Published online Nov 15, 2020. doi: 10.4251/wjgo.v12.i11.1311
Peer-review started: July 31, 2020
First decision: September 17, 2020
Revised: September 28, 2020
Accepted: October 19, 2020
Article in press: October 19, 2020
Published online: November 15, 2020
Processing time: 103 Days and 20.7 Hours
Colorectal cancer (CRC) is an important disease worldwide, accounting for the second highest number of cancer-related deaths and the third highest number of new cancer cases. The blood test is a simple and minimally invasive diagnostic test. However, there is currently no blood test that can accurately diagnose CRC.
To develop a comprehensive, spontaneous, minimally invasive, label-free, blood-based CRC screening technique based on Raman spectroscopy.
We used Raman spectra recorded using 184 serum samples obtained from patients undergoing colonoscopies. Patients with malignant tumor histories as well as those with cancers in organs other than the large intestine were excluded. Consequently, the specific diseases of 184 patients were CRC (12), rectal neuroendocrine tumor (2), colorectal adenoma (68), colorectal hyperplastic polyp (18), and others (84). We used the 1064-nm wavelength laser for excitation. The power of the laser was set to 200 mW.
Use of the recorded Raman spectra as training data allowed the construction of a boosted tree CRC prediction model based on machine learning. Therefore, the generalized R2 values for CRC, adenomas, hyperplastic polyps, and neuroendocrine tumors were 0.9982, 0.9630, 0.9962, and 0.9986, respectively.
For machine learning using Raman spectral data, a highly accurate CRC prediction model with a high R2 value was constructed. We are currently planning studies to demonstrate the accuracy of this model with a large amount of additional data.
Core Tip: We developed a comprehensive, spontaneous, minimally invasive, label-free, blood-based colorectal cancer (CRC) screening technique based on Raman spectroscopy. We used Raman spectra recorded using 184 serum samples obtained from patients undergoing colonoscopies. Use of the recorded Raman spectra as training data allowed the construction of a boosted tree CRC prediction model based on machine learning. The generalized R2 values for CRC was 0.9982. For machine learning using Raman spectral data, we are currently working on the construction of a more accurate CRC prediction model with a large amount of additional data.
- Citation: Ito H, Uragami N, Miyazaki T, Yang W, Issha K, Matsuo K, Kimura S, Arai Y, Tokunaga H, Okada S, Kawamura M, Yokoyama N, Kushima M, Inoue H, Fukagai T, Kamijo Y. Highly accurate colorectal cancer prediction model based on Raman spectroscopy using patient serum. World J Gastrointest Oncol 2020; 12(11): 1311-1324
- URL: https://www.wjgnet.com/1948-5204/full/v12/i11/1311.htm
- DOI: https://dx.doi.org/10.4251/wjgo.v12.i11.1311
Colorectal cancer (CRC) is an important disease worldwide. According to Globocan 2018 (http://gco.iarc.fr/today/data/factsheets/cancers/39-All-cancers-fact-sheet.pdf), among all cancers, CRC accounted for the second highest number of deaths and the third highest number of new cases[1]. The blood test is a simple minimally invasive diagnostic test. However, presently, no blood test method can accurately diagnose CRC. Tumor marker tests such as those for carcinoembryonic antigen[2], carbohydrate antigen 19-9 (CA 19-9)[3], CA72-4[4], and CA125[5] are minimally invasive tests for CRC patients and can be performed in many medical institutions. However, these conventional tumor markers have low-detection sensitivity for CRC[6]; they are mostly used for prognostic[7] and recurrence predictions[8] rather than early diagnosis. The presence of cell-free DNA[9] and microRNA (miRNA)[10] has been reported in the blood of CRC patients; however, these tests have not yet become common practice. Circulating cancer cells[11] can be detected in the blood of CRC patients, but it is unclear whether the detection of circulating cancer cells is useful for the early diagnosis of CRC[12].
Raman spectroscopy is a non-destructive method[13] used to analyze the components contained in a sample. This technique can analyze samples in various states (gases, liquids, and solids) without labeling. Research has also been conducted using Raman spectroscopy to diagnose cancer using a blood sample. Raman spectroscopy is useful for the diagnosis of colorectal[14], gastric[15], esophageal[16], pancreatic[17], lung[18], breast[19], prostate[20,21], and bladder[22] cancers. Lin et al[14] reported the results of analysis of serum obtained from 38 CRC patients and 45 volunteers by gold nanoparticle-based surface-enhanced Raman spectroscopy, which had diagnostic sensitivity and specificity of 97.4% and 100%, respectively. However, the effectiveness of Raman spectroscopy in cancer diagnosis has not yet been evaluated. We recorded the highly sensitive surface-enhanced Raman scattering spectra of human serum samples using a silver nanocomplex biochip[23,24]. There were significant differences in the scattered light intensities of Raman shifts, attributed to specific molecular bonds, between the serum samples of cancer patients with stomach or colon cancer and those with benign disease. However, the procedure was complicated, and the detection of substances was limited by the fact that specific silver nanoparticles should be used. Thus, we decided to develop another comprehensive, label-free Raman technique to detect known and unknown substances in the serum. The subsequent preliminary study showed that our Raman spectroscopy system could detect spontaneous Raman scattering spectra from untreated human serum samples within 1 min[25]. Hence, we considered that serum analysis based on Raman spectroscopy could provide a rapid cancer diagnosis.
In this study, we confirmed the correlation between the Raman scattering spectra of the serum samples collected before examination and the endoscopic diagnosis of patients who underwent colonoscopies. Additionally, we constructed a model that predicts cancer with increased accuracy based on machine learning.
This study included patients who underwent colonoscopies at the Showa University Koto Toyosu Hospital (Tokyo, Japan) between September 2018 and September 2019. Patients were excluded if they were < 20-years-old, > 80-years-old, had a history of malignancy, or had malignant diseases in organs other than the colon. The protocol in this study complied with the Declaration of Helsinki and the Clinical Trial Act in Japan. The study protocol was reviewed and approved (No. 18T5005) by the Institutional Review Board of Showa University Koto Toyosu Hospital. All participants provided written consent for their participation in this study. The study protocol was registered in the University Hospital Medical Information Network clinical trial registry (UMIN-CTR, No. UMIN000034306).
Based on previous clinical research at the Showa University Koto Toyosu Hospital, at least 150 patients were required to capture > 3 CRC patients. Therefore, 184 patients were recruited for the study (110 men and 74 women, aged between 20 and 80 years). The median and average ages were 57 and 56.9 years, respectively (Table 1).
| Parameters | n = 184 |
| Sex | |
| Male | 110 |
| Female | 74 |
| Age in yr, mean (range) | 56.9 (20-80) |
| Gastroscopy | 100 |
| Computed tomography | 51 |
| Ultrasonography | 5 |
| Magnetic resonance imaging | 5 |
In addition to colonoscopy, gastroscopy, computed tomography, ultrasonography, and magnetic resonance imaging were performed in 100, 51, 5, and 5 cases, respectively.
