Published online Dec 15, 2021. doi: 10.4251/wjgo.v13.i12.2101
Peer-review started: May 11, 2021
First decision: June 12, 2021
Revised: June 24, 2021
Accepted: September 14, 2021
Article in press: September 14, 2021
Published online: December 15, 2021
Processing time: 217 Days and 14.1 Hours
Colorectal cancer (CRC) ranks third in terms of incidence and second in mortality worldwide. In CRC, the silencing of mismatch repair genes, including the mutL homolog 1 (hMLH1) has been linked to microsatellite instability (MSI), the lengthening or shortening of microsatellite repeats. Very limited data have been presented so far on the link of hMLH1 methylation and MSI in Southeast Asia populations with sporadic CRC, and on its clinical significance.
To investigate the significance of the MSI status and hMLH1 methylation in CRC Filipino patients.
Fifty-four sporadic CRC patients with complete clinical data were included in this study. Genomic DNA from CRC tumor biopsies and their normal tissue counterparts were profiled for MSI by high resolution melting (HRM) analysis using the Bethesda Panel of Markers (BAT25, BAT26, D2S123, D5S346, and D17S250). hMLH1 methylation screening was performed using bisulfite conversion and methylation specific polymerase chain reaction. Statistical analysis was conducted to calculate their associations to clinicopathological characteristics and survival relevance (Kaplan-Meier curves and the log-rank test).
hMLH1 methylation was observed in 9% and 35% of CRC and normal samples, respectively. Higher incidence of consistently methylated hMLH1 found in both normal and CRC was noticed for relation to location of tumor (P < 0.05). As for MSI status, D2S123 the most common unstable microsatellite and MSI-high (MSI-H) was the most common MSI profile, counted for 46% and 50% of normal and CRC tissues, respectively. The presence of MSI-low (MSI-L) and microsatellite stable (MSS) was 43% and 11% for normal, and 31% and 19% for CRC samples. The mean month of patients’ survival was shorter in patients whose normal and tumor tissues had methylated compared to those with unmethylated hMLH1 and with MSI-H compared to those with MSI-L/MSS (P < 0.05). This was supported by significant difference in Kaplan-Meier with log-rank analysis. This data indicated that hMLH1 methylation and high MSI status have prognostic value.
This study showed the clinical significance of hMLH1 methylation and MSI status in sporadic CRC Filipino patients, especially in the normal part of the tumor.
Core Tip: Colorectal cancer (CRC) ranks third in terms of incidence and second in mortality worldwide. In CRC, the silencing of mismatch repair genes, including the mutL homolog 1 (hMLH1) has been linked to microsatellite instability (MSI). This study investigated the status of hMLH1 methylation and MSI in normal and tumor tissues of Filipinos sporadic CRC patients and their clinical significances.
- Citation: Cabral LKD, Mapua CA, Natividad FF, Sukowati CHC, Cortez ER, Enriquez MLD. MutL homolog 1 methylation and microsatellite instability in sporadic colorectal tumors among Filipinos. World J Gastrointest Oncol 2021; 13(12): 2101-2113
- URL: https://www.wjgnet.com/1948-5204/full/v13/i12/2101.htm
- DOI: https://dx.doi.org/10.4251/wjgo.v13.i12.2101
Colorectal cancer (CRC) occurs when malignant tumors form in the lining of the large intestine, which includes the ascending, transverse, and descending colon, and the rectum. CRC ranks third in terms of incidence and second in mortality worldwide, for both sexes. Over 1.9 million new CRC cases and 935000 deaths were estimated to occur in 2020, accounting for about 1/10 cancer cases and deaths[1]. In the Philippines, CRC is currently the third leading site of malignancy. The incidence of CRC had almost doubled from 2010 to 2015 with survival rates under 50%[2,3]. In terms of mortality rate, Filipinos (and Chinese) ethnicity had significantly decreased risk of death compared with Caucasians[4].
CRC, like many other cancers, is a malignancy caused by DNA changes causing abnormal behavior of cells. DNA deletions, duplications, substitutions, mutations, and rearrangements can either activate or inactivate a gene or several genes and con
In an inherited CRC, the hereditary non-polyposis CRC (HNPCC), germline mutation is involved in one or two of the mutator or mismatch repair (MMR) genes, including the mutL homolog 1 (hMLH1). Mutations in hMLH1 have been linked to microsatellite instability (MSI), the lengthening or shortening of microsatellite repeats (sequences with 1-6 repeating nucleotides). When instability is left unrepaired, it may accumulate as mutations and can affect other genes that have microsatellite repeats in their coding regions[8,9]. MSI is found in 85%-90% of HNPCC patients[10].
Recent studies demonstrated that in a subset of sporadic CRC, DNA methylation, the transfer of a covalent methyl group to the C5 position of the cytosine to form 5-methylcytosine by DNA methyltransferases, may cause the loss of function of a MMR gene. Likewise, it leads to the accumulation of MSI[11,12]. Tumors with methylated hMLH1 and high levels of MSI present with a distinct characteristic from other CRC tumors. CRC with methylated hMLH1 had a delayed onset and was associated with female gender[13]. This linked hMLH1 and MSI in the development of sporadic CRC, suggesting that the two molecular profiles are closely related[14].
So far, very limited data have been presented on the link of hMLH1 methylation and MSI in Southeast Asia populations with sporadic CRC. This study presents the detection and characterization of MSI status and hMLH1 methylation in CRC and its paired non-tumoral adjacent tissues in Filipino patients.
Fifty-four sporadic CRC patients with complete clinical data were included in this study. Diagnosis of CRC was based on the presence of malignancy in the initial biopsy. Patients should be Filipino by descent with no family history of cancer. From each patient, paired tumor and its corresponding normal tissue were obtained from surgical resection at the St. Luke’s Medical Center, Quezon City, Philippines. Normal tissues were collected approximately 6 inches away from the margin of the tumor. Upon pathological confirmation, fresh frozen sections were stored in -80°C. The project of the Colorectal Cancer Study Group was approved by the Institutional Ethics Review Board of St. Luke’s Medical Center (Project Code No. 06-015). All patients enrolled in the study signed an Informed Consent Form allowing the use of their tissues and clinical data in the Colorectal Cancer (CRC) Databank of St. Luke’s Medical Center.
CRC cell lines SW480 (ATCC® CCL-228) and SW48 (ATCC® CCL-23) were purchased from American Type Culture Collection (ATCC) as controls for MSI and hMLH1 methylation. SW480 is a CRC cell line that has stable microsatellite and unmethylated hMLH1, while SW48 is high MSI and methylated hMLH1. Lymphocytes from patients who underwent colonoscopy and who were found to be free of cancer and polyp were used as additional controls for stable microsatellite.
Genomic DNA (gDNA) extraction was performed using the QIAamp® DNA Mini Kit (Qiagen), according to the manufacturer's instructions. Briefly, tissues were finely minced and put in lysis solution and proteinase K until completely lysed. After DNA precipitation with ethanol, DNA extract was washed twice and eluted with appro
gDNA from normal and tumor specimens was subjected to bisulfite treatment to differentiate methylated cytosines from unmethylated ones. Bisulfite chemically modifies non-methylated cytosines into uracil, which is then converted to thymidine in polymerase chain reaction (PCR) cycles.
