Published online Nov 15, 2019. doi: 10.4251/wjgo.v11.i11.998
Peer-review started: March 14, 2019
First decision: May 9, 2019
Revised: July 26, 2019
Accepted: August 27, 2019
Article in press: August 27, 2019
Published online: November 15, 2019
Processing time: 246 Days and 13.2 Hours
Toll-like receptors (TLRs) are the first line of host defense, and are involved in Helicobacter pylori (H. pylori) recognition and activation of both inflammatory and carcinogenic processes. The presence of single nucleotide polymorphisms (SNPs) in genes that activate the immune response may modulate the risk of precancerous lesions and gastric cancer (GC). Among them, Toll-like receptor 9 (TLR9) polymorphisms have emerged with a risk factor of infectious diseases and cancer, however the studies are still inconclusive.
To evaluate whether TLR9 rs5743836 and rs187084 SNPs contribute to the risk of gastric carcinogenesis, and its influence on mRNA expression.
A case-control study was conducted to evaluate two TLR9 SNPs (TLR9-1237 TC-rs5743836 and TLR9-1486 CT-rs187084) in chronic gastritis (CG) and GC patients. A total of 609 DNA samples of peripheral blood [248 CG, 161 GC, and 200 samples from healthy individuals (C)] were genotyped by polymerase chain reaction-restriction fragment length polymorphism. All samples were tested for the H. pylori infection using Hpx1 and Hpx2 primers. Quantitative polymerase chain reaction by TaqMan® assay was used to quantify TLR9 mRNA from fresh gastric tissues (48 GC, 26 CG, and 14 C).
For TLR9-1237, the TC + CC or CC genotypes were associated with a higher risk of GC than C [recessive model odds ratio (OR) = 5.01, 95% confidence interval (CI): 2.52-9.94, P < 0.0001], and the CG (recessive model OR =4.63; 95%CI: 2.44-8.79, P < 0.0001) groups. For TLR9-1486, an association between the CT + TT genotypes and increased risk of both GC (dominant model OR = 2.72, 95%CI: 1.57-4.72, P < 0.0001) and CG (dominant model OR = 1.79, 95%CI: 1.15-2.79, P = 0.0094) was observed when compared to the C group. Moreover, the presence of TLR9-1237 TC/CC + TLR9-1486 CC genotypes potentiate the risk for this neoplasm (OR = 18.57; 95%CI: 5.06-68.15, P < 0.0001). The TLR9 mRNA level was significantly higher in the GC group (RQ = 9.24, P < 0.0001) in relation to the CG group (RQ = 1.55, P = 0.0010) and normal mucosa (RQ = 1.0). When the samples were grouped according to the polymorphic genotypes and the presence of H. pylori infection, an influence of TLR9-1237 TC + CC polymorphic genotypes (P = 0.0083) and H. pylori infection (P < 0.0001) was observed on the upregulation of mRNA expression.
Our findings show that TLR9 rs5743836 and rs187084 polymorphisms are associated with a higher risk of carcinogenesis gastric, and that TLR9 mRNA levels can be modulated by TLR9-1237 TC + CC variant genotypes and H. pylori infection.
Core tip: This study investigated the influence of Toll-like receptor 9 (TLR9) polymorphisms on mRNA and Helicobacter pylori (H. pylori) infection in gastric cancer samples, and the association of these single nucleotide polymorphisms with the risk of developing this neoplasm. Increased expression of TLR9 in tumor tissue was observed compared with chronic gastritis and normal tissue. Moreover, when samples were grouped according to H. pylori presence, an upregulation of TLR9 mRNA expression was observed. Thus, both H. pylori infection as well as functional TLR9 polymorphisms may change gene expression levels, accentuating inflammation and aggravating the development of gastric cancer.
- Citation: Susi MD, Lourenço Caroline M, Rasmussen LT, Payão SLM, Rossi AFT, Silva AE, Oliveira-Cucolo JG. Toll-like receptor 9 polymorphisms and Helicobacter pylori influence gene expression and risk of gastric carcinogenesis in the Brazilian population.. World J Gastrointest Oncol 2019; 11(11): 998-1010
- URL: https://www.wjgnet.com/1948-5204/full/v11/i11/998.htm
- DOI: https://dx.doi.org/10.4251/wjgo.v11.i11.998
Toll-like receptors (TLRs) are a class of transmembrane receptors known as pattern recognition receptors. They consist of a family of eleven human proteins (TLR1 to TLR11) and are composed of extracellular, transmembrane, and intracellular regions[1]. The extracellular region consists of ten to thirty leucine-rich repeats, which detect conserved molecular patterns (PAMPs) shared by most microorganisms, including the Helicobacter pylori (H. pylori) bacterium, widely known as a class I carcinogen in gastric diseases[1,2]. Thus, the pattern of the host’s immune response beyond genetic and environmental factors is essential for understanding the pathology of gastric cancer (GC)[3].
H. pylori infection disrupts gastric homeostasis within the local mucosa and promotes the production of multiple inflammatory cytokines[4]. The inflammatory process can progress through a multi-step process, developing from chronic gastritis (CG) to gastric atrophy, intestinal metaplasia, dysplasia, and finally to GC[2].
After microorganism infection, TLRs are the first line of host defense. They are important components of innate immunity and are involved in the activation of the adaptive immune response[5]. Thus, during H. pylori infection, TLRs on gastric epithelial and immune cells recognize many PAMPs. Toll-like receptors 2 and 4 recognize lipopolysaccharides, Toll-like receptor 5 recognizes flagellin, and Toll-like receptor 9 (TLR9) recognizes unmethylated CpG oligonucleotides, which are abundant in bacterial DNA[6]. Based on the recognition and activation of these receptor signaling pathways, the inflammatory process and the presence of genetic polymorphisms may modulate the risk of developing precancerous gastric lesions and GC[5].
The activation of TLR9 requires a complex pathway, since it is located in the intracellular compartment of endosomes. It is therefore believed that unmethylated CpG oligonucleotides are transferred to the intracellular compartment through non-specific endocytosis[7].
Some TLR9 polymorphisms (rs352139, rs187084, rs41308230, and rs5743844) have been associated with infection-related diseases, such as tuberculosis, malaria, and lupus, as well as with liver cancer, GC, and colorectal cancer[5,7-11].
TLR9-1237 TC (rs5743836), which replaces tyrosine with cytosine within the proximal promoter region, has had its variant allele TLR9-1237 C associated with the development of premalignant gastric diseases induced by H. pylori[10,12]. However, another study on an Asian population did not detect an association between a risk for GC and H. pylori infection[9].
In the case of TLR9-1486 C/T (rs187084), which is located in the promoter region, it is not yet clear which alleles (C or T) are associated with susceptibility to disease. Some studies have indicated an association between the CT or CC genotypes and a greater risk of cervical cancer[12,13]. Conversely, a recent study found a protective association between the CT genotype and the risk of severe bronchiolitis[14].
Given inconsistent results, as well as the importance of these receptors in the immune response and the susceptibility to inflammatory diseases and cancer, new studies are needed. Thus, the aim of this study was to evaluate whether the TLR9-1237 TC (rs5743836) and TLR9-1486 CT (rs187084) polymorphisms (alone and in combination) are associated with a risk of CG and GC. In addition, we also evaluate the influence of both polymorphisms and H. pylori infection on TLR9 mRNA expression. Our results show that both single nucleotide polymorphisms (SNPs) already play a major role in the early stages of the gastric carcinogenic cascade.
Written informed consent for the collection of biological material (peripheral blood and gastric tissues) and access to medical records for research purposes was obtained from all individuals included in the study. The Research Ethics Committee of Sacred Heart University in Bauru, São Paulo, Brazil approved this study (Registration NO. 382.514).