The primary diagnoses of the 184 patients recruited in this study were: CRC in 12 cases (3, tumor-node-metastasis [TNM] stage 0; 1, TNM stage I; 2, TNM stage II; 5, TNM stage III; 1, TNM stage IV), rectal carcinoids in 2 cases, colorectal adenomas in 68 cases, colon hyperplastic polyps in 18 cases, other diseases in 54 cases (1, leiomyoma; 9, nonspecific colitis; 2, ulcerative colitis; 24, colon diverticulum; 3, nonspecific ileitis; and 15, internal hemorrhoid), and no specific finding in 30 cases (Table 2). One CRC patient was also diagnosed with colorectal adenoma and another with ulcerative colitis. Sixteen of sixty-eight patients with colorectal adenoma were also diagnosed with colorectal hyperplastic polyps. All cancers and adenomas were histopathologically diagnosed by at least two qualified clinical pathologists. Polyps were diagnosed endoscopically as hyperplastic polyps are not usually treated.
| Main diagnosis | n = 184 |
| Cancer | 12 |
| Colon, TNM stage 0-II | 5 |
| Colon, TNM stage III or IV | 3 |
| Rectum, TNM stage III or IV | 3 |
| Colon and rectum, double, TNM stage IIA | 1 |
| Malignant potential tumor | 2 |
| Rectal neuroendocrine tumor | 2 |
| Adenoma | 68 |
| Hyperplastic polyp | 18 |
| Other diseases | 54 |
| Leiomyoma | 1 |
| Nonspecific colitis | 9 |
| Ulcerative colitis | 2 |
| Colon diverticulum | 24 |
| Nonspecific ileitis | 3 |
| Internal hemorrhoids | 15 |
| No specific findings | 30 |
Blood samples were collected prior to endoscopic examinations. Serum samples were obtained by centrifuging blood samples for 5 min (1500 × g). The extracted serum samples were dispensed into 2.0 mL hyperplastic polypropylene microtubes (Biosphere® plus; Sarstedt Ag & Co. Kg, Sarstedtstraße, Nümbrecht, Germany), which were free from DNA, DNase/RNase, polymerase chain reaction inhibitor, adenosine triphosphate, and pyrogens/endotoxins. The specimens were preserved at -80 °C in an ultralow temperature freezer (MDF-C8V1; Panasonic Corporation, Osaka, Japan).
A Nomadic Raman microscope with a computer-controlled electrical stage running Pathologic System Software Version 1.0.1.0 (BaySpec Inc., San Jose, CA, United States) was used for analysis. The details of the Raman microscope used in this study have been described in a previous paper[25], and the outline is given below. A wavelength of 1064 nm was selected as the excitation laser. The power of the laser was set to 200 mW. A 20 × magnifying objective lens with a correction collar with near-infrared microscopy (LCPLN20XIR; Olympus Corporation, Tokyo, Japan) and a 2048 × 64 pixel thermoelectric cooled indium gallium arsenide, charge-couple device (CCD) detector, with a spectral range of 100-3200 cm-1 (grating 4 cm-1) were used to record the spectra. A CCD camera with 1392 × 1040 colors and a maximum acquisition rate of 30 frames per second (Lw135R; Lumenera Corporation, Capella Court, Ottawa, ON, Canada) was used for focusing before each Raman scattering spectral acquisition (Figure 1). The dark background noise of the CCD camera was acquired in the form of a spectrum in the absence of a sample and was subtracted from all spectra acquired in which the sample was present. Baseline correction was performed for each spectrum with the Pathologic System Software (BaySpec Inc.). Moreover, some figures in this paper were created using RaspWin Ver 8.0.1 (HT SoftLab) and Adobe Illustrator CS6 Version 16.2.0 (Adobe Systems Incorporated, San Jose, CA, United States).
An overview of the measurement and analysis processes is summarized in Online resource 10. The cryopreserved serum was thawed immediately before the measurement and was set at 25 °C. We manually placed a drop of the serum stock solution on the tip of a thin stainless-steel tube. The serum was irradiated with a 1064-nm wavelength laser three times for 15 s, and the average value was recorded as the Raman scattering spectra. A new droplet was then prepared, and the same measurement was repeated three times for each serum sample.
