Published online Feb 15, 2019. doi: 10.4251/wjgo.v11.i2.91
- This article has been corrected.
- See: World J Gastrointest Oncol. Jul 15, 2022; 14(7): 1372-1374
Peer-review started: November 14, 2018
First decision: December 7, 2018
Revised: December 22, 2018
Accepted: January 8, 2019
Article in press: January 8, 2019
Published online: February 15, 2019
Processing time: 96 Days and 22.2 Hours
Long non-coding RNAs (lncRNAs) are a kind of single-stranded RNA of more than 200 nucleotides in length and have no protein-coding function. Amounting studies have indicated that lncRNAs could play a vital role in the initiation and development of cancers, including gastric cancer (GC). Considering the crucial functions of lncRNAs, the identification and exploration of novel lncRNAs in GC is necessary.
To explore the role of novel lncRNA LINC02532 in GC.
The upregulated LINC02532 was identified by processing the GC RNA-Seq data from The Cancer Genome Atlas. The qRT-PCR assay was performed to confirm the expression levels in GC cell lines and tissues. Cell proliferation, migration, and invasion were evaluated by the cell counting kit-8, colony formation, wound healing, and Transwell assays. The miRNAs downregulated in GC and sponged by LINC02532 were identified from and predicted by the data from the Firehose and RNA22 software programs, respectively. The miRNA downstream target genes were obtained from the TargetScan, miRDB, and DIANA online tools. Gene functional enrichment analysis was carried out using the Database for Annotation, Visualization, and Integrated Discovery software in the categories of cellular components, biological processes, molecular functions, and KEGG pathways.
The qRT-PCR assay demonstrated that the LINC02532 expression level was significantly upregulated in the GC cell lines and 52 paired tissues. Kaplan-Meier survival analysis based on The Cancer Genome Atlas data showed that patients with higher LINC02532 expression had poorer prognosis than those with lower LINC02532 expression. The correlation analysis between expression and clinicopathological features revealed that high expression of LINC02532 was associated with a high TNM stage (P = 0.008) and poor differentiation grade (P = 0.023). Functional experiments showed that LINC02532 promoted GC cell proliferation, migration, and invasion. According to the bioinformatics analysis, LINC02532 may act as a ceRNA by sponging downregulated miR-129-5p and miR-490-5p. Target genes of the two miRNAs were selected for further functional enrichment analysis. Importantly, KEGG pathway analysis showed that the genes were mainly involved in transcriptional misregulation in cancer, cell cycle, and TGF-beta, mTOR, and p53 signaling pathways.
The present study suggested that LINC02532 acted as an oncogene in GC and may be a promising target for therapy and prognosis management of GC.
Core tip: Our study identified a novel long non-coding RNA LINC02532 and found that it was significantly overexpressed in gastric cancer (GC). Kaplan-Meier survival analysis showed that patients with higher LINC02532 expression had poorer prognosis than those with lower LINC02532 expression. The correlation analysis between expression and clinicopathological features revealed that the high expression of LINC02532 was associated with a high TNM stage and poor differentiation grade. Functional assays supported the finding that LINC02532 promoted GC cell proliferation, migration, and invasion. According to the bioinformatics analysis, LINC02532 may sponge downregulated miR-129-5p and miR-490-5p and participate in transcriptional misregulation in cancer, cell cycle, and TGF-beta, mTOR, and p53 signaling pathways.
- Citation: Zhang C, Ma MH, Liang Y, Wu KZ, Dai DQ. Novel long non-coding RNA LINC02532 promotes gastric cancer cell proliferation, migration, and invasion in vitro. World J Gastrointest Oncol 2019; 11(2): 91-101
- URL: https://www.wjgnet.com/1948-5204/full/v11/i2/91.htm
- DOI: https://dx.doi.org/10.4251/wjgo.v11.i2.91
Gastric cancer (GC) is a common cancer in incidence and mortality worldwide, with 1033701 new cases and 782685 deaths in 2018[1]. These numbers have seen consistent growth because most GC patients are diagnosed at late stages and endure poor outcomes. The 5-year overall survival rate could exceed 90% if patients were diagnosed at early stages of the disease and treated appropriately[2,3]. In light of improved prognosis resulting from early diagnosis, it is urgently needed to identify the molecular mechanisms associated with the initiation and development of GC and discover crucial biomarkers for diagnosis and treatment.
Long non-coding RNAs (lncRNAs, 200 nucleotides in length) are a series of single-stranded RNA molecules, which have no protein-coding functions[4]. However, studies have shown that deregulated lncRNAs could participate in vital biological processes of various carcinomas, including GC[5-7]. The competitive endogenous RNA (ceRNA) hypothesis supported that lncRNAs, which harbor microRNA-response elements, can sponge microRNAs (miRNAs) from target mRNAs sharing common microRNA-response elements[8]. For example, LINC00483 acts as ceRNA to sponge miR-30a-3p and regulates proliferation and apoptosis in GC[9]. LncRNA-CDC6 can sponge miR-215 to promote the proliferation and metastasis of breast cancer[10]. All this evidence indicates that lncRNA sponges are closely related to cancer tumorigenesis. Despite all of that, the exact role that lncRNAs play in GC tumorigenesis remains vague. Thus, it is essential to explore the lncRNA features that can be used as diagnostic biomarkers and therapy targets in GC.
In this study, we downloaded the RNA-Seq data from The Cancer Genome Atlas (TCGA) database. After data processing, we first identified a novel lncRNA LINC02532 whose expression levels and biological features in cancers have not yet been reported. We found that LINC02532 was significantly overexpressed in GC cell lines and tissues. Functional assays showed that LINC02532 promoted GC cells proliferation, migration, and invasion. Using the bioinformatics analysis, we predicted that LINC02532 may act as ceRNA to sponge miR-129-5p and miR-490-5p. In addition, reliable target genes of each miRNA were identified. Further gene ontology and KEGG pathway enrichment analyses of the target genes were performed and their results showed that these genes were involved in transcriptional misregulation in cancer, cell cycle, and TGF-beta, mTOR, and p53 signaling pathways. In conclusion, our study found that LINC02532 participates in the tumorigenesis of GC as an oncogene and could be useful in the therapy and prognosis management of GC.
The RNA-Seq data for 375 GC and 32 normal tissues and patients’ clinical information data were downloaded from the TCGA[11] database (update to 2018.03. https://cancergenome.nih.gov/). The data for GC mature miRNA sequencing were obtained from the Firebrowse[12] (http://firebrowse.org/) website, which provides integrated data from the TCGA database. RNA22 tools[13] (https://cm.jefferson.edu/rna22/) were selected for predicting targeted miRNAs, which may be sponged by LINC02532. Differentially expressed genes and miRNAs were identified at the cut-off criterion of fold change > 2.0 and P < 0.05.
