Published online Jul 15, 2018. doi: 10.4251/wjgo.v10.i7.145
Peer-review started: March 8, 2018
First decision: March 19, 2018
Revised: April 22, 2018
Accepted: June 8, 2018
Article in press: June 9, 2018
Published online: July 15, 2018
Processing time: 129 Days and 6.5 Hours
Colorectal cancer (CRC) treatment has become more personalised, incorporating a combination of the individual patient risk assessment, gene testing, and chemotherapy with surgery for optimal care. The improvement of staging with high-resolution imaging has allowed more selective treatments, optimising survival outcomes. The next step is to identify biomarkers that can inform clinicians of expected prognosis and offer the most beneficial treatment, while reducing unnecessary morbidity for the patient. The search for biomarkers in CRC has been of significant interest, with questions remaining on their impact and applicability. The study of biomarkers can be broadly divided into metabolic, molecular, microRNA, epithelial-to-mesenchymal-transition (EMT), and imaging classes. Although numerous molecules have claimed to impact prognosis and treatment, their clinical application has been limited. Furthermore, routine testing of prognostic markers with no demonstrable influence on response to treatment is a questionable practice, as it increases cost and can adversely affect expectations of treatment. In this review we focus on recent developments and emerging biomarkers with potential utility for clinical translation in CRC. We examine and critically appraise novel imaging and molecular-based approaches; evaluate the promising array of microRNAs, analyze metabolic profiles, and highlight key findings for biomarker potential in the EMT pathway.
Core tip: Biomarkers are an emerging field that can potentially guide the diagnosis, prognosis, and treatment course in rectal cancer. Here, the current definitions, classifications, recent developments and emerging biomarkers with potential utility for clinical translation in colorectal cancer are reviewed by international experts for a better understanding in surgery.
- Citation: Chand M, Keller DS, Mirnezami R, Bullock M, Bhangu A, Moran B, Tekkis PP, Brown G, Mirnezami A, Berho M. Novel biomarkers for patient stratification in colorectal cancer: A review of definitions, emerging concepts, and data. World J Gastrointest Oncol 2018; 10(7): 145-158
- URL: https://www.wjgnet.com/1948-5204/full/v10/i7/145.htm
- DOI: https://dx.doi.org/10.4251/wjgo.v10.i7.145
Colorectal cancer (CRC) is one of the most common types of cancer and cancer related deaths worldwide, with more than a third of the incidence involving the rectum[1,2]. Historically, rectal cancer was associated with the worst oncological outcomes[3]. The choice of treatment for rectal cancer was traditionally based upon the histologic type of malignancy, stage of the disease, the tumour-node-metastasis (TNM) staging system, and circumferential resection margin (CRM) status[2,4]. These variables provide clinical utility, help determine the need for neoadjuvant chemoradiotherapy (CRT) in patients with a threatened or involved CRM, post-operative adjuvant treatment in stage III disease, and are prognostic of oncological outcome. Nevertheless, they provide an incomplete picture, as many patients with predicted early-stage disease harbour lymph node and systemic micrometastases, which can ultimately result in local and/or distant disease recurrence. Administration of neoadjuvant CRT is also sub-optimal as this treatment modality has many side effects, some of which are fatal, while others impair quality of life (QOL). Response to CRT is also unpredictable; up to 30% of patients will have a complete pathological response (pCR = tumour regression grade 1, TRG1), and could have omitted surgery altogether[5,6]. In 10% of cases however, no reduction in tumour volume is achieved, (tumour regression grade 5, TRG5); patients get no benefit from CRT, but are exposed to its side effects and may also experience cancer progression from delay to surgery[7]. These observations underscore the limitations of current methods for accurate stratification of patients with rectal cancer, and highlight the pressing need to identify biomarkers indicative of aggressive disease and/or response to CRT, in order to avoid patient under- or over-treatment.
With the advent of the “holy plane”, standards for utilising chemoradiation, the application of minimally invasive surgery, and multidisciplinary tumour boards to guide care, the diagnosis, staging and management of rectal cancer has improved significantly in the past 25 years[8-18]. However, considerable variation still exists in management and outcomes, and recurrence continues to be a problem, with 5-year survival rates stubbornly below 60% in most European countries[19]. To further improve outcomes, there is a paradigm shift in the methods of diagnosis, staging, determining the patient’s prognosis, and developing a personalized therapeutic course using advances in molecular biology, genetics, biochemistry, imaging, and the individual patient’s personal risk assessment, neoadjuvant chemoradiotherapy, and adjuvant chemotherapy with surgery to optimise care[20].
The routine evaluation of microsatellite instability (MSI) and KRAS/NRAS/BRAF mutational status in clinical practice, for risk stratification in stage II CRC and to determine the utility of monoclonal antibody-based adjuvant therapy, such as panitumumab or cetuximab, in metastatic disease, provides a clear proof-of-concept that more tailored therapeutic strategies can be translated to improve patient care through identification of biomarkers with functional activity. In this review, we explore the recent developments and emerging biomarkers with potential utility for clinical translation in CRC. We examine and critically appraise both novel imaging and molecular pathology based approaches; evaluating the promising array of microRNAs with biomarker potential; examining the developing techniques and studies analysing metabolic profiles, and highlight key findings in the biomarker potential in the epithelial-to-mesenchymal-transition (EMT) pathway.
From the Biomarkers Definitions Working Group, the formal definition of a biomarker is a tumour characteristic that can be objectively measured and evaluated as an indicator(s) of normal biological or pathogenic processes, or pharmacologic responses to a therapeutic intervention that identify increased or decreased risk of patient benefit or harm[21,22]. Biomarkers can take multiple forms when used to detect or confirm presence of disease or to identify affected individuals[23]. Table 1 shows the categorisation of biomarkers. Most biomarkers applicable in CRC are prognostic - providing information about the likelihood of a condition, disease recurrence or progression; or predictive - providing information about the likelihood to respond to specific treatments. A cause of confusion around biomarkers has been the loose application of their definition and application. Distinguishing between predictive and prognostic biomarkers- which may not be mutually exclusive- has been another source of confusion in patient stratification and developing treatment strategies[23]. Another source of confusion is the inconsistent terminology previously used, restricting the scope of biomarkers to describing biological molecules or monitoring the treatment response. The current definition laid out by Cancer Research United Kingdom provides a standardised vocabulary for investigators, explicitly stating, “molecular, histologic, radiographic or physiologic characteristics are examples of biomarkers”[24]. With this progression, biomarkers may be used in a variety of situations and serve a number of purposes - as a diagnostic tool; for risk-stratification and staging of disease; as an estimator of prognosis; and, for prediction of disease response. The study of such biomarkers can be broadly divided into metabolic; miRNA; EMT; and imaging biomarkers. This review describes the current status of biomarkers in CRC within this framework.
| Biomarker type | Objective |
| Diagnostic biomarker | These aim to identify the type of cancer, e.g., PSA, CEA. They may also be used to monitor or detect disease recurrence |
| Pharmacological biomarker | These are used to measure response to a specific drug treatment. They are based on accurate pharmokinetic data and measure treatment response in early drug trials, e.g., drug therapy to angiogenesis |
| Predictive biomarker | These are used to identify individuals who will most likely show a survival benefit to a specific targeted treatment, e.g., improvement in local recurrence risk following treatment for circumferential resection margin involvement |
| Prognostic biomarker | These indicate the progress of disease and to estimate the risk of disease recurrence for example. They are used to estimate survival outcome and are independent of treatment strategy, e.g., nodal disease |
| Risk/predisposition biomarker | These aim to identify individuals who are at significant risk of developing tumours, e.g., MLH1 gene |
| Screening biomarker | These are used to identify disease at an early stage, e.g., PSA |
| Surrogate response biomarker | These can be used as an alternative to a clinically meaningful endpoint. Therefore there must be correlation with a clinical endpoint, e.g., CEA |
The search for molecular markers in CRC has been of significant recent interest. Extensive research has revealed that CRC develops through three major pathways: (1) chromosomal abnormalities that lead to mutations of oncogenes and tumour suppressor genes (classic pathway), characterised by the adenoma-carcinoma progression; (2) the microsatellite instability pathway that results from defects in the DNA repair system; and (3) the methylation pathway characterized by the epigenetic (post cellular division) methylation of numerous genes (methylator pathway). Hundreds of molecules involved in the chromosomal instability pathway have been associated with prognosis, however, only 1 single marker- the epidermal growth factor receptor (EGFR) pathway-has successfully proven clinical utility to date, largely due to the complexity and redundancy of cellular pathways, as well as the lack of therapies that can target the different biomarkers.
The EGFR pathway is the most clinically relevant molecule involved in the chromosomal instability pathway, and the EGFR serves as the main target for treatment in locally advanced CRC. However, this treatment is only useful for patients with wild-type KRAS (wtKRAS)[25]. Abnormal activation of the EGFR signalling pathways in CRC is mainly associated with three mutations in the mitogen-activated protein kinase and phosphatidylinositol-3-kinase (PI3K) pathways - KRAS, NRAS, and BRAF; these three mutations are reported to occur in more than half of all CRC cases[26]. Mutation of some of the components of the EGFR pathway, specifically BRAF V600E, KRAS (exon 2, 3, 4), and NRAS mutation (exon 2, 3, 4) cause the malignant cells to become resistant to anti-EGFR therapy; thus, patients should not be treated with either cetuximab or panitumumab. As a result, all patients with metastatic CRC should have investigation of KRAS/NRAS and BRAF mutation status prior to the start of treatment. KRAS/NRAS and BRAF mutational status may be performed by a variety of techniques, detailed discussion of the different methodologies is out of the scope of this review, however it is essential to emphasize that several technical factors including tissue fixation and tumour volume amongst others may affect the accuracy of the test results leading to erroneous information with the consequent impact on the decision making process. Furthermore, any tumour molecular analysis should be performed only by a certified laboratory that can prove competency and proficiency to perform testing.