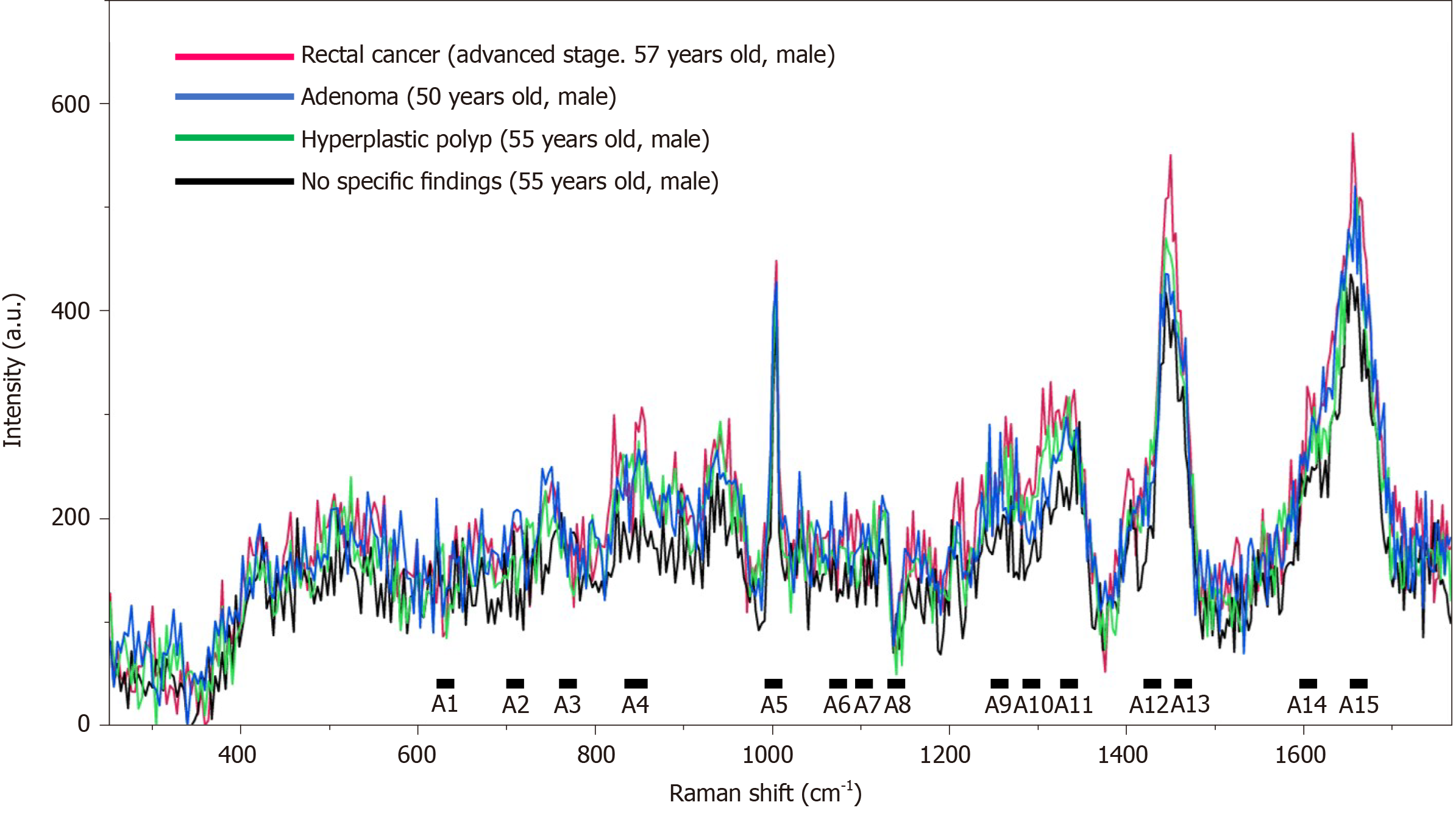
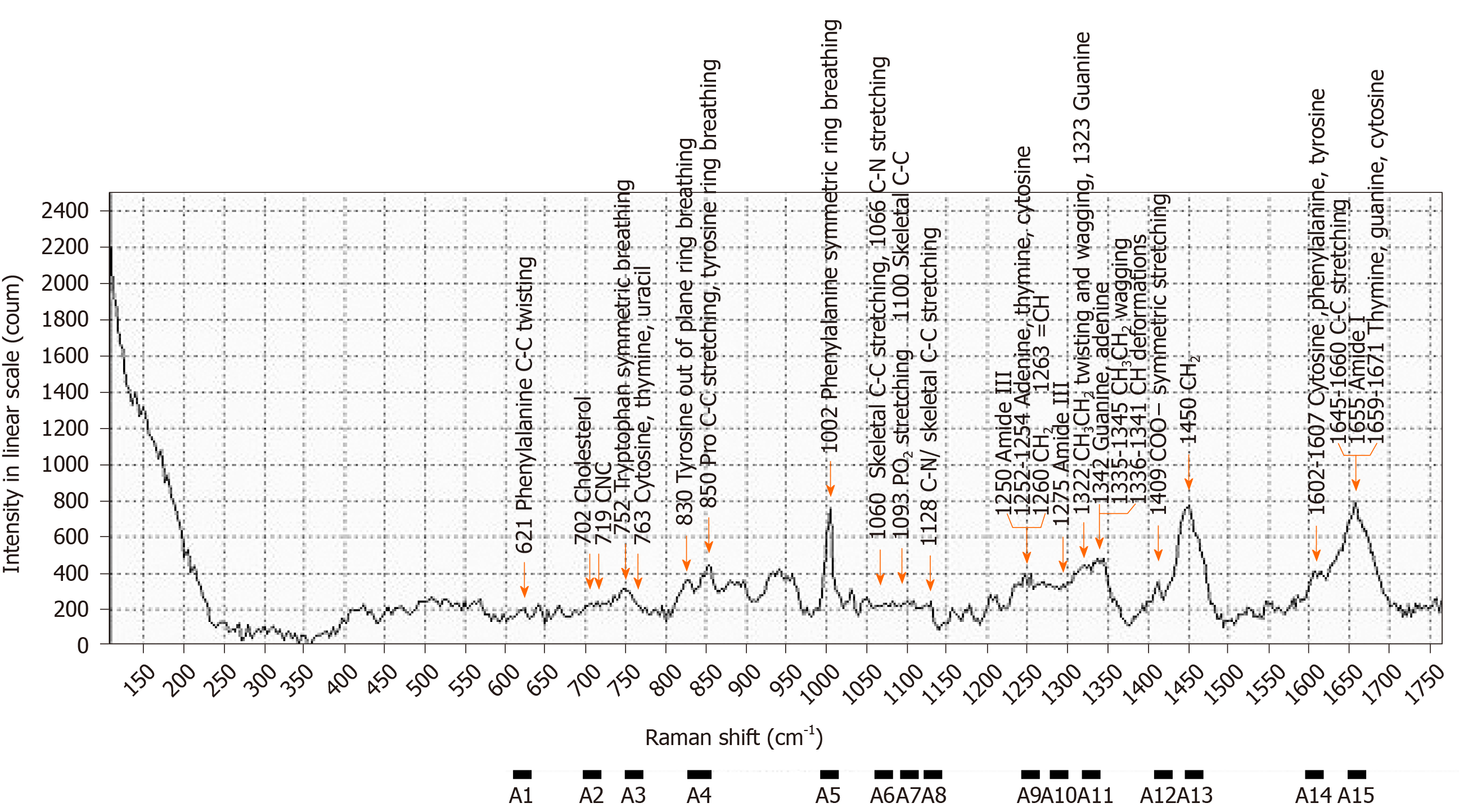
The scattered light intensity (15 range, A1-A15) of the Raman shift related to nucleic acids[26-34], proteins[28,30-32,34,35], and lipids[27,28,30,32,34,36-41] in serum was extracted from the obtained Raman spectra (Figures 2 and 3, Table 3). The average values of the three extracted scattered light intensities from each spectrum were analyzed as training data by the boosted tree model[42] with the use of JMP® Pro 14.3.0 (SAS Institute Inc., Cary, NC, United States). The average values for the three extracted scattered light intensities among the patient groups were evaluated for normality using the Tukey test. Intergroup differences in non-normally distributed data were compared using the Steel-Dwass nonparametric test. All analyses were performed using JMP® Pro 14.3.0 (SAS Institute Inc.).
| Raman shift, cm-1 | Assignments | |||
| Nucleic acids | Proteins | Lipids | ||
| A1 | 611-631 | Phenylalanine | ||
| A2 | 700-720 | Cholesterol, C-N-C | ||
| A3 | 751-771 | DNA, pyrimidines | Tryptophan | |
| A4 | 830-859 | Tyrosine, pro C-C | ||
| A5 | 993-1013 | Phenylalanine | ||
| A6 | 1060-1080 | Skeletal C-C, C-N | Skeletal C-C | |
| A7 | 1091-1111 | PO2 stretching, DNA | Skeletal C-C | |
| A8 | 1123-1143 | Skeletal C-C, C-N | Skeletal C-C | |
| A9 | 1244-1264 | Adenine, thymine, cytosine | Amide III | =CH, CH2 |
| A10 | 1275-1295 | Amide III | ||
| A11 | 1322-1343 | Guanine, adenine | CH3CH2 | CH |
| A12 | 1408-1428 | COO- | ||
| A13 | 1448-1468 | CH2 | CH2 | |
| A14 | 1596-1616 | Cytosine | Phenylalanine, tyrosine | |
| A15 | 1647-1667 | Thymine, guanine, cytosine | Amide I | C=C |
The relevant definitions were: Entropy R-squared = 1 − Loglike(model)/Loglike(0); Generalized R-squared = {1 − [L(0)/L(model)](2/n)}/[1 − L(0)(2/n)]; Mean Logp = ∑−Log[ρ(j)]/n; Root-mean-square error = √∑[y(j) − ρ(j)]²/n; Mean absolute deviation = ∑|y(j) − ρ(j)|/n; and Misclassification rate = ∑[ρ(j) ≠ ρMax]/n (Supplementary Table 1). The detailed conditions for the analysis based on the boosted tree model were: Number of layers = 200 (maximum number of layers to include in the final tree is 200); Splits per tree = 3 (number of splits for each layer is 3); Learning rate = 0.1; Overfit penalty = 0.0001; Minimum size split = 5; Row sampling rate = 1; and Column sampling rate = 1 (Supplementary Table 2).
Raman spectra of all serum samples were recorded, and the highest values of the scattered light intensity, ranging from A1 to A15, were extracted (Figures 2 and 3). The boosted tree model was used to predict CRC, and a highly accurate model was constructed based on a generalized R2 value of 0.9977 and an entropy R2 value of 0.9982 (Supplementary Table 3). Similarly, the boosted tree model was used to predict colorectal adenomas and hyperplastic polyps, and a highly accurate adenoma prediction model was constructed based on a generalized R2 value of 0.9269, and an entropy R2 value of 0.9630 (Supplementary Table 4). Furthermore, a highly accurate hyperplastic polyp prediction model was constructed based on a generalized R2 value of 0.9947 and an entropy R2 value of 0.9962 (Supplementary Table 5). The boosted tree model was used to predict rectal neuroendocrine tumors based on data from two patients, and a highly accurate rectal neuroendocrine tumor prediction model was also constructed based on a generalized R2 value of 0.9985 and an entropy R2 value of 0.9986 (Supplementary Table 6).
