Published online Nov 15, 2021. doi: 10.4251/wjgo.v13.i11.1799
Peer-review started: March 1, 2021
First decision: June 23, 2021
Revised: July 6, 2021
Accepted: September 7, 2021
Article in press: September 7, 2021
Published online: November 15, 2021
Processing time: 255 Days and 16.8 Hours
Gastrointestinal tumors are among the most common cancer types, and early detection is paramount to improve their management. Cell-free DNA (cfDNA) liquid biopsy raises significant hopes for non-invasive early detection.
To describe current applications of this technology for gastrointestinal cancer detection and screening.
A systematic review of the literature was performed across the PubMed database. Articles reporting the use of cfDNA liquid biopsy in the screening or diagnosis of gastrointestinal cancers were included in the analysis.
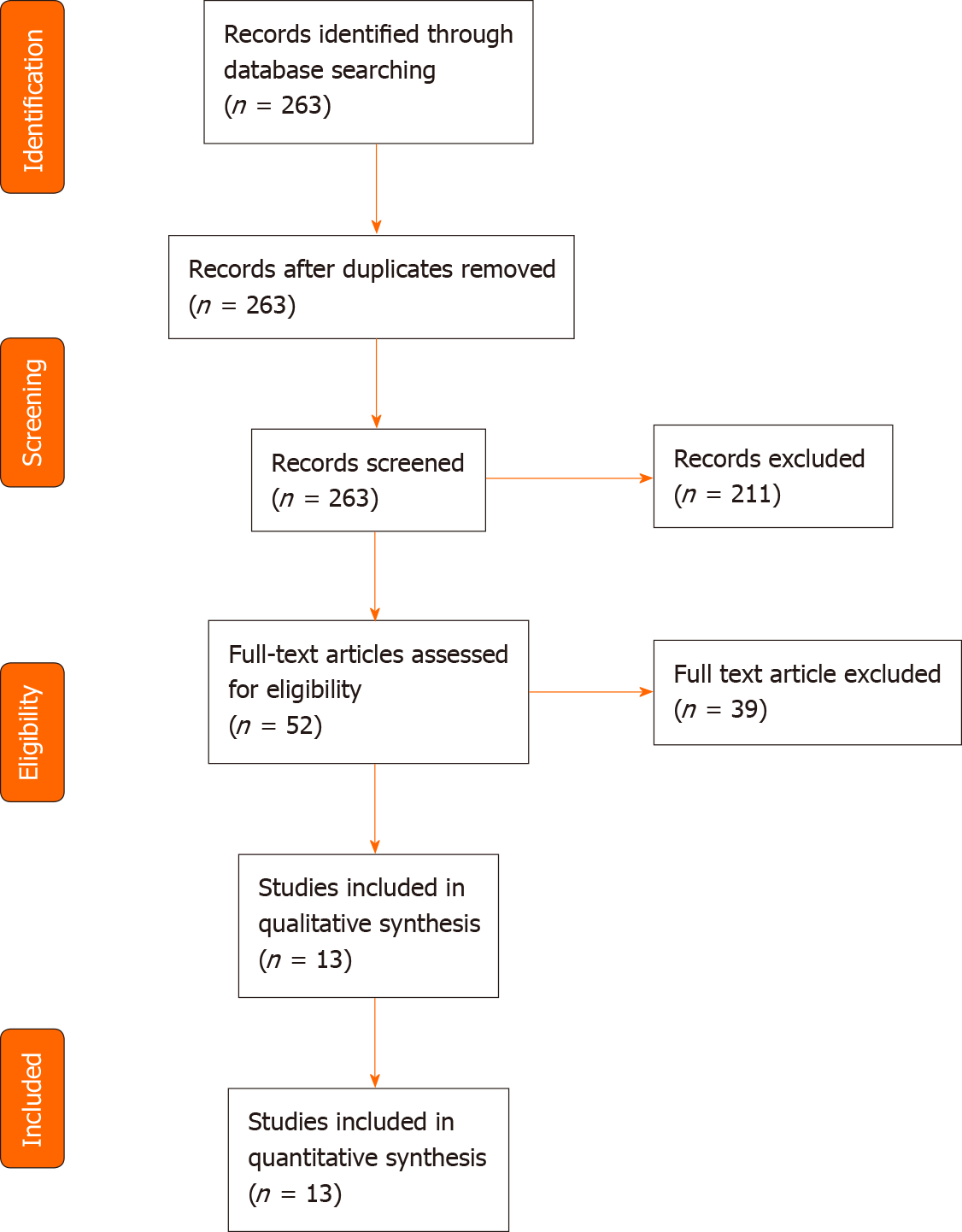
A total of 263 articles were screened for eligibility, of which 13 articles were included. Studies investigated colorectal cancer (5 studies), pancreatic cancer (2 studies), hepatocellular carcinoma (3 studies), and multi-cancer detection (3 studies), including gastric, oesophageal, or bile duct cancer, representing a total of 4824 patients. Test sensitivities ranged from 71% to 100%, and specificities ranged from 67.4% to 100%. Pre-cancerous lesions detection was less performant with a sensitivity of 16.9% and a 100% specificity in one study. Another study using a large biobank demonstrated a 94.9% sensitivity in detecting cancer up to 4 years before clinical symptoms, with a 61% accuracy in tissue-of-origin identification.
cfDNA liquid biopsy seems capable of detecting gastrointestinal cancers at an early stage of development in a non-invasive and repeatable manner and screening simultaneously for multiple cancer types in a single blood sample. Further trials in clinically relevant settings are required to determine the exact place of this technology in gastrointestinal cancer screening and diagnosis strategies.
Core Tip: Liquid biopsy cell-free DNA represents a promising non-invasive method for detecting various gastrointestinal cancers at an early stage of development. The current literature suggests a high-performance profile for this technology and the potential to improve the global course of gastrointestinal cancers currently diagnosed at an advanced stage, such as pancreatic cancer. Prospective validation studies in relevant clinical settings are required to determine the applicability and added value of these new diagnostic and screening tests in global cancer care.
- Citation: Uhe I, Hagen ME, Ris F, Meyer J, Toso C, Douissard J. Cell-free DNA liquid biopsy for early detection of gastrointestinal cancers: A systematic review. World J Gastrointest Oncol 2021; 13(11): 1799-1812
- URL: https://www.wjgnet.com/1948-5204/full/v13/i11/1799.htm
- DOI: https://dx.doi.org/10.4251/wjgo.v13.i11.1799
Tumors developing from the gastrointestinal tract are among the most common cancer types, colorectal and stomach cancer, counting for 19.5% and 11.1% respectively worldwide in 2020[1]. Risk factors notably include smoking, obesity, poor diet, genetic factors, and infections with hepatitis B virus or Helicobacter pylori bacteria[2]. Early detection and diagnosis represent a crucial component to allow effective treatment and improve survival. Nowadays, different screening strategies have been developed, such as colonoscopy for colorectal cancer or blood testing for alpha-fetoprotein (AFP) or magnetic resonance imaging in high-risk patients for liver cancer, but other types of tumors often lack screening strategies and non-invasive testing. For instance, so far, no efficient screening methods are available for pancreatic cancer; most patients experience their first symptoms at advanced and metastatic stages, explaining the 5-year survival rate of only 5% to 10%[3].
