Published online Oct 15, 2021. doi: 10.4251/wjgo.v13.i10.1244
Peer-review started: February 16, 2021
First decision: March 29, 2021
Revised: April 6, 2021
Accepted: August 2, 2021
Article in press: August 2, 2021
Published online: October 15, 2021
Processing time: 238 Days and 18.6 Hours
Gastric cancer represents a common and highly fatal malignancy, and thus a pathophysiology-based reconsideration is necessary, given the absence of efficient therapeutic regimens. In this regard, emerging data reveal a significant role of autophagy in gastric oncogenesis, progression, metastasis and chemoresistance. Although autophagy comprises a normal primordial process, ensuring cellular homeostasis under energy depletion and stress conditions, alterations at any stage of the complex regulatory system could stimulate a tumorigenic and promoting cascade. Among others, Helicobacter pylori infection induces a variety of signaling molecules modifying autophagy, during acute infection or after chronic autophagy degeneration. Subsequently, defective autophagy allows malignant transformation and upon cancer establishment, an overactive autophagy is stimulated. This overexpressed autophagy provides energy supplies and resistance mechanisms to gastric cancer cells against hosts defenses and anticancer treatment. This review interprets the implicated autophagic pathways in normal cells and in gastric cancer to illuminate the potential preventive, therapeutic and prognostic benefits of understanding and intervening autophagy.
Core Tip: Autophagy comprises a substantial normal cellular function, implicated in benign and malignant diseases. Its complex regulatory system can be affected at any stage by endogenous and environmental factors. Helicobacter pylori expresses and stimulates a wide range of autophagy modulators, thus promoting or inhibiting autophagy to yield a survival benefit and cause damage. Concerning gastric cancer, the dysregulated autophagic process facilitates tumor generation, progress and resistance to chemotherapy, through various mechanisms. The current review attempts to decrypt and simplify those mechanisms and the respective pathways, to explain their promising potential for the development of therapeutic agents and prognostic tools.
- Citation: Papaefthymiou A, Christodoulidis G, Koffas A, Doulberis M, Polyzos SA, Manolakis A, Potamianos S, Kapsoritakis A, Kountouras J. Role of autophagy in gastric carcinogenesis. World J Gastrointest Oncol 2021; 13(10): 1244-1262
- URL: https://www.wjgnet.com/1948-5204/full/v13/i10/1244.htm
- DOI: https://dx.doi.org/10.4251/wjgo.v13.i10.1244
Gastric cancer (GC) represents a global health burden, being fifth in annual worldwide incidence among malignancies, with more than one million new diagnoses every year[1]. A total of 1089103 GC patients were identified globally in 2020, and 768793 of them died from this malignancy, making it the third major cause of cancer-related death[2]. Anatomy of the stomach and paucity of early symptoms often delay diagnosis, especially in regions without screening programs in place. Patients with advanced or metastatic disease are not eligible for surgery and receive conventional chemotherapy, which may be associated with prolonged survival and improved quality of life[3]. Palliative measures can prolong survival, depending on the regimen and tumor’s genotypic characteristics; despite chemotherapy remaining the principal therapeutic option, the median overall survival of stage IV GC patients appears to be between 9-11 months[4,5]. In patients with localized or locally advanced disease, the prognosis remains dismal even after surgery and adjuvant chemotherapy, with more than 60% of patients relapsing, especially within 2 years post-surgery[6]. The latter underscores the necessity for further optimization, based on the conception of pathophysiological pathways of gastric oncogenesis[7-9].
Among other pathophysiological pathway mechanisms implicated in GC, autophagy appears to play a crucial role in gastric tumorigenesis, progression, metastasis and prognosis[10,11]. Autophagy (derived from the Greek term “αυτο-φαγία” meaning self-eating) was initially introduced in medical terminology in the middle of the 19th century to describe an endogenous, microenvironmental reaction of animal models to starvation. However, the decryption of implicated mechanisms required nearly a century to take place[12]. In 2016, the Japanese cell biologist, Ohsumi Y, was awarded with the Nobel Prize in Physiology and Medicine for his contribution to the comprehension and description of autophagy, through interpretation of genetic loci and pathways regulating this primitive cellular function[13].
Autophagy is a complex biological process that is essential for maintaining cellular homeostasis and cell survival[14,15]. It is an intracellular self-digestion pathway for the maintenance of homeostasis. This can be found as an important catabolic pathway in all eukaryotic cells and can be further activated by a variety of stimuli, including deprivation of nutrients, infection, hormones, oxidative stress and endoplasmic reticulum (ER) stress[16,17]. It is characterized by the formation of autophagosomes with double-membrane structure, which fuse with lysosomes to degrade cytosolic proteins, damaged or excess organelles, protein aggregates and invasive microbes[14,18].
Specifically, autophagy comprises a baseline recycling process of the cytoplasmic material to preserve natural homeostasis and unhindered power supply. Besides, energy deprivation, tissue stress and injury trigger more intense autophagy, to provide an interim nutritional reservoir, degrade the accumulated harmful molecules or stimulate the autophagy-related type II programmed cell death[19,20]. In this regard, a dysregulated autophagic process, either defective or overactive, is implicated in both benign and malignant diseases. In particular, specific mutations of autophagy-related genes (ATG) have been incriminated for susceptibility to infectious, autoimmune, autoinflammatory, metabolic, cardiovascular and neurodegenerative disorders as well as for predisposition to breast, ovarian, liver and gastrointestinal carcinogenesis, progression and chemotherapy resistance[21,22].
Regarding tumorigenesis, autophagy seems to play a dual role based on the cancer type, stage and genetic background of the patient. Initially, autophagy acts as a tumor suppressor by decreasing oncogenic protein p62 and assists in eliminating impaired organelles or DNA to impede further cellular damage and malignancy development[23,24]. Subsequently, autophagy can protect tumors from damages triggered by nutrient shortage, radiation and chemotherapeutics and strengthen tumor immune escape, metabolism and growth, resulting in drug resistance[23,25,26].
The increasing research on GC and autophagy interaction warrant a clinical approach to illuminate and simplify this relationship and the resulting feasible potential. This review summarizes the main principles of autophagy, its effect on gastric carcinogenesis and the respective impact on treatment and prognosis.
As aforementioned, autophagy comprises a substantial safety barrier of normal cells, providing a seamless functionality and integrity of cellular and tissue functions[27,28]. The autophagy-stimulating factors can define a possible substrate selectivity, and the final receiver of the under degraded molecules is in any case the lysosome.
The process of autophagy can be characterized by the following phases: (1) initiation; (2) vesicle nucleation; (3) vesicle elongation and maturation; (4) vesicle fusion, and (5) cargo degradation[29]. Each phase of autophagy is regulated by a diversity of ATGs, such as ATG5, ATG7, ATG12, ATG16L1 and their complexes[30].
There are three subtypes of autophagy depending on the mechanisms transporting the substrates in the lysosomes: macro-autophagy, micro-autophagy and chaperone-mediated autophagy (CMA). Macro-autophagy (the principal form of autophagy) is developed in response to stress and targets the release and degradation of every kind of aggregated protein and dysfunctional organelle by inducing a specific form of cytosolic vesicles, the autophagosomes[30]; the core machinery of autophagosome formation requires more than 40 largely conserved ATG[31]. Micro-autophagy, a non-selective procedure, is activated commonly to ingest senescent endoplasmic membranes by direct engulfment in the lysosome membrane[32]; it is the only constitutive autophagy mechanism that triggers small cytoplasmic debris engulfment into the lysosome for degradation[31]. CMA is a main route for the elimination of cellular aberrant proteins and can provide a cytoprotective effect[33]; it is an intracellular catabolic pathway that mediates the degradation of soluble cytosolic proteins in lysosomes[34].
In contrast to macro-autophagy and micro-autophagy, CMA protein targets are recognized one-by-one by the cytosolic chaperone heat shock-cognate protein Hsc70, which along with its modulatory co-chaperones transports substrates to the lysosome’s surface[35]. CMA selectivity is conferred by a specific sequence existing in all CMA target proteins[36]. Specific amino-acid sequences, KFERQ, are recognized by Hsc70 and tethered by cytosolic vectors[34-36]. These vectors are ligated to the complementary lysosome receptors LAMP-2A and transport the substrate proteins, enzymes, transcriptional factors and nucleosomes to recycling[35].
The most common and intensively studied subtype of autophagy is the mentioned macro-autophagy acting both selectively and non-selectively, depending on the triggering event, microenvironmental conditions and targeting cargo[37-39]. An isolated phagophore membrane, originating from the ER, matures around the targeted substances to form a closed vesicle, the autophagosome. Upon generation, autophagosome fuses with the lysosome to form the autophagolysosome. After the cargo’s disposal at the lysosome, the endogenous hydrolases degrade all included molecules, and the products are subsequently released in the cytoplasm as raw material for further cellular functions[40].
The autophagic pathway is regulated by complex molecular signals. The initial stimulus inducing autophagy is energy depletion, which modifies the expression of two mainstay molecule regulators of autophagy inception: mammalian target of rapamycin complex 1 (mTORC1) and adenosine monophosphate-activated protein kinase (AMPK). These proteins act oppositely with the former inhibiting autophagy initiation. In this regard, mTORC1 and mTORC2 have a common central kinase, mTOR, a conserved serine/threonine protein kinase, which plays important roles in multiple biological processes involved in cell growth, such as regulating autophagy[41]. mTORC1, a primary regulator of autophagy, acts as a mediator of normal cellular processes, such as proliferation, metabolism and protein synthesis and is regulated by a plethora of molecules. mTORC1 exhibits a major role in controlling cell growth and cellular metabolism by integrating different external and internal signals, including growth factors, amino acids, glucose and energy status[42].
Its dysregulation contributes to several pathologies including malignancies, diabetes mellitus and cardiovascular diseases[43]. Likewise, suppression of mTORC1 leads to activation of a critical molecule, Unc-51-like autophagy activating kinase 1 (ULK1), which translocates to the ER for the initiation of autophagy[43]. In this regard, under nutrient deficiency, hypoxia, inflammatory and/or infectious conditions, autophagy is initiated by the ULK1 complex[44]. In contrast, upregulated mTORC1, under normal conditions, inhibits the autophagy promoter ULK1 through phosphorylation[44].
Moreover, several signaling pathways, such as the intracellular phosphatidylinositol kinase (PI3K), mitogen activated kinase-like protein, AMPK, p53 and phosphatase and tensin homolog, can control mTORC1 and autophagy. More specifically, hormonal and cytokine signals activate PI3K and stimulate the generation of the Akt/PKB complex, which activates mTORC1 and directly inhibits AMPK. Conversely, tissue stress, hypoxia, low glycose, and increased AMP/ATP ratio induce AMPK expression, which triggers autophagy directly or through mTORC1 phosphorylation inactivation[10]. Thereafter, the autophagy cascade is upregulated through a primary ubiquitin complex (Ulk1, FIP200, ATG13) generation[45], which catalyzes the accumulation of PI3KIII, Beclin1, Vps34, Vps15 and ATG14L molecules to synthesize phosphatidylinositol 3 phosphate. In particular, the recruitment of an ATG14-containing Class III PI3K complex to specific sites in the ER plays a critical role in the induction of autophagy during starvation, a process in which the PI3K effectors WIPI1/2 are thought to play a major role[46]. Phosphatidylinositol 3 phosphate stimulates the formation of the phagophore membrane, and further genetic interactions and molecular activations compose a nodal cluster of ATG12-ATG5-ATG16L to guide the maturation of the phagophore membrane[47].
In parallel, light chain 3-I (LC3-I) proteins are catalyzed to LC3-II and are incorporated into the autophagosome membrane as receptors, facilitating its conjunction with lysosomes[48,49]. Moreover, after ligation with specific cytosolic proteins, such as p62, LC3-II recognizes and opsonizes the targets’ loci to transfer them in the autophagosomes[50]. More specifically, during autophagy, LC3-I, the cytosolic form of LC3 (a microtubule-associated protein that is constitutively expressed in mammalian tissues) binds to phosphatidylethanolamine to form LC3-II, which is transported to autophagosome membranes[51]. When autophagosomes are fused to lysosomes to form autophagosomes, LC3-II in autophagosomes is degraded[51]. Therefore, the relative ratio of LC3-I/LC3-II expression can be used to monitor autophagy progression[18].
Except for the stem regulators of autophagy, a plethora of further environmental, genetic and cytoplasmic stimuli could modify this cascade at any stage. Environmental factors, diet and microbiota preserve a perpetual impact on DNA methylation or histone modification, thus causing epigenetic changes. These alterations and further stressors are capable of up- or downregulating the expression of specific transcriptional factors or gene adjusters, such as activator of transcription (STAT) 3 and microRNAs (miR), to modify autophagy with respect to cellular needs[52]. Speci
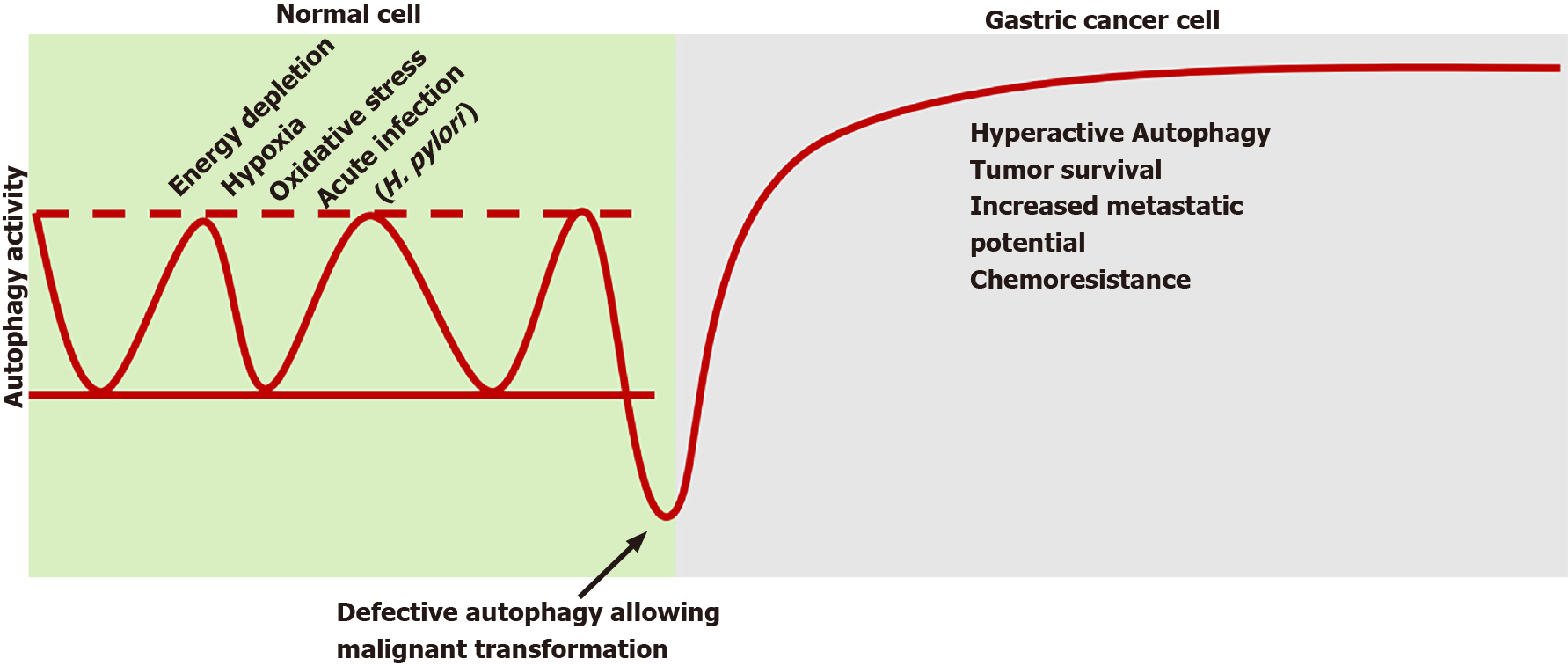
It is well accepted that autophagy plays a critical role in cancers[54]. Autophagy appears to play a dual role in cancer (Figure 1). First, it can eliminate the injured cellular constituents or whole cells. Second, the energy or nutrients as a result of degradation can provide malignant cells nutrition or be recycled for apoptosis-associated protein synthesis of apoptosis relative proteins[55]. Specifically, autophagy participates in oncogenesis and tumor progression via two main pillars: a defective pro-death and an excessive pro-survival action.