First, a case-control study on CG, GC, and healthy individuals (C) was performed on a total of 609 DNA samples obtained from peripheral blood, which were genotyped for TLR9 polymorphisms (rs187084 and rs5743836). All of the samples included were obtained from patients with gastric complaints who were treated in the Gastroenterology Department of the Clinical Hospital of Marília Medical School in Marília-São Paulo or in the State Hospital of Bauru, São Paulo, Brazil. These patients received upper digestive endoscopies between January 2010 and March 2015. None of the subjects received chemotherapy drugs, radiotherapy, antibiotics, anti-inflammatory agents, or proton pump inhibitors before 30 days of endoscopy.
The CG group consisted of 248 patients (110 men and 138 women; mean age: 43.10 ± 21.79 years, range: 10-90 years) with a confirmed histopathological diagnosis of CG as per the Sidney System[15], while the GC group included 161 patients (111 men and 50 women; mean age: 63.33 ± 14.45 years, range: 23-90 years) with a confirmed histopathological diagnosis of GC as per Lauren’s classification[16]. The control group (C) consisted of 200 healthy individuals free of gastric diseases (97 men and 103 women; mean age: 49.58 ± 21.01 years, range: 10-90 years) who underwent gastric endoscopy and were confirmed negative for any gastric disease, as well as for H. pylori infection.
In addition, biopsies of fresh gastric tissues were also collected for quantification of TLR9 mRNA. Biopsies were mainly collected from the gastric antrum and corpus regions during endoscopic evaluation or gastric surgery of 48 patients with CG (29 men and 19 women; mean age: 53.10 ± 9.41 years, range: 41-84 years), 26 patients with GC (19 men and 7 women; mean age: 62.77 ± 14.20 years, range: 33-88 years), and 14 individuals who were H. pylori-negative and free of gastric diseases as tested by histopathology (9 men and 5 women; mean age: 49.58 ± 21.01 years, range: 10-92 years).
DNA from peripheral blood was extracted following the protocol by[17], while DNA from fresh gastric tissue was extracted using the QIAamp® tissue kit (Qiagen, Hilden, Germany) according to the manufacturer’s instructions and stored at -20 °C. The polymerase chain reaction-restriction fragment length polymorphism technique was used to identify the TLR9-1486 CT (rs187084) and TLR9-1237 TC (rs5743836) SNPs in all three groups. Table 1 summarizes the polymerase chain reaction (PCR) conditions, sets of primers, enzymes used in each assay, minor allele frequency (MAF), and location for both polymorphisms. Approximately 10% of the samples were processed in duplicate for quality control. The presence of H. pylori was detected using PCR where a 150 bp fragment was amplified from genomic DNA samples using Hpx1 and Hpx2 primers, as described in a previous study[18].
| Genes | Location | MAF | Primers, 5’-3’ | Cycles | T melting | Enzyme | Genotypes, bp |
| TLR9-1237 T/C (rs5743836) | Chromosome 3: 52226766 (intron variant) | 0.1725 (C) | F: TGGGAGCAGAGACATAATGGA | 35 | 60 °C | BstNI | CC = 60 + 48 + 27 |
| R: CTGCTTGCAGTTGACTGTGT | CT = 108 + 60 + 48 + 27 | ||||||
| TT = 108 + 27 | |||||||
| TLR9-1486 C/T (rs187084) | Chromosome 3: 52227015 (intron variant) | 0.3776 (C) | F: TCCCAGCAGCAACAATTCATTA | 30 | 60 °C | AflII | CC = 499 |
| R: CTGCTTGCAGTTGACTGTGT | TC = 499 + 327 + 172 | ||||||
| TT = 327 + 172 |
Total RNA was extracted using a RNeasy Mini Kit (Qiagen) according to the manufacturer’s protocol. RNA concentrations and quality were measured using the NanoDrop 2000 spectrophotometer (ThermoFisher Scientific, Waltham, MA, United States). The RNA samples were stored at 80°C and used for reverse transcription. cDNA was synthesized from 2.5 μg of total RNA using random primers and a High Capacity cDNA Archive Kit (Applied Biosystems, Foster City, CA, United States) according to the manufacturer’s instructions.
The relative expression levels of TLR9 mRNA were measured by real-time quantitative PCR based on the TaqMan® System (Life Technologies, Carlsbad, CA, United States) within the StepOne Plus Real-Time PCR system 2.2.3 (Applied Biosystems) using a specific TaqMan probe for TLR9 (H00002973_m1) and two reference genes, GUSB (Hs00187320_m1) and TBP (Hs00187332_m1) which were tested in a previous study[19]. A pool of normal H. pylori-negative gastric tissue samples was used as a calibrator. All reactions were performed in triplicate and included a negative reaction control. To determine the amplification efficacy of the probe, a standard curve was constructed with serial dilutions of cDNA samples (pool of pure cDNA and 1:5, 1:25, 1:125, and 1:625 dilutions). Relative quantification (RQ) was calculated using the comparative CT method (2(-∆∆Ct))[20], while using a pool of the normal mucosa samples as calibrator and reference genes for normalization. The data were expressed as median values.
We evaluated MAF and the Hardy-Weinberg equilibrium (HWE) using OEGE software[21]. Odds ratios (ORs) and 95% confidence intervals (CIs) were calculated for the polymorphisms under study using multiple logistic regression models adjusted for age, gender, and H. pylori infection. ORs were calculated using the SNPStat program, which considered the dominant model (major allele homozygotes vs heterozygotes + minor allele homozygotes), the recessive model (major allele homozygotes + heterozygotes vs minor allele homozygotes), and the log-additive model (major allele homozygotes vs heterozygotes vs minor allele homozygotes for the two polymorphisms). The haplotype frequencies of TLR9 were inferred using the Haploview program, version 4.0. Data regarding the relative expression of mRNA were expressed as a median to determine possible associations between the differences in relative gene expression in each group. The distribution of continuous data was evaluated using the D’Agostino and Pearson omnibus normality test. Wilcoxon Signed Rank test was used to compare the expression values between the CG and GC groups in relation to normal mucosa. The Mann-Whitney test was used to analyze the influence of the factors as variant genotypes and H. pylori infection on TLR9 mRNA expression. Statistical analyses were performed using the GraphPad Prism 5 software, version 5.01, and the SPSS program, version 11.5. A probability level (P) of < 0.05 was considered significant.
The rs5743836 and rs187084 SNPs were found not to be in HWE in the GC (P < 0.0001), CG (P < 0.0001), and C (P = 0.0008) groups. The results of the genotype and allele frequencies for the TLR9-1237 TC and TLR9-1486 CT polymorphisms in all three groups (GC vs C, CG vs C, and GC vs CG) are shown in Table 2.