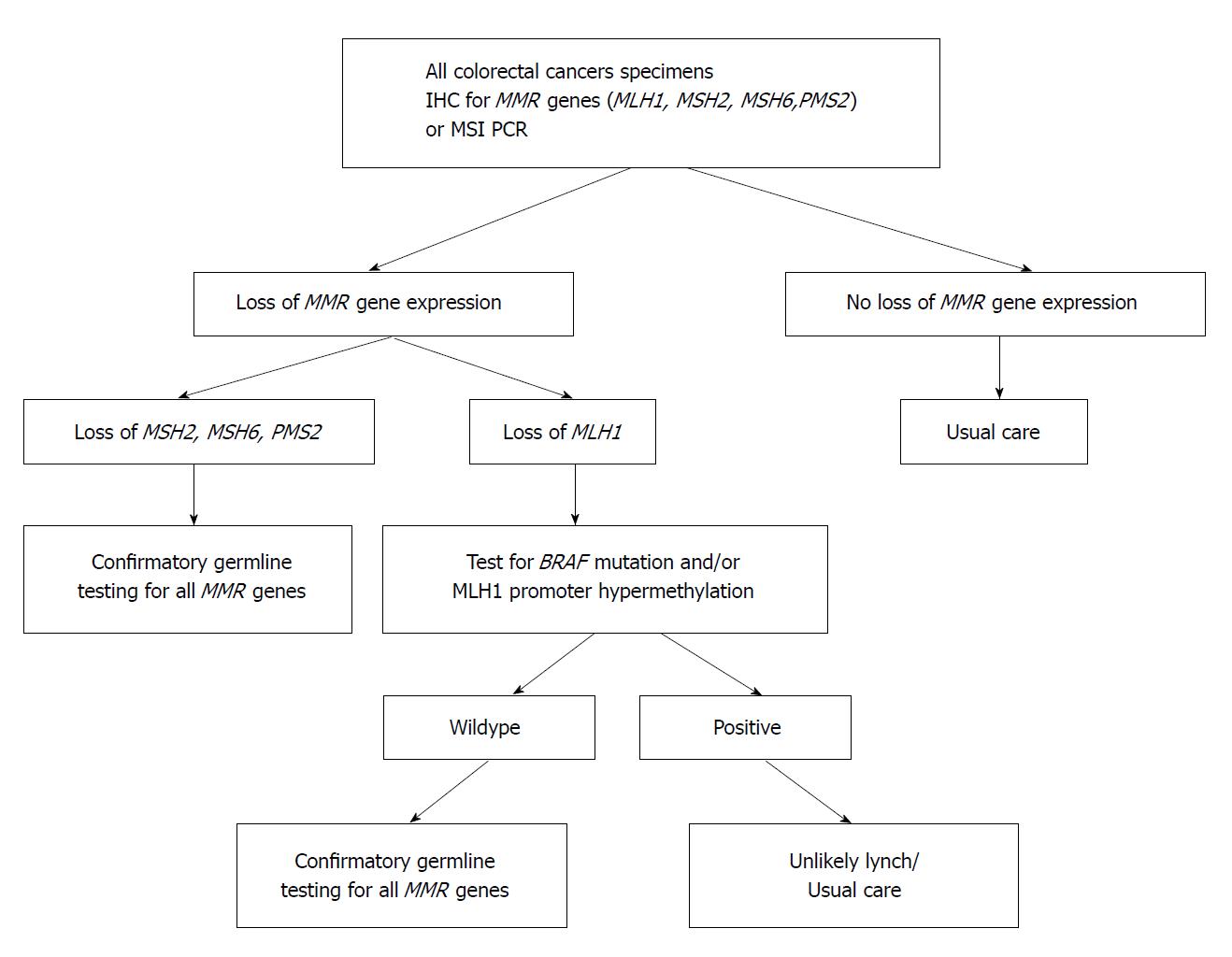
Microsatellite instability status (MSI) (high or low) is the primary molecular marker for stratification of stage II CRC. In node negative CRC, patients that are MSI-high have better outcomes than MSI-low tumours; therefore, adjuvant chemotherapy is usually not indicated in MSI-high tumours. MSI-high tumours arise in the setting of a defective DNA repair machinery, although several proteins have been implicated in DNA repair, abnormalities in MSH2, MSH6, PMS2 and MLH1 are the most commonly described. MSI-high tumours may be the result of an inherited mutation of the DNA repair genes (Lynch syndrome) or, more commonly, the abnormal epigenetic methylation of the MLH1 promoter gene (sporadic MSI-high CRC). Analysis of the DNA repair system may be directly investigated by the tissue expression of MSH2, MSH6, PMS2 and MLH1 by immunohistochemistry, or alternatively by determination of microsatellite status by PCR.
The CpG Island Methylator Phenotype (methylator) pathway has been associated with a constellation of clinical (elderly patients, female, right-sided colon tumours) and histological features (poorly differentiated tumours and advanced stage disease). This pattern seen in approximately 15%-20% of CRCs, and involves atypical methylation of the mismatch repair gene MLH1. The precursor lesions in CIMP cancers are serrated polyps, not adenomatous lesions, with the initial mutation occurring most often in the BRAF oncogene[27]. BRAF mutations transform normal mucosa to aberrant crypt foci, hyperplastic, or sessile serrated polyps (SSP). With promoter methylation, loss of p16 occurs, allowing cells to progress to advanced polyps[28]. Increasing activity leads to methylation of MLH1, silencing transcription. Loss of MLH1 results in MMR deficiency and the MSI-H CRC phenotype. This is clinically important for diagnosis and therapeutic planning. An estimated 85% of MMR deficiency CRC is due to methylation of the MLH1 promoter region. BRAF can be used to distinguish between MLH1 promoter methylation and Lynch syndrome as the cause of CRC. A positive BRAF mutation is associated with the methylator pathway, and indicates MLH1 down-regulation through somatic methylation of the gene’s promoter region, not through a germline mutation. BRAF mutations are rare in Lynch Syndrome-related CRC. On the converse, MLH1 promotor methylation in the absence of a BRAF mutation is consistent with Lynch Syndrome. Figure 1 shows a clinical algorithm for testing MMR deficiency. Several promising new therapies aimed at demethylation of genes are being developed.
In recent years the majority of molecular profiling approaches applied to the study of rectal cancer have focused on macromolecules (DNA, RNA, protein). While these avenues of research continue to offer significant insights into rectal cancer development and progression[29,30], it is widely accepted that a macromolecular, “bottom up” view of system activity cannot provide all the answers to facilitate precision approaches for rectal cancer diagnosis, prognosis and therapeutic personalisation[31]. Metabonomics (metabolomics/metabolic profiling) offers a dynamic “top down” view of system activity and is defined as the systematic, time-dependent measurement of metabolic shifts occurring in response to drugs, environmental stimuli or disease[32-34]. This approach provides rich micromolecular data downstream of the genome and proteome, offering a genuine functional “snapshot” of system activity[33].
The basic concept of altered cancer metabolism is well described across a variety of cancer subtypes[35-38]; the Warburg effect[39] is central to our understanding of cancer metabolism and glycolytic flux forms the basis for [18F]-fluorodeoxyglucose enhanced positron emission tomography (FDG-PET) solid tumour imaging[40]. Current and next-generation nuclear magnetic resonance (NMR) spectroscopy and mass spectrometry (MS)-based profiling platforms offer a means of interrogating the cancer metabolome in unprecedented detail and moving beyond the Warburg phenomenon to identify an entirely new pool of disease-relevant biomolecular data. These profiling approaches are likely to have three main areas of application in rectal cancer phenotyping: (1) to identify novel metabolic fingerprints for accurate and ultra-fast tumour tissue diagnosis, staging and grading; (2) to develop metabolite-based models for prediction of response to chemo and/or radiotherapy; and (3) to devise novel next-generation targeted therapies designed to disrupt specific metabolic pathways implicated in rectal cancer.
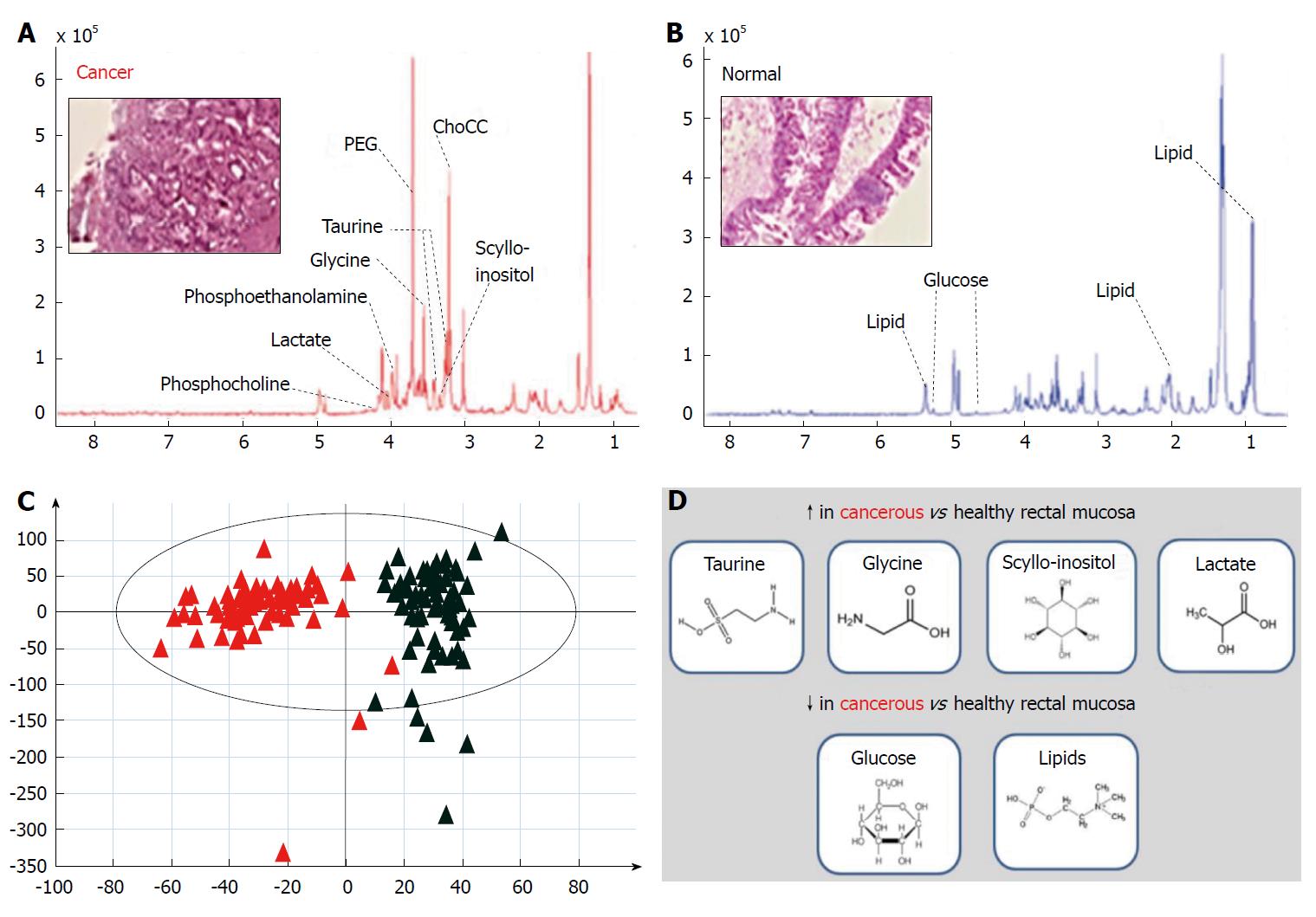
NMR spectroscopy techniques are highly versatile and have been developed and applied for metabolic profiling of liquid-state and solid-state systems[41,42]. The technique of HR-MAS NMR has been introduced more recently to overcome spectral line-broadening effects seen with conventional NMR analysis of solids[41]. This approach allows acquisition of tissue-specific high-resolution spectra, which in combination with chemometric data treatment methods have the capacity to identify novel molecular signatures within rectal cancer tissue[43]. Recent work in this area has demonstrated increased abundance of taurine, glycine, lactate and scyllo-inositol in cancerous relative to healthy rectal mucosa, with a relative reduction in abundance observed for lipids and glucose[44] (Figure 2). These findings can be used to determine tissue status (cancerous or healthy) by entirely biochemical means, and have also revealed strong differences in metabolite profiles according to tumour stage[44]. From a pharmaco-therapeutic perspective these discoveries offer the chance to develop novel anti-cancer agents; for example, taurine (2-aminoethane sulphonic acid), a common beta-amino acid has a known role in a number of fundamental physiological functions including cellular osmoregulation, cell-membrane stabilization and protein assembly[45]. Exploiting this finding by disrupting taurine handling within the rectal cancer microenvironment may offer a means of developing next-generation targeted agents for rectal cancer down-staging[46].
Mass spectrometry approaches have shown recent promise in the development of metabolite-based biomarker discovery for prediction of response to chemoradiotherapy. Crotti et al[47] described novel peptidomic methodology in an analysis of samples of serum collected pre- and post-CRT subjected to matrix-assisted laser desorption/ionisation-time of flight (MALDI-TOF) mass spectrometry. A comparison of pre-treatment serum fingerprints from responders [Mandard tumour regression grade (TRG) 1 and 2] and non-responders (Mandard TRG 3-5) identified three peptides (m/z 1082.552, m/z 1098.537 and 1104.538) that were capable of robust class separation. Kim and colleagues also used a MALDI-based approach, but specifically sought to evaluate the abundance of low-mass ions (< m/z 1000) in serum samples acquired from 73 patients with locally advanced rectal cancer, prior to CRT[48]. A panel of nine low-mass ions were found to have discriminatory capacity, with hypoxanthine (HX; m/z 137.08) and phosphoenolpyruvic acid (PEP; m/z 169.04) highlighted as the most significant. Lower levels of HX and higher levels of PEP were shown to strongly correlate with improved response to CRT (TRG 1, 2). These studies indicate the exciting potential for the development of a circulating biomarker panel to predict chemoradiosensitivity prior to commencing therapy.
