Published online Feb 15, 2020. doi: 10.4251/wjgo.v12.i2.124
Peer-review started: September 13, 2019
First decision: October 18, 2019
Revised: October 30, 2019
Accepted: November 29, 2019
Article in press: November 29, 2019
Published online: February 15, 2020
Processing time: 155 Days and 0.4 Hours
Colorectal cancer (CRC) is a global problem affecting millions of people worldwide. This disease is unique because of its slow progress that makes it preventable and often curable. CRC symptoms usually emerge only at advanced stages of the disease, consequently its early detection can be achieved only through active population screening, which markedly reduces mortality due to this cancer. CRC screening tests that employ non-invasively detectable biomarkers are currently being actively developed and, in most cases, samples of either stool or blood are used. However, alternative biological substances that can be collected non-invasively (colorectal mucus, urine, saliva, exhaled air) have now emerged as new sources of diagnostic biomarkers. The main categories of currently explored CRC biomarkers are: (1) Proteins (comprising widely used haemoglobin); (2) DNA (including mutations and methylation markers); (3) RNA (in particular microRNAs); (4) Low molecular weight metabolites (comprising volatile organic compounds) detectable by metabolomic techniques; and (5) Shifts in gut microbiome composition. Numerous tests for early CRC detection employing such non-invasive biomarkers have been proposed and clinically studied. While some of these studies generated promising early results, very few of the proposed tests have been transformed into clinically validated diagnostic/screening techniques. Such DNA-based tests as Food and Drug Administration-approved multitarget stool test (marketed as Cologuard®) or blood test for methylated septin 9 (marketed as Epi proColon® 2.0 CE) show good diagnostic performance but remain too expensive and technically complex to become effective CRC screening tools. It can be concluded that, despite its deficiencies, the protein (haemoglobin) detection-based faecal immunochemical test (FIT) today presents the most cost-effective option for non-invasive CRC screening. The combination of non-invasive FIT and confirmatory invasive colonoscopy is the current strategy of choice for CRC screening. However, continuing intense research in the area promises the emergence of new superior non-invasive CRC screening tests that will allow the development of improved disease prevention strategies.
Core tip: Numerous biomarkers detectable in non-invasively collected samples of stool, colorectal mucus, blood, urine, saliva and exhaled air have been investigated to develop new tests for colorectal cancer (CRC) early detection and screening. Promising results are often reported, but it is difficult to achieve the right balance between technical complexity, cost and diagnostic performance of the new tests. Today the combination of non-invasive faecal immunochemical test and confirmatory invasive colonoscopy remains the CRC screening strategy of choice. However, on-going intense research promises the emergence of new superior non-invasive screening tests that will allow the development of improved prevention strategies for these malignancies.
- Citation: Loktionov A. Biomarkers for detecting colorectal cancer non-invasively: DNA, RNA or proteins? World J Gastrointest Oncol 2020; 12(2): 124-148
- URL: https://www.wjgnet.com/1948-5204/full/v12/i2/124.htm
- DOI: https://dx.doi.org/10.4251/wjgo.v12.i2.124
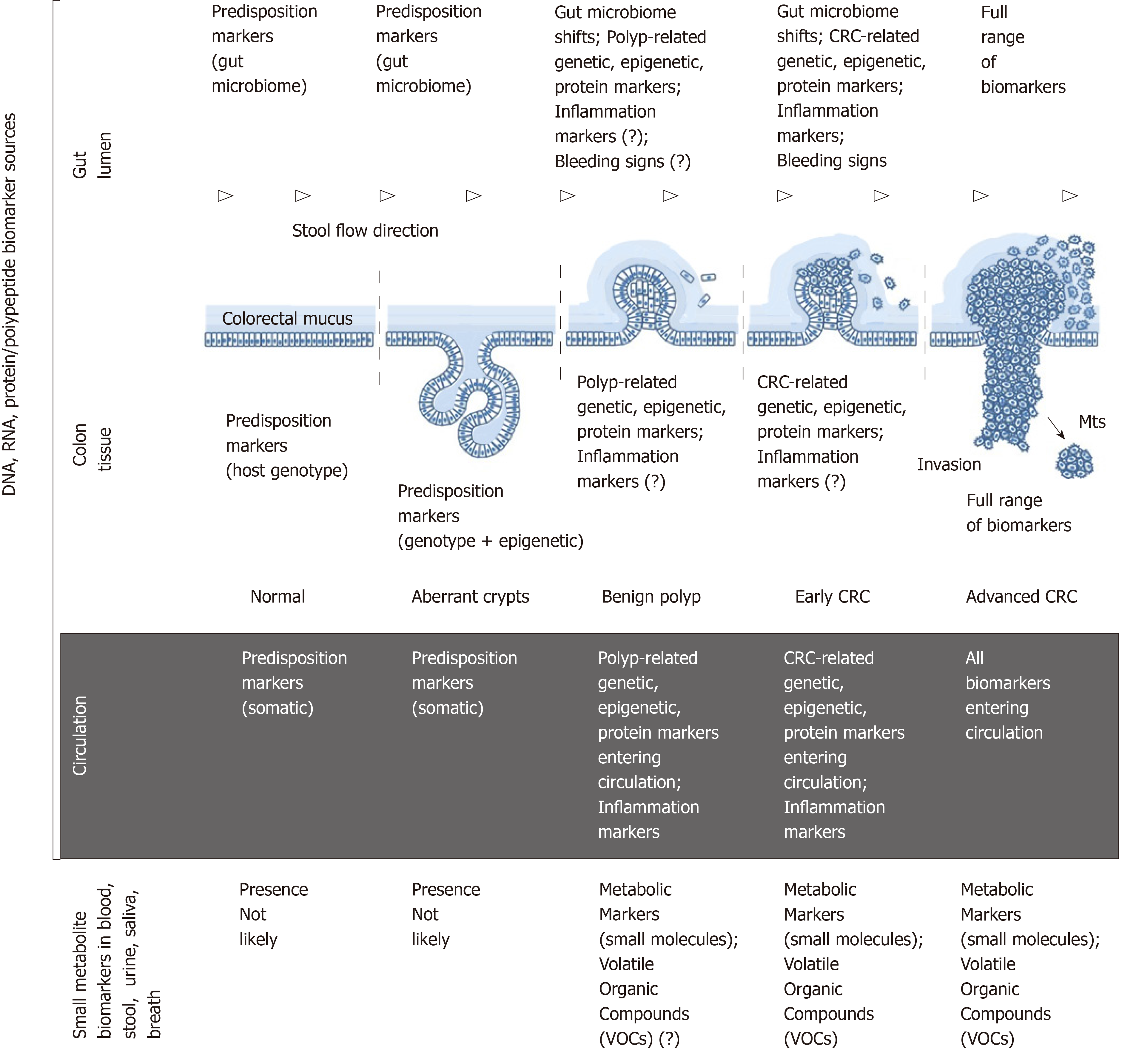
Colorectal cancer (CRC) is currently the third most frequently diagnosed cancer worldwide. The global incidence for 2018 is estimated at 1801000 new cases, and the number of CRC-related deaths for this period is 861700[1]. Although the highest CRC incidence continues to be observed in economically developed Western countries, it is now rapidly increasing in other parts of the world[2]. Sporadic CRC development can take decades and is in most cases characterised by a slow progression from aberrant crypt formation in the colonic mucosa to benign polyps that may give rise to early cancer, then gradually evolving to invasive and metastasising advanced neoplasms (Figure 1)[2-4]. These pathogenetic features make CRC one of the most preventable and often curable malignancies. However, disease curability entirely depends on its early detection, which is not straightforward as clinical symptoms usually emerge only when CRC is already advanced. The latter factor warrants the necessity of active population screening for CRC, and it has been well proven that screening saves lives[2].
Full colonoscopy is regarded as the gold standard diagnostic technique for colorectal tumour detection[5], and it has become a very popular method for primary CRC screening[6-8] in the United States. One apparent reason for this trend is that diagnostic colonoscopy is usually combined with the simultaneous removal of detected polyps and functions as both a diagnostic and preventive procedure clearly reducing mortality from CRC[9]. Nonetheless, colonoscopy is an expensive and invasive technique that requires unpleasant bowel preparation and occasionally causes serious complications[10]. Moreover, its sensitivity is not perfect, with polyps sometimes missed[11], the latter problem often depending on the operator’s skills[12]. Although colonoscopy as the final (confirmatory) diagnostic step is undisputable, its use in primary CRC screening remains questionable as indiscriminate application of this method inevitably results in frequent negative outcomes and a large health economic burden[13]. In theory, the global introduction of non-invasive tests employing biomarker analysis to select patients that really require endoscopy could dramatically reduce the numbers of unnecessary colonoscopies. Unfortunately, none of the existing non-invasive tests successfully combine high diagnostic sensitivity and specificity with technical simplicity and low cost, the key characteristics of an ideal screening modality. This paper provides a brief overview of the current state of the area encompassing biomarker-based non-invasive tests for CRC detection.
CRC development is an extraordinarily complex process driven by multiple genetic, epigenetic, metabolic and immune alterations at the host level and influenced by numerous environmental factors[4,14,15]. Despite intense research, precise mechanisms of CRC development remain largely obscure[4,14,15]. Genome-targeting investigations, especially genome-wide association studies, have revealed a highly complex pathogenetic landscape comprising multiple alternative cascades of molecular events that may eventually result in cancer[4,16]. This complexity leads some investigators to a hardly satisfactory conclusion that “each patient’s CRC is genetically and epigenetically unique”[4]. Nevertheless, colorectal tumours frequently have common molecular patterns that are diagnostically relevant and will be considered below.
The series of morphological events accompanying CRC development is presented in Figure 1. This sequence involves numerous associations with various types of biomolecules that can be characterised as biomarkers. The ideal biomarkers for CRC can be defined as substances that satisfy the following criteria: “(1) Are measured easily and inexpensively to identify a patient’s cancer; (2) Identify a patient’s prognosis to improve treatment outcome; and (3) Predict a patient’s response to a specific treatment”[15]. This paper is focused only on the first category, i.e., diagnostic biomarkers of CRC that can be sampled and tested non-invasively.
Figure 1 outlines the main sources of CRC biomarkers in relation to disease stages. From the morphological point of view, it is obvious that (1) colon tissue; (2) gut lumen; (3) blood/lymph circulation are the main sources of CRC-associated DNA, RNA and protein/polypeptide biomarkers associated with the host; (4) moreover, specific pattern shifts in small metabolite molecules derived from CRC-affected metabolic pathways constitute an additional group of post-metabolic markers that can be analysed by metabolomics techniques[17,18]; and (5) CRC-associated gut microbiome changes[19] deserve to be considered as a separate category of diagnostic markers of non-human origin.
Colonic epithelium is the site of neoplastic growth initiation. After that CRC progresses within the colonic wall until advanced stages of the disease, hence pre-malignant and malignant colon tissues are certainly the richest biomarker sources[4]. However, invasive biopsies are required for sampling tissue. Therefore, CRC markers detectable in tissue samples are not discussed here.
Colonic epithelium is the key element of the gastrointestinal barrier between host tissues and microbiota-rich colon contents. Until recently it was presumed that all host cells exfoliated or migrated from the surface of the colonic epithelium were immediately incorporated in the faecal matter. According to this simplistic notion, it seemed to be logical that analysing naturally excreted stool samples constitutes the only perfectly non-invasive approach to investigating CRC biomarkers. It should, however, be stressed that stool is a complex mixture of microbiota-dominated faecal matter and occasional fragments of colorectal mucus secreted by goblet cells of the colonic epithelium. While the prevailing faecal component of stool entirely belongs to the environment, colorectal mucus is host-derived. The two-layered structure and functional significance of the mucus overlaying colonic epithelium have been elucidated only during the last decade[20,21], and it is now clear that colorectal mucus rather than faecal matter is the main receptacle of all cells and biomolecules released from either normal or malignant epithelium[22,23]. Intrarectal collection of colorectal mucus had demonstrated high informativeness of this substance[22,23], which was shown to accept CRC-generated malignant colonocytes exfoliated from tumour surface and transport them distally alongside stool flow without incorporating them into faeces (Figure 1)[20,23]. Biomarker-rich colorectal mucus essentially serves as a border between well oxygenated colonic epithelium and anaerobic gut lumen. Our group has recently developed a simple technique for non-invasive sampling of this mucus[24-26], the analysis of which may constitute a very convenient alternative to stool-based tests.
Blood-derived biomarker analysis is another area of significant interest in the context of CRC detection since blood collection is regarded as a practically non-invasive procedure. It is evident that a wide range of CRC-associated biomarkers can be detected in the circulating blood and lymph of patients with these malignancies, but lymph collection cannot be performed with minimal invasiveness. For this reason, only biomarkers measurable in blood will be discussed below. In the modern literature the term “liquid biopsy” is often applied to this group of biomarker-based techniques[27]. Nevertheless, despite the easiness of blood sampling and the availability of numerous analytical techniques for biomarker detection in human plasma or serum, the presence of cancer biomarkers in blood may or may not be associated with CRC. Malignancies of other sites should always be excluded if this approach is considered for CRC screening.
The use of metabolomics for revealing CRC-specific changes in patterns of low molecular weight metabolites has recently become another area of active exploration[28]. This new approach can potentially employ a wider range of biological samples comprising blood, stool, colorectal mucus, urine, saliva and exhaled breath, thus bringing about additional diagnostic options.
Recent research has revealed that specific changes in gut microbiome composition may be associated with the development of CRC[19]. In this context stool samples are usually investigated quantitatively for the presence of particular types of bacteria.
The limited choice of sample sources for non-invasive testing creates obvious problems. Collecting gut-derived samples looks preferable, but stool samples, albeit containing cells and molecules originating from the colonic mucosa (i.e., colorectal mucus fragments), are usually dominated by the presence of abundant microbiota-rich faecal matter that often interferes with analytical procedures employed for host-related biomarker detection. A recently described analysis of non-invasively collected colorectal mucus presents a very interesting alternative; however, this approach is new and requires further testing. On the other hand, blood collection is very straightforward and easy to standardise, but molecular changes detectable in blood (or plasma/serum) samples are not necessarily gut-specific. Finally, although the use of easily collectable materials (urine, saliva or exhaled air) is extremely attractive, the presence of CRC-specific biomarkers in such samples remains to be adequately explored. The sources of biological material characterised above may contain several types of diagnostic biomarkers that are discussed in the next section.