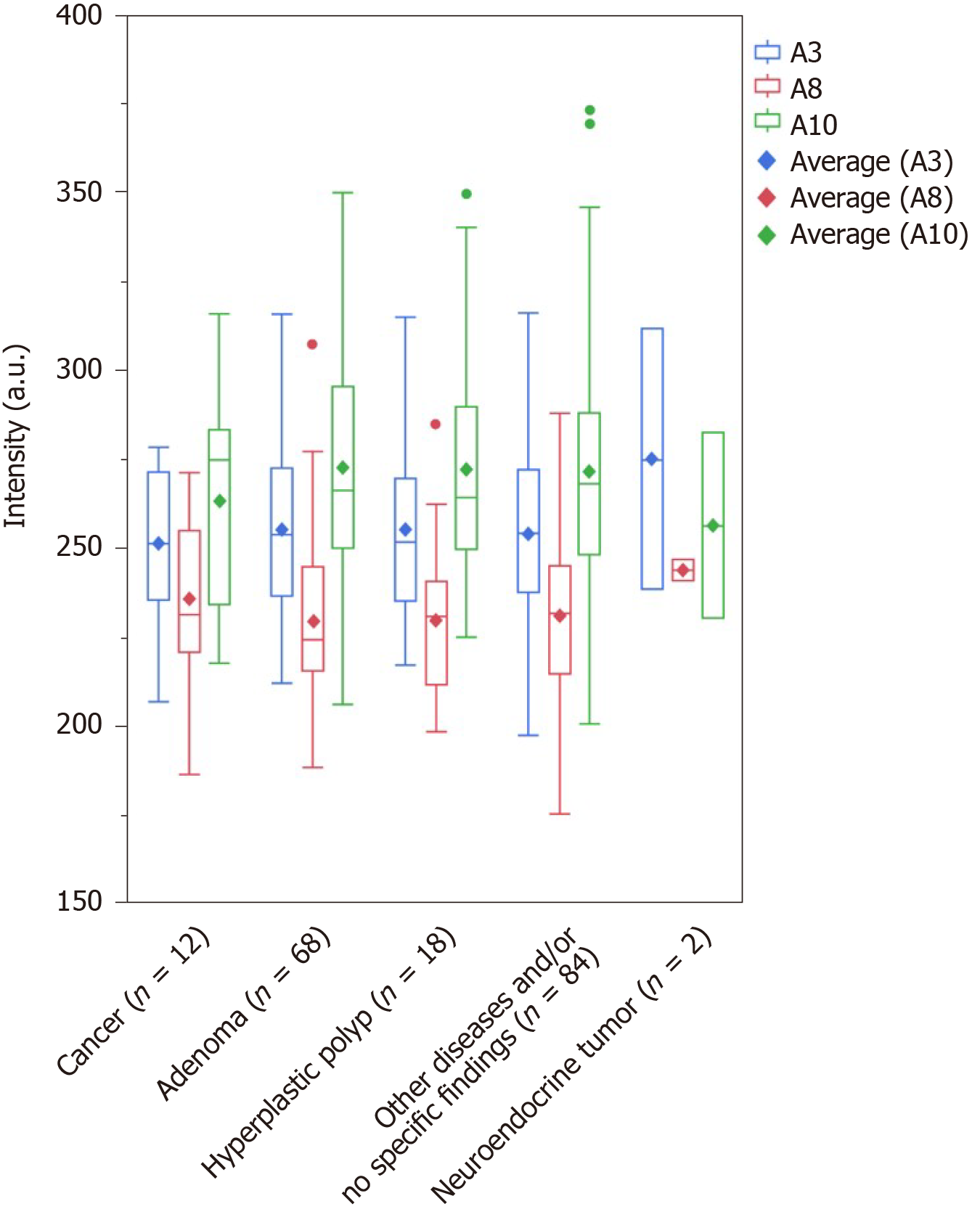
The major Raman shifts[17-19,21-37,43-45] that contributed to the prediction of the presence of CRC (effect > 0.1) were higher in the order of A10 (1275-1295 cm-1; amide III), A8 (1123-1143 cm-1; C-N and skeletal C-C), and A3 (751-771 cm-1; DNA, pyrimidines [cytosine, thymine, uracil], and tryptophan). The major Raman shifts that contributed to the prediction of the presence of colorectal adenoma (effect > 0.1) were higher in the order of A4 (830-859 cm-1; tyrosine and pro C-C), and A10 (1275-1295 cm-1; amide III). The major Raman shifts that contributed to the prediction of colorectal polyps (effect > 0.1) were higher in the order of A7 (1091-1111 cm-1; PO2 stretching and skeletal C-C), A1 (611-631 cm-1; phenylalanine), and A4 (830-859 cm-1; tyrosine and pro C-C). A10 (1275-1295 cm-1; amide III) was the Raman shift that affected the prediction of the presence of cancer and adenomas. A4 (830-859 cm-1; tyrosine and pro C-C) was the Raman shift that influenced the prediction of both adenomas and polyps. The only Raman shift that influenced the prediction of rectal neuroendocrine tumors was A11, which was different from those for the prediction of cancer, adenomas, and polyps (Table 4, Supplementary Tables 3-6). We investigated the maximum scattered light intensities of the Raman shifts of A10, A8, and A3 that contributed to the prediction of the presence of CRC. Compared with the group with adenomas, hyperplastic polyps, and no specific findings and/or other diseases, the mean scattered light intensity of A8 tended to be higher, and the mean scattered light intensities of A3 and A10 tended to be lower in the cancer samples (Figure 4). However, there was no significant difference based on the Steel-Dwass test (Supplementary Table 7).
| Cancer | Adenoma | Hyperplastic polyp | NET | ||
| Measure | Training | ||||
| Entropy R-square | 0.9977 | 0.9269 | 0.9947 | 0.9985 | |
| Generalized R-square | 0.9982 | 0.9630 | 0.9962 | 0.9986 | |
| Raman shift, cm-1 | Effect | ||||
| A1 | 611-631 | 0.0176 | 0.0285 | 0.2404 | 0.0148 |
| A2 | 700-720 | 0.0191 | 0.0636 | 0.0476 | 0.0062 |
| A3 | 751-771 | 0.1716 | 0.0479 | 0.0274 | 0.0182 |
| A4 | 830-859 | 0.0580 | 0.2008 | 0.1936 | 0.0246 |
| A5 | 993-1013 | 0.0084 | 0.0478 | 0.0054 | 0.0324 |
| A6 | 1060-1080 | 0.0052 | 0.0555 | 0.0407 | 0.0000 |
| A7 | 1091-1111 | 0.0686 | 0.0370 | 0.2886 | 0.0180 |
| A8 | 1123-1143 | 0.1873 | 0.0722 | 0.0563 | 0.0412 |
| A9 | 1244-1264 | 0.0481 | 0.0548 | 0.0129 | 0.0022 |
| A10 | 1275-1295 | 0.2291 | 0.1146 | 0.0224 | 0.0000 |
| A11 | 1322-1343 | 0.0505 | 0.0463 | 0.0149 | 0.7714 |
| A12 | 1408-1428 | 0.0663 | 0.0551 | 0.0132 | 0.0662 |
| A13 | 1448-1468 | 0.0089 | 0.0066 | 0.0041 | 0.0000 |
| A14 | 1596-1616 | 0.0145 | 0.0758 | 0.0293 | 0.0049 |
| A15 | 1647-1667 | 0.0470 | 0.0936 | 0.0033 | 0.0000 |
Numerous studies have been performed to diagnose cancer using blood tests. However, tumor markers, free DNA[9], miRNA[10], and circulating cancer cells[11] have been the main targets of blood-based CRC diagnostic techniques. To date, high-precision technologies have not been developed. Additionally, standard blood-based procedures for cancer diagnosis have not yet been established. Raman spectroscopy is an analytical method that can quickly evaluate the components of unlabeled samples that have not been pre-treated[46]. However, the measurement of Raman spectra is strongly inhibited by autofluorescence[47], and thus it is difficult to analyze a biological sample with high accuracy. Furthermore, given that the detection sensitivity of label-free, spontaneous Raman spectroscopy is lower than other labeling techniques, it is difficult to detect minute quantities of target substances. Correspondingly, it is necessary to enhance scattered light using various methods, including the use of surface-enhanced Raman spectroscopy. In fact, most Raman spectroscopic analyses of blood performed to date have utilized surface-enhanced Raman spectroscopy[19,20,48-50]. Some studies have used small sample sizes for blood-based diagnoses of CRC with Raman spectroscopy. Almost all of these studies have been based on the surface-enhanced Raman scattering technique[14,51-54]. Principal component analysis (PCA)[51], partial least squares[52,53], linear discriminant analysis (LDA)[52], and PCA-LDA[14,54] were used for evaluation, and respective diagnostic sensitivities and specificities of 86.4%-100% and 80%-100% were reported[14,51-54]. However, the surface-enhanced Raman scattering technique is highly sensitive. Furthermore, target substances are limited by the designed specifications of colloids based on precious metals, such as gold and silver. Therefore, important unknown substances that can be used for the detection of cancer may escape when surface-enhanced Raman scattering techniques are used. In fact, significant molecular bonds present in the serum and involved in cancer detection have varied[14,51-54]. Furthermore, the Raman scattered light enhancement technique does not have the advantages of Raman spectroscopy. For example, it is not label-free, more complex, and its response is slower. It would therefore be ideal if diagnosis could be established at high accuracy using ordinary, spontaneous Raman spectroscopy. Our label-free, spontaneous Raman spectroscopy system could detect known and unknown substances in a comprehensive manner. In this study, a highly accurate CRC prediction model with a generalized R2 value that exceeded 0.99 was constructed. We used a near-infrared laser as the excitation light source that was hardly affected by autofluorescence. Furthermore, a microtube was developed for serum measurements. Therefore, the Raman spectra of the serum could be obtained without a surface-enhanced Raman technique.