MicroRNAs (miRNA) are highly conserved, short, non-coding nucleotide segments that regulate gene expression post-transcriptionally through repressing translation or targeting mRNAs for degradation[49]. miRNA genes account for between 2%-5% of the human genome and are commonly clustered within introns[50]. Each miRNA is estimated to interact with multiple mRNA targets and, as a consequence, thus, these sequences may regulate more than 30% of all human genes[51,52]. Oncogenes and tumour-suppressor genes are being discovered under miRNA control, with the majority of miRNA genes found within cancer-associated genomic regions[53,54]. In CRC, abnormally expressed miRNAs disrupt cellular signal transduction and cell survival pathways, such as Wnt/β-catenin, EGFR, and p53, linking miRNA to known events in the pathway of malignant transformation[55].
Accumulating evidence suggests that miRNAs may also have powerful clinical applications. miRNA expression profiles are capable of discriminating tumours of different developmental origin[56]. Furthermore, the expression of individual miRNAs may be used to predict patient survival, tumour stage, the presence of lymph node metastases and the response to therapy in CRC[55,57,58].
Three studies have specifically examined the utility of miRNA expression signatures in predicting chemoradiotherapy response in rectal cancer[59-61]. Della Vittoria Scarpati et al[59] examined miRNA expression in fresh-frozen pre-treatment tumour specimens from 38 patients with locally advanced (T3/T4 Node +ve) rectal cancer and compared miRNA profiles in patients with complete (Mandard TRG 1; n = 9) and incomplete (Mandard TRG > 1; n = 29) pathological responses to a standardised neoadjuvant chemoradiotherapy regime consisting of capecitabine, oxaliplatin and 45 Gy of pelvic conformal radiotherapy. Thirteen significantly differentially expressed miRNAs were subsequently validated using high sensitivity TaqMan® qRT-PCR, of which 2; miR-622 and miR-630, were found to predict chemoradiotherapy response with 100% sensitivity and specificity[59].
A similar analysis of 20 patients undergoing combined radiotherapy and capecitabine/5-FU chemotherapy compared “responders”, namely those displaying a positive response to treatment (Mandard TRG 1 and 2) with “non-responders” (Mandard TRG 3-5). TaqMan Low Density Arrays identified a miRNA signature consisting of 8 miRNAs capable of correctly classifying 90% (9/10) of responders and 90% (9/10) of non-responders[60].
A third study, which used formalin fixed rather than fresh rectal cancer specimens identified a miRNA signature consisting of just 3 miRNAs (miR-153, miR-16 and miR-590-5p), capable of distinguishing patients with complete and incomplete responses to therapy, however the value of this data is unclear as patient demographics, tumour characteristics, study end-points and the neo-adjuvant treatment strategy were not clearly described[61].
As profiling methodology and the definition of tumour regression vary between these 3 studies, inter-study comparisons are of limited value; however it is important to note that no overlap is observed between the miRNA signatures described. This suggests that an miRNA based “therapy-response” prediction tool is some way from becoming a reality however; other studies have clearly established that miRNAs do play a role in regulating the tissue response to neoadjuvant therapy in CRC[62-64]. Perhaps by focusing on the contribution of miRNAs within the biological pathways that govern resistance and/or sensitivity to neo-adjuvant therapy in rectal cancer, more clinically pertinent data will emerge on the role of miRNA as a potential biomarker in cancer treatment strategies[65].
The term “liquid biopsy” in cancer arose when circulating tumor cells (CTC) were proposed as alternatives to conventional tissue biopsy in breast cancer for prognosis and evaluation of treatment responses[66]. The theory has continued to grow experimentally and has gained particular traction in CRC. The clinical applications of liquid biopsy in CRC continue to grow, including detecting premalignant and early-stage cancers, identification of aggressive phenotypes and high-risk patients, assessing tumor heterogeneity, residual, and recurrent disease, and monitoring treatment response[67]. In colon cancers, liquid biopsies may hold prognostic information beyond the nodal status for determining whether to administer adjuvant chemotherapy, while in rectal cancer, liquid biopsy may have roles for both primary disease evaluation and monitoring treatment response[68]. Possible sources of liquid biopsies include blood, urine, saliva, and stool, which contain cancer-derived subcellular components, such as circulating tumor DNA (ctDNA) and circulating miRNAs.
Tumour-tissue remains the “gold standard”, but the advent of ctDNA analysis from blood samples has promise as a non-invasive biomarkers. Studies have reported a direct relationship between ctDNA levels and tumor burden, stage, vascularity, cellular turnover, and response to therapy[69-71]. It can enable efficient temporal assessment of disease status, response to intervention, and early detection of recurrence superior to current strategies, such as CEA[72]. ctDNA can monitor and recognize high-risk individuals, as the plasma tumour DNA levels are significantly higher in patients with increased advanced/stage IV disease, recurrence, or metastasis[73,74]. ctDNA may be sensitive to detect with early, presumably curable CRC from common mutations, which could have implication for diagnostic testing[75]. Meta-analysis has demonstrated high overall sensitivity and specificity for detecting the KRAS oncogene mutation in CRC, showing it may be a viable alternative to tissue analysis for the detection of KRAS mutations and subsequent therapeutic planning[75]. Further, comparative analysis between CTCs and ctDNA in metastatic CRC has shown strong concordance between ctDNA and tissue for RAS, BRAF, and ERBB2 mutations (84.6%) and greater detectability than CTCs with a smaller amount of blood sampling[76]. ctDNA may hold specific promise as a biomarker to guide therapy in post-operative locally advanced rectal cancer, but further studies are needed for validation[77]. There are limitations to ctDNA as a biomarker. Although ctDNA targets offer a high specificity, it is scarce in circulating biofluids- representing less than 1% of the total circulating free DNA and may be inadequate as clinically applicable diagnostic biomarkers. The best source of ctDNA is still uncertain and the size of the DNA released from dead cancer cells is longer than that of non-neoplastic DNA[70,78]. Large scale controlled trials are needed for validation.
miRNA is an alternate for liquid biopsy. miRNAs have features making them ideal candidates for development as disease-specific biomarkers, and may offer superior sensitivity and specificity compared with ctDNA for diagnosing CRC[79]. miRNAs are generally stable in blood and other body fluids due to their small size and their ability to escape from RNase-mediated degradation. miRNA expression levels are different in tumour compared to normal colon tissues[80]. miRNA are actively secreted from living cells, while most ctDNA is dependent on release from apoptotic or necrotic cells[81,82]. miRNA-based diagnostic markers and panels have been identified for early detection, risk of recurrence at the time of diagnosis, complement to CEA for identification of distant metastasis, and stratification of patients with poor prognosis and greater likelihood of metastasis to the lymph nodes, liver, and peritoneum[80,83-88]. These miRNAs are detailed in Table 2. While a promising tool for “precision medicine”, there are limitations of circulating miRNAs as biomarkers in CRC. The existing studies use relatively small sample sizes, are retrospective in design, and utilized non-standardized sampling procedures. Larger, controlled studies are needed in order to validate the best purification method and clinical use of circulating miRNAs in CRC.
| Expression level | Diagnostic biomarker | Prognostic biomarker (malignant potential, tumor recurrence) | Predictive biomarker (chemosensitivity) |
| High | miR-92a, miR-141, let-7a, miR-1229, miR-1246, miR-150, miR-21, miR-223, miR-23a, miR-378 | miR-141, miR-320, miR-596, miR-203 | miR-106a, miR-484, miR-130b |
| Low | miR-15a, miR-103, miR-148a, miR451 |
An example of a blood sample-based diagnostic biomarker that could make a clinical impact is methylated Septin 9 (mSEPT9), which is validated to distinguish CRC from normal blood using real-time PCR[89]. This non-invasive, blood-based tool for CRC could improve screening and surveillance compliance over colonoscopy and other screening methods[90]. While monitoring of mSEPT9 may hold promise for CRC screening, a larger study population and more prospective studies are needed to validate mSEPT9 as a diagnostic biomarker in CRC.
EMT is a physiological process resulting in transformation of stable epithelial cells into mobile mesenchymal cells[91]. While EMT is a normal process during human development, it has also been shown to occur in carcinogenesis[92]. In this situation, the resulting abnormal mesenchymal cells, which evade the influence of normal cellular control mechanisms, display an aggressive and invasive phenotype. These cells are increasingly linked to formation of micro-metastases, and causation of resistance to the effects of radiotherapy.
Down-regulation of membranous E-cadherin is the classical finding of EMT. This results in loss of intercellular epithelial junctional complexes, promoting migration of cells[93-95]. The microRNA-200 family has been identified as a key post-transcriptional regulator of this process, through its targeting of E-cadherin transcriptional receptors[96]. Subsequent escape from growth factor control, with uncontrolled proliferation, results from the EMT process[94,95]. An end consequence of this pathway is tumour budding, defined as the presence of single cells or small cell clusters at the invasive front of tumour growth[97]. Tumour budding is highly likely to be associated to EMT at the poorly differentiated invasive front[97-100].
There is increasing evidence linking EMT to chemoresistance in ovarian, pancreatic and breast cancer cell lines[101-104], and in human lung cancer specimens[105]. Emerging evidence is also relating EMT to response to chemoradiotherapy in CRC. This initially arose from testing chemoresistance in colorectal cell lines[106-108]. However newer human evidence is relating EMT as an independent biomarker of tumour budding, lymph node metastases, and radioresistance[109]. The largest of these demonstrated that, in 103 patients with advanced rectal cancer, an EMT phenotype was associated with nonresponse to neoadjuvant therapy and reduced cancer specific survival[110]. More evidence from human rectal cancer tissue is urgently needed to assess its potential as a biomarker.
A genetic predisposition to loss of E-cadherin and subsequent EMT may be causative, meaning that pre-treatment biopsy analysis presents a window for intervention. Radiotherapy may also be a traumatic triggering stimulus which forces some cells into an EMT phenotype, meaning other methods for patient selection may be required; overlap in causation is likely.
The biological action of metformin down-regulates the EMT transcription factors and up regulations E-cadherin[110]. Its low toxicity profile makes it a feasible option in EMT prevention attempts, with subsequent improvements in response to neoadjuvant therapies[111,112]. Additionally, cyclo-oxygenase (COX) inhibitors have shown potential to prevent EMT by reducing vimentin expression and increasing cell surface E-cadherin expression in cell line models[113]. However, due to their serious associated cardiovascular side-effects, the particular COX agent and dose require optimisation before widescale use[113,114]. The potential role of post-transcriptional microRNA-200 regulation presents a further potential therapeutic target[96].
The concept of an imaging biomarker is relatively new, but one which is becoming an increasingly important component of many phase II/III clinical trials as a surrogate endpoint. Imaging biomarkers may allow objective assessment of the tumour response to therapy and/or non-invasively detect early disease. Currently, the imaging techniques that seek to quantify treatment response in CRC can be broadly divided into those which measure tumour size and those which measure tumour activity. Whilst size criteria are the more commonly used biomarkers to assess radiological response in clinical trials because of their association with survival outcomes, it is the functional imaging techniques which are feted as having the greatest potential in uncovering the underlying biological processes which lead to cancer.