The story of non-invasively detectable CRC markers started due to a 1967 publication by Greegor, describing his observation of the frequent presence of occult blood in stool samples collected from patients with CRC[29]. That important discovery resulted in the development and prolonged use of the haemoglobin-recognising faecal occult blood test (FOBT) as the only non-invasive test for CRC detection. The situation had changed considerably in 1992, when a publication by Sidransky et al[30] described K-ras gene mutation detection in stool samples obtained from CRC patients and shifted the focus of attention to molecular markers. The area of CRC biomarker research has since exponentially expanded with thousands of papers published, but many initially promising findings failed to transform into clinically relevant diagnostic approaches. The purpose of this paper is to briefly outline the present status of non-invasive biomarkers proposed for detecting asymptomatic CRC. Only the most impressive and clinically relevant observations related to the main groups of these biomarkers (proteins/polypeptides, DNA, RNA, small metabolites, microbiome changes) are highlighted in the text below. However, numerous other markers that demonstrated promise in the context of CRC detection are presented in comprehensive Tables 1, 2, 3, 4 and 5. As it was impossible to cover all relevant studies, restrictions had to be applied when the Tables were prepared. Publications describing very small studies or reporting negative results were omitted. Likewise, only papers related to CRC, but not adenoma detection, were included since in most cases diagnostic sensitivity of biomarker tests for adenomatous polyps correlates with that for CRC. In addition, the necessity of non-invasive detection of colorectal polyps is still a debatable question, as the proportion of adenomas likely to progress to malignancy is relatively small, whereas the vast majority of these lesions (especially small polyps) never give rise to CRC[134,135].
| Study setting | Sample type | Marker type | Biomarker(s) | Sensitivity (or its range) | Specificity (or its range) | Ref. |
| Screening (reviewed) | Stool | Protein | Haemoglobin (FIT) | 66.0%-74.0% | 84.0%-95.0% | [31] |
| Case-control (reviewed) | Stool | Protein | M2-PK | 68.0%-93.0% | 70.0%-97.5% | [32,33] |
| Case-control | Stool | Protein | MMP 9 | 89.30% | 91.20% | [34] |
| Case-control | Stool | Protein panel | Complement C3, Lactotransferrin, Haemoglobin subunit α1 and Haptoglobin | 71.00% | 95.00% | [35] |
| Case-control | Serum | Protein | CA11-19 | 98.00% | 84.00% | [36] |
| Case-control | Serum | Protein (cytokine) | MIC-1 (GDF15) | 43.80% | 96.70% | [36] |
| Case-control | Serum | Protein (cytokine) | IL-6 | 28.0%-89.5% | 46.0%-94.0% | [37] |
| Case-control | Serum | Protein (cytokine) | IL-8 | 70.00% | 91.00% | [36] |
| Case-control | Serum | Protein (cytokine) | Growth-related gene product β1 | 56.10% | 95.30% | [36] |
| Case-control | Serum | Protein | Cyr61 | 83.00% | 97.00% | [38] |
| Case-control | Serum | Protein | Β6-integrin | 69.80% | 100.00% | [39] |
| Case-control (reviewed) | Serum | Protein | TIMP-1 | 52.0%-85.0% | 60.0%-95.0% | [40] |
| Case-control | Serum | Protein | RBP4 | 74.90% | 81.70% | [36] |
| Case-control | Serum | Protein | THBS2 | 64.90% | 87.10% | [36] |
| Case-control | Serum | Protein | TFF3 | 74.20% | 94.80% | [36] |
| Case-control | Serum | Protein | COL3A1 | 98.80% | 69.10% | [36] |
| Case-control | Serum | Protein | COL10A1 | 63.00% | 85.00% | [36] |
| Case-control | Serum | Protein | AZGP1 | 55.80% | 85.00% | [36] |
| Case-control | Serum | Protein | Angiopoietin-2 | 79.30% | 82.40% | [36] |
| Case-control | Serum | Protein | Kininogen | 63.60% | 65.90% | [36] |
| Case-control | Plasma | Protein | Melanotransferrin | 48.20% | 92.50% | [36] |
| Case-control | Serum | Protein panel | RBP4 and CEA | 80.80% | 91.20% | [36] |
| Case-control | Serum | Protein panel | TFF3 and CEA | 89.40% | 87.80% | [41] |
| Case-control | Serum | Protein panel | sDC-SIGN and sDC-SIGNR | 98.70% | 94.80% | [42] |
| Case-control | Serum | Protein panel | IGFBP-3 and CEA | 75.00% | 90.00% | [43] |
| Case-control | Serum | Protein panel | AZGP1, CEA and CA19-9 | 67.50% | 82.50% | [36] |
| Case-control | Serum | Protein panel | IGFBP2, DKK3 and PKM2 | 73.00% | 95.00% | [36] |
| Case-control | Plasma | Protein panel | BAG4, IL6ST, VWF, EGFR and CD44 | 73.00% | 90.00% | [44] |
| Case-control, prospective | Serum | Protein panel | CEA, hs-CRP, CYFra21-1 and Ferritin | 60.0%-70.0% | 81.0%-89.0% | [45] |
Protein biomarkers considered in CRC early detection and screening are listed in Table 1. Historically, the use of haemoglobin detection in stool for non-invasive CRC detection can be regarded as the most popular approach in terms of population screening. Indeed, the traditional guaiac FOBT was almost exclusively employed for this purpose for several decades, and was attractive due to its simplicity and low cost. Although this test has insufficient sensitivity, it can be credited for saving many human lives[2,136,137]. Nevertheless, the outdated FOBT is now being replaced by the faecal immunochemical test (FIT) characterised by a much higher sensitivity. In a recent comprehensive review on FIT, Gies et al[31] discussed numerous studies of varying sizes and reported sensitivities between 66% and 74% and specificity levels between 84% and 95% when numbers of analysed CRC cases and controls were over 50. Table 1 also shows that M2-pyruvate kinase (M2-PK) is a relatively well-studied stool marker of CRC[32,33]; however, FIT performs better and remains considerably more popular. Other stool tests, including metalloproteinase 9 (MMP9)[34] and multimarker protein panels (see Table 1) have been investigated, but these tests have not been clinically accepted so far. It is also intriguing that in a recent small study, our group compared 24 protein biomarkers in non-invasively collected samples of colorectal mucus and concluded that haemoglobin, tissue inhibitor of metalloproteinase 1, M2-PK, peptidyl arginine deiminase 1, C-reactive protein and MMP9 could reliably detect CRC[138].
Blood (or plasma/serum) testing for CRC-associated proteins has been employed by many research groups (Table 1), but most of those studies produced relatively modest results. Among single protein markers detectable in the serum only CA11-19 marker protein[36], cysteine-rich 61 protein of the CCN family (Cyr 61)[38], B6-integrin[39] and trefoil factor 3 (TFF3)[36] can be regarded as promising. A number of protein panels were also examined; however, analysing multiple proteins is usually more technically complex and expensive. Impressive test sensitivity and specificity values (98.7% and 94.8%, respectively) were reported for combined testing for lectins DC-SIGN and DC-SIGNR by Jiang et al[42] in 2014, but these results remain to be confirmed in larger studies. Although blood collection is simple and easy to standardise, protein biomarkers of CRC present in stool or colorectal mucus currently look more diagnostically reliable than those detectable in blood.
An additional advantage of using protein biomarkers for CRC detection is defined by the fact that their immunochemical detection can be easily presented as point of care (POC) tests, which are already available for FIT[139].
This sub-section briefly discusses studies on CRC detection using DNA and mRNA markers that are listed in Table 2.
| Study setting | Sample type | Marker type | Biomarker(s) | Sensitivity (or its range) | Specificity (or its range) | Ref. |
| Screening | Stool | DNA mutation panel | 3 K-ras mutations, 10 APC mutations, 8 p53 mutations, microsatellite instability marker BAT-26 and long DNA marker | 51.60% | 94.40% | [46] |
| Case-control | Stool | Panel including DNA mutation, DNA methylation, DNA amount and protein testing | K-ras mutation, methylation of Vimentin (VIM), BMP3, NDRG4 and TFPI2 genes, DNA measurement by β-actin assessment and HemoQuant test for haemoglobin | 78.0%-85.0% | 85.0%-90.0% | [47] |
| Screening | Stool | Panel including DNA mutation, DNA methylation, DNA amount and protein testing | K-ras mutation, BMP3 and NDRG4 promoter methylation, DNA measurement by β-actin assessment and test for haemoglobin (FIT) | 92.30% | 86.60% | [48] |
| Case-control | Stool | Methylated DNA | BMP3 gene | 51.0%-84.0% | 90.0%-100.0% | [49] |
| Case-control | Stool | Methylated DNA | CDKN2A gene | 20.0%-40.0% | 84.0%-100.0% | [49] |
| Case-control | Stool | Methylated DNA | ECAD gene | 65.20% | 88.00% | [49] |
| Case-control | Stool | Methylated DNA | FBN1 gene | 72.00% | 93.30% | [49] |
| Case-control | Stool | Methylated DNA | GATA 4/5 gene promoter | 42.9%-71.0% | 84.0%-95.0% | [49,50] |
| Case-control | Stool | Methylated DNA | HLTF gene | 20.0%-37.5% | 90.0%-92.6% | [49] |
| Case-control | Stool | Methylated DNA | HIC1 gene | 42.30% | 98.00% | [49] |
| Case-control | Stool | Methylated DNA | HPP1 gene | 71.20% | 57.10% | [49] |
| Case-control | Stool | Methylated DNA | ING1b gene | 73.70% | 95.00% | [49] |
| Case-control | Stool | Methylated DNA | ITGA4 gene | 40.00% | 96.80% | [49] |
| Case-control | Stool | Methylated DNA | MGMT gene | 33.9-55.1% | 52.0%-100.0% | [49] |
| Case-control | Stool | Methylated DNA | NDRG4 gene promoter | 53.0%-92.0% | 89.1%-100.0% | [49-51] |
| Case-control | Stool | Methylated DNA | P16INK4A gene | 71.70% | 86.00% | [49] |
| Case-control | Stool | Methylated DNA | PHACTR3 gene | 55.0%-66.0% | 95.0%-100.0% | [49] |
| Case-control | Stool | Methylated DNA | RASSF2 gene | 45.30% | 94.70% | [49] |
| Case-control | Stool | Methylated DNA | SDC2 gene | 81.10% | 93.30% | [52] |
| Case-control | Stool | Methylated DNA | SEPT9 gene | 20.0%-84.8% | 80.0%-94.5% | [49] |
| Case-control | Stool | Methylated DNA | SFRP1 gene | 26.4%-89.0% | 86.0%-95.5% | [49] |
| Case-control | Stool | Methylated DNA | SFRP2 gene | 32.1%-94.2% | 54.0%-100.0% | [49,51] |
| Case-control | Stool | Methylated DNA | SPG20 gene | 80.2%-89.0% | 99.0%-100.0% | [49,51] |
| Case-control | Stool | Methylated DNA | SNCA gene | 83.90% | 75.00% | [49] |
| Case-control | Stool | Methylated DNA | TFPI2 gene | 63.3%-92.0% | 79.0%-100.0% | [49-51] |
| Case-control | Stool | Methylated DNA | TP53 gene | 56.30% | 100.00% | [49] |
| Case-control | Stool | Methylated DNA | Vimentin (VIM) gene | 32.6%-86.0% | 82.0%-100.0% | [49-51] |
| Case-control | Stool | Methylated DNA | WIF1 gene | 19.3%-60.4% | 96.7%-99.4% | [49] |
| Case-control | Stool | Methylated DNA | XAF1 gene | 55.90% | 52.00% | [49] |
| Case-control | Stool | Methylated DNA panel | BMP3 and NDRG4 genes | 98.00% | 90.00% | [49] |
| Case-control | Stool | Methylated DNA panel | MGMT and XAF1 genes | 73.50% | 52.00% | [49] |
| Case-control | Stool | Methylated DNA panel | MGMT-B and SFRP2 genes | 88.30% | 91.20% | [49] |
| Case-control | Stool | Methylated DNA panel | RASSF1A and SFRP2 genes | 75.00% | 89.40% | [51] |
| Case-control | Stool | Methylated DNA panel | SNCA and FNB1 genes | 84.30% | 93.30% | [53] |
| Case-control | Stool | Methylated DNA panel | Vimentin (VIM) and SFRP2 genes | 92.50% | 91.20% | [53] |
| Case-control | Stool | Methylated DNA panel | AGTR1, WNT2 and SLIT2 genes | 74.0%-78.0% | 88.0%-89.0% | [49,50] |
| Case-control | Stool | Methylated DNA panel | ECAD, MGMT and P16INK4A genes | 72.00% | 88.00% | [49] |
| Case-control | Stool | Methylated DNA panel | ITGA4, SFRP2 and P16INK4A genes | 70.00% | 96.80% | [49] |
| Case-control | Stool | Methylated DNA panel | MGMT, CDKN2A and hMTH1 genes | 55.00% | 63.00% | [49] |
| Case-control | Stool | Methylated DNA panel | MGMT, MLH1 and Vimentin (VIM) genes | 75.00% | 86.50% | [49,51] |
| Case-control | Stool | Methylated DNA panel | SFRP2, HPP1 and MGMT genes | 93.70% | 77.10% | [49] |
| Case-control | Stool | Methylated DNA panel | WIF-1, ALX-4 and Vimentin (VIM) genes | 25.00% | 98.00% | [49] |
| Case-control | Stool | Methylated DNA panel | Vimentin (VIM), OMSR and TFPI2 genes | 86.70% | 87.60% | [49] |
| Case-control | Stool | Methylated DNA panel | SFRP2, GATA4/5, NRDG4 and Vimentin (VIM) genes | 96.40% | 65.00% | [49] |
| Case-control | Stool | Human DNA content | Total human DNA content | 66.00% | 89.80% | [54] |
| Case-control | Bowel Lavage Fluid | Methylated DNA panel | miR-124-3, LOC386758 and SFRP1 genes | 82.00% | 79.00% | [55] |
| Case-control | Intrarectally collected colorectal mucus | Human DNA content | Total human DNA content | 60.40% | 94.80% | [56] |
| Case-control | Serum/plasma | Methylated DNA | ALX4 gene | 23.0%-90.7% | 72.5%-100.0% | [57] |
| Case-control | Serum/plasma | Methylated DNA | APC gene | 57.0%-86.5% | 86.0%-92.1% | [57] |
| Case-control | Plasma | Methylated DNA | CDH1 (E-cadherin) gene | 60.00% | 84.00% | [55] |
| Case-control | Serum/plasma | Methylated DNA | SDC2 gene | 87.0%-90.7% | 72.5%-95.2% | [36,57] |
| Case-control | Serum/plasma | Methylated DNA | SEPT9 gene | 47.1-95.6% | 81.0%-96.7% | [36,57-62] |
| Case-control | Serum/plasma | Methylated DNA | SFRP2 gene | 54.0%-69.4% | 40.0%-98.7% | [57,63] |
| Case-control | Plasma | Methylated DNA | THBD (Thrombomodulin) gene | 70.70% | 80.30% | [51] |
| Case-control | Serum/plasma | Methylated DNA | TPEF gene | 65.0%-81.0% | 69.0%-90.0% | [57] |
| Case-control | Serum/plasma | Methylated DNA | VIM (Vimentin) gene | 59.0%-90.7% | 72.5%-93.0% | [57] |
| Case-control | Plasma | Hypomethylated DNA | LINE-1 transposable DNA element | 65.80% | 90.00% | [36] |
| Case-control | Serum/plasma | Methylated DNA panel | IKFZ and BCAT1 genes | 62.1%-95.0% | 92.0%-95.0% | [36,57] |
| Case-control | Serum | Methylated DNA panel | SEPT9 and SDC2 genes | 86.50% | 92.10% | [64] |
| Case-control | Serum/plasma | Methylated DNA panel | APC, MGMT, RASSF2A and WIF-1 genes | 86.50% | 92.10% | [57] |
| Case-control | Plasma | Methylated DNA panel | ALX4, BMP3, NPTX2, RARB, SDC2, SEPT9 and VIM genes | 90.70% | 72.50% | [63] |
| Case-control | Serum | ALU115 DNA content | Free ALU115 DNA content | 69.20% | 99.10% | [36] |
| Case-control | Serum | DNA integrity | ALU247/115 DNA integrity index | 73.10% | 97.30% | [36] |
| Case-control | Serum | Free DNA content | ALU-based cell-free DNA | 64.50% | 98.90% | [36] |
| Case-control | Whole blood | mRNA expression | TSPAN8 gene | 83.60% | 58.20% | [36] |
| Case-control | Whole blood | mRNA expression | LGALS gene | 82.10% | 61.20% | [36] |
| Case-control | Whole blood | mRNA expression | COL1A2 gene | 73.10% | 59.70% | [36] |
| Case-control | Whole blood | mRNA expression | CEACAM6 gene | 65.70% | 61.20% | [36] |
| Case-control | Whole blood or serum | mRNA expression | SALL4 gene | 85.9%-96.1% | 85.7%-95.0% | [65,66] |
| Case-control | Whole blood | mRNA expression panel | TSPAN8 and LGALS4 genes | 92.50% | 67.20% | [36] |
| Case-control (CRC and high-risk adenomas in the case group) | Whole blood | mRNA expression panel | LGALS4, CEACAM6, TSPAN8 and Col1A2 genes | 75.00% | 87.00% | [67] |
| Case-control | Whole blood | mRNA expression panel | CEA, EpCAM, CK19, MUC1, EGFR and C-Met genes | 87.00% | 85.00% | [68] |
| Case-control | Whole blood | Long non-coding RNA expression | NEAT1 variant 1 | 69.00% | 79% | [36] |
| Case-control | Whole blood | Long non-coding RNA expression | NEAT1 variant 2 | 70.00% | 96.00% | [36] |
| Case-control | Serum | Long non-coding RNA expression | BLACAT1 | 83.30% | 76.70% | [69] |
| Case-control | Plasma | Long non-coding RNA expression panel | ATB and CCAT1 | 82.00% | 75.00% | [70] |
| Case-control | Plasma | Long non-coding RNA expression panel | 91H, PVT-1 and MEG3 | 82.80% | 78.60% | [71] |
| Case-control | Serum | Long non-coding RNA expression panel | LOC285194, RP11-462C24.1 and Nbla12061 | 68.30% | 86.90% | [72] |
Gene mutations, especially those of K-Ras and APC genes, were the first CRC-associated genetic markers assessed with the purpose of developing new non-invasive modalities for CRC early detection and screening. Regrettably, it soon became clear that using gene mutations alone does not achieve satisfactory levels of diagnostic sensitivity. One demonstrative study evaluating this approach in a representative colonoscopy screening group concluded that the sensitivity of a panel comprising 21 DNA alterations (point mutations in K-ras, APC and p53 genes, microsatellite instability marker BAT-26 deletions and long DNA assay) was only slightly above 50%[46].