Four human GC cell lines (SGC-7901, MGC-803, AGS, and MKN-45) and a normal gastric epithelium cell line (GES-1) purchased from the Chinese Academy of Sciences (Shanghai, China), were cultivated in RPMI 1640 medium (HyClone, Logan, UT, United States) supplemented with 10% fetal bovine serum (FBS; Gibco, Grand Island, NY, United States), 100 mg/mL streptomycin, and 100 U/mL penicillin. All cells were cultured at 37 °C in a humidified incubator with 5% CO2.
A total of 52 paired GC tissues and corresponding adjacent normal tissues were collected from the Fourth Affiliated Hospital of China Medical University. All patients did not receive preoperative chemo- or radio-therapy and collected tissues were promptly stored at -80 °C. The study was authorized by the Research Ethics Committee of the Fourth Affiliated Hospital of China Medical University. The GC patients were confirmed by histopathology and signed the informed consent.
Total RNA was isolated from tissues and cells using Trizol reagent (Invitrogen, Carlsbad, CA, United States). RNA reverse transcription was operated using the PrimeScriptTM RT reagent Kit (Takara, Otsu, Shiga, Japan). The quantitative real-time PCR (qRT-PCR) was performed using the SYBR Premix Ex Taq II (Takara) on an ABI 7500 Fast Real-Time PCR system (Applied Biosystems). The primers used for qRT-PCR were as follows: LINC02532, Forward - 5′-TCCTGGTGCCACTGAGACTGTG-3′; Reverse - 5′- AGCCTCCATGATTCCTGCCTCTAG-3′; GAPDH, Forward - 5′- AGCCACATCGCTCAGACAC-3′; Reverse - 5′-GCCCAATACGACCAAATCC-3′. The relative gene expression was calculated by the 2-ΔΔCT method, and all of the operations were performed according to the manufacturer's instructions.
Three small interfering RNA (siRNA) against LINC02532 and negative control siRNA (si-NC) were synthesized and purified by GenePharma (Shanghai, China). The sequences are as follows (5′ to 3′): si-LINC02532 #1, sense: GGAACACAUAUAGCUGGAATT, antisense: UUCCAGCUAUAUGUGUUCCTT; si-LINC02532 #2, sense: GCAGGAAUCAUGGAGGCUUTT, antisense: AAGCCUCCAUGAUUCCUGCTT; si-LINC02532 #3, sense: GCCAGUAUAAGCAAGAGUATT, antisense: UACUCUUGCUUAUACUGGCTT; si-NC, sense: UUCUCCGAACGUGUCACGUTT, antisense: ACGUGACACGUUCGGAGAATT. The siRNAs were transfected into SGC-7901 and MGC-803 cells by the Lipofectamine 3000 Reagent (Invitrogen). After transfection, cells were harvested for detection of the interference effect and further functional assays.
Cell counting kit-8 (CCK-8) and colony formation assays were performed to assess the cell proliferation capacity. After a 24-h transfection with siRNA, the SGC-7901 and MGC-803 cells were seeded on 96-well plates (5000 cells/well) and then cultured in a humidified incubator at 37 °C with 5% CO2. At 0, 24, 48, and 72 h, 10 μL of the cell counting kit-8 solution (Dojindo Laboratories, Japan) were added to each well. After incubating for 4 h, the absorbance of each well at 450 nm was measured using a spectrophotometer. For colony formation assays, cells were seeded on 6-well plates (2000 cells/well). After culturing for 1 wk, the cell colonies were fixed with 4% paraformaldehyde and then stained with a 0.1% crystal violet solution. Visible colonies were counted and corresponding photographs were taken.
The transfected cells were seeded on 6-well plates and cultured to approximately 90% density. Then, a straight wound was drawn with a 100-μL pipette tip on the confluent monolayer. The cells continued to be cultured with serum-free medium for 48 h and photographed by an inverted microscope.
Invasion and migration assays were performed in 24-well plates with inserts (8 µmol/L pore size, Corning, New York, NY, United States) with or without Matrigel (Solarbio, Beijing), respectively. A total of 2.0 × 104 cells with 200 μL of serum-free medium were added into each upper chamber, and 500 μL of medium with 10% FBS were added into each well. After incubation for 48 h, the cells migrated to and invaded the lower chamber surfaces and were fixed and stained as described above. Cells per field under the inverted microscope were counted and photographed.
The functional and pathway enrichment analyses were performed using the Database for Annotation, Visualization, and Integrated Discovery (https://david.ncifcrf.gov/) tool, which provides systematic functional annotation to comprehend the potential biological gene functions[14]. The analysis was carried out in the categories of cellular components, molecular functions, biological processes, and KEGG pathways.
The statistical comparisons were performed by paired and two-tailed Student’s t test or chi-squared test. The Kaplan-Meier method and log-rank test were utilized for the survival analysis. All data were analyzed using the SPSS version 19.0 (Chicago, IL, United States), where P < 0.05 was considered to be statistically significant. All experiments were performed three times.