Considering the first axis, baseline autophagy in normal cells provides a protective effect on tissue homeostasis by supervising inflammation, injury and genetic instability to suppress potential derogations[19]. Considering the second axis, the possible defective repairing and tissue imbalance trigger the autophagic cell death to ensure the engraving of premalignant foci. Preclinical studies revealed that suppressing autophagy, through respective gene inhibition, promoted oxidative stress, genetic instability, p62 protein accumulation and finally carcinogenesis[56].
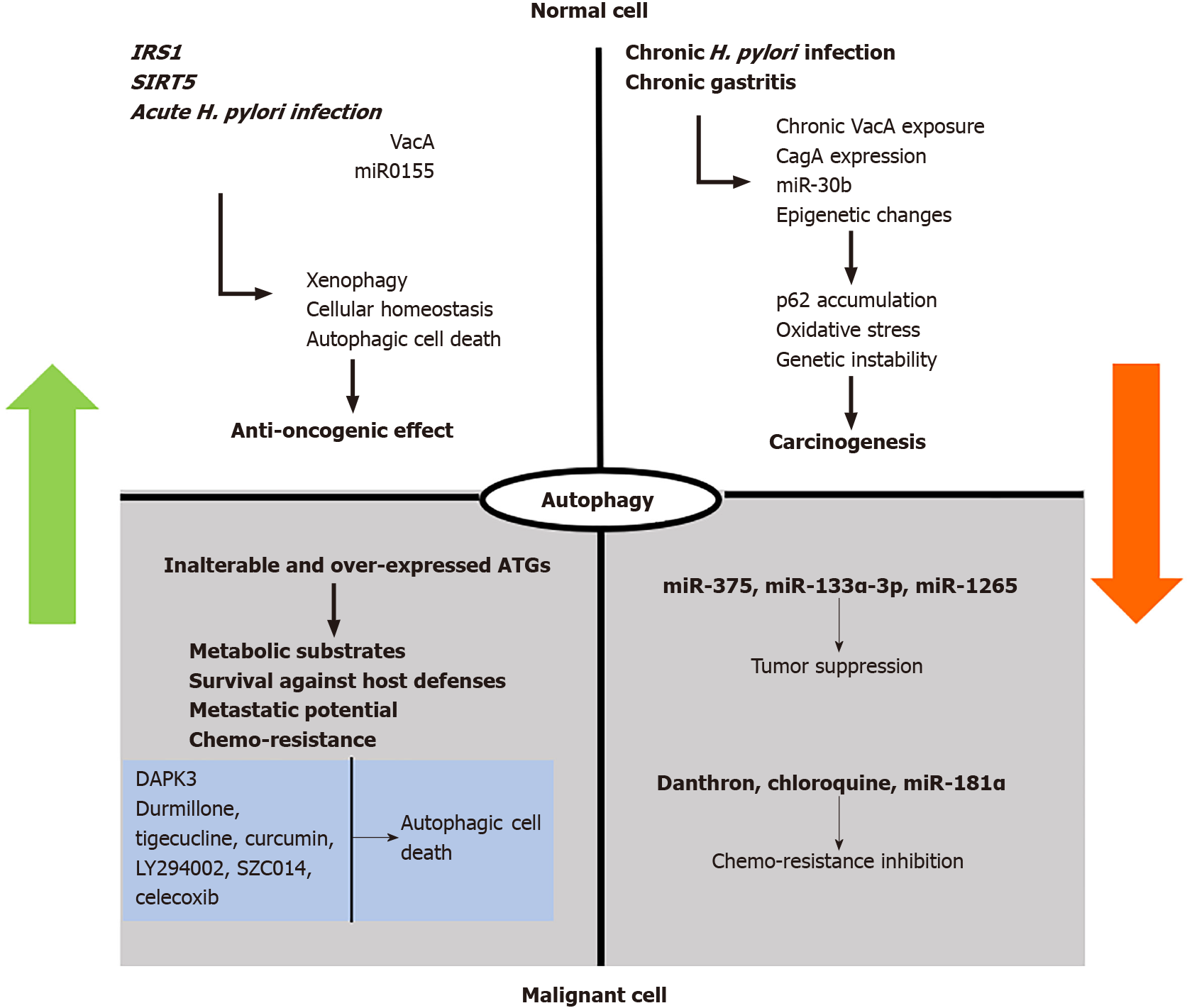
Recently, IRS1, an oncogene implicated in a wide variety of cancers, was suggested to promote gastric carcinogenesis, though the existence of a single nucleotide polymorphism of this gene increased baseline autophagy, therefore inhibiting tumor progression[57]. The IRS1 appears to be a specific biomarker for GC[57]. Recent data also indicate that sirtuins, particularly sirtuin 5, which regulate autophagy and apoptosis in tumor cells, seem to be critical regulators of autophagy and apoptosis in GC cell lines by providing the balance of autophagy and apoptosis[58].
Undergoing malignant transformation, tumor cells modulate autophagy to obtain survival benefit in the stressful microenvironment. In contrast to other genetic positions, autophagy genes remain generally inalterable in the majority of cancers[59]. The increased baseline autophagy in cancer cells provides substrates and energy to replenish hypoxia and under irrigation. Concerning the gastric acidic environment, cancer cells overexpress acid-sensing ion channels, which induce ATG5-mediated autophagy to facilitate tumor survival[60]. The respective overactive pathway in GC cells can be affected by intracellular miR-375, miR-133a-3p and miR-1265 to reduce cellular proliferation and metastatic potential[61,62].
Autophagy plays a significant role in tumor chemotherapy. For example, danthron, 1, 8-dihydroxyanthrquinone, isolated from Pheum palmatum, exhibited a suppressive effect on cancer cell autophagy, thereby suggesting a relative chemotherapeutic benefit[63]. Nevertheless, autophagy induction is still undeniably considered as a potential strategy for cancer prevention, resulting in type II programmed cell death. A relative study suggested that durmillone, a compound isolated from stems of Millettia pachyloba Drake, displayed the best activity among flavonoids that induced autophagy, in both HeLa and MCF-7 cells. A significant upregulation in expression of LC3-II, Beclin1 and Atg7 was detected after intervention of durmillone. Likewise, durmillone induced apoptosis, in dose-dependent manner[64].
Besides, autophagy adjustment after exposure to conventional chemotherapy generates chemoresistance of malignant cells, occasionally reversed after autophagy inhibitors[10]. Chemotherapy administration, such as cisplatin, induces MALAT1 expression, which antagonizes miR-23b-3p in autophagosome inhibition, thus providing chemoresistance. miR-181a reverses this action, thus providing an adjuvant option for future therapeutics[61,62].
Nevertheless, upregulated autophagy is not necessarily helpful for malignant aggregates, but it largely depends on the stimuli. In particular, Li et al[65] documented that death-associated protein kinase 3, after phosphorylating ULK1, induces intratumor autophagic cell death and provides a positive death-associated protein kinase 3 feedback. Interestingly, metastatic GC lesions presented defective death-associated protein kinase 3 expression, thus promising a potential future prognostic marker and a therapeutic target[65].
Helicobacter pylori (H. pylori) infection (H. pylori-I) induces a plethora of autophagy-modifying molecules to yield its survival in a host microenvironment.
The H. pylori-related vacuolating cytotoxin (VacA), a vital bacterial virulence factor, is a secreted toxin that inserts into host cell membranes, forming a chloride channel. VacA appears to induce more severe pathologies including GC[66], though the mechanisms underlying VacA effects remain insufficient. It is initially recognized by the host defensive mechanisms, thus triggering an autophagy over reaction and autophagosome generation to ensure direct H. pylori clearance and chronicity avoidance[67]. Nevertheless, the continuous exposure to VacA provokes tolerance of autophagic reaction and inhibition of autophagosome maturation, thus promoting carcinogenesis by favoring the accumulation of oxidative substances and tumorigenic molecules, such as p62 and nuclear factor-erythroid 2-related factor 2 and the reduction of nuclear factor κB[68,69]. A potential mechanism of VacA-mediated autophagy disruption consists of its direct effect on the lysosomal calcium channels to production of deformational, dysfunctional and attenuated vesicles, thus providing a survival benefit of H. pylori against natural defense and antibiotic treatment[70].
Specifically, VacA impairs host endolysosomal trafficking, thus inducing accumulation of dysfunctional lysosomes and autophagosomes[69]. H. pylori-associated VacA usurps the lysosomal calcium transient receptor potential membrane channel mucolipin 1 (TRPML1; also recognized as ML), thereby reducing lysosomal and autophagic killing to promote an intracellular niche that permits H. pylori survival[71]. In contrast, a molecule directed against TRPML1 inverted the VacA toxic effects on endolysosomal trafficking, thereby contributing to the clearance of intracellular H. pylori. Thus, TRPML1 might represent a therapeutic target for chronic H. pylori infection[71].
Beyond VacA, H. pylori induces defective autophagy or inhibition by means of H. pylori-related cytotoxin-associated gene A (CagA) resulting in gastric oncogenesis[72]. H. pylori-related CagA protein was reported to negatively regulate autophagy via the c-Met-PI3k/Akt-mTOR signaling pathway and promote inflammatory process[72,73]. Likewise, some data suggest that H. pylori-related CagA may represent the Trojan Horse of H. pylori resistance to autophagy[74]. Moreover, H. pylori may promote inflammatory process via H. pylori-related CagA-dependent activation of the mentioned mTORC[75]. Besides, H. pylori produces outer membrane vesicles (OMVs) that contain various virulence factors; H. pylori OMVs may amplify its virulence[76,77]. H. pylori-related OMVs can induce autophagy[78], relying on the nucleotide-binding oligomerization domain-1-receptor interacting serine/threonine kinase 2 signaling pathway, which is crucial for the induction of autophagy and the production of interleukin 8[78,79]. Likewise, H. pylori OMVs stimulate autophagosome formation, which is not dependent on VacA[78], and by inducing interleukin 8 production and nuclear factor κB activation may contribute to gastric pathologies[80].
H. pylori secretory protein HP0175, an inducer of apoptosis in gastric epithelial cells[81], can also regulate PKR-like ER kinase, which in turn activates the transcription of ATF4 and CHOP, resulting in the induction of autophagy in gastric epithelial cells[82]. Likewise, HP0175 can upregulate the expression of ATGs independently to functional VacA during acute infection[82]. Moreover, HP0175 appears to stimulate epidermal growth factor receptor-dependent vascular endothelial growth factor production in the GC cell line, thereby possibly contributing to gastric cancer development and/or progression[83,84]. Similarly, Bravo et al[85] recently described the virulent role of H. pylori-expressed gamma glutamyl transpeptidase, which suppresses the late stages of autophagic defenses to establish permanent H. pylori-I, by inhibiting cathepsin B activity in the lysosomes.
H. pylori-induced prolonged inflammatory process may progress to gastric oncogenesis[86,87]. Autophagy seems to play a role in H. pylori-associated gastritis, and long-term H. pylori-I has been reported to promote GC development and progression by dramatically impairing autophagy[88]. In this regard, autophagy seems to be implicated in H. pylori-related intestinal metaplasia; after infected parietal cell death, chief cells decrease the expression of the zymogen granule maturation transcription factor Mist1, thus triggering the autophagic degradation of zymogen granules to dematurate and reprogram the chief cell phenotype[89]. Additional data indicate that, as chronic gastritis progresses to atrophy, ATG16L1 gene expression becomes defective, thus resulting into incomplete autophagosome formation[11]. Moreover, H. pylori-I is associated with raised levels of p62 in gastric intestinal metaplasia and reduced levels of Rad51 in gastric dysplasia, thus providing a mechanism into the connection of H. pylori-I, autophagy, DNA injury and gastric oncogenesis[90].
Specifically, several genes appear to play principal roles in autophagy-induced oncogenesis such as p62[91], and chronic H. pylori-I provokes the loss of autophagy and consequent accumulation of autophagic substrate p62, leading to ubiquitination of Rad51, thereby suppressing capability of the DNA damage repair[90]. DNA injury has been connected to autophagy[92], and the Rad51 repair protein catalyzes homologous DNA strand pairing and exchange, whereas its genetic polymorphism may be a critical predictor for gastric malignancy of H. pylori positive patients[93]. On the other hand, Mills and Sansom[89] indicated that autophagic activity, measured by ATG5, LC3A and LC3B expression, had an increasing tendency during progression to intestinal metaplasia, followed by a gradual reduction in this rate upon metaplasia establish
Moreover, H. pylori has been incriminated for epigenetic inactivation of autophagy regulating genes, such as microtubule-associated protein 1 light chain 3 variant 1, thus inhibiting autophagic cell death and provoking cellular proliferation and invasiveness in vitro, implying a potential carcinogenic pathway in vivo[94]. Data from cultures of H. pylori-related GC stem cells indicate an overexpression of autophagy markers, reflecting intensive baseline autophagy to assist their survival in harsh malignant environments[95]. The resultant suppressive autophagy allows the intracellular oxidative stress and nucleic acid alterations to induce malignant mutations. Recently, Piao et al[96] revealed that H. pylori-I modifies the function of the signal transducer and STAT3 by phosphorylating its Ser727. Thus a perpetual mitophagy is mediated and promotes gastritis and carcinogenesis[96].
However, preclinical studies reported a protective role of autophagy against H. pylori-I, as the knockdown of the ATG16L1 gene of autophagy resulted in increased vulnerability of rodents in H. pylori-I and propensity to chronic infection[69]. Additionally, miR-155 triggers the autophagy-related H. pylori clearance, and miR-99b suppresses the oncogenic role of H. pylori-I[61,62]. Nevertheless, the success of autophagy-mediated H. pylori eradication is the exception, as it escapes initial defenses and reprograms the baseline autophagy to H. pylori beneficial diversion, thus yielding bacterial survival and chronic gastritis.