Reduction in tumour size has been shown to be a useful biomarker[115]. This can be measured in one-, two- or three-dimensions by various routine imaging techniques such as CT and MRI[116]. However, the two commonly used criteria - WHO[117] and RECIST[118] (Table 1); have contrasting characteristics, in particular in the technique used to measure tumour size - only one dimension using RECIST criteria. Further limitations to using size measurements have been deciding on what degree of tumour bulk reduction constitutes a significant clinical response. An example of this is has been shown by Morgan et al[119], who investigated the effect of a VEGF receptor inhibitor on colorectal metastases, whereby significant size reduction was not met with an equally significant overall response (< 10%). However the novel MRI-based tumour regression grade (mrTRG), which stratifies response on the degree of fibrosis visualised in the tumour following chemoradiotherapy, has been shown to be a useful clinical tool[120]. The degree of fibrosis seen on MRI following CRT on a scale analogous to histopathological tumour regression grade (TRG)[121] - tumour signal that has been completely replaced by radiological evidence of fibrosis is defined as radiological complete response (mrTRG1-2)[122]. These findings have been validated in a prospectively enrolled, multicentre study[123] and used to influence treatment decisions in particular “deferral of surgery” programs. In the above study, multivariate analysis showed mrTRG hazard ratios (HR) were independently significant for overall and disease-free survival. Using fibrosis as a radiological feature is not limited to measuring tumour size but can be used to quantify other prognostic factors such as extramural venous invasion (EMVI), for example[120]. A further study using prospectively collected data on EMVI response to neo-adjuvant chemoradiotherapy showed hazard ratio of 2.37 for DFS in tumours which had undergone more than 50% fibrosis of tumour signal in extramural vasculature[124].
These techniques involve analysis of images to quantify the functional activity of tumours. The most common example of this is positron emission tomograhy (PET) with Fluorodeoxyglucose (18-FDG), which relies on the principle of a differential glycolytic rate seen in tumour cells. Using the glucose analogue 18-FDG gives an assessment of tumour metabolism[125,126] by quantification of standard uptake values (SUV). However as timing of the scans from administration of the 18-FDG and subsequent clearance rates may vary between centres and patients, comparisons and standardisation of technique has been difficult. It is also important to note that until now, there has been no validation of response.
Dynamic contrast-enhanced (DCE) CT/MRI provides a detailed assessment of tumour bloodflow through acquisition of data as specific contrast material passes through the vasculature. DCE-CT has the potential to identify angiogenesis and has been shown to be able to distinguish from diverticular disease as well as detect early liver metastases[127,128]. Although reports have identified a correlation between tumour blood flow, the development of metastases, and decreased survival outcomes[129,130], this has not been translated to widespread clinical application. Vascular endothelial growth factor (VEGF) is upregulated in up to 78% of CRCs[131,132] and is a potential target for functional imaging techniques. Bevacizmab is an anti-VEGF-A monoclonal antibody and DCE-MRI has been used in rectal cancer to evaluate treatment response using conjugation with a radioclueotide[133-135]. The analysis in DCE-MRI uses two compartments of plasma and extravascular-extracellular space to compare contrast agent - Ktrans is the constant which is used to depict the bloodflow. Several studies have validated Ktrans with expression of growth factors, such as VEGF and immunohistochemical confirmation of vessel architecture[136-139]. Reduction in Ktrans using Vatalanib (tyrosine kinase inhibitor which target VEGF receptor-2) for metastatic CRC with liver disease have shown promising results in the phase I/II setting[119,140] but not been translated to survival benefit in phase IIItrials.
Diffusion weighted imaging (DWI) assesses the movement of water molecules within cells using diffusion-weighted gradients to T2 sequences. Quantitative analysis is possible by calculation of the apparent diffusion coefficients (ADC), which are inversely correlated with tumour cellularity. DWI has been effective in detecting small liver metastases and differentiation from inflammatory lesion[141-143], as well as detecting lymph node metastases[144], but application has been limited to mainly experimental work.
The interest in biomarkers relating to rectal cancer is clearly increasing. They form a new aspect of clinical and laboratory research which help translate these concepts to more meaningful applications in patient management. Much of the current literature is still in its embryonic stage, but as more results from clinical trials using biomarker endpoints and outcome measures become available, there will be a better understanding by clinicians of their potential, with possible future application to improve the predictive and prognosis of rectal cancer.
Manuscript source: Invited manuscript
Specialty type: Oncology
Country of origin: United Kingdom
Peer-review report classification
Grade A (Excellent): 0
Grade B (Very good): B, B, B, B
Grade C (Good): C
Grade D (Fair): 0
Grade E (Poor): 0
P- Reviewer: Horesh N, Ocker M, Seow-Choen F, Sipos F, Wang SK S- Editor: Ji FF L- Editor: A E- Editor: Tan WW
| 1. | American Cancer Society. Key Statistics for Colorectal Cancer 2017. Available from: http://www.cancer.org/cancer/colonandrectumcancer/detailedguide/colorectal-cancer-key-statistics. |
| 2. | NCCN Clinical Practice Guidelines in Oncology (NCCN Guidelines®). Rectal Cancer, Version 3, 2017. Available from: https://www.nccn.org/professionals/physician_gls/PDF/rectal.pdf. |
| 3. | Wolmark N, Wieand HS, Rockette HE, Fisher B, Glass A, Lawrence W, Lerner H, Cruz AB, Volk H, Shibata H. The prognostic significance of tumor location and bowel obstruction in Dukes B and C colorectal cancer. Findings from the NSABP clinical trials. Ann Surg. 1983;198:743-752. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 123] [Cited by in RCA: 120] [Article Influence: 2.9] [Reference Citation Analysis (0)] |
| 4. | National Institute for Health and Care Excellence. Colorectal cancer: diagnosis and management. Available from: https://www.nice.org.uk/guidance/cg131. |
| 5. | García-Aguilar J, Hernandez de Anda E, Sirivongs P, Lee SH, Madoff RD, Rothenberger DA. A pathologic complete response to preoperative chemoradiation is associated with lower local recurrence and improved survival in rectal cancer patients treated by mesorectal excision. Dis Colon Rectum. 2003;46:298-304. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 308] [Cited by in RCA: 298] [Article Influence: 13.5] [Reference Citation Analysis (0)] |
| 6. | Maas M, Nelemans PJ, Valentini V, Das P, Rödel C, Kuo LJ, Calvo FA, García-Aguilar J, Glynne-Jones R, Haustermans K. Long-term outcome in patients with a pathological complete response after chemoradiation for rectal cancer: a pooled analysis of individual patient data. Lancet Oncol. 2010;11:835-844. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1189] [Cited by in RCA: 1460] [Article Influence: 97.3] [Reference Citation Analysis (0)] |
| 7. | Guillem JG, Díaz-González JA, Minsky BD, Valentini V, Jeong SY, Rodriguez-Bigas MA, Coco C, Leon R, Hernandez-Lizoain JL, Aristu JJ. cT3N0 rectal cancer: potential overtreatment with preoperative chemoradiotherapy is warranted. J Clin Oncol. 2008;26:368-373. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 165] [Cited by in RCA: 156] [Article Influence: 9.2] [Reference Citation Analysis (0)] |
| 8. | Guren MG, Kørner H, Pfeffer F, Myklebust TÅ, Eriksen MT, Edna TH, Larsen SG, Knudsen KO, Nesbakken A, Wasmuth HH, Vonen B, Hofsli E, Færden AE, Brændengen M, Dahl O, Steigen SE, Johansen MJ, Lindsetmo RO, Drolsum A, Tollåli G, Dørum LM, Møller B, Wibe A. Nationwide improvement of rectal cancer treatment outcomes in Norway, 1993-2010. Acta Oncol. 2015;54:1714-1722. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 57] [Cited by in RCA: 71] [Article Influence: 7.1] [Reference Citation Analysis (0)] |
| 9. | Wibe A, Møller B, Norstein J, Carlsen E, Wiig JN, Heald RJ, Langmark F, Myrvold HE, Søreide O; Norwegian Rectal Cancer Group. A national strategic change in treatment policy for rectal cancer--implementation of total mesorectal excision as routine treatment in Norway. A national audit. Dis Colon Rectum. 2002;45:857-866. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 437] [Cited by in RCA: 434] [Article Influence: 18.9] [Reference Citation Analysis (0)] |
| 10. | Kapiteijn E, Marijnen CA, Nagtegaal ID, Putter H, Steup WH, Wiggers T, Rutten HJ, Pahlman L, Glimelius B, van Krieken JH. Preoperative radiotherapy combined with total mesorectal excision for resectable rectal cancer. N Engl J Med. 2001;345:638-646. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 3104] [Cited by in RCA: 3118] [Article Influence: 129.9] [Reference Citation Analysis (0)] |
| 11. | Sauer R, Becker H, Hohenberger W, Rödel C, Wittekind C, Fietkau R, Martus P, Tschmelitsch J, Hager E, Hess CF. Preoperative versus postoperative chemoradiotherapy for rectal cancer. N Engl J Med. 2004;351:1731-1740. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 4342] [Cited by in RCA: 4462] [Article Influence: 212.5] [Reference Citation Analysis (1)] |
| 12. | Heald RJ, Moran BJ, Ryall RD, Sexton R, MacFarlane JK. Rectal cancer: the Basingstoke experience of total mesorectal excision, 1978-1997. Arch Surg. 1998;133:894-899. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1037] [Cited by in RCA: 1055] [Article Influence: 39.1] [Reference Citation Analysis (0)] |
| 13. | MERCURY Study Group. Diagnostic accuracy of preoperative magnetic resonance imaging in predicting curative resection of rectal cancer: prospective observational study. BMJ. 2006;333:779. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 666] [Cited by in RCA: 659] [Article Influence: 34.7] [Reference Citation Analysis (0)] |
| 14. | Fleshman J, Sargent DJ, Green E, Anvari M, Stryker SJ, Beart RW Jr, Hellinger M, Flanagan R Jr, Peters W, Nelson H; Clinical Outcomes of Surgical Therapy Study Group. Laparoscopic colectomy for cancer is not inferior to open surgery based on 5-year data from the COST Study Group trial. Ann Surg. 2007;246:655-662; discussion 662-664. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 789] [Cited by in RCA: 801] [Article Influence: 44.5] [Reference Citation Analysis (0)] |
| 15. | Jayne DG, Thorpe HC, Copeland J, Quirke P, Brown JM, Guillou PJ. Five-year follow-up of the Medical Research Council CLASICC trial of laparoscopically assisted versus open surgery for colorectal cancer. Br J Surg. 2010;97:1638-1645. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 830] [Cited by in RCA: 737] [Article Influence: 49.1] [Reference Citation Analysis (0)] |