| Polymorp-hisms | Models | Genotypes/ Alleles | C, n = 200 | Cases | ||||
| GC, n = 161 | OR (95%CI), P value | CG, n = 248 | OR (95%CI), P value | |||||
| TLR9-1237 T/C (rs5743836) | Dominant | TT | 148 (74%) | 78 (48.5%) | 1.00 | 188 (75.8%) | 1.00 | |
| TC + CC | 52 (26%) | 83 (51.5%) | 3.03 (1.95-4.71), < 0.0001 | 60 (24.2%) | 0.91 (0.59-1.40), 0.6600 | |||
| Recessive | TT + TC | 188 (94%) | 122 (75.8%) | 1.00 | 232 (93.5%) | 1.00 | ||
| CC | 12 (6%) | 39 (24.2%) | 5.01 (2.52-9.94), < 0.0001 | 16 (6.5%) | 1.08 (0.50-2.34), 0.8400 | |||
| Log-additive | 2.37 (1.74-3.23), < 0.0001 | 0.96 (0.70-1.32), 0.8100 | ||||||
| Alleles | T | 336 (0.84) | 200 (0.62) | 1.00 | 420 (0.85) | 1.00 | ||
| C | 64 (0.16) | 122 (0.38) | 3.20 (2.25-4.54), < 0.0001 | 76 (0.15) | 0.95 (0.66-1.36), 0.7822 | |||
| GC vs CG | Dominant | 3.46 (2.23-5.37), < 0.0001 | ||||||
| OR (95%CI), P | Recessive | 4.63 (2.44-8.79), < 0.0001 | ||||||
| Log-additive | 2.46 (1.82-3.34), < 0.0001 | |||||||
| Alleles | 3.37 (2.41-4.70), < 0.0001 | |||||||
| TLR9-1486 C/T (rs187084) | Dominant | CC | 58 (29%) | 21 (13%) | 1.00 | 46 (18.6%) | 1.00 | |
| CT + TT | 142 (71%) | 140 (87%) | 2.72 (1.57-4.72), < 0.0001 | 202 (81.5%) | 1.79 (1.15-2.79), 0.0094 | |||
| Recessive | CC + CT | 164 (82%) | 122 (75.8%) | 1.00 | 182 (73.4%) | 1.00 | ||
| TT | 36 (18%) | 39 (24.2%) | 1.46 (0.87-2.43), 0.1500 | 66 (26.6%) | 1.65 (1.05-2.61), 0.0300 | |||
| Log-additive | 1.70 (1.23-2.37), 0.0013 | 1.52 (1.15-2.02), 0.0030 | ||||||
| Alleles | C | 222 (0.56) | 143 (0.44) | 1.00 | 228 (0.46) | 1.00 | ||
| T | 178 (0.44) | 179 (0.56) | 1.56 (1.16-2.09), 0.0035 | 268 (0.54) | 1.46 (1.12-1.91), 0.0048 | |||
| GC vs CG | Dominant | 1.15 (0.72-1.84), 0.5700 | ||||||
| OR (95%CI), P | Recessive | 0.69 (0.39-1.24), 0.2100 | ||||||
| Log-additive | 0.95 (0.69-1.31), 0.7500 | |||||||
| Alleles | 0.93 (0.70-1.24), 0.6670 | |||||||
For TLR9-1237 TC, both TC + CC and solely CC genotype were associated with a risk of GC in all three evaluated models, whether compared to each other or relative to the C group dominant model (OR = 3.03, 95%CI: 1.95-4.71, P < 0.0001), recessive model (OR = 5.01, 95%CI: 2.52-9.94, P < 0.0001), and log-additive model (OR = 2.37, 95%CI: 1.74-3.23, P < 0.0001)], as well as compared to the CG group [dominant model (OR = 3.46, 95%CI: 2.23-5.37, P < 0.0001), recessive model (OR = 4.63, 95%CI: 2.44-8.79, P < 0.0001), and log-additive model (OR = 2.46, 95%CI: 1.82-3.34, P < 0.0001)]. However no association was found between CG vs C [dominant model (OR = 0.91, 95%CI: 0.59-1.40, P = 0.6600), recessive model (OR = 1.08, 95%CI: 0.50-2.34, P = 0.8400), and log-additive model (OR = 0.96, 95%CI: 0.70-1.32, P = 0.8100)] (Table 2).
For TLR9-1486 CT, the CT + TT polymorphic genotypes were also associated with risk of GC in the dominant model (OR = 2.72, 95%CI: 1.57-4.72), P < 0.0001) and in the log-additive model (OR = 1.70, 95%CI: 1.23-2.37, P = 0.0013) relative to the C group. Additionally, between the CG and C groups, we observed a higher risk associated for the CT + TT and also the TT genotype in the three evaluated models [dominant model (OR = 1.79, 95%CI: 1.15-2.79, P = 0.0094), recessive model (OR = 1.65, 95%CI: 1.05-2.61, P = 0.0300), and log-additive model (OR = 1.52, 95%CI: 1.15-2.02, P = 0.0030)]. However, no statistically significant difference was observed between the two case groups (Table 2).
The TLR9 haplotype analysis (Table 3) revealed a higher frequency of the two wild alleles (T-C haplotype) in the C group than in the GC group (47.7 and 35.2, respectively; P ≤ 0.0001), or in the CG group (48.4 and 40.5, respectively; P = 0.0178). Similarly, a higher frequency of T-C haplotype was also found in the CG group than in the GC group (39.7 and 24.9, respectively; P ≤ 0.0001). However, the opposite was observed for the variant haplotype C-T, which was more frequent in the GC group than in the C group (18.7 and 8.2, respectively; P ≤ 0.0001), as well as in relation to the CG group (18.3 and 9.0, respectively; P ≤ 0.0001).
| Haplotypes | GC, % | C, % | X2 | P value | CG, % | C, % | X2 | P value | GC, % | CG, % | X2 | P value |
| TLR9-1237/-1486 | ||||||||||||
| T-C | 35.2 | 47.7 | 38.284 | ≤ 0.0001 | 40.5 | 48.4 | 5.617 | 0.0178 | 24.9 | 39.7 | 19.098 | ≤ 0.0001 |
| T-T | 36.9 | 36.3 | 0.024 | 0.8772 | 44.2 | 35.6 | 6.778 | 0.0092 | 37.3 | 45.0 | 4.838 | 0.0278 |
| C-T | 18.7 | 8.2 | 17.656 | ≤ 0.0001 | 9.8 | 8.9 | 0.233 | 0.6290 | 18.3 | 9.0 | 15.335 | ≤ 0.0001 |
| C-C | 19.2 | 7.8 | 20.475 | ≤ 0.0001 | 5.5 | 7.1 | 1.000 | 0.3158 | 19.6 | 6.3 | 33.560 | ≤ 0.0001 |
In another statistical analysis, when we evaluated the different combinations between the genotypes of the two polymorphisms (TLR9-1237 TC + TLR9-1486 CT), OR was found to be higher in the two case groups relative to the control group, as well as in the GC group relative to the CG group (Table 4). The highest OR values were observed for the combination TLR9-1237 TC/CC + TLR9-1486 CC when the GC group was compared to the C group (OR = 18.57, CI: 5.06-68.15, P < 0.0001), and when the GC group was compared to the CG group (OR = 10.08, CI: 2.96-34.24, P = 0.0002). Elevated OR values were also observed for the combination TLR9-1237 TC/CC + TLR9-1486 CT/TT when the GC group was compared to the C group (OR = 10.79, CI: 4.50-25.83, P < 0.0001).