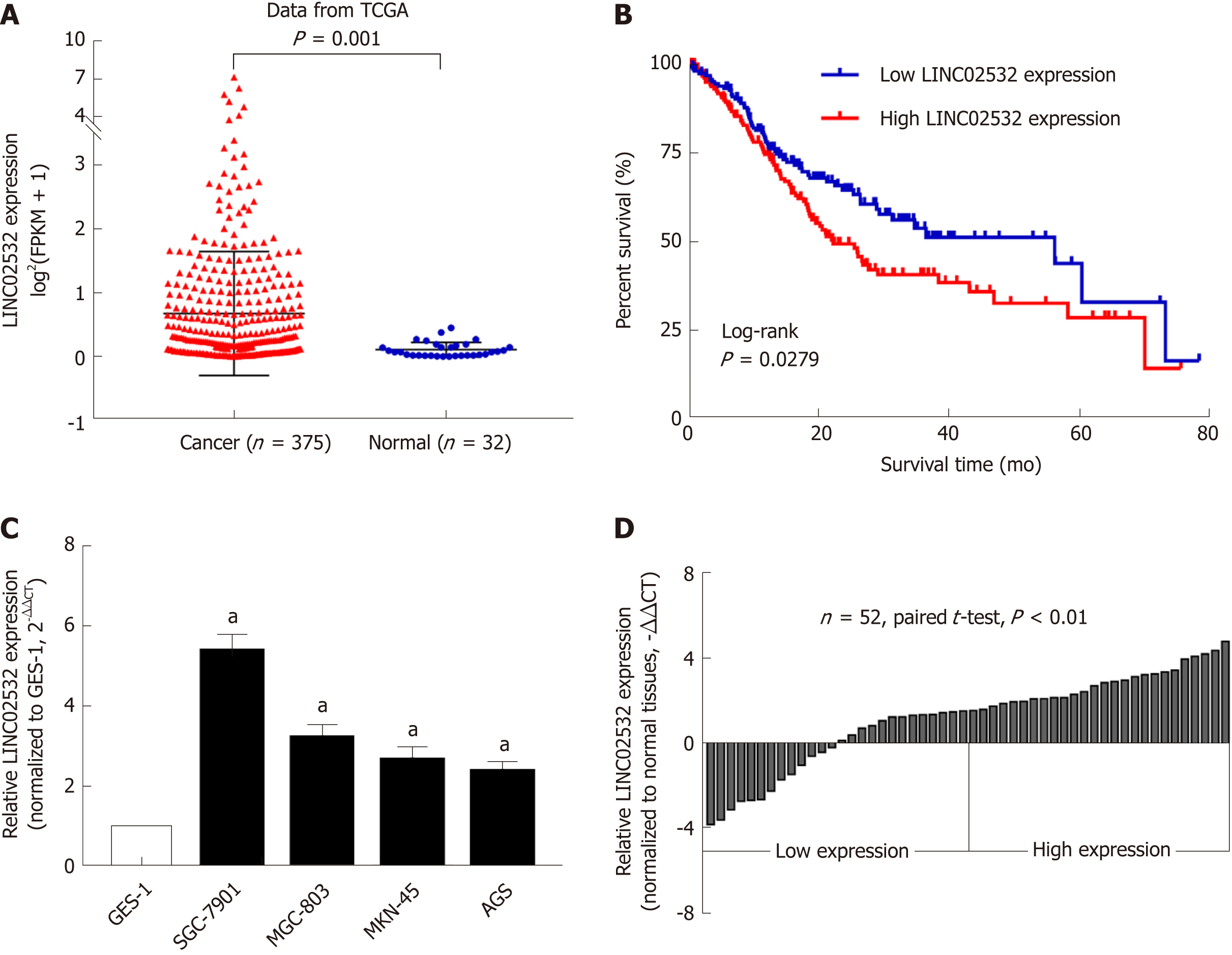
After analyzing the GC RNA-Seq data from the TCGA database, we noted that the expression of LINC02532 in tumor tissues was significantly higher than in normal tissues (Figure 1A). We then studied the association between LINC02532 expression and the overall survival of GC patients. A total of 347 patients with detailed information and overall survival time less than 80 mo were divided into two groups, according to the median expression value. The Kaplan-Meier survival curve showed that the group with higher LINC02532 expression had poorer prognosis than the group with lower LINC02532 expression (Figure 1B). Further qRT-PCR results from 52 GC patients and cell lines also indicated that LINC02532 was upregulated in GC (Figure 1C and D). To investigate whether the expression level of LINC02532 was associated with GC development and progression, we analyzed the correlation between expression and clinicopathological features. The results showed that a high expression of LINC02532 was associated with high TNM stage and poor differentiation (Table 1).
| Variable | LINC02532 expression | Total samples | P value | |
| Low, n, % | High, n, % | |||
| Age | ||||
| < 60 | 12 (23.1) | 15 (28.8) | 27 | 0.405 |
| ≥ 60 | 14 (26.9) | 11 (21.2) | 25 | |
| Sex | ||||
| Male | 16 (30.8) | 17 (32.7) | 33 | 0.773 |
| Female | 10 (19.2) | 9 (17.3) | 19 | |
| Tumor invasion | ||||
| T1 + T2 | 13 (25.0) | 11 (21.2) | 24 | 0.578 |
| T3 + T4 | 13 (25.0) | 15 (28.8) | 28 | |
| TNM stage | ||||
| I + II | 13 (25.0) | 4 (7.7) | 17 | 0.008a |
| III + IV | 13 (25.0) | 22 (42.3) | 35 | |
| Lymph node metastasis | ||||
| No | 9 (17.3) | 8 (15.4) | 17 | 0.768 |
| Yes | 17 (32.7) | 18 (34.6) | 35 | |
| Differentiation grade | ||||
| Well and moderate | 20 (38.5) | 12 (23.1) | 32 | 0.023a |
| Poor | 6 (11.5) | 14 (26.9) | 20 | |
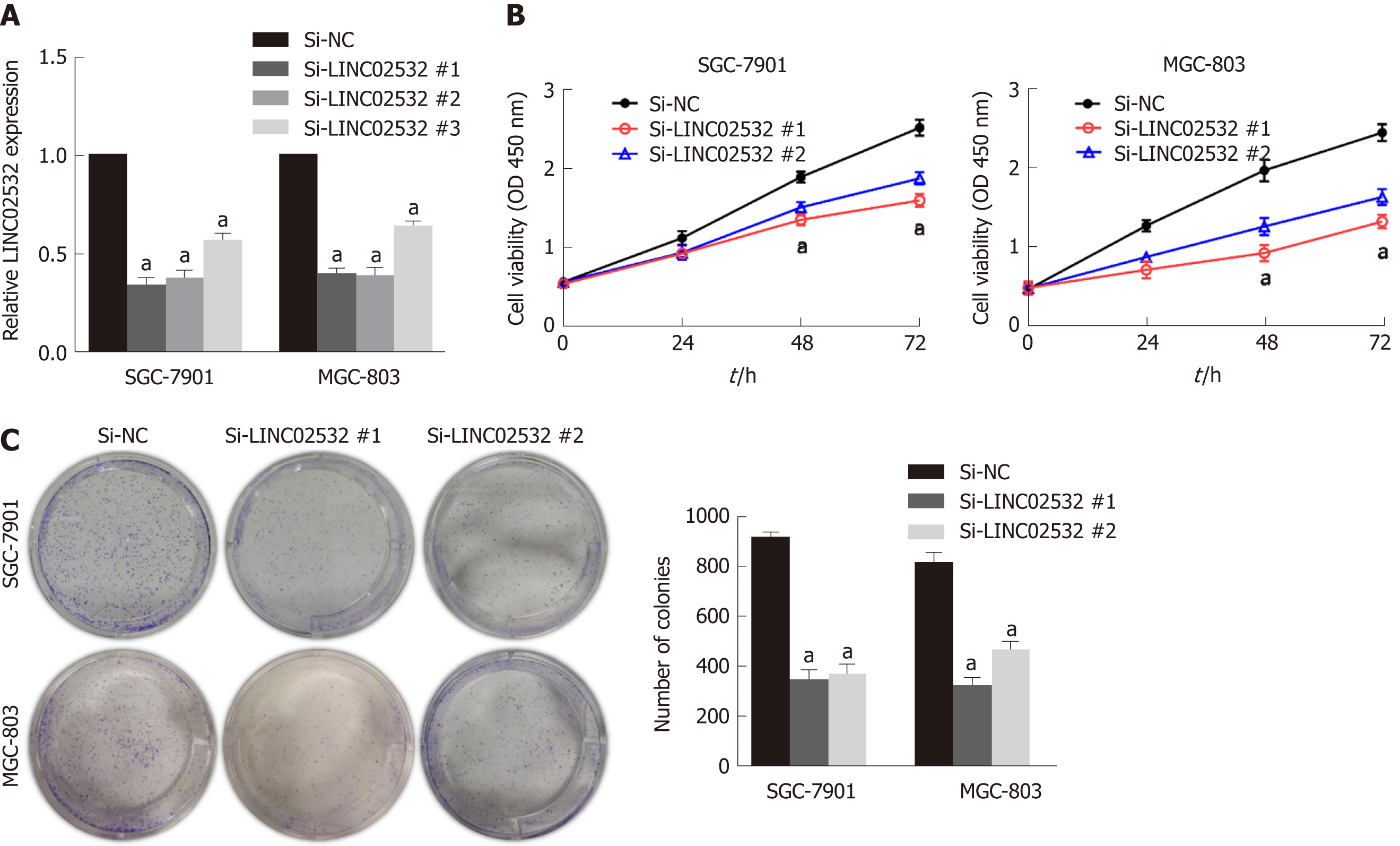
To study the role of LINC02532 in vitro, three kinds of siRNA against LINC02532 were transfected into SGC-7901 and MGC-803 cell lines. The qRT-PCR results revealed that LINC02532 expression was downregulated more significantly by siRNA #1 and #2 (Figure 2A). Accordingly, siRNA #1 and #2 were selected for further functional assays. The cell counting kit-8 assay showed that LINC02532 knockdown effectively restrained GC cell proliferation (Figure 2B). In addition, the colony formation assay showed that the cell proliferation rate was decreased after the LINC02532 knockdown (Figure 2C). All these findings suggested that LINC02532 could promote GC cell proliferation.