Bisulfite treatment of the DNA sample was performed using EZ DNA Methylation Lightning Kit (Zymo Research) according to the manufacturer’s suggestion. Briefly, 200-500 ng gDNA was incubated in the conversion reagent and then treated with binding buffer in a spin column. Converted DNA was then subjected to desulphonation and clean-up using washing buffer. DNA (approximately 10 μL) was eluted and collected for methylation specific PCR (MS-PCR)[15]. Reaction was carried out using Qiagen Taq Core Kit in a reaction volume of 25 μL with 10x PCR Buffer, 2.5 mmol/L of MgCl2, 50 pmol of primer, 1.25 mmol/L of dNTPs and 1.25 units of Taq DNA polymerase added to 1.5-2 μL of converted DNA. Primer sets used for hMLH1 MS-PCR are taken from published work of Fox et al[15].
MSI test was carried out using PCR for five markers from the Bethesda panel that included BAT25, BAT26, D2S123, D5S346, and D17S250, as in a previous study[16]. PCR conditions of each marker were optimized and validated in our earlier study on HNPCC (Evangelista & Enriquez, unpublished work).
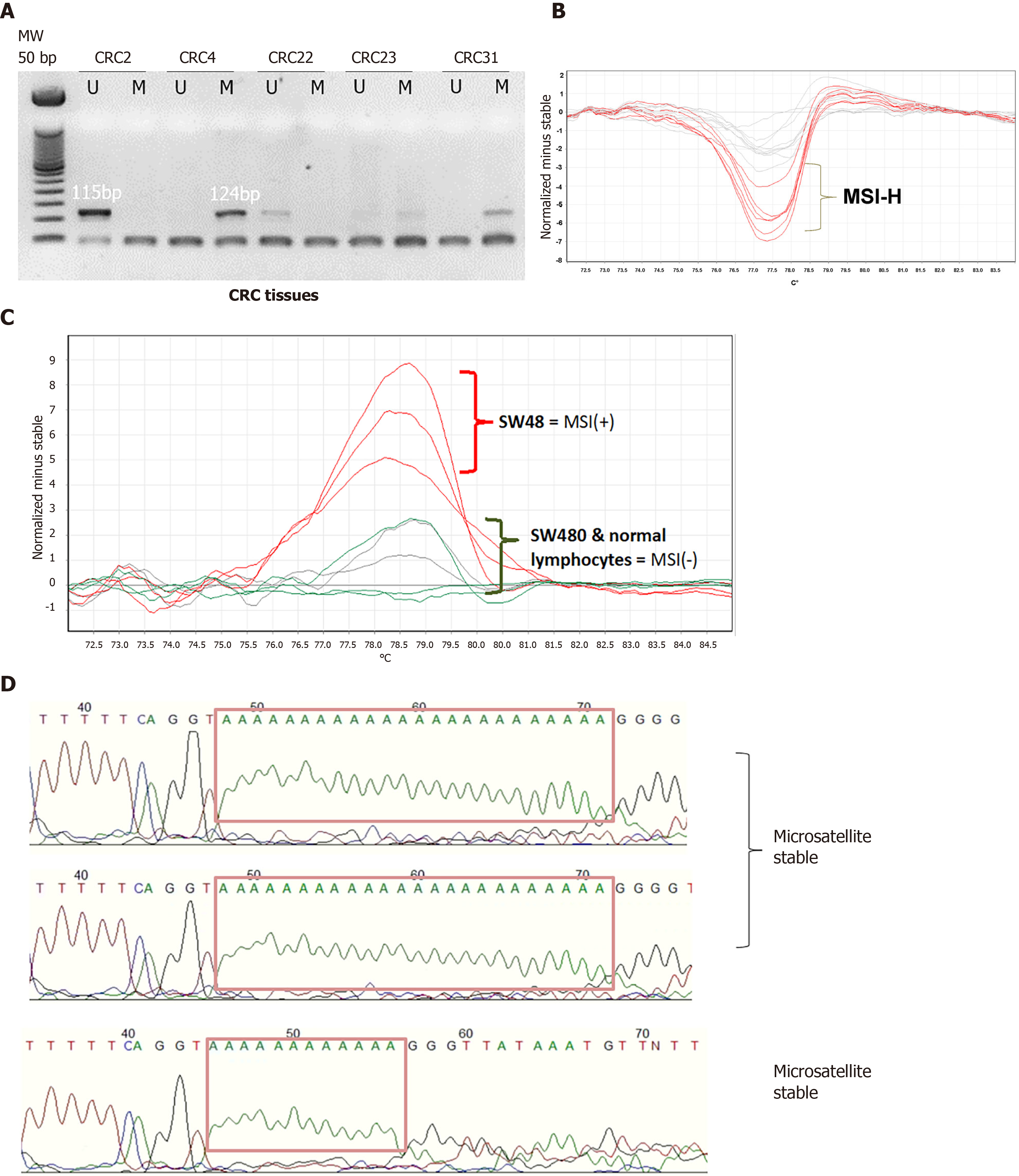
In brief, PCR was performed using Qiagen Taq PCR core kit with EvaGreen dye (Biotium) in a 25 μL reaction volume containing 50 ng of gDNA. All reactions were done in triplicates. PCR and high resolution melting (HRM) analysis were carried out using Rotor-Gene™ 6000 (Qiagen) system with data collected over the range from 55 °C to 95 °C with ramp rising at 0.1 °C/s[17]. Melting curve was analyzed using Rotor-Gene Q (RGQ) Scanning Software version 2.0.2 (Qiagen). Raw melting-curve data were normalized by manual adjustment of linear regions before (pre-; 100% fluorescence) and after (post-; 0% fluorescence) the melting transition. The melting curve of DNA from SW480 cells which exhibits microsatellite stable (MSS) phenotype[18] was used as the normal/stable control. The RGQ software assigned the profile of each sample in reference to the stable control. The confidence percentage (cut-off), was optimized by analyzing non-cancer patients’ DNA, with values not lower than 60%. Therefore, the confidence value of ≥ 60% was regarded as MSS, while any confidence value of < 60% was regarded as unstable.
MSI score was defined as MSI-high (MSI-H) where HRM instability was observed in ≥ 2 markers; MSI-low (MSI-L) where instability was only in one marker; and MSS if no instability was observed in any of the markers. For validation, amplified PCR products of the stable and unstable control were subjected to Sanger sequencing to verify their microsatellite repeats.
Statistical analysis was constructed using software GraphPrism version 5.01 (GraphPad Software, Inc., La Jolla, CA, United States). Associations between clini
This study analyzed 54 Filipino CRC patients with comparable male to female ratio (30M:24F). The age mean was 56.9 ± 11.8 years old, with 5 patients under 40 years old and 49 patients above 40 years old. Regarding tumor parameters, CRC was mostly found in the distal part of the colon (38; 70%), followed by the rectum (10; 19%) and proximal part of the colon (6; 11%). By histology, moderately-differentiated grade CRC was noticed in 44 (81%), poorly-differentiated in 8 (15%), and well-differentiated in 2 (4%) patients. By disease stage, stage 1 CRC was observed in 6 (11%), stage 2 in 14 (26%), stage 3 in 30 (56%), and stage in 4 (7%) of patients. None of the patients had family history of CRC.