These past few years, researchers have focused their attention on a new promising diagnostic method, liquid biopsy, which uses biomarkers such as circulating cell tumor, RNA fragments, or cell-free DNA (cfDNA). Unlike tissue samples obtained by invasive methods like needle biopsies or endoscopies, biomarkers can be detected in body fluids, mostly blood[4], and address limitations of tissues biopsies not only in diagnosis and screening, but also in diagnosis and screening the treatment response and follow-up[5-7]. Among liquid biopsy options, cfDNA raises the most significant hopes in early cancer detection. Historically, it was first reported in 1948 by Mandel et al among healthy patients. In 1977, Leon et al described elevated levels of cfDNA in the serum of cancerous patients for the first time[4,8,9]. CfDNA is continuously released in the bloodstream through different mechanisms such as apoptosis, necrosis, and active secretion by the tumor cell. When originating from a cancer cell, cfDNA is called circulating tumor DNA (ctDNA)[4]. Concentration levels seem to correlate with the cancer stage and size; advanced-stage cancer patients show a higher concentration of cfDNA[8,9]. While cfDNA quantification in the bloodstream might indicate the presence or absence of cancer, sequencing and analyzing the mutation patterns of this cfDNA goes one step further: mutational profiling might give the researchers clues on the tumor’s tissue of origin, providing information to target further specific investigations[9]. Recent progress in genomic technology also provides highly sensitive detection of low-prevalence mutations, even in high signal-to-noise configurations, thus theoretically enabling very early cancer diagnosis. The ability to run repeatable, non-invasive, multi-cancer early detection tests would bring significant advantages in the global care of frequently hardly reachable cancer locations, such as gastrointestinal cancers.
The present systematic review of the literature aims to describe the current state of developing cfDNA liquid biopsies as a means of early gastrointestinal cancer detection and screening.
A systematic review of the literature was performed following the PRISMA guidelines[10]. All articles written in English from January 2010 to January 2021 were searched on January 19th, 2021, through the PubMed database using the following research algorithm: (liquid biopsy OR cfdna) AND (multiple OR gastrointestinal OR colon OR colorectal OR gastric OR oesophag* OR liver OR hepatocellular OR pancreatic) AND (cancer OR tumor OR tumour) AND (screening OR diagnos* OR detect*) AND early AND (blood OR venous OR plasma) NOT review.
After a first selection based on titles for screening, eligible articles were selected based on abstract analysis. Then, full-text analysis of the eligible articles searched for criteria of the finally included articles. Two investigators (I Uhe, J Douissard) independently assessed the articles for eligibility and inclusion. Discordances in study inclusions were solved by re-evaluation between the two reviewers.
All relevant articles reporting human studies investigating cfDNA liquid biopsy as a screening method or diagnosis method for newly discovered untreated primary gastrointestinal cancers were included. Studies investigating multiple cancer screening, including gastrointestinal but not limited to them, were also included. Excluded articles were studies investigating cfDNA as a follow-up method after cancer treatment, minimal residual disease detection, studies investigating cfDNA as a prognosis method only, reviews, meta-analyses, theoretical papers, and biological studies not reporting clinical outcomes. Studies reporting cancer patients who were already treated, surgically or medically, have also been excluded. To improve the present review’s clinical relevance, only the total number of participants in the papers’ validations cohorts were considered. If available, test performances were reported in terms of sensitivity (Se), specificity (Sp), positive and negative predictive values, or area under the curve (AUC).
A total of 263 articles were identified through the PubMed search. Two articles were not written in English, 11 were not original publications, and 119 did not involve cfDNA. Thirty-five articles did not mention gastrointestinal cancer, and 44 did not investigate cfDNA as a screening or diagnosis method, leaving 52 articles. After full-text reading, thirteen studies were ultimately included for analysis, representing a total of 4824 patients (Table 1, Figure 1). The largest study included blood samples from 1194 participants[11], while the smallest study included samples of 130 participants[12]. Six studies took place in China[11,13-17], three in the United States[9,18,19], and four in Europe[12,20-22]. Five were multicentric[9,11,16,18,19], four monocentric[13,14,17,22] and four studies did not mention the information. Five studies focused on colorectal cancer (CRC)[9,12,17,20,22], three on various cancer types[14,19,21] of which two included gastric cancers[14,19], three on hepatocellular carcinoma (HCC)[11,15,16] and two on pancreatic ductal adenocarcinoma (PDAC)[13,18]. All studies compared cancer and non-cancer individuals. Five of them also included in their analysis a group of patients with pre-cancerous lesions, such as colorectal adenoma or hyperplasia, liver cirrhosis, or chronic hepatitis B virus infection[11,12,15,16,22] (Table 2).
