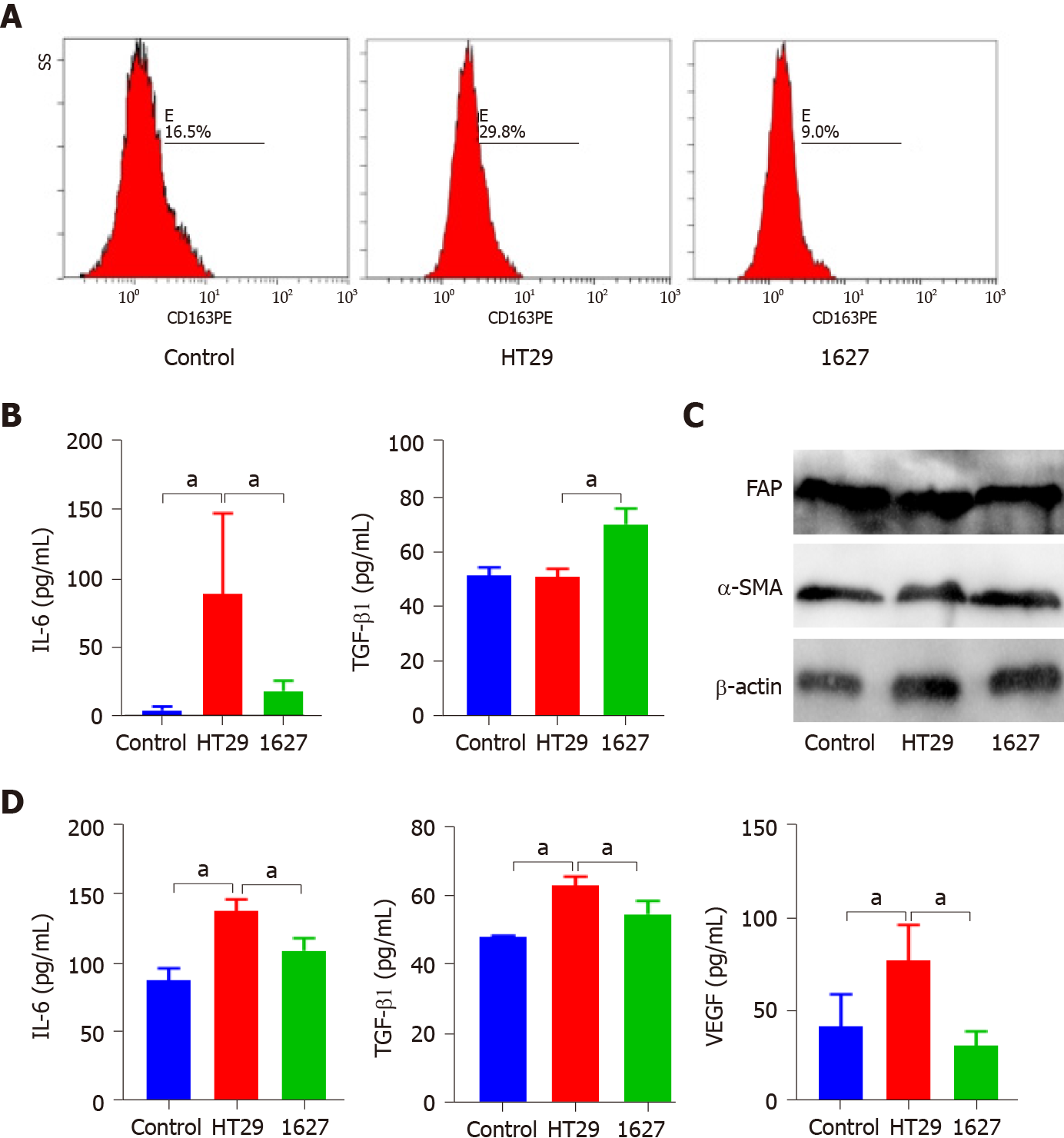
The exosomes derived from HT29 and 1627 cells were cocultured with macrophages. Flow cytometry revealed that the exosomes of the HT29 cell group promoted the expression of CD163 in macrophages compared with the control group and 1627 cell group (P < 0.05) (Figure 4A). ELISA indicated that IL-6 secreted by macrophages in the HT29 cell group was markedly elevated (P < 0.05), whereas TGF-β1 was decreased (P < 0.05) (Figure 4B). Western blot analysis demonstrated that there was no significant difference in the expression of fibroblast FAP and α-SMA in the control, HT29 cell, and 1627 cell groups (Figure 4C). Conversely, the levels of IL-6, TGF-β1, and VEGF secreted by fibroblasts in the 1627 cell group decreased, compared with the HT29 cell group (P < 0.05) (Figure 4D). Exosomes derived from BRAFV600E mutant CRC cells promoted polarization of macrophages to M2 subtype and enhanced the secretory function of fibroblasts.