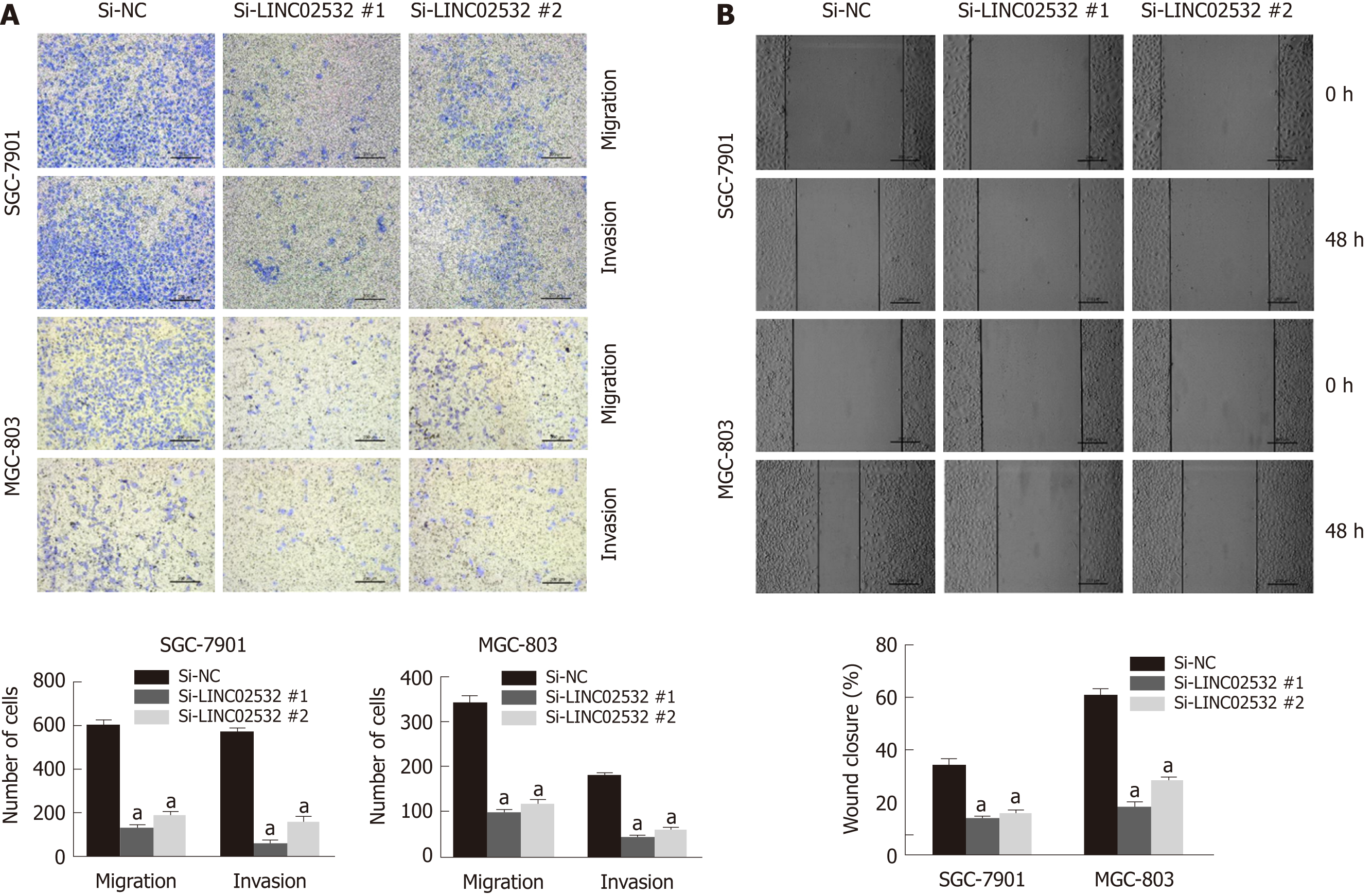
We further performed Transwell and wound healing assays to investigate the effect of LINC02532 on GC cell migration and invasion. The results showed that LINC02532 knockdown attenuated the migration and invasion ability of SGC-7901 and MGC-803 cells (Figure 3A and B), indicating that LINC02532 promoted cell migration and invasion in GC.