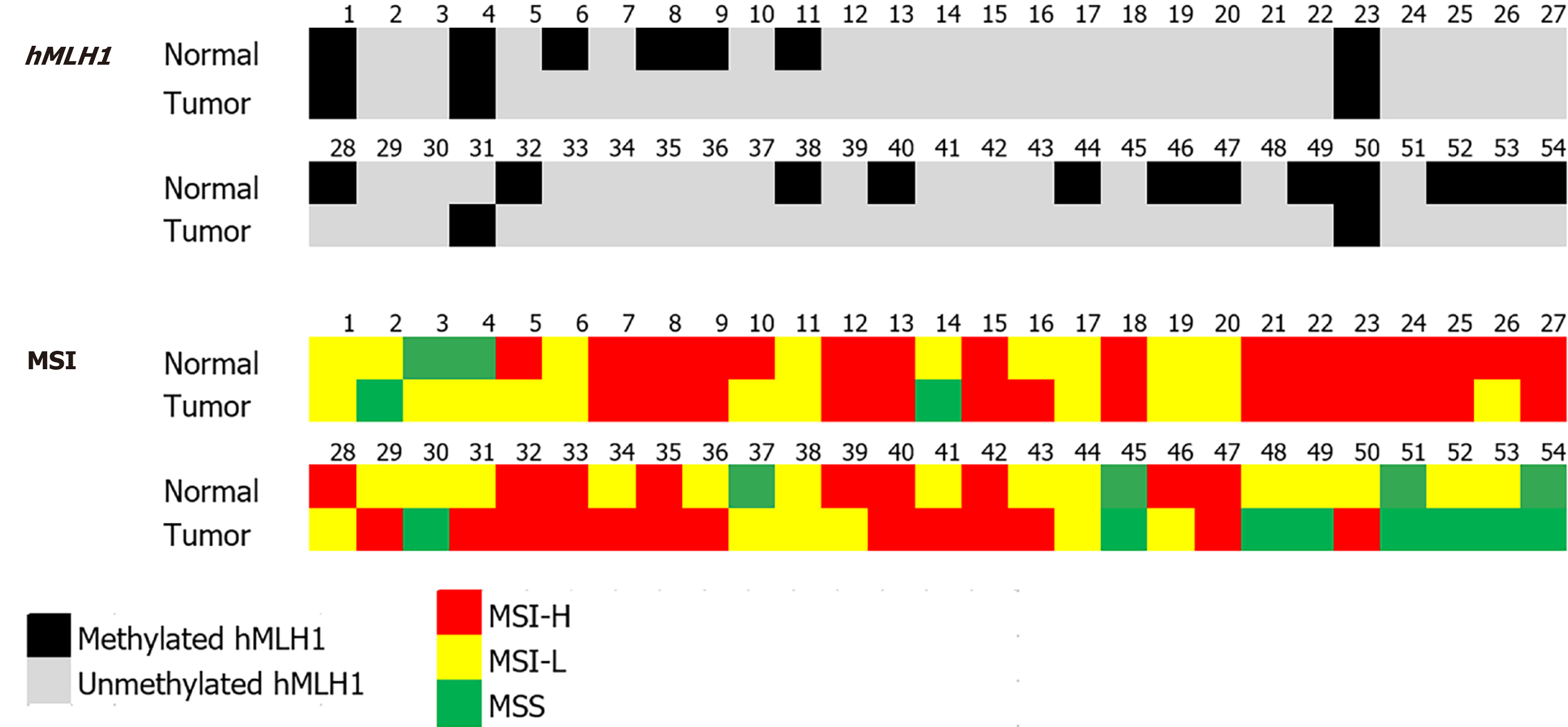
We analyzed the hMLH1 DNA methylation by MS-PCR. From 54 paired CRC and its normal adjacent tissue, most of the samples were unmethylated (91% and 65%, respectively). Representative MS-PCR gel electrophoresis is shown in Figure 1. hMLH1 methylation was noticed only in 5 (9%) and 19 (35%) of CRC and normal samples, respectively. It is interesting to note that the methylated hMLH1 in tumor tissues was accompanied by methylation in its paired normal tissues (4/5, 80%). Only 1 sample showed tumoral hMLH1 methylation without methylation in its normal tissue (Figure 2).
The distribution of hMLH1 methylation status in patients’ demography and tumor parameters is shown in Table 1 (left panel). In normal tissues, the incidence of hMLH1 methylation was slightly higher in female compared to male (42% vs 30%), and in tumors located in the proximal colon compared to distal/rectum location (67% vs 31%). The same pattern was found for hMLH1 methylation in tumor tissues. Methylation was higher in females compared to males (13% vs 7%), and in tumors located in the proximal colon compared to those in distal colon or rectum (33% vs 6%, P = 0.031).
| Normal tissue (%) | Tumor tissue(%) | Normal/tumor (%) | |||||||||
| M | U | M | U | M/M | U/U | M/U or U/M | |||||
| Freq | 19 | 35 | P value | 5 | 49 | P value | 4 | 34 | 16 | P value | |
| Gender | |||||||||||
| Male | 30 | 9 (30) | 21 (70) | NS | 2 (7) | 28 (93) | NS1 | 1 (3) | 20 (67) | 9 (30) | NS |
| Female | 24 | 10 (42) | 14 (58) | 3 (12) | 21 (88) | 3 (13) | 14 (58) | 7 (29) | |||
| Age | |||||||||||
| ≤ 40 | 5 | 3 (60) | 2 (40) | NS1 | 0 (0) | 5 (100) | NS1 | 0 (0) | 2 (40) | 3 (60) | NS |
| > 40 | 49 | 16 (33) | 33 (67) | 5 (10) | 44 (90) | 4 (8) | 32 (65) | 13 (27) | |||
| Location of tumor | |||||||||||
| Proximal | 6 | 4 (67) | 2 (33) | NS1 | 2 (33) | 4 (67) | 0.031 | 2 (33) | 2 (33) | 2 (33) | 0.029 |
| Distal/rectum | 48 | 15 (31) | 33 (69) | 3 (6) | 45 (94) | 2 (4) | 32 (67) | 14 (29) | |||
| Tumor grade | |||||||||||
| Poor | 8 | 5 (62) | 3 (38) | NS | 1 (12) | 7 (88) | NS | 1 (12) | 3 (38) | 4 (50) | NS |
| Moderate | 44 | 13 (30) | 31 (70) | 4 (9) | 40 (91) | 3 (7) | 30 (68) | 11 (25) | |||
| Well | 2 | 1 (50) | 1 (50) | 0 (0) | 2 (100) | 0 (0) | 1 (50) | 1 (50) | |||
| Tumor stage | |||||||||||
| I-II | 20 | 6 (30) | 14 (70) | NS | 2 (10) | 18 (90) | NS1 | 2 (10) | 14 (70) | 4 (20) | NS |
| III-IV | 34 | 13 (38) | 21 (62) | 3 (9) | 31 (91) | 2 (6) | 20 (59) | 12 (35) | |||
The signature of hMLH1 status in paired normal and tumor tissue is presented in Table 1 (right panel). Although the incidence was not statistically significant, a higher incidence of methylated hMLH1 in both normal and tumor tissues (M/M) was noticed in females, in > 40 years old patients, and in poorly differentiated cancer. A significant difference was found for the tumor location (P = 0.029), where M/M signature was noticed in 33% of proximal tumors, while U/U was in 67% of distally located or rectal tumors.