This study demonstrated that BRAFV600E mutant CRC generates a unique immune microenvironment. Compared with BRAF wild type CRC, there was more angiogenesis and lymphoangiogenesis in the microenvironment. Meanwhile, the polarization of macrophages to M2 subtype was more obvious, and the immunosuppression was more prominent. Exosomes derived from BRAFV600E mutant CRC cells could induce this change. Further analysis indicated that exosomes derived from BRAFV600E mutant CRC cells were rich in certain lncRNAs and mRNAs, which might link to these alterations. Our findings suggested that for this particular CRC, it might be worthwhile to try to investigate the TME.
It is currently argued that BRAFV600E mutation is of vital significance in predicting the prognosis of CRC patients[22]. Several studies have indicated that BRAFV600E mutation is not only correlated to poor tissue differentiation, but also to gender, advanced stage, high T stage, right colon, lymph node metastasis, mucous tissue, and high levels of platelets and CEA[22,23]. Conversely, our results were inconsistent with those reported in the literature. We recognized that BRAFV600E mutation was only associated with poor tissue differentiation and increased preoperative CA19-9 levels, which might have been caused by the small sample size.
However, we analyzed the TME of such patients. The results demonstrated that the MVD and MLVD in BRAFV600E mutant CRC tissues were higher than those in BRAF wild type tissues, and the number of CD163+ M2 macrophages increased substantially. In papillary thyroid carcinoma, BRAFV600E mutation upregulates the expression of VEGF-A in cancer cells and promotes angiogenesis, and increases the expression of VEGF-C in cancer cells and promotes lymphangiogenesis, whereas silencing BRAF gene can reverse these effects[24]. Under normal circumstances, HCT116 colon cancer cells can activate the VEGF signaling pathway, promote the proliferation, migration, and angiogenesis of vascular endothelial cells, and facilitate tumor metastasis by releasing exosomes[25]. Exosomes derived from CRC cells can also promote TAMs to release VEGF-C, enhance proliferation of lymphatic endothelial cells, and induce lymphangiogenesis[26]. However, our results indicate that BRAFV600E mutation promotes angiogenesis and lymphoangiogenesis in the microenvironment.
Simultaneously, BRAFV600E mutation affected cellular components in the microenvironment. We identified that there was no difference in the content of CD68+ macrophages in BRAFV600E mutant and BRAF wild-type CRC tissues, and the number of M2 macrophages was markedly higher than that of BRAF wild-type tissues. This is in agreement with the results reported in the literature. Compared with BRAF wild type tumor, there is no difference in CD68+ macrophages in BRAFV600E mutant thyroid cancer, whereas M2 subtype macrophages increase[27]. Furthermore, TAMs have been positively related to lymph node and peritoneal metastasis in ovarian cancer, gastric cancer, and other tumors[28-30]. CAFs play an essential role in tumor development and metastasis. Our findings indicated no difference in the content of CAFs in CRC tissues of BRAFV600E mutant and BRAF wild-type CRC. We hypothesized that BRAFV600E mutant CRC might promote lymph node and peritoneal metastasis through other mechanisms, but the specific mechanism needs further exploration. This harsh microenvironment may lead to a worse prognosis and even result in resistance to traditional treatments.
Recent research has drawn a link between exosomes and intercellular communi
To explore the effect of exosomes secreted by BRAFV600E mutant cells on the surrounding immune microenvironment, we cocultured exosomes derived from two colon cancer cell lines (HT29 and 1627) with HUVECs or HLECs. The exosomes derived from BRAFV600E mutant CRC cells promoted proliferation, migration, and tube formation of endothelial cells, and induced angiogenesis and lymphoangiogenesis. Silencing BRAF gene generated corresponding inhibitory effects. BRAFV600E mutation can promote the expression of matrix metalloproteinase-2 and VEGF-A in malignant melanoma cells, mediate angiogenesis, and enhance the invasiveness of tumor cells. However, BRAF gene deletion leads to a lack of VEGF-A, inhibiting angiogenesis and minimizing the permeability between endothelial cells[33]. Additional research has also proposed that BRAFV600E mutant thyroid cancer cells can promote angiogenesis and lymphoangiogenesis by releasing VEGF-A and VEGF-C into the TME. Zelboraf can reduce the contents of these factors in the TME and inhibit angiogenesis and lymphoangiogenesis, thereby minimizing distant metastasis.