| 16. | Jeong SY, Park JW, Nam BH, Kim S, Kang SB, Lim SB, Choi HS, Kim DW, Chang HJ, Kim DY. Open versus laparoscopic surgery for mid-rectal or low-rectal cancer after neoadjuvant chemoradiotherapy (COREAN trial): survival outcomes of an open-label, non-inferiority, randomised controlled trial. Lancet Oncol. 2014;15:767-774. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 561] [Cited by in RCA: 647] [Article Influence: 58.8] [Reference Citation Analysis (0)] |
| 17. | Color II Study Group. Buunen M, Bonjer HJ, Hop WC, Haglind E, Kurlberg G, Rosenberg J, Lacy AM, Cuesta MA, D’Hoore A, Fürst A, Lange JF, Jess P, Bulut O, Poornoroozy P, Jensen KJ, Christensen MM, Lundhus E, Ovesen H, Birch D, Iesalnieks I, Jäger C, Kreis M, van riet Y, van der Harst E, Gerhards MF, Bemelman WA, Hansson BM, Neijenhuis PA, Prins HA, Balague C, Targarona E, Luján Mompeán JA, Franco Osorio JD, Garcia Molina FJ, Skullman S, Läckberg Z, Kressner U, Matthiessen P, Kim SH, Poza AA. COLOR II. A randomized clinical trial comparing laparoscopic and open surgery for rectal cancer. Dan Med Bull. 2009;56:89-91. [PubMed] |
| 18. | Lacy AM, Delgado S, Castells A, Prins HA, Arroyo V, Ibarzabal A, Pique JM. The long-term results of a randomized clinical trial of laparoscopy-assisted versus open surgery for colon cancer. Ann Surg. 2008;248:1-7. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 407] [Cited by in RCA: 436] [Article Influence: 25.6] [Reference Citation Analysis (0)] |
| 19. | Allemani C, Rachet B, Weir HK, Richardson LC, Lepage C, Faivre J, Gatta G, Capocaccia R, Sant M, Baili P. Colorectal cancer survival in the USA and Europe: a CONCORD high-resolution study. BMJ Open. 2013;3:e003055. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 68] [Cited by in RCA: 64] [Article Influence: 5.3] [Reference Citation Analysis (0)] |
| 20. | Schmoll HJ, Van Cutsem E, Stein A, Valentini V, Glimelius B, Haustermans K, Nordlinger B, van de Velde CJ, Balmana J, Regula J. ESMO Consensus Guidelines for management of patients with colon and rectal cancer. a personalized approach to clinical decision making. Ann Oncol. 2012;23:2479-2516. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1035] [Cited by in RCA: 1107] [Article Influence: 85.2] [Reference Citation Analysis (1)] |
| 21. | Biomarkers Definitions Working Group. Biomarkers and surrogate endpoints: preferred definitions and conceptual framework. Clin Pharmacol Ther. 2001;69:89-95. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 4510] [Cited by in RCA: 4130] [Article Influence: 172.1] [Reference Citation Analysis (0)] |
| 22. | Strimbu K, Tavel JA. What are biomarkers? Curr Opin HIV AIDS. 2010;5:463-466. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 1599] [Cited by in RCA: 1209] [Article Influence: 80.6] [Reference Citation Analysis (0)] |
| 23. | FDA-NIH Biomarker Working Group. BEST (Biomarkers, Endpoints, and other Tools) Resource. Available from: https://www.ncbi.nlm.nih.gov/books/NBK326791/. |
| 24. | Lioumi M, Newell D. CR-UK biomarker roadmaps. Clin Cancer Res. 2010;16:B33-B33. [DOI] [Full Text] |
| 25. | Plano D, Alcolea V, Sanmartín C, Sharma AK. Methods of selecting combination therapy for colorectal cancer patients: a patent evaluation of US20160025730A1. Expert Opin Ther Pat. 2017;27:527-538. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 3] [Cited by in RCA: 3] [Article Influence: 0.4] [Reference Citation Analysis (0)] |
| 26. | De Roock W, Claes B, Bernasconi D, De Schutter J, Biesmans B, Fountzilas G, Kalogeras KT, Kotoula V, Papamichael D, Laurent-Puig P. Effects of KRAS, BRAF, NRAS, and PIK3CA mutations on the efficacy of cetuximab plus chemotherapy in chemotherapy-refractory metastatic colorectal cancer: a retrospective consortium analysis. Lancet Oncol. 2010;11:753-762. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1491] [Cited by in RCA: 1656] [Article Influence: 110.4] [Reference Citation Analysis (1)] |
| 27. | Kambara T, Simms LA, Whitehall VL, Spring KJ, Wynter CV, Walsh MD, Barker MA, Arnold S, McGivern A, Matsubara N. BRAF mutation is associated with DNA methylation in serrated polyps and cancers of the colorectum. Gut. 2004;53:1137-1144. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 536] [Cited by in RCA: 567] [Article Influence: 27.0] [Reference Citation Analysis (1)] |
| 28. | Bettington M, Walker N, Clouston A, Brown I, Leggett B, Whitehall V. The serrated pathway to colorectal carcinoma: current concepts and challenges. Histopathology. 2013;62:367-386. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 309] [Cited by in RCA: 351] [Article Influence: 29.3] [Reference Citation Analysis (0)] |
| 29. | Dineen S. Biology of rectal cancer--the rationale for targeted therapy. Crit Rev Oncog. 2012;17:383-392. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 2] [Cited by in RCA: 2] [Article Influence: 0.2] [Reference Citation Analysis (0)] |
| 30. | Cunningham D, Atkin W, Lenz HJ, Lynch HT, Minsky B, Nordlinger B, Starling N. Colorectal cancer. Lancet. 2010;375:1030-1047. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1208] [Cited by in RCA: 1177] [Article Influence: 78.5] [Reference Citation Analysis (0)] |
| 31. | Mirnezami R, Nicholson J, Darzi A. Preparing for precision medicine. N Engl J Med. 2012;366:489-491. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 484] [Cited by in RCA: 429] [Article Influence: 33.0] [Reference Citation Analysis (0)] |
| 32. | Nicholson JK, Lindon JC, Holmes E. ‘Metabonomics’: understanding the metabolic responses of living systems to pathophysiological stimuli via multivariate statistical analysis of biological NMR spectroscopic data. Xenobiotica. 1999;29:1181-1189. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 2918] [Cited by in RCA: 2723] [Article Influence: 104.7] [Reference Citation Analysis (0)] |
| 33. | Mirnezami R, Kinross JM, Vorkas PA, Goldin R, Holmes E, Nicholson J, Darzi A. Implementation of molecular phenotyping approaches in the personalized surgical patient journey. Ann Surg. 2012;255:881-889. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 32] [Cited by in RCA: 27] [Article Influence: 2.1] [Reference Citation Analysis (0)] |
| 34. | Nicholson JK, Holmes E, Kinross JM, Darzi AW, Takats Z, Lindon JC. Metabolic phenotyping in clinical and surgical environments. Nature. 2012;491:384-392. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 361] [Cited by in RCA: 371] [Article Influence: 28.5] [Reference Citation Analysis (0)] |
| 35. | Sitter B, Bathen TF, Singstad TE, Fjøsne HE, Lundgren S, Halgunset J, Gribbestad IS. Quantification of metabolites in breast cancer patients with different clinical prognosis using HR MAS MR spectroscopy. NMR Biomed. 2010;23:424-431. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 107] [Cited by in RCA: 107] [Article Influence: 7.1] [Reference Citation Analysis (0)] |
| 36. | Chen W, Zu Y, Huang Q, Chen F, Wang G, Lan W, Bai C, Lu S, Yue Y, Deng F. Study on metabonomic characteristics of human lung cancer using high resolution magic-angle spinning 1H NMR spectroscopy and multivariate data analysis. Magn Reson Med. 2011;66:1531-1540. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 34] [Cited by in RCA: 32] [Article Influence: 2.3] [Reference Citation Analysis (0)] |
| 37. | Kobayashi T, Nishiumi S, Ikeda A, Yoshie T, Sakai A, Matsubara A, Izumi Y, Tsumura H, Tsuda M, Nishisaki H. A novel serum metabolomics-based diagnostic approach to pancreatic cancer. Cancer Epidemiol Biomarkers Prev. 2013;22:571-579. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 125] [Cited by in RCA: 134] [Article Influence: 11.2] [Reference Citation Analysis (0)] |
| 38. | Zhang T, Wu X, Ke C, Yin M, Li Z, Fan L, Zhang W, Zhang H, Zhao F, Zhou X. Identification of potential biomarkers for ovarian cancer by urinary metabolomic profiling. J Proteome Res. 2013;12:505-512. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 114] [Cited by in RCA: 138] [Article Influence: 10.6] [Reference Citation Analysis (0)] |
| 39. | Warburg O. On the origin of cancer cells. Science. 1956;123:309-314. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 9117] [Cited by in RCA: 9927] [Article Influence: 143.9] [Reference Citation Analysis (0)] |
| 40. | Reivich M, Alavi A. Positron emission tomographic studies of local cerebral glucose metabolism in humans in physiological and pathophysiological conditions. Adv Metab Disord. 1983;10:135-176. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 24] [Cited by in RCA: 24] [Article Influence: 0.6] [Reference Citation Analysis (0)] |
| 41. | Beckonert O, Coen M, Keun HC, Wang Y, Ebbels TM, Holmes E, Lindon JC, Nicholson JK. High-resolution magic-angle-spinning NMR spectroscopy for metabolic profiling of intact tissues. Nat Protoc. 2010;5:1019-1032. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 291] [Cited by in RCA: 265] [Article Influence: 17.7] [Reference Citation Analysis (0)] |
| 42. | Beckonert O, Keun HC, Ebbels TM, Bundy J, Holmes E, Lindon JC, Nicholson JK. Metabolic profiling, metabolomic and metabonomic procedures for NMR spectroscopy of urine, plasma, serum and tissue extracts. Nat Protoc. 2007;2:2692-2703. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1534] [Cited by in RCA: 1561] [Article Influence: 91.8] [Reference Citation Analysis (0)] |
| 43. | Trygg J, Holmes E, Lundstedt T. Chemometrics in metabonomics. J Proteome Res. 2007;6:469-479. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 987] [Cited by in RCA: 898] [Article Influence: 49.9] [Reference Citation Analysis (0)] |
| 44. | Mirnezami R, Jiménez B, Li JV, Kinross JM, Veselkov K, Goldin RD, Holmes E, Nicholson JK, Darzi A. Rapid diagnosis and staging of colorectal cancer via high-resolution magic angle spinning nuclear magnetic resonance (HR-MAS NMR) spectroscopy of intact tissue biopsies. Ann Surg. 2014;259:1138-1149. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 56] [Cited by in RCA: 53] [Article Influence: 4.8] [Reference Citation Analysis (0)] |
| 45. | Jiménez B, Mirnezami R, Kinross J, Cloarec O, Keun HC, Holmes E, Goldin RD, Ziprin P, Darzi A, Nicholson JK. 1H HR-MAS NMR spectroscopy of tumor-induced local metabolic “field-effects” enables colorectal cancer staging and prognostication. J Proteome Res. 2013;12:959-968. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 88] [Cited by in RCA: 92] [Article Influence: 7.7] [Reference Citation Analysis (0)] |
| 46. | Gupta RC, Win T, Bittner S. Taurine analogues; a new class of therapeutics: retrospect and prospects. Curr Med Chem. 2005;12:2021-2039. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 65] [Cited by in RCA: 67] [Article Influence: 3.4] [Reference Citation Analysis (0)] |
| 47. | Crotti S, Enzo MV, Bedin C, Pucciarelli S, Maretto I, Del Bianco P, Traldi P, Tasciotti E, Ferrari M, Rizzolio F. Clinical predictive circulating peptides in rectal cancer patients treated with neoadjuvant chemoradiotherapy. J Cell Physiol. 2015;230:1822-1828. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 15] [Cited by in RCA: 17] [Article Influence: 1.7] [Reference Citation Analysis (0)] |
| 48. | Kim K, Yeo SG, Yoo BC. Identification of hypoxanthine and phosphoenolpyruvic Acid as serum markers of chemoradiotherapy response in locally advanced rectal cancer. Cancer Res Treat. 2015;47:78-89. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 23] [Cited by in RCA: 25] [Article Influence: 2.5] [Reference Citation Analysis (0)] |
| 49. | Bartel DP. MicroRNAs: genomics, biogenesis, mechanism, and function. Cell. 2004;116:281-297. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 25833] [Cited by in RCA: 27840] [Article Influence: 1325.7] [Reference Citation Analysis (0)] |