MSI status was assessed using HRM of the Bethesda panel that included BAT25, BAT26, D2S123, D5S346, and D17S250. As controls, cell lines SW480 and SW48 were used to represent a MSS and MSI-H, respectively. MSS feature was also checked in lymphocytes DNA from a normal individual. A representative HRM analysis of BAT26 gene and its direct Sanger sequencing are shown in Figure 1.
From the Bethesda panel markers, the microsatellite with the highest instability was D2S123 and this was found in 32 (59%) and 33 (61%) samples of normal and tumor, respectively. On the other hand, BAT26 instability was observed only in 1 (2%) sample of each normal and tumor tissues. In normal tissues, the instability of BAT25, D17S250, and D5S346, were 2 (4%), 14 (26%), and 29 (54%), respectively. Among the CRC tissues, instability was found in BAT25, BAT26, D17S250, and D5S346 accounting for 5 (20%), 12 (22%), and 25 (46%), respectively. There was no statistical difference in the instability of these markers between normal and tumor tissues.
The distribution of MSI status in patients’ demography and tumor parameter is shown in Table 2 (left panel). In normal tissue samples, 25 (46%), 23 (43%), and 6 (11%) samples were MSI-H, MSI-L, and MSS, respectively. In female, the highest percentage was for MSI-H (54%), followed by MSI-L (33%), and MSS (13%). Similar pattern was observed in tumoral tissues with the following rates: MSI-H with 27 (50%), MSI-L with 17 (31%), and MSS with 10 (19%). Instability rates of microsatellites in tumors tissues in the female group were the exactly the same as in the normal tissues.
| Normal tissue (%) | Tumor tissue (%) | Normal/tumor (%) | |||||||||||||
| MSI-H | MSI-L | MSS | P value | MSI-H | MSI-L | MSS | P value | H/H | L/L | S/S | L/S or S/L | L/H or H/L | P value | ||
| Freq | 25 | 23 | 6 | 27 | 17 | 10 | 19 | 8 | 3 | 10 | 14 | ||||
| Gender | |||||||||||||||
| Male | 30 | 12 (40) | 15 (50) | 3 (10) | NS | 14 (47) | 9 (30) | 7 (23) | NS | 9 (30) | 4 (13) | 1 (3) | 8 (27) | 8 (27) | NS |
| Female | 24 | 13 (54) | 8 (33) | 3 (13) | 13 (54) | 8 (33) | 3 (13) | 10 (42) | 4 (17) | 2 (8) | 2 (8) | 6 (25) | |||
| Age | |||||||||||||||
| ≤ 40 | 5 | 3 (60) | 2 (40) | 0 (0) | NS | 2 (40) | 1 (20) | 2 (40) | NS | 2 (40) | 0 (0) | 0 (0) | 2 (40) | 1 (20) | NS |
| > 40 | 49 | 22 (45) | 21 (43) | 6 (12) | 25 (51) | 16 (33) | 8 (16) | 17 (35) | 8 (16) | 3 (6) | 8 (16) | 13 (27) | |||
| Location of tumor | |||||||||||||||
| Proximal | 6 | 4 (67) | 2 (33) | 0 (0) | NS | 4 (67) | 2 (33) | 0 (0) | NS | 4 (67) | 2 (33) | 0 (0) | 0 (0) | 0 (0) | NS |
| Distal/rectum | 48 | 21 (44) | 21 (44) | 6 (12) | 23 (48) | 15 (31) | 10 (21) | 15 (31) | 6 (13) | 3 (6) | 10 (21) | 14 (29) | |||
| Tumor grade | |||||||||||||||
| Poor | 8 | 5 (62) | 3 (38) | 0 (0) | NS | 5 (63) | 2 (25) | 1 (12) | NS | 4 (50) | 1 (13) | 0 (0) | 1 (13) | 2 (25) | NS |
| Moderate | 44 | 19 (43) | 19 (43) | 6 (14) | 21 (48) | 15 (34) | 8 (18) | 14 (32) | 7 (16) | 3 (7) | 8 (18) | 12 (27) | |||
| Well | 2 | 1 (50) | 1 (50) | 0 (0) | 1 (50) | 0 (0) | 1 (50) | 1 (50) | 0 (0) | 0 (0) | 1 (50) | 0 (0) | NS | ||
| Tumor stage | |||||||||||||||
| I-II | 20 | 9 (45) | 9 (45) | 2 (10) | NS | 8 (40) | 10 (50) | 2 (10) | NS | 7 (35) | 6 (30) | 0 (0) | 4 (20) | 3 (15) | NS |
| III-IV | 34 | 16 (47) | 14 (41) | 4 (12) | 19 (56) | 7 (21) | 8 (23) | 12 (35) | 2 (6) | 3 (9) | 6 (18) | 11(32) | |||
The signature of MSI status in paired normal and tumor tissue is presented in Table 2 (right panel). Analysis of data using showed that the MSI status between normal and tumor tissues was not statistically significant. Nonetheless, a slightly higher incidence of MSI-H in hMLH1 was observed in both normal and tumor tissues (H/H) among females, similar to methylated hMLH1.
Table 3 shows the MSI and methylation status of each sample according to the paired groupings described previously. There was no statistically significant correlation between MSI and hMLH1 methylation status of all the sample pairs. Thirteen paired samples (24%) were unmethylated with MSI-H (H/H and U/U). The clinicopathological profile of this cancer group showed that 11 samples came from the distal colon/rectum (85%), 12 (92%) were > 40 years old, 10 (77%) were moderately differentiated, and 8 (62%) were stage III/IV cancers.
| hMLH1 methylation (%) | |||||||
| M/M | M/U | U/M | U/U | Total | P value | ||
| MSI status | H/H | 1 (5) | 5 (26) | 0 (0) | 13 (68) | 19 | NS |
| L/L | 1 (13) | 4 (50) | 0 (0) | 3 (37) | 8 | ||
| S/S | 0 (0) | 1 (33) | 0 (0) | 2 (67) | 3 | ||
| L/S or S/L | 1 (10) | 3 (30) | 0 (0) | 6 (60) | 10 | ||
| L/H or H/L | 1 (7) | 2 (14) | 1 (7) | 10 (71) | 14 | ||
| Total | 4 (7) | 15 (28) | 1 (2) | 34 (63) | 54 | ||
To determine the prognostic values of both hMLH1 methylation and MSI status, we performed Kaplan-Meier analysis on patient survival for independent groups. The survival was defined as death event (in months) post-surgery. Patients were followed up until 60 mo after surgery.