The expression of VEGF-A, bFGF, TGF-β, and VEGF-C proteins in exosomes derived from BRAFV600E mutant CRC cells was also increased. Tight junction proteins ZO-1, claudin-5, and occludin participate in the formation of the endothelial barrier, affect cell permeability, and play an vital role in regulating the proliferation, migration, and tube formation of endothelial cells[34-36]. This study found that exosomes derived from BRAFV600E mutant CRC cells could inhibit the expression of ZO-1, claudin-5, and occludin in HLECs and ZO-1 in HUVECs. Due to the low concentrations of claudin-5 and occludin in HUVECs, this study failed to detect the effect of silencing BRAF gene on the expression of claudin-5 and occludin proteins in HUVECs. Despite that inhibition of ZO-1, claudin-5, and occludin can restrain the growth of endothelial cells, it may not be enough to resist BRAFV600E mutation for the promotion of vascularization (namely, increased expression of VEGF-A, bFGF, TGF-β, and VEGF-C proteins). Our study suggests that exosomes derived from BRAFV600E mutant CRC cells promote the expression of VEGF-A, bFGF, TGF-β, and VEGF-C to facilitate angiogenesis and lymphangiogenesis.
We cocultured exosomes derived from HT29 and 1627 cells with macrophages and found that exosomes derived from BRAFV600E mutant CRC cells enhanced the polarization of macrophages to M2 subtype. M1 subtype macrophages secrete IL-6, and M2 subtype macrophages secrete TGF-β1[37]. IL-6 is highly expressed in diverse malignant TMEs, and it can promote tumor invasion, distant metastasis, and angiogenesis, and participate in tumor resistance[38]. Exosomes derived from BRAFV600E mutant colon cancer cells can promote the secretion of IL-6 by macrophages and after silencing the BRAF gene, exosomes can inhibit the secretion of IL-6 by macrophages. Some researchers have discovered that exosomes secreted by hepatoma promote the secretion of IL-6 by macrophages, whereas exosomes secreted by melatonin-treated hepatocellular carcinoma can inhibit the secretion of IL-6 by macrophages[39]. TGF-β1 inhibits tumor proliferation and induces apoptosis in the early stage of tumor development. Conversely, when in the advanced stage, it promotes the development and metastasis of the tumor by promoting epithelial–mesenchymal transition, regulating the microenvironment and the immune system[40,41]. Some studies have found that BRAFV600E mutant CRC-derived exosomes can inhibit macrophages from secreting more TGF-β1. Therefore, we speculated that exosomes derived from BRAFV600E mutant CRC cells could promote the polarization of macrophages to M2 subtype, increase the secretion of IL-6, and reduce the secretion of TGF-β1, thereby facilitating distant metastasis.
The cell component with the highest content in the microenvironment is CAFs, which highly express α-SMA and FAP, and secrete IL-6, TGF-β1, and VEGF. Meanwhile, they assist tumor cells in immune escape, and promote angiogenesis, tumor invasion, and metastasis[42,43]. The present study indicated that after exosomes derived from BRAFV600E mutant CRC cells were cocultured with fibroblasts, the expression of α-SMA and FAP in fibroblasts did not change markedly. Instead, they promoted the secretion of IL-6 and TGF- β1 and VEGF by CAFs. It was suggested that BRAFV600E mutation had little effect on the number of CAFs, mainly affecting their function. Some researchers have found that exosomes derived from hepatocellular carcinoma can promote the differentiation of hepatic astrocytes into CAFs. The activated CAFs secrete cytokines VEGF, TGF-β, and IL-6, and promote angiogenesis and liver metastasis[44,45].
We selected only one BRAFV600E mutant CRC cell line, and only performed in vitro experiments. The differentially expressed lncRNAs were not verified. Nevertheless, our research demonstrated that BRAFV600E mutant CRC had a unique immune microenvironment, which might be induced by the release of exosomes rich in certain lncRNAs. Therefore, in the future, we can consider to reshape the immune microenvironment, combined with traditional treatment, to treat this specific type of CRC.