The illumination of the interaction between H. pylori and autophagy motivated researchers to investigate the impact of autophagy regulators on H. pylori eradication in order to ameliorate the increasing antibiotic resistance. In this regard, Hu et al[97] at a preclinical level suggested that vitamin D supplementation facilitated H. pylori eradication, even in cases of antibiotic-resistant strains, by recovering the deactivated autophagic function of lysosomes[97]. In this respect, recent data indicate the occurrence of a new antibacterial signaling pathway of vitamin D3 via the activation of PDIA3/STAT3 - MCOLN3 - Ca2+ axis, to reactivate the lysosomal acidification and degradation function of autolysosomes, which is the main signaling pathway for the antibacterial action of vitamin D3 both in cells and animals and possibly in humans[98]. Moreover, simvastatin treatment in H. pylori-infected macrophages promoted the maturation of the phagophore membrane to autophagosomes and their fusion with lysosomes, thus yielding H. pylori clearance[99].
The most studied clinical impact of autophagy on GC consists of the provided drug resistance, due to its increased baseline activity. Cisplatin treatment induces an endogenous protective reaction of GC cells by triggering resistance via ATG5 stimulation[100]. In this regard, supplementation treatment with autophagy inhibitors, such as miR-199a-5p/30a, miR-181a or miR-23b-3p, may prove a promising adjuvant approach to sensitize malignant cells to conventional therapy[101-103]. Moreover, GC stem cells develop resistance mechanisms through autophagy, suppressed by chloroquine co-administration with 5-fluorouracil through inhibition of the Notch-1 pathway[104]. To date, a plethora of autophagy regulators have been studied concerning their impact on cancer therapeutics. Autophagy inhibitors, such as chloroquine and 3-methyladenine, improved the outcomes of conventional chemotherapy probably by sensitizing malignant cells, thus yielding increased survival compared to chemotherapy used as monotherapy[105,106]. Preclinical studies of autophagy promoters suggested that tigecycline, curcumin, LY294002, SZC014 and celecoxib could stimulate type II programmed cell death in GC cells[105-110]. Similarly, in vivo co-administration of everolimus, an mTOR agonist, with cisplatin prolonged survival in subjects with advanced GC through disease stabilization, although no regression was achieved[106,111].
Additionally, the role of autophagy in metastatic potential of GC cells is also important. In this regard, STRT1 triggers autophagy, by deacetylating ATGs to modify epithelial-to-mesenchymal transition and thus lymph node and/or distal metastasis in GC[112,113]. Upon insertion in the systematic circulation and migration to distal sites, malignant cells experience potent metabolic, hypoxic and immune stress. Subsequently, the tumor interim autophagy serves as the necessary nutritional substrate for cancer cell survival, promotes neo-angiogenesis and neutralizes the toxic effect of host defensive mechanisms[114,115].
Recent data indicate that TDB, a chemically synthesized derivative of benzoxazole, inhibits the proliferation of human GC MGC-803 cells by inducing autophagy and apoptosis exerted via the PI3K/AKT/mTOR pathway. Inhibiting autophagy also increased apoptosis. Furthermore, TDB showed good antitumor activity in vivo, thereby providing a potential new targeted drug for the treatment of GC[116]. Moreover, the mentioned autophagy-related Beclin 1 protein might be a potential marker of gastric carcinogenesis, aggressiveness and prognostic prediction. Beclin 1-related autophagy seems to promote GC at an early clinal stage and thus appears to be a novel target of gene therapy in GC[117,118]. Likewise, recent data suggest that quantification of autophagic activity in GC tissues could provide a prognostic tool for short- and long-term survival.
More specifically, Qiu et al[119], after retrospectively analyzing 208 ATGs, identified the transcriptome calculation of four of them, GRID2, ATG4D, GABARAPL2 and CXCR4, could be incorporated in a prognostic model. Increased expression of these genes determined a high-risk group with diminished survival rates compared to the low-risk group. After evaluation of additional confounders, a nomogram was suggested to predict the overall 3 year and 5 year survival[119]. Other investigators, by introducing the Kaplan–Meier plotter online database to assess the value of ATG gene expression levels in overall survival prediction in GC patients with different clinical stage, differentiation, gender, HER2 status and therapeutic strategy showed the following: increased levels of ATG3, ATG4C, ATG5 and ATG10 mRNA were correlated with prolonged overall survival, whereas high levels ATG4B, ATG7, ATG12, ATG16L1 and TECPR1 mRNA were correlated with negative overall survival in patients with GC. Therefore, individual ATG estimation by using Kaplan-Meier plotter analysis may offer a guide to clinical therapeutic strategy by means of individualizing gene therapy for GC patients[55].
Autophagy constitutes a primary, fundamental cellular function to preserve natural homeostasis, regulated by complex mechanisms. Although it provides an anti-oncogenic shield in normal cells, a plethora of modifying stimuli, such as H. pylori-I, inhibit its action thus facilitating malignant transformation. Once malignancy is established, excessive autophagy favors optimal conditions for cancer survival, progression and treatment resistance. Figure 2 summarizes this stimuli-dependent role of autophagy in normal and malignant gastric cells.
The comprehension of autophagy pathway and the respective regulatory mechanisms provides an invaluable opportunity to decrypt further factors implicated in gastric carcinogenesis but also to apply this knowledge in clinical practice. The autophagy cascade provides a wide range of molecule biomarkers that could serve as components for prognostic stratification and a target of novel treatment strategies, especially considering cases with advanced non-operable disease.
Manuscript source: Invited manuscript
Specialty type: Oncology
Country/Territory of origin: Greece
Peer-review report’s scientific quality classification
Grade A (Excellent): 0
Grade B (Very good): B
Grade C (Good): 0
Grade D (Fair): 0
Grade E (Poor): 0
P-Reviewer: Liu XM S-Editor: Gao CC L-Editor: Filipodia P-Editor: Guo X
| 1. | Bray F, Ferlay J, Soerjomataram I, Siegel RL, Torre LA, Jemal A. Global cancer statistics 2018: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J Clin. 2018;68:394-424. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 53206] [Cited by in RCA: 55806] [Article Influence: 7972.3] [Reference Citation Analysis (132)] |
| 2. | Yang L, Ying X, Liu S, Lyu G, Xu Z, Zhang X, Li H, Li Q, Wang N, Ji J. Gastric cancer: Epidemiology, risk factors and prevention strategies. Chin J Cancer Res. 2020;32:695-704. [RCA] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 38] [Cited by in RCA: 145] [Article Influence: 29.0] [Reference Citation Analysis (0)] |
| 3. | Sjöstedt S, Pieper R. Gastric cancer. Factors influencing longterm survival and postoperative mortality. Acta Chir Scand Suppl. 1986;530:25-29. [PubMed] |
| 4. | Boku N, Yamamoto S, Fukuda H, Shirao K, Doi T, Sawaki A, Koizumi W, Saito H, Yamaguchi K, Takiuchi H, Nasu J, Ohtsu A; Gastrointestinal Oncology Study Group of the Japan Clinical Oncology Group. Fluorouracil vs combination of irinotecan plus cisplatin vs S-1 in metastatic gastric cancer: a randomised phase 3 study. Lancet Oncol. 2009;10:1063-1069. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 449] [Cited by in RCA: 475] [Article Influence: 29.7] [Reference Citation Analysis (0)] |
| 5. | Zhang F, Huang X, Song Y, Gao P, Zhou C, Guo Z, Shi J, Wu Z, Wang Z. Conversion Surgery for Stage IV Gastric Cancer. Front Oncol. 2019;9:1158. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 16] [Cited by in RCA: 28] [Article Influence: 4.7] [Reference Citation Analysis (0)] |
| 6. | Liu D, Lu M, Li J, Yang Z, Feng Q, Zhou M, Zhang Z, Shen L. The patterns and timing of recurrence after curative resection for gastric cancer in China. World J Surg Oncol. 2016;14:305. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 52] [Cited by in RCA: 95] [Article Influence: 10.6] [Reference Citation Analysis (0)] |
| 7. | Van Cutsem E, Moiseyenko VM, Tjulandin S, Majlis A, Constenla M, Boni C, Rodrigues A, Fodor M, Chao Y, Voznyi E, Risse ML, Ajani JA; V325 Study Group. Phase III study of docetaxel and cisplatin plus fluorouracil compared with cisplatin and fluorouracil as first-line therapy for advanced gastric cancer: a report of the V325 Study Group. J Clin Oncol. 2006;24:4991-4997. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1331] [Cited by in RCA: 1457] [Article Influence: 76.7] [Reference Citation Analysis (0)] |
| 8. | Shah MA, Bang YJ, Lordick F, Alsina M, Chen M, Hack SP, Bruey JM, Smith D, McCaffery I, Shames DS, Phan S, Cunningham D. Effect of Fluorouracil, Leucovorin, and Oxaliplatin With or Without Onartuzumab in HER2-Negative, MET-Positive Gastroesophageal Adenocarcinoma: The METGastric Randomized Clinical Trial. JAMA Oncol. 2017;3:620-627. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 247] [Cited by in RCA: 251] [Article Influence: 31.4] [Reference Citation Analysis (0)] |
| 9. | Fuchs CS, Shitara K, Di Bartolomeo M, Lonardi S, Al-Batran SE, Van Cutsem E, Ilson DH, Alsina M, Chau I, Lacy J, Ducreux M, Mendez GA, Alavez AM, Takahari D, Mansoor W, Enzinger PC, Gorbounova V, Wainberg ZA, Hegewisch-Becker S, Ferry D, Lin J, Carlesi R, Das M, Shah MA; RAINFALL Study Group. Ramucirumab with cisplatin and fluoropyrimidine as first-line therapy in patients with metastatic gastric or junctional adenocarcinoma (RAINFALL): a double-blind, randomised, placebo-controlled, phase 3 trial. Lancet Oncol. 2019;20:420-435. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 170] [Cited by in RCA: 230] [Article Influence: 38.3] [Reference Citation Analysis (0)] |
| 10. | Cao Y, Luo Y, Zou J, Ouyang J, Cai Z, Zeng X, Ling H, Zeng T. Autophagy and its role in gastric cancer. Clin Chim Acta. 2019;489:10-20. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 71] [Cited by in RCA: 123] [Article Influence: 17.6] [Reference Citation Analysis (0)] |
| 11. | Zhang F, Chen C, Hu J, Su R, Zhang J, Han Z, Chen H, Li Y. Molecular mechanism of Helicobacter pylori-induced autophagy in gastric cancer. Oncol Lett. 2019;18:6221-6227. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 14] [Cited by in RCA: 23] [Article Influence: 3.8] [Reference Citation Analysis (0)] |
| 12. | Ktistakis NT. In praise of M. Anselmier who first used the term "autophagie" in 1859. Autophagy. 2017;13:2015-2017. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 17] [Cited by in RCA: 27] [Article Influence: 3.4] [Reference Citation Analysis (0)] |
| 13. | Tsukada M, Ohsumi Y. Isolation and characterization of autophagy-defective mutants of Saccharomyces cerevisiae. FEBS Lett. 1993;333:169-174. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1305] [Cited by in RCA: 1387] [Article Influence: 43.3] [Reference Citation Analysis (0)] |
| 14. | Colletti M, Ceglie D, Di Giannatale A, Nazio F. Autophagy and Exosomes Relationship in Cancer: Friends or Foes? Front Cell Dev Biol. 2020;8:614178. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 10] [Cited by in RCA: 38] [Article Influence: 9.5] [Reference Citation Analysis (0)] |
| 15. | Mizushima N. Autophagy: process and function. Genes Dev. 2007;21:2861-2873. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 2692] [Cited by in RCA: 3071] [Article Influence: 170.6] [Reference Citation Analysis (0)] |
| 16. | Glick D, Barth S, Macleod KF. Autophagy: cellular and molecular mechanisms. J Pathol. 2010;221:3-12. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 2980] [Cited by in RCA: 2786] [Article Influence: 185.7] [Reference Citation Analysis (0)] |