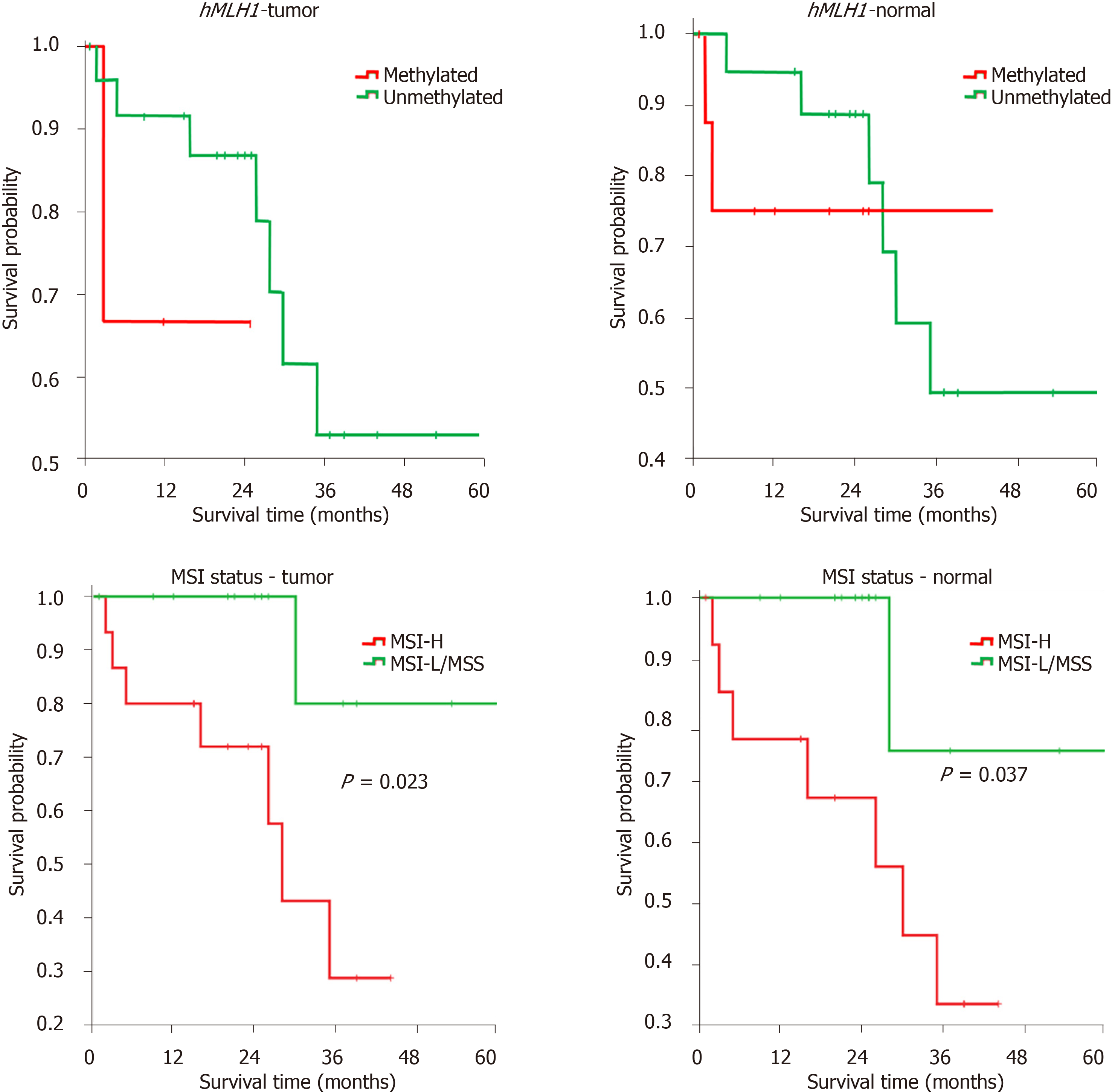
Our results show that the mean survival was relatively shorter in patients with methylated compared to those with unmethylated hMLH1, both in tumor [18 (6-29) mo vs 44 (34-54) mo] and in normal tissues [34 (21-46) mo vs 44 (33-54) mo], expressed as mean (95% confidence interval). A significant difference was also observed in tumor tissues with MSI-H compared to those with MSI-L/MSS [27 (19-36) mo vs 56 (44-67) mo]. For the MSI status in normal tissues, again a shorter survival was found for MSI-H compared to MSI-L/MSS [27 (18-36) mo vs 54 (39-68) mo].
Kaplan-Meier analysis was performed to associate the hMLH1 methylation and MSI status with patients’ survival in both cohorts. In concordance with the mean survival above, we found the association of MSI-H status in both normal and tumor tissues with shorter probability of survival (P < 0.05), compared to that of MSI-L/MSS (Figure 3). Our data indicated that hMLH1 methylation and high MSI status might have a prognostic value.
CRC is a classic example of disease progression accompanied by molecular changes. Its carcinogenesis is composed of stage-specific molecular changes in gene expression and the accumulation of mutations. A mutation in the MMR genes may lead to accu
DNA methylation has been linked to tumor suppressor gene silencing[19], including the silencing of hMLH1 MMR gene[20]. In a subset of sporadic CRC, the loss of function of hMLH1 gene led to the accumulation of instability in microsatellite regions of the DNA[11,12].
In this study, there was a notable higher incidence of methylation of hMLH1 in normal tissue as compared to its paired tumor tissue (35% vs 9%). This is in agreement with other studies showing elevated hMLH1 methylation in the normal gastric epithelia in patients with stomach cancer as compared to those of non-cancer patients[21]. Methylation in the normal mucosa was proposed as a marker of risk for the development of CRC[22]. As demonstrated recently, the acceleration of ageing-related DNA methylation drift in normal mucosa was correlated with an earlier age of diagnosis[23].
hMLH1 methylation in non-neoplastic epithelia was shown also to exhibit MSI[24]. It is interesting to note that the methylated hMLH1 in tumor was accompanied by methylation in its paired normal tissue. This finding warrants further investigation on the clinical utility of hMLH1 methylation in normal tissue as a useful marker not only in CRC but in also in other gastrointestinal cancers.
However, in this study, a high incidence of non-methylated hMLH1 in MSI-H samples (both normal and tumor) suggests that hMLH1methylation did not bring about MSI. We assumed, at least in our cohort, that unstable microsatellites can be attributed to methylation in other MMR genes, such as hSMH2[25,26] and mutation in hSMH6[27]. It was also noted that MSI may also result from other defects in the component of base excision repair of the proteins (DNA glycosylase, AP endonuclease, DNA polymerase, etc.)[28]. MSI tumors were also grouped to Type A (≤ 6 bp change) and Type B (≥ 8 bp change). In a report on MMR gene knock-out animals, no Type B MSI was observed, suggesting that a kind of MSI group may involve malignancies apart from MMR deficiency[29].
Significant differences for hMLH1 methylation were found for the location of the tumor. Colon is divided into two anatomical regions: the proximal/right colon and the distal/left colon and each with very unique features[30-32]. This may be partially attributed to their difference in embryological development and physiological circumstances. The proximal colon develops from the hindgut while the distal colon originates from the midgut[33]. Proximal and distal colon are also different in the genetic patterns[34].