Our study showed that, compared with wild type BRAF, BRAFV600E mutation led to more angiogenesis and lymphoangiogenesis in the microenvironment. Meanwhile, the polarization of macrophages to M2 subtype was more obvious, and the immunosuppression was more prominent. Further analysis indicated that exosomes derived from BRAFV600E mutant CRC cells were rich in certain lncRNAs and mRNAs, which might be linked to these alterations. This provides a hypothesis for finding new therapeutic strategies for BRAFV600E mutant CRC.
BRAFV600E gene mutation accounts for approximately 10% of patients with metastatic colorectal cancer (CRC). Compared with CRC patients with wild-type BRAF, patients with BRAFV600E mutant CRC are prone to peritoneal metastasis and distant lymph node metastasis with a poor prognosis. Previous findings suggest that BRAFV600E mutation affects the tumor microenvironment (TME).
BRAFV600E mutation is involved in the formation of the immunosuppressive microenvironment in thyroid cancer. However, the influence and the related mechanism of BRAFV600E mutation in CRC on the surrounding immune microenvironment are not clear.
The study aimed to determine the influence of BRAFV600E mutation in CRC on the surrounding immune microenvironment, elucidating whether BRAFV600E mutant CRC cell-derived exosomes participate in the formation of an immunosuppressive microenvironment.
CRC patients were divided into either a control group or a treatment group. The formation of microvessels and microlymphatic vessels and M2 subtype macrophages in tumor tissues were detected by immunohistochemistry. Screening and functional analysis of exosomal long noncoding RNAs (lncRNAs) were performed by transcriptomics. The proliferation and migration of human umbilical vein endothelial cells (HUVECs) and human lymphatic endothelial cells (HLECs) were detected by CCK-8 assays and scratch test, respectively. The tube-forming ability of endothelial cells was assessed by tube formation assay. The macrophage subtypes were obtained by flow cytometry. The expression of vascular endothelial growth factor (VEGF)-A, basic fibroblast growth factor (bFGF), transforming growth factor (TGF)-β1, VEGF-C, claudin-5, occludin, zonula occludens (ZO)-1, fibroblast activation protein (FAP), and α-smooth muscle actin was assessed by Western blot analysis. The levels of cytokines interleukin (IL)-6, TGF-β1, and VEGF were assessed by ELISA.
BRAFV600E mutation was positively correlated with a poor prognosis in CRC (P < 0.01). Microvascular density and microlymphatic vessel density in BRAFV600E mutant CRC tissues were higher than those in BRAF wild-type CRC (P < 0.05). The number of CD163+ M2 macrophages in BRAFV600E mutant CRC tumor tissue was markedly increased (P < 0.05). Compared with exosomes from CRC cells with BRAF gene silencing, the expression of 13 lncRNAs and 192 mRNAs in the BRAFV600E mutant CRC cell exosomes was upregulated, and the expression of 22 lncRNAs and 236 mRNAs was downregulated (P < 0.05). The biological functions and signaling pathways predicted by differential lncRNA target genes and differential mRNA were closely related to angiogenesis, tumor cell proliferation, differentiation, metabolism, and changes in the microenvironment. The proliferation, migration, and tube formation ability of HUVECs and HLECs induced by exosomes in the 1627 cell group (HT29 cells with BRAF gene silencing) was greatly reduced compared with the HT29 cell group (P < 0.05). Compared with the HT29 cell group, the expression levels of VEGF-A, bFGF, TGF-β1, and VEGF-C in the exosomes derived from 1627 cells were reduced. The expression of ZO-1 in HUVECs, and claudin-5, occludin, and ZO-1 in HLECs of the 1627 cell group was higher. Compared with the 1627 cell group, the exosomes of the HT29 group promoted the expression of CD163 in macrophages (P < 0.05). IL-6 secretion by macrophages in the HT29 cell group was markedly elevated (P < 0.05), whereas TGF-β1 was decreased (P < 0.05). The levels of IL-6, TGF-β1, and VEGF secreted by fibroblasts in the 1627 cell group decreased, compared with the HT29 group (P < 0.05).
BRAFV600E mutant CRC cells can reach the TME by releasing exosomal lncRNAs, inducing the formation of an immunosuppressive microenvironment.
The study will provide a novel therapeutic strategy for BRAFV600E mutant CRC.
Provenance and peer review: Unsolicited article; Externally peer reviewed.