| 17. | Chen D, Zheng K, Wu H, Zhang X, Ye W, Tan X, Xiong Y. Lin28a attenuates cerebral ischemia/reperfusion injury through regulating Sirt3-induced autophagy. Brain Res Bull. 2021;170:39-48. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 7] [Cited by in RCA: 6] [Article Influence: 1.5] [Reference Citation Analysis (0)] |
| 18. | Klionsky DJ, Abdelmohsen K, Abe A, Abedin MJ, Abeliovich H, Acevedo Arozena A, Adachi H, Adams CM, Adams PD, Adeli K, Adhihetty PJ, Adler SG, Agam G, Agarwal R, Aghi MK, Agnello M, Agostinis P, Aguilar PV, Aguirre-Ghiso J, Airoldi EM, Ait-Si-Ali S, Akematsu T, Akporiaye ET, Al-Rubeai M, Albaiceta GM, Albanese C, Albani D, Albert ML, Aldudo J, Algül H, Alirezaei M, Alloza I, Almasan A, Almonte-Beceril M, Alnemri ES, Alonso C, Altan-Bonnet N, Altieri DC, Alvarez S, Alvarez-Erviti L, Alves S, Amadoro G, Amano A, Amantini C, Ambrosio S, Amelio I, Amer AO, Amessou M, Amon A, An Z, Anania FA, Andersen SU, Andley UP, Andreadi CK, Andrieu-Abadie N, Anel A, Ann DK, Anoopkumar-Dukie S, Antonioli M, Aoki H, Apostolova N, Aquila S, Aquilano K, Araki K, Arama E, Aranda A, Araya J, Arcaro A, Arias E, Arimoto H, Ariosa AR, Armstrong JL, Arnould T, Arsov I, Asanuma K, Askanas V, Asselin E, Atarashi R, Atherton SS, Atkin JD, Attardi LD, Auberger P, Auburger G, Aurelian L, Autelli R, Avagliano L, Avantaggiati ML, Avrahami L, Awale S, Azad N, Bachetti T, Backer JM, Bae DH, Bae JS, Bae ON, Bae SH, Baehrecke EH, Baek SH, Baghdiguian S, Bagniewska-Zadworna A, Bai H, Bai J, Bai XY, Bailly Y, Balaji KN, Balduini W, Ballabio A, Balzan R, Banerjee R, Bánhegyi G, Bao H, Barbeau B, Barrachina MD, Barreiro E, Bartel B, Bartolomé A, Bassham DC, Bassi MT, Bast RC Jr, Basu A, Batista MT, Batoko H, Battino M, Bauckman K, Baumgarner BL, Bayer KU, Beale R, Beaulieu JF, Beck GR Jr, Becker C, Beckham JD, Bédard PA, Bednarski PJ, Begley TJ, Behl C, Behrends C, Behrens GM, Behrns KE, Bejarano E, Belaid A, Belleudi F, Bénard G, Berchem G, Bergamaschi D, Bergami M, Berkhout B, Berliocchi L, Bernard A, Bernard M, Bernassola F, Bertolotti A, Bess AS, Besteiro S, Bettuzzi S, Bhalla S, Bhattacharyya S, Bhutia SK, Biagosch C, Bianchi MW, Biard-Piechaczyk M, Billes V, Bincoletto C, Bingol B, Bird SW, Bitoun M, Bjedov I, Blackstone C, Blanc L, Blanco GA, Blomhoff HK, Boada-Romero E, Böckler S, Boes M, Boesze-Battaglia K, Boise LH, Bolino A, Boman A, Bonaldo P, Bordi M, Bosch J, Botana LM, Botti J, Bou G, Bouché M, Bouchecareilh M, Boucher MJ, Boulton ME, Bouret SG, Boya P, Boyer-Guittaut M, Bozhkov PV, Brady N, Braga VM, Brancolini C, Braus GH, Bravo-San Pedro JM, Brennan LA, Bresnick EH, Brest P, Bridges D, Bringer MA, Brini M, Brito GC, Brodin B, Brookes PS, Brown EJ, Brown K, Broxmeyer HE, Bruhat A, Brum PC, Brumell JH, Brunetti-Pierri N, Bryson-Richardson RJ, Buch S, Buchan AM, Budak H, Bulavin DV, Bultman SJ, Bultynck G, Bumbasirevic V, Burelle Y, Burke RE, Burmeister M, Bütikofer P, Caberlotto L, Cadwell K, Cahova M, Cai D, Cai J, Cai Q, Calatayud S, Camougrand N, Campanella M, Campbell GR, Campbell M, Campello S, Candau R, Caniggia I, Cantoni L, Cao L, Caplan AB, Caraglia M, Cardinali C, Cardoso SM, Carew JS, Carleton LA, Carlin CR, Carloni S, Carlsson SR, Carmona-Gutierrez D, Carneiro LA, Carnevali O, Carra S, Carrier A, Carroll B, Casas C, Casas J, Cassinelli G, Castets P, Castro-Obregon S, Cavallini G, Ceccherini I, Cecconi F, Cederbaum AI, Ceña V, Cenci S, Cerella C, Cervia D, Cetrullo S, Chaachouay H, Chae HJ, Chagin AS, Chai CY, Chakrabarti G, Chamilos G, Chan EY, Chan MT, Chandra D, Chandra P, Chang CP, Chang RC, Chang TY, Chatham JC, Chatterjee S, Chauhan S, Che Y, Cheetham ME, Cheluvappa R, Chen CJ, Chen G, Chen GC, Chen H, Chen JW, Chen JK, Chen M, Chen P, Chen Q, Chen SD, Chen S, Chen SS, Chen W, Chen WJ, Chen WQ, Chen X, Chen YH, Chen YG, Chen Y, Chen YJ, Chen YQ, Chen Z, Cheng A, Cheng CH, Cheng H, Cheong H, Cherry S, Chesney J, Cheung CH, Chevet E, Chi HC, Chi SG, Chiacchiera F, Chiang HL, Chiarelli R, Chiariello M, Chieppa M, Chin LS, Chiong M, Chiu GN, Cho DH, Cho SG, Cho WC, Cho YY, Cho YS, Choi AM, Choi EJ, Choi EK, Choi J, Choi ME, Choi SI, Chou TF, Chouaib S, Choubey D, Choubey V, Chow KC, Chowdhury K, Chu CT, Chuang TH, Chun T, Chung H, Chung T, Chung YL, Chwae YJ, Cianfanelli V, Ciarcia R, Ciechomska IA, Ciriolo MR, Cirone M, Claerhout S, Clague MJ, Clària J, Clarke PG, Clarke R, Clementi E, Cleyrat C, Cnop M, Coccia EM, Cocco T, Codogno P, Coers J, Cohen EE, Colecchia D, Coletto L, Coll NS, Colucci-Guyon E, Comincini S, Condello M, Cook KL, Coombs GH, Cooper CD, Cooper JM, Coppens I, Corasaniti MT, Corazzari M, Corbalan R, Corcelle-Termeau E, Cordero MD, Corral-Ramos C, Corti O, Cossarizza A, Costelli P, Costes S, Cotman SL, Coto-Montes A, Cottet S, Couve E, Covey LR, Cowart LA, Cox JS, Coxon FP, Coyne CB, Cragg MS, Craven RJ, Crepaldi T, Crespo JL, Criollo A, Crippa V, Cruz MT, Cuervo AM, Cuezva JM, Cui T, Cutillas PR, Czaja MJ, Czyzyk-Krzeska MF, Dagda RK, Dahmen U, Dai C, Dai W, Dai Y, Dalby KN, Dalla Valle L, Dalmasso G, D'Amelio M, Damme M, Darfeuille-Michaud A, Dargemont C, Darley-Usmar VM, Dasarathy S, Dasgupta B, Dash S, Dass CR, Davey HM, Davids LM, Dávila D, Davis RJ, Dawson TM, Dawson VL, Daza P, de Belleroche J, de Figueiredo P, de Figueiredo RC, de la Fuente J, De Martino L, De Matteis A, De Meyer GR, De Milito A, De Santi M, de Souza W, De Tata V, De Zio D, Debnath J, Dechant R, Decuypere JP, Deegan S, Dehay B, Del Bello B, Del Re DP, Delage-Mourroux R, Delbridge LM, Deldicque L, Delorme-Axford E, Deng Y, Dengjel J, Denizot M, Dent P, Der CJ, Deretic V, Derrien B, Deutsch E, Devarenne TP, Devenish RJ, Di Bartolomeo S, Di Daniele N, Di Domenico F, Di Nardo A, Di Paola S, Di Pietro A, Di Renzo L, DiAntonio A, Díaz-Araya G, Díaz-Laviada I, Diaz-Meco MT, Diaz-Nido J, Dickey CA, Dickson RC, Diederich M, Digard P, Dikic I, Dinesh-Kumar SP, Ding C, Ding WX, Ding Z, Dini L, Distler JH, Diwan A, Djavaheri-Mergny M, Dmytruk K, Dobson RC, Doetsch V, Dokladny K, Dokudovskaya S, Donadelli M, Dong XC, Dong X, Dong Z, Donohue TM Jr, Doran KS, D'Orazi G, Dorn GW 2nd, Dosenko V, Dridi S, Drucker L, Du J, Du LL, Du L, du Toit A, Dua P, Duan L, Duann P, Dubey VK, Duchen MR, Duchosal MA, Duez H, Dugail I, Dumit VI, Duncan MC, Dunlop EA, Dunn WA Jr, Dupont N, Dupuis L, Durán RV, Durcan TM, Duvezin-Caubet S, Duvvuri U, Eapen V, Ebrahimi-Fakhari D, Echard A, Eckhart L, Edelstein CL, Edinger AL, Eichinger L, Eisenberg T, Eisenberg-Lerner A, Eissa NT, El-Deiry WS, El-Khoury V, Elazar Z, Eldar-Finkelman H, Elliott CJ, Emanuele E, Emmenegger U, Engedal N, Engelbrecht AM, Engelender S, Enserink JM, Erdmann R, Erenpreisa J, Eri R, Eriksen JL, Erman A, Escalante R, Eskelinen EL, Espert L, Esteban-Martínez L, Evans TJ, Fabri M, Fabrias G, Fabrizi C, Facchiano A, Færgeman NJ, Faggioni A, Fairlie WD, Fan C, Fan D, Fan J, Fang S, Fanto M, Fanzani A, Farkas T, Faure M, Favier FB, Fearnhead H, Federici M, Fei E, Felizardo TC, Feng H, Feng Y, Ferguson TA, Fernández ÁF, Fernandez-Barrena MG, Fernandez-Checa JC, Fernández-López A, Fernandez-Zapico ME, Feron O, Ferraro E, Ferreira-Halder CV, Fesus L, Feuer R, Fiesel FC, Filippi-Chiela EC, Filomeni G, Fimia GM, Fingert JH, Finkbeiner S, Finkel T, Fiorito F, Fisher PB, Flajolet M, Flamigni F, Florey O, Florio S, Floto RA, Folini M, Follo C, Fon EA, Fornai F, Fortunato F, Fraldi A, Franco R, Francois A, François A, Frankel LB, Fraser ID, Frey N, Freyssenet DG, Frezza C, Friedman SL, Frigo DE, Fu D, Fuentes JM, Fueyo J, Fujitani Y, Fujiwara Y, Fujiya M, Fukuda M, Fulda S, Fusco C, Gabryel B, Gaestel M, Gailly P, Gajewska M, Galadari S, Galili G, Galindo I, Galindo MF, Galliciotti G, Galluzzi L, Galy V, Gammoh N, Gandy S, Ganesan AK, Ganesan S, Ganley IG, Gannagé M, Gao FB, Gao F, Gao JX, García Nannig L, García Véscovi E, Garcia-Macía M, Garcia-Ruiz C, Garg AD, Garg PK, Gargini R, Gassen NC, Gatica D, Gatti E, Gavard J, Gavathiotis E, Ge L, Ge P, Ge S, Gean PW, Gelmetti V, Genazzani AA, Geng J, Genschik P, Gerner L, Gestwicki JE, Gewirtz DA, Ghavami S, Ghigo E, Ghosh D, Giammarioli AM, Giampieri F, Giampietri C, Giatromanolaki A, Gibbings DJ, Gibellini L, Gibson SB, Ginet V, Giordano A, Giorgini F, Giovannetti E, Girardin SE, Gispert S, Giuliano S, Gladson CL, Glavic A, Gleave M, Godefroy N, Gogal RM Jr, Gokulan K, Goldman GH, Goletti D, Goligorsky MS, Gomes AV, Gomes LC, Gomez H, Gomez-Manzano C, Gómez-Sánchez R, Gonçalves DA, Goncu E, Gong Q, Gongora C, Gonzalez CB, Gonzalez-Alegre P, Gonzalez-Cabo P, González-Polo RA, Goping IS, Gorbea C, Gorbunov NV, Goring DR, Gorman AM, Gorski SM, Goruppi S, Goto-Yamada S, Gotor C, Gottlieb RA, Gozes I, Gozuacik D, Graba Y, Graef M, Granato GE, Grant GD, Grant S, Gravina GL, Green DR, Greenhough A, Greenwood MT, Grimaldi B, Gros F, Grose C, Groulx JF, Gruber F, Grumati P, Grune T, Guan JL, Guan KL, Guerra B, Guillen C, Gulshan K, Gunst J, Guo C, Guo L, Guo M, Guo W, Guo XG, Gust AA, Gustafsson ÅB, Gutierrez E, Gutierrez MG, Gwak HS, Haas A, Haber JE, Hadano S, Hagedorn M, Hahn DR, Halayko AJ, Hamacher-Brady A, Hamada K, Hamai A, Hamann A, Hamasaki M, Hamer I, Hamid Q, Hammond EM, Han F, Han W, Handa JT, Hanover JA, Hansen M, Harada M, Harhaji-Trajkovic L, Harper JW, Harrath AH, Harris AL, Harris J, Hasler U, Hasselblatt P, Hasui K, Hawley RG, Hawley TS, He C, He CY, He F, He G, He RR, He XH, He YW, He YY, Heath JK, Hébert MJ, Heinzen RA, Helgason GV, Hensel M, Henske EP, Her C, Herman PK, Hernández A, Hernandez C, Hernández-Tiedra S, Hetz C, Hiesinger PR, Higaki K, Hilfiker S, Hill BG, Hill JA, Hill WD, Hino K, Hofius D, Hofman P, Höglinger GU, Höhfeld J, Holz MK, Hong Y, Hood DA, Hoozemans JJ, Hoppe T, Hsu C, Hsu CY, Hsu LC, Hu D, Hu G, Hu HM, Hu H, Hu MC, Hu YC, Hu ZW, Hua F, Hua Y, Huang C, Huang HL, Huang KH, Huang KY, Huang S, Huang WP, Huang YR, Huang Y, Huber TB, Huebbe P, Huh WK, Hulmi JJ, Hur GM, Hurley JH, Husak Z, Hussain SN, Hussain S, Hwang JJ, Hwang S, Hwang TI, Ichihara A, Imai Y, Imbriano C, Inomata M, Into T, Iovane V, Iovanna JL, Iozzo RV, Ip NY, Irazoqui JE, Iribarren P, Isaka Y, Isakovic AJ, Ischiropoulos H, Isenberg JS, Ishaq M, Ishida H, Ishii I, Ishmael JE, Isidoro C, Isobe K, Isono E, Issazadeh-Navikas S, Itahana K, Itakura E, Ivanov AI, Iyer AK, Izquierdo JM, Izumi Y, Izzo V, Jäättelä M, Jaber N, Jackson DJ, Jackson WT, Jacob TG, Jacques TS, Jagannath C, Jain A, Jana NR, Jang BK, Jani A, Janji B, Jannig PR, Jansson PJ, Jean S, Jendrach M, Jeon JH, Jessen N, Jeung EB, Jia K, Jia L, Jiang H, Jiang L, Jiang T, Jiang X, Jiang Y, Jiménez A, Jin C, Jin H, Jin L, Jin M, Jin S, Jinwal UK, Jo EK, Johansen T, Johnson DE, Johnson GV, Johnson JD, Jonasch E, Jones C, Joosten LA, Jordan J, Joseph AM, Joseph B, Joubert AM, Ju D, Ju J, Juan HF, Juenemann K, Juhász G, Jung HS, Jung JU, Jung YK, Jungbluth H, Justice MJ, Jutten B, Kaakoush NO, Kaarniranta K, Kaasik A, Kabuta T, Kaeffer B, Kågedal K, Kahana A, Kajimura S, Kakhlon O, Kalia M, Kalvakolanu DV, Kamada Y, Kambas K, Kaminskyy VO, Kampinga HH, Kandouz M, Kang C, Kang R, Kang TC, Kanki T, Kanneganti TD, Kanno H, Kanthasamy AG, Kantorow M, Kaparakis-Liaskos M, Kapuy O, Karantza V, Karim MR, Karmakar P, Kaser A, Kaushik S, Kawula T, Kaynar AM, Ke PY, Ke ZJ, Kehrl JH, Keller KE, Kemper JK, Kenworthy AK, Kepp O, Kern A, Kesari S, Kessel D, Ketteler R, Kettelhut Ido C, Khambu B, Khan MM, Khandelwal VK, Khare S, Kiang JG, Kiger AA, Kihara A, Kim AL, Kim CH, Kim DR, Kim DH, Kim EK, Kim HY, Kim HR, Kim JS, Kim JH, Kim JC, Kim KW, Kim MD, Kim MM, Kim PK, Kim SW, Kim SY, Kim YS, Kim Y, Kimchi A, Kimmelman AC, Kimura T, King JS, Kirkegaard K, Kirkin V, Kirshenbaum LA, Kishi S, Kitajima Y, Kitamoto K, Kitaoka Y, Kitazato K, Kley RA, Klimecki WT, Klinkenberg M, Klucken J, Knævelsrud H, Knecht E, Knuppertz L, Ko JL, Kobayashi S, Koch JC, Koechlin-Ramonatxo C, Koenig U, Koh YH, Köhler K, Kohlwein SD, Koike M, Komatsu M, Kominami E, Kong D, Kong HJ, Konstantakou EG, Kopp BT, Korcsmaros T, Korhonen L, Korolchuk VI, Koshkina NV, Kou Y, Koukourakis MI, Koumenis C, Kovács AL, Kovács T, Kovacs WJ, Koya D, Kraft C, Krainc D, Kramer H, Kravic-Stevovic T, Krek W, Kretz-Remy C, Krick R, Krishnamurthy M, Kriston-Vizi J, Kroemer G, Kruer MC, Kruger R, Ktistakis NT, Kuchitsu K, Kuhn C, Kumar AP, Kumar A, Kumar