Since methylation is affected by environmental exposure, the difference might also be related to function. The proximal colon is the site where breakdown of complex carbohydrates occurs while the undigested dietary proteins are broken down in the distal colon. This process of digestion in the distal colon generates by-products such as ammonia, phenols, indoles and sulfurs, that may inhibit DNA methyltransferase activity[35]. Among African-American race, higher vegetable intake diet was shown being associated with greater odds for high hMLH1 methylation[36]. Because methylation is less inhibited in proximal region, this may explain the higher incidence of methylated normal and tumor tissue (M/M), in contrast to high percentage of unmethylation in distal region.
In this study, we observed that from both the average of survival data and log-rank test of Kaplan-Meier analysis, hMLH1 methylation in normal tissues and MSI status in both normal and CRC tissues had prognostic value. Unmethylated hMLH1 and MSI-L/MSS status were positively associated with better patients’ survival. Our data may support earlier findings where expression of hMLH1 is considered an independent prognostic and predictive factor stage II-III CRC in Chinese population[37]. Data from this study, however, in contrast with previous studies showing the association between MSI-H with improved overall and disease-free survival[38,39], showed that population genetics and the disease stages might influence the prognosis of CRC patients.
Although there was no statistical difference, the incidence of MSI and hMLH1 methylation showed a similar trend in association with sex and grade. MSI and hMLH1 methylation were more common in females and in poorly differentiated tumors. This non-significant correlation may be attributed to the small sample size, thus studies using larger number of samples that are well distributed in terms of tumor clinicopathological parameters will be needed. Moreover, significance of both markers in relation to patients' prognosis, especially for Southeast Asian population, needs to be validated. Investigation of the methylation status of two or more MMR genes and their correlation with MSI status is also recommended, since this study only looked into one MMR gene out of the six. A recent data from metastatic CRC showed that a novel epigenetic signature of eight hypermethylated genes was able to identify CRC with poor prognosis. These genes were characterized with CpG-island high methylator and MSI-like phenotype[40].
The establishment of molecular biomarkers in diseases is essential for an effective management and treatment protocol for cancer patients. For the precision treatment in the future, especially for immunotherapy, it was shown that patients with MSI tumors exhibited significant response to anti-PD-1 inhibitors after failure of conventional therapy[41,42]. The role of MSI and hMLH1 methylation is shedding light in the management of sporadic CRC in clinical practice in other countries[43,44] but not yet in the Philippines.
To conclude, we showed the clinical significance of hMLH1 methylation and MSI status in sporadic CRC Filipino patients, especially in the normal part of the organ. This study is one of the few attempts to establish the molecular profile of Filipino CRC patients in the local setting, to highlight the importance of epigenetic modification. Understanding the diverse molecular and genetic key players involved in cancer development will ultimately translate to improvement in patient care.
A distinct molecular signature marks a particular subset of sporadic colorectal cancer (CRC). It involves the mismatch repair (MMR) genes silencing due to DNA methylation, leading to microsatellite instability (MSI).
To improve the management of the CRC patients based on their distinct molecular subtypes.
To examine the association of mutL homolog 1 (hMLH1) methylation (MMR gene) and the MSI phenotype in relation to cancer characteristics and patient survival among Filipino sporadic CRC patients.
Paired tissues (normal and tumor) from sporadic CRC patients was screened for hMLH1 methylation using methylation specific polymerase chain reaction. Subsequent MSI typing was done by high resolution melting analysis.
The results of this study showed that hMLH1 methylation was mostly noticed in proximal tumors. Low overall survival was observed in methylated hMLH1 and MSI tumors.
The epigenetic silencing of hMLH1 as well as MSI may present a distinct pattern of CRC in Filipino patients.
This is an initial attempt to characterize sporadic CRC in Filipino population. It may shed light in understanding molecular epigenetic modification in CRC as well as its role in tumor development and management.
The members of the Colorectal Cancer Study Group of the St. Luke's Medical Center actively and consistently participated in the study.
Provenance and peer review: Invited article; Externally peer reviewed.
Corresponding Author's Membership in Professional Societies: The Philippine Society for Cell Biology.
Specialty type: Oncology
Country/Territory of origin: Philippines
Peer-review report’s scientific quality classification
Grade A (Excellent): 0
Grade B (Very good): 0
Grade C (Good): C
Grade D (Fair): 0
Grade E (Poor): 0
P-Reviewer: Lei XH S-Editor: Gao CC L-Editor: A P-Editor: Yu HG
| 1. | Sung H, Ferlay J, Siegel RL, Laversanne M, Soerjomataram I, Jemal A, Bray F. Global Cancer Statistics 2020: GLOBOCAN Estimates of Incidence and Mortality Worldwide for 36 Cancers in 185 Countries. CA Cancer J Clin. 2021;71:209-249. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 75126] [Cited by in RCA: 64671] [Article Influence: 16167.8] [Reference Citation Analysis (176)] |
| 2. | Ting FIL, Sacdalan DBL, Tampo MMT, Apellido RT, Monroy HJ 3rd, Sacdalan MDP, Sacdalan DL; written on behalf of the University of the Philippines, Philippine General Hospital Colorectal Polyp and Cancer Study Group. Treatment Outcomes of Patients With Colorectal Cancer Enrolled in a Comprehensive Benefits Program of the National Insurance System in the Philippines: Data From the Pilot Site. JCO Glob Oncol. 2020;6:35-46. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 5] [Cited by in RCA: 10] [Article Influence: 2.0] [Reference Citation Analysis (0)] |
| 3. | Laudico AV, Mirasol-Lumague MR, Mapua CA, Uy GB, Toral JA, Medina VM, Pukkala E. Cancer incidence and survival in Metro Manila and Rizal province, Philippines. Jpn J Clin Oncol. 2010;40:603-612. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 20] [Cited by in RCA: 22] [Article Influence: 1.5] [Reference Citation Analysis (0)] |
| 4. | Le H, Ziogas A, Taylor TH, Lipkin SM, Zell JA. Survival of distinct Asian groups among colorectal cancer cases in California. Cancer. 2009;115:259-270. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 14] [Cited by in RCA: 17] [Article Influence: 1.1] [Reference Citation Analysis (0)] |
| 5. | Srivastava S, Verma M, Henson DE. Biomarkers for early detection of colon cancer. Clin Cancer Res. 2001;7:1118-1126. [PubMed] |