The relatively disappointing diagnostic performance of mutation-based assays stimulated the search for CRC-related epigenetic changes, in particular aberrant hypermethylation of CpG islands usually located in gene promoter regions[140]. Gene-specific DNA methylation in stool was extensively investigated (Table 2), and several genes, including BMP3, NDGR4, septin 9 (SEPT9), SFRP2, SPG20, TFPI2 and vimentin (VIM) were shown to have diagnostic sensitivities between 50% and 92% at specificities between 80% and 100% for CRC detection (see recent reviews by Liu et al[49], Lam et al[50] and Rasmussen et al[51]). However, the reproducibility of these results was often problematic, and attempts to combine multiple methylated genes in panels were undertaken to increase assay reliability. It is remarkable that high CRC detection sensitivity and specificity values could be achieved by combining methylation testing for BMP3 and NDRG4[49] or VIM and SFRP2[53] genes, but these results need to be corroborated. The ColosureTM test detecting methylated VIM in stool was the first methylation-based commercial test for CRC[141]. This diagnostic product was marketed in the USA but has recently been replaced by a more efficient multimarker Cologuard® test considered later in this sub-section.
Table 2 demonstrates that in the context of CRC diagnostics, DNA methylation markers detectable in blood attract at least as much attention as similar markers in stool. Although investigations of different groups often produce conflicting results, it is now apparent that SEPT9 methylation detection is the best studied option amongst these blood tests[57]. This test has recently been commercialised and regulated for clinical application as Epi proColon® 2.0 CE[142], but its use appears to be limited to opportunistic CRC screening[57]. Moreover, DNA methylation analysis in biological samples is relatively laborious (especially for multimarker panels) and difficult to present in POC format. These factors limit diagnostic potential of this approach. In addition, Table 2 shows that samples of stool, blood, bowel lavage fluid and colorectal mucus were also tested for total and ALU-based DNA quantification, DNA integrity assessment, examination of gene expression and long non-coding RNA expression. However, none of these assays could provide sufficiently high values for diagnostic sensitivity and specificity.
It is now becoming clear that tests involving DNA markers tend to perform better only when markers of different types are combined. Long-term research projects led by a United States company, Exact Sciences, allowed the design of a multitarget stool test that demonstrated high levels of sensitivity and specificity for CRC detection. An early version of this test that included K-ras mutation, methylation of VIM, BMP3, NDRG4 and TFPI2 genes, DNA measurement by β-actin assessment and the HemoQuant test for haemoglobin achieved diagnostic sensitivity between 78% and 85% at specificity between 85% and 90% in a case-control study[47]. It is remarkable that this test performed significantly better when directly compared with the test for methylated SEPT9 in plasma (similar to Epi proColon)[143]. The multitarget test was then simplified, and its final version includes only determination of K-ras mutation, BMP3 and NDRG4 promoter methylation, DNA measurement by β-actin assessment and FIT. Screening application of this test in a large study produced CRC detection sensitivity of 92.3% at a specificity of 86.6%[48], which makes this assay the best among all available tests involving DNA markers. The test was approved by the United States Food and Drug Administration in 2014 and is now marketed as Cologuard®. However, this test, which can be regarded as an enhanced version of FIT, requires stool collection, remains technically complex, with a multistep analytical procedure required[144], and is very expensive at over $600.
MicroRNAs (a sub-class of small non-coding RNA molecules) were discovered and characterised during the last decade of the XX century. Since that time, it was established that microRNAs are important regulators of gene expression intimately involved in the pathogenesis of many diseases including cancer[145]. As many of them are associated with the presence of colorectal tumours, it was suggested that microRNA determination in stool or blood samples may provide a new diagnostic modality for CRC early detection and screening[73]. MicroRNA variants investigated as potential CRC markers are listed in Table 3. Several published studies that used stool sample analysis highlight miR-21 as the best-studied marker of this type, but do not show outstanding sensitivity and specificity values[73]. MiR-451 and miR-223 detectable in stool produced high sensitivity and specificity values in a small study[75]; however, these markers looked less impressive in other studies, when combined with other microRNAs[73,76]. It is impossible to exclude that these discrepancies may be associated with either technical problems or different ethnic composition of the studied patient groups since clinical studies providing material for microRNA analyses were performed mostly in East Asia.
| Study setting | Sample type | Marker type | Biomarker(s) and detection methods | Sensitivity (or its range) | Specificity (or its range) | Ref. |
| Case-control | Stool | MicroRNA | miR-18a, upregulated | 61.00% | 69.00% | [73] |
| Case-control | Stool | MicroRNA | miR-20a, upregulated | 55.00% | 82.00% | [73] |
| Case-control | Stool | MicroRNA | miR-21, upregulated | 56.0%-86.0% | 73.0%-81.1% | [73,74] |
| Case-control | Stool | MicroRNA | miR-92a, upregulated | 72.00% | 73.00% | [73] |
| Case-control | Stool | MicroRNA | miR-106a, upregulated | 34.00% | 97.00% | [73] |
| Case-control | Stool | MicroRNA | miR-135b, upregulated | 78.00% | 68.00% | [73] |
| Case-control | Stool | MicroRNA | miR-144*, upregulated | 74.00% | 87.00% | [73] |
| Case-control | Stool | MicroRNA | miR-221, upregulated | 62.00% | 74.00% | [73] |
| Case-control | Stool | MicroRNA | miR-223, upregulated | 77.00% | 96.00% | [75] |
| Case-control | Stool | MicroRNA | miR-451, upregulated | 88.00% | 100.00% | [75] |
| Case-control | Stool | MicroRNA panel | miR-223 and mir-92a, both upregulated | 97.00% | 75.00% | [73] |
| Case-control | Stool | MicroRNA panel | miR-17-93 cluster and miR-135b, all upregulated | 74.00% | 79.00% | [73] |
| Case-control | Stool | MicroRNA panel | miR-144-5p, miR-451a and miR-20b-5p, all upregulated | 66.00% | 95.00% | [76] |
| Case-control | Plasma | MicroRNA | miR-17-3p, upregulated | 64.00% | 70.00% | [73,77] |
| Case-control | Plasma | MicroRNA | miR-18a, upregulated | 73.10% | 79.10% | [77] |
| Case-control | Plasma | MicroRNA | miR-20a, upregulated | 46.00% | 73.40% | [73,77] |
| Case-control | Serum/plasma | MicroRNA | miR-21, upregulated | 65.0%-91.4% | 74.4%-95.0% | [73,77-79] |
| Case-control | Plasma | MicroRNA | miR-24, downregulated | 78.40% | 83.80% | [77] |
| Case-control | Plasma | MicroRNA | miR-29a, upregulated | 69.00% | 89.10% | [77] |
| Case-control | Serum/plasma | MicroRNA | miR-29b, downregulated | 61.4%-77.0% | 72.5%-75.0% | [77] |
| Case-control | Plasma | MicroRNA | miR-92, upregulated | 89.00% | 70.00% | [77] |
| Case-control | Serum/plasma | MicroRNA | miR-92a, upregulated | 65.5%-84.0% | 71.2%-82.5% | [73,77] |
| Case-control | Plasma | MicroRNA | miR-96, upregulated | 65.40% | 73.30% | [73,77] |
| Case-control | Plasma | MicroRNA | miR-106a, upregulated | 74.00% | 44.40% | [77] |
| Case-control | Serum | MicroRNA | miR-139-3p, downregulated | 96.60% | 97.80% | [80] |
| Case-control | Serum | MicroRNA | miR-139a-5p, upregulated | 76.70% | 88.00% | [81] |
| Case-control | Plasma | MicroRNA | miR-155, upregulated | 58.20% | 95.00% | [73] |
| Case-control | Plasma | MicroRNA | miR-182, upregulated | 78.00% | 91.00% | [82] |
| Case-control | Serum | MicroRNA | miR-194, downregulated | 72.00% | 80.00% | [77] |
| Case-control | Serum | MicroRNA | miR-196b, upregulated | 63.00% | 87.40% | [84] |
| Case-control | Plasma | MicroRNA | miR-200c, upregulated | 64.10% | 73.30% | [77] |
| Case-control | Serum | MicroRNA | miR-210, upregulated | 74.6%-88.6% | 73.5%-90.1% | [77,79] |
| Case-control | Plasma | MicroRNA | miR-221, upregulated | 86.00% | 41.00% | [73,77] |
| Case-control | Plasma | MicroRNA | miR-320a, downregulated | 92.80% | 73.10% | [77] |
| Case-control | Serum | MicroRNA | miR-338-5p, upregulated | 76.30% | 92.50% | [84] |
| Case-control | Serum | MicroRNA | miR-372, upregulated | 81.90% | 73.30% | [77] |
| Case-control | Serum | MicroRNA | miR-375, downregulated | 76.90% | 64.60% | [77] |
| Case-control | Plasma | MicroRNA | miR-423-5p, downregulated | 91.90% | 70.80% | [77] |
| Case-control | Plasma | MicroRNA | miR-506, upregulated | 76.80% | 60.70% | [85] |
| Case-control | Plasma | MicroRNA | miR-601, downregulated | 69.20% | 72.40% | [77] |
| Case-control | Plasma | MicroRNA | miR-760, downregulated | 80.00% | 72.40% | [77] |
| Case-control | Serum | MicroRNA | miR-1290, upregulated | 70.10% | 91.20% | [86] |
| Case-control | Plasma | MicroRNA | miR-4316, upregulated | 76.80% | 75.00% | [85] |
| Case-control | Plasma | MicroRNA panel | miR-19a and miR-19b, both upregulated | 78.60% | 77.40% | [77] |
| Case-control | Serum | MicroRNA panel | miR-21 and miR-92a, both upregulated | 68.00% | 91.20% | [73,77] |
| Case-control | Plasma | MicroRNA panel | miR-29a and miR-92a, both upregulated | 83.00% | 84.70% | [73,77] |
| Case-control | Plasma | MicroRNA panel | miR-200c and miR-18a, both upregulated | 84.60% | 75.60% | [36,77] |
| Case-control | Plasma | MicroRNA panel | miR-223 and miR-92a, both upregulated | 76.00% | 71.00% | [73] |
| Case-control | Plasma | MicroRNA panel | miR-320d, downregulated; miR-1290, upregulated | 81.20% | 90.70% | [87] |
| Case-control | Plasma | MicroRNA panel | miR-431 and miR-139-p3, both upregulated | 91.00% | 57.00% | [77] |
| Case-control | Plasma | MicroRNA panel | miR-601 and miR-760, both downregulated | 83.30% | 69.10% | [73,77] |
| Case-control | Plasma | MicroRNA panel | miR-19a, miR-19b and miR-15b, all upregulated | 78.60% | 79.20% | [77] |
| Case-control | Plasma | MicroRNA panel | miR-24, miR-320a and miR-423-5p, all downregulated | 92.80% | 70.80% | [36,77] |
| Case-control | Plasma | MicroRNA panel | miR-144-3p, miR-425-5p and miR-1260b, all downregulated | 93.80% | 91.30% | [88] |
| Case-control | Serum | MicroRNA panel | miR-145, downregulated; miR-106a and miR-17-3p, upregulated | 78.50% | 82.80% | [73,77] |
| Case-control | Plasma | MicroRNA panel | miR-409-3p, upregulated; miR-7 and miR-93, downregulated | 82.00% | 89.00% | [73,77] |
| Case-control | Plasma | MicroRNA panel | miR-18a, miR-21, miR-22 and miR-25, all upregulated | 67.00% | 90.00% | [89] |
| Case-control | Serum | MicroRNA panel | miR-23a-3p, miR-27a-3p, miR-142-5p and miR-376c-3p, all upregulated | 89.00% | 81% | [36] |
| Case-control | Plasma | MicroRNA panel | miR-29a, miR-92a, upregulated; miR-601, miR-760, downregulated | 83.30% | 93.10% | [77] |
| Case-control | Serum | MicroRNA panel | miR-21, miR-29, miR-92, miR-125, miR-223, all upregulated | 84.70% | 98.70% | [78] |
| Case-control | Plasma | MicroRNA panel | miR-19a, miR-19b, miR-15b, miR-29a, miR-335, miR-18a, all upregulated | 91.00% | 90.00% | [90] |
| Case-control | Plasma | MicroRNA panel | miR-21, let-7g, upregulated, mir-31, mir-92a, miR-181b, miR-203, downregulated | 96.00% | 81.00% | [73] |
| Case-control | Plasma | MicroRNA panel | miR-103a-3p, miR-127-3p, miR-151a-5p, miR-17-5p, miR-181a-3p, miR-18a-5p, miR-18b-5p, all upregulated | 76.90% | 86.70% | [91] |
| Case-control | Plasma | Exosomal MicroRNA panel | miR-27a, miR-130a, both upregulated | 82.50% | 75.00% | [92] |
| Case-control | Saliva | MicroRNA | miR-21, upregulated | 97.00% | 91.00% | [93] |
Table 3 also indicates that microRNA markers of CRC were intensely investigated in blood. Hitherto most of these studies produced modest or inconsistent results. Again, miR-21 was assessed by many groups, and conflicting results were published. Although very high diagnostic sensitivity (96.6%) and specificity (97.8%) values were reported by Ng et al[80] for miR-139-3p, which was shown to be downregulated in the serum of CRC patients, this finding remains to be confirmed. Combinations of microRNA markers detectable in plasma or serum were also tested as diagnostic panels. Among these panels (Table 3), combinations of downregulated miR-144-3p, miR-425-5p and miR-1260b[88] and upregulated miR-19a, miR-19b, miR-15b, miR-29a, miR-335 and miR-18a[90] demonstrated sensitivity and specificity levels exceeding 90%.
In addition, it should be noted that a recent small study has revealed that quantification of miR-21 in saliva samples resulted in CRC detection with 97% sensitivity and 91% specificity[93]. However, these highly intriguing results remain to be corroborated.
Although microRNAs constitute a group of promising CRC biomarkers, further research in this relatively new area is needed to establish clinically valid diagnostic techniques using these markers. The relative technical complexity of laboratory procedures used in microRNA analysis (RNA extraction, reverse transcription and qPCR analysis) and the necessity of careful assay optimisation and standardisation[146] should also be taken into account when the diagnostic potential of this interesting approach is considered.