In this study, our system could accurately predict the presence of CRC, adenomas, and hyperplastic polyps. Even though our system could also predict the presence of rectal neuroendocrine tumors, the number of patients with rectal neuroendocrine tumors in this study was only two. Accordingly, findings should be confirmed with additional future studies. There is a possibility that our less invasive blood-based test could be used for screening CRC. In addition, this technique can comprehensively detect all known and unknown molecules contained in a sample. Therefore, by constructing an optimal algorithm based on the Raman spectrum, there is a possibility that it can be applied to the diagnosis of various diseases, including congenital, genetic, and metabolic diseases.
The Raman shifts (effect > 0.1) in our CRC prediction model were in the ranges of 1275-1295 cm-1 (amide III), 1123-1143 cm-1 (C-N and skeletal C-C), and 751-771 cm-1 [DNA, pyrimidines (cytosine, thymidine, uracil), and tryptophan] (Table 3). Additionally, the scattered light intensities of 1275-1295 cm-1 (amide III), 1123-1143 cm-1 (C-N and skeletal C-C), and 751-771 cm-1 [DNA, pyrimidines (cytosine, thymidine, uracil), and tryptophan] of the sera obtained from CRC patients were respectively low, high, and low. Compared with the results of our study, Feng et al[52] reported the exact opposite result with a surface-enhanced Raman scattering technique. Hong et al[51] also reported that the scattered light of the serum from CRC patients was relatively high intensity at 1275-1295 cm-1 (amide III). Other studies reported no difference regarding these Raman shifts[14,53,54]. These discrepancies may be attributed to differences between spontaneous Raman spectra and surface-enhanced Raman spectra or to the inaccuracies of the analyzed results owing to the small data sizes. In this study, only the disease was used as an index, and the effects of age, sex, presence or absence of co-morbidity, total protein concentration, albumin concentration, and other biological factors were not considered. We need to carefully analyze more samples after matching these factors to enhance our CRC prediction model.
The Raman shift that had a strong effect on the prediction of the presence of colorectal adenomas and hyperplastic polyps was partially consistent with the Raman shift used for the prediction of CRC. For the prediction of colorectal adenomas, effects larger than 0.01 were in the range of 830-859 cm-1 (tyrosine and pro C-C) and 1275-1295 cm-1 (amide III). Neither our or previous studies have shown a clear link between colorectal adenomas and serum tyrosine levels. The “adenoma-carcinoma sequence”[55] was estimated. Accordingly, it has been suggested that abnormalities in Wnt signaling in colorectal adenomas were associated with the development of CRC[56,57]. In this study, the amide III intensity level was a significant factor for CRC and adenomas. Furthermore, the amide III intensity level was low in cancer and high in adenomas. Thus, serum amide III levels may be used to assess the risk of CRC. For the prediction of hyperplastic colon polyps, effects were larger in the ranges of 1091-1111 cm-1 (PO2 stretching and skeletal C-C), 611-631 cm-1 (phenylalanine), and 830-859 cm-1 (tyrosine and pro C-C). The Raman shift at 830-859 cm-1 (tyrosine and pro C-C) was also the appropriate Raman shift for the prediction of colorectal adenomas. This overlap may be associated with the transition of hyperplastic colorectal polyps to colorectal adenomas and may be predictive for the development of colorectal adenomas[58]. In this study, all polyps were diagnosed endoscopically. Therefore, the possibility of adenomas cannot be ruled out.
The limitations of this study were as follows. First, the number of subjects was small; the total number of subjects was 184, of which only 12 were CRC patients. Additionally, since there were only two samples from patients with neuroendocrine tumors, we could not give definitive results for neuroendocrine tumors in this study. In the future, verification with more subjects must be performed. Second, this study did not consider the effects of biological factors other than diseases such as age, sex, presence or absence of co-morbidities, and total protein or albumin concentrations. We need to revalidate the technology in this study with prospective trials tailored to the subject’s background. Third, the patients analyzed in this study received inconsistent clinical examinations that may have been associated with various degrees of accuracies regarding clinical diagnoses. They may have also had malignancies of organs other than the colon. Fourth, it was not clear whether our technology was able to detect high-risk states of carcinogenesis or the cancer cells themselves after the onset of carcinogenesis. If the moieties detected by our technology are derived from cancer cells, this technology can be used to determine the therapeutic effects of cancer.
In summary, we could present a model for diagnosing CRC with serum and machine learning in this study. However, the clinical usefulness of this model is still undecided. We plan to continue our research in the future to improve the accuracy of our results. First, we will include larger cohorts. Second, we will increase the number of target cancer types. Third, blood will be collected twice before and after the treatment, and the Raman spectra of the serum will be compared. From these results, we believe that the significance of our label-free, spontaneous Raman spectroscopy technology in cancer treatment will become clearer. Regarding the Raman spectral analysis, a comprehensive machine learning method capable of analyzing the entire spectrum will be designed and implemented.
We studied a minimally invasive and accurate diagnostic method of CRC, which is the second most common cancer-related death worldwide. We evaluated that analysis of Raman spectrum of serum by machine learning could be an excellent diagnostic method for CRC. Since Raman spectroscopy is greatly affected by autofluorescence, it is technically difficult to analyze biological samples by Raman spectroscopy. We have succeeded in recording the Raman spectrum of untreated serum with high accuracy by using a near-infrared laser, which is less affected by autofluorescence, as the excitation light source. Then, using the recorded Raman spectra as data, we constructed a CRC prediction model by "Boosted Tree Model" which is a kind of machine learning. Although this model may predict CRC with high accuracy, we should analyze more clinical data to confirm the clinical usefulness of this model.
Colorectal cancer (CRC) is an important disease worldwide. Among all cancers, CRC accounts for the second highest number of deaths and the third highest number of new cases. The blood test is a simple minimally invasive diagnostic test. However, presently, no blood test method can accurately diagnose cancer.
We have tried to develop a comprehensive, spontaneous, minimally invasive, label-free, blood-based CRC screening technique based on Raman spectroscopy.
We used Raman spectra recorded using 184 serum samples obtained from patients (CRC in 12 patients, rectal neuroendocrine tumor in 2 patients, colorectal adenoma in 68 patients, colorectal hyperplastic polyp in 18 patients, and others in 84 patients) undergoing colonoscopies.