| Risk genotype | Groups | OR (95%CI), P value | |||||
| TLR9-1237 | TLR9-1486 | C, n = 200 | GC, n = 161 | CG, n = 248 | GC × C | CG × C | GC × CG |
| TT | CC | 50 | 7 | 38 | 1.00 | 1.00 | 1.00 |
| TC/CC | CC | 5 | 13 | 7 | 18.57 (5.06-68.15), < 0.0001 | 1.842 (0.54-6.25), 0.3665 | 10.08 (2.96-34.24). 0.0002 |
| TT | CT/TT | 98 | 70 | 150 | 5.102 (2.18-11.92), < 0.0001 | 7.737(4.72-12.66), < 0.0001 | 2.533 (1.07-5.95), 0.0307 |
| TC/CC | CT/TT | 47 | 71 | 53 | 10.79 (4.50-25.83), < 0.0001 | 1.484 (0.83-1.64), 0.1911 | 7.272 (3.01-17.56), < 0.0001 |
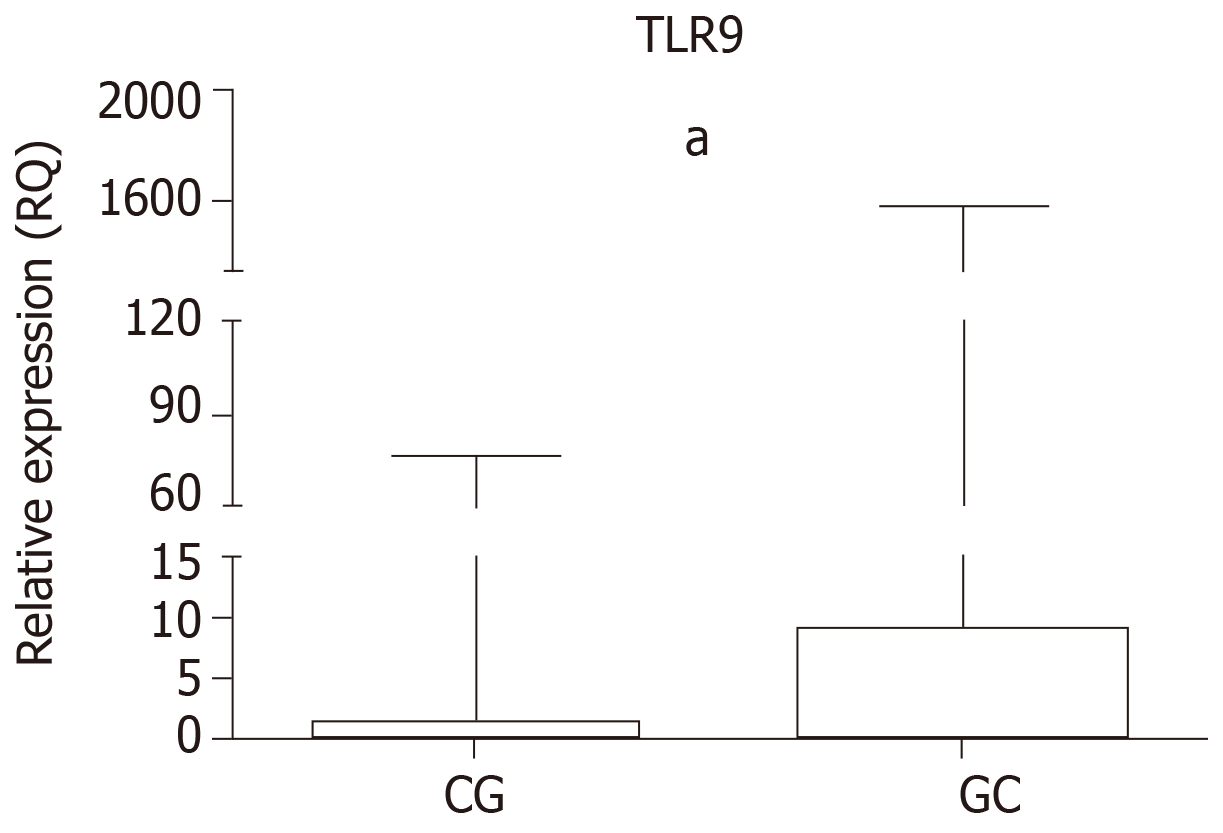
The relative expression of TLR9 in the GC and CG groups is shown in Table 5 and Figure 1. Higher mRNA expression was observed in the GC group (median RQ = 9.24, P < 0.0001) and the CG group (median RQ = 1.55, P = 0.0010) relative to normal mucosa, as was significant upregulation in the GC group relative to the CG group (P < 0.0001).
| Variables | Cases | |
| GC, n = 23 | CG, n = 48 | |
| Median | 9.24 | 1.55 |
| Range | 3.64-24.06 | 1.04-3.87 |
| P value | < 0.00011 | |
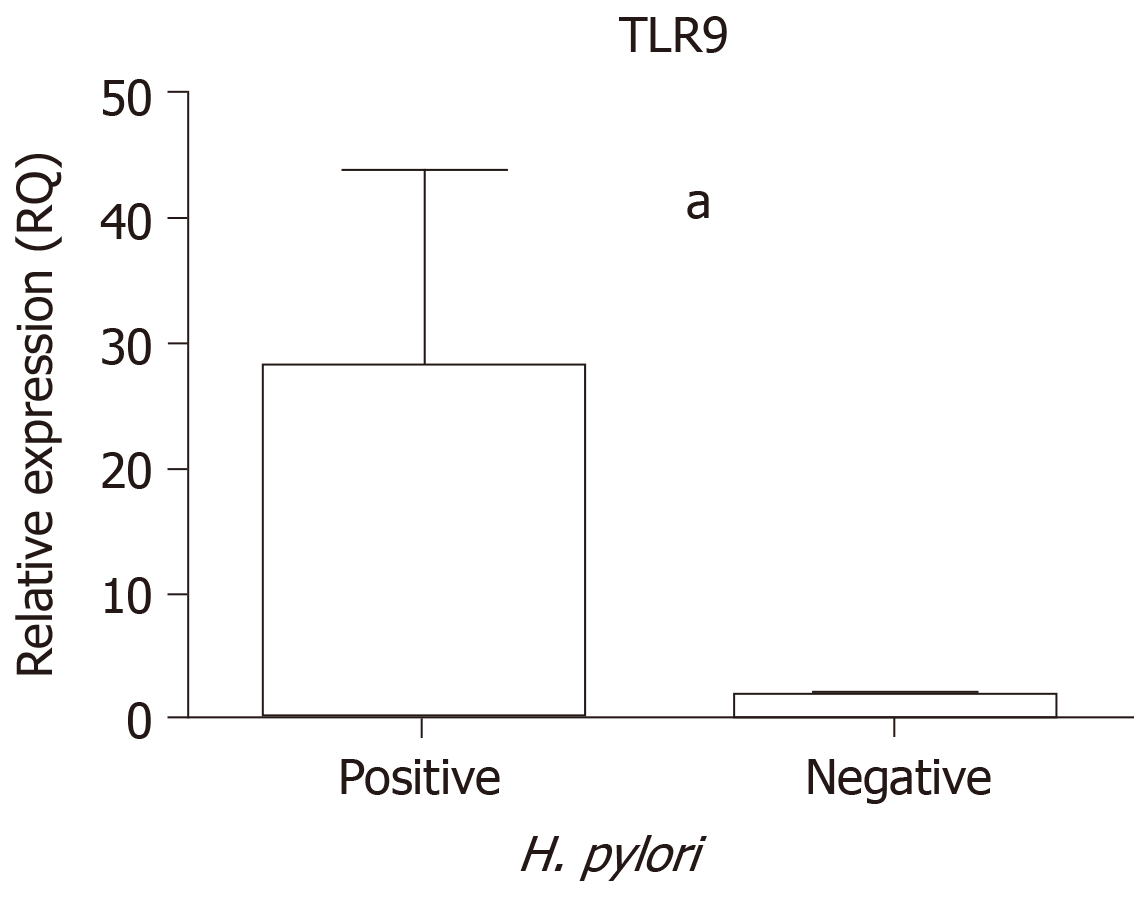
In addition, an analysis was performed to determine the influence of H. pylori infection on TLR9 mRNA levels. The GC and CG tissue samples were considered together to compare the positive and negative H. pylori samples, and significantly higher TLR9 expression was demonstrated among samples that were H. pylori-positive (median RQ = 2.85) than among H. pylori-negative samples (median RQ = 0.86, P < 0.0001; Figure 2).