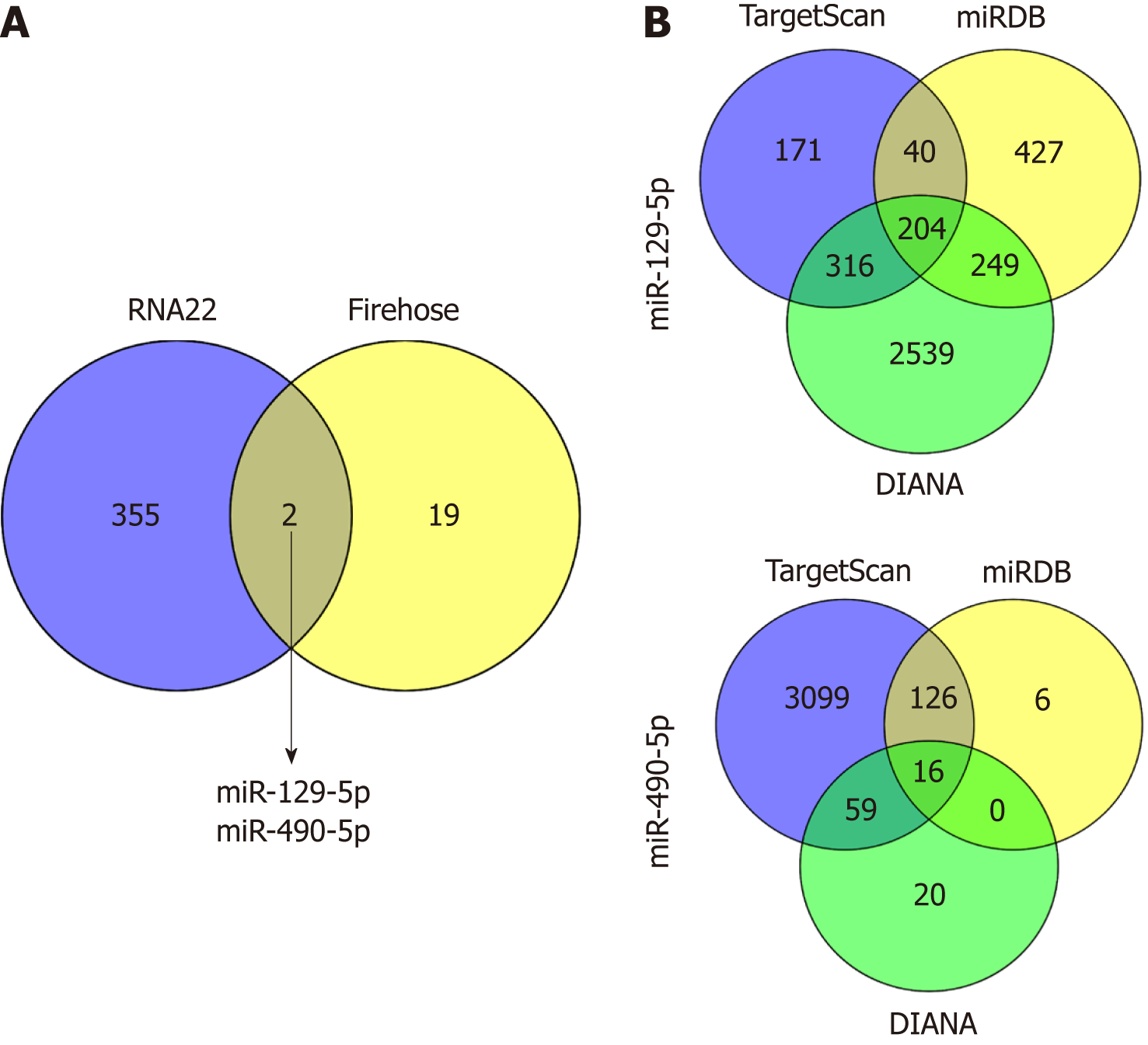
To further explore the molecular mechanism, we hypothesized that LINC02532 may promote GC cell proliferation, migration, and invasion by sponging miRNAs. First, we predicted the subcellular locations of LINC02532 using the online software iLoc-LncRNA[15]. The results suggested that the cytoplasm was the LINC02532 subcellular location with a probability score of about 0.98, which greatly supported the theory that LINC02532 may function as a ceRNA. Furthermore, we found that there were two miRNAs (miR-129-5p and miR-490-5p) that were both potential targets of LINC02532 and were downregulated in GC (Figure 4A). The possible miRNA target genes were obtained by the TargetScan[16], miRDB[17], and DIANA[18] software programs (Figure 4B). In total, 218 target genes (two duplicates were removed) were acquired.
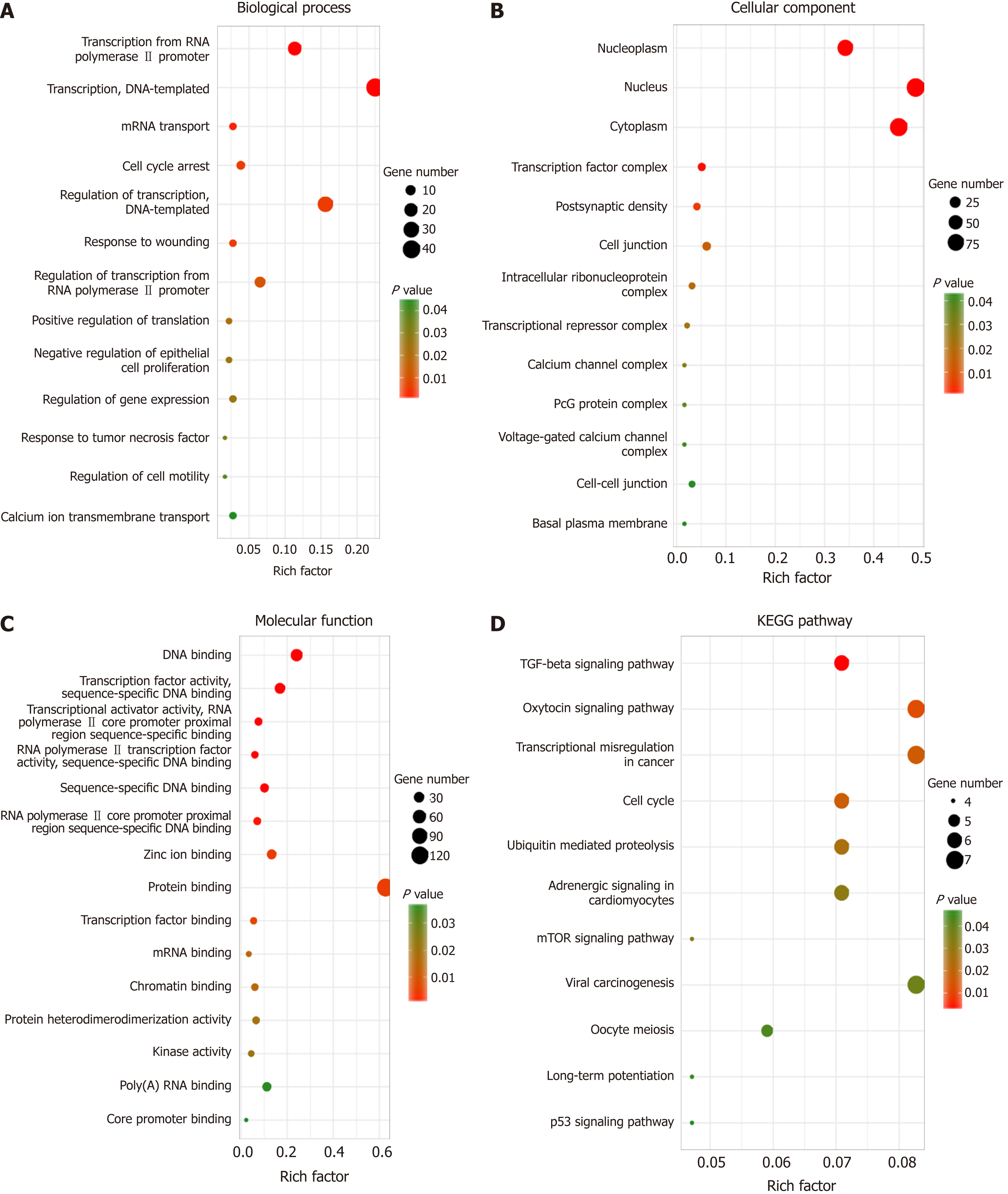
Identifying the function of these target genes may reveal the novel molecular roles of LINC02532 in GC. Thus, functional enrichment analysis was performed on 218 target genes. The biological process analysis indicated that the genes were enriched during the cell cycle arrest phase, in response to wounding, tumor necrosis factors, and regulation of cell motility. Regarding molecular function, these genes mainly participated in protein binding, transcription factor binding, and kinase activity. KEGG pathway analysis showed that the genes were significantly enriched in transcriptional misregulation in cancer, cell cycle, and TGF-beta, mTOR, and p53 signaling pathways (Figure 5).