| Ref. | Year | Country | Mono/multicentric | Type of cancer | Total number of patients in validation cohort | Type of groups analyzed |
| Li et al[13] | 2020 | China | Monocentric | PDAC | 208 | Cancer vs healthy |
| Chen et al[14] | 2020 | China | Monocentric | Gastric, esophagus, colorectal, lung or liver | 418 | Cancer diagnosed vs healthy; Pre-diagnosed patients vs healthy |
| Guler et al[18] | 2020 | United States | Multicentric | PDAC | 228 | Cancer vs healthy |
| Junca et al[12] | 2020 | France | NA | Colorectal | 130 | Cancer vs healthy vs advanced-adenoma vs non-advanced adenoma and/or hyperplastic polyp(s) |
| Tao et al[15] | 2020 | China | NA | HCC | 175 | HBV-related HCC vs cancer-free HBV patients |
| Cristiano et al[19] | 2019 | United States | Multicentric | Breast, colorectal, lung, ovarian, pancreatic, gastric, bile duct | 423 | Cancer vs healthy |
| Li et al[17] | 2019 | China | Monocentric | Colorectal | 140 | Cancer vs healthy |
| Qu et al[16] | 2019 | China | Multicentric | HCC | 331 | HBsAg1 positive without cancer based on screening with serum AFP and ultrasonography |
| Cai et al[11] | 2019 | China | Multicentric | HCC | 1194 | Cancer vs healthy vs 392 LC/HB vs BLL |
| Wan et al[9] | 2019 | United States | Multicentric | Colorectal | 817 | Cancer vs healthy |
| Jensen et al[20] | 2019 | Denmark | NA | Colorectal | 234 | Cancer vs healthy |
| Nunes et al[21] | 2018 | Portugal | NA | Breast, colorectal, lung | 356 | Cancer vs healthy |
| Perrone et al[22] | 2014 | Italy | Monocentric | Colorectal | 170 | Cancer vs healthy vs premalignant lesion (adenoma/hyperplasia) |
| Ref. | Total patients in validation cohort | Nbr patient cancer group | Nbr patient healthy group | Nbr patient additional group 1 | Nbr patient in aditionnal group 2 |
| Li et al[13] | 208 | 101 | 107 | - | - |
| Chen et al[14] | 418 | 113 | 2071 | 98 pre-diagnosed patients | - |
| Guler et al[18] | 228 | 23 | 205 | - | - |
| Junca et al[12] | 130 | 20 | 40 | 39 advance adenoma | 31 non-advance adenoma |
| Tao et al[15] | 175 | 89 | 86 | - | - |
| Cristiano et al[19] | 423 | 208 | 215 | - | - |
| Li et al[17] | 140 | 74 | 66 | - | - |
| Qu et al[16] | 331 | - | - | HBsAg (+) | - |
| Cai et al[11] | 1194 | 809 | 256 | 129 LC/CHB | - |
| Wan et al[9] | 817 | 546 | 271 | - | - |
| Jensen et al[20] | 234 | 143 | 91 | - | - |
| Nunes et al[21] | 356 | 253 | 103 | - | - |
| Perrone et al[22] | 170 | 34 | 63 | 73 adenoma/hyperplasia | - |
The risk of bias of included studies was determined using the ROBINS-I tool (2016)[23]. Except for one study with an overall low risk of bias[16], all included studies were at moderate risk (Table 3).
| Ref. | Entry | Judgement | Support for judgement | |
| Li et al[13] | A | Bias due to confounding | Low risk | No confounding factors |
| B | Bias in selection of participants into the study | No information | No information about the start of follow up and intervention for the participants | |
| C | Bias in classification of interventions | No information | No information about the start of follow up and intervention for the participants | |
| D | Bias due to deviationsfrom intended interventions | Low risk | No deviations from the planned interventions | |
| E | Bias due to missing data | Low risk | All data were reported | |
| F | Bias in measurement of outcomes | Low risk | Comparable methods of outcome assessment in the groups, intervention received in each group unlikely to influence the outcome measure, any error in measuring the outcome is unrelated to intervention | |
| G | Bias in selection of the reported result | Moderate risk | No pre-registered protocol available; outcome measurements and analyses consistent with a priori plan | |
| Chen et al[14] | A | Bias due to confounding | Low risk | No confounding factors |
| B | Bias in selection of participants into the study | Low risk | Information provided about the start of follow up and intervention for the participants | |
| C | Bias in classification of interventions | Low risk | Information provided about the start of follow up and intervention for the participants | |
| D | Bias due to deviationsfrom intended interventions | Low risk | No deviations from the planned interventions | |
| E | Bias due to missing data | Low risk | All data were reported | |
| F | Bias in measurement of outcomes | Low risk | Comparable methods of outcome assessment in the groups, intervention received in each group unlikely to influence the outcome measure, any error in measuring the outcome is unrelated to intervention | |
| G | Bias in selection of the reported result | Moderate risk | No pre-registered protocol available; outcome measurements and analyses consistent with a priori plan | |
| Guler et al[18] | A | Bias due to confounding | Low risk | No confounding factors |
| B | Bias in selection of participants into the study | No information | No information about the start of follow up and intervention for the participants | |
| C | Bias in classification of interventions | No information | No information about the start of follow up and intervention for the participants | |
| D | Bias due to deviationsfrom intended interventions | Low risk | No deviations from the planned interventions | |
| E | Bias due to missing data | Low risk | All data were reported | |
| F | Bias in measurement of outcomes | Low risk | Comparable methods of outcome assessment in the groups, intervention received in each group unlikely to influence the outcome measure, any error in measuring the outcome is unrelated to intervention | |
| G | Bias in selection of the reported result | Moderate risk | No pre-registered protocol available; outcome measurements and analyses consistent with a priori plan | |
| Junca et al[12] | A | Bias due to confounding | Low risk | No confounding factors |
| B | Bias in selection of participants into the study | No information | No information about the start of follow up and intervention for the participants | |
| C | Bias in classification of interventions | No information | No information about the start of follow up and intervention for the participants | |
| D | Bias due to deviationsfrom intended interventions | Low risk | No deviations from the planned interventions | |
| E | Bias due to missing data | Low risk | All data were reported | |
| F | Bias in measurement of outcomes | Low risk | Comparable methods of outcome assessment in the groups, intervention received in each group unlikely to influence the outcome measure, any error in measuring the outcome is unrelated to intervention | |
| G | Bias in selection of the reported result | Moderate risk | No pre-registered protocol available; outcome measurements and analysesconsistent with a priori plan | |
| Tao et al[15] | A | Bias due to confounding | Low risk | No confounding factors |
| B | Bias in selection of participants into the study | Low risk | Information provided about the start of follow up and intervention for the participants in the supplementary materials | |
| C | Bias in classification of interventions | Low risk | Information provided about the start of follow up and intervention for the participants in the supplementary materials | |
| D | Bias due to deviationsfrom intended interventions | Low risk | No deviations from the planned interventions | |
| E | Bias due to missing data | Low risk | All data were reported | |
| F | Bias in measurement of outcomes | Low risk | Comparable methods of outcome assessment in the groups, intervention received in each group unlikely to influence the outcome measure, any error in measuring the outcome is unrelated to intervention | |
| G | Bias in selection of the reported result | Moderate risk | No pre-registered protocol available; outcome measurements and analyses consistent with a priori plan | |
| Cristiano et al[19] | A | Bias due to confounding | Low risk | No confounding factors |
| B | Bias in selection of participants into the study | No information | No information about the start of follow up and intervention for the participants | |
| C | Bias in classification of interventions | No information | No information about the start of follow up and intervention for the participants | |