We used Raman spectra recorded using 184 serum samples. We used 1064-nm wavelength laser for excitation.
Use of the recorded Raman spectra as training data allowed the construction of a boosted tree CRC prediction model based on machine learning. Therefore, the generalized R2 values for CRC, adenomas, hyperplastic polyps, and neuroendocrine tumors were 0.9982, 0.9630, 0.9962, and 0.9986, respectively.
We could show a diagnostic model of machine learning using Raman spectral data, a highly accurate CRC prediction with a high R2 value.
We are currently planning studies to demonstrate the clinical usefulness of this model with a vast volume of additional data.
We are grateful to the patients who donated blood. We are also grateful to the clinical staff at Showa University, Koto Toyosu Hospital. We would like to thank Ms. Iono A and Mr. Oi H (SAS Institute Inc.) for their dedicated support in machine learning.
Manuscript source: Invited manuscript
Specialty type: Oncology
Country/Territory of origin: Japan
Peer-review report’s scientific quality classification
Grade A (Excellent): 0
Grade B (Very good): 0
Grade C (Good): C, C
Grade D (Fair): 0
Grade E (Poor): 0
P-Reviewer: Chong CS, Kuo SH S-Editor: Chen XF L-Editor: Filipodia P-Editor: Li JH
| 1. | Ciccolallo L, Capocaccia R, Coleman MP, Berrino F, Coebergh JW, Damhuis RA, Faivre J, Martinez-Garcia C, Møller H, Ponz de Leon M, Launoy G, Raverdy N, Williams EM, Gatta G. Survival differences between European and US patients with colorectal cancer: role of stage at diagnosis and surgery. Gut. 2005;54:268-273. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 94] [Cited by in RCA: 94] [Article Influence: 4.7] [Reference Citation Analysis (0)] |
| 2. | Nicolini A, Caciagli M, Zampieri F, Ciampalini G, Carpi A, Spisni R, Colizzi C. Usefulness of CEA, TPA, GICA, CA 72.4, and CA 195 in the Diagnosis of primary colorectal cancer and at its relapse. Cancer Detect Prev. 1995;19:183-195. [PubMed] |
| 3. | Morita S, Nomura T, Fukushima Y, Morimoto T, Hiraoka N, Shibata N. Does serum CA19-9 play a practical role in the management of patients with colorectal cancer? Dis Colon Rectum. 2004;47:227-232. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 31] [Cited by in RCA: 33] [Article Influence: 1.6] [Reference Citation Analysis (0)] |
| 4. | Filella X, Molina R, Mengual PJ, Jo J, Ballesta AM. Significance of CA72.4 in patients with colorectal cancer. Comparison with CEA and CA19.9. J Nucl Biol Med. 1991;35:158-161. [PubMed] |
| 5. | Gao Y, Wang J, Zhou Y, Sheng S, Qian SY, Huo X. Evaluation of Serum CEA, CA19-9, CA72-4, CA125 and Ferritin as Diagnostic Markers and Factors of Clinical Parameters for Colorectal Cancer. Sci Rep. 2018;8:2732. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 94] [Cited by in RCA: 200] [Article Influence: 28.6] [Reference Citation Analysis (1)] |
| 6. | Shinkins B, Nicholson BD, Primrose J, Perera R, James T, Pugh S, Mant D. The diagnostic accuracy of a single CEA blood test in detecting colorectal cancer recurrence: Results from the FACS trial. PLoS One. 2017;12:e0171810. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 36] [Cited by in RCA: 49] [Article Influence: 6.1] [Reference Citation Analysis (0)] |
| 7. | Kanellos I, Zacharakis E, Kanellos D, Pramateftakis MG, Betsis D. Prognostic significance of CEA levels and positive cytology in peritoneal washings in patients with colorectal cancer. Colorectal Dis. 2006;8:436-440. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 23] [Cited by in RCA: 25] [Article Influence: 1.3] [Reference Citation Analysis (0)] |
| 8. | Bhatti I, Patel M, Dennison AR, Thomas MW, Garcea G. Utility of postoperative CEA for surveillance of recurrence after resection of primary colorectal cancer. Int J Surg. 2015;16:123-128. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 36] [Cited by in RCA: 48] [Article Influence: 4.8] [Reference Citation Analysis (0)] |
| 9. | Hamfjord J, Guren TK, Dajani O, Johansen JS, Glimelius B, Sorbye H, Pfeiffer P, Lingjærde OC, Tveit KM, Kure EH, Pallisgaard N, Spindler KG. Total circulating cell-free DNA as a prognostic biomarker in metastatic colorectal cancer before first-line oxaliplatin-based chemotherapy. Ann Oncol. 2019;30:1088-1095. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 60] [Cited by in RCA: 64] [Article Influence: 10.7] [Reference Citation Analysis (0)] |
| 10. | Desmond BJ, Dennett ER, Danielson KM. Circulating Extracellular Vesicle MicroRNA as Diagnostic Biomarkers in Early Colorectal Cancer-A Review. Cancers. 12:52. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 34] [Cited by in RCA: 50] [Article Influence: 8.3] [Reference Citation Analysis (0)] |
| 11. | Yang C, Zou K, Zheng L, Xiong B. Prognostic and clinicopathological significance of circulating tumor cells detected by RT-PCR in non-metastatic colorectal cancer: a meta-analysis and systematic review. BMC Cancer. 2017;17:725. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 31] [Cited by in RCA: 44] [Article Influence: 5.5] [Reference Citation Analysis (0)] |
| 12. | Negin BP, Cohen SJ. Circulating tumor cells in colorectal cancer: past, present, and future challenges. Curr Treat Options Oncol. 2010;11:1-13. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 65] [Cited by in RCA: 78] [Article Influence: 5.2] [Reference Citation Analysis (0)] |
| 13. | Edwards HG, Newton EM, Wynn-Williams DD, Lewis-Smith RI. Non-destructive analysis of pigments and other organic compounds in lichens using Fourier-transform Raman spectroscopy: a study of Antarctic epilithic lichens. Spectrochim Acta A Mol Biomol Spectrosc. 2003;59:2301-2309. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 35] [Cited by in RCA: 17] [Article Influence: 0.8] [Reference Citation Analysis (0)] |
| 14. | Lin D, Feng S, Pan J, Chen Y, Lin J, Chen G, Xie S, Zeng H, Chen R. Colorectal cancer detection by gold nanoparticle based surface-enhanced Raman spectroscopy of blood serum and statistical analysis. Opt Express. 2011;19:13565-13577. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 207] [Cited by in RCA: 153] [Article Influence: 10.9] [Reference Citation Analysis (0)] |
| 15. | Chen Y, Chen G, Zheng X, He C, Feng S, Chen Y, Lin X, Chen R, Zeng H. Discrimination of gastric cancer from normal by serum RNA based on surface-enhanced Raman spectroscopy (SERS) and multivariate analysis. Med Phys. 2012;39:5664-5668. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 24] [Cited by in RCA: 23] [Article Influence: 1.8] [Reference Citation Analysis (0)] |
| 16. | Li X, Yang T, Li S, Wang D, Guan D. Detecting Esophageal Cancer Using Surface-Enhanced Raman Spectroscopy (SERS) of Serum Coupled with Hierarchical Cluster Analysis and Principal Component Analysis. Appl Spectrosc. 2015;69:1334-1341. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 19] [Cited by in RCA: 16] [Article Influence: 1.6] [Reference Citation Analysis (0)] |