Specialty type: Oncology
Country/Territory of origin: China
Peer-review report’s scientific quality classification
Grade A (Excellent): 0
Grade B (Very good): B
Grade C (Good): 0
Grade D (Fair): 0
Grade E (Poor): 0
P-Reviewer: Saber A S-Editor: Gao CC L-Editor: Wang TQ P-Editor: Gao CC
| 1. | Siegel RL, Miller KD, Jemal A. Cancer statistics, 2019. CA Cancer J Clin. 2019;69:7-34. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 13300] [Cited by in RCA: 15468] [Article Influence: 2578.0] [Reference Citation Analysis (2)] |
| 2. | Zhou J, Zheng R, Zhang S, Zeng H, Wang S, Chen R, Sun K, Li M, Gu J, Zhuang G, Wei W. Colorectal cancer burden and trends: Comparison between China and major burden countries in the world. Chin J Cancer Res. 2021;33:1-10. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 17] [Cited by in RCA: 61] [Article Influence: 15.3] [Reference Citation Analysis (0)] |
| 3. | Fiskus W, Mitsiades N. B-Raf Inhibition in the Clinic: Present and Future. Annu Rev Med. 2016;67:29-43. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 48] [Cited by in RCA: 52] [Article Influence: 5.8] [Reference Citation Analysis (0)] |
| 4. | Ursem C, Atreya CE, Van Loon K. Emerging treatment options for BRAF-mutant colorectal cancer. Gastrointest Cancer. 2018;8:13-23. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 21] [Cited by in RCA: 31] [Article Influence: 4.4] [Reference Citation Analysis (0)] |
| 5. | Taieb J, Lapeyre-Prost A, Laurent Puig P, Zaanan A. Exploring the best treatment options for BRAF-mutant metastatic colon cancer. Br J Cancer. 2019;121:434-442. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 47] [Cited by in RCA: 60] [Article Influence: 10.0] [Reference Citation Analysis (0)] |
| 6. | Davies H, Bignell GR, Cox C, Stephens P, Edkins S, Clegg S, Teague J, Woffendin H, Garnett MJ, Bottomley W, Davis N, Dicks E, Ewing R, Floyd Y, Gray K, Hall S, Hawes R, Hughes J, Kosmidou V, Menzies A, Mould C, Parker A, Stevens C, Watt S, Hooper S, Wilson R, Jayatilake H, Gusterson BA, Cooper C, Shipley J, Hargrave D, Pritchard-Jones K, Maitland N, Chenevix-Trench G, Riggins GJ, Bigner DD, Palmieri G, Cossu A, Flanagan A, Nicholson A, Ho JW, Leung SY, Yuen ST, Weber BL, Seigler HF, Darrow TL, Paterson H, Marais R, Marshall CJ, Wooster R, Stratton MR, Futreal PA. Mutations of the BRAF gene in human cancer. Nature. 2002;417:949-954. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 7459] [Cited by in RCA: 7634] [Article Influence: 331.9] [Reference Citation Analysis (0)] |
| 7. | Fedorenko IV, Paraiso KH, Smalley KS. Acquired and intrinsic BRAF inhibitor resistance in BRAF V600E mutant melanoma. Biochem Pharmacol. 2011;82:201-209. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 127] [Cited by in RCA: 136] [Article Influence: 9.7] [Reference Citation Analysis (0)] |
| 8. | Bhatt KV, Spofford LS, Aram G, McMullen M, Pumiglia K, Aplin AE. Adhesion control of cyclin D1 and p27Kip1 Levels is deregulated in melanoma cells through BRAF-MEK-ERK signaling. Oncogene. 2005;24:3459-3471. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 113] [Cited by in RCA: 109] [Article Influence: 5.5] [Reference Citation Analysis (0)] |
| 9. | Jin Z, Sinicrope FA. Advances in the therapy of BRAFV600E metastatic colorectal cancer. Expert Rev Anticancer Ther. 2019;19:823-829. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 3] [Cited by in RCA: 2] [Article Influence: 0.3] [Reference Citation Analysis (0)] |
| 10. | Biller LH, Schrag D. Diagnosis and Treatment of Metastatic Colorectal Cancer: A Review. JAMA. 2021;325:669-685. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 398] [Cited by in RCA: 1425] [Article Influence: 356.3] [Reference Citation Analysis (0)] |
| 11. | Wong V, Lee M, Wong R, Tie J, Shapiro J, Desai J, Nott L, Steel S, Burge M, Ma B, Khattak A, Hong W, Gibbs P. BRAFV600E Mutations Arising from a Left-Side Primary in Metastatic Colorectal Cancer: Are They a Distinct Subset? Target Oncol. 2021;16:227-236. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 2] [Cited by in RCA: 2] [Article Influence: 0.5] [Reference Citation Analysis (0)] |