Metabolomics is a new discipline that focuses on evaluating a wide variety of endogenous metabolites produced by the organism[17,18,28]. These metabolites can serve as late stage biomarkers of either normal physiological or pathophysiological events, and cancer metabolome is defined as the entire suite of low molecular weight (< 1500 Da) cancer-specific metabolites[17]. Interestingly, some of these metabolites are VOC-s that are present in the gas phase of various excreted biological materials and can potentially be used for detecting malignancies including CRC[99]. The outcomes of metabolomic studies on CRC detection are summarised in Table 4. Remarkably, very impressive results (with CRC detection sensitivity reaching 97% at 99% specificity) were achieved by Sonoda et al[97], when dog scent judgment was applied to faeces and exhaled breath samples for discriminating between CRC patients and controls. Unfortunately, it is not realistic to expect that this natural phenomenon could constitute a reliable diagnostic tool. Hence, advanced Electronic Nose technologies are being developed and tested for CRC detection (Table 4) alongside widely used combinations of gas chromatography (GC) and mass spectrometry (MS)[18,94,99]. The latter approach, albeit regarded as the technical gold standard, is complex, costly and unsuitable for population screening. This point is especially important because most of the numerous studies applying metabolomic approaches to detecting CRC-related metabolites (non-VOC-s) in biological substances use various versions of MS (Table 4). Although some of the studies listed in Table 4 produced sensitivity and specificity values above 90% for CRC detection[102,109,113,116,125], cost and complexity issues remain major obstacles to the introduction of these assays into routine clinical practice. In this context, the use of electronic noses sensing CRC-associated VOC-s appears to be more promising, especially in view of CRC detection sensitivity and specificity both reaching 95% in a recent study by Zonta et al[98].
| Study setting | Sample type | Marker type | Biomarker(s) and detection methods | Sensitivity (or its range) | Specificity (or its range) | Ref. |
| Case-control | Stool | VOCs | Hydrogen sulphide, Dimethylsulphide, Dimethyldisulphide, mlz 90 - detected by selected ion flow tube (SIFT) mass spectrometry (MS) | 72.00% | 78.00% | [94] |
| Case-control | Stool | VOCs | Propan-2-ol, 3-methylbutanoic acid - detected by gas chromatography (GC) and MS | 87.90% | 84.60% | [95] |
| Case-control | Stool | VOCs | Methyl mercaptan (increased) and hydrogen (decreased) – detected by GC | 90.00% | 57.70% | [96] |
| Case-control | Stool | VOCs | Pattern recognition technique - canine scent judgment | 97.00% | 99.00% | [97] |
| Case-control | Stool | VOCs | Pattern recognition technique (eNose Cyranose® 320) | 85.00% | 87.00% | [94] |
| Case-control | Stool | VOCs | Pattern recognition technique (SCENT A1) | 95.00% | 95.00% | [98] |
| Case-control | Urine | VOCs | Ion mobility spectroscopy technology (FAIMS) | 88.00% | 60.00% | [99] |
| Case-control | Urine | VOCs | Ion mobility spectroscopy technology (FAIMS) | 63.00% | 63.00% | [100] |
| Case-control | Urine | VOCs | Pattern recognition technique (eNose applied) | 78.00% | 79.00% | [99] |
| Case-control | Breath | VOCs | Pattern recognition technique - canine scent judgment | 91.00% | 99.00% | [97] |
| Case-control | Breath | VOCs | Acetone (increased), ethyl acetate (increased), ethanol (decreased) and 4-methyl octane (decreased) detected by GC-MS | 85.00% | 94.00% | [99] |
| Case-control | Breath | VOCs | Nonanal, decanal, 4-methyl-pentanone, 2-methylbutane, 4-methyloctane, 4-methylundecane, 2-methylpentane, methylcyclopentane, cycloxehane, methylcyclohexane, trimethyldecane-1,2-pentadiene, 1,3-dimethylbenzene, 1,4-dimethylbenzene – detected by GC-MS | 86.00% | 83.00% | [99] |
| Case-control | Stool | Magnetic resonance spectra | Magnetic resonance spectra patterns | 85.20% | 86.90% | [101] |
| Case-control | Stool | Small metabolites | Acetate – detected by proton magnetic resonance spectroscopy (PMRS) | 94.70% | 92.30% | [102] |
| Case-control | Stool | Small metabolites | Succinate – detected by PMRS | 91.20% | 93.50% | [102] |
| Case-control | Serum | Aromatic carboxylic acids | Benzoic acid – detected by CE-time of flight (TOF) MS | 89.00% | 82.00% | [103] |
| Case-control | Serum | Fatty acids | GTA-446 – detected by flow injection analysis MS | 83.30% | 84.80% | [104] |
| Case-control | Plasma | Amino acid metabolites | L-kynurenine – detected by high-performance liquid chromatography (HPLC) | 85.20% | 100.00% | [105] |
| Case-control | Plasma | Fatty acids | Decanoic acid – detected by CE-TOFMS | 87.80% | 80.00% | [106] |
| Case-control | Serum | Multiple metabolites | 38 metabolites detected by GC-MS | 85.00% | 86.00% | [107] |
| Case-control | Serum | Phospholipids (sphingomyelins and phosphatidylcho-lines) | SM (34:1), PC (34:1), PC (34:2), PC (36:4), PC (36:2), PC (36:3) - detected by MS | ♂77.3%; ♀80.8% | ♂92.4%; ♀85.9% | [108] |
| Case-control | Serum | Unsaturated free fatty acids (panel) | C16:1, C18:3, C20:4, C22:6, all downregulated – detected by MS | 93.80% | 92.20% | [109] |
| Case-control | Serum | Amino acids (panel) | 8 amino acids – detected by LC-MS/MS | 65.00% | 95.00% | [110] |
| Case-control | Serum | Amino acids, fatty acids, carbohydrates | 13 metabolites – detected by LC-MS/MS | 96.00% | 80.00% | [111] |
| Case-control | Serum | Metabolite panel | 2-hydroxy-butyrate, aspartic acid, kynurenine, cystamine – detected by GC-MS | 83.10% | 81.00% | [112] |
| Case-control | Serum | Lipid metabolites (panel) | Palmitic amide, oleamide, hexadecaneodioic acid, octadecanoic acid, eicosatrienoic acid, LPC(18:2), LPC(20:4), LPC(22:6), myristic acid, LPC(16:0) – detected by ion cyclotron resonance MS | 98.10% | 100.00% | [113] |
| Case-control | Serum | Panel of hydroxylated polyunsaturated ultra long-chain fatty acids | C28H46O4, C28H48O4 and C28H50O4, all downregulated – detected by LC-MS/MS and nuclear MR | 75.00% | 90.00% | [114] |
| Case-control | Serum | Multiple metabolites (panel) | 11,14-eicosadienoic acid, 12a-hydroxy-3-oxocholadienic acid, 12-ketodeoxycholic acid, 12-keto-tetrahydro-leukotriene B4, 13-cis-retinoic acid, 1b-hydrocholic acid, 1-methylhistamine, 1-monopalmitin, 2,3-dihydroxybutanoic acid, 24-hydroxycalcitriol – detected by GC-TOFMS and UPLC-QTOFMS | 83.70% | 91.70% | [115] |
| Case-control | Plasma | Amino acids, fatty acids, carbohydrates | 8 metabolites – detected by CT-TQMS | 99.30% | 93.80% | [116] |
| Case-control | Plasma | Choline-containing phospholipids (panel) | Total saturated lysophosphatidyl-cholines (LPCs), 18:2 LPC and sphingosylphosphorylcholine – detected by LC-MS/MS | 88.30% | 80.00% | [117] |
| Case-control | Plasma | Choline-containing phospholipids (panel) | Total saturated lysophosphatidyl-cholines (LPCs) and the difference between 18:2 LPC and 18:1 LPC – detected by LC-MS | 82.00% | 93.00% | [118] |
| Case-control | Dried blood | Amino acids and acylcarnitines (panel) | C16, Arg, C4/C8, C5/C3, Val, Phe/Tyr, Ala, C4/C3 – detected by direct infusion MS | 81.20% | 83.90% | [119] |
| Case-control | Urine | Polyamines | N1, N12-diacetylspermine – detected by ELISA | 75.80% | 96.00% | [120] |
| Case-control | Urine | Polyamines and amino acid metabolites | N1, N12-diacetylspermine and kynurenine – detected by LC-MS | 80.00% | 80.00% | [121] |
| Case-control | Urine | Amino acids and acetoacetate (panel) | Alanine, glutamine, aspartic acid and acetoacetate – detected by PMRS | 87.50% | 91.30% | [122] |
| Case-control | Urine | Nucleosides (panel) | 5-hydroxymethyluracil and 8-oxo-7,8-dihydroguanine – detected by UPLC-MS/MS | 78.60% | 75.00% | [123] |
| Case-control | Urine | Nucleosides (panel) | Cytidine, 3-methylcitidine, 1-methyladenosine, 2-deoxyguanosine, adenosine, inosine – detected by HPLC-MS/MS | 69.00% | 98.00% | [124] |
| Case-control | Urine | Metabolite panel | Citrate, Hippurate, p-cresol, 2-aminobutyrate, myristate, putrescine and kynurenate - detected by UPLC-QTOFMS | 97.50% | 100% | [125] |
| Case-control | Urine | Nucleosides (panel) | Adenosine, N4-acetylcytidine, cytidine, guanosine, inosine, 1-methyladenosine, 1-methylguanosine, 1-methylinosine, 2-methylguanosine, 2,2-methylguanosine, N6-methyladenosine, uridine, 3-methyluridine+5-methyluridine, pseudouridine – detected by reverse phase HPLC | 76.90% | 90.40% | [126] |
| Case-control | Urine | Nucleosides (panel) | Adenosine, N4-acetylcytidine, cytidine, guanosine, inosine, 1-methyladenosine, 1-methylguanosine, 1-methylinosine, 2-methylguanosine, 2,2-methylguanosine, N6-methyladenosine, 5-methyluridine, pseudouridine, uridine – detected by column switching HPLC | 71.00% | 96.00% | [127] |
The structure of the gastrointestinal tract engenders permanent interactions between its epithelial tissue and luminal microbiota, thus significant microbial impact in colorectal carcinogenesis appears to be likelier than in any other neoplasia. Steadily accumulating evidence indicates a pivotal role for the gut microbiome in influencing the development of CRC[19]. It is now believed that bacterial effects predisposing to CRC include impacts in gut surface barrier disruption, induction of colonic inflammation, direct genotoxic action against epithelial cells and dysbiosis leading to CRC-promoting shifts in gut microflora composition and the colonic microenvironment[19,147]. These advances prompted interest in evaluating gut microbiome shifts as possible diagnostic markers for CRC[148]. The results of several recent studies (presented in Table 5) show that alterations in gut microbiome composition can potentially serve as non-invasive diagnostic markers for this disease. One remarkable common feature of all the studies listed in Table 5 is the obligatory presence of Fusobacterium nucleatum (F. nucleatum) as one of the components of all tested panels. Indeed, F. nucleatum, an anaerobic oral commensal, is now identified as a pathogenetic factor contributing to multiple disorders comprising among others inflammatory bowel disease and CRC[19,148,149]. This interesting diagnostic approach is being actively investigated; however, further studies are necessary to firmly establish the value of the gut microbiome in non-invasive CRC detection.
| Study setting | Sample type | Marker type | Biomarker(s) | Sensitivity (or its range) | Specificity (or its range) | Ref. |
| Case-control | Stool | Bacterial | Fusobacterium nucleatum | 54.0%-92.8% | 79.8%-91.0% | [128-131] |
| Case-control | Stool | Bacterial | clbA-positive bacteria | 56.4% | 81.5% | [131] |
| Case-control | Stool | Bacterial panel | Fusobacterium nucleatum, Bacteroides clarus, Roseburia intestinalis and Clostridium hathewayi | 92.8% | 79.8% | [130] |
| Case-control | Stool | Bacterial panel | clbA-positive bacteria and Fusobacterium nucleatum | 84.6% | 63.1% | [131] |
| Case-control | Stool | Bacterial panel | Ratio of Fusobacterium nucleatum to Bifidobacterium | 84.6% | 92.3% | [132] |
| Case-control | Stool | Bacterial panel | Combination of ratios of Fusobacterium nucleatum to Bifidobacterium and Fusobacterium nucleatum to Faecalibacterium prausnitzii | 90.0% | 90.2% | [132] |
| Case-control (CRC and adenomatous polyps in the case group) | Stool | Bacterial panel | Fusobacterium nucleatum, Enterococcus faecalis, Streptococcus bovis, Enterotoxigenic Bacteroides fragilis, and Porphyromonas spp | 91.4% | 93.5% | [133] |
The existing plethora of potential non-invasive approaches to CRC detection briefly reviewed in this paper looks impressive in terms of numbers, but often disappointing in terms of outcome. Most of the published results clearly fail to transform into diagnostic or screening tests that would be highly sensitive and specific, simple to perform and not associated with excessive cost. As a matter of fact, the choice of available biomarker-based tests practically used for CRC screening remains strictly limited. Today FIT is by far the most popular option[2,9,31] owing to its relative simplicity and affordability. The recently introduced and widely advertised multitarget Cologuard® stool test or Epi proColon test targeting SEPT9 methylation in plasma, albeit approved for clinical use, are technically complex and prohibitively expensive. Comparative studies addressing the health economics of CRC screening have demonstrated that the multitarget stool test, being more cost-effective that no screening, is significantly less cost-effective when compared to the FIT or invasive endoscopic testing[150-152]. Likewise, methylated SEPT9 detection in plasma samples[153] is clearly less cost-effective than the FIT. Considering a unit cost of $8 for the FIT (sampling kit and analysis only), Lansdorp-Vogelaar et al[154] concluded that a biomarker-based test that detects CRC with higher levels of sensitivity and specificity (up to 100%) should never be more expensive than $57 to be cost-effective. These estimates seem to indicate that in practical terms the FIT is currently the most cost-effective test for non-invasive CRC screening. Other authors argue that a highly specific non-invasive biomarker with an improved sensitivity for advanced adenomas (that progress to CRC) would probably be cost-effective at higher threshold costs[155], but the $600 price tag currently attached to Cologuard® is obviously excessive.
In any case, it is apparent that the FIT is not a perfect screening test. Its specificity reaching 95% is sufficiently high to be deemed satisfactory, but the sensitivity of this test remains relatively modest[31]. There is, however, an opinion that repeated FIT testing with one-year intervals may compensate for the lack of sensitivity[12]. Moreover, accurate identification of individuals with different levels of CRC risk could lead to creating objective approaches to risk stratification and personalised screening[12,155,156].
The effectiveness of a screening strategy is defined not only by screening test performance characteristics, but also by screening participant adherence[12]. One additional practical problem in CRC screening programmes employing faecal tests is insufficient screening uptake[157,158] that often results from participants’ reluctance to collect stool samples[159,160]. The use of non-invasively collected colorectal mucus samples[24,138] in FIT-like tests can help solve this problem, but this new approach remains to be thoroughly evaluated, and this will require large comparative randomised trials that usually take several years to complete[155]. The existing combination of the FIT and confirmatory colonoscopy is the strategy of choice today, and its further optimisation is currently regarded as the main factor in improving CRC screening effectiveness.
The present strong position of the FIT as the test of choice for non-invasive CRC screening will certainly be temporary as this test has one intrinsic deficiency that is impossible to eliminate. The FIT detects blood, which is shed but not produced by tumours, and bleeding may not occur in some CRC patients. For this reason, FIT sensitivity will never approach 100%, and it is likely that this target will become achievable only when a screening test employing CRC-specific biomarker(s) is developed. As no single biomarker detectable in all colorectal tumours has been identified so far, multitarget strategies combining either multiple markers of the same type or different assays (such as Cologuard®) emerge as CRC screening options advocated by some experts. However, these complex assays usually require sophisticated laboratory equipment and are laborious and expensive. Although future technological advances can help in eliminating these deficiencies, the search for more reliable and easily detectable single CRC biomarkers should continue.
It can be expected that rapid progress in cancer biomarker research accompanied by accelerated development of new non-invasive tests promises forthcoming breakthroughs in CRC screening and prevention of this disease.