| 17. | Wang G, Lipert RJ, Jain M, Kaur S, Chakraboty S, Torres MP, Batra SK, Brand RE, Porter MD. Detection of the potential pancreatic cancer marker MUC4 in serum using surface-enhanced Raman scattering. Anal Chem. 2011;83:2554-2561. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 212] [Cited by in RCA: 157] [Article Influence: 11.2] [Reference Citation Analysis (0)] |
| 18. | Zhang K, Liu X, Man B, Yang C, Zhang C, Liu M, Zhang Y, Liu L, Chen C. Label-free and stable serum analysis based on Ag-NPs/PSi surface-enhanced Raman scattering for noninvasive lung cancer detection. Biomed Opt Express. 2018;9:4345-4358. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 32] [Cited by in RCA: 34] [Article Influence: 4.9] [Reference Citation Analysis (0)] |
| 19. | Vargas-Obieta E, Martínez-Espinosa JC, Martínez-Zerega BE, Jave-Suárez LF, Aguilar-Lemarroy A, González-Solís JL. Breast cancer detection based on serum sample surface enhanced Raman spectroscopy. Lasers Med Sci. 2016;31:1317-1324. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 42] [Cited by in RCA: 51] [Article Influence: 5.7] [Reference Citation Analysis (0)] |
| 20. | Chen N, Rong M, Shao X, Zhang H, Liu S, Dong B, Xue W, Wang T, Li T, Pan J. Surface-enhanced Raman spectroscopy of serum accurately detects prostate cancer in patients with prostate-specific antigen levels of 4-10 ng/mL. Int J Nanomedicine. 2017;12:5399-5407. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 34] [Cited by in RCA: 45] [Article Influence: 5.6] [Reference Citation Analysis (0)] |
| 21. | Shao X, Pan J, Wang Y, Zhu Y, Xu F, Shangguan X, Dong B, Sha J, Chen N, Chen Z, Wang T, Liu S, Xue W. Evaluation of expressed prostatic secretion and serum using surface-enhanced Raman spectroscopy for the noninvasive detection of prostate cancer, a preliminary study. Nanomedicine. 2017;13:1051-1059. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 24] [Cited by in RCA: 26] [Article Influence: 2.9] [Reference Citation Analysis (0)] |
| 22. | Li S, Li L, Zeng Q, Zhang Y, Guo Z, Liu Z, Jin M, Su C, Lin L, Xu J, Liu S. Characterization and noninvasive diagnosis of bladder cancer with serum surface enhanced Raman spectroscopy and genetic algorithms. Sci Rep. 2015;5:9582. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 74] [Cited by in RCA: 56] [Article Influence: 5.6] [Reference Citation Analysis (0)] |
| 23. | Ito H, Inoue H, Hasegawa K, Hasegawa Y, Shimizu T, Kimura S, Onimaru M, Ikeda H, Kudo SE. Use of surface-enhanced Raman scattering for detection of cancer-related serum-constituents in gastrointestinal cancer patients. Nanomedicine. 2014;10:599-608. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 29] [Cited by in RCA: 32] [Article Influence: 2.7] [Reference Citation Analysis (0)] |
| 24. | Ito H, Hasegawa K, Hasegawa Y, Nishimaki T, Hosomichi K, Kimura S, Ohba M, Yao H, Onimaru M, Inoue I, Inoue H. Silver Nanoscale Hexagonal Column Chips for Detecting Cell-free DNA and Circulating Nucleosomes in Cancer Patients. Sci Rep. 2015;5:10455. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 18] [Cited by in RCA: 19] [Article Influence: 1.9] [Reference Citation Analysis (0)] |
| 25. | Ito H, Uragami N, Miyazaki T, Yokoyama N, Inoue H. Raman spectroscopic evaluation of human serum using metal plate and 785- and 1064-nm excitation lasers. PLoS One. 2019;14:e0211986. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 3] [Cited by in RCA: 3] [Article Influence: 0.5] [Reference Citation Analysis (2)] |
| 26. | Sajan D, Binoy J, Pradeep B, Venkata Krishna K, Kartha VB, Hubert Joe I, Jayakumar VS. NIR-FT Raman and infrared spectra and ab initio computations of glycinium oxalate. Spectrochim Acta A Mol Biomol Spectrosc. 2004;60:173-180. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 183] [Cited by in RCA: 132] [Article Influence: 6.3] [Reference Citation Analysis (0)] |
| 27. | Notingher I, Green C, Dyer C, Perkins E, Hopkins N, Lindsay C, Hench LL. Discrimination between ricin and sulphur mustard toxicity in vitro using Raman spectroscopy. J R Soc Interface. 2004;1:79-90. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 188] [Cited by in RCA: 183] [Article Influence: 9.6] [Reference Citation Analysis (0)] |
| 28. | Shetty G, Kendall C, Shepherd N, Stone N, Barr H. Raman spectroscopy: elucidation of biochemical changes in carcinogenesis of oesophagus. Br J Cancer. 2006;94:1460-1464. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 278] [Cited by in RCA: 270] [Article Influence: 14.2] [Reference Citation Analysis (0)] |
| 29. | Farquharson S, Gift A, Shende C, Inscore F, Ordway B, Farquharson C, Murren J. Surface-enhanced Raman spectral measurements of 5-fluorouracil in saliva. Molecules. 2008;13:2608-2627. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 57] [Cited by in RCA: 49] [Article Influence: 2.9] [Reference Citation Analysis (0)] |
| 30. | Chan JW, Taylor DS, Zwerdling T, Lane SM, Ihara K, Huser T. Micro-Raman spectroscopy detects individual neoplastic and normal hematopoietic cells. Biophys J. 2006;90:648-656. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 511] [Cited by in RCA: 416] [Article Influence: 21.9] [Reference Citation Analysis (0)] |
| 31. | Malini R, Venkatakrishna K, Kurien J, Pai KM, Rao L, Kartha VB, Krishna CM. Discrimination of normal, inflammatory, premalignant, and malignant oral tissue: a Raman spectroscopy study. Biopolymers. 2006;81:179-193. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 209] [Cited by in RCA: 186] [Article Influence: 9.8] [Reference Citation Analysis (0)] |
| 32. | Stone N, Kendall C, Smith J, Crow P, Barr H. Raman spectroscopy for identification of epithelial cancers. Faraday Discuss. 2004;126:141-157. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 467] [Cited by in RCA: 460] [Article Influence: 21.9] [Reference Citation Analysis (0)] |
| 33. | Ruiz-Chica AJ, Medina MA, Sánchez-Jiménez F, Ramírez FJ. Characterization by Raman spectroscopy of conformational changes on guanine-cytosine and adenine-thymine oligonucleotides induced by aminooxy analogues of spermidine. J Raman Spectrosc. 2004;35:93-100. [DOI] [Full Text] |
| 34. | De Gelder J, De Gussem K, Vandenabeele P, Moens L. Reference database of Raman spectra of biological molecules. J Raman Spectrosc. 2007;38:1133-1147. [DOI] [Full Text] |
| 35. | Nijssen A, Bakker Schut TC, Heule F, Caspers PJ, Hayes DP, Neumann MH, Puppels GJ. Discriminating basal cell carcinoma from its surrounding tissue by Raman spectroscopy. J Invest Dermatol. 2002;119:64-69. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 262] [Cited by in RCA: 201] [Article Influence: 8.7] [Reference Citation Analysis (0)] |
| 36. | Cheng WT, Liu MT, Liu HN, Lin SY. Micro-Raman spectroscopy used to identify and grade human skin pilomatrixoma. Microsc Res Tech. 2005;68:75-79. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 201] [Cited by in RCA: 227] [Article Influence: 11.9] [Reference Citation Analysis (0)] |
| 37. | Jyothi Lakshmi R, Kartha VB, Murali Krishna C, R Solomon JG, Ullas G, Uma Devi P. Tissue Raman spectroscopy for the study of radiation damage: brain irradiation of mice. Radiat Res. 2002;157:175-182. [PubMed] [DOI] [Full Text] |