Studies have demonstrated that lncRNAs are dysregulated and participate in the oncogenesis and progression of GC[19-21], which suggests that lncRNAs can be promising targets in the diagnosis and treatment of GC. In the present study, we downloaded and analyzed the GC RNA-Seq data from the TCGA database and discovered that LINC02532 was significantly upregulated in the GC tissues. However, the role of LINC02532 in cancers, including GC, is still unclear, which attracted our interest for further exploration.
First, we performed the qRT-PCR assay and confirmed that LINC02532 was overexpressed in the GC cell lines and 56 paired tissues. The association analysis between the LINC02532 expression and clinicopathological features found that high expression levels are significantly associated with the advanced TNM stage and poor differentiation. The Kaplan-Meier survival analysis based on the GC clinical data from the TCGA database indicated that patients with high expression levels of LINC02532 had a poorer prognosis than those with low expression levels. Furthermore, the functional assays suggested that LINC02532 promoted GC cells proliferation, migration, and invasion in vitro. All this evidence demonstrated that LINC02532 may play a vital role in the initiation and development of GC.
The functions of lncRNAs are diverse and closely related to their location in the cells. The lncRNAs that are located in the nucleus mainly regulate gene expression through modulating the activity of transcription factors, while those located in the cytoplasm participate in the regulation of mRNA by sponging the miRNAs[22,23]. Thus, it is necessary to identify the subcellular locations of lncRNAs before exploring their potential molecular mechanisms. According to the prediction based on the RNALocate database, we found that the probability score of LNC02532 being located in the cytoplasm reached up to 0.98. Thus, we predicted the miRNAs that were both potential targets of LINC02532 and were downregulated in GC based on the Firehose database. As a result, two miRNAs (miR-129-5p and miR-490-5p) were screened out. Previous studies confirmed that the miR-129-5p acted as a tumor suppressor by targeting ADAM9[24] and COL1A1[25] in GC. Chen et al[26] found that miR-129-5p inhibits GC cell proliferation by regulating the glyco-metabolism process. Hou et al[27] demonstrated that miR-490-5p was downregulated and inhibited GC cell proliferation by regulating the ERK signaling pathway. In summary, these findings are in accordance with our hypothesis and suggest the crucial role of miR-129-5p and miR-490-5p in GC.
Furthermore, we predicted the target genes of miR-129-5p and miR-490-5p. In order to improve the reliability of prediction, we used three prediction software programs with different algorithms and selected consistent results. Finally, functional enrichment analysis was performed on the common target genes. The KEGG pathway analysis showed that the genes were significantly enriched in transcriptional misregulation in cancer, cell cycle, and TGF-beta, mTOR, and p53 signaling pathways. Several studies have reported that the TGF-beta[28], mTOR[29,30], and p53[31] signaling pathways are closely associated with the initiation and development of GC. All these results support that LINC02532 may play a key role in the molecular mechanism of GC tumorigenesis.
Above all, the present study found that novel LINC02532 acts as an oncogene to promote the tumorigenesis of GC and could be a promising target in the diagnosis, therapy, and prognosis management of GC.
Long non-coding RNAs (lncRNAs) are a class of non-coding and single-stranded RNAs, with the length of more than 200 nucleotides. Although lncRNAs have no protein-coding capacity, they take part in the regulation of gene expression at the post-transcriptional level. Recent studies have found that lncRNAs are involved in the initiation and development of cancers, including gastric cancer (GC). To identify and explore the potential role of lncRNAs in GC could contribute to finding a promising biomarker in the diagnosis, therapy, and prognosis management of GC.
GC is a malignant digestive system tumor with high incidence and mortality in the world. Considering the actual mechanisms of lncRNAs in GC remain unclear, it is urgent to identify and explore the potential role of lncRNAs. The finding of novel lncRNA could contribute to studying the initiation and development mechanisms of GC.
In the study, we aimed to identify a novel lncRNA and explore its biological effect in GC. Once its actual role is confirmed, it could be a promising molecule in studying the diagnosis, therapy, and prognosis management of GC. The potential microRNAs (miRNAs) that LINC02532 may sponged were also predicted by bioinformatics. The results are useful for further exploring the mechanisms of competing endogenous RNAs (ceRNA).
We first identified differentially expressed genes by processing the RNA-Seq or mature miRNAs sequencing data of GC downloaded from The Cancer Genome Atlas or Firebrowse website. The RNA-Seq data supported that LINC02532 was upregulated and its expression level in GC cells and tissues was confirmed by qRT-PCR assay. The functional assays indicated that LINC02532 promoted GC cells proliferation, migration, and invasion in vitro. RNA22 tool was selected for predicting target miRNAs, which may be sponged by LINC02532. The miRNA target genes were obtained by the TargetScan, miRDB, and DIANA software. The gene functional enrichment analysis of the common target genes was performed by the Database for Annotation, Visualization, and Integrated Discovery tool.