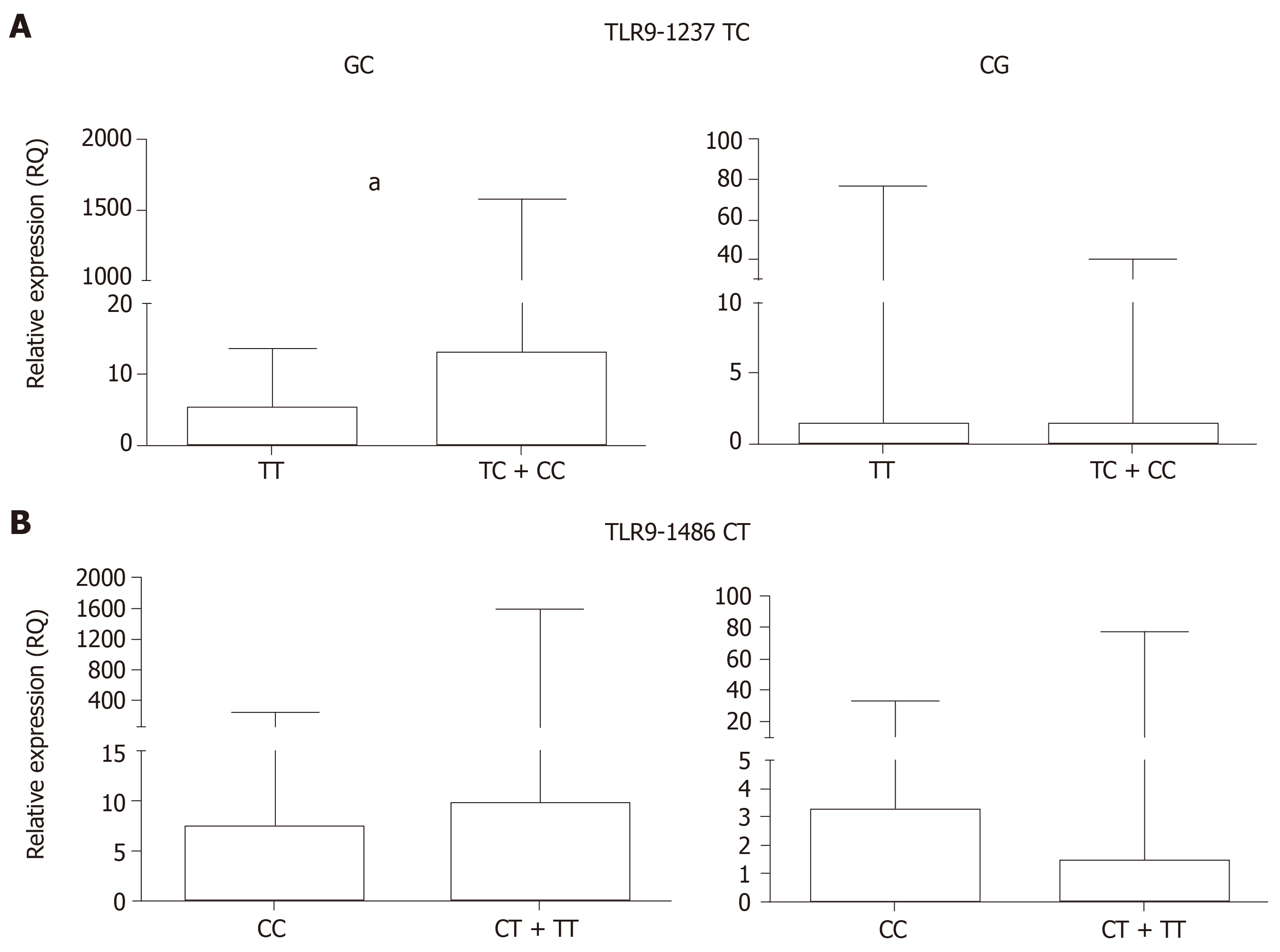
Posteriorly, the GC and CG samples were grouped according to the polymorphic genotypes for TLR9-1237T/C and -1486 C/T in order to assess whether these polymorphisms affect TLR9 mRNA expression levels (Table 6 and Figure 3).
| Groups | TLR9-1237T/C (rs5743836) | P value | TLR9-1486 C/T (rs187084) | P value | |||
| TT | TC/CC | CC | CT / TT | ||||
| Upregulated cases | Upregulated cases | ||||||
| GC (n = 26) | Median | 5.24 | 13.01 | 0.0083 | 7.37 | 9.90 | 0.8763 |
| Range | 1.72-9.27 | 7.55-189.00 | 5.74-14.16 | 2.49-53-75 | |||
| CG (n = 48) | Median | 1.47 | 1.45 | 0.8442 | 3.26 | 1.50 | 0.1515 |
| Range | 0.77-4.19 | 0.75-3.82 | 1.39-8.46 | 0.75-3.00 | |||
In the GC group, individuals carrying the TLR9-1237 TC + CC polymorphic genotype exhibited higher mRNA expression levels (RQ = 13.01) than the TT wild-type genotype carriers (RQ = 5.24, P = 0.0083). However, in the CG group, differences between the TC + CC and TT wild-type genotypes were not statistically significant (P = 0.8442). For TLR9-1486 CT, in both the GC and the CG groups, when the individuals were grouped according to T allele carriers (CT + TT) and as CC wild-type homozygous, the differences were not significant (P = 0.8763 and 0.1515, respectively).
Our findings show that the functional polymorphisms TLR9-1237 T/C- (rs5743836) and TLR9-1486 C/T (rs187084) receptors are associated with an increased risk of developing premalignant and malignant gastric diseases in this Brazilian population. Therefore, they may act as a potential factor in the progression of gastric carcinogenesis. TLR9 mRNA expression levels are upregulated in GC tissues, and are modulated by both H. pylori infection and the presence of TLR9-1237 TC + CC variant genotypes.
In our analyses, the case and control groups were not found to be in HWE; however, this disequilibrium does not invalidate the association observed in this study, since the groups are part of the same population. It is important to note that this theorem assumes an ideal population in which allele and genotype frequencies remain constant from generation to generation in the absence of other evolutionary influences. In genes such as those involved in the control of inflammatory and immunity diseases or cancer, this imbalance in HWE may appear to be associated with the selection of a particular allele, which allows for more effective protection or response[21].
The first meta-analysis to investigate the association of these two TLR9 polymorphisms with the susceptibility to cancer evaluated 11 studies on various ethnic groups, and failed to demonstrate any influence of rs187084 or rs5743836 SNPs. This study did, however, indicate that rs352140, in the TLR9 2848 C/T gene, was associated with an increased cancer risk in Caucasians[22]. One study in a Mexican population also did not find an association of TLR9 1237 T/C SNP with gastroduodenal diseases, yet found that the TLR9 2848 G allele increased the risk of duodenal ulcers and H. pylori infection[23]. Thus, despite that both cited studies differed from our TLR9 1237 T/C SNP results, it is necessary to investigate TLR9 2848 C/T (rs352140) in our Brazilian population in future analyses.
Another study analyzed the influence of three TLR9 polymorphisms on multiple cancers, and in the case of the TLR9-1237 TC (rs5743836) SNP, found a protective effect of the C variant allele on digestive system tumors and breast cancer. In contrast to the SNP TLR9-1486 CT (rs187084), there was a high risk for C wild allele carriers in cancer development, particularly in the case of cervical cancer in both the Chinese[13,24] and Caucasian populations[25]. Tian et al[13] stratified the population by race to eliminate heterogeneity, and demonstrated for the rs187084 SNP that the influence of heterogeneity decreased when stratified only among the Chinese population. These results are inconsistent with those found in the present study, and thus reflect the complexity of studies on genetic associations with predisposition to diseases, since it involves several factors, such as ethnic origin, lifestyle, gene-environment interactions, and sample size[14,26-28].
On the other hand, Gębura et al[29] found results similar to ours in patients with rheumatoid arthritis. They evaluated certain SNPs on TLRs in a Caucasian population, and were able to demonstrate a positive association between the TLR9-1237 C and TLR9-1486 T variants and susceptibility to the development of rheumatoid arthritis. However, TLR9-1486 T was found to be associated with the disease only among women. Similarly, in cervical cancer and pre-neoplastic cervical lesions, Martínez-Campos et al[26] found a positive association between the TLR9-1486 TT genotype and the diseases, but did not detect any association between the TLR9-1237 TC SNP and the lesions evaluated.
In a Brazilian population, a single study that assessed various SNPs, among them the TLR9-1486 C/T, reported a protective association between the TLR9-1486 C allele or CT genotype and the severity of bronchiolitis caused by rhinovirus, with a higher expectation of intensive care hospitalization[14].