D, Kumar R, Kumar S, Kundu M, Kung HJ, Kuno A, Kuo SH, Kuret J, Kurz T, Kwok T, Kwon TK, Kwon YT, Kyrmizi I, La Spada AR, Lafont F, Lahm T, Lakkaraju A, Lam T, Lamark T, Lancel S, Landowski TH, Lane DJ, Lane JD, Lanzi C, Lapaquette P, Lapierre LR, Laporte J, Laukkarinen J, Laurie GW, Lavandero S, Lavie L, LaVoie MJ, Law BY, Law HK, Law KB, Layfield R, Lazo PA, Le Cam L, Le Roch KG, Le Stunff H, Leardkamolkarn V, Lecuit M, Lee BH, Lee CH, Lee EF, Lee GM, Lee HJ, Lee H, Lee JK, Lee J, Lee JH, Lee M, Lee MS, Lee PJ, Lee SW, Lee SJ, Lee SY, Lee SH, Lee SS, Lee S, Lee YR, Lee YJ, Lee YH, Leeuwenburgh C, Lefort S, Legouis R, Lei J, Lei QY, Leib DA, Leibowitz G, Lekli I, Lemaire SD, Lemasters JJ, Lemberg MK, Lemoine A, Leng S, Lenz G, Lenzi P, Lerman LO, Lettieri Barbato D, Leu JI, Leung HY, Levine B, Lewis PA, Lezoualc'h F, Li C, Li F, Li FJ, Li J, Li K, Li L, Li M, Li Q, Li R, Li S, Li W, Li X, Li Y, Lian J, Liang C, Liang Q, Liao Y, Liberal J, Liberski PP, Lie P, Lieberman AP, Lim HJ, Lim KL, Lim K, Lima RT, Lin CS, Lin CF, Lin F, Lin FC, Lin K, Lin KH, Lin PH, Lin T, Lin WW, Lin YS, Lin Y, Linden R, Lindholm D, Lindqvist LM, Lingor P, Linkermann A, Liotta LA, Lipinski MM, Lira VA, Lisanti MP, Liton PB, Liu B, Liu C, Liu CF, Liu F, Liu HJ, Liu J, Liu JJ, Liu JL, Liu K, Liu L, Liu Q, Liu RY, Liu S, Liu W, Liu XD, Liu X, Liu XH, Liu Y, Liu Z, Liuzzi JP, Lizard G, Ljujic M, Lodhi IJ, Logue SE, Lokeshwar BL, Long YC, Lonial S, Loos B, López-Otín C, López-Vicario C, Lorente M, Lorenzi PL, Lõrincz P, Los M, Lotze MT, Lovat PE, Lu B, Lu J, Lu Q, Lu SM, Lu S, Lu Y, Luciano F, Luckhart S, Lucocq JM, Ludovico P, Lugea A, Lukacs NW, Lum JJ, Lund AH, Luo H, Luo J, Luo S, Luparello C, Lyons T, Ma J, Ma Y, Ma Z, Machado J, Machado-Santelli GM, Macian F, MacIntosh GC, MacKeigan JP, Macleod KF, MacMicking JD, MacMillan-Crow LA, Madeo F, Madesh M, Madrigal-Matute J, Maeda A, Maeda T, Maegawa G, Maellaro E, Maes H, Magariños M, Maiese K, Maiti TK, Maiuri L, Maiuri MC, Maki CG, Malli R, Malorni W, Maloyan A, Mami-Chouaib F, Man N, Mancias JD, Mandelkow EM, Mandell MA, Manfredi AA, Manié SN, Manzoni C, Mao K, Mao Z, Mao ZW, Marambaud P, Marconi AM, Marelja Z, Marfe G, Margeta M, Margittai E, Mari M, Mariani FV, Marin C, Marinelli S, Mariño G, Markovic I, Marquez R, Martelli AM, Martens S, Martin KR, Martin SJ, Martin S, Martin-Acebes MA, Martín-Sanz P, Martinand-Mari C, Martinet W, Martinez J, Martinez-Lopez N, Martinez-Outschoorn U, Martínez-Velázquez M, Martinez-Vicente M, Martins WK, Mashima H, Mastrianni JA, Matarese G, Matarrese P, Mateo R, Matoba S, Matsumoto N, Matsushita T, Matsuura A, Matsuzawa T, Mattson MP, Matus S, Maugeri N, Mauvezin C, Mayer A, Maysinger D, Mazzolini GD, McBrayer MK, McCall K, McCormick C, McInerney GM, McIver SC, McKenna S, McMahon JJ, McNeish IA, Mechta-Grigoriou F, Medema JP, Medina DL, Megyeri K, Mehrpour M, Mehta JL, Mei Y, Meier UC, Meijer AJ, Meléndez A, Melino G, Melino S, de Melo EJ, Mena MA, Meneghini MD, Menendez JA, Menezes R, Meng L, Meng LH, Meng S, Menghini R, Menko AS, Menna-Barreto RF, Menon MB, Meraz-Ríos MA, Merla G, Merlini L, Merlot AM, Meryk A, Meschini S, Meyer JN, Mi MT, Miao CY, Micale L, Michaeli S, Michiels C, Migliaccio AR, Mihailidou AS, Mijaljica D, Mikoshiba K, Milan E, Miller-Fleming L, Mills GB, Mills IG, Minakaki G, Minassian BA, Ming XF, Minibayeva F, Minina EA, Mintern JD, Minucci S, Miranda-Vizuete A, Mitchell CH, Miyamoto S, Miyazawa K, Mizushima N, Mnich K, Mograbi B, Mohseni S, Moita LF, Molinari M, Møller AB, Mollereau B, Mollinedo F, Mongillo M, Monick MM, Montagnaro S, Montell C, Moore DJ, Moore MN, Mora-Rodriguez R, Moreira PI, Morel E, Morelli MB, Moreno S, Morgan MJ, Moris A, Moriyasu Y, Morrison JL, Morrison LA, Morselli E, Moscat J, Moseley PL, Mostowy S, Motori E, Mottet D, Mottram JC, Moussa CE, Mpakou VE, Mukhtar H, Mulcahy Levy JM, Muller S, Muñoz-Moreno R, Muñoz-Pinedo C, Münz C, Murphy ME, Murray JT, Murthy A, Mysorekar IU, Nabi IR, Nabissi M, Nader GA, Nagahara Y, Nagai Y, Nagata K, Nagelkerke A, Nagy P, Naidu SR, Nair S, Nakano H, Nakatogawa H, Nanjundan M, Napolitano G, Naqvi NI, Nardacci R, Narendra DP, Narita M, Nascimbeni AC, Natarajan R, Navegantes LC, Nawrocki ST, Nazarko TY, Nazarko VY, Neill T, Neri LM, Netea MG, Netea-Maier RT, Neves BM, Ney PA, Nezis IP, Nguyen HT, Nguyen HP, Nicot AS, Nilsen H, Nilsson P, Nishimura M, Nishino I, Niso-Santano M, Niu H, Nixon RA, Njar VC, Noda T, Noegel AA, Nolte EM, Norberg E, Norga KK, Noureini SK, Notomi S, Notterpek L, Nowikovsky K, Nukina N, Nürnberger T, O'Donnell VB, O'Donovan T, O'Dwyer PJ, Oehme I, Oeste CL, Ogawa M, Ogretmen B, Ogura Y, Oh YJ, Ohmuraya M, Ohshima T, Ojha R, Okamoto K, Okazaki T, Oliver FJ, Ollinger K, Olsson S, Orban DP, Ordonez P, Orhon I, Orosz L, O'Rourke EJ, Orozco H, Ortega AL, Ortona E, Osellame LD, Oshima J, Oshima S, Osiewacz HD, Otomo T, Otsu K, Ou JH, Outeiro TF, Ouyang DY, Ouyang H, Overholtzer M, Ozbun MA, Ozdinler PH, Ozpolat B, Pacelli C, Paganetti P, Page G, Pages G, Pagnini U, Pajak B, Pak SC, Pakos-Zebrucka K, Pakpour N, Palková Z, Palladino F, Pallauf K, Pallet N, Palmieri M, Paludan SR, Palumbo C, Palumbo S, Pampliega O, Pan H, Pan W, Panaretakis T, Pandey A, Pantazopoulou A, Papackova Z, Papademetrio DL, Papassideri I, Papini A, Parajuli N, Pardo J, Parekh VV, Parenti G, Park JI, Park J, Park OK, Parker R, Parlato R, Parys JB, Parzych KR, Pasquet JM, Pasquier B, Pasumarthi KB, Patschan D, Patterson C, Pattingre S, Pattison S, Pause A, Pavenstädt H, Pavone F, Pedrozo Z, Peña FJ, Peñalva MA, Pende M, Peng J, Penna F, Penninger JM, Pensalfini A, Pepe S, Pereira GJ, Pereira PC, Pérez-de la Cruz V, Pérez-Pérez ME, Pérez-Rodríguez D, Pérez-Sala D, Perier C, Perl A, Perlmutter DH, Perrotta I, Pervaiz S, Pesonen M, Pessin JE, Peters GJ, Petersen M, Petrache I, Petrof BJ, Petrovski G, Phang JM, Piacentini M, Pierdominici M, Pierre P, Pierrefite-Carle V, Pietrocola F, Pimentel-Muiños FX, Pinar M, Pineda B, Pinkas-Kramarski R, Pinti M, Pinton P, Piperdi B, Piret JM, Platanias LC, Platta HW, Plowey ED, Pöggeler S, Poirot M, Polčic P, Poletti A, Poon AH, Popelka H, Popova B, Poprawa I, Poulose SM, Poulton J, Powers SK, Powers T, Pozuelo-Rubio M, Prak K, Prange R, Prescott M, Priault M, Prince S, Proia RL, Proikas-Cezanne T, Prokisch H, Promponas VJ, Przyklenk K, Puertollano R, Pugazhenthi S, Puglielli L, Pujol A, Puyal J, Pyeon D, Qi X, Qian WB, Qin ZH, Qiu Y, Qu Z, Quadrilatero J, Quinn F, Raben N, Rabinowich H, Radogna F, Ragusa MJ, Rahmani M, Raina K, Ramanadham S, Ramesh R, Rami A, Randall-Demllo S, Randow F, Rao H, Rao VA, Rasmussen BB, Rasse TM, Ratovitski EA, Rautou PE, Ray SK, Razani B, Reed BH, Reggiori F, Rehm M, Reichert AS, Rein T, Reiner DJ, Reits E, Ren J, Ren X, Renna M, Reusch JE, Revuelta JL, Reyes L, Rezaie AR, Richards RI, Richardson DR, Richetta C, Riehle MA, Rihn BH, Rikihisa Y, Riley BE, Rimbach G, Rippo MR, Ritis K, Rizzi F, Rizzo E, Roach PJ, Robbins J, Roberge M, Roca G, Roccheri MC, Rocha S, Rodrigues CM, Rodríguez CI, de Cordoba SR, Rodriguez-Muela N, Roelofs J, Rogov VV, Rohn TT, Rohrer B, Romanelli D, Romani L, Romano PS, Roncero MI, Rosa JL, Rosello A, Rosen KV, Rosenstiel P, Rost-Roszkowska M, Roth KA, Roué G, Rouis M, Rouschop KM, Ruan DT, Ruano D, Rubinsztein DC, Rucker EB 3rd, Rudich A, Rudolf E, Rudolf R, Ruegg MA, Ruiz-Roldan C, Ruparelia AA, Rusmini P, Russ DW, Russo GL, Russo G, Russo R, Rusten TE, Ryabovol V, Ryan KM, Ryter SW, Sabatini DM, Sacher M, Sachse C, Sack MN, Sadoshima J, Saftig P, Sagi-Eisenberg R, Sahni S, Saikumar P, Saito T, Saitoh T, Sakakura K, Sakoh-Nakatogawa M, Sakuraba Y, Salazar-Roa M, Salomoni P, Saluja AK, Salvaterra PM, Salvioli R, Samali A, Sanchez AM, Sánchez-Alcázar JA, Sanchez-Prieto R, Sandri M, Sanjuan MA, Santaguida S, Santambrogio L, Santoni G, Dos Santos CN, Saran S, Sardiello M, Sargent G, Sarkar P, Sarkar S, Sarrias MR, Sarwal MM, Sasakawa C, Sasaki M, Sass M, Sato K, Sato M, Satriano J, Savaraj N, Saveljeva S, Schaefer L, Schaible UE, Scharl M, Schatzl HM, Schekman R, Scheper W, Schiavi A, Schipper HM, Schmeisser H, Schmidt J, Schmitz I, Schneider BE, Schneider EM, Schneider JL, Schon EA, Schönenberger MJ, Schönthal AH, Schorderet DF, Schröder B, Schuck S, Schulze RJ, Schwarten M, Schwarz TL, Sciarretta S, Scotto K, Scovassi AI, Screaton RA, Screen M, Seca H, Sedej S, Segatori L, Segev N, Seglen PO, Seguí-Simarro JM, Segura-Aguilar J, Seki E, Sell C, Seiliez I, Semenkovich CF, Semenza GL, Sen U, Serra AL, Serrano-Puebla A, Sesaki H, Setoguchi T, Settembre C, Shacka JJ, Shajahan-Haq AN, Shapiro IM, Sharma S, She H, Shen CK, Shen CC, Shen HM, Shen S, Shen W, Sheng R, Sheng X, Sheng ZH, Shepherd TG, Shi J, Shi Q, Shi Y, Shibutani S, Shibuya K, Shidoji Y, Shieh JJ, Shih CM, Shimada Y, Shimizu S, Shin DW, Shinohara ML, Shintani M, Shintani T, Shioi T, Shirabe K, Shiri-Sverdlov R, Shirihai O, Shore GC, Shu CW, Shukla D, Sibirny AA, Sica V, Sigurdson CJ, Sigurdsson EM, Sijwali PS, Sikorska B, Silveira WA, Silvente-Poirot S, Silverman GA, Simak J, Simmet T, Simon AK, Simon HU, Simone C, Simons M, Simonsen A, Singh R, Singh SV, Singh SK, Sinha D, Sinha S, Sinicrope FA, Sirko A, Sirohi K, Sishi BJ, Sittler A, Siu PM, Sivridis E, Skwarska A, Slack R, Slaninová I, Slavov N, Smaili SS, Smalley KS, Smith DR, Soenen SJ, Soleimanpour SA, Solhaug A, Somasundaram K, Son JH, Sonawane A, Song C, Song F, Song HK, Song JX, Song W, Soo KY, Sood AK, Soong TW, Soontornniyomkij V, Sorice M, Sotgia F, Soto-Pantoja DR, Sotthibundhu A, Sousa MJ, Spaink HP, Span PN, Spang A, Sparks JD, Speck PG, Spector SA, Spies CD, Springer W, Clair DS, Stacchiotti A, Staels B, Stang MT, Starczynowski DT, Starokadomskyy P, Steegborn C, Steele JW, Stefanis L, Steffan J, Stellrecht CM, Stenmark H, Stepkowski TM, Stern ST, Stevens C, Stockwell BR, Stoka V, Storchova Z, Stork B, Stratoulias V, Stravopodis DJ, Strnad P, Strohecker AM, Ström AL, Stromhaug P, Stulik J, Su YX, Su Z, Subauste CS, Subramaniam S, Sue CM, Suh SW, Sui X, Sukseree S, Sulzer D, Sun FL, Sun J, Sun SY, Sun Y, Sundaramoorthy V, Sung J, Suzuki H, Suzuki K, Suzuki N, Suzuki T, Suzuki YJ, Swanson MS, Swanton C, Swärd K, Swarup G, Sweeney ST, Sylvester PW, Szatmari Z, Szegezdi E, Szlosarek PW, Taegtmeyer H, Tafani M, Taillebourg E, Tait SW, Takacs-Vellai K, Takahashi Y, Takáts S, Takemura G, Takigawa N, Talbot NJ, Tamagno E, Tamburini J, Tan CP, Tan L, Tan ML, Tan M, Tan YJ, Tanaka K, Tanaka M, Tang D, Tang G, Tanida I, Tanji K, Tannous BA, Tapia JA, Tasset-Cuevas I, Tatar M, Tavassoly I, Tavernarakis N, Taylor A, Taylor GS, Taylor GA, Taylor JP, Taylor MJ, Tchetina EV, Tee AR, Teixeira-Clerc F, Telang S, Tencomnao T, Teng BB, Teng RJ, Terro F, Tettamanti G, Theiss AL, Theron AE, Thomas KJ, Thomé MP, Thomes PG, Thorburn A, Thorner J, Thum T, Thumm M, Thurston TL, Tian L, Till A, Ting JP, Titorenko VI, Toker L, Toldo S, Tooze SA, Topisirovic I, Torgersen ML, Torosantucci L, Torriglia A, Torrisi MR, Tournier C, Towns R, Trajkovic V, Travassos LH, Triola G, Tripathi DN, Trisciuoglio D, Troncoso R, Trougakos IP, Truttmann AC, Tsai KJ, Tschan MP, Tseng YH, Tsukuba T, Tsung A, Tsvetkov AS, Tu S, Tuan HY, Tucci M, Tumbarello DA, Turk B, Turk V, Turner RF, Tveita AA, Tyagi SC, Ubukata M, Uchiyama Y, Udelnow A, Ueno T, Umekawa M, Umemiya-Shirafuji R, Underwood BR, Ungermann C, Ureshino RP, Ushioda R, Uversky VN, Uzcátegui NL, Vaccari T, Vaccaro MI, Váchová L, Vakifahmetoglu-Norberg H, Valdor R, Valente EM, Vallette F, Valverde AM, Van den Berghe G, Van Den Bosch L, van den Brink GR, van der Goot FG, van der Klei IJ, van der Laan LJ, van Doorn WG, van Egmond M, van Golen KL, Van Kaer L, van Lookeren Campagne M, Vandenabeele P, Vandenberghe W, Vanhorebeek I, Varela-Nieto I, Vasconcelos MH, Vasko R, Vavvas DG, Vega-Naredo I, Velasco G, Velentzas AD, Velentzas PD, Vellai T, Vellenga E, Vendelbo MH, Venkatachalam K, Ventura N, Ventura S, Veras PS, Verdier M, Vertessy BG, Viale A, Vidal M, Vieira HL, Vierstra RD, Vigneswaran N, Vij N, Vila M, Villar M, Villar VH, Villarroya J, Vindis C, Viola G, Viscomi MT, Vitale G, Vogl DT, Voitsekhovskaja OV, von Haefen C, von Schwarzenberg K, Voth DE, Vouret-Craviari