| 12. | Galon J, Bruni D. Tumor Immunology and Tumor Evolution: Intertwined Histories. Immunity. 2020;52:55-81. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 211] [Cited by in RCA: 382] [Article Influence: 76.4] [Reference Citation Analysis (0)] |
| 13. | Means C, Clayburgh DR, Maloney L, Sauer D, Taylor MH, Shindo ML, Coussens LM, Tsujikawa T. Tumor immune microenvironment characteristics of papillary thyroid carcinoma are associated with histopathological aggressiveness and BRAF mutation status. Head Neck. 2019;41:2636-2646. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 12] [Cited by in RCA: 20] [Article Influence: 3.3] [Reference Citation Analysis (0)] |
| 14. | Denton AE, Roberts EW, Fearon DT. Stromal Cells in the Tumor Microenvironment. Adv Exp Med Biol. 2018;1060:99-114. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 122] [Cited by in RCA: 194] [Article Influence: 27.7] [Reference Citation Analysis (0)] |
| 15. | Erreni M, Mantovani A, Allavena P. Tumor-associated Macrophages (TAM) and Inflammation in Colorectal Cancer. Cancer Microenviron. 2011;4:141-154. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 225] [Cited by in RCA: 262] [Article Influence: 17.5] [Reference Citation Analysis (1)] |
| 16. | Herrera M, Herrera A, Domínguez G, Silva J, García V, García JM, Gómez I, Soldevilla B, Muñoz C, Provencio M, Campos-Martin Y, García de Herreros A, Casal I, Bonilla F, Peña C. Cancer-associated fibroblast and M2 macrophage markers together predict outcome in colorectal cancer patients. Cancer Sci. 2013;104:437-444. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 187] [Cited by in RCA: 244] [Article Influence: 20.3] [Reference Citation Analysis (0)] |
| 17. | Ping Q, Yan R, Cheng X, Wang W, Zhong Y, Hou Z, Shi Y, Wang C, Li R. Cancer-associated fibroblasts: overview, progress, challenges, and directions. Cancer Gene Ther. 2021;28:984-999. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 84] [Cited by in RCA: 174] [Article Influence: 43.5] [Reference Citation Analysis (0)] |
| 18. | Bracci L, Lozupone F, Parolini I. The role of exosomes in colorectal cancer disease progression and response to therapy. Cytokine Growth Factor Rev. 2020;51:84-91. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 13] [Cited by in RCA: 18] [Article Influence: 3.6] [Reference Citation Analysis (0)] |
| 19. | Galamb O, Barták BK, Kalmár A, Nagy ZB, Szigeti KA, Tulassay Z, Igaz P, Molnár B. Diagnostic and prognostic potential of tissue and circulating long non-coding RNAs in colorectal tumors. World J Gastroenterol. 2019;25:5026-5048. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in CrossRef: 83] [Cited by in RCA: 83] [Article Influence: 13.8] [Reference Citation Analysis (0)] |
| 20. | Sun Z, Liu J, Chen C, Zhou Q, Yang S, Wang G, Song J, Li Z, Zhang Z, Xu J, Sun X, Chang Y, Yuan W. The Biological Effect and Clinical Application of Long Noncoding RNAs in Colorectal Cancer. Cell Physiol Biochem. 2018;46:431-441. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 23] [Cited by in RCA: 28] [Article Influence: 4.0] [Reference Citation Analysis (0)] |
| 21. | Liang ZX, Liu HS, Wang FW, Xiong L, Zhou C, Hu T, He XW, Wu XJ, Xie D, Wu XR, Lan P. LncRNA RPPH1 promotes colorectal cancer metastasis by interacting with TUBB3 and by promoting exosomes-mediated macrophage M2 polarization. Cell Death Dis. 2019;10:829. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 125] [Cited by in RCA: 260] [Article Influence: 43.3] [Reference Citation Analysis (0)] |
| 22. | Wang J, Shen J, Huang C, Cao M, Shen L. Clinicopathological Significance of BRAFV600E Mutation in Colorectal Cancer: An Updated Meta-Analysis. J Cancer. 2019;10:2332-2341. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 17] [Cited by in RCA: 26] [Article Influence: 4.3] [Reference Citation Analysis (0)] |