| 50. | Rodriguez A, Griffiths-Jones S, Ashurst JL, Bradley A. Identification of mammalian microRNA host genes and transcription units. Genome Res. 2004;14:1902-1910. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1501] [Cited by in RCA: 1456] [Article Influence: 69.3] [Reference Citation Analysis (0)] |
| 51. | Garzon R, Fabbri M, Cimmino A, Calin GA, Croce CM. MicroRNA expression and function in cancer. Trends Mol Med. 2006;12:580-587. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 550] [Cited by in RCA: 582] [Article Influence: 30.6] [Reference Citation Analysis (0)] |
| 52. | Lewis BP, Burge CB, Bartel DP. Conserved seed pairing, often flanked by adenosines, indicates that thousands of human genes are microRNA targets. Cell. 2005;120:15-20. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 8847] [Cited by in RCA: 9285] [Article Influence: 464.3] [Reference Citation Analysis (0)] |
| 53. | Iorio MV, Croce CM. MicroRNAs in cancer: small molecules with a huge impact. J Clin Oncol. 2009;27:5848-5856. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 727] [Cited by in RCA: 789] [Article Influence: 49.3] [Reference Citation Analysis (0)] |
| 54. | Calin GA, Sevignani C, Dumitru CD, Hyslop T, Noch E, Yendamuri S, Shimizu M, Rattan S, Bullrich F, Negrini M. Human microRNA genes are frequently located at fragile sites and genomic regions involved in cancers. Proc Natl Acad Sci USA. 2004;101:2999-3004. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 2908] [Cited by in RCA: 3110] [Article Influence: 148.1] [Reference Citation Analysis (0)] |
| 55. | Slaby O, Svoboda M, Michalek J, Vyzula R. MicroRNAs in colorectal cancer: translation of molecular biology into clinical application. Mol Cancer. 2009;8:102. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 251] [Cited by in RCA: 290] [Article Influence: 18.1] [Reference Citation Analysis (0)] |
| 56. | Lu J, Getz G, Miska EA, Alvarez-Saavedra E, Lamb J, Peck D, Sweet-Cordero A, Ebert BL, Mak RH, Ferrando AA. MicroRNA expression profiles classify human cancers. Nature. 2005;435:834-838. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 7723] [Cited by in RCA: 7369] [Article Influence: 368.5] [Reference Citation Analysis (0)] |
| 57. | Slaby O, Svoboda M, Fabian P, Smerdova T, Knoflickova D, Bednarikova M, Nenutil R, Vyzula R. Altered expression of miR-21, miR-31, miR-143 and miR-145 is related to clinicopathologic features of colorectal cancer. Oncology. 2007;72:397-402. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 475] [Cited by in RCA: 543] [Article Influence: 31.9] [Reference Citation Analysis (0)] |
| 58. | Schetter AJ, Leung SY, Sohn JJ, Zanetti KA, Bowman ED, Yanaihara N, Yuen ST, Chan TL, Kwong DL, Au GK. MicroRNA expression profiles associated with prognosis and therapeutic outcome in colon adenocarcinoma. JAMA. 2008;299:425-436. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 1037] [Cited by in RCA: 1196] [Article Influence: 70.4] [Reference Citation Analysis (0)] |
| 59. | Della Vittoria Scarpati G, Falcetta F, Carlomagno C, Ubezio P, Marchini S, De Stefano A, Singh VK, D’Incalci M, De Placido S, Pepe S. A specific miRNA signature correlates with complete pathological response to neoadjuvant chemoradiotherapy in locally advanced rectal cancer. Int J Radiat Oncol Biol Phys. 2012;83:1113-1119. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 116] [Cited by in RCA: 133] [Article Influence: 9.5] [Reference Citation Analysis (0)] |
| 60. | Svoboda M, Sana J, Fabian P, Kocakova I, Gombosova J, Nekvindova J, Radova L, Vyzula R, Slaby O. MicroRNA expression profile associated with response to neoadjuvant chemoradiotherapy in locally advanced rectal cancer patients. Radiat Oncol. 2012;7:195. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 92] [Cited by in RCA: 103] [Article Influence: 7.9] [Reference Citation Analysis (0)] |
| 61. | Kheirelseid EA, Miller N, Chang KH, Curran C, Hennessey E, Sheehan M, Newell J, Lemetre C, Balls G, Kerin MJ. miRNA expressions in rectal cancer as predictors of response to neoadjuvant chemoradiation therapy. Int J Colorectal Dis. 2013;28:247-260. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 52] [Cited by in RCA: 59] [Article Influence: 4.9] [Reference Citation Analysis (0)] |
| 62. | Svoboda M, Izakovicova Holla L, Sefr R, Vrtkova I, Kocakova I, Tichy B, Dvorak J. Micro-RNAs miR125b and miR137 are frequently upregulated in response to capecitabine chemoradiotherapy of rectal cancer. Int J Oncol. 2008;33:541-547. [PubMed] |
| 63. | Rossi L, Bonmassar E, Faraoni I. Modification of miR gene expression pattern in human colon cancer cells following exposure to 5-fluorouracil in vitro. Pharmacol Res. 2007;56:248-253. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 131] [Cited by in RCA: 138] [Article Influence: 7.7] [Reference Citation Analysis (0)] |
| 64. | Nakajima G, Hayashi K, Xi Y, Kudo K, Uchida K, Takasaki K, Yamamoto M, Ju J. Non-coding MicroRNAs hsa-let-7g and hsa-miR-181b are Associated with Chemoresponse to S-1 in Colon Cancer. Cancer Genomics Proteomics. 2006;3:317-324. [PubMed] |
| 65. | Esquela-Kerscher A, Slack FJ. Oncomirs - microRNAs with a role in cancer. Nat Rev Cancer. 2006;6:259-269. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 5384] [Cited by in RCA: 5607] [Article Influence: 295.1] [Reference Citation Analysis (0)] |
| 66. | Lianidou ES, Mavroudis D, Sotiropoulou G, Agelaki S, Pantel K. What’s new on circulating tumor cells? A meeting report. Breast Cancer Res. 2010;12:307. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 27] [Cited by in RCA: 35] [Article Influence: 2.3] [Reference Citation Analysis (0)] |
| 67. | Shigeyasu K, Toden S, Zumwalt TJ, Okugawa Y, Goel A. Emerging Role of MicroRNAs as Liquid Biopsy Biomarkers in Gastrointestinal Cancers. Clin Cancer Res. 2017;23:2391-2399. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 75] [Cited by in RCA: 106] [Article Influence: 13.3] [Reference Citation Analysis (0)] |
| 68. | Nordgård O, Tjensvoll K, Gilje B, Søreide K. Circulating tumour cells and DNA as liquid biopsies in gastrointestinal cancer. Br J Surg. 2018;105:e110-e120. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 36] [Cited by in RCA: 44] [Article Influence: 6.3] [Reference Citation Analysis (0)] |
| 69. | Bettegowda C, Sausen M, Leary RJ, Kinde I, Wang Y, Agrawal N, Bartlett BR, Wang H, Luber B, Alani RM. Detection of circulating tumor DNA in early- and late-stage human malignancies. Sci Transl Med. 2014;6:224ra24. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 2770] [Cited by in RCA: 3549] [Article Influence: 322.6] [Reference Citation Analysis (0)] |
| 70. | Jahr S, Hentze H, Englisch S, Hardt D, Fackelmayer FO, Hesch RD, Knippers R. DNA fragments in the blood plasma of cancer patients: quantitations and evidence for their origin from apoptotic and necrotic cells. Cancer Res. 2001;61:1659-1665. [PubMed] |
| 71. | Bhangu JS, Taghizadeh H, Braunschmid T, Bachleitner-Hofmann T, Mannhalter C. Circulating cell-free DNA in plasma of colorectal cancer patients - A potential biomarker for tumor burden. Surg Oncol. 2017;26:395-401. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 15] [Cited by in RCA: 20] [Article Influence: 2.5] [Reference Citation Analysis (0)] |
| 72. | Reinert T, Schøler LV, Thomsen R, Tobiasen H, Vang S, Nordentoft I, Lamy P, Kannerup AS, Mortensen FV, Stribolt K. Analysis of circulating tumour DNA to monitor disease burden following colorectal cancer surgery. Gut. 2016;65:625-634. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 277] [Cited by in RCA: 341] [Article Influence: 37.9] [Reference Citation Analysis (0)] |
| 73. | Frattini M, Gallino G, Signoroni S, Balestra D, Battaglia L, Sozzi G, Leo E, Pilotti S, Pierotti MA. Quantitative analysis of plasma DNA in colorectal cancer patients: a novel prognostic tool. Ann N Y Acad Sci. 2006;1075:185-190. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 66] [Cited by in RCA: 74] [Article Influence: 4.1] [Reference Citation Analysis (0)] |
| 74. | Schwarzenbach H, Stoehlmacher J, Pantel K, Goekkurt E. Detection and monitoring of cell-free DNA in blood of patients with colorectal cancer. Ann N Y Acad Sci. 2008;1137:190-196. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 131] [Cited by in RCA: 128] [Article Influence: 7.5] [Reference Citation Analysis (0)] |
| 75. | Tang M, Deng Z, Li B, Peng Y, Song M, Liu J. Circulating tumor DNA is effective for detection of KRAS mutation in colorectal cancer: a meta-analysis. Int J Biol Markers. 2017;32:e421-e427. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 11] [Cited by in RCA: 11] [Article Influence: 1.4] [Reference Citation Analysis (0)] |
| 76. | Germano G, Mauri G, Siravegna G, Dive C, Pierce J, Di Nicolantonio F, D’Incalci M, Bardelli A, Siena S, Sartore-Bianchi A. Parallel Evaluation of Circulating Tumor DNA and Circulating Tumor Cells in Metastatic Colorectal Cancer. Clin Colorectal Cancer. 2018;17:80-83. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 35] [Cited by in RCA: 39] [Article Influence: 5.6] [Reference Citation Analysis (0)] |
| 77. | Tie J, Semira C, Gibbs P. Circulating tumor DNA as a biomarker to guide therapy in post-operative locally advanced rectal cancer: the best option? Expert Rev Mol Diagn. 2018;18:1-3. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 6] [Cited by in RCA: 6] [Article Influence: 0.8] [Reference Citation Analysis (0)] |
| 78. | Shapiro B, Chakrabarty M, Cohn EM, Leon SA. Determination of circulating DNA levels in patients with benign or malignant gastrointestinal disease. Cancer. 1983;51:2116-2120. [RCA] [PubMed] [DOI] [Full Text] [Cited by in RCA: 1] [Reference Citation Analysis (0)] |
| 79. | Ganepola GA, Nizin J, Rutledge JR, Chang DH. Use of blood-based biomarkers for early diagnosis and surveillance of colorectal cancer. World J Gastrointest Oncol. 2014;6:83-97. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in CrossRef: 58] [Cited by in RCA: 62] [Article Influence: 5.6] [Reference Citation Analysis (0)] |
| 80. | Zanutto S, Pizzamiglio S, Ghilotti M, Bertan C, Ravagnani F, Perrone F, Leo E, Pilotti S, Verderio P, Gariboldi M. Circulating miR-378 in plasma: a reliable, haemolysis-independent biomarker for colorectal cancer. Br J Cancer. 2014;110:1001-1007. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 97] [Cited by in RCA: 108] [Article Influence: 9.8] [Reference Citation Analysis (0)] |
| 81. | Diaz LA Jr, Bardelli A. Liquid biopsies: genotyping circulating tumor DNA. J Clin Oncol. 2014;32:579-586. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1413] [Cited by in RCA: 1686] [Article Influence: 153.3] [Reference Citation Analysis (0)] |
| 82. | Cortez MA, Bueso-Ramos C, Ferdin J, Lopez-Berestein G, Sood AK, Calin GA. MicroRNAs in body fluids--the mix of hormones and biomarkers. Nat Rev Clin Oncol. 2011;8:467-477. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1009] [Cited by in RCA: 1130] [Article Influence: 80.7] [Reference Citation Analysis (0)] |
| 83. | Ogata-Kawata H, Izumiya M, Kurioka D, Honma Y, Yamada Y, Furuta K, Gunji T, Ohta H, Okamoto H, Sonoda H. Circulating exosomal microRNAs as biomarkers of colon cancer. PLoS One. 2014;9:e92921. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 557] [Cited by in RCA: 647] [Article Influence: 58.8] [Reference Citation Analysis (1)] |