V, Vuori K, Vyas JM, Waeber C, Walker CL, Walker MJ, Walter J, Wan L, Wan X, Wang B, Wang C, Wang CY, Wang D, Wang F, Wang G, Wang HJ, Wang H, Wang HG, Wang HD, Wang J, Wang M, Wang MQ, Wang PY, Wang P, Wang RC, Wang S, Wang TF, Wang X, Wang XJ, Wang XW, Wang Y, Wang YJ, Wang YT, Wang ZN, Wappner P, Ward C, Ward DM, Warnes G, Watada H, Watanabe Y, Watase K, Weaver TE, Weekes CD, Wei J, Weide T, Weihl CC, Weindl G, Weis SN, Wen L, Wen X, Wen Y, Westermann B, Weyand CM, White AR, White E, Whitton JL, Whitworth AJ, Wiels J, Wild F, Wildenberg ME, Wileman T, Wilkinson DS, Wilkinson S, Willbold D, Williams C, Williams K, Williamson PR, Winklhofer KF, Witkin SS, Wohlgemuth SE, Wollert T, Wolvetang EJ, Wong E, Wong GW, Wong RW, Wong VK, Woodcock EA, Wright KL, Wu C, Wu D, Wu GS, Wu J, Wu M, Wu S, Wu WK, Wu Y, Wu Z, Xavier CP, Xavier RJ, Xia GX, Xia T, Xia W, Xia Y, Xiao H, Xiao J, Xiao S, Xiao W, Xie CM, Xie Z, Xilouri M, Xiong Y, Xu C, Xu F, Xu H, Xu J, Xu L, Xu X, Xu Y, Xu ZX, Xu Z, Xue Y, Yamada T, Yamamoto A, Yamanaka K, Yamashina S, Yamashiro S, Yan B, Yan X, Yan Z, Yanagi Y, Yang DS, Yang JM, Yang L, Yang M, Yang PM, Yang P, Yang Q, Yang W, Yang WY, Yang X, Yang Y, Yang Z, Yao MC, Yao PJ, Yao X, Yao Z, Yasui LS, Ye M, Yedvobnick B, Yeganeh B, Yeh ES, Yeyati PL, Yi F, Yi L, Yin XM, Yip CK, Yoo YM, Yoo YH, Yoon SY, Yoshida K, Yoshimori T, Young KH, Yu H, Yu JJ, Yu JT, Yu J, Yu L, Yu WH, Yu XF, Yu Z, Yuan J, Yuan ZM, Yue BY, Yue J, Yue Z, Zacks DN, Zacksenhaus E, Zaffaroni N, Zaglia T, Zakeri Z, Zecchini V, Zeng J, Zeng M, Zeng Q, Zervos AS, Zhang DD, Zhang F, Zhang G, Zhang GC, Zhang H, Zhang J, Zhang JP, Zhang L, Zhang MY, Zhang X, Zhang XD, Zhang Y, Zhao M, Zhao WL, Zhao X, Zhao YG, Zhao Y, Zhao YX, Zhao Z, Zhao ZJ, Zheng D, Zheng XL, Zheng X, Zhivotovsky B, Zhong Q, Zhou GZ, Zhou G, Zhou H, Zhou SF, Zhou XJ, Zhu H, Zhu WG, Zhu W, Zhu XF, Zhu Y, Zhuang SM, Zhuang X, Ziparo E, Zois CE, Zoladek T, Zong WX, Zorzano A, Zughaier SM. Guidelines for the use and interpretation of assays for monitoring autophagy (3rd edition). Autophagy. 2016;12:1-222. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 4248] [Cited by in RCA: 4238] [Article Influence: 470.9] [Reference Citation Analysis (0)] |
| 19. | Qian HR, Yang Y. Functional role of autophagy in gastric cancer. Oncotarget. 2016;7:17641-17651. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 63] [Cited by in RCA: 82] [Article Influence: 10.3] [Reference Citation Analysis (0)] |
| 20. | Kelekar A. Introduction to the Review Series Autophagy in Higher Eukaryotes- A matter of survival or death. Autophagy. 2008;4:555-556. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 17] [Cited by in RCA: 20] [Article Influence: 1.2] [Reference Citation Analysis (0)] |
| 21. | Mizushima N, Levine B. Autophagy in Human Diseases. N Engl J Med. 2020;383:1564-1576. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 420] [Cited by in RCA: 752] [Article Influence: 150.4] [Reference Citation Analysis (0)] |
| 22. | Choi AM, Ryter SW, Levine B. Autophagy in human health and disease. N Engl J Med. 2013;368:651-662. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1792] [Cited by in RCA: 1912] [Article Influence: 159.3] [Reference Citation Analysis (0)] |
| 23. | Qiu YH, Zhang TS, Wang XW, Wang MY, Zhao WX, Zhou HM, Zhang CH, Cai ML, Chen XF, Zhao WL, Shao RG. Mitochondria autophagy: a potential target for cancer therapy. J Drug Target. 2021;29:576-591. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 4] [Cited by in RCA: 28] [Article Influence: 7.0] [Reference Citation Analysis (0)] |
| 24. | Guo JY, Xia B, White E. Autophagy-mediated tumor promotion. Cell. 2013;155:1216-1219. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 304] [Cited by in RCA: 395] [Article Influence: 35.9] [Reference Citation Analysis (0)] |
| 25. | White E. Deconvoluting the context-dependent role for autophagy in cancer. Nat Rev Cancer. 2012;12:401-410. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1226] [Cited by in RCA: 1379] [Article Influence: 106.1] [Reference Citation Analysis (0)] |
| 26. | Ashrafi G, Schwarz TL. The pathways of mitophagy for quality control and clearance of mitochondria. Cell Death Differ. 2013;20:31-42. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1172] [Cited by in RCA: 1397] [Article Influence: 116.4] [Reference Citation Analysis (0)] |
| 27. | Budini M, Buratti E, Morselli E, Criollo A. Autophagy and Its Impact on Neurodegenerative Diseases: New Roles for TDP-43 and C9orf72. Front Mol Neurosci. 2017;10:170. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 53] [Cited by in RCA: 75] [Article Influence: 9.4] [Reference Citation Analysis (0)] |
| 28. | Ricci V. Relationship between VacA Toxin and Host Cell Autophagy in Helicobacter pylori Infection of the Human Stomach: A Few Answers, Many Questions. Toxins (Basel). 2016;8. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 34] [Cited by in RCA: 48] [Article Influence: 5.3] [Reference Citation Analysis (0)] |
| 29. | Levy JMM, Towers CG, Thorburn A. Targeting autophagy in cancer. Nat Rev Cancer. 2017;17:528-542. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1382] [Cited by in RCA: 1946] [Article Influence: 243.3] [Reference Citation Analysis (0)] |
| 30. | Levine B, Kroemer G. Biological Functions of Autophagy Genes: A Disease Perspective. Cell. 2019;176:11-42. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1024] [Cited by in RCA: 2059] [Article Influence: 343.2] [Reference Citation Analysis (0)] |
| 31. | Luo S, Li X, Zhang Y, Fu Y, Fan B, Zhu C, Chen Z. Cargo Recognition and Function of Selective Autophagy Receptors in Plants. Int J Mol Sci. 2021;22. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 7] [Cited by in RCA: 18] [Article Influence: 4.5] [Reference Citation Analysis (0)] |
| 32. | Li WW, Li J, Bao JK. Microautophagy: lesser-known self-eating. Cell Mol Life Sci. 2012;69:1125-1136. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 568] [Cited by in RCA: 536] [Article Influence: 41.2] [Reference Citation Analysis (0)] |
| 33. | Handa K, Kanno H, Matsuda M, Sugaya T, Murakami T, Prudnikova M, Ozawa H, Itoi E. Chaperone-Mediated Autophagy after Spinal Cord Injury. J Neurotrauma. 2020;37:1687-1695. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 3] [Cited by in RCA: 14] [Article Influence: 2.8] [Reference Citation Analysis (0)] |
| 34. | Kaushik S, Bandyopadhyay U, Sridhar S, Kiffin R, Martinez-Vicente M, Kon M, Orenstein SJ, Wong E, Cuervo AM. Chaperone-mediated autophagy at a glance. J Cell Sci. 2011;124:495-499. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 144] [Cited by in RCA: 160] [Article Influence: 11.4] [Reference Citation Analysis (0)] |
| 35. | Kaushik S, Cuervo AM. Chaperone-mediated autophagy: a unique way to enter the lysosome world. Trends Cell Biol. 2012;22:407-417. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 571] [Cited by in RCA: 635] [Article Influence: 48.8] [Reference Citation Analysis (0)] |
| 36. | Cuervo AM, Wong E. Chaperone-mediated autophagy: roles in disease and aging. Cell Res. 2014;24:92-104. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 550] [Cited by in RCA: 632] [Article Influence: 52.7] [Reference Citation Analysis (0)] |
| 37. | Pietrocola F, Malik SA, Mariño G, Vacchelli E, Senovilla L, Chaba K, Niso-Santano M, Maiuri MC, Madeo F, Kroemer G. Coffee induces autophagy in vivo. Cell Cycle. 2014;13:1987-1994. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 35] [Cited by in RCA: 45] [Article Influence: 4.1] [Reference Citation Analysis (0)] |
| 38. | Amaravadi RK, Kimmelman AC, Debnath J. Targeting Autophagy in Cancer: Recent Advances and Future Directions. Cancer Discov. 2019;9:1167-1181. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 337] [Cited by in RCA: 670] [Article Influence: 111.7] [Reference Citation Analysis (0)] |
| 39. | Abdrakhmanov A, Gogvadze V, Zhivotovsky B. To Eat or to Die: Deciphering Selective Forms of Autophagy. Trends Biochem Sci. 2020;45:347-364. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 50] [Cited by in RCA: 71] [Article Influence: 14.2] [Reference Citation Analysis (0)] |
| 40. | White E, Mehnert JM, Chan CS. Autophagy, Metabolism, and Cancer. Clin Cancer Res. 2015;21:5037-5046. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 379] [Cited by in RCA: 521] [Article Influence: 57.9] [Reference Citation Analysis (0)] |
| 41. | Murugan AK. mTOR: Role in cancer, metastasis and drug resistance. Semin Cancer Biol. 2019;59:92-111. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 138] [Cited by in RCA: 326] [Article Influence: 54.3] [Reference Citation Analysis (0)] |
| 42. | Yin Z, Pascual C, Klionsky DJ. Autophagy: machinery and regulation. Microb Cell. 2016;3:588-596. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 353] [Cited by in RCA: 440] [Article Influence: 48.9] [Reference Citation Analysis (0)] |
| 43. | Chappell WH, Steelman LS, Long JM, Kempf RC, Abrams SL, Franklin RA, Bäsecke J, Stivala F, Donia M, Fagone P, Malaponte G, Mazzarino MC, Nicoletti F, Libra M, Maksimovic-Ivanic D, Mijatovic S, Montalto G, Cervello M, Laidler P, Milella M, Tafuri A, Bonati A, Evangelisti C, Cocco L, Martelli AM, McCubrey JA. Ras/Raf/MEK/ERK and PI3K/PTEN/Akt/mTOR inhibitors: rationale and importance to inhibiting these pathways in human health. Oncotarget. 2011;2:135-164. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 417] [Cited by in RCA: 461] [Article Influence: 32.9] [Reference Citation Analysis (0)] |
| 44. | König P, Svrlanska A, Read C, Feichtinger S, Stamminger T. The Autophagy-Initiating Protein Kinase ULK1 Phosphorylates Human Cytomegalovirus Tegument Protein pp28 and Regulates Efficient Virus Release. J Virol. 2021;95. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 7] [Cited by in RCA: 10] [Article Influence: 2.5] [Reference Citation Analysis (0)] |
| 45. | Ganley IG, Lam du H, Wang J, Ding X, Chen S, Jiang X. ULK1.ATG13.FIP200 complex mediates mTOR signaling and is essential for autophagy. J Biol Chem. 2009;284:12297-12305. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1000] [Cited by in RCA: 1169] [Article Influence: 73.1] [Reference Citation Analysis (0)] |
| 46. | Roberts R, Ktistakis NT. Omegasomes: PI3P platforms that manufacture autophagosomes. Essays Biochem. 2013;55:17-27. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 53] [Cited by in RCA: 58] [Article Influence: 4.8] [Reference Citation Analysis (0)] |
| 47. | Lin MG, Hurley JH. Structure and function of the ULK1 complex in autophagy. Curr Opin Cell Biol. 2016;39:61-68. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 146] [Cited by in RCA: 189] [Article Influence: 21.0] [Reference Citation Analysis (0)] |
| 48. | Ichimura Y, Kirisako T, Takao T, Satomi Y, Shimonishi Y, Ishihara N, Mizushima N, Tanida I, Kominami E, Ohsumi M, Noda T, Ohsumi Y. A ubiquitin-like system mediates protein lipidation. Nature. 2000;408:488-492. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1457] [Cited by in RCA: 1606] [Article Influence: 64.2] [Reference Citation Analysis (0)] |
| 49. | Mizushima N, Noda T, Yoshimori T, Tanaka Y, Ishii T, George MD, Klionsky DJ, Ohsumi M, Ohsumi Y. A protein conjugation system essential for autophagy. Nature. 1998;395:395-398. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1205] [Cited by in RCA: 1292] [Article Influence: 47.9] [Reference Citation Analysis (0)] |
| 50. | Pankiv S, Clausen TH, Lamark T, Brech A, Bruun JA, Outzen H, Øvervatn A, Bjørkøy G, Johansen T. p62/SQSTM1 binds directly to Atg8/LC3 to facilitate degradation of ubiquitinated protein aggregates by autophagy. J Biol Chem. 2007;282:24131-24145. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 3082] [Cited by in RCA: 3627] [Article Influence: 201.5] [Reference Citation Analysis (0)] |
| 51. | Tanida I, Ueno T, Kominami E. LC3 and Autophagy. Methods Mol Biol. 2008;445:77-88. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 967] [Cited by in RCA: 1353] [Article Influence: 79.6] [Reference Citation Analysis (0)] |