Manuscript source: Invited manuscript
Specialty type: Oncology
Country of origin: United Kingdom
Peer-review report classification
Grade A (Excellent): A, A
Grade B (Very good): B
Grade C (Good): 0
Grade D (Fair): D, D
Grade E (Poor): 0
P-Reviewer: Cao ZF, Fan RY, Kadiyska T, Shenoy S, Yamada SL S-Editor: Dou Y L-Editor: Webster JR E-Editor: Liu MY
| 1. | Ferlay J, Colombet M, Soerjomataram I, Mathers C, Parkin DM, Piñeros M, Znaor A, Bray F. Estimating the global cancer incidence and mortality in 2018: GLOBOCAN sources and methods. Int J Cancer. 2019;144:1941-1953. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 3585] [Cited by in RCA: 4888] [Article Influence: 698.3] [Reference Citation Analysis (1)] |
| 2. | Brenner H, Chen C. The colorectal cancer epidemic: challenges and opportunities for primary, secondary and tertiary prevention. Br J Cancer. 2018;119:785-792. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 149] [Cited by in RCA: 193] [Article Influence: 27.6] [Reference Citation Analysis (0)] |
| 3. | Grady WM, Markowitz SD. The molecular pathogenesis of colorectal cancer and its potential application to colorectal cancer screening. Dig Dis Sci. 2015;60:762-772. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 105] [Cited by in RCA: 129] [Article Influence: 12.9] [Reference Citation Analysis (0)] |
| 4. | Carethers JM, Jung BH. Genetics and Genetic Biomarkers in Sporadic Colorectal Cancer. Gastroenterology. 2015;149:1177-1190.e3. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 254] [Cited by in RCA: 340] [Article Influence: 34.0] [Reference Citation Analysis (0)] |
| 5. | Hazewinkel Y, Dekker E. Colonoscopy: basic principles and novel techniques. Nat Rev Gastroenterol Hepatol. 2011;8:554-564. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 39] [Cited by in RCA: 45] [Article Influence: 3.2] [Reference Citation Analysis (0)] |
| 6. | Hoff G, Dominitz JA. Contrasting US and European approaches to colorectal cancer screening: which is best? Gut. 2010;59:407-414. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 51] [Cited by in RCA: 50] [Article Influence: 3.3] [Reference Citation Analysis (0)] |
| 7. | Lieberman DA, Williams JL, Holub JL, Morris CD, Logan JR, Eisen GM, Carney P. Colonoscopy utilization and outcomes 2000 to 2011. Gastrointest Endosc. 2014;80:133-143. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 86] [Cited by in RCA: 102] [Article Influence: 9.3] [Reference Citation Analysis (0)] |
| 8. | Young GP, Rabeneck L, Winawer SJ. The Global Paradigm Shift in Screening for Colorectal Cancer. Gastroenterology. 2019;156:843-851.e2. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 49] [Cited by in RCA: 72] [Article Influence: 12.0] [Reference Citation Analysis (0)] |
| 9. | Zauber AG, Winawer SJ, O'Brien MJ, Lansdorp-Vogelaar I, van Ballegooijen M, Hankey BF, Shi W, Bond JH, Schapiro M, Panish JF, Stewart ET, Waye JD. Colonoscopic polypectomy and long-term prevention of colorectal-cancer deaths. N Engl J Med. 2012;366:687-696. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1952] [Cited by in RCA: 2282] [Article Influence: 175.5] [Reference Citation Analysis (1)] |
| 10. | Lieberman D. Colon cancer screening and surveillance controversies. Curr Opin Gastroenterol. 2009;25:422-427. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 27] [Cited by in RCA: 25] [Article Influence: 1.6] [Reference Citation Analysis (0)] |
| 11. | Zhao S, Wang S, Pan P, Xia T, Chang X, Yang X, Guo L, Meng Q, Yang F, Qian W, Xu Z, Wang Y, Wang Z, Gu L, Wang R, Jia F, Yao J, Li Z, Bai Y. Magnitude, Risk Factors, and Factors Associated With Adenoma Miss Rate of Tandem Colonoscopy: A Systematic Review and Meta-analysis. Gastroenterology. 2019;156:1661-1674.e11. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 205] [Cited by in RCA: 369] [Article Influence: 61.5] [Reference Citation Analysis (0)] |
| 12. | Ladabaum U, Dominitz JA, Kahi C, Schoen RE. Strategies for Colorectal Cancer Screening. Gastroenterology. 2019;. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 198] [Cited by in RCA: 408] [Article Influence: 81.6] [Reference Citation Analysis (0)] |
| 13. | Corte CJ, Leong RW. Improving the utility of colonoscopy: Recent advances in practice. J Gastroenterol Hepatol. 2016;31:32-44. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 8] [Cited by in RCA: 12] [Article Influence: 1.3] [Reference Citation Analysis (0)] |
| 14. | Hanahan D, Weinberg RA. Hallmarks of cancer: the next generation. Cell. 2011;144:646-674. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 51728] [Cited by in RCA: 47052] [Article Influence: 3360.9] [Reference Citation Analysis (5)] |
| 15. | Okugawa Y, Grady WM, Goel A. Epigenetic Alterations in Colorectal Cancer: Emerging Biomarkers. Gastroenterology. 2015;149:1204-1225.e12. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 521] [Cited by in RCA: 565] [Article Influence: 56.5] [Reference Citation Analysis (1)] |
| 16. | Vogelstein B, Papadopoulos N, Velculescu VE, Zhou S, Diaz LA, Kinzler KW. Cancer genome landscapes. Science. 2013;339:1546-1558. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 5906] [Cited by in RCA: 5591] [Article Influence: 465.9] [Reference Citation Analysis (0)] |
| 17. | Aboud OA, Weiss RH. New opportunities from the cancer metabolome. Clin Chem. 2013;59:138-146. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 70] [Cited by in RCA: 79] [Article Influence: 6.1] [Reference Citation Analysis (0)] |
| 18. | Liu X, Locasale JW. Metabolomics: A Primer. Trends Biochem Sci. 2017;42:274-284. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 184] [Cited by in RCA: 274] [Article Influence: 34.3] [Reference Citation Analysis (0)] |
| 19. | Chen J, Pitmon E, Wang K. Microbiome, inflammation and colorectal cancer. Semin Immunol. 2017;32:43-53. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 140] [Cited by in RCA: 195] [Article Influence: 24.4] [Reference Citation Analysis (1)] |
| 20. | Johansson ME, Larsson JM, Hansson GC. The two mucus layers of colon are organized by the MUC2 mucin, whereas the outer layer is a legislator of host-microbial interactions. Proc Natl Acad Sci USA. 2011;108 Suppl 1:4659-4665. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 848] [Cited by in RCA: 1015] [Article Influence: 67.7] [Reference Citation Analysis (0)] |
| 21. | Johansson ME, Ambort D, Pelaseyed T, Schütte A, Gustafsson JK, Ermund A, Subramani DB, Holmén-Larsson JM, Thomsson KA, Bergström JH, van der Post S, Rodriguez-Piñeiro AM, Sjövall H, Bäckström M, Hansson GC. Composition and functional role of the mucus layers in the intestine. Cell Mol Life Sci. 2011;68:3635-3641. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 396] [Cited by in RCA: 384] [Article Influence: 27.4] [Reference Citation Analysis (0)] |
| 22. | Loktionov A. Cell exfoliation in the human colon: myth, reality and implications for colorectal cancer screening. Int J Cancer. 2007;120:2281-2289. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 63] [Cited by in RCA: 71] [Article Influence: 3.9] [Reference Citation Analysis (0)] |
| 23. | Loktionov A, Bandaletova T, Llewelyn AH, Dion C, Lywood HG, Lywood RC, Rockall TA, Stebbing JF, Broughton M, Caffarey S, Marks CG. Colorectal cancer detection by measuring DNA from exfoliated colonocytes obtained by direct contact with rectal mucosa. Int J Oncol. 2009;34:301-311. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1] [Cited by in RCA: 1] [Article Influence: 0.1] [Reference Citation Analysis (0)] |
| 24. | Loktionov A, Chhaya V, Bandaletova T, Poullis A. Assessment of cytology and mucin 2 in colorectal mucus collected from patients with inflammatory bowel disease: Results of a pilot trial. J Gastroenterol Hepatol. 2016;31:326-333. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 8] [Cited by in RCA: 10] [Article Influence: 1.1] [Reference Citation Analysis (0)] |
| 25. | Bandaletova T, Chhaya V, Poullis A, Loktionov A. Colorectal mucus non-invasively collected from patients with inflammatory bowel disease and its suitability for diagnostic cytology. APMIS. 2016;124:160-168. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 9] [Cited by in RCA: 11] [Article Influence: 1.1] [Reference Citation Analysis (0)] |
| 26. | Loktionov A, Chhaya V, Bandaletova T, Poullis A. Inflammatory bowel disease detection and monitoring by measuring biomarkers in non-invasively collected colorectal mucus. J Gastroenterol Hepatol. 2017;32:992-1002. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 13] [Cited by in RCA: 13] [Article Influence: 1.6] [Reference Citation Analysis (0)] |
| 27. | Yamada T, Matsuda A, Koizumi M, Shinji S, Takahashi G, Iwai T, Takeda K, Ueda K, Yokoyama Y, Hara K, Hotta M, Matsumoto S, Yoshida H. Liquid Biopsy for the Management of Patients with Colorectal Cancer. Digestion. 2019;99:39-45. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 46] [Cited by in RCA: 57] [Article Influence: 9.5] [Reference Citation Analysis (0)] |
| 28. | Erben V, Bhardwaj M, Schrotz-King P, Brenner H. Metabolomics Biomarkers for Detection of Colorectal Neoplasms: A Systematic Review. Cancers (Basel). 2018;10:246. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 38] [Cited by in RCA: 49] [Article Influence: 7.0] [Reference Citation Analysis (0)] |
| 29. | Greegor DH. Diagnosis of large-bowel cancer in the asymptomatic patient. JAMA. 1967;201:943-945. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 115] [Cited by in RCA: 96] [Article Influence: 1.7] [Reference Citation Analysis (0)] |
| 30. | Sidransky D, Tokino T, Hamilton SR, Kinzler KW, Levin B, Frost P, Vogelstein B. Identification of ras oncogene mutations in the stool of patients with curable colorectal tumors. Science. 1992;256:102-105. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 513] [Cited by in RCA: 475] [Article Influence: 14.4] [Reference Citation Analysis (0)] |
| 31. | Gies A, Bhardwaj M, Stock C, Schrotz-King P, Brenner H. Quantitative fecal immunochemical tests for colorectal cancer screening. Int J Cancer. 2018;143:234-244. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 31] [Cited by in RCA: 34] [Article Influence: 4.9] [Reference Citation Analysis (2)] |
| 32. | Uppara M, Adaba F, Askari A, Clark S, Hanna G, Athanasiou T, Faiz O. A systematic review and meta-analysis of the diagnostic accuracy of pyruvate kinase M2 isoenzymatic assay in diagnosing colorectal cancer. World J Surg Oncol. 2015;13:48. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 30] [Cited by in RCA: 37] [Article Influence: 3.7] [Reference Citation Analysis (1)] |
| 33. | Sithambaram S, Hilmi I, Goh KL. The Diagnostic Accuracy of the M2 Pyruvate Kinase Quick Stool Test--A Rapid Office Based Assay Test for the Detection of Colorectal Cancer. PLoS One. 2015;10:e0131616. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 17] [Cited by in RCA: 23] [Article Influence: 2.3] [Reference Citation Analysis (1)] |
| 34. | Annaházi A, Ábrahám S, Farkas K, Rosztóczy A, Inczefi O, Földesi I, Szűcs M, Rutka M, Theodorou V, Eutamene H, Bueno L, Lázár G, Wittmann T, Molnár T, Róka R. A pilot study on faecal MMP-9: a new noninvasive diagnostic marker of colorectal cancer. Br J Cancer. 2016;114:787-792. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 40] [Cited by in RCA: 40] [Article Influence: 4.4] [Reference Citation Analysis (0)] |
| 35. | Bosch LJW, de Wit M, Pham TV, Coupé VMH, Hiemstra AC, Piersma SR, Oudgenoeg G, Scheffer GL, Mongera S, Sive Droste JT, Oort FA, van Turenhout ST, Larbi IB, Louwagie J, van Criekinge W, van der Hulst RWM, Mulder CJJ, Carvalho B, Fijneman RJA, Jimenez CR, Meijer GA. Novel Stool-Based Protein Biomarkers for Improved Colorectal Cancer Screening: A Case-Control Study. Ann Intern Med. 2017;167:855-866. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 31] [Cited by in RCA: 45] [Article Influence: 5.6] [Reference Citation Analysis (1)] |
| 36. | Nikolaou S, Qiu S, Fiorentino F, Rasheed S, Tekkis P, Kontovounisios C. Systematic review of blood diagnostic markers in colorectal cancer. Tech Coloproctol. 2018;22:481-498. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 84] [Cited by in RCA: 96] [Article Influence: 13.7] [Reference Citation Analysis (4)] |
| 37. | Xu J, Ye Y, Zhang H, Szmitkowski M, Mäkinen MJ, Li P, Xia D, Yang J, Wu Y, Wu H. Diagnostic and Prognostic Value of Serum Interleukin-6 in Colorectal Cancer. Medicine (Baltimore). 2016;95:e2502. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 70] [Cited by in RCA: 86] [Article Influence: 9.6] [Reference Citation Analysis (0)] |
| 38. | Song YF, Xu ZB, Zhu XJ, Tao X, Liu JL, Gao FL, Wu CL, Song B, Lin Q. Serum Cyr61 as a potential biomarker for diagnosis of colorectal cancer. Clin Transl Oncol. 2017;19:519-524. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 17] [Cited by in RCA: 28] [Article Influence: 3.1] [Reference Citation Analysis (2)] |
| 39. | Bengs S, Becker E, Busenhart P, Spalinger MR, Raselli T, Kasper S, Lang S, Atrott K, Mamie C, Vavricka SR, von Boehmer L, Knuth A, Tuomisto A, Mäkinen MJ, Hruz P, Turina M, Rickenbacher A, Petrowsky H, Weber A, Frei P, Halama M, Jenkins G, Sheppard D, Croner RS, Christoph J, Britzen-Laurent N, Naschberger E, Schellerer V, Stürzl M, Fried M, Rogler G, Scharl M. β6 -integrin serves as a novel serum tumor marker for colorectal carcinoma. Int J Cancer. 2019;145:678-685. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 18] [Cited by in RCA: 31] [Article Influence: 5.2] [Reference Citation Analysis (1)] |
| 40. | Meng C, Yin X, Liu J, Tang K, Tang H, Liao J. TIMP-1 is a novel serum biomarker for the diagnosis of colorectal cancer: A meta-analysis. PLoS One. 2018;13:e0207039. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 20] [Cited by in RCA: 38] [Article Influence: 5.4] [Reference Citation Analysis (1)] |
| 41. | Xie H, Guo JH, An WM, Tian ST, Yu HP, Yang XL, Wang HM, Guo Z. Diagnostic value evaluation of trefoil factors family 3 for the early detection of colorectal cancer. World J Gastroenterol. 2017;23:2159-2167. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in CrossRef: 9] [Cited by in RCA: 10] [Article Influence: 1.3] [Reference Citation Analysis (1)] |
| 42. | Jiang Y, Zhang C, Chen K, Chen Z, Sun Z, Zhang Z, Ding D, Ren S, Zuo Y. The clinical significance of DC-SIGN and DC-SIGNR, which are novel markers expressed in human colon cancer. PLoS One. 2014;9:e114748. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 27] [Cited by in RCA: 36] [Article Influence: 3.3] [Reference Citation Analysis (1)] |