| 6. | Mármol I, Sánchez-de-Diego C, Pradilla Dieste A, Cerrada E, Rodriguez Yoldi MJ. Colorectal Carcinoma: A General Overview and Future Perspectives in Colorectal Cancer. Int J Mol Sci. 2017;18. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 520] [Cited by in RCA: 907] [Article Influence: 113.4] [Reference Citation Analysis (2)] |
| 7. | Fearon ER, Vogelstein B. A genetic model for colorectal tumorigenesis. Cell. 1990;61:759-767. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 8087] [Cited by in RCA: 8006] [Article Influence: 228.7] [Reference Citation Analysis (1)] |
| 8. | Potter JD. Colorectal cancer: molecules and populations. J Natl Cancer Inst. 1999;91:916-932. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 573] [Cited by in RCA: 542] [Article Influence: 20.8] [Reference Citation Analysis (0)] |
| 9. | Imai K, Yamamoto H. Carcinogenesis and microsatellite instability: the interrelationship between genetics and epigenetics. Carcinogenesis. 2008;29:673-680. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 300] [Cited by in RCA: 324] [Article Influence: 19.1] [Reference Citation Analysis (0)] |
| 10. | Bianchi F, Galizia E, Catalani R, Belvederesi L, Ferretti C, Corradini F, Cellerino R. CAT25 is a mononucleotide marker to identify HNPCC patients. J Mol Diagn. 2009;11:248-252. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 32] [Cited by in RCA: 34] [Article Influence: 2.1] [Reference Citation Analysis (0)] |
| 11. | Berg KD, Glaser CL, Thompson RE, Hamilton SR, Griffin CA, Eshleman JR. Detection of microsatellite instability by fluorescence multiplex polymerase chain reaction. J Mol Diagn. 2000;2:20-28. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 114] [Cited by in RCA: 119] [Article Influence: 4.8] [Reference Citation Analysis (0)] |
| 12. | Samowitz WS, Curtin K, Ma KN, Schaffer D, Coleman LW, Leppert M, Slattery ML. Microsatellite instability in sporadic colon cancer is associated with an improved prognosis at the population level. Cancer Epidemiol Biomarkers Prev. 2001;10:917-923. [PubMed] |
| 13. | Malkhosyan SR, Yamamoto H, Piao Z, Perucho M. Late onset and high incidence of colon cancer of the mutator phenotype with hypermethylated hMLH1 gene in women. Gastroenterology. 2000;119:598. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 50] [Cited by in RCA: 49] [Article Influence: 2.0] [Reference Citation Analysis (0)] |
| 14. | Poynter JN, Siegmund KD, Weisenberger DJ, Long TI, Thibodeau SN, Lindor N, Young J, Jenkins MA, Hopper JL, Baron JA, Buchanan D, Casey G, Levine AJ, Le Marchand L, Gallinger S, Bapat B, Potter JD, Newcomb PA, Haile RW, Laird PW; Colon Cancer Family Registry Investigators. Molecular characterization of MSI-H colorectal cancer by MLHI promoter methylation, immunohistochemistry, and mismatch repair germline mutation screening. Cancer Epidemiol Biomarkers Prev. 2008;17:3208-3215. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 184] [Cited by in RCA: 190] [Article Influence: 11.2] [Reference Citation Analysis (0)] |
| 15. | Fox EJ, Leahy DT, Geraghty R, Mulcahy HE, Fennelly D, Hyland JM, O'Donoghue DP, Sheahan K. Mutually exclusive promoter hypermethylation patterns of hMLH1 and O6-methylguanine DNA methyltransferase in colorectal cancer. J Mol Diagn. 2006;8:68-75. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 29] [Cited by in RCA: 33] [Article Influence: 1.7] [Reference Citation Analysis (0)] |
| 16. | Loukola A, Eklin K, Laiho P, Salovaara R, Kristo P, Järvinen H, Mecklin JP, Launonen V, Aaltonen LA. Microsatellite marker analysis in screening for hereditary nonpolyposis colorectal cancer (HNPCC). Cancer Res. 2001;61:4545-4549. [PubMed] |
| 17. | Janavicius R, Matiukaite D, Jakubauskas A, Griskevicius L. Microsatellite instability detection by high-resolution melting analysis. Clin Chem. 2010;56:1750-1757. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 27] [Cited by in RCA: 29] [Article Influence: 1.9] [Reference Citation Analysis (0)] |
| 18. | Brennetot C, Buhard O, Jourdan F, Flejou JF, Duval A, Hamelin R. Mononucleotide repeats BAT-26 and BAT-25 accurately detect MSI-H tumors and predict tumor content: implications for population screening. Int J Cancer. 2005;113:446-450. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 39] [Cited by in RCA: 48] [Article Influence: 2.4] [Reference Citation Analysis (0)] |
| 19. | Wong JJ, Hawkins NJ, Ward RL. Colorectal cancer: a model for epigenetic tumorigenesis. Gut. 2007;56:140-148. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 125] [Cited by in RCA: 123] [Article Influence: 6.8] [Reference Citation Analysis (0)] |
| 20. | Yamashita K, Dai T, Dai Y, Yamamoto F, Perucho M. Genetics supersedes epigenetics in colon cancer phenotype. Cancer Cell. 2003;4:121-131. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 143] [Cited by in RCA: 144] [Article Influence: 6.5] [Reference Citation Analysis (0)] |
| 21. | Waki T, Tamura G, Tsuchiya T, Sato K, Nishizuka S, Motoyama T. Promoter methylation status of E-cadherin, hMLH1, and p16 genes in nonneoplastic gastric epithelia. Am J Pathol. 2002;161:399-403. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 112] [Cited by in RCA: 114] [Article Influence: 5.0] [Reference Citation Analysis (0)] |
| 22. | Kawakami K, Ruszkiewicz A, Bennett G, Moore J, Grieu F, Watanabe G, Iacopetta B. DNA hypermethylation in the normal colonic mucosa of patients with colorectal cancer. Br J Cancer. 2006;94:593-598. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 103] [Cited by in RCA: 111] [Article Influence: 5.8] [Reference Citation Analysis (0)] |
| 23. | Joo JE, Clendenning M, Wong EM, Rosty C, Mahmood K, Georgeson P, Winship IM, Preston SG, Win AK, Dugué PA, Jayasekara H, English D, Macrae FA, Hopper JL, Jenkins MA, Milne RL, Giles GG, Southey MC, Buchanan DD. DNA Methylation Signatures and the Contribution of Age-Associated Methylomic Drift to Carcinogenesis in Early-Onset Colorectal Cancer. Cancers (Basel). 2021;13. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 18] [Cited by in RCA: 19] [Article Influence: 4.8] [Reference Citation Analysis (0)] |
| 24. | Sakata K, Tamura G, Endoh Y, Ohmura K, Ogata S, Motoyama T. Hypermethylation of the hMLH1 gene promoter in solitary and multiple gastric cancers with microsatellite instability. Br J Cancer. 2002;86:564-567. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 31] [Cited by in RCA: 35] [Article Influence: 1.5] [Reference Citation Analysis (0)] |
| 25. | Morán A, Ortega P, de Juan C, Fernández-Marcelo T, Frías C, Sánchez-Pernaute A, Torres AJ, Díaz-Rubio E, Iniesta P, Benito M. Differential colorectal carcinogenesis: Molecular basis and clinical relevance. World J Gastrointest Oncol. 2010;2:151-158. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in CrossRef: 38] [Cited by in RCA: 43] [Article Influence: 2.9] [Reference Citation Analysis (0)] |