Few studies have investigated the association between TLR9 polymorphisms and the risk of GC, and most of those have focused on the Asian populations [9,10,27,30]. Liu et al[30] failed to demonstrate an association between TLR9-1486 CT SNPs and GC risk; meanwhile, Wang et al[9] were able to demonstrate an association between both the CT and CC genotypes of TLR9-1486 and a higher risk of GC, as well as an association with the presence of H. pylori. The authors suggest that TLR9-1486 C carriers are associated with an increased risk and poor prognosis of gastric carcinoma in the Chinese population. However, they could not find any individuals with the C allele for the TLR9-1237 T/C polymorphism, and all individuals were genotyped as TLR9-1237 TT. Conversely, Ng et al[10] found an association between the C variant allele of TLR9-1237 and the development of H. pylori-induced premalignant gastric lesions.
We were able to demonstrate that variant haplotypes (C-T) for the TLR9 -1237 and -1486 SNPs were more frequent in the GC and CG groups than the control group. Moreover, the combination of two risk alleles,homozygous or heterozygous (TLR9-1237 TC/CC + TLR9-1486 CT/TT) also conferred higher ORs, a result which suggests a synergistic or addictive effect of these variant alleles and the risk of gastric lesions. Ashton et al[31] performed the haplotype analysis for TLR9 (TLR9-1237 TC/CC + TLR9-1486 CT/TT) in endometrial cancer, but found a higher presence of the C-C combination in the case group than in the C group. In this population, the TLR9-1486 C allele was considered the least frequent variant. Another study that evaluated the frequency of four SNPs on TLR9 (rs187084, rs352139, rs5743836, and rs352140) found a significantly higher frequency of CATT in patients with lupus erythematous relative to control groups[32].
Despite this variability of the effect of wild-type and polymorphic alleles on the risk of inflammatory diseases and cancer, Omar et al[33] were able to demonstrate the synergistic effect of the TLR9 TTAG (-1486 T, -1237 T, + 1174 A, + 2848 G) haplotype on promoter activity. This TTAG haplotype was significantly associated with protection against malaria, and these findings were strongly supported by the result of luciferase reporter gene expression assay, which showed a significantly higher promoter activity for the TTA haplotype relative to the CCG, CTG, and TCG haplotypes. Therefore, the presence of the wild-type or variant alleles may influence the levels of TLR9 mRNA.
When we analyzed the TLR9 mRNA expression, we also found significantly higher levels of TLR9 mRNA in GC and CG tissues, as well as upregulation of TLR9 expression in the H. pylori-positive samples, regardless of the type of lesion (cancer or gastritis). Moreover, we observed that carriers of the TLR9-1237 TC + CC genotypes also exhibit higher TLR9 expression levels than the TT genotype, a finding which supports the hypothesis that infection by H. pylori triggers inflammatory responses mediated by TLR receptors, the expression of which may be affected by the occurrence of polymorphisms in their promoter regions.
The TLR9-1237 TC (rs5743836) SNP is located in the promoter region, and an in silico study found that the C variant allele creates an alternative NF-κB binding site, which may be functionally relevant. It is believed to enhance the transcriptional activation of TLR9 and potentially affect CpG-DNA activation of pro-inflammatory cytokines, consequently exacerbating the inflammatory response[10]. Moreover, functional studies have demonstrated that C variant allele carriers exhibit significantly higher luciferase activity, indicating that rs5743836 regulates TLR9 transcriptional activity[10].
Mutations in the promoter region have been predicted to affect the expression or stability of regulatory regions[34], a theory which would explain the higher expression levels of TLR9 mRNA in GC tissues, particularly for TLR9-1237 C allele carriers.
The presence of H. pylori in the gastric mucosa is a preponderant factor for carcinogenesis. The vast majority of these host colonies are free-living, but it is known that this bacterium can bind strongly to gastric epithelial cells and induce humoral and cellular immune responses, and may also cause ultra-structural changes in these cells[28]. However, H. pylori lipopolysaccharide is not a good immune system activator, so the creation of an alternative NF-κB binding site generated by the presence of TLR9-1237 C variant allele would cause a greater inflammatory response and facilitate the carcinogenic process[35].
Regarding TLR9-1486 C/T (rs187084), an in silico analysis showed that this SNP created a putative Sp1 binding site, which may be functionally relevant[36]. Recently, Martínez-Campos et al[26] found both upregulation of TLR9 mRNA in peripheral blood mononuclear cells and a higher concentration of TLR9 protein in the sera of patients diagnosed with uterine cervical cancer and human papillomavirus. However, no significant difference was observed in TLR9 expression levels among CC, TC, and TT carriers of TLR9-1486 SNP, a finding which was likely due to the limited sample size.
In conclusion, our results demonstrate that TLR9-1237 TC + CC and TLR9-1486 TC + TT, whether alone or in combination, are associated with risk of gastric carcinogenesis in the Brazilian population. Moreover, the results suggest that upregulation of TLR9 mRNA levels in premalignant and malignant gastric tissues may be influenced by TLR9-1237 TC + CC variant genotypes. The pattern of the host’s immune response, along with genetic and environmental factors, are essential for understanding the pathology of GC. Thus, polymorphisms in genes that affects its expression, such as TLR9, could have an effect on the development and clinical manifestation of disease.
Gastric cancer (GC) is one of the most prevalent cancers worldwide, with high rates of incidence and death. It ranks sixth in the world and is the fifth leading cause of cancer-related deaths worldwide. In Brazil, GC is the fourth most frequent type of cancer in men, and the sixth in women, with an estimated incidence of 21,290 new cases in 2018. The main risk factor associated with this type of cancer is the infection by the bacterium Helicobacter pylori (H. pylori). Toll-like receptors are the first line of host defense, and are involved in H. pylori recognition and activation of the inflammatory and carcinogenic process. The presence of single nucleotide polymorphisms (SNPs) in genes that activate the immune response may modulate the risk of precancerous lesions and GC, among them of which is TLR9 polymorphisms.
Polymorphisms in toll-like receptors genes have emerged with a risk factor of infectious diseases and cancer. However, the literature presents conflicting results. Therefore, several studies are needed to assess and confirm the real role among factors that influence changes in immune inflammatory processes related to GC, especially when it is a mixed population such as the Brazilian population.
Considering the inconsistent results and the importance of these receptors in the immune response and to susceptibility to inflammatory diseases and cancer, new studies are needed. Thus, the aim of this study was to evaluate whether the TLR9-1237 TC (rs5743836) and TLR9-1486 CT (rs187084) polymorphisms (alone and in combination) are associated with a risk of chronic gastritis (CG) and GC. In addition, we also evaluate the influence of both polymorphisms and H. pylori infection on TLR9 mRNA expression. The results may highlight important polymorphisms that act on gastric carcinogenesis.
A case-control study was conducted to evaluate two TLR9 SNPs (TLR9-1237 TC-rs5743836 and TLR9-1486 CT-rs187084) in CG and GC patients. A total of 609 DNA samples of peripheral blood [248 CG, 161 GC, and 200 samples from healthy individuals (C)] were genotyped by polymerase chain reaction-restriction fragment length polymorphism. All samples were tested for the H. pylori infection using Hpx1 and Hpx2 primers. Quantitative polymerase chain reaction by TaqMan® assay was used to quantify TLR9 mRNA from fresh gastric tissues (48 GC, 26 CG, and 14 C).
The data showed that for TLR9-1237, the TC + CC or CC genotypes were associated with a higher risk of GC than the C and CG groups. For TLR9-1486, an association between the CT + TT genotypes and increased risk of both GC and CG was observed when compared to the C group. Moreover, the presence of TLR9-1237 TC/CC + TLR9-1486 CC genotypes potentiates the risk for this neoplasm. The TLR9 mRNA level was significantly higher in the GC group in relation to the CG group and normal mucosa. When the samples were grouped according to the polymorphic genotypes and the presence of H. pylori infection, an influence of TLR9-1237 TC + CC polymorphic genotypes and H. pylori infection was observed on the upregulation of mRNA expression.