| 43. | Hou YL, Luo P, Ji GY, Chen H. Clinical significance of serum IGFBP-3 in colorectal cancer. J Clin Lab Anal. 2019;33:e22912. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 7] [Cited by in RCA: 19] [Article Influence: 3.2] [Reference Citation Analysis (0)] |
| 44. | Rho JH, Ladd JJ, Li CI, Potter JD, Zhang Y, Shelley D, Shibata D, Coppola D, Yamada H, Toyoda H, Tada T, Kumada T, Brenner DE, Hanash SM, Lampe PD. Protein and glycomic plasma markers for early detection of adenoma and colon cancer. Gut. 2018;67:473-484. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 42] [Cited by in RCA: 58] [Article Influence: 8.3] [Reference Citation Analysis (0)] |
| 45. | Wilhelmsen M, Christensen IJ, Rasmussen L, Jørgensen LN, Madsen MR, Vilandt J, Hillig T, Klaerke M, Nielsen KT, Laurberg S, Brünner N, Gawel S, Yang X, Davis G, Heijboer A, Martens F, Nielsen HJ. Detection of colorectal neoplasia: Combination of eight blood-based, cancer-associated protein biomarkers. Int J Cancer. 2017;140:1436-1446. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 29] [Cited by in RCA: 39] [Article Influence: 4.9] [Reference Citation Analysis (0)] |
| 46. | Imperiale TF, Ransohoff DF, Itzkowitz SH, Turnbull BA, Ross ME; Colorectal Cancer Study Group. Fecal DNA versus fecal occult blood for colorectal-cancer screening in an average-risk population. N Engl J Med. 2004;351:2704-2714. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 548] [Cited by in RCA: 525] [Article Influence: 25.0] [Reference Citation Analysis (3)] |
| 47. | Ahlquist DA, Zou H, Domanico M, Mahoney DW, Yab TC, Taylor WR, Butz ML, Thibodeau SN, Rabeneck L, Paszat LF, Kinzler KW, Vogelstein B, Bjerregaard NC, Laurberg S, Sørensen HT, Berger BM, Lidgard GP. Next-generation stool DNA test accurately detects colorectal cancer and large adenomas. Gastroenterology. 2012;142:248-56; quiz e25-6. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 223] [Cited by in RCA: 232] [Article Influence: 17.8] [Reference Citation Analysis (1)] |
| 48. | Imperiale TF, Ransohoff DF, Itzkowitz SH, Levin TR, Lavin P, Lidgard GP, Ahlquist DA, Berger BM. Multitarget stool DNA testing for colorectal-cancer screening. N Engl J Med. 2014;370:1287-1297. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1015] [Cited by in RCA: 1235] [Article Influence: 112.3] [Reference Citation Analysis (1)] |
| 49. | Liu R, Su X, Long Y, Zhou D, Zhang X, Ye Z, Ma J, Tang T, Wang F, He C. A systematic review and quantitative assessment of methylation biomarkers in fecal DNA and colorectal cancer and its precursor, colorectal adenoma. Mutat Res. 2019;779:45-57. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 18] [Cited by in RCA: 30] [Article Influence: 5.0] [Reference Citation Analysis (0)] |
| 50. | Lam K, Pan K, Linnekamp JF, Medema JP, Kandimalla R. DNA methylation based biomarkers in colorectal cancer: A systematic review. Biochim Biophys Acta. 2016;1866:106-120. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 52] [Cited by in RCA: 67] [Article Influence: 7.4] [Reference Citation Analysis (0)] |
| 51. | Rasmussen SL, Krarup HB, Sunesen KG, Pedersen IS, Madsen PH, Thorlacius-Ussing O. Hypermethylated DNA as a biomarker for colorectal cancer: a systematic review. Colorectal Dis. 2016;18:549-561. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 65] [Cited by in RCA: 63] [Article Influence: 7.0] [Reference Citation Analysis (2)] |
| 52. | Niu F, Wen J, Fu X, Li C, Zhao R, Wu S, Yu H, Liu X, Zhao X, Liu S, Wang X, Wang J, Zou H. Stool DNA Test of Methylated Syndecan-2 for the Early Detection of Colorectal Neoplasia. Cancer Epidemiol Biomarkers Prev. 2017;26:1411-1419. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 49] [Cited by in RCA: 75] [Article Influence: 9.4] [Reference Citation Analysis (0)] |
| 53. | Xiao Z, Li B, Wang G, Zhu W, Wang Z, Lin J, Xu A, Wang X. Validation of methylation-sensitive high-resolution melting (MS-HRM) for the detection of stool DNA methylation in colorectal neoplasms. Clin Chim Acta. 2014;431:154-163. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 20] [Cited by in RCA: 26] [Article Influence: 2.4] [Reference Citation Analysis (0)] |
| 54. | Teixeira Y, Lima JM, Souza ML, Aguiar P, Silva TD, Forones NM. HUMAN DNA QUANTIFICATION IN THE STOOLS OF PATIENTS WITH COLORECTAL CANCER. Arq Gastroenterol. 2015;52:293-298. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 8] [Cited by in RCA: 12] [Article Influence: 1.3] [Reference Citation Analysis (0)] |
| 55. | Harada T, Yamamoto E, Yamano HO, Nojima M, Maruyama R, Kumegawa K, Ashida M, Yoshikawa K, Kimura T, Harada E, Takagi R, Tanaka Y, Aoki H, Nishizono M, Nakaoka M, Tsuyada A, Niinuma T, Kai M, Shimoda K, Shinomura Y, Sugai T, Imai K, Suzuki H. Analysis of DNA methylation in bowel lavage fluid for detection of colorectal cancer. Cancer Prev Res (Phila). 2014;7:1002-1010. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 31] [Cited by in RCA: 38] [Article Influence: 3.5] [Reference Citation Analysis (1)] |
| 56. | Loktionov A, Ferrett CG, Gibson JJS, Bandaletova T, Dion C, Llewelyn AH, Lywood HGG, Lywood RCG, George BD, Mortensen NJ. A case-control study of colorectal cancer detection by quantification of DNA isolated from directly collected exfoliated colonocytes. Int J Cancer. 2010;126:1910-1919. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 11] [Cited by in RCA: 11] [Article Influence: 0.7] [Reference Citation Analysis (0)] |
| 57. | Worm Ørntoft MB. Review of Blood-Based Colorectal Cancer Screening: How Far Are Circulating Cell-Free DNA Methylation Markers From Clinical Implementation? Clin Colorectal Cancer. 2018;17:e415-e433. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 35] [Cited by in RCA: 51] [Article Influence: 7.3] [Reference Citation Analysis (1)] |
| 58. | Zhang M, He Y, Zhang X, Zhang M, Kong L. A pooled analysis of the diagnostic efficacy of plasmic methylated septin-9 as a novel biomarker for colorectal cancer. Biomed Rep. 2017;7:353-360. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 12] [Cited by in RCA: 18] [Article Influence: 2.3] [Reference Citation Analysis (0)] |
| 59. | Fu B, Yan P, Zhang S, Lu Y, Pan L, Tang W, Chen S, Chen S, Zhang A, Liu W. Cell-Free Circulating Methylated SEPT9 for Noninvasive Diagnosis and Monitoring of Colorectal Cancer. Dis Markers. 2018;2018:6437104. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 36] [Cited by in RCA: 60] [Article Influence: 8.6] [Reference Citation Analysis (0)] |
| 60. | Xie L, Jiang X, Li Q, Sun Z, Quan W, Duan Y, Li D, Chen T. Diagnostic Value of Methylated Septin9 for Colorectal Cancer Detection. Front Oncol. 2018;8:247. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 29] [Cited by in RCA: 54] [Article Influence: 7.7] [Reference Citation Analysis (0)] |
| 61. | Li H, Wang Z, Zhao G, Ma Y, Chen Y, Xue Q, Zheng M, Fei S. Performance of a MethyLight assay for methylated SFRP2 DNA detection in colorectal cancer tissue and serum. Int J Biol Markers. 2019;34:54-59. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 16] [Cited by in RCA: 24] [Article Influence: 4.0] [Reference Citation Analysis (0)] |
| 62. | Sun J, Fei F, Zhang M, Li Y, Zhang X, Zhu S, Zhang S. The role of mSEPT9 in screening, diagnosis, and recurrence monitoring of colorectal cancer. BMC Cancer. 2019;19:450. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 98] [Cited by in RCA: 97] [Article Influence: 16.2] [Reference Citation Analysis (0)] |
| 63. | Rasmussen SL, Krarup HB, Sunesen KG, Johansen MB, Stender MT, Pedersen IS, Madsen PH, Thorlacius-Ussing O. Hypermethylated DNA, a circulating biomarker for colorectal cancer detection. PLoS One. 2017;12:e0180809. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 46] [Cited by in RCA: 72] [Article Influence: 9.0] [Reference Citation Analysis (0)] |
| 64. | Chen Y, Wang Z, Zhao G, Sun C, Ma Y, Zhang L, Zheng M, Li H. Performance of a Novel Blood-Based Early Colorectal Cancer Screening Assay in Remaining Serum after the Blood Biochemical Test. Dis Markers. 2019;2019:5232780. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 9] [Cited by in RCA: 24] [Article Influence: 4.0] [Reference Citation Analysis (0)] |
| 65. | Ardalan Khales S, Abbaszadegan MR, Abdollahi A, Raeisossadati R, Tousi MF, Forghanifard MM. SALL4 as a new biomarker for early colorectal cancers. J Cancer Res Clin Oncol. 2015;141:229-235. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 60] [Cited by in RCA: 56] [Article Influence: 5.6] [Reference Citation Analysis (0)] |
| 66. | Wu HK, Liu C, Fan XX, Wang H, Zhou L. Spalt-like transcription factor 4 as a potential diagnostic and prognostic marker of colorectal cancer. Cancer Biomark. 2017;20:191-198. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 8] [Cited by in RCA: 13] [Article Influence: 1.9] [Reference Citation Analysis (0)] |
| 67. | Rodia MT, Solmi R, Pasini F, Nardi E, Mattei G, Ugolini G, Ricciardiello L, Strippoli P, Miglio R, Lauriola M. LGALS4, CEACAM6, TSPAN8, and COL1A2: Blood Markers for Colorectal Cancer-Validation in a Cohort of Subjects With Positive Fecal Immunochemical Test Result. Clin Colorectal Cancer. 2018;17:e217-e228. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 15] [Cited by in RCA: 26] [Article Influence: 3.3] [Reference Citation Analysis (0)] |
| 68. | Shou X, Li Y, Hu W, Ye T, Wang G, Xu F, Sui M, Xu Y. Six-gene Assay as a new biomarker in the blood of patients with colorectal cancer: establishment and clinical validation. Mol Oncol. 2019;13:781-791. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 9] [Cited by in RCA: 12] [Article Influence: 2.0] [Reference Citation Analysis (1)] |
| 69. | Dai M, Chen X, Mo S, Li J, Huang Z, Huang S, Xu J, He B, Zou Y, Chen J, Dai S. Meta-signature LncRNAs serve as novel biomarkers for colorectal cancer: integrated bioinformatics analysis, experimental validation and diagnostic evaluation. Sci Rep. 2017;7:46572. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 44] [Cited by in RCA: 55] [Article Influence: 6.9] [Reference Citation Analysis (0)] |
| 70. | Abedini P, Fattahi A, Agah S, Talebi A, Beygi AH, Amini SM, Mirzaei A, Akbari A. Expression analysis of circulating plasma long noncoding RNAs in colorectal cancer: The relevance of lncRNAs ATB and CCAT1 as potential clinical hallmarks. J Cell Physiol. 2019;234:22028-22033. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 29] [Cited by in RCA: 44] [Article Influence: 7.3] [Reference Citation Analysis (0)] |
| 71. | Liu H, Ye D, Chen A, Tan D, Zhang W, Jiang W, Wang M, Zhang X. A pilot study of new promising non-coding RNA diagnostic biomarkers for early-stage colorectal cancers. Clin Chem Lab Med. 2019;57:1073-1083. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 22] [Cited by in RCA: 39] [Article Influence: 7.8] [Reference Citation Analysis (0)] |
| 72. | Wang C, Yu J, Han Y, Li L, Li J, Li T, Qi P. Long non-coding RNAs LOC285194, RP11-462C24.1 and Nbla12061 in serum provide a new approach for distinguishing patients with colorectal cancer from healthy controls. Oncotarget. 2016;7:70769-70778. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 29] [Cited by in RCA: 37] [Article Influence: 5.3] [Reference Citation Analysis (0)] |
| 73. | Slaby O. Non-coding RNAs as Biomarkers for Colorectal Cancer Screening and Early Detection. Adv Exp Med Biol. 2016;937:153-170. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 28] [Cited by in RCA: 46] [Article Influence: 5.8] [Reference Citation Analysis (0)] |
| 74. | Bastaminejad S, Taherikalani M, Ghanbari R, Akbari A, Shabab N, Saidijam M. Investigation of MicroRNA-21 Expression Levels in Serum and Stool as a Potential Non-Invasive Biomarker for Diagnosis of Colorectal Cancer. Iran Biomed J. 2017;21:106-113. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 54] [Cited by in RCA: 53] [Article Influence: 6.6] [Reference Citation Analysis (0)] |
| 75. | Phua LC, Chue XP, Koh PK, Cheah PY, Chan EC, Ho HK. Global fecal microRNA profiling in the identification of biomarkers for colorectal cancer screening among Asians. Oncol Rep. 2014;32:97-104. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 49] [Cited by in RCA: 53] [Article Influence: 4.8] [Reference Citation Analysis (0)] |
| 76. | Wu CW, Cao X, Berger CK, Foote PH, Mahoney DW, Simonson JA, Anderson BW, Yab TC, Taylor WR, Boardman LA, Kisiel JB, Ahlquist DA. Novel Approach to Fecal Occult Blood Testing by Assay of Erythrocyte-Specific microRNA Markers. Dig Dis Sci. 2017;62:1985-1994. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 21] [Cited by in RCA: 30] [Article Influence: 3.8] [Reference Citation Analysis (0)] |
| 77. | Carter JV, Galbraith NJ, Yang D, Burton JF, Walker SP, Galandiuk S. Blood-based microRNAs as biomarkers for the diagnosis of colorectal cancer: a systematic review and meta-analysis. Br J Cancer. 2017;116:762-774. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 76] [Cited by in RCA: 117] [Article Influence: 14.6] [Reference Citation Analysis (0)] |
| 78. | Chen B, Xia Z, Deng YN, Yang Y, Zhang P, Zhu H, Xu N, Liang S. Emerging microRNA biomarkers for colorectal cancer diagnosis and prognosis. Open Biol. 2019;9:180212. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 110] [Cited by in RCA: 100] [Article Influence: 16.7] [Reference Citation Analysis (0)] |
| 79. | Sabry D, El-Deek SEM, Maher M, El-Baz MAH, El-Bader HM, Amer E, Hassan EA, Fathy W, El-Deek HEM. Role of miRNA-210, miRNA-21 and miRNA-126 as diagnostic biomarkers in colorectal carcinoma: impact of HIF-1α-VEGF signaling pathway. Mol Cell Biochem. 2019;454:177-189. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 61] [Cited by in RCA: 102] [Article Influence: 14.6] [Reference Citation Analysis (0)] |
| 80. | Ng L, Wan TM, Man JH, Chow AK, Iyer D, Chen G, Yau TC, Lo OS, Foo DC, Poon JT, Leung WK, Pang RW, Law WL. Identification of serum miR-139-3p as a non-invasive biomarker for colorectal cancer. Oncotarget. 2017;8:27393-27400. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 43] [Cited by in RCA: 52] [Article Influence: 7.4] [Reference Citation Analysis (0)] |
| 81. | Wang Q, Zhang H, Shen X, Ju S. Serum microRNA-135a-5p as an auxiliary diagnostic biomarker for colorectal cancer. Ann Clin Biochem. 2017;54:76-85. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 23] [Cited by in RCA: 23] [Article Influence: 2.6] [Reference Citation Analysis (0)] |
| 82. | Liu X, Xu T, Hu X, Chen X, Zeng K, Sun L, Wang S. Elevated circulating miR-182 acts as a diagnostic biomarker for early colorectal cancer. Cancer Manag Res. 2018;10:857-865. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 24] [Cited by in RCA: 47] [Article Influence: 6.7] [Reference Citation Analysis (0)] |