| 84. | Liu GH, Zhou ZG, Chen R, Wang MJ, Zhou B, Li Y, Sun XF. Serum miR-21 and miR-92a as biomarkers in the diagnosis and prognosis of colorectal cancer. Tumour Biol. 2013;34:2175-2181. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 143] [Cited by in RCA: 170] [Article Influence: 14.2] [Reference Citation Analysis (0)] |
| 85. | Toiyama Y, Takahashi M, Hur K, Nagasaka T, Tanaka K, Inoue Y, Kusunoki M, Boland CR, Goel A. Serum miR-21 as a diagnostic and prognostic biomarker in colorectal cancer. J Natl Cancer Inst. 2013;105:849-859. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 345] [Cited by in RCA: 402] [Article Influence: 33.5] [Reference Citation Analysis (0)] |
| 86. | Shivapurkar N, Weiner LM, Marshall JL, Madhavan S, Deslattes Mays A, Juhl H, Wellstein A. Recurrence of early stage colon cancer predicted by expression pattern of circulating microRNAs. PLoS One. 2014;9:e84686. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 39] [Cited by in RCA: 45] [Article Influence: 4.1] [Reference Citation Analysis (0)] |
| 87. | Hur K, Toiyama Y, Okugawa Y, Ide S, Imaoka H, Boland CR, Goel A. Circulating microRNA-203 predicts prognosis and metastasis in human colorectal cancer. Gut. 2017;66:654-665. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 129] [Cited by in RCA: 161] [Article Influence: 20.1] [Reference Citation Analysis (0)] |
| 88. | Cheng H, Zhang L, Cogdell DE, Zheng H, Schetter AJ, Nykter M, Harris CC, Chen K, Hamilton SR, Zhang W. Circulating plasma MiR-141 is a novel biomarker for metastatic colon cancer and predicts poor prognosis. PLoS One. 2011;6:e17745. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 303] [Cited by in RCA: 340] [Article Influence: 24.3] [Reference Citation Analysis (0)] |
| 89. | Lofton-Day C, Model F, Devos T, Tetzner R, Distler J, Schuster M, Song X, Lesche R, Liebenberg V, Ebert M. DNA methylation biomarkers for blood-based colorectal cancer screening. Clin Chem. 2008;54:414-423. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 340] [Cited by in RCA: 375] [Article Influence: 20.8] [Reference Citation Analysis (0)] |
| 90. | Adler A, Geiger S, Keil A, Bias H, Schatz P, deVos T, Dhein J, Zimmermann M, Tauber R, Wiedenmann B. Improving compliance to colorectal cancer screening using blood and stool based tests in patients refusing screening colonoscopy in Germany. BMC Gastroenterol. 2014;14:183. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 111] [Cited by in RCA: 164] [Article Influence: 14.9] [Reference Citation Analysis (0)] |
| 91. | Bhangu A, Wood G, Mirnezami A, Darzi A, Tekkis P, Goldin R. Epithelial mesenchymal transition in colorectal cancer: Seminal role in promoting disease progression and resistance to neoadjuvant therapy. Surg Oncol. 2012;21:316-323. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 41] [Cited by in RCA: 43] [Article Influence: 3.3] [Reference Citation Analysis (0)] |
| 92. | Kalluri R, Weinberg RA. The basics of epithelial-mesenchymal transition. J Clin Invest. 2009;119:1420-1428. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 6575] [Cited by in RCA: 7906] [Article Influence: 494.1] [Reference Citation Analysis (0)] |
| 93. | Peinado H, Portillo F, Cano A. Transcriptional regulation of cadherins during development and carcinogenesis. Int J Dev Biol. 2004;48:365-375. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 419] [Cited by in RCA: 434] [Article Influence: 21.7] [Reference Citation Analysis (0)] |
| 94. | Thiery JP, Sleeman JP. Complex networks orchestrate epithelial-mesenchymal transitions. Nat Rev Mol Cell Biol. 2006;7:131-142. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 2893] [Cited by in RCA: 3087] [Article Influence: 162.5] [Reference Citation Analysis (0)] |
| 95. | Reinacher-Schick A, Baldus SE, Romdhana B, Landsberg S, Zapatka M, Mönig SP, Hölscher AH, Dienes HP, Schmiegel W, Schwarte-Waldhoff I. Loss of Smad4 correlates with loss of the invasion suppressor E-cadherin in advanced colorectal carcinomas. J Pathol. 2004;202:412-420. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 40] [Cited by in RCA: 46] [Article Influence: 2.2] [Reference Citation Analysis (0)] |
| 96. | Hur K, Toiyama Y, Takahashi M, Balaguer F, Nagasaka T, Koike J, Hemmi H, Koi M, Boland CR, Goel A. MicroRNA-200c modulates epithelial-to-mesenchymal transition (EMT) in human colorectal cancer metastasis. Gut. 2013;62:1315-1326. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 390] [Cited by in RCA: 452] [Article Influence: 37.7] [Reference Citation Analysis (0)] |
| 97. | Zlobec I, Lugli A. Epithelial mesenchymal transition and tumor budding in aggressive colorectal cancer: tumor budding as oncotarget. Oncotarget. 2010;1:651-661. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 209] [Cited by in RCA: 256] [Article Influence: 18.3] [Reference Citation Analysis (0)] |
| 98. | Kevans D, Wang LM, Sheahan K, Hyland J, O’Donoghue D, Mulcahy H, O’Sullivan J. Epithelial-mesenchymal transition (EMT) protein expression in a cohort of stage II colorectal cancer patients with characterized tumor budding and mismatch repair protein status. Int J Surg Pathol. 2011;19:751-760. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 46] [Cited by in RCA: 56] [Article Influence: 4.0] [Reference Citation Analysis (0)] |
| 99. | Nemolato S, Restivo A, Cabras T, Coni P, Zorcolo L, Orrù G, Fanari M, Cau F, Gerosa C, Fanni D. Thymosin β 4 in colorectal cancer is localized predominantly at the invasion front in tumor cells undergoing epithelial mesenchymal transition. Cancer Biol Ther. 2012;13:191-197. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 29] [Cited by in RCA: 38] [Article Influence: 2.9] [Reference Citation Analysis (0)] |
| 100. | Zlobec I, Lugli A. Invasive front of colorectal cancer: dynamic interface of pro/anti-tumor factors. World J Gastroenterol. 2009;15:5898-5906. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in CrossRef: 89] [Cited by in RCA: 100] [Article Influence: 6.3] [Reference Citation Analysis (2)] |
| 101. | Kurrey NK, Jalgaonkar SP, Joglekar AV, Ghanate AD, Chaskar PD, Doiphode RY, Bapat SA. Snail and slug mediate radioresistance and chemoresistance by antagonizing p53-mediated apoptosis and acquiring a stem-like phenotype in ovarian cancer cells. Stem Cells. 2009;27:2059-2068. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 461] [Cited by in RCA: 525] [Article Influence: 32.8] [Reference Citation Analysis (0)] |
| 102. | Du Z, Qin R, Wei C, Wang M, Shi C, Tian R, Peng C. Pancreatic cancer cells resistant to chemoradiotherapy rich in “stem-cell-like” tumor cells. Dig Dis Sci. 2011;56:741-750. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 90] [Cited by in RCA: 103] [Article Influence: 7.4] [Reference Citation Analysis (0)] |
| 103. | Zhang S, Wang X, Osunkoya AO, Iqbal S, Wang Y, Chen Z, Müller S, Chen Z, Josson S, Coleman IM. EPLIN downregulation promotes epithelial-mesenchymal transition in prostate cancer cells and correlates with clinical lymph node metastasis. Oncogene. 2011;30:4941-4952. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 67] [Cited by in RCA: 68] [Article Influence: 4.9] [Reference Citation Analysis (0)] |
| 104. | Nicolini A, Ferrari P, Fini M, Borsari V, Fallahi P, Antonelli A, Berti P, Carpi A, Miccoli P. Stem cells: their role in breast cancer development and resistance to treatment. Curr Pharm Biotechnol. 2011;12:196-205. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 28] [Cited by in RCA: 32] [Article Influence: 2.3] [Reference Citation Analysis (0)] |
| 105. | Shintani Y, Okimura A, Sato K, Nakagiri T, Kadota Y, Inoue M, Sawabata N, Minami M, Ikeda N, Kawahara K. Epithelial to mesenchymal transition is a determinant of sensitivity to chemoradiotherapy in non-small cell lung cancer. Ann Thorac Surg. 2011;92:1794-804; discussion 1804. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 117] [Cited by in RCA: 131] [Article Influence: 9.4] [Reference Citation Analysis (0)] |
| 106. | Hoshino H, Miyoshi N, Nagai K, Tomimaru Y, Nagano H, Sekimoto M, Doki Y, Mori M, Ishii H. Epithelial-mesenchymal transition with expression of SNAI1-induced chemoresistance in colorectal cancer. Biochem Biophys Res Commun. 2009;390:1061-1065. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 54] [Cited by in RCA: 61] [Article Influence: 3.8] [Reference Citation Analysis (0)] |
| 107. | Papageorgis P, Cheng K, Ozturk S, Gong Y, Lambert AW, Abdolmaleky HM, Zhou JR, Thiagalingam S. Smad4 inactivation promotes malignancy and drug resistance of colon cancer. Cancer Res. 2011;71:998-1008. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 177] [Cited by in RCA: 175] [Article Influence: 12.5] [Reference Citation Analysis (0)] |
| 108. | Yang AD, Fan F, Camp ER, van Buren G, Liu W, Somcio R, Gray MJ, Cheng H, Hoff PM, Ellis LM. Chronic oxaliplatin resistance induces epithelial-to-mesenchymal transition in colorectal cancer cell lines. Clin Cancer Res. 2006;12:4147-4153. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 366] [Cited by in RCA: 435] [Article Influence: 24.2] [Reference Citation Analysis (0)] |
| 109. | Kawamoto A, Yokoe T, Tanaka K, Saigusa S, Toiyama Y, Yasuda H, Inoue Y, Miki C, Kusunoki M. Radiation induces epithelial-mesenchymal transition in colorectal cancer cells. Oncol Rep. 2012;27:51-57. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 24] [Cited by in RCA: 67] [Article Influence: 4.8] [Reference Citation Analysis (0)] |
| 110. | Bhangu A, Wood G, Brown G, Darzi A, Tekkis P, Goldin R. The role of epithelial mesenchymal transition and resistance to neoadjuvant therapy in locally advanced rectal cancer. Colorectal Dis. 2014;16:O133-O143. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 38] [Cited by in RCA: 44] [Article Influence: 4.0] [Reference Citation Analysis (0)] |
| 111. | Vazquez-Martin A, Oliveras-Ferraros C, Cufí S, Del Barco S, Martin-Castillo B, Menendez JA. Metformin regulates breast cancer stem cell ontogeny by transcriptional regulation of the epithelial-mesenchymal transition (EMT) status. Cell Cycle. 2010;9:3807-3814. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 136] [Cited by in RCA: 152] [Article Influence: 10.1] [Reference Citation Analysis (0)] |
| 112. | Jiralerspong S, Palla SL, Giordano SH, Meric-Bernstam F, Liedtke C, Barnett CM, Hsu L, Hung MC, Hortobagyi GN, Gonzalez-Angulo AM. Metformin and pathologic complete responses to neoadjuvant chemotherapy in diabetic patients with breast cancer. J Clin Oncol. 2009;27:3297-3302. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 627] [Cited by in RCA: 670] [Article Influence: 41.9] [Reference Citation Analysis (0)] |
| 113. | Adhim Z, Matsuoka T, Bito T, Shigemura K, Lee KM, Kawabata M, Fujisawa M, Nibu K, Shirakawa T. In vitro and in vivo inhibitory effect of three Cox-2 inhibitors and epithelial-to-mesenchymal transition in human bladder cancer cell lines. Br J Cancer. 2011;105:393-402. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 46] [Cited by in RCA: 52] [Article Influence: 3.7] [Reference Citation Analysis (0)] |
| 114. | Baron JA, Sandler RS, Bresalier RS, Quan H, Riddell R, Lanas A, Bolognese JA, Oxenius B, Horgan K, Loftus S. A randomized trial of rofecoxib for the chemoprevention of colorectal adenomas. Gastroenterology. 2006;131:1674-1682. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 330] [Cited by in RCA: 313] [Article Influence: 16.5] [Reference Citation Analysis (0)] |