| 52. | Huang T, Song X, Yang Y, Wan X, Alvarez AA, Sastry N, Feng H, Hu B, Cheng SY. Autophagy and Hallmarks of Cancer. Crit Rev Oncog. 2018;23:247-267. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 76] [Cited by in RCA: 82] [Article Influence: 11.7] [Reference Citation Analysis (0)] |
| 53. | Dehhaghi M, Kazemi Shariat Panahi H, Heng B, Guillemin GJ. The Gut Microbiota, Kynurenine Pathway, and Immune System Interaction in the Development of Brain Cancer. Front Cell Dev Biol. 2020;8:562812. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 18] [Cited by in RCA: 58] [Article Influence: 11.6] [Reference Citation Analysis (0)] |
| 54. | Levine B, Kroemer G. Autophagy in the pathogenesis of disease. Cell. 2008;132:27-42. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 5765] [Cited by in RCA: 5746] [Article Influence: 338.0] [Reference Citation Analysis (1)] |
| 55. | Wu M, Chen B, Pan X, Su J. Prognostic Value of Autophagy-related Proteins in Human Gastric Cancer. Cancer Manag Res. 2020;12:13527-13540. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 2] [Cited by in RCA: 7] [Article Influence: 1.4] [Reference Citation Analysis (0)] |
| 56. | Marinković M, Šprung M, Buljubašić M, Novak I. Autophagy Modulation in Cancer: Current Knowledge on Action and Therapy. Oxid Med Cell Longev. 2018;2018:8023821. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 107] [Cited by in RCA: 150] [Article Influence: 21.4] [Reference Citation Analysis (0)] |
| 57. | Zheng W, Wu C, Wu X, Cai Y, Liu B, Wang C. Genetic variants of autophagy-related genes in the PI3K/Akt/mTOR pathway and risk of gastric cancer in the Chinese population. Gene. 2021;769:145190. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 12] [Cited by in RCA: 10] [Article Influence: 2.5] [Reference Citation Analysis (0)] |
| 58. | Gu W, Qian Q, Xu Y, Xu X, Zhang L, He S, Li D. SIRT5 regulates autophagy and apoptosis in gastric cancer cells. J Int Med Res. 2021;49:300060520986355. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 6] [Cited by in RCA: 25] [Article Influence: 6.3] [Reference Citation Analysis (0)] |
| 59. | Lebovitz CB, Robertson AG, Goya R, Jones SJ, Morin RD, Marra MA, Gorski SM. Cross-cancer profiling of molecular alterations within the human autophagy interaction network. Autophagy. 2015;11:1668-1687. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 91] [Cited by in RCA: 112] [Article Influence: 12.4] [Reference Citation Analysis (0)] |
| 60. | Zhang Q, Wu S, Zhu J, Chai D, Gan H. Down-regulation of ASIC1 suppressed gastric cancer via inhibiting autophagy. Gene. 2017;608:79-85. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 15] [Cited by in RCA: 19] [Article Influence: 2.4] [Reference Citation Analysis (0)] |
| 61. | Pourhanifeh MH, Vosough M, Mahjoubin-Tehran M, Hashemipour M, Nejati M, Abbasi-Kolli M, Sahebkar A, Mirzaei H. Autophagy-related microRNAs: Possible regulatory roles and therapeutic potential in and gastrointestinal cancers. Pharmacol Res. 2020;161:105133. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 56] [Cited by in RCA: 57] [Article Influence: 11.4] [Reference Citation Analysis (0)] |
| 62. | YiRen H, YingCong Y, Sunwu Y, Keqin L, Xiaochun T, Senrui C, Ende C, XiZhou L, Yanfan C. Long noncoding RNA MALAT1 regulates autophagy associated chemoresistance via miR-23b-3p sequestration in gastric cancer. Mol Cancer. 2017;16:174. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 189] [Cited by in RCA: 277] [Article Influence: 34.6] [Reference Citation Analysis (0)] |
| 63. | Chen H, Zhao C, He R, Zhou M, Liu Y, Guo X, Wang M, Zhu F, Qin R, Li X. Danthron suppresses autophagy and sensitizes pancreatic cancer cells to doxorubicin. Toxicol In Vitro. 2019;54:345-353. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 22] [Cited by in RCA: 28] [Article Influence: 4.0] [Reference Citation Analysis (0)] |
| 64. | Yan W, Yang J, Tang H, Xue L, Chen K, Wang L, Zhao M, Tang M, Peng A, Long C, Chen X, Ye H, Chen L. Flavonoids from the stems of Millettia pachyloba Drake mediate cytotoxic activity through apoptosis and autophagy in cancer cells. J Adv Res. 2019;20:117-127. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 17] [Cited by in RCA: 18] [Article Influence: 3.0] [Reference Citation Analysis (0)] |
| 65. | Li GM, Li L, Li MQ, Chen X, Su Q, Deng ZJ, Liu HB, Li B, Zhang WH, Jia YX, Wang WJ, Ma JY, Zhang HL, Xie D, Zhu XF, He YL, Guan XY, Bi J. DAPK3 inhibits gastric cancer progression via activation of ULK1-dependent autophagy. Cell Death Differ. 2021;28:952-967. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 25] [Cited by in RCA: 54] [Article Influence: 10.8] [Reference Citation Analysis (0)] |
| 66. | Foegeding NJ, Caston RR, McClain MS, Ohi MD, Cover TL. An Overview of Helicobacter pylori VacA Toxin Biology. Toxins (Basel). 2016;8. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 109] [Cited by in RCA: 160] [Article Influence: 17.8] [Reference Citation Analysis (0)] |
| 67. | Terebiznik MR, Raju D, Vázquez CL, Torbricki K, Kulkarni R, Blanke SR, Yoshimori T, Colombo MI, Jones NL. Effect of Helicobacter pylori's vacuolating cytotoxin on the autophagy pathway in gastric epithelial cells. Autophagy. 2009;5:370-379. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 151] [Cited by in RCA: 161] [Article Influence: 10.1] [Reference Citation Analysis (0)] |
| 68. | Tang B, Li N, Gu J, Zhuang Y, Li Q, Wang HG, Fang Y, Yu B, Zhang JY, Xie QH, Chen L, Jiang XJ, Xiao B, Zou QM, Mao XH. Compromised autophagy by MIR30B benefits the intracellular survival of Helicobacter pylori. Autophagy. 2012;8:1045-1057. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 99] [Cited by in RCA: 112] [Article Influence: 8.6] [Reference Citation Analysis (0)] |
| 69. | Raju D, Hussey S, Ang M, Terebiznik MR, Sibony M, Galindo-Mata E, Gupta V, Blanke SR, Delgado A, Romero-Gallo J, Ramjeet MS, Mascarenhas H, Peek RM, Correa P, Streutker C, Hold G, Kunstmann E, Yoshimori T, Silverberg MS, Girardin SE, Philpott DJ, El Omar E, Jones NL. Vacuolating cytotoxin and variants in Atg16L1 that disrupt autophagy promote Helicobacter pylori infection in humans. Gastroenterology. 2012;142:1160-1171. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 154] [Cited by in RCA: 182] [Article Influence: 14.0] [Reference Citation Analysis (0)] |
| 70. | Capurro MI, Prashar A, Jones NL. MCOLN1/TRPML1 inhibition - a novel strategy used by Helicobacter pylori to escape autophagic killing and antibiotic eradication therapy in vivo. Autophagy. 2020;16:169-170. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 10] [Cited by in RCA: 14] [Article Influence: 2.3] [Reference Citation Analysis (0)] |
| 71. | Capurro MI, Greenfield LK, Prashar A, Xia S, Abdullah M, Wong H, Zhong XZ, Bertaux-Skeirik N, Chakrabarti J, Siddiqui I, O'Brien C, Dong X, Robinson L, Peek RM Jr, Philpott DJ, Zavros Y, Helmrath M, Jones NL. VacA generates a protective intracellular reservoir for Helicobacter pylori that is eliminated by activation of the lysosomal calcium channel TRPML1. Nat Microbiol. 2019;4:1411-1423. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 46] [Cited by in RCA: 83] [Article Influence: 13.8] [Reference Citation Analysis (0)] |
| 72. | Eslami M, Yousefi B, Kokhaei P, Arabkari V, Ghasemian A. Current information on the association of Helicobacter pylori with autophagy and gastric cancer. J Cell Physiol. 2019;. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 26] [Cited by in RCA: 36] [Article Influence: 6.0] [Reference Citation Analysis (0)] |
| 73. | Li N, Tang B, Jia YP, Zhu P, Zhuang Y, Fang Y, Li Q, Wang K, Zhang WJ, Guo G, Wang TJ, Feng YJ, Qiao B, Mao XH, Zou QM. Helicobacter pylori CagA Protein Negatively Regulates Autophagy and Promotes Inflammatory Response via c-Met-PI3K/Akt-mTOR Signaling Pathway. Front Cell Infect Microbiol. 2017;7:417. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 67] [Cited by in RCA: 94] [Article Influence: 11.8] [Reference Citation Analysis (0)] |
| 74. | Deen NS, Huang SJ, Gong L, Kwok T, Devenish RJ. The impact of autophagic processes on the intracellular fate of Helicobacter pylori: more tricks from an enigmatic pathogen? Autophagy. 2013;9:639-652. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 39] [Cited by in RCA: 48] [Article Influence: 4.0] [Reference Citation Analysis (0)] |
| 75. | Feng GJ, Chen Y, Li K. Helicobacter pylori promote inflammation and host defense through the cagA-dependent activation of mTORC1. J Cell Physiol. 2020;235:10094-10108. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 8] [Cited by in RCA: 14] [Article Influence: 2.8] [Reference Citation Analysis (0)] |
| 76. | Cullen TW, Giles DK, Wolf LN, Ecobichon C, Boneca IG, Trent MS. Helicobacter pylori vs the host: remodeling of the bacterial outer membrane is required for survival in the gastric mucosa. PLoS Pathog. 2011;7:e1002454. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 134] [Cited by in RCA: 161] [Article Influence: 11.5] [Reference Citation Analysis (0)] |
| 77. | Olofsson A, Vallström A, Petzold K, Tegtmeyer N, Schleucher J, Carlsson S, Haas R, Backert S, Wai SN, Gröbner G, Arnqvist A. Biochemical and functional characterization of Helicobacter pylori vesicles. Mol Microbiol. 2010;77:1539-1555. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 143] [Cited by in RCA: 174] [Article Influence: 11.6] [Reference Citation Analysis (0)] |
| 78. | Parker H, Keenan JI. Composition and function of Helicobacter pylori outer membrane vesicles. Microbes Infect. 2012;14:9-16. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 37] [Cited by in RCA: 46] [Article Influence: 3.3] [Reference Citation Analysis (0)] |
| 79. | Kaparakis M, Turnbull L, Carneiro L, Firth S, Coleman HA, Parkington HC, Le Bourhis L, Karrar A, Viala J, Mak J, Hutton ML, Davies JK, Crack PJ, Hertzog PJ, Philpott DJ, Girardin SE, Whitchurch CB, Ferrero RL. Bacterial membrane vesicles deliver peptidoglycan to NOD1 in epithelial cells. Cell Microbiol. 2010;12:372-385. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 279] [Cited by in RCA: 342] [Article Influence: 21.4] [Reference Citation Analysis (0)] |
| 80. | Choi MS, Ze EY, Park JY, Shin TS, Kim JG. Helicobacter pylori-derived outer membrane vesicles stimulate interleukin 8 secretion through nuclear factor kappa B activation. Korean J Intern Med. 2021;36:854-867. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 7] [Cited by in RCA: 20] [Article Influence: 5.0] [Reference Citation Analysis (0)] |
| 81. | Basak C, Pathak SK, Bhattacharyya A, Pathak S, Basu J, Kundu M. The secreted peptidyl prolyl cis,trans-isomerase HP0175 of Helicobacter pylori induces apoptosis of gastric epithelial cells in a TLR4- and apoptosis signal-regulating kinase 1-dependent manner. J Immunol. 2005;174:5672-5680. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 66] [Cited by in RCA: 70] [Article Influence: 3.5] [Reference Citation Analysis (0)] |
| 82. | Halder P, Datta C, Kumar R, Sharma AK, Basu J, Kundu M. The secreted antigen, HP0175, of Helicobacter pylori links the unfolded protein response (UPR) to autophagy in gastric epithelial cells. Cell Microbiol. 2015;17:714-729. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 26] [Cited by in RCA: 31] [Article Influence: 3.1] [Reference Citation Analysis (0)] |
| 83. | Song H, Wang T, Tian L, Bai S, Chen L, Zuo Y, Xue Y. Macrophages on the Peritoneum are involved in Gastric Cancer Peritoneal Metastasis. J Cancer. 2019;10:5377-5387. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 17] [Cited by in RCA: 31] [Article Influence: 5.2] [Reference Citation Analysis (0)] |
| 84. | Basu S, Pathak SK, Chatterjee G, Pathak S, Basu J, Kundu M. Helicobacter pylori protein HP0175 transactivates epidermal growth factor receptor through TLR4 in gastric epithelial cells. J Biol Chem. 2008;283:32369-32376. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 42] [Cited by in RCA: 49] [Article Influence: 2.9] [Reference Citation Analysis (0)] |
| 85. | Bravo J, Díaz P, Corvalán AH, Quest AFG. A Novel Role for Helicobacter pylori Gamma-Glutamyltranspeptidase in Regulating Autophagy and Bacterial Internalization in Human Gastric Cells. Cancers (Basel). 2019;11. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 9] [Cited by in RCA: 13] [Article Influence: 2.2] [Reference Citation Analysis (0)] |