| 83. | Xu C, Gu L. The diagnostic effect of serum miR-196b as biomarker in colorectal cancer. Biomed Rep. 2017;6:39-45. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 9] [Cited by in RCA: 15] [Article Influence: 1.7] [Reference Citation Analysis (0)] |
| 84. | Bilegsaikhan E, Liu HN, Shen XZ, Liu TT. Circulating miR-338-5p is a potential diagnostic biomarker in colorectal cancer. J Dig Dis. 2018;19:404-410. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 17] [Cited by in RCA: 28] [Article Influence: 4.0] [Reference Citation Analysis (0)] |
| 85. | Krawczyk P, Powrózek T, Olesiński T, Dmitruk A, Dziwota J, Kowalski D, Milanowski J. Evaluation of miR-506 and miR-4316 expression in early and non-invasive diagnosis of colorectal cancer. Int J Colorectal Dis. 2017;32:1057-1060. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 31] [Cited by in RCA: 43] [Article Influence: 5.4] [Reference Citation Analysis (0)] |
| 86. | Imaoka H, Toiyama Y, Fujikawa H, Hiro J, Saigusa S, Tanaka K, Inoue Y, Mohri Y, Mori T, Kato T, Toden S, Goel A, Kusunoki M. Circulating microRNA-1290 as a novel diagnostic and prognostic biomarker in human colorectal cancer. Ann Oncol. 2016;27:1879-1886. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 85] [Cited by in RCA: 122] [Article Influence: 13.6] [Reference Citation Analysis (0)] |
| 87. | Liu X, Xu X, Pan B, He B, Chen X, Zeng K, Xu M, Pan Y, Sun H, Xu T, Hu X, Wang S. Circulating miR-1290 and miR-320d as Novel Diagnostic Biomarkers of Human Colorectal Cancer. J Cancer. 2019;10:43-50. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 30] [Cited by in RCA: 53] [Article Influence: 8.8] [Reference Citation Analysis (0)] |
| 88. | Lundström B, Lindqvist B, Söderbergh H, Wentzel T, Hallmans G. Nephroangiography in Wegener's granulumatosis. A comparison with panarteritis nodosa. Acta Radiol Diagn (Stockh). 1975;16:641-653. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 19] [Cited by in RCA: 36] [Article Influence: 0.7] [Reference Citation Analysis (0)] |
| 89. | Wikberg ML, Myte R, Palmqvist R, van Guelpen B, Ljuslinder I. Plasma miRNA can detect colorectal cancer, but how early? Cancer Med. 2018;7:1697-1705. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 23] [Cited by in RCA: 37] [Article Influence: 5.3] [Reference Citation Analysis (0)] |
| 90. | Herreros-Villanueva M, Duran-Sanchon S, Martín AC, Pérez-Palacios R, Vila-Navarro E, Marcuello M, Diaz-Centeno M, Cubiella J, Diez MS, Bujanda L, Lanas A, Jover R, Hernández V, Quintero E, José Lozano J, García-Cougil M, Martínez-Arranz I, Castells A, Gironella M, Arroyo R. Plasma MicroRNA Signature Validation for Early Detection of Colorectal Cancer. Clin Transl Gastroenterol. 2019;10:e00003. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 61] [Cited by in RCA: 64] [Article Influence: 10.7] [Reference Citation Analysis (0)] |
| 91. | Zhang H, Zhu M, Shan X, Zhou X, Wang T, Zhang J, Tao J, Cheng W, Chen G, Li J, Liu P, Wang Q, Zhu W. A panel of seven-miRNA signature in plasma as potential biomarker for colorectal cancer diagnosis. Gene. 2019;687:246-254. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 57] [Cited by in RCA: 98] [Article Influence: 14.0] [Reference Citation Analysis (0)] |
| 92. | Liu X, Pan B, Sun L, Chen X, Zeng K, Hu X, Xu T, Xu M, Wang S. Circulating Exosomal miR-27a and miR-130a Act as Novel Diagnostic and Prognostic Biomarkers of Colorectal Cancer. Cancer Epidemiol Biomarkers Prev. 2018;27:746-754. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 63] [Cited by in RCA: 104] [Article Influence: 14.9] [Reference Citation Analysis (0)] |
| 93. | Sazanov AA, Kiselyova EV, Zakharenko AA, Romanov MN, Zaraysky MI. Plasma and saliva miR-21 expression in colorectal cancer patients. J Appl Genet. 2017;58:231-237. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 49] [Cited by in RCA: 60] [Article Influence: 6.7] [Reference Citation Analysis (0)] |
| 94. | Bosch S, Berkhout DJ, Ben Larbi I, de Meij TG, de Boer NK. Fecal volatile organic compounds for early detection of colorectal cancer: where are we now? J Cancer Res Clin Oncol. 2019;145:223-234. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 33] [Cited by in RCA: 29] [Article Influence: 4.8] [Reference Citation Analysis (1)] |
| 95. | Bond A, Greenwood R, Lewis S, Corfe B, Sarkar S, O'Toole P, Rooney P, Burkitt M, Hold G, Probert C. Volatile organic compounds emitted from faeces as a biomarker for colorectal cancer. Aliment Pharmacol Ther. 2019;49:1005-1012. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 40] [Cited by in RCA: 57] [Article Influence: 9.5] [Reference Citation Analysis (0)] |
| 96. | Ishibe A, Ota M, Takeshita A, Tsuboi H, Kizuka S, Oka H, Suwa Y, Suzuki S, Nakagawa K, Suwa H, Momiyama M, Watanabe J, Taguri M, Kunisaki C, Endo I. Detection of gas components as a novel diagnostic method for colorectal cancer. Ann Gastroenterol Surg. 2018;2:147-153. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 17] [Cited by in RCA: 20] [Article Influence: 2.9] [Reference Citation Analysis (0)] |
| 97. | Sonoda H, Kohnoe S, Yamazato T, Satoh Y, Morizono G, Shikata K, Morita M, Watanabe A, Morita M, Kakeji Y, Inoue F, Maehara Y. Colorectal cancer screening with odour material by canine scent detection. Gut. 2011;60:814-819. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 179] [Cited by in RCA: 180] [Article Influence: 12.9] [Reference Citation Analysis (0)] |
| 98. | Zonta G, Anania G, Feo C, Gaiardo A, Gherardi S, Giberti A, Guidi V, Landini N, Palmonari C, Ricci L, de Togni A, Malagù C. Use of gas sensors and FOBT for the early detection of colorectal cancer. Sens Actuators B. 2018;262:884-891. [DOI] [Full Text] |
| 99. | Di Lena M, Porcelli F, Altomare DF. Volatile organic compounds as new biomarkers for colorectal cancer: a review. Colorectal Dis. 2016;18:654-663. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 66] [Cited by in RCA: 75] [Article Influence: 8.3] [Reference Citation Analysis (0)] |
| 100. | Widlak MM, Neal M, Daulton E, Thomas CL, Tomkins C, Singh B, Harmston C, Wicaksono A, Evans C, Smith S, Savage RS, Covington JA, Arasaradnam RP. Risk stratification of symptomatic patients suspected of colorectal cancer using faecal and urinary markers. Colorectal Dis. 2018;20:O335-O342. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 47] [Cited by in RCA: 59] [Article Influence: 8.4] [Reference Citation Analysis (0)] |
| 101. | Bezabeh T, Somorjai R, Dolenko B, Bryskina N, Levin B, Bernstein CN, Jeyarajah E, Steinhart AH, Rubin DT, Smith IC. Detecting colorectal cancer by 1H magnetic resonance spectroscopy of fecal extracts. NMR Biomed. 2009;22:593-600. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 46] [Cited by in RCA: 46] [Article Influence: 2.9] [Reference Citation Analysis (0)] |
| 102. | Lin Y, Ma C, Liu C, Wang Z, Yang J, Liu X, Shen Z, Wu R. NMR-based fecal metabolomics fingerprinting as predictors of earlier diagnosis in patients with colorectal cancer. Oncotarget. 2016;7:29454-29464. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 85] [Cited by in RCA: 97] [Article Influence: 10.8] [Reference Citation Analysis (0)] |
| 103. | Uchiyama K, Yagi N, Mizushima K, Higashimura Y, Hirai Y, Okayama T, Yoshida N, Katada K, Kamada K, Handa O, Ishikawa T, Takagi T, Konishi H, Kuriu Y, Nakanishi M, Otsuji E, Itoh Y, Naito Y. Serum metabolomics analysis for early detection of colorectal cancer. J Gastroenterol. 2017;52:677-694. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 80] [Cited by in RCA: 68] [Article Influence: 8.5] [Reference Citation Analysis (0)] |
| 104. | Hata T, Takemasa I, Takahashi H, Haraguchi N, Nishimura J, Hata T, Mizushima T, Doki Y, Mori M. Downregulation of serum metabolite GTA-446 as a novel potential marker for early detection of colorectal cancer. Br J Cancer. 2017;117:227-232. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 10] [Cited by in RCA: 12] [Article Influence: 1.5] [Reference Citation Analysis (0)] |
| 105. | Cavia-Saiz M, Muñiz Rodríguez P, Llorente Ayala B, García-González M, Coma-Del Corral MJ, García Girón C. The role of plasma IDO activity as a diagnostic marker of patients with colorectal cancer. Mol Biol Rep. 2014;41:2275-2279. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 21] [Cited by in RCA: 20] [Article Influence: 1.8] [Reference Citation Analysis (0)] |
| 106. | Crotti S, Agnoletto E, Cancemi G, Di Marco V, Traldi P, Pucciarelli S, Nitti D, Agostini M. Altered plasma levels of decanoic acid in colorectal cancer as a new diagnostic biomarker. Anal Bioanal Chem. 2016;408:6321-6328. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 28] [Cited by in RCA: 35] [Article Influence: 3.9] [Reference Citation Analysis (0)] |
| 107. | Farshidfar F, Weljie AM, Kopciuk KA, Hilsden R, McGregor SE, Buie WD, MacLean A, Vogel HJ, Bathe OF. A validated metabolomic signature for colorectal cancer: exploration of the clinical value of metabolomics. Br J Cancer. 2016;115:848-857. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 86] [Cited by in RCA: 113] [Article Influence: 12.6] [Reference Citation Analysis (0)] |
| 108. | Guo Y, Ren J, Li X, Liu X, Liu N, Wang Y, Li Z. Simultaneous Quantification of Serum Multi-Phospholipids as Potential Biomarkers for Differentiating Different Pathophysiological states of lung, stomach, intestine, and pancreas. J Cancer. 2017;8:2191-2204. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 13] [Cited by in RCA: 21] [Article Influence: 2.6] [Reference Citation Analysis (0)] |
| 109. | Zhang Y, He C, Qiu L, Wang Y, Qin X, Liu Y, Li Z. Serum Unsaturated Free Fatty Acids: A Potential Biomarker Panel for Early-Stage Detection of Colorectal Cancer. J Cancer. 2016;7:477-483. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 27] [Cited by in RCA: 33] [Article Influence: 3.7] [Reference Citation Analysis (0)] |
| 110. | Gu H, Du J, Carnevale Neto F, Carroll PA, Turner SJ, Chiorean EG, Eisenman RN, Raftery D. Metabolomics method to comprehensively analyze amino acids in different domains. Analyst. 2015;140:2726-2734. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 36] [Cited by in RCA: 36] [Article Influence: 3.6] [Reference Citation Analysis (0)] |
| 111. | Zhu J, Djukovic D, Deng L, Gu H, Himmati F, Chiorean EG, Raftery D. Colorectal cancer detection using targeted serum metabolic profiling. J Proteome Res. 2014;13:4120-4130. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 147] [Cited by in RCA: 170] [Article Influence: 15.5] [Reference Citation Analysis (0)] |
| 112. | Nishiumi S, Kobayashi T, Ikeda A, Yoshie T, Kibi M, Izumi Y, Okuno T, Hayashi N, Kawano S, Takenawa T, Azuma T, Yoshida M. A novel serum metabolomics-based diagnostic approach for colorectal cancer. PLoS One. 2012;7:e40459. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 207] [Cited by in RCA: 202] [Article Influence: 15.5] [Reference Citation Analysis (0)] |
| 113. | Li F, Qin X, Chen H, Qiu L, Guo Y, Liu H, Chen G, Song G, Wang X, Li F, Guo S, Wang B, Li Z. Lipid profiling for early diagnosis and progression of colorectal cancer using direct-infusion electrospray ionization Fourier transform ion cyclotron resonance mass spectrometry. Rapid Commun Mass Spectrom. 2013;27:24-34. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 89] [Cited by in RCA: 84] [Article Influence: 7.0] [Reference Citation Analysis (0)] |
| 114. | Ritchie SA, Ahiahonu PW, Jayasinghe D, Heath D, Liu J, Lu Y, Jin W, Kavianpour A, Yamazaki Y, Khan AM, Hossain M, Su-Myat KK, Wood PL, Krenitsky K, Takemasa I, Miyake M, Sekimoto M, Monden M, Matsubara H, Nomura F, Goodenowe DB. Reduced levels of hydroxylated, polyunsaturated ultra long-chain fatty acids in the serum of colorectal cancer patients: implications for early screening and detection. BMC Med. 2010;8:13. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 109] [Cited by in RCA: 113] [Article Influence: 7.5] [Reference Citation Analysis (0)] |
| 115. | Tan B, Qiu Y, Zou X, Chen T, Xie G, Cheng Y, Dong T, Zhao L, Feng B, Hu X, Xu LX, Zhao A, Zhang M, Cai G, Cai S, Zhou Z, Zheng M, Zhang Y, Jia W. Metabonomics identifies serum metabolite markers of colorectal cancer. J Proteome Res. 2013;12:3000-3009. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 164] [Cited by in RCA: 163] [Article Influence: 13.6] [Reference Citation Analysis (0)] |
| 116. | Nishiumi S, Kobayashi T, Kawana S, Unno Y, Sakai T, Okamoto K, Yamada Y, Sudo K, Yamaji T, Saito Y, Kanemitsu Y, Okita NT, Saito H, Tsugane S, Azuma T, Ojima N, Yoshida M. Investigations in the possibility of early detection of colorectal cancer by gas chromatography/triple-quadrupole mass spectrometry. Oncotarget. 2017;8:17115-17126. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 40] [Cited by in RCA: 58] [Article Influence: 7.3] [Reference Citation Analysis (0)] |
| 117. | Li S, Guo B, Song J, Deng X, Cong Y, Li P, Zhao K, Liu L, Xiao G, Xu F, Ye Y, Zhao Z, Yu M, Xu Y, Sang J, Zhang J. Plasma choline-containing phospholipids: potential biomarkers for colorectal cancer progression. Metabolomics. 2013;9:202-212. [DOI] [Full Text] |
| 118. | Zhao Z, Xiao Y, Elson P, Tan H, Plummer SJ, Berk M, Aung PP, Lavery IC, Achkar JP, Li L, Casey G, Xu Y. Plasma lysophosphatidylcholine levels: potential biomarkers for colorectal cancer. J Clin Oncol. 2007;25:2696-2701. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 139] [Cited by in RCA: 177] [Article Influence: 9.8] [Reference Citation Analysis (0)] |
| 119. | Jing Y, Wu X, Gao P, Fang Z, Wu J, Wang Q, Li C, Zhu Z, Cao Y. Rapid differentiating colorectal cancer and colorectal polyp using dried blood spot mass spectrometry metabolomic approach. IUBMB Life. 2017;69:347-354. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 31] [Cited by in RCA: 26] [Article Influence: 3.3] [Reference Citation Analysis (0)] |
| 120. | Hiramatsu K, Takahashi K, Yamaguchi T, Matsumoto H, Miyamoto H, Tanaka S, Tanaka C, Tamamori Y, Imajo M, Kawaguchi M, Toi M, Mori T, Kawakita M. N(1),N(12)-Diacetylspermine as a sensitive and specific novel marker for early- and late-stage colorectal and breast cancers. Clin Cancer Res. 2005;11:2986-2990. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 58] [Cited by in RCA: 61] [Article Influence: 3.1] [Reference Citation Analysis (0)] |
| 121. | Deng L, Ismond K, Liu Z, Constable J, Wang H, Alatise OI, Weiser MR, Kingham TP, Chang D. Urinary Metabolomics to Identify a Unique Biomarker Panel for Detecting Colorectal Cancer: A Multicenter Study. Cancer Epidemiol Biomarkers Prev. 2019;28:1283-1291. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 33] [Cited by in RCA: 33] [Article Influence: 5.5] [Reference Citation Analysis (0)] |