We identified that LINC02532 was upregulated in GC and promoted GC cell proliferation, migration, and invasion in vitro. The results of statistical analysis showed that a high expression of LINC02532 was associated with high TNM stage and poor differentiation. Besides, Kaplan-Meier survival analysis based on The Cancer Genome Atlas data indicated that patients with higher LINC02532 expression had poorer prognosis than those with lower LINC02532 expression. Furthermore, we found that LINC02532 may act as a ceRNA by sponging miR-129-5p and miR-490-5p. The target genes of the two miRNAs were selected for gene functional enrichment analysis and the results supported the potential oncogene role of LINC02532 in GC.
We found that novel LINC02532 was significantly upregulated in GC, and it may act as an oncogene to promote the tumorigenesis of GC. The present study revealed that LINC02532 could be a promising target in the diagnosis, therapy, and prognosis management of GC.
The present study was well designed by combining bioinformatics prediction with experimental evidence. Furthermore, we will further focus on the mechanisms of LINC02532, such as the ceRNA hypothesis.
Manuscript source: Unsolicited manuscript
Specialty type: Oncology
Country of origin: China
Peer-review report classification
Grade A (Excellent): 0
Grade B (Very good): B, B
Grade C (Good): 0
Grade D (Fair): 0
Grade E (Poor): 0
P- Reviewer: Li C, Muhammad JS S- Editor: Ji FF L- Editor: Filipodia E- Editor: Bian YN
| 1. | Bray F, Ferlay J, Soerjomataram I, Siegel RL, Torre LA, Jemal A. Global cancer statistics 2018: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J Clin. 2018;68:394-424. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 53206] [Cited by in RCA: 55806] [Article Influence: 7972.3] [Reference Citation Analysis (132)] |
| 2. | Suzuki H, Oda I, Abe S, Sekiguchi M, Mori G, Nonaka S, Yoshinaga S, Saito Y. High rate of 5-year survival among patients with early gastric cancer undergoing curative endoscopic submucosal dissection. Gastric Cancer. 2016;19:198-205. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 135] [Cited by in RCA: 188] [Article Influence: 20.9] [Reference Citation Analysis (0)] |
| 3. | Yu H, Yang AM, Lu XH, Zhou WX, Yao F, Fei GJ, Guo T, Yao LQ, He LP, Wang BM. Magnifying narrow-band imaging endoscopy is superior in diagnosis of early gastric cancer. World J Gastroenterol. 2015;21:9156-9162. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in CrossRef: 30] [Cited by in RCA: 39] [Article Influence: 3.9] [Reference Citation Analysis (1)] |
| 4. | Kapranov P, Cheng J, Dike S, Nix DA, Duttagupta R, Willingham AT, Stadler PF, Hertel J, Hackermüller J, Hofacker IL, Bell I, Cheung E, Drenkow J, Dumais E, Patel S, Helt G, Ganesh M, Ghosh S, Piccolboni A, Sementchenko V, Tammana H, Gingeras TR. RNA maps reveal new RNA classes and a possible function for pervasive transcription. Science. 2007;316:1484-1488. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1800] [Cited by in RCA: 1923] [Article Influence: 106.8] [Reference Citation Analysis (0)] |
| 5. | Yan K, Tian J, Shi W, Xia H, Zhu Y. LncRNA SNHG6 is Associated with Poor Prognosis of Gastric Cancer and Promotes Cell Proliferation and EMT through Epigenetically Silencing p27 and Sponging miR-101-3p. Cell Physiol Biochem. 2017;42:999-1012. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 111] [Cited by in RCA: 145] [Article Influence: 18.1] [Reference Citation Analysis (0)] |
| 6. | Li J, Li Z, Zheng W, Li X, Wang Z, Cui Y, Jiang X. LncRNA-ATB: An indispensable cancer-related long noncoding RNA. Cell Prolif. 2017;50. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 112] [Cited by in RCA: 151] [Article Influence: 18.9] [Reference Citation Analysis (0)] |
| 7. | Chen X, Chen Z, Yu S, Nie F, Yan S, Ma P, Chen Q, Wei C, Fu H, Xu T, Ren S, Sun M, Wang Z. Long Noncoding RNA LINC01234 Functions as a Competing Endogenous RNA to Regulate CBFB Expression by Sponging miR-204-5p in Gastric Cancer. Clin Cancer Res. 2018;24:2002-2014. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 133] [Cited by in RCA: 174] [Article Influence: 24.9] [Reference Citation Analysis (0)] |
| 8. | Karreth FA, Pandolfi PP. ceRNA cross-talk in cancer: when ce-bling rivalries go awry. Cancer Discov. 2013;3:1113-1121. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 504] [Cited by in RCA: 681] [Article Influence: 56.8] [Reference Citation Analysis (0)] |
| 9. | Li D, Yang M, Liao A, Zeng B, Liu D, Yao Y, Hu G, Chen X, Feng Z, Du Y, Zhou Y, He J, Nie Y. Linc00483 as ceRNA regulates proliferation and apoptosis through activating MAPKs in gastric cancer. J Cell Mol Med. 2018;. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 34] [Cited by in RCA: 46] [Article Influence: 6.6] [Reference Citation Analysis (0)] |
| 10. | Kong X, Duan Y, Sang Y, Li Y, Zhang H, Liang Y, Liu Y, Zhang N, Yang Q. LncRNA-CDC6 promotes breast cancer progression and function as ceRNA to target CDC6 by sponging microRNA-215. J Cell Physiol. 2018;. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 93] [Cited by in RCA: 227] [Article Influence: 32.4] [Reference Citation Analysis (0)] |
| 11. | Wang Z, Jensen MA, Zenklusen JC. A Practical Guide to The Cancer Genome Atlas (TCGA). Methods Mol Biol. 2016;1418:111-141. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 219] [Cited by in RCA: 433] [Article Influence: 48.1] [Reference Citation Analysis (0)] |
| 12. | Deng M, Brägelmann J, Kryukov I, Saraiva-Agostinho N, Perner S. FirebrowseR: an R client to the Broad Institute's Firehose Pipeline. Database (Oxford). 2017;2017. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 96] [Cited by in RCA: 122] [Article Influence: 15.3] [Reference Citation Analysis (0)] |
| 13. | Miranda KC, Huynh T, Tay Y, Ang YS, Tam WL, Thomson AM, Lim B, Rigoutsos I. A pattern-based method for the identification of MicroRNA binding sites and their corresponding heteroduplexes. Cell. 2006;126:1203-1217. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1440] [Cited by in RCA: 1547] [Article Influence: 81.4] [Reference Citation Analysis (0)] |