| 86. | Butcher LD, den Hartog G, Ernst PB, Crowe SE. Oxidative Stress Resulting From Helicobacter pylori Infection Contributes to Gastric Carcinogenesis. Cell Mol Gastroenterol Hepatol. 2017;3:316-322. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 105] [Cited by in RCA: 163] [Article Influence: 20.4] [Reference Citation Analysis (0)] |
| 87. | Han H, Lim JW, Kim H. Astaxanthin Inhibits Helicobacter pylori-induced Inflammatory and Oncogenic Responses in Gastric Mucosal Tissues of Mice. J Cancer Prev. 2020;25:244-251. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 12] [Cited by in RCA: 17] [Article Influence: 3.4] [Reference Citation Analysis (0)] |
| 88. | Tsugawa H, Suzuki H, Saya H, Hatakeyama M, Hirayama T, Hirata K, Nagano O, Matsuzaki J, Hibi T. Reactive oxygen species-induced autophagic degradation of Helicobacter pylori CagA is specifically suppressed in cancer stem-like cells. Cell Host Microbe. 2012;12:764-777. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 163] [Cited by in RCA: 177] [Article Influence: 14.8] [Reference Citation Analysis (0)] |
| 89. | Mills JC, Sansom OJ. Reserve stem cells: Differentiated cells reprogram to fuel repair, metaplasia, and neoplasia in the adult gastrointestinal tract. Sci Signal. 2015;8:re8. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 102] [Cited by in RCA: 112] [Article Influence: 11.2] [Reference Citation Analysis (0)] |
| 90. | Xie C, Li N, Wang H, He C, Hu Y, Peng C, Ouyang Y, Wang D, Xie Y, Chen J, Shu X, Zhu Y, Lu N. Inhibition of autophagy aggravates DNA damage response and gastric tumorigenesis via Rad51 ubiquitination in response to H. pylori infection. Gut Microbes. 2020;11:1567-1589. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 28] [Cited by in RCA: 53] [Article Influence: 10.6] [Reference Citation Analysis (0)] |
| 91. | Usman RM, Razzaq F, Akbar A, Farooqui AA, Iftikhar A, Latif A, Hassan H, Zhao J, Carew JS, Nawrocki ST, Anwer F. Role and mechanism of autophagy-regulating factors in tumorigenesis and drug resistance. Asia Pac J Clin Oncol. 2021;17:193-208. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 67] [Cited by in RCA: 78] [Article Influence: 19.5] [Reference Citation Analysis (0)] |
| 92. | Wang Y, Zhu WG, Zhao Y. Autophagy substrate SQSTM1/p62 regulates chromatin ubiquitination during the DNA damage response. Autophagy. 2017;13:212-213. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 32] [Cited by in RCA: 44] [Article Influence: 4.9] [Reference Citation Analysis (0)] |
| 93. | Trang TT, Nagashima H, Uchida T, Mahachai V, Vilaichone RK, Tshering L, Binh TT, Yamaoka Y. RAD51 G135C genetic polymorphism and their potential role in gastric cancer induced by Helicobacter pylori infection in Bhutan. Epidemiol Infect. 2016;144:234-240. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 5] [Cited by in RCA: 5] [Article Influence: 0.5] [Reference Citation Analysis (0)] |
| 94. | Camilo V, Sugiyama T, Touati E. Pathogenesis of Helicobacter pylori infection. Helicobacter. 2017;22 Suppl 1. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 62] [Cited by in RCA: 110] [Article Influence: 13.8] [Reference Citation Analysis (3)] |
| 95. | Courtois S, Haykal M, Bodineau C, Sifré E, Azzi-Martin L, Ménard A, Mégraud F, Lehours P, Durán RV, Varon C, Bessède E. Autophagy induced by Helicobacter pylori infection is necessary for gastric cancer stem cell emergence. Gastric Cancer. 2021;24:133-144. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 15] [Cited by in RCA: 25] [Article Influence: 6.3] [Reference Citation Analysis (0)] |
| 96. | Piao JY, Kim SJ, Kim DH, Park JH, Park SA, Han HJ, Na HK, Yoon K, Lee HN, Kim N, Hahm KB, Surh YJ. Helicobacter pylori infection induces STAT3 phosphorylation on Ser727 and autophagy in human gastric epithelial cells and mouse stomach. Sci Rep. 2020;10:15711. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 11] [Cited by in RCA: 26] [Article Influence: 5.2] [Reference Citation Analysis (0)] |
| 97. | Hu W, Zhang L, Li MX, Shen J, Liu XD, Xiao ZG, Wu DL, Ho IHT, Wu JCY, Cheung CKY, Zhang YC, Lau AHY, Ashktorab H, Smoot DT, Fang EF, Chan MTV, Gin T, Gong W, Wu WKK, Cho CH. Vitamin D3 activates the autolysosomal degradation function against Helicobacter pylori through the PDIA3 receptor in gastric epithelial cells. Autophagy. 2019;15:707-725. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 71] [Cited by in RCA: 114] [Article Influence: 19.0] [Reference Citation Analysis (0)] |
| 98. | Hu W, Zhang L, Wu WKK, Cho CH. Involvement of autophagy in antibacterial actions of vitamin D in Helicobacter pylori infection: abridged secondary publication. Hong Kong Med J. 2020;26 Suppl 4:26-28. [PubMed] |
| 99. | Liao WC, Huang MZ, Wang ML, Lin CJ, Lu TL, Lo HR, Pan YJ, Sun YC, Kao MC, Lim HJ, Lai CH. Statin Decreases Helicobacter pylori Burden in Macrophages by Promoting Autophagy. Front Cell Infect Microbiol. 2016;6:203. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 24] [Cited by in RCA: 43] [Article Influence: 5.4] [Reference Citation Analysis (0)] |
| 100. | Xu N, Zhang J, Shen C, Luo Y, Xia L, Xue F, Xia Q. Cisplatin-induced downregulation of miR-199a-5p increases drug resistance by activating autophagy in HCC cell. Biochem Biophys Res Commun. 2012;423:826-831. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 146] [Cited by in RCA: 155] [Article Influence: 11.9] [Reference Citation Analysis (0)] |
| 101. | An Y, Zhang Z, Shang Y, Jiang X, Dong J, Yu P, Nie Y, Zhao Q. miR-23b-3p regulates the chemoresistance of gastric cancer cells by targeting ATG12 and HMGB2. Cell Death Dis. 2015;6:e1766. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 141] [Cited by in RCA: 183] [Article Influence: 18.3] [Reference Citation Analysis (0)] |
| 102. | Zhao J, Nie Y, Wang H, Lin Y. MiR-181a suppresses autophagy and sensitizes gastric cancer cells to cisplatin. Gene. 2016;576:828-833. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 75] [Cited by in RCA: 91] [Article Influence: 9.1] [Reference Citation Analysis (0)] |
| 103. | Ge J, Chen Z, Huang J, Chen J, Yuan W, Deng Z. Upregulation of autophagy-related gene-5 (ATG-5) is associated with chemoresistance in human gastric cancer. PLoS One. 2014;9:e110293. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 79] [Cited by in RCA: 90] [Article Influence: 8.2] [Reference Citation Analysis (0)] |
| 104. | Li LQ, Pan D, Zhang SW, -Y-Xie D, Zheng XL, Chen H. Autophagy regulates chemoresistance of gastric cancer stem cells via the Notch signaling pathway. Eur Rev Med Pharmacol Sci. 2018;22:3402-3407. [RCA] [PubMed] [DOI] [Full Text] [Cited by in RCA: 23] [Reference Citation Analysis (0)] |
| 105. | Rojas-Puentes LL, Gonzalez-Pinedo M, Crismatt A, Ortega-Gomez A, Gamboa-Vignolle C, Nuñez-Gomez R, Dorantes-Gallareta Y, Arce-Salinas C, Arrieta O. Phase II randomized, double-blind, placebo-controlled study of whole-brain irradiation with concomitant chloroquine for brain metastases. Radiat Oncol. 2013;8:209. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 85] [Cited by in RCA: 106] [Article Influence: 8.8] [Reference Citation Analysis (0)] |
| 106. | Zhou H, Yuan M, Yu Q, Zhou X, Min W, Gao D. Autophagy regulation and its role in gastric cancer and colorectal cancer. Cancer Biomark. 2016;17:1-10. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 54] [Cited by in RCA: 65] [Article Influence: 8.1] [Reference Citation Analysis (0)] |
| 107. | Liu M, Li CM, Chen ZF, Ji R, Guo QH, Li Q, Zhang HL, Zhou YN. Celecoxib regulates apoptosis and autophagy via the PI3K/Akt signaling pathway in SGC-7901 gastric cancer cells. Int J Mol Med. 2014;33:1451-1458. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 46] [Cited by in RCA: 64] [Article Influence: 5.8] [Reference Citation Analysis (0)] |
| 108. | Zhao K, Zhu BS, Gong W, Zhu ML, Gao ZT, Wu YY, Chen Q, Yang XD, Xing CG. SN50 enhances the effects of LY294002 on cell death induction in gastric cancer cell line SGC7901. Arch Med Sci. 2013;9:990-998. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 8] [Cited by in RCA: 14] [Article Influence: 1.2] [Reference Citation Analysis (0)] |
| 109. | Rui LX, Shu SY, Jun WJ, Mo CZ, Wu SZ, Min LS, Yuan L, Yong PJ, Cheng SZ, Sheng WS, Yao TZ. The dual induction of apoptosis and autophagy by SZC014, a synthetic oleanolic acid derivative, in gastric cancer cells via NF-κB pathway. Tumour Biol. 2016;37:5133-5144. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 20] [Cited by in RCA: 23] [Article Influence: 2.3] [Reference Citation Analysis (0)] |
| 110. | Tang C, Yang L, Jiang X, Xu C, Wang M, Wang Q, Zhou Z, Xiang Z, Cui H. Antibiotic drug tigecycline inhibited cell proliferation and induced autophagy in gastric cancer cells. Biochem Biophys Res Commun. 2014;446:105-112. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 38] [Cited by in RCA: 52] [Article Influence: 4.7] [Reference Citation Analysis (0)] |
| 111. | Shen YC, Li CP, Yen CJ, Hsu C, Lin YL, Lin ZZ, Chen LT, Su WC, Chao Y, Yeh KH, Cheng AL. Phase II multicentered study of low-dose everolimus plus cisplatin and weekly 24-hour infusion of high-dose 5-fluorouracil and leucovorin as first-line treatment for patients with advanced gastric cancer. Oncology. 2014;87:104-113. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 9] [Cited by in RCA: 12] [Article Influence: 1.1] [Reference Citation Analysis (0)] |
| 112. | Zhang L, Wang X, Chen P. MiR-204 down regulates SIRT1 and reverts SIRT1-induced epithelial-mesenchymal transition, anoikis resistance and invasion in gastric cancer cells. BMC Cancer. 2013;13:290. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 101] [Cited by in RCA: 126] [Article Influence: 10.5] [Reference Citation Analysis (0)] |
| 113. | Huang R, Xu Y, Wan W, Shou X, Qian J, You Z, Liu B, Chang C, Zhou T, Lippincott-Schwartz J, Liu W. Deacetylation of nuclear LC3 drives autophagy initiation under starvation. Mol Cell. 2015;57:456-466. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 470] [Cited by in RCA: 519] [Article Influence: 51.9] [Reference Citation Analysis (0)] |
| 114. | Lock R, Debnath J. Extracellular matrix regulation of autophagy. Curr Opin Cell Biol. 2008;20:583-588. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 137] [Cited by in RCA: 128] [Article Influence: 7.5] [Reference Citation Analysis (0)] |
| 115. | Ding YP, Yang XD, Wu Y, Xing CG. Autophagy promotes the survival and development of tumors by participating in the formation of vasculogenic mimicry. Oncol Rep. 2014;31:2321-2327. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 17] [Cited by in RCA: 18] [Article Influence: 1.6] [Reference Citation Analysis (0)] |
| 116. | Xiao M, Lin C, Yang Z, Tian S, Huang Y, Fu J. Compound TDB (Tricyclic decyl benzoxazole) induces autophagy-dependent apoptosis in the gastric cancer cell line MGC-803 by regulating PI3K/AKT/mTOR. Am J Transl Res. 2021;13:73-87. [PubMed] |
| 117. | Masuda GO, Yashiro M, Kitayama K, Miki Y, Kasashima H, Kinoshita H, Morisaki T, Fukuoka T, Hasegawa T, Sakurai K, Toyokawa T, Kubo N, Tanaka H, Muguruma K, Masaichi O, Hirakawa K. Clinicopathological Correlations of Autophagy-related Proteins LC3, Beclin 1 and p62 in Gastric Cancer. Anticancer Res. 2016;36:129-136. [PubMed] |
| 118. | Zheng HC, Zhao S, Xue H, Zhao EH, Jiang HM, Hao CL. The Roles of Beclin 1 Expression in Gastric Cancer: A Marker for Carcinogenesis, Aggressive Behaviors and Favorable Prognosis, and a Target of Gene Therapy. Front Oncol. 2020;10:613679. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 8] [Cited by in RCA: 21] [Article Influence: 4.2] [Reference Citation Analysis (0)] |
| 119. | Qiu J, Sun M, Wang Y, Chen B. Identification and validation of an individualized autophagy-clinical prognostic index in gastric cancer patients. Cancer Cell Int. 2020;20:178. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 28] [Cited by in RCA: 45] [Article Influence: 9.0] [Reference Citation Analysis (0)] |