| D | Bias due to deviationsfrom intended interventions | Low risk | No deviations from the planned interventions | |
| E | Bias due to missing data | Low risk | All data were reported | |
| F | Bias in measurement of outcomes | Low risk | Comparable methods of outcome assessment in the groups, intervention received in each group unlikely to influence the outcome measure, any error in measuring the outcome is unrelated to intervention | |
| G | Bias in selection of the reported result | Moderate risk | No pre-registered protocol available; outcome measurements and analyses consistent with a priori plan | |
| Li et al[17] | A | Bias due to confounding | Low risk | No confounding factors |
| B | Bias in selection of participants into the study | No information | No information about the start of follow up and intervention for the participants | |
| C | Bias in classification of interventions | No information | No information about the start of follow up and intervention for the participants | |
| D | Bias due to deviationsfrom intended interventions | Low risk | No deviations from the planned interventions | |
| E | Bias due to missing data | Low risk | All data were reported | |
| F | Bias in measurement of outcomes | Low risk | Comparable methods of outcome assessment in the groups, intervention received in each group unlikely to influence the outcome measure, any error in measuring the outcome is unrelated to intervention | |
| G | Bias in selection of the reported result | Moderate risk | No pre-registered protocol available; outcome measurements and analyses consistent with a priori plan | |
| Qu et al[16] | A | Bias due to confounding | Low risk | No confounding factors |
| B | Bias in selection of participants into the study | Low risk | Information provided about the start of follow up and intervention for the participants | |
| C | Bias in classification of interventions | Low risk | Information provided about the start of follow up and intervention for the participants | |
| D | Bias due to deviationsfrom intended interventions | Low risk | No deviations from the planned interventions | |
| E | Bias due to missing data | Low risk | All data were reported | |
| F | Bias in measurement of outcomes | Low risk | Comparable methods of outcome assessment in the groups, intervention received in each group unlikely to influence the outcome measure, any error in measuring the outcome is unrelated to intervention | |
| G | Bias in selection of the reported result | Low risk | Pre-registered protocol available (NCC201709011) | |
| Cai et al[11] | A | Bias due to confounding | Low risk | No confounding factors |
| B | Bias in selection of participants into the study | Low risk | Information provided about the start of follow up and intervention for the participants | |
| C | Bias in classification of interventions | Low risk | Information provided about the start of follow up and intervention for the participants | |
| D | Bias due to deviationsfrom intended interventions | Low risk | No deviations from the planned interventions | |
| E | Bias due to missing data | Low risk | All data were reported | |
| F | Bias in measurement of outcomes | Low risk | Comparable methods of outcome assessment in the groups, intervention received in each group unlikely to influence the outcome measure, any error in measuring the outcome is unrelated to intervention | |
| G | Bias in selection of the reported result | Moderate risk | No pre-registered protocol available; outcome measurements and analyses consistent with a priori plan | |
| Wan et al[9] | A | Bias due to confounding | Low risk | No confounding factors |
| B | Bias in selection of participants into the study | No information | No information about the start of follow up and intervention for the participants | |
| C | Bias in classification of interventions | No information | No information about the start of follow up and intervention for the participants | |
| D | Bias due to deviationsfrom intended interventions | Low risk | No deviations from the planned interventions | |
| E | Bias due to missing data | Low risk | All data were reported | |
| F | Bias in measurement of outcomes | Low risk | Comparable methods of outcome assessment in the groups, intervention received in each group unlikely to influence the outcome measure, any error in measuring the outcome is unrelated to intervention | |
| G | Bias in selection of the reported result | Moderate risk | No pre-registered protocol available; outcome measurements and analyses consistent with a priori plan | |
| Jensen et al[20] | A | Bias due to confounding | Low risk | No confounding factors |
| B | Bias in selection of participants into the study | Low risk | Information provided about the start of follow up and intervention for the participants | |
| C | Bias in classification of interventions | Low risk | Information provided about the start of follow up and intervention for the participants | |
| D | Bias due to deviationsfrom intended interventions | Low risk | No deviations from the planned interventions | |
| E | Bias due to missing data | Low risk | All data were reported | |
| F | Bias in measurement of outcomes | Low risk | Comparable methods of outcome assessment in the groups, intervention received in each group unlikely to influence the outcome measure, any error in measuring the outcome is unrelated to intervention | |
| G | Bias in selection of the reported result | Moderate risk | No pre-registered protocol available; outcome measurements and analyses consistent with a priori plan | |
| Nunes et al[21] | A | Bias due to confounding | Low risk | No confounding factors |
| B | Bias in selection of participants into the study | No information | No information about the start of follow up and intervention for the participants | |
| C | Bias in classification of interventions | No information | No information about the start of follow up and intervention for the participants | |
| D | Bias due to deviationsfrom intended interventions | Low risk | No deviations from the planned interventions | |
| E | Bias due to missing data | Low risk | All data were reported | |
| F | Bias in measurement of outcomes | Low risk | Comparable methods of outcome assessment in the groups, intervention received in each group unlikely to influence the outcome measure, any error in measuring the outcome is unrelated to intervention | |
| G | Bias in selection of the reported result | Moderate risk | No pre-registered protocol available; outcome measurements and analyses consistent with a priori plan | |
| Perrone et al[22] | A | Bias due to confounding | Low risk | No confounding factors |
| B | Bias in selection of participants into the study | Low risk | Information provided about the start of follow up and intervention for the participants | |
| C | Bias in classification of interventions | Low risk | Information provided about the start of follow up and intervention for the participants | |
| D | Bias due to deviationsfrom intended interventions | Low risk | No deviations from the planned interventions | |
| E | Bias due to missing data | Low risk | All data were reported | |
| F | Bias in measurement of outcomes | Low risk | Comparable methods of outcome assessment in the groups, intervention received in each group unlikely to influence the outcome measure, any error in measuring the outcome is unrelated to intervention | |
| G | Bias in selection of the reported result | Moderate risk | No pre-registered protocol available; outcome measurements and analyses consistent with a priori plan |
All studies collected cfDNA from plasma samples. Kits used for cfDNA extraction from plasma samples can be found in Table 4. The QIAamp circulating nucleic acid kit was the most employed, a spin column-based kit (n = 7/13). A large majority of studies used next-generation sequencing (NGS) (n = 9/13), two used real-time polymerase chain reaction (RT-PCR), one digital droplet PCR, and one multiplex methylation-specific PCR. Various mutational patterns and genomic profiling strategies were investigated (Table 4). Most studies focused on methylation variations (n = 7/13), while others investigated specific mutation locations such as KRAS and BRAF or more complex mutational patterns.