| 115. | Benjamin RS, Choi H, Macapinlac HA, Burgess MA, Patel SR, Chen LL, Podoloff DA, Charnsangavej C. We should desist using RECIST, at least in GIST. J Clin Oncol. 2007;25:1760-1764. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 434] [Cited by in RCA: 410] [Article Influence: 22.8] [Reference Citation Analysis (0)] |
| 116. | O’Connor JP, Jackson A, Asselin MC, Buckley DL, Parker GJ, Jayson GC. Quantitative imaging biomarkers in the clinical development of targeted therapeutics: current and future perspectives. Lancet Oncol. 2008;9:766-776. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 132] [Cited by in RCA: 118] [Article Influence: 6.9] [Reference Citation Analysis (0)] |
| 117. | Hoogstraten B, Miller AB, Staquet M, Winkler A. WHO Handbook for Reporting Results of Cancer Treatment. Geneva: World Health Organization 1979; . |
| 118. | Therasse P, Arbuck SG, Eisenhauer EA, Wanders J, Kaplan RS, Rubinstein L, Verweij J, Van Glabbeke M, van Oosterom AT, Christian MC. New guidelines to evaluate the response to treatment in solid tumors. European Organization for Research and Treatment of Cancer, National Cancer Institute of the United States, National Cancer Institute of Canada. J Natl Cancer Inst. 2000;92:205-216. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 12751] [Cited by in RCA: 13076] [Article Influence: 523.0] [Reference Citation Analysis (0)] |
| 119. | Morgan B, Thomas AL, Drevs J, Hennig J, Buchert M, Jivan A, Horsfield MA, Mross K, Ball HA, Lee L. Dynamic contrast-enhanced magnetic resonance imaging as a biomarker for the pharmacological response of PTK787/ZK 222584, an inhibitor of the vascular endothelial growth factor receptor tyrosine kinases, in patients with advanced colorectal cancer and liver metastases: results from two phase I studies. J Clin Oncol. 2003;21:3955-3964. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 544] [Cited by in RCA: 496] [Article Influence: 22.5] [Reference Citation Analysis (0)] |
| 120. | Patel UB, Blomqvist LK, Taylor F, George C, Guthrie A, Bees N, Brown G. MRI after treatment of locally advanced rectal cancer: how to report tumor response--the MERCURY experience. AJR Am J Roentgenol. 2012;199:W486-W495. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 126] [Cited by in RCA: 157] [Article Influence: 12.1] [Reference Citation Analysis (0)] |
| 121. | Mandard AM, Dalibard F, Mandard JC, Marnay J, Henry-Amar M, Petiot JF, Roussel A, Jacob JH, Segol P, Samama G. Pathologic assessment of tumor regression after preoperative chemoradiotherapy of esophageal carcinoma. Clinicopathologic correlations. Cancer. 1994;73:2680-2686. [RCA] [PubMed] [DOI] [Full Text] [Cited by in RCA: 25] [Reference Citation Analysis (0)] |
| 122. | Patel UB, Brown G, Rutten H, West N, Sebag-Montefiore D, Glynne-Jones R, Rullier E, Peeters M, Van Cutsem E, Ricci S. Comparison of magnetic resonance imaging and histopathological response to chemoradiotherapy in locally advanced rectal cancer. Ann Surg Oncol. 2012;19:2842-2852. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 150] [Cited by in RCA: 173] [Article Influence: 13.3] [Reference Citation Analysis (0)] |
| 123. | Patel UB, Taylor F, Blomqvist L, George C, Evans H, Tekkis P, Quirke P, Sebag-Montefiore D, Moran B, Heald R. Magnetic resonance imaging-detected tumor response for locally advanced rectal cancer predicts survival outcomes: MERCURY experience. J Clin Oncol. 2011;29:3753-3760. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 431] [Cited by in RCA: 501] [Article Influence: 35.8] [Reference Citation Analysis (0)] |
| 124. | Chand M, Swift RI, Tekkis PP, Chau I, Brown G. Extramural venous invasion is a potential imaging predictive biomarker of neoadjuvant treatment in rectal cancer. Br J Cancer. 2014;110:19-25. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 102] [Cited by in RCA: 121] [Article Influence: 10.1] [Reference Citation Analysis (0)] |
| 125. | Kelloff GJ, Hoffman JM, Johnson B, Scher HI, Siegel BA, Cheng EY, Cheson BD, O’shaughnessy J, Guyton KZ, Mankoff DA. Progress and promise of FDG-PET imaging for cancer patient management and oncologic drug development. Clin Cancer Res. 2005;11:2785-2808. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 490] [Cited by in RCA: 474] [Article Influence: 23.7] [Reference Citation Analysis (0)] |
| 126. | Tarantola G, Zito F, Gerundini P. PET instrumentation and reconstruction algorithms in whole-body applications. J Nucl Med. 2003;44:756-769. [PubMed] |
| 127. | Leggett DA, Kelley BB, Bunce IH, Miles KA. Colorectal cancer: diagnostic potential of CT measurements of hepatic perfusion and implications for contrast enhancement protocols. Radiology. 1997;205:716-720. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 93] [Cited by in RCA: 92] [Article Influence: 3.3] [Reference Citation Analysis (0)] |
| 128. | Goh V, Halligan S, Taylor SA, Burling D, Bassett P, Bartram CI. Differentiation between diverticulitis and colorectal cancer: quantitative CT perfusion measurements versus morphologic criteria--initial experience. Radiology. 2007;242:456-462. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 100] [Cited by in RCA: 76] [Article Influence: 4.2] [Reference Citation Analysis (0)] |
| 129. | Goh V, Halligan S, Wellsted DM, Bartram CI. Can perfusion CT assessment of primary colorectal adenocarcinoma blood flow at staging predict for subsequent metastatic disease? A pilot study. Eur Radiol. 2009;19:79-89. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 71] [Cited by in RCA: 68] [Article Influence: 4.0] [Reference Citation Analysis (0)] |
| 130. | Hayano K, Shuto K, Koda K, Yanagawa N, Okazumi S, Matsubara H. Quantitative measurement of blood flow using perfusion CT for assessing clinicopathologic features and prognosis in patients with rectal cancer. Dis Colon Rectum. 2009;52:1624-1629. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 54] [Cited by in RCA: 48] [Article Influence: 3.0] [Reference Citation Analysis (0)] |
| 131. | Abdou AG, Aiad H, Asaad N, Abd El-Wahed M, Serag El-Dien M. Immunohistochemical evaluation of vascular endothelial growth factor (VEGF) in colorectal carcinoma. J Egypt Natl Canc Inst. 2006;18:311-322. [PubMed] |
| 132. | Cao D, Hou M, Guan YS, Jiang M, Yang Y, Gou HF. Expression of HIF-1alpha and VEGF in colorectal cancer: association with clinical outcomes and prognostic implications. BMC Cancer. 2009;9:432. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 134] [Cited by in RCA: 159] [Article Influence: 9.9] [Reference Citation Analysis (0)] |
| 133. | Scheer MG, Stollman TH, Boerman OC, Verrijp K, Sweep FC, Leenders WP, Ruers TJ, Oyen WJ. Imaging liver metastases of colorectal cancer patients with radiolabelled bevacizumab: Lack of correlation with VEGF-A expression. Eur J Cancer. 2008;44:1835-1840. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 45] [Cited by in RCA: 43] [Article Influence: 2.5] [Reference Citation Analysis (0)] |
| 134. | Stollman TH, Scheer MG, Leenders WP, Verrijp KC, Soede AC, Oyen WJ, Ruers TJ, Boerman OC. Specific imaging of VEGF-A expression with radiolabeled anti-VEGF monoclonal antibody. Int J Cancer. 2008;122:2310-2314. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 51] [Cited by in RCA: 49] [Article Influence: 2.9] [Reference Citation Analysis (0)] |
| 135. | Willett CG, Boucher Y, di Tomaso E, Duda DG, Munn LL, Tong RT, Chung DC, Sahani DV, Kalva SP, Kozin SV. Direct evidence that the VEGF-specific antibody bevacizumab has antivascular effects in human rectal cancer. Nat Med. 2004;10:145-147. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1468] [Cited by in RCA: 1443] [Article Influence: 68.7] [Reference Citation Analysis (0)] |
| 136. | Lankester KJ, Taylor JN, Stirling JJ, Boxall J, d’Arcy JA, Collins DJ, Walker-Samuel S, Leach MO, Rustin GJ, Padhani AR. Dynamic MRI for imaging tumor microvasculature: comparison of susceptibility and relaxivity techniques in pelvic tumors. J Magn Reson Imaging. 2007;25:796-805. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 38] [Cited by in RCA: 37] [Article Influence: 2.1] [Reference Citation Analysis (0)] |
| 137. | Tuncbilek N, Karakas HM, Altaner S. Dynamic MRI in indirect estimation of microvessel density, histologic grade, and prognosis in colorectal adenocarcinomas. Abdom Imaging. 2004;29:166-172. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 58] [Cited by in RCA: 59] [Article Influence: 2.8] [Reference Citation Analysis (0)] |
| 138. | Atkin G, Taylor NJ, Daley FM, Stirling JJ, Richman P, Glynne-Jones R, d’Arcy JA, Collins DJ, Padhani AR. Dynamic contrast-enhanced magnetic resonance imaging is a poor measure of rectal cancer angiogenesis. Br J Surg. 2006;93:992-1000. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 54] [Cited by in RCA: 58] [Article Influence: 3.1] [Reference Citation Analysis (0)] |
| 139. | George ML, Dzik-Jurasz AS, Padhani AR, Brown G, Tait DM, Eccles SA, Swift RI. Non-invasive methods of assessing angiogenesis and their value in predicting response to treatment in colorectal cancer. Br J Surg. 2001;88:1628-1636. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 194] [Cited by in RCA: 191] [Article Influence: 8.0] [Reference Citation Analysis (0)] |
| 140. | Thomas AL, Morgan B, Horsfield MA, Higginson A, Kay A, Lee L, Masson E, Puccio-Pick M, Laurent D, Steward WP. Phase I study of the safety, tolerability, pharmacokinetics, and pharmacodynamics of PTK787/ZK 222584 administered twice daily in patients with advanced cancer. J Clin Oncol. 2005;23:4162-4171. [PubMed] |
| 141. | Koh DM, Brown G, Riddell AM, Scurr E, Collins DJ, Allen SD, Chau I, Cunningham D, deSouza NM, Leach MO. Detection of colorectal hepatic metastases using MnDPDP MR imaging and diffusion-weighted imaging (DWI) alone and in combination. Eur Radiol. 2008;18:903-910. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 119] [Cited by in RCA: 100] [Article Influence: 5.9] [Reference Citation Analysis (0)] |
| 142. | Taouli B, Chouli M, Martin AJ, Qayyum A, Coakley FV, Vilgrain V. Chronic hepatitis: role of diffusion-weighted imaging and diffusion tensor imaging for the diagnosis of liver fibrosis and inflammation. J Magn Reson Imaging. 2008;28:89-95. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 158] [Cited by in RCA: 171] [Article Influence: 10.1] [Reference Citation Analysis (0)] |
| 143. | Ichikawa T, Erturk SM, Motosugi U, Sou H, Iino H, Araki T, Fujii H. High-B-value diffusion-weighted MRI in colorectal cancer. AJR Am J Roentgenol. 2006;187:181-184. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 175] [Cited by in RCA: 170] [Article Influence: 8.9] [Reference Citation Analysis (0)] |
| 144. | Kim JK, Kim KA, Park BW, Kim N, Cho KS. Feasibility of diffusion-weighted imaging in the differentiation of metastatic from nonmetastatic lymph nodes: early experience. J Magn Reson Imaging. 2008;28:714-719. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 142] [Cited by in RCA: 137] [Article Influence: 8.1] [Reference Citation Analysis (0)] |
| 145. | Kawaguchi T, Komatsu S, Ichikawa D, Tsujiura M, Takeshita H, Hirajima S, Miyamae M, Okajima W, Ohashi T, Imamura T. Circulating MicroRNAs: A Next-Generation Clinical Biomarker for Digestive System Cancers. Int J Mol Sci. 2016;17. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 58] [Cited by in RCA: 67] [Article Influence: 7.4] [Reference Citation Analysis (0)] |