Our findings highlight that the functional polymorphisms of the TLR9-1237 T/C (rs5743836) and TLR9-1486 C/T (rs187084) receptors are associated with an increased risk of developing premalignant and malignant gastric diseases in this Brazilian population, and therefore may act as a potential factor in the progression of gastric carcinogenesis. TLR9 mRNA expression levels are upregulated in GC tissues and are modulated by both H. pylori infection and the presence of TLR9-1237 TC + CC-variant genotypes. The pattern of the host’s immune response, along with genetic and environmental factors, are essential for understanding the pathology of GC. Thus, polymorphisms in genes that affect its expression, such as TLR9, could have an effect on the development and clinical manifestation of disease.
Considering that most cases of GC have a good prognosis and are treatable when diagnosed at an early stage, it is of the utmost importance to establish molecular markers capable of identifying risk groups and providing early diagnosis in individuals with increased risk of developing this neoplasm. Overall, our results indicate that the TLR9 gene plays an important role in gastric carcinogenesis, highlighting the importance of the TLR9-1237 T/C (rs5743836) polymorphism in increasing gene expression and H. pylori infection, possibly triggering a stronger inflammatory response, which in turn enhances the risk of tumor progression. In the future, it would be important to investigate another polymorphism in the TLR9 gene [TLR9-2848 C/T (rs352140)], described in the literature as associated with cancer, but not yet analyzed in our Brazilian population.
The authors are grateful to Joice Matos Biselli and Profa Dra Lilian Castiglioni for her support in the statistical analysis.
Manuscript source: Invited manuscript
Specialty type: Oncology
Country of origin: Brazil
Peer-review report classification
Grade A (Excellent): 0
Grade B (Very good): 0
Grade C (Good): C, C
Grade D (Fair): 0
Grade E (Poor): 0
P-Reviewer: Romo-Gonzalez C, Islek A S-Editor: Tang JZ L-Editor: Filipodia E-Editor: Qi LL
| 1. | Varga MG, Peek RM. DNA Transfer and Toll-like Receptor Modulation by Helicobacter pylori. Curr Top Microbiol Immunol. 2017;400:169-193. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 14] [Cited by in RCA: 20] [Article Influence: 2.5] [Reference Citation Analysis (0)] |
| 2. | Correa P. The biological model of gastric carcinogenesis. IARC Sci Publ. 2004;301-310. [PubMed] |
| 3. | Song M, Rabkin CS, Camargo MC. Gastric Cancer: an Evolving Disease. Curr Treat Options Gastroenterol. 2018;16:561-569. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 25] [Cited by in RCA: 32] [Article Influence: 4.6] [Reference Citation Analysis (0)] |
| 4. | Cadamuro AC, Rossi AF, Maniezzo NM, Silva AE. Helicobacter pylori infection: host immune response, implications on gene expression and microRNAs. World J Gastroenterol. 2014;20:1424-1437. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in CrossRef: 80] [Cited by in RCA: 88] [Article Influence: 8.0] [Reference Citation Analysis (2)] |
| 5. | Wang TR, Peng JC, Qiao YQ, Zhu MM, Zhao D, Shen J, Ran ZH. Helicobacter pylori regulates TLR4 and TLR9 during gastric carcinogenesis. Int J Clin Exp Pathol. 2014;7:6950-6955. [PubMed] |
| 6. | Fukata M, Abreu MT. Role of Toll-like receptors in gastrointestinal malignancies. Oncogene. 2008;27:234-243. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 148] [Cited by in RCA: 140] [Article Influence: 8.2] [Reference Citation Analysis (0)] |
| 7. | Fűri I, Sipos F, Germann TM, Kalmár A, Tulassay Z, Molnár B, Műzes G. Epithelial toll-like receptor 9 signaling in colorectal inflammation and cancer: clinico-pathogenic aspects. World J Gastroenterol. 2013;19:4119-4126. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in CrossRef: 34] [Cited by in RCA: 32] [Article Influence: 2.7] [Reference Citation Analysis (0)] |
| 8. | Lopes JA, Borges-Canha M, Pimentel-Nunes P. Innate immunity and hepatocarcinoma: Can toll-like receptors open the door to oncogenesis? World J Hepatol. 2016;8:162-182. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 19] [Cited by in RCA: 24] [Article Influence: 2.7] [Reference Citation Analysis (0)] |
| 9. | Wang X, Xue L, Yang Y, Xu L, Zhang G. TLR9 promoter polymorphism is associated with both an increased susceptibility to gastric carcinoma and poor prognosis. PLoS One. 2013;8:e65731. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 25] [Cited by in RCA: 26] [Article Influence: 2.2] [Reference Citation Analysis (0)] |
| 10. | Ng MT, Van't Hof R, Crockett JC, Hope ME, Berry S, Thomson J, McLean MH, McColl KE, El-Omar EM, Hold GL. Increase in NF-kappaB binding affinity of the variant C allele of the toll-like receptor 9 -1237T/C polymorphism is associated with Helicobacter pylori-induced gastric disease. Infect Immun. 2010;78:1345-1352. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 74] [Cited by in RCA: 92] [Article Influence: 5.8] [Reference Citation Analysis (0)] |
| 11. | Messaritakis I, Stogiannitsi M, Koulouridi A, Sfakianaki M, Voutsina A, Sotiriou A, Athanasakis E, Xynos E, Mavroudis D, Tzardi M, Souglakos J. Evaluation of the detection of Toll-like receptors (TLRs) in cancer development and progression in patients with colorectal cancer. PLoS One. 2018;13:e0197327. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 21] [Cited by in RCA: 37] [Article Influence: 5.3] [Reference Citation Analysis (0)] |
| 12. | Chen X, Wang S, Liu L, Chen Z, Qiang F, Kan Y, Shen Y, Wu J, Shen H, Hu Z. A genetic variant in the promoter region of Toll-like receptor 9 and cervical cancer susceptibility. DNA Cell Biol. 2012;31:766-771. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 33] [Cited by in RCA: 35] [Article Influence: 2.5] [Reference Citation Analysis (0)] |
| 13. | Tian S, Zhang L, Yang T, Wei X, Zhang L, Yu Y, Li Y, Cao D, Yang X. The Associations between Toll-Like Receptor 9 Gene Polymorphisms and Cervical Cancer Susceptibility. Mediators Inflamm. 2018;2018:9127146. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 9] [Cited by in RCA: 11] [Article Influence: 1.6] [Reference Citation Analysis (0)] |
| 14. | Alvarez AE, Marson FAL, Bertuzzo CS, Bastos JCS, Baracat ECE, Brandão MB, Tresoldi AT, das Neves Romaneli MT, Almeida CCB, de Oliveira T, Schlodtmann PG, Corrêa E, de Miranda MLF, Dos Reis MC, De Pieri JV, Arns CW, Ribeiro JD. Association between single nucleotide polymorphisms in TLR4, TLR2, TLR9, VDR, NOS2 and CCL5 genes with acute viral bronchiolitis. Gene. 2018;645:7-17. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 18] [Cited by in RCA: 22] [Article Influence: 2.8] [Reference Citation Analysis (0)] |
| 15. | Dixon MF, Genta RM, Yardley JH, Correa P. Classification and grading of gastritis. The updated Sydney System. International Workshop on the Histopathology of Gastritis, Houston 1994. Am J Surg Pathol. 1996;20:1161-1181. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 3221] [Cited by in RCA: 3543] [Article Influence: 122.2] [Reference Citation Analysis (3)] |
| 16. | Lauren P. The two histological main types of gastric carcinoma: diffuse and so-called intestinal-type carcinoma. an attempt at a histo-clinical classification. Acta Pathol Microbiol Scand. 1965;64:31-49. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 4011] [Cited by in RCA: 4310] [Article Influence: 148.6] [Reference Citation Analysis (0)] |