| Ref. | Source of cfDNA | Focus in cfDNA | Extraction method (used kit) | Sequencing method | Sequencing method details |
| Li et al[13] | Plasma | Methylated markers | QIAamp Circulating Nucleic Acid Kit (Qiagen, 55114) | NGS | Illumina HiSeq 2000 platform |
| Chen et al[14] | Plasma | Cancer-specific methylation signatures | QIAamp Circulating Nucleic Acid kit (Qiagen, 55114) | NGS | APA Library Quantification Kit for Illumina (KK4844) and sequenced on an Illumina NextSeq 500 |
| Guler et al[18] | Plasma | 5hmC modifications | QIAamp Circulating Nucleic Acid Kit (QIAGEN, Germantown, MD) | NGS | NextSeq550 instrument with version 2 reagent chemistry (Illumina, San Diego, CA). |
| Junca et al[12] | Plasma | KRAS and BRAF mutational status | QIAamp Circulating Nucleic Acid kit (Qiagen, Hilden, Germany) | RT-PCR | Q24 PyroMark system (Qiagen, Hilden, Germany) |
| Tao et al[15] | Plasma | Somatic copy number aberration | QIAamp CirculatingNucleic Acid Kit (Qiagen) | NGS | Next generation sequencing (Illumina) |
| Cristiano et al[19] | Plasma | Fragmentation size | Qiagen Circulating Nucleic Acids Kit (Qiagen GmbH) | NGS | NEBNext DNA Library Prep Kit for Illumina |
| Li et al[17] | Plasma | Aberrant DNA hypermethylation of CpGislands | DNeasy Blood & TissueKit (Qiagen) | NGS | Methylated CpG tandem ampli-fication and sequencing |
| Qu et al[16] | Plasma | Specific mutations | ARCHITECT i2000SR Chemical luminescence immunity analyzer | NGS | Next generation sequencing |
| Cai et al[11] | Plasma | 5hmC modifications | NA | NGS | 5hmC-Seal |
| Wan et al[9] | Plasma | cfDNA mutations patterns | MagMAX cfDNA Isolation Kit | NGS | Illumina NovaSeq 6000 Sequencing System |
| Jensen et al[20] | Plasma | Tumour-specific DNA methylation | Gentra Puregene Tissue Kit (Qiagen) | DD-PCR | Bisulfite sequencing and methylation-specific droplet digital PCR |
| Nunes et al[21] | Plasma | Aberrant DNA methylation | QIAamp MinElute ccfDNA (Qiagen, Hilden, Germany) | qMSP | qMSP |
| Perrone et al[22] | Plasma | KRAS mutated cfDNA | Qiamp DNA Blood Extraction Kit (Qiagen) | RT-PCR | RT-PCR |
Overall test performances for each cancer subgroup are described in Table 5.
| Ref. | Group of validation cohorts | Sensitivity | Specificity | Positive predictive value | Negative predictive value | AUC | |
| PDCA | Li et al[13] | Cancer vs healthy | 93.2 | 95.2 | NA | NA | 0.943 |
| Chen et al[14] | Cancer vs healthy | NA | NA | NA | NA | 0.921 | |
| HCC | Guler et al[18] | HBV-related HCC vs cancer-free HBV group 1 | 18 | 97.4 | NA | NA | 0.92 |
| HBV-related HCC vs cancer-free HBV group 2 | 29 | 95.6 | NA | NA | 0.81 | ||
| Junca et al[12] | HCC vs cancer-free HBV | 100 | 94 | 17 | 100 | NA | |
| Tao et al[15] | HCC vs healthy | 82.7 | 76.4 | NA | NA | 0.884 | |
| HCC vs high risk (HBV and cirrhosis) | 82.7 | 67.4 | NA | NA | 0.846 | ||
| Various cancer types | Cristiano et al[19] | Pre-diagnosis vs healthy | 84.9 | 96.1 | NA | NA | NA |
| Post-diagnosis vs healthy | 87.5 | 96.1 | |||||
| Li et al[17] | All cancer vs healthy | 80 | 95 | NA | NA | 0.94 | |
| 73 | 98 | ||||||
| Gastric cancer vs healthy | 81 | 95 | |||||
| 81 | 98 | ||||||
| Colorectal cancer vs healthy | 81 | 95 | |||||
| 70 | 98 | ||||||
| Bile duct cancer vs healthy | 88 | 95 | |||||
| 81 | 98 | ||||||
| Pancreatic cancer vs healthy | 71 | 95 | |||||
| 65 | 98 | ||||||
| Qu et al[16] | All cancer vs healthy | 74.2 | 73.5 | 87.1 | 52.1 | NA | |
| Colorectal cancer vs healthy | 78.4 | 69.9 | 48.3 | 90 | |||
| Colorectal | Cai et al[11] | Cancer/adenoma vs healthy | 16.9 | 100 | 100 | 59.2 | NA |
| Wan et al[9] | Cancer vs healthy | 74 | 90 | NA | NA | 0.887 | |
| Jensen et al[20] | Cancer vs healthy | 85 | 85 | NA | Na | 0.92 | |
| Nunes et al[21] | Cancer vs healthy | 85 | 99 | NA | NA | NA | |
| Perrone et al[22] | Cancer vs healthy | NA | NA | NA | NA | 0.709 | |
| Adenomas vs healthy | NA | NA | NA | NA | 0.535 |
Clinically relevant sensitivities and specificities to detect colorectal adenocarcinoma were achieved in three studies[9,20,21], Li et al[17] and Jensen et al[20] focusing on tumor-specific methylations. In contrast, Wan et al[9] investigated complex cfDNA mutational patterns using a machine-learning-based model. Sensitivities ranged from 74% to 85%, while specificities ranged from 85% to 99%. In a fourth study, Perrone et al[22] reported an AUC of 0.709 when discriminating CRC from healthy patients. However, for premalignant lesions, the performance was lower, with an AUC of 0.535[22]. Similarly, investigating adenomas and adenocarcinomas through cfDNA KRAS and BRAF mutations, Junca et al[12] found a mean sensitivity of 16.9% for a 100% specificity reflecting a still lower sensitivity in premalignant lesions detection but allowing a high level of precision.
Examining methylation patterns in cfDNA, Li et al[13] described eight methylation markers in patients suffering from PDAC; SIX3, TRIM73, MAPT, FAM150A, EPB41L3, MIR663, LOC100130148, and LOC100128977. These markers identified PDAC patients efficiently, with a sensitivity of 93.2% and a specificity of 95.2% (AUC = 0.943). By investigating 5-hydroxymethylcytosine (5hmC) changes in circulating cfDNA, Guler et al[18] achieved similar performance with an AUC of 0.921.
Cai et al[11] found promising results using a mutational pattern of 32 gene markers to discriminate HCC patients from healthy individuals, with a sensitivity and specificity of 82.7% and 76.4%, respectively. Furthermore, when comparing HCC patients with cancer-free high-risk patients (chronic hepatitis B or liver cirrhosis), the model performed similarly with an 82.7% sensitivity and 67.4% specificity[11].