| 23. | Caputo F, Santini C, Bardasi C, Cerma K, Casadei-Gardini A, Spallanzani A, Andrikou K, Cascinu S, Gelsomino F. BRAF-Mutated Colorectal Cancer: Clinical and Molecular Insights. Int J Mol Sci. 2019;20. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 76] [Cited by in RCA: 108] [Article Influence: 18.0] [Reference Citation Analysis (0)] |
| 24. | Husain A, Hu N, Sadow PM, Nucera C. Expression of angiogenic switch, cachexia and inflammation factors at the crossroad in undifferentiated thyroid carcinoma with BRAF(V600E). Cancer Lett. 2016;380:577-585. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 9] [Cited by in RCA: 11] [Article Influence: 1.1] [Reference Citation Analysis (0)] |
| 25. | Hu HY, Yu CH, Zhang HH, Zhang SZ, Yu WY, Yang Y, Chen Q. Exosomal miR-1229 derived from colorectal cancer cells promotes angiogenesis by targeting HIPK2. Int J Biol Macromol. 2019;132:470-477. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 98] [Cited by in RCA: 94] [Article Influence: 15.7] [Reference Citation Analysis (0)] |
| 26. | Sun B, Zhou Y, Fang Y, Li Z, Gu X, Xiang J. Colorectal cancer exosomes induce lymphatic network remodeling in lymph nodes. Int J Cancer. 2019;145:1648-1659. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 45] [Cited by in RCA: 73] [Article Influence: 12.2] [Reference Citation Analysis (0)] |
| 27. | Angell TE, Lechner MG, Jang JK, Correa AJ, LoPresti JS, Epstein AL. BRAF V600E in papillary thyroid carcinoma is associated with increased programmed death ligand 1 expression and suppressive immune cell infiltration. Thyroid. 2014;24:1385-1393. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 94] [Cited by in RCA: 103] [Article Influence: 9.4] [Reference Citation Analysis (0)] |
| 28. | Wang R, Zhang T, Ma Z, Wang Y, Cheng Z, Xu H, Li W, Wang X. The interaction of coagulation factor XII and monocyte/macrophages mediating peritoneal metastasis of epithelial ovarian cancer. Gynecol Oncol. 2010;117:460-466. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 27] [Cited by in RCA: 27] [Article Influence: 1.8] [Reference Citation Analysis (0)] |
| 29. | Yamagata Y, Tomioka H, Sakamoto K, Sato K, Harada H, Ikeda T, Kayamori K. CD163-Positive Macrophages Within the Tumor Stroma Are Associated With Lymphangiogenesis and Lymph Node Metastasis in Oral Squamous Cell Carcinoma. J Oral Maxillofac Surg. 2017;75:2144-2153. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 32] [Cited by in RCA: 46] [Article Influence: 5.8] [Reference Citation Analysis (0)] |
| 30. | Go Y, Tanaka H, Tokumoto M, Sakurai K, Toyokawa T, Kubo N, Muguruma K, Maeda K, Ohira M, Hirakawa K. Tumor-Associated Macrophages Extend Along Lymphatic Flow in the Pre-metastatic Lymph Nodes of Human Gastric Cancer. Ann Surg Oncol. 2016;23 Suppl 2:S230-S235. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 17] [Cited by in RCA: 21] [Article Influence: 2.1] [Reference Citation Analysis (0)] |
| 31. | Kok VC, Yu CC. Cancer-Derived Exosomes: Their Role in Cancer Biology and Biomarker Development. Int J Nanomedicine. 2020;15:8019-8036. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 300] [Cited by in RCA: 293] [Article Influence: 58.6] [Reference Citation Analysis (0)] |
| 32. | Gao T, Liu X, He B, Nie Z, Zhu C, Zhang P, Wang S. Exosomal lncRNA 91H is associated with poor development in colorectal cancer by modifying HNRNPK expression. Cancer Cell Int. 2018;18:11. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 61] [Cited by in RCA: 91] [Article Influence: 13.0] [Reference Citation Analysis (0)] |
| 33. | Declercq M, Treps L. BRAF, A gatekeeper controlling endothelial permeability. FEBS J. 2019;286:2273-2276. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 2] [Cited by in RCA: 3] [Article Influence: 0.5] [Reference Citation Analysis (0)] |
| 34. | Tornavaca O, Chia M, Dufton N, Almagro LO, Conway DE, Randi AM, Schwartz MA, Matter K, Balda MS. ZO-1 controls endothelial adherens junctions, cell-cell tension, angiogenesis, and barrier formation. J Cell Biol. 2015;208:821-838. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 308] [Cited by in RCA: 418] [Article Influence: 41.8] [Reference Citation Analysis (0)] |