| 14. | Huang da W, Sherman BT, Lempicki RA. Bioinformatics enrichment tools: paths toward the comprehensive functional analysis of large gene lists. Nucleic Acids Res. 2009;37:1-13. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 10350] [Cited by in RCA: 11166] [Article Influence: 656.8] [Reference Citation Analysis (0)] |
| 15. | Su ZD, Huang Y, Zhang ZY, Zhao YW, Wang D, Chen W, Chou KC, Lin H. iLoc-lncRNA: predict the subcellular location of lncRNAs by incorporating octamer composition into general PseKNC. Bioinformatics. 2018;34:4196-4204. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 98] [Cited by in RCA: 151] [Article Influence: 25.2] [Reference Citation Analysis (0)] |
| 16. | Agarwal V, Bell GW, Nam JW, Bartel DP. Predicting effective microRNA target sites in mammalian mRNAs. Elife. 2015;4. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 4013] [Cited by in RCA: 5411] [Article Influence: 541.1] [Reference Citation Analysis (0)] |
| 17. | Wong N, Wang X. miRDB: an online resource for microRNA target prediction and functional annotations. Nucleic Acids Res. 2015;43:D146-D152. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 1247] [Cited by in RCA: 1419] [Article Influence: 129.0] [Reference Citation Analysis (0)] |
| 18. | Paraskevopoulou MD, Georgakilas G, Kostoulas N, Vlachos IS, Vergoulis T, Reczko M, Filippidis C, Dalamagas T, Hatzigeorgiou AG. DIANA-microT web server v5.0: service integration into miRNA functional analysis workflows. Nucleic Acids Res. 2013;41:W169-W173. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 749] [Cited by in RCA: 914] [Article Influence: 76.2] [Reference Citation Analysis (0)] |
| 19. | Huang Y, Zhang J, Hou L, Wang G, Liu H, Zhang R, Chen X, Zhu J. LncRNA AK023391 promotes tumorigenesis and invasion of gastric cancer through activation of the PI3K/Akt signaling pathway. J Exp Clin Cancer Res. 2017;36:194. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 103] [Cited by in RCA: 164] [Article Influence: 20.5] [Reference Citation Analysis (0)] |
| 20. | Sun TT, He J, Liang Q, Ren LL, Yan TT, Yu TC, Tang JY, Bao YJ, Hu Y, Lin Y, Sun D, Chen YX, Hong J, Chen H, Zou W, Fang JY. LncRNA GClnc1 Promotes Gastric Carcinogenesis and May Act as a Modular Scaffold of WDR5 and KAT2A Complexes to Specify the Histone Modification Pattern. Cancer Discov. 2016;6:784-801. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 308] [Cited by in RCA: 313] [Article Influence: 34.8] [Reference Citation Analysis (0)] |
| 21. | Lü MH, Tang B, Zeng S, Hu CJ, Xie R, Wu YY, Wang SM, He FT, Yang SM. Long noncoding RNA BC032469, a novel competing endogenous RNA, upregulates hTERT expression by sponging miR-1207-5p and promotes proliferation in gastric cancer. Oncogene. 2016;35:3524-3534. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 128] [Cited by in RCA: 151] [Article Influence: 15.1] [Reference Citation Analysis (0)] |
| 22. | Moran VA, Perera RJ, Khalil AM. Emerging functional and mechanistic paradigms of mammalian long non-coding RNAs. Nucleic Acids Res. 2012;40:6391-6400. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 463] [Cited by in RCA: 525] [Article Influence: 40.4] [Reference Citation Analysis (0)] |
| 23. | Geisler S, Coller J. RNA in unexpected places: long non-coding RNA functions in diverse cellular contexts. Nat Rev Mol Cell Biol. 2013;14:699-712. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1015] [Cited by in RCA: 1154] [Article Influence: 96.2] [Reference Citation Analysis (0)] |
| 24. | Liu Q, Jiang J, Fu Y, Liu T, Yu Y, Zhang X. MiR-129-5p functions as a tumor suppressor in gastric cancer progression through targeting ADAM9. Biomed Pharmacother. 2018;105:420-427. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 22] [Cited by in RCA: 31] [Article Influence: 4.4] [Reference Citation Analysis (0)] |
| 25. | Wang Q, Yu J. MiR-129-5p suppresses gastric cancer cell invasion and proliferation by inhibiting COL1A1. Biochem Cell Biol. 2018;96:19-25. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 74] [Cited by in RCA: 103] [Article Influence: 12.9] [Reference Citation Analysis (0)] |
| 26. | Chen D, Wang H, Chen J, Li Z, Li S, Hu Z, Huang S, Zhao Y, He X. MicroRNA-129-5p Regulates Glycolysis and Cell Proliferation by Targeting the Glucose Transporter SLC2A3 in Gastric Cancer Cells. Front Pharmacol. 2018;9:502. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 35] [Cited by in RCA: 61] [Article Influence: 8.7] [Reference Citation Analysis (0)] |
| 27. | Hou DZ, Dai JS, Deng ZB, Yan DG. Overexpression of microRNA-490-5p targeting CDK1 suppresses proliferation and cell cycle progression of gastric cancer cells by inhibiting ERK signaling pathway. Journal of Modern Oncology. 2017;25:2565-2572. [DOI] [Full Text] |
| 28. | Mishra L, Katuri V, Evans S. The role of PRAJA and ELF in TGF-beta signaling and gastric cancer. Cancer Biol Ther. 2005;4:694-699. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 23] [Cited by in RCA: 24] [Article Influence: 1.2] [Reference Citation Analysis (0)] |
| 29. | Du W, Wang S, Zhou Q, Li X, Chu J, Chang Z, Tao Q, Ng EK, Fang J, Sung JJ, Yu J. ADAMTS9 is a functional tumor suppressor through inhibiting AKT/mTOR pathway and associated with poor survival in gastric cancer. Oncogene. 2013;32:3319-3328. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 73] [Cited by in RCA: 90] [Article Influence: 6.9] [Reference Citation Analysis (0)] |
| 30. | Du C, Li DQ, Li N, Chen L, Li SS, Yang Y, Hou MX, Xie MJ, Zheng ZD. DDX5 promotes gastric cancer cell proliferation in vitro and in vivo through mTOR signaling pathway. Sci Rep. 2017;7:42876. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 43] [Cited by in RCA: 47] [Article Influence: 5.9] [Reference Citation Analysis (0)] |
| 31. | Fu H, Wang C, Yang D, Wei Z, Xu J, Hu Z, Zhang Y, Wang W, Yan R, Cai Q. Curcumin regulates proliferation, autophagy, and apoptosis in gastric cancer cells by affecting PI3K and P53 signaling. J Cell Physiol. 2018;233:4634-4642. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 83] [Cited by in RCA: 126] [Article Influence: 18.0] [Reference Citation Analysis (0)] |