| 122. | Wang Z, Lin Y, Liang J, Huang Y, Ma C, Liu X, Yang J. NMR-based metabolomic techniques identify potential urinary biomarkers for early colorectal cancer detection. Oncotarget. 2017;8:105819-105831. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 36] [Cited by in RCA: 45] [Article Influence: 5.6] [Reference Citation Analysis (0)] |
| 123. | Rozalski R, Gackowski D, Siomek-Gorecka A, Starczak M, Modrzejewska M, Banaszkiewicz Z, Olinski R. Urinary 5-hydroxymethyluracil and 8-oxo-7,8-dihydroguanine as potential biomarkers in patients with colorectal cancer. Biomarkers. 2015;20:287-291. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 30] [Cited by in RCA: 34] [Article Influence: 3.4] [Reference Citation Analysis (0)] |
| 124. | Hsu WY, Chen CJ, Huang YC, Tsai FJ, Jeng LB, Lai CC. Urinary nucleosides as biomarkers of breast, colon, lung, and gastric cancer in Taiwanese. PLoS One. 2013;8:e81701. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 39] [Cited by in RCA: 48] [Article Influence: 4.0] [Reference Citation Analysis (0)] |
| 125. | Cheng Y, Xie G, Chen T, Qiu Y, Zou X, Zheng M, Tan B, Feng B, Dong T, He P, Zhao L, Zhao A, Xu LX, Zhang Y, Jia W. Distinct urinary metabolic profile of human colorectal cancer. J Proteome Res. 2012;11:1354-1363. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 177] [Cited by in RCA: 160] [Article Influence: 12.3] [Reference Citation Analysis (1)] |
| 126. | Feng B, Zheng MH, Zheng YF, Lu AG, Li JW, Wang ML, Ma JJ, Xu GW, Liu BY, Zhu ZG. Normal and modified urinary nucleosides represent novel biomarkers for colorectal cancer diagnosis and surgery monitoring. J Gastroenterol Hepatol. 2005;20:1913-1919. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 56] [Cited by in RCA: 60] [Article Influence: 3.0] [Reference Citation Analysis (0)] |
| 127. | Zheng YF, Yang J, Zhao XJ, Feng B, Kong HW, Chen YJ, Lv S, Zheng MH, Xu GW. Urinary nucleosides as biological markers for patients with colorectal cancer. World J Gastroenterol. 2005;11:3871-3876. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in CrossRef: 55] [Cited by in RCA: 55] [Article Influence: 2.8] [Reference Citation Analysis (0)] |
| 128. | Suehiro Y, Sakai K, Nishioka M, Hashimoto S, Takami T, Higaki S, Shindo Y, Hazama S, Oka M, Nagano H, Sakaida I, Yamasaki T. Highly sensitive stool DNA testing of Fusobacterium nucleatum as a marker for detection of colorectal tumours in a Japanese population. Ann Clin Biochem. 2017;54:86-91. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 48] [Cited by in RCA: 57] [Article Influence: 6.3] [Reference Citation Analysis (0)] |
| 129. | Wong SH, Kwong TNY, Chow TC, Luk AKC, Dai RZW, Nakatsu G, Lam TYT, Zhang L, Wu JCY, Chan FKL, Ng SSM, Wong MCS, Ng SC, Wu WKK, Yu J, Sung JJY. Quantitation of faecal Fusobacterium improves faecal immunochemical test in detecting advanced colorectal neoplasia. Gut. 2017;66:1441-1448. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 157] [Cited by in RCA: 224] [Article Influence: 28.0] [Reference Citation Analysis (0)] |
| 130. | Liang Q, Chiu J, Chen Y, Huang Y, Higashimori A, Fang J, Brim H, Ashktorab H, Ng SC, Ng SSM, Zheng S, Chan FKL, Sung JJY, Yu J. Fecal Bacteria Act as Novel Biomarkers for Noninvasive Diagnosis of Colorectal Cancer. Clin Cancer Res. 2017;23:2061-2070. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 165] [Cited by in RCA: 229] [Article Influence: 25.4] [Reference Citation Analysis (0)] |
| 131. | Eklöf V, Löfgren-Burström A, Zingmark C, Edin S, Larsson P, Karling P, Alexeyev O, Rutegård J, Wikberg ML, Palmqvist R. Cancer-associated fecal microbial markers in colorectal cancer detection. Int J Cancer. 2017;141:2528-2536. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 95] [Cited by in RCA: 139] [Article Influence: 17.4] [Reference Citation Analysis (0)] |
| 132. | Guo S, Li L, Xu B, Li M, Zeng Q, Xiao H, Xue Y, Wu Y, Wang Y, Liu W, Zhang G. A Simple and Novel Fecal Biomarker for Colorectal Cancer: Ratio of Fusobacterium Nucleatum to Probiotics Populations, Based on Their Antagonistic Effect. Clin Chem. 2018;64:1327-1337. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 67] [Cited by in RCA: 110] [Article Influence: 15.7] [Reference Citation Analysis (0)] |
| 133. | Rezasoltani S, Sharafkhah M, Asadzadeh Aghdaei H, Nazemalhosseini Mojarad E, Dabiri H, Akhavan Sepahi A, Modarressi MH, Feizabadi MM, Zali MR. Applying simple linear combination, multiple logistic and factor analysis methods for candidate fecal bacteria as novel biomarkers for early detection of adenomatous polyps and colon cancer. J Microbiol Methods. 2018;155:82-88. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 42] [Cited by in RCA: 40] [Article Influence: 5.7] [Reference Citation Analysis (0)] |
| 134. | Sievers CK, Grady WM, Halberg RB, Pickhardt PJ. New insights into the earliest stages of colorectal tumorigenesis. Expert Rev Gastroenterol Hepatol. 2017;11:723-729. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 23] [Cited by in RCA: 31] [Article Influence: 3.9] [Reference Citation Analysis (0)] |
| 135. | Pickhardt PJ, Pooler BD, Kim DH, Hassan C, Matkowskyj KA, Halberg RB. The Natural History of Colorectal Polyps: Overview of Predictive Static and Dynamic Features. Gastroenterol Clin North Am. 2018;47:515-536. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 29] [Cited by in RCA: 52] [Article Influence: 7.4] [Reference Citation Analysis (0)] |
| 136. | Levin B, Lieberman DA, McFarland B, Smith RA, Brooks D, Andrews KS, Dash C, Giardiello FM, Glick S, Levin TR, Pickhardt P, Rex DK, Thorson A, Winawer SJ; American Cancer Society Colorectal Cancer Advisory Group; US Multi-Society Task Force; American College of Radiology Colon Cancer Committee. Screening and surveillance for the early detection of colorectal cancer and adenomatous polyps, 2008: a joint guideline from the American Cancer Society, the US Multi-Society Task Force on Colorectal Cancer, and the American College of Radiology. CA Cancer J Clin. 2008;58:130-160. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1169] [Cited by in RCA: 1201] [Article Influence: 70.6] [Reference Citation Analysis (0)] |
| 137. | Wolf AMD, Fontham ETH, Church TR, Flowers CR, Guerra CE, LaMonte SJ, Etzioni R, McKenna MT, Oeffinger KC, Shih YT, Walter LC, Andrews KS, Brawley OW, Brooks D, Fedewa SA, Manassaram-Baptiste D, Siegel RL, Wender RC, Smith RA. Colorectal cancer screening for average-risk adults: 2018 guideline update from the American Cancer Society. CA Cancer J Clin. 2018;68:250-281. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 945] [Cited by in RCA: 1309] [Article Influence: 187.0] [Reference Citation Analysis (0)] |
| 138. | Loktionov A, Soubieres A, Bandaletova T, Mathur J, Poullis A. Colorectal cancer detection by biomarker quantification in noninvasively collected colorectal mucus: preliminary comparison of 24 protein biomarkers. Eur J Gastroenterol Hepatol. 2019;31:1220-1227. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 18] [Cited by in RCA: 9] [Article Influence: 1.5] [Reference Citation Analysis (0)] |
| 139. | Huddy JR, Ni MZ, Markar SR, Hanna GB. Point-of-care testing in the diagnosis of gastrointestinal cancers: current technology and future directions. World J Gastroenterol. 2015;21:4111-4120. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in CrossRef: 18] [Cited by in RCA: 18] [Article Influence: 1.8] [Reference Citation Analysis (0)] |
| 140. | Baylin SB, Jones PA. A decade of exploring the cancer epigenome - biological and translational implications. Nat Rev Cancer. 2011;11:726-734. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 2049] [Cited by in RCA: 2104] [Article Influence: 150.3] [Reference Citation Analysis (0)] |
| 141. | Ned RM, Melillo S, Marrone M. Fecal DNA testing for Colorectal Cancer Screening: the ColoSure™ test. PLoS Curr. 2011;3:RRN1220. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 58] [Cited by in RCA: 73] [Article Influence: 5.2] [Reference Citation Analysis (0)] |
| 142. | Lamb YN, Dhillon S. Epi proColon® 2.0 CE: A Blood-Based Screening Test for Colorectal Cancer. Mol Diagn Ther. 2017;21:225-232. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 111] [Cited by in RCA: 139] [Article Influence: 17.4] [Reference Citation Analysis (0)] |
| 143. | Ahlquist DA, Taylor WR, Mahoney DW, Zou H, Domanico M, Thibodeau SN, Boardman LA, Berger BM, Lidgard GP. The stool DNA test is more accurate than the plasma septin 9 test in detecting colorectal neoplasia. Clin Gastroenterol Hepatol. 2012;10:272-7.e1. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 112] [Cited by in RCA: 131] [Article Influence: 10.1] [Reference Citation Analysis (1)] |
| 144. | Lidgard GP, Domanico MJ, Bruinsma JJ, Light J, Gagrat ZD, Oldham-Haltom RL, Fourrier KD, Allawi H, Yab TC, Taylor WR, Simonson JA, Devens M, Heigh RI, Ahlquist DA, Berger BM. Clinical performance of an automated stool DNA assay for detection of colorectal neoplasia. Clin Gastroenterol Hepatol. 2013;11:1313-1318. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 99] [Cited by in RCA: 114] [Article Influence: 9.5] [Reference Citation Analysis (1)] |
| 145. | Romano G, Veneziano D, Acunzo M, Croce CM. Small non-coding RNA and cancer. Carcinogenesis. 2017;38:485-491. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 346] [Cited by in RCA: 324] [Article Influence: 40.5] [Reference Citation Analysis (0)] |
| 146. | McDonald JS, Milosevic D, Reddi HV, Grebe SK, Algeciras-Schimnich A. Analysis of circulating microRNA: preanalytical and analytical challenges. Clin Chem. 2011;57:833-840. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 492] [Cited by in RCA: 520] [Article Influence: 37.1] [Reference Citation Analysis (0)] |
| 147. | Louis P, Hold GL, Flint HJ. The gut microbiota, bacterial metabolites and colorectal cancer. Nat Rev Microbiol. 2014;12:661-672. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1596] [Cited by in RCA: 1958] [Article Influence: 178.0] [Reference Citation Analysis (0)] |
| 148. | Amitay EL, Krilaviciute A, Brenner H. Systematic review: Gut microbiota in fecal samples and detection of colorectal neoplasms. Gut Microbes. 2018;9:293-307. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 17] [Cited by in RCA: 32] [Article Influence: 4.6] [Reference Citation Analysis (0)] |
| 149. | Han YW. Fusobacterium nucleatum: a commensal-turned pathogen. Curr Opin Microbiol. 2015;23:141-147. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 378] [Cited by in RCA: 550] [Article Influence: 55.0] [Reference Citation Analysis (0)] |
| 150. | Bailey JR, Aggarwal A, Imperiale TF. Colorectal Cancer Screening: Stool DNA and Other Noninvasive Modalities. Gut Liver. 2016;10:204-211. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 37] [Cited by in RCA: 48] [Article Influence: 5.3] [Reference Citation Analysis (0)] |
| 151. | Ladabaum U, Mannalithara A. Comparative Effectiveness and Cost Effectiveness of a Multitarget Stool DNA Test to Screen for Colorectal Neoplasia. Gastroenterology. 2016;151:427-439.e6. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 89] [Cited by in RCA: 121] [Article Influence: 13.4] [Reference Citation Analysis (0)] |
| 152. | Naber SK, Knudsen AB, Zauber AG, Rutter CM, Fischer SE, Pabiniak CJ, Soto B, Kuntz KM, Lansdorp-Vogelaar I. Cost-effectiveness of a multitarget stool DNA test for colorectal cancer screening of Medicare beneficiaries. PLoS One. 2019;14:e0220234. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 41] [Cited by in RCA: 40] [Article Influence: 6.7] [Reference Citation Analysis (0)] |
| 153. | Ladabaum U, Alvarez-Osorio L, Rösch T, Brueggenjuergen B. Cost-effectiveness of colorectal cancer screening in Germany: current endoscopic and fecal testing strategies versus plasma methylated Septin 9 DNA. Endosc Int Open. 2014;2:E96-E104. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 28] [Cited by in RCA: 39] [Article Influence: 3.5] [Reference Citation Analysis (0)] |
| 154. | Lansdorp-Vogelaar I, Goede SL, Bosch LJW, Melotte V, Carvalho B, van Engeland M, Meijer GA, de Koning HJ, van Ballegooijen M. Cost-effectiveness of High-performance Biomarker Tests vs Fecal Immunochemical Test for Noninvasive Colorectal Cancer Screening. Clin Gastroenterol Hepatol. 2018;16:504-512.e11. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 29] [Cited by in RCA: 38] [Article Influence: 5.4] [Reference Citation Analysis (0)] |
| 155. | Imperiale TF, Kahi CJ. Cost-effectiveness of Future Biomarkers for Colorectal Cancer Screening: Quantified Futility or Call for Innovation? Clin Gastroenterol Hepatol. 2018;16:483-485. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 4] [Cited by in RCA: 4] [Article Influence: 0.6] [Reference Citation Analysis (0)] |
| 156. | McFerran E, O'Mahony JF, Fallis R, McVicar D, Zauber AG, Kee F. Evaluation of the Effectiveness and Cost-Effectiveness of Personalized Surveillance After Colorectal Adenomatous Polypectomy. Epidemiol Rev. 2017;39:148-160. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 22] [Cited by in RCA: 20] [Article Influence: 2.5] [Reference Citation Analysis (0)] |
| 157. | Lo SH, Halloran S, Snowball J, Seaman H, Wardle J, von Wagner C. Colorectal cancer screening uptake over three biennial invitation rounds in the English bowel cancer screening programme. Gut. 2015;64:282-291. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 93] [Cited by in RCA: 106] [Article Influence: 10.6] [Reference Citation Analysis (0)] |
| 158. | Hirst Y, Stoffel S, Baio G, McGregor L, von Wagner C. Uptake of the English Bowel (Colorectal) Cancer Screening Programme: an update 5 years after the full roll-out. Eur J Cancer. 2018;103:267-273. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 36] [Cited by in RCA: 47] [Article Influence: 6.7] [Reference Citation Analysis (0)] |
| 159. | Palmer CK, Thomas MC, von Wagner C, Raine R. Reasons for non-uptake and subsequent participation in the NHS Bowel Cancer Screening Programme: a qualitative study. Br J Cancer. 2014;110:1705-1711. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 100] [Cited by in RCA: 105] [Article Influence: 9.5] [Reference Citation Analysis (0)] |
| 160. | Reynolds LM, Bissett IP, Consedine NS. Emotional predictors of bowel screening: the avoidance-promoting role of fear, embarrassment, and disgust. BMC Cancer. 2018;18:518. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 18] [Cited by in RCA: 34] [Article Influence: 4.9] [Reference Citation Analysis (0)] |