| 35. | Chidiac R, Zhang Y, Tessier S, Faubert D, Delisle C, Gratton JP. Comparative Phosphoproteomics Analysis of VEGF and Angiopoietin-1 Signaling Reveals ZO-1 as a Critical Regulator of Endothelial Cell Proliferation. Mol Cell Proteomics. 2016;15:1511-1525. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 13] [Cited by in RCA: 21] [Article Influence: 2.3] [Reference Citation Analysis (0)] |
| 36. | Zhang H, Zhang S, Zhang J, Liu D, Wei J, Fang W, Zhao W, Chen Y, Shang D. ZO-1 expression is suppressed by GM-CSF via miR-96/ERG in brain microvascular endothelial cells. J Cereb Blood Flow Metab. 2018;38:809-822. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 22] [Cited by in RCA: 27] [Article Influence: 3.9] [Reference Citation Analysis (0)] |
| 37. | Xu YW, Xing RX, Zhang WH, Li L, Wu Y, Hu J, Wang C, Luo QL, Shen JL, Chen X. Toxoplasma ROP16I/III ameliorated inflammatory bowel diseases via inducing M2 phenotype of macrophages. World J Gastroenterol. 2019;25:6634-6652. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in CrossRef: 13] [Cited by in RCA: 27] [Article Influence: 4.5] [Reference Citation Analysis (0)] |
| 38. | Choy EH, De Benedetti F, Takeuchi T, Hashizume M, John MR, Kishimoto T. Translating IL-6 biology into effective treatments. Nat Rev Rheumatol. 2020;16:335-345. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 201] [Cited by in RCA: 437] [Article Influence: 87.4] [Reference Citation Analysis (0)] |
| 39. | Cheng L, Liu J, Liu Q, Liu Y, Fan L, Wang F, Yu H, Li Y, Bu L, Li X, Wei W, Wang H, Sun G. Exosomes from Melatonin Treated Hepatocellularcarcinoma Cells Alter the Immunosupression Status through STAT3 Pathway in Macrophages. Int J Biol Sci. 2017;13:723-734. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 63] [Cited by in RCA: 91] [Article Influence: 11.4] [Reference Citation Analysis (0)] |
| 40. | Liu S, Ren J, Ten Dijke P. Targeting TGFβ signal transduction for cancer therapy. Signal Transduct Target Ther. 2021;6:8. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 132] [Cited by in RCA: 239] [Article Influence: 59.8] [Reference Citation Analysis (0)] |
| 41. | Derynck R, Turley SJ, Akhurst RJ. TGFβ biology in cancer progression and immunotherapy. Nat Rev Clin Oncol. 2021;18:9-34. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 188] [Cited by in RCA: 587] [Article Influence: 117.4] [Reference Citation Analysis (0)] |
| 42. | Piersma B, Hayward MK, Weaver VM. Fibrosis and cancer: A strained relationship. Biochim Biophys Acta Rev Cancer. 2020;1873:188356. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 147] [Cited by in RCA: 408] [Article Influence: 81.6] [Reference Citation Analysis (0)] |
| 43. | Lim H, Koh M, Jin H, Bae M, Lee SY, Kim KM, Jung J, Kim HJ, Park SY, Kim HS, Moon WK, Hwang S, Cho NH, Moon A. Cancer-associated fibroblasts induce an aggressive phenotypic shift in non-malignant breast epithelial cells via interleukin-8 and S100A8. J Cell Physiol. 2021;236:7014-7032. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 15] [Cited by in RCA: 15] [Article Influence: 3.8] [Reference Citation Analysis (0)] |
| 44. | Zhou Y, Ren H, Dai B, Li J, Shang L, Huang J, Shi X. Hepatocellular carcinoma-derived exosomal miRNA-21 contributes to tumor progression by converting hepatocyte stellate cells to cancer-associated fibroblasts. J Exp Clin Cancer Res. 2018;37:324. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 329] [Cited by in RCA: 353] [Article Influence: 50.4] [Reference Citation Analysis (0)] |
| 45. | Fang T, Lv H, Lv G, Li T, Wang C, Han Q, Yu L, Su B, Guo L, Huang S, Cao D, Tang L, Tang S, Wu M, Yang W, Wang H. Tumor-derived exosomal miR-1247-3p induces cancer-associated fibroblast activation to foster lung metastasis of liver cancer. Nat Commun. 2018;9:191. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 407] [Cited by in RCA: 747] [Article Influence: 106.7] [Reference Citation Analysis (0)] |