| 17. | Miller SA, Dykes DD, Polesky HF. A simple salting out procedure for extracting DNA from human nucleated cells. Nucleic Acids Res. 1988;16:1215. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 13387] [Cited by in RCA: 14440] [Article Influence: 390.3] [Reference Citation Analysis (0)] |
| 18. | Rasmussen LT, Labio RW, Gatti LL, Silva LC, Queiroz VF, Smith Mde A, Payão SL. Helicobacter pylori detection in gastric biopsies, saliva and dental plaque of Brazilian dyspeptic patients. Mem Inst Oswaldo Cruz. 2010;105:326-330. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 27] [Cited by in RCA: 32] [Article Influence: 2.1] [Reference Citation Analysis (0)] |
| 19. | Zabaglia LM, Sallas ML, Santos MPD, Orcini WA, Peruquetti RL, Constantino DH, Chen E, Smith MAC, Payão SM, Rasmussen LT. Expression of miRNA-146a, miRNA-155, IL-2, and TNF-α in inflammatory response to Helicobacter pylori infection associated with cancer progression. Ann Hum Genet. 2018;82:135-142. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 12] [Cited by in RCA: 14] [Article Influence: 1.8] [Reference Citation Analysis (0)] |
| 20. | Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) Method. Methods. 2001;25:402-408. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 117419] [Cited by in RCA: 133250] [Article Influence: 5552.1] [Reference Citation Analysis (1)] |
| 21. | Rodriguez S, Gaunt TR, Day IN. Hardy-Weinberg equilibrium testing of biological ascertainment for Mendelian randomization studies. Am J Epidemiol. 2009;169:505-514. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 804] [Cited by in RCA: 787] [Article Influence: 49.2] [Reference Citation Analysis (0)] |
| 22. | Zhang L, Qin H, Guan X, Zhang K, Liu Z. The TLR9 gene polymorphisms and the risk of cancer: evidence from a meta-analysis. PLoS One. 2013;8:e71785. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 21] [Cited by in RCA: 24] [Article Influence: 2.0] [Reference Citation Analysis (0)] |
| 23. | Trejo-de la O A, Torres J, Sánchez-Zauco N, Pérez-Rodríguez M, Camorlinga-Ponce M, Flores-Luna L, Lazcano-Ponce E, Maldonado-Bernal C. Polymorphisms in TLR9 but not in TLR5 increase the risk for duodenal ulcer and alter cytokine expression in the gastric mucosa. Innate Immun. 2015;21:706-713. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 18] [Cited by in RCA: 20] [Article Influence: 2.0] [Reference Citation Analysis (0)] |
| 24. | Mu X, Zhao J, Yuan X, Zhao X, Yao K, Liu Y, Zhao X. Gene Polymorphisms of Toll-Like Receptor 9 -1486T/C and 2848G/A in Cervical Cancer Risk. Int J Gynecol Cancer. 2015;25:1173-1178. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 13] [Cited by in RCA: 12] [Article Influence: 1.2] [Reference Citation Analysis (0)] |
| 25. | Wan GX, Cao YW, Li WQ, Li YC, Zhang WJ, Li F. Associations between TLR9 polymorphisms and cancer risk: evidence from an updated meta-analysis of 25,685 subjects. Asian Pac J Cancer Prev. 2014;15:8279-8285. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 12] [Cited by in RCA: 16] [Article Influence: 1.6] [Reference Citation Analysis (0)] |
| 26. | Martínez-Campos C, Bahena-Román M, Torres-Poveda K, Burguete-García AI, Madrid-Marina V. TLR9 gene polymorphism -1486T/C (rs187084) is associated with uterine cervical neoplasm in Mexican female population. J Cancer Res Clin Oncol. 2017;143:2437-2445. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 15] [Cited by in RCA: 10] [Article Influence: 1.3] [Reference Citation Analysis (1)] |
| 27. | Liu F, Lu W, Qian Q, Qi W, Hu J, Feng B. Frequency of TLR 2, 4, and 9 gene polymorphisms in Chinese population and their susceptibility to type 2 diabetes and coronary artery disease. J Biomed Biotechnol. 2012;2012:373945. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 31] [Cited by in RCA: 40] [Article Influence: 3.1] [Reference Citation Analysis (0)] |
| 28. | Mommersteeg MC, Yu J, Peppelenbosch MP, Fuhler GM. Genetic host factors in Helicobacter pylori-induced carcinogenesis: Emerging new paradigms. Biochim Biophys Acta Rev Cancer. 2018;1869:42-52. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 41] [Cited by in RCA: 39] [Article Influence: 4.9] [Reference Citation Analysis (0)] |
| 29. | Gębura K, Świerkot J, Wysoczańska B, Korman L, Nowak B, Wiland P, Bogunia-Kubik K. Polymorphisms within Genes Involved in Regulation of the NF-κB Pathway in Patients with Rheumatoid Arthritis. Int J Mol Sci. 2017;18. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 26] [Cited by in RCA: 25] [Article Influence: 3.1] [Reference Citation Analysis (0)] |
| 30. | Liu S, Wang X, Shi Y, Han L, Zhao Z, Zhao C, Luo B. Toll-like receptor gene polymorphisms and susceptibility to Epstein-Barr virus-associated and -negative gastric carcinoma in Northern China. Saudi J Gastroenterol. 2015;21:95-103. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 18] [Cited by in RCA: 18] [Article Influence: 1.8] [Reference Citation Analysis (0)] |
| 31. | Ashton KA, Proietto A, Otton G, Symonds I, McEvoy M, Attia J, Scott RJ. Toll-like receptor (TLR) and nucleosome-binding oligomerization domain (NOD) gene polymorphisms and endometrial cancer risk. BMC Cancer. 2010;10:382. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 40] [Cited by in RCA: 49] [Article Influence: 3.3] [Reference Citation Analysis (0)] |
| 32. | Zhang J, Zhu Q, Meng F, Lei H, Zhao Y. Association study of TLR-9 polymorphisms and systemic lupus erythematosus in northern Chinese Han population. Gene. 2014;533:385-388. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 33] [Cited by in RCA: 38] [Article Influence: 3.2] [Reference Citation Analysis (0)] |
| 33. | Omar AH, Yasunami M, Yamazaki A, Shibata H, Ofori MF, Akanmori BD, Shuaibu MN, Kikuchi M, Hirayama K. Toll-like receptor 9 (TLR9) polymorphism associated with symptomatic malaria: a cohort study. Malar J. 2012;11:168. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 40] [Cited by in RCA: 42] [Article Influence: 3.2] [Reference Citation Analysis (0)] |
| 34. | Lemos B, Bettencourt BR, Meiklejohn CD, Hartl DL. Evolution of proteins and gene expression levels are coupled in Drosophila and are independently associated with mRNA abundance, protein length, and number of protein-protein interactions. Mol Biol Evol. 2005;22:1345-1354. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 200] [Cited by in RCA: 211] [Article Influence: 10.6] [Reference Citation Analysis (0)] |
| 35. | Carvalho A, Osório NS, Saraiva M, Cunha C, Almeida AJ, Teixeira-Coelho M, Ludovico P, Pedrosa J, Pitzurra L, Aversa F, Romani L, Castro AG, Rodrigues F. The C allele of rs5743836 polymorphism in the human TLR9 promoter links IL-6 and TLR9 up-regulation and confers increased B-cell proliferation. PLoS One. 2011;6:e28256. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 31] [Cited by in RCA: 35] [Article Influence: 2.5] [Reference Citation Analysis (0)] |
| 36. | Hamann L, Hamprecht A, Gomma A, Schumann RR. Rapid and inexpensive real-time PCR for genotyping functional polymorphisms within the Toll-like receptor -2, -4, and -9 genes. J Immunol Methods. 2004;285:281-291. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 50] [Cited by in RCA: 52] [Article Influence: 2.5] [Reference Citation Analysis (0)] |