| 38. | Huang Z, McWilliams A, Lui H, McLean DI, Lam S, Zeng H. Near-infrared Raman spectroscopy for optical diagnosis of lung cancer. Int J Cancer. 2003;107:1047-1052. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 554] [Cited by in RCA: 488] [Article Influence: 23.2] [Reference Citation Analysis (0)] |
| 39. | Krafft C, Neudert L, Simat T, Salzer R. Near infrared Raman spectra of human brain lipids. Spectrochim Acta A Mol Biomol Spectrosc. 2005;61:1529-1535. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 408] [Cited by in RCA: 395] [Article Influence: 19.8] [Reference Citation Analysis (0)] |
| 40. | Koljenović S, Schut TB, Vincent A, Kros JM, Puppels GJ. Detection of meningioma in dura mater by Raman spectroscopy. Anal Chem. 2005;77:7958-7965. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 107] [Cited by in RCA: 113] [Article Influence: 6.3] [Reference Citation Analysis (0)] |
| 41. | Patil CA, Kalkman J, Faber DJ, Nyman JS, van Leeuwen TG, Mahadevan-Jansen A. Integrated system for combined Raman spectroscopy-spectral domain optical coherence tomography. J Biomed Opt. 2011;16:011007. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 48] [Cited by in RCA: 32] [Article Influence: 2.3] [Reference Citation Analysis (0)] |
| 42. | Hastie T, Tibshirani R, Friedman J. The elements of statistical learning: data mining, inference, and prediction. 2nd ed. Berlin: Springer Science+Business Media, 2009: 1-745. |
| 43. | Ó Faoláin E, Hunter MB, Byrne JM, Kelehan P, McNamara M, Byrne HJ, Lyng FM. A study examining the effects of tissue processing on human tissue sections using vibrational spectroscopy. Vib Spectrosc. 2005;38:121-127. [DOI] [Full Text] |
| 44. | Short MA, Lui H, McLean D, Zeng H, Alajlan A, Chen XK. Changes in nuclei and peritumoral collagen within nodular basal cell carcinomas via confocal micro-Raman spectroscopy. J Biomed Opt. 2006;11:34004. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 46] [Cited by in RCA: 24] [Article Influence: 1.3] [Reference Citation Analysis (0)] |
| 45. | Lau DP, Huang Z, Lui H, Anderson DW, Berean K, Morrison MD, Shen L, Zeng H. Raman spectroscopy for optical diagnosis in the larynx: preliminary findings. Lasers Surg Med. 2005;37:192-200. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 74] [Cited by in RCA: 57] [Article Influence: 3.0] [Reference Citation Analysis (0)] |
| 46. | Gardiner DJ, Graves PR. Practical Raman spectroscopy. Berlin: Springer, 1989: 1-154. |
| 47. | Huang Z, Lui H, McLean DI, Korbelik M, Zeng H. Raman spectroscopy in combination with background near-infrared autofluorescence enhances the in vivo assessment of malignant tissues. Photochem Photobiol. 2005;81:1219-1226. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 90] [Cited by in RCA: 74] [Article Influence: 3.9] [Reference Citation Analysis (0)] |
| 48. | Xue L, Yan B, Li Y, Tan Y, Luo X, Wang M. Surface-enhanced Raman spectroscopy of blood serum based on gold nanoparticles for tumor stages detection and histologic grades classification of oral squamous cell carcinoma. Int J Nanomedicine. 2018;13:4977-4986. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 35] [Cited by in RCA: 41] [Article Influence: 5.9] [Reference Citation Analysis (0)] |
| 49. | Tan Y, Yan B, Xue L, Li Y, Luo X, Ji P. Surface-enhanced Raman spectroscopy of blood serum based on gold nanoparticles for the diagnosis of the oral squamous cell carcinoma. Lipids Health Dis. 2017;16:73. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 53] [Cited by in RCA: 42] [Article Influence: 5.3] [Reference Citation Analysis (0)] |
| 50. | Xiao R, Zhang X, Rong Z, Xiu B, Yang X, Wang C, Hao W, Zhang Q, Liu Z, Duan C, Zhao K, Guo X, Fan Y, Zhao Y, Johnson H, Huang Y, Feng X, Xu X, Zhang H, Wang S. Non-invasive detection of hepatocellular carcinoma serum metabolic profile through surface-enhanced Raman spectroscopy. Nanomedicine. 2016;12:2475-2484. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 39] [Cited by in RCA: 43] [Article Influence: 4.8] [Reference Citation Analysis (0)] |
| 51. | Hong Y, Li Y, Huang L, He W, Wang S, Wang C, Zhou G, Chen Y, Zhou X, Huang Y, Huang W, Gong T, Zhou Z. Label-free diagnosis for colorectal cancer through coffee ring-assisted surface-enhanced Raman spectroscopy on blood serum. J Biophotonics. 2020;13:e201960176. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 41] [Cited by in RCA: 49] [Article Influence: 9.8] [Reference Citation Analysis (0)] |
| 52. | Feng S, Wang W, Tai IT, Chen G, Chen R, Zeng H. Label-free surface-enhanced Raman spectroscopy for detection of colorectal cancer and precursor lesions using blood plasma. Biomed Opt Express. 2015;6:3494-3502. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 42] [Cited by in RCA: 40] [Article Influence: 4.0] [Reference Citation Analysis (0)] |
| 53. | Wang J, Lin D, Lin J, Yu Y, Huang Z, Chen Y, Lin J, Feng S, Li B, Liu N, Chen R. Label-free detection of serum proteins using surface-enhanced Raman spectroscopy for colorectal cancer screening. J Biomed Opt. 2014;19:087003. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 65] [Cited by in RCA: 61] [Article Influence: 5.5] [Reference Citation Analysis (0)] |
| 54. | Chen Y, Chen G, Feng S, Pan J, Zheng X, Su Y, Chen Y, Huang Z, Lin X, Lan F, Chen R, Zeng H. Label-free serum ribonucleic acid analysis for colorectal cancer detection by surface-enhanced Raman spectroscopy and multivariate analysis. J Biomed Opt. 2012;17:067003. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 47] [Cited by in RCA: 35] [Article Influence: 2.7] [Reference Citation Analysis (0)] |
| 55. | Hill MJ, Morson BC, Bussey HJ. Aetiology of adenoma--carcinoma sequence in large bowel. Lancet. 1978;1:245-247. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 277] [Cited by in RCA: 259] [Article Influence: 5.5] [Reference Citation Analysis (0)] |
| 56. | Prasetyanti PR, Zimberlin CD, Bots M, Vermeulen L, Melo Fde S, Medema JP. Regulation of stem cell self-renewal and differentiation by Wnt and Notch are conserved throughout the adenoma-carcinoma sequence in the colon. Mol Cancer. 2013;12:126. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 39] [Cited by in RCA: 52] [Article Influence: 4.3] [Reference Citation Analysis (0)] |
| 57. | Murakami T, Mitomi H, Saito T, Takahashi M, Sakamoto N, Fukui N, Yao T, Watanabe S. Distinct WNT/β-catenin signaling activation in the serrated neoplasia pathway and the adenoma-carcinoma sequence of the colorectum. Mod Pathol. 2015;28:146-158. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 41] [Cited by in RCA: 50] [Article Influence: 5.0] [Reference Citation Analysis (0)] |
| 58. | Mohammadi M, Kristensen MH, Nielsen HJ, Bonde JH, Holck S. Qualities of sessile serrated adenoma/polyp/lesion and its borderline variant in the context of synchronous colorectal carcinoma. J Clin Pathol. 2012;65:924-927. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 18] [Cited by in RCA: 19] [Article Influence: 1.5] [Reference Citation Analysis (0)] |