Comparing HCC patients with cancer-free asymptomatic HBV patients based on cfDNA mutational pattern of specific locations, Qu et al[16] achieved a sensitivity and specificity of 100% and 94%, respectively. Further, using somatic copy number aberration in cfDNA as an alternative to methylation or specific mutations analysis, Tao et al[15] investigated the possibility of discriminating HBV-related HCC from cancer-free chronic HBV patients. Their predictive model performed appropriately, showing a high level of precision in two validation cohorts, with an AUC of 0.92 and 0.81.
Nunes et al[21] investigated the possibility to diagnose lung, breast, and colorectal cancer patients simultaneously from healthy individuals by detecting aberrant methylations on specific locations. They achieved an overall specificity of 73.5% and a sensitivity of 74.2%. For colorectal cancer, specificity was 69.9%, and sensitivity was 78.4%[21].
With a comparable strategy targeting five cancers (gastric, oesophageal, lung, liver, and colorectal), Chen et al[14] demonstrated the potential ability of cfDNA liquid biopsy to achieve multicancer detection several years before the actual diagnosis. Based on blood samples from a large biobank, they analyzed samples from 3 groups. The post-diagnosis group included patients with a newly discovered and untreated malignancy at the time of sampling. The pre-diagnosis group included patients with no known malignancy at the sampling time but who developed cancer within four years after sampling (pre-diagnosis). Finally, the control group included healthy individuals who were still free of malignant disease four years after sampling. Their model achieved an overall detection specificity of 96% when comparing healthy individuals to pre-diagnosis and post-diagnosis groups. Overall sensitivity was 87.5% for the post-diagnosis group, ranging from 75% in colorectal cancer to 96% in lung cancer. It reached 94.9% in the pre-diagnosis group, ranging from 91% in oesophageal cancer to 100% in liver cancer[14].
In contrast to these two studies focused on cfDNA methylations, Cristiano et al[19] explored a multi-cancer detection model analyzing cfDNA fragmentation patterns, including gastric, bile duct, colorectal and pancreatic cancers. Their model reached an overall detection sensitivity of 80% for a specificity of 95%, or a sensitivity of 73% for a specificity of 98%, and a global AUC of 0.94. Furthermore, enhanced by a machine-learning algorithm, they were able to identify the tissue of origin of cancer samples with a 61% accuracy[19]. Detailed performances per cancer type of this model can be found in Table 3.
Liquid biopsy appears as a promising non-invasive method for the initial screening and diagnosis of various gastrointestinal cancers. High levels of sensitivity and specificity described in the included studies seem within acceptable ranges for eventual clinical use. In the case of HCC, cfDNA tests demonstrated better detection performances when compared to the standard surveillance of high-risk patients combining AFP dosage and ultra-sound monitoring. It also appears to be a viable solution regarding the challenge of pancreatic cancer screening; due to the paucity of symptoms in the early phases and the absence of acceptable screening strategies even for high-risk groups, this type of cancer remains frequently detected at metastatic or locally advanced and unresectable stages. Conversely, colorectal cancer is one of the few cancers with a standardized and efficient large-scale screening strategy based on the colonoscopy and the fecal occult blood test. Still, there is room for improved and more cost-effective strategies. Of note, cfDNA liquid biopsy’s ability to detect several cancer types simultaneously appears as a potential paradigm shift in global cancer care, and studies investigating such application achieved a high level of performance. Further, as demonstrated by Chen et al[13], this technology bears the potential to predict cancer several years before the onset of clinical symptoms and identify or direct investigations towards specific tissues of origin.
The central role of early cancer detection in improving oncologic and public-health outcomes is well established. However, it is a challenge for liquid biopsy since smaller and earlier-stage tumors tend to release lower levels of ctDNA[24]. The signal-to-noise ratio of ctDNA is thus meager compared to non-cancer-derived cfDNA, with a detection percentage ranging from 0 to 11.7%[25,26]. The extraction method plays a critical role in improving detection performance. Different procedures have been developed, the more widespread being column-based, polymer-based, phenol-chloroform, or magnet-based[9,27]. These methods are efficient and allow to reach a high DNA concentration but remain expensive and time-consuming[9,27]. In this context, some authors proposed plasma processing methods without the need for DNA extraction. Breitbach et al[28] notably used quantitative RT-PCR to measure cfDNA concentration in plasma. Not only did the method showed great feasibility with higher levels of cfDNA found among cancer patients, but it also proved to be more time effective and more efficient than the eluate of the QIAamp DNA Blood Mini Kit, for example, with levels of cfDNA in unpurified plasma 2.79 fold higher[28].
Regarding the sequencing method, some authors focused their attention on specific mutations while others analyzed the whole genome searching for non-specific mutational patterns, most of them using NGS methods. Different factors can explain the apparent predominance of NGS over other PCR methods such as RT-PCR in the published studies. Although more technically demanding and expensive, NGS is a hypothesis-free approach that carries a higher discovery power of new mutational patterns, in addition to a higher sensitivity to rare variants[29,30]. Further, its superior multiplex capabilities tend to improve the workflow when studying a large number of locations and samples. These high throughput and detection sensitivity capabilities might be valuable in a screening configuration for early cancer detection, which deals with lower levels of mutation than advanced stage cancers and aims at testing a high volume of patients.
As the field is at an early stage of clinical exploration, there is still a high variability in trial designs and reporting methods, thus undermining the global quality of tests’ performance analysis. Of note, biocomputational trials based on biobank samples often report higher levels of sensitivity and specificity but are less likely to translate into clinically relevant performances as prospective trials would. Applicability to real-life clinical applications is thus the most awaited step to achieve for the scientific validation of this technology, and upcoming clinical trials will need to address many questions, such as the appropriate balance between sensitivity and specificity in a screening purpose, the timing of screening tests, patient selection, socio-economic parameters and dealing with the uncertainty around tissues of origin in positive tests.
Liquid biopsy cfDNA represents an efficient, non-invasive, and promising method for detecting various gastrointestinal cancers at an early stage of development. These tools could improve the global prognosis of cancers currently diagnosed at an advanced stage due to the lack of effective screening and diagnostic methods, such as pancreatic cancer. Allowing early detection of several types of cancers and reducing the burden of multiple screening tests, cfDNA liquid biopsies could change the course of gastrointestinal cancers care for a significant number of patients and induce a paradigm shift in cancer-related public health policies, provided that they can demonstrate their clinical relevance in future studies.
Liquid biopsy cell-free DNA (cfDNA) represents a promising non-invasive method for detecting various gastrointestinal cancers at an early stage of development.
Various and recent literature is available on this topic, with exponentially growing interest.
To review the current state of development of cfDNA liquid biopsy in the field of gastrointestinal cancer early detection.
A systematic review of the literature according to the PRISMA guidelines.