| 26. | Zhang H, Fu WL, Huang Q. Mapping of the methylation pattern of the hMSH2 promoter in colon cancer, using bisulfite genomic sequencing. J Carcinog. 2006;5:22. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 7] [Cited by in RCA: 7] [Article Influence: 0.4] [Reference Citation Analysis (0)] |
| 27. | Kawaguchi M, Banno K, Yanokura M, Kobayashi Y, Kishimi A, Ogawa S, Kisu I, Nomura H, Hirasawa A, Susumu N, Aoki D. Analysis of candidate target genes for mononucleotide repeat mutation in microsatellite instability-high (MSI-H) endometrial cancer. Int J Oncol. 2009;35:977-982. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 4] [Cited by in RCA: 14] [Article Influence: 0.9] [Reference Citation Analysis (0)] |
| 28. | Brim H, Mokarram P, Naghibalhossaini F, Saberi-Firoozi M, Al-Mandhari M, Al-Mawaly K, Al-Mjeni R, Al-Sayegh A, Raeburn S, Lee E, Giardiello F, Smoot DT, Vilkin A, Boland CR, Goel A, Hafezi M, Nouraie M, Ashktorab H. Impact of BRAF, MLH1 on the incidence of microsatellite instability high colorectal cancer in populations based study. Mol Cancer. 2008;7:68. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 48] [Cited by in RCA: 63] [Article Influence: 3.7] [Reference Citation Analysis (0)] |
| 29. | Oda S, Maehara Y, Ikeda Y, Oki E, Egashira A, Okamura Y, Takahashi I, Kakeji Y, Sumiyoshi Y, Miyashita K, Yamada Y, Zhao Y, Hattori H, Taguchi K, Ikeuchi T, Tsuzuki T, Sekiguchi M, Karran P, Yoshida MA. Two modes of microsatellite instability in human cancer: differential connection of defective DNA mismatch repair to dinucleotide repeat instability. Nucleic Acids Res. 2005;33:1628-1636. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 48] [Cited by in RCA: 52] [Article Influence: 2.6] [Reference Citation Analysis (0)] |
| 30. | Bufill JA. Colorectal cancer: evidence for distinct genetic categories based on proximal or distal tumor location. Ann Intern Med. 1990;113:779-788. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 505] [Cited by in RCA: 550] [Article Influence: 15.7] [Reference Citation Analysis (0)] |
| 31. | Irving MH, Catchpole B. ABC of colorectal diseases. Anatomy and physiology of the colon, rectum, and anus. BMJ. 1992;304:1106-1108. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 13] [Cited by in RCA: 11] [Article Influence: 0.3] [Reference Citation Analysis (0)] |
| 32. | Minoo P, Zlobec I, Peterson M, Terracciano L, Lugli A. Characterization of rectal, proximal and distal colon cancers based on clinicopathological, molecular and protein profiles. Int J Oncol. 2010;37:707-718. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 115] [Cited by in RCA: 145] [Article Influence: 9.7] [Reference Citation Analysis (0)] |
| 33. | Szmulowicz UM, Hull TL. Colonic Physiology. In: Beck DE, Roberts PL, Saclarides TJ, Senagore AJ, Stamos MJ, Wexner SD. The ASCRS Textbook of Colon and Rectal Surgery. New York, NY: Springer, 2011: 23-39. [DOI] [Full Text] |
| 34. | Li FY, Lai MD. Colorectal cancer, one entity or three. J Zhejiang Univ Sci B. 2009;10:219-229. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 172] [Cited by in RCA: 184] [Article Influence: 11.5] [Reference Citation Analysis (0)] |
| 35. | Lim U, Song MA. Dietary and lifestyle factors of DNA methylation. Methods Mol Biol. 2012;863:359-376. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 147] [Cited by in RCA: 154] [Article Influence: 11.8] [Reference Citation Analysis (0)] |
| 36. | Busch EL, Galanko JA, Sandler RS, Goel A, Keku TO. Lifestyle Factors, Colorectal Tumor Methylation, and Survival Among African Americans and European Americans. Sci Rep. 2018;8:9470. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 5] [Cited by in RCA: 6] [Article Influence: 0.9] [Reference Citation Analysis (0)] |
| 37. | Wang SM, Jiang B, Deng Y, Huang SL, Fang MZ, Wang Y. Clinical significance of MLH1/MSH2 for stage II/III sporadic colorectal cancer. World J Gastrointest Oncol. 2019;11:1065-1080. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in CrossRef: 11] [Cited by in RCA: 18] [Article Influence: 3.0] [Reference Citation Analysis (0)] |
| 38. | Toh JWT, Phan K, Reza F, Chapuis P, Spring KJ. Rate of dissemination and prognosis in early and advanced stage colorectal cancer based on microsatellite instability status: systematic review and meta-analysis. Int J Colorectal Dis. 2021;36:1573-1596. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 11] [Cited by in RCA: 24] [Article Influence: 6.0] [Reference Citation Analysis (0)] |
| 39. | Popat S, Hubner R, Houlston RS. Systematic review of microsatellite instability and colorectal cancer prognosis. J Clin Oncol. 2005;23:609-618. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1267] [Cited by in RCA: 1347] [Article Influence: 67.4] [Reference Citation Analysis (1)] |
| 40. | Condelli V, Calice G, Cassano A, Basso M, Rodriquenz MG, Zupa A, Maddalena F, Crispo F, Pietrafesa M, Aieta M, Sgambato A, Tortora G, Zoppoli P, Landriscina M. Novel Epigenetic Eight-Gene Signature Predictive of Poor Prognosis and MSI-Like Phenotype in Human Metastatic Colorectal Carcinomas. Cancers (Basel). 2021;13. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 6] [Cited by in RCA: 22] [Article Influence: 5.5] [Reference Citation Analysis (0)] |
| 41. | Huang Z, Chen X, Liu C, Cui L. The Clinical Significance of Microsatellite Instability in Precision Treatment. Methods Mol Biol. 2020;2204:33-38. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 3] [Cited by in RCA: 3] [Article Influence: 0.6] [Reference Citation Analysis (0)] |
| 42. | Diao Z, Han Y, Chen Y, Zhang R, Li J. The clinical utility of microsatellite instability in colorectal cancer. Crit Rev Oncol Hematol. 2021;157:103171. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 14] [Cited by in RCA: 43] [Article Influence: 8.6] [Reference Citation Analysis (0)] |
| 43. | García-Alfonso P, García-Carbonero R, García-Foncillas J, Pérez-Segura P, Salazar R, Vera R, Ramón Y Cajal S, Hernández-Losa J, Landolfi S, Musulén E, Cuatrecasas M, Navarro S. Update of the recommendations for the determination of biomarkers in colorectal carcinoma: National Consensus of the Spanish Society of Medical Oncology and the Spanish Society of Pathology. Clin Transl Oncol. 2020;22:1976-1991. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 8] [Cited by in RCA: 11] [Article Influence: 2.2] [Reference Citation Analysis (0)] |
| 44. | Sepulveda AR, Hamilton SR, Allegra CJ, Grody W, Cushman-Vokoun AM, Funkhouser WK, Kopetz SE, Lieu C, Lindor NM, Minsky BD, Monzon FA, Sargent DJ, Singh VM, Willis J, Clark J, Colasacco C, Rumble RB, Temple-Smolkin R, Ventura CB, Nowak JA. Molecular Biomarkers for the Evaluation of Colorectal Cancer: Guideline From the American Society for Clinical Pathology, College of American Pathologists, Association for Molecular Pathology, and the American Society of Clinical Oncology. J Clin Oncol. 2017;35:1453-1486. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 198] [Cited by in RCA: 238] [Article Influence: 29.8] [Reference Citation Analysis (0)] |