The current literature suggests a high-performance profile for this technology and the potential to improve the global course of gastrointestinal cancers currently diagnosed at an advanced stage, such as pancreatic cancer.
cfDNA liquid biopsy showed high potential in the diagnosis of early gastrointestinal cancers and simultaneous screening of multiple cancer types.
Further trials in clinically relevant settings are required to determine the exact place of this technology in future diagnosis strategies.
Provenance and peer review: Invited article; Externally peer reviewed.
Specialty type: Oncology
Country/Territory of origin: Switzerland
Peer-review report’s scientific quality classification
Grade A (Excellent): 0
Grade B (Very good): 0
Grade C (Good): C
Grade D (Fair): 0
Grade E (Poor): 0
P-Reviewer: Luglio G S-Editor: Zhang H L-Editor: A P-Editor: Li X
| 1. | Cancer today. [cited 1 February 2021]. Available from: http://gco.iarc.fr/today/home. |
| 2. | Dizdar Ö, Kılıçkap S. Global Epidemiology of Gastrointestinal Cancers. In: Yalcin S, Philip PA. Textbook of Gastrointestinal Oncology. Switzerland: Springer International Publishing, 2019: 1-12. [DOI] [Full Text] |
| 3. | Pancreatic Cancer Prognosis. [cited 1 February 2021]. Available from: https://www.hopkinsmedicine.org/health/conditions-and-diseases/pancreatic-cancer/pancreatic-cancer-prognosis. |
| 4. | Alberti LR, Garcia DP, Coelho DL, De Lima DC, Petroianu A. How to improve colon cancer screening rates. World J Gastrointest Oncol. 2015;7:484-491. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in CrossRef: 14] [Cited by in RCA: 14] [Article Influence: 1.4] [Reference Citation Analysis (0)] |
| 5. | Banys-Paluchowski M, Krawczyk N, Fehm T. Liquid Biopsy in Breast Cancer. Geburtshilfe Frauenheilkd. 2020;80:1093-1104. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 13] [Cited by in RCA: 13] [Article Influence: 2.6] [Reference Citation Analysis (0)] |
| 6. | De Rubis G, Rajeev Krishnan S, Bebawy M. Liquid Biopsies in Cancer Diagnosis, Monitoring, and Prognosis. Trends Pharmacol Sci. 2019;40:172-186. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 252] [Cited by in RCA: 381] [Article Influence: 63.5] [Reference Citation Analysis (0)] |
| 7. | He HJ, Stein EV, Konigshofer Y, Forbes T, Tomson FL, Garlick R, Yamada E, Godfrey T, Abe T, Tamura K, Borges M, Goggins M, Elmore S, Gulley ML, Larson JL, Ringel L, Haynes BC, Karlovich C, Williams PM, Garnett A, Ståhlberg A, Filges S, Sorbara L, Young MR, Srivastava S, Cole KD. Multilaboratory Assessment of a New Reference Material for Quality Assurance of Cell-Free Tumor DNA Measurements. J Mol Diagn. 2019;21:658-676. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 9] [Cited by in RCA: 12] [Article Influence: 2.0] [Reference Citation Analysis (0)] |
| 8. | Huang Z, Gu B. Circulating tumor DNA: a resuscitative gold mine? Ann Transl Med. 2015;3:253. [RCA] [PubMed] [DOI] [Full Text] [Cited by in RCA: 3] [Reference Citation Analysis (0)] |
| 9. | Wan JCM, Massie C, Garcia-Corbacho J, Mouliere F, Brenton JD, Caldas C, Pacey S, Baird R, Rosenfeld N. Liquid biopsies come of age: towards implementation of circulating tumour DNA. Nat Rev Cancer. 2017;17:223-238. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1634] [Cited by in RCA: 1767] [Article Influence: 220.9] [Reference Citation Analysis (0)] |
| 10. | PRISMA. [cited 28 February 2021]. Available from: http://prisma-statement.org/PRISMAStatement/. |
| 11. | Cai J, Chen L, Zhang Z, Zhang X, Lu X, Liu W, Shi G, Ge Y, Gao P, Yang Y, Ke A, Xiao L, Dong R, Zhu Y, Yang X, Wang J, Zhu T, Yang D, Huang X, Sui C, Qiu S, Shen F, Sun H, Zhou W, Zhou J, Nie J, Zeng C, Stroup EK, Chiu BC, Lau WY, He C, Wang H, Zhang W, Fan J. Genome-wide mapping of 5-hydroxymethylcytosines in circulating cell-free DNA as a non-invasive approach for early detection of hepatocellular carcinoma. Gut. 2019;68:2195-2205. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 206] [Cited by in RCA: 199] [Article Influence: 33.2] [Reference Citation Analysis (0)] |
| 12. | Junca A, Tachon G, Evrard C, Villalva C, Frouin E, Karayan-Tapon L, Tougeron D. Detection of Colorectal Cancer and Advanced Adenoma by Liquid Biopsy (Decalib Study): The ddPCR Challenge. Cancers (Basel). 2020;12. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 22] [Cited by in RCA: 18] [Article Influence: 3.6] [Reference Citation Analysis (0)] |
| 13. | Li S, Wang L, Zhao Q, Wang Z, Lu S, Kang Y, Jin G, Tian J. Genome-Wide Analysis of Cell-Free DNA Methylation Profiling for the Early Diagnosis of Pancreatic Cancer. Front Genet. 2020;11:596078. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 12] [Cited by in RCA: 27] [Article Influence: 5.4] [Reference Citation Analysis (0)] |
| 14. | Chen X, Gole J, Gore A, He Q, Lu M, Min J, Yuan Z, Yang X, Jiang Y, Zhang T, Suo C, Li X, Cheng L, Zhang Z, Niu H, Li Z, Xie Z, Shi H, Zhang X, Fan M, Wang X, Yang Y, Dang J, McConnell C, Zhang J, Wang J, Yu S, Ye W, Gao Y, Zhang K, Liu R, Jin L. Non-invasive early detection of cancer four years before conventional diagnosis using a blood test. Nat Commun. 2020;11:3475. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 416] [Cited by in RCA: 373] [Article Influence: 74.6] [Reference Citation Analysis (0)] |
| 15. | Tao K, Bian Z, Zhang Q, Guo X, Yin C, Wang Y, Zhou K, Wan S, Shi M, Bao D, Yang C, Xing J. Machine learning-based genome-wide interrogation of somatic copy number aberrations in circulating tumor DNA for early detection of hepatocellular carcinoma. EBioMedicine. 2020;56:102811. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 21] [Cited by in RCA: 48] [Article Influence: 9.6] [Reference Citation Analysis (0)] |
| 16. | Qu C, Wang Y, Wang P, Chen K, Wang M, Zeng H, Lu J, Song Q, Diplas BH, Tan D, Fan C, Guo Q, Zhu Z, Yin H, Jiang L, Chen X, Zhao H, He H, Li G, Bi X, Zhao X, Chen T, Tang H, Lv C, Wang D, Chen W, Zhou J, Cai J, Wang X, Wang S, Yan H, Zeng YX, Cavenee WK, Jiao Y. Detection of early-stage hepatocellular carcinoma in asymptomatic HBsAg-seropositive individuals by liquid biopsy. Proc Natl Acad Sci U S A. 2019;116:6308-6312. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 136] [Cited by in RCA: 138] [Article Influence: 23.0] [Reference Citation Analysis (0)] |
| 17. | Li J, Zhou X, Liu X, Ren J, Wang J, Wang W, Zheng Y, Shi X, Sun T, Li Z, Kang A, Tang F, Wen L, Fu W. Detection of Colorectal Cancer in Circulating Cell-Free DNA by Methylated CpG Tandem Amplification and Sequencing. Clin Chem. 2019;65:916-926. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 17] [Cited by in RCA: 27] [Article Influence: 4.5] [Reference Citation Analysis (0)] |
| 18. | Guler GD, Ning Y, Ku CJ, Phillips T, McCarthy E, Ellison CK, Bergamaschi A, Collin F, Lloyd P, Scott A, Antoine M, Wang W, Chau K, Ashworth A, Quake SR, Levy S. Detection of early stage pancreatic cancer using 5-hydroxymethylcytosine signatures in circulating cell free DNA. Nat Commun. 2020;11:5270. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 89] [Cited by in RCA: 115] [Article Influence: 23.0] [Reference Citation Analysis (0)] |
| 19. | Cristiano S, Leal A, Phallen J, Fiksel J, Adleff V, Bruhm DC, Jensen SØ, Medina JE, Hruban C, White JR, Palsgrove DN, Niknafs N, Anagnostou V, Forde P, Naidoo J, Marrone K, Brahmer J, Woodward BD, Husain H, van Rooijen KL, Ørntoft MW, Madsen AH, van de Velde CJH, Verheij M, Cats A, Punt CJA, Vink GR, van Grieken NCT, Koopman M, Fijneman RJA, Johansen JS, Nielsen HJ, Meijer GA, Andersen CL, Scharpf RB, Velculescu VE. Genome-wide cell-free DNA fragmentation in patients with cancer. Nature. 2019;570:385-389. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 797] [Cited by in RCA: 858] [Article Influence: 143.0] [Reference Citation Analysis (0)] |
| 20. | Jensen SØ, Øgaard N, Ørntoft MW, Rasmussen MH, Bramsen JB, Kristensen H, Mouritzen P, Madsen MR, Madsen AH, Sunesen KG, Iversen LH, Laurberg S, Christensen IJ, Nielsen HJ, Andersen CL. Novel DNA methylation biomarkers show high sensitivity and specificity for blood-based detection of colorectal cancer-a clinical biomarker discovery and validation study. Clin Epigenetics. 2019;11:158. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 50] [Cited by in RCA: 96] [Article Influence: 16.0] [Reference Citation Analysis (0)] |
| 21. | Nunes SP, Moreira-Barbosa C, Salta S, Palma de Sousa S, Pousa I, Oliveira J, Soares M, Rego L, Dias T, Rodrigues J, Antunes L, Henrique R, Jerónimo C. Cell-Free DNA Methylation of Selected Genes Allows for Early Detection of the Major Cancers in Women. Cancers (Basel). 2018;10. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 29] [Cited by in RCA: 53] [Article Influence: 7.6] [Reference Citation Analysis (0)] |
| 22. | Perrone F, Lampis A, Bertan C, Verderio P, Ciniselli CM, Pizzamiglio S, Frattini M, Nucifora M, Molinari F, Gallino G, Gariboldi M, Meroni E, Leo E, Pierotti MA, Pilotti S. Circulating free DNA in a screening program for early colorectal cancer detection. Tumori. 2014;100:115-121. [RCA] [PubMed] [DOI] [Full Text] [Cited by in RCA: 22] [Reference Citation Analysis (0)] |
| 23. | ROBINS-I tool. [cited 27 June 2021]. Available from: https://methods.cochrane.org/methods-cochrane/robins-i-tool. |
| 24. | Fiala C, Diamandis EP. Utility of circulating tumor DNA in cancer diagnostics with emphasis on early detection. BMC Med. 2018;16:166. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 138] [Cited by in RCA: 187] [Article Influence: 26.7] [Reference Citation Analysis (0)] |
| 25. | Newman AM, Bratman SV, To J, Wynne JF, Eclov NC, Modlin LA, Liu CL, Neal JW, Wakelee HA, Merritt RE, Shrager JB, Loo BW Jr, Alizadeh AA, Diehn M. An ultrasensitive method for quantitating circulating tumor DNA with broad patient coverage. Nat Med. 2014;20:548-554. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 1341] [Cited by in RCA: 1643] [Article Influence: 149.4] [Reference Citation Analysis (0)] |
| 26. | Lu JL, Liang ZY. Circulating free DNA in the era of precision oncology: Pre- and post-analytical concerns. Chronic Dis Transl Med. 2016;2:223-230. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 18] [Cited by in RCA: 25] [Article Influence: 2.8] [Reference Citation Analysis (0)] |
| 27. | Jin CE, Koo B, Lee TY, Han K, Lim SB, Park IJ, Shin Y. Simple and Low-Cost Sampling of Cell-Free Nucleic Acids from Blood Plasma for Rapid and Sensitive Detection of Circulating Tumor DNA. Adv Sci (Weinh). 2018;5:1800614. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 47] [Cited by in RCA: 52] [Article Influence: 7.4] [Reference Citation Analysis (0)] |
| 28. | Breitbach S, Tug S, Helmig S, Zahn D, Kubiak T, Michal M, Gori T, Ehlert T, Beiter T, Simon P. Direct quantification of cell-free, circulating DNA from unpurified plasma. PLoS One. 2014;9:e87838. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 95] [Cited by in RCA: 107] [Article Influence: 9.7] [Reference Citation Analysis (0)] |
| 29. | Parilla M, Ritterhouse LL. Beyond the Variants: Mutational Patterns in Next-Generation Sequencing Data for Cancer Precision Medicine. Front Cell Dev Biol. 2020;8:370. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 2] [Cited by in RCA: 6] [Article Influence: 1.2] [Reference Citation Analysis (0)] |
| 30. | Federici G, Soddu S. Variants of uncertain significance in the era of high-throughput genome sequencing: a lesson from breast and ovary cancers. J Exp Clin Cancer Res. 2020;39:46. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 56] [Cited by in RCA: 117] [Article Influence: 23.4] [Reference Citation Analysis (0)] |