Copyright
©The Author(s) 2025.
World J Gastrointest Oncol. Apr 15, 2025; 17(4): 103113
Published online Apr 15, 2025. doi: 10.4251/wjgo.v17.i4.103113
Published online Apr 15, 2025. doi: 10.4251/wjgo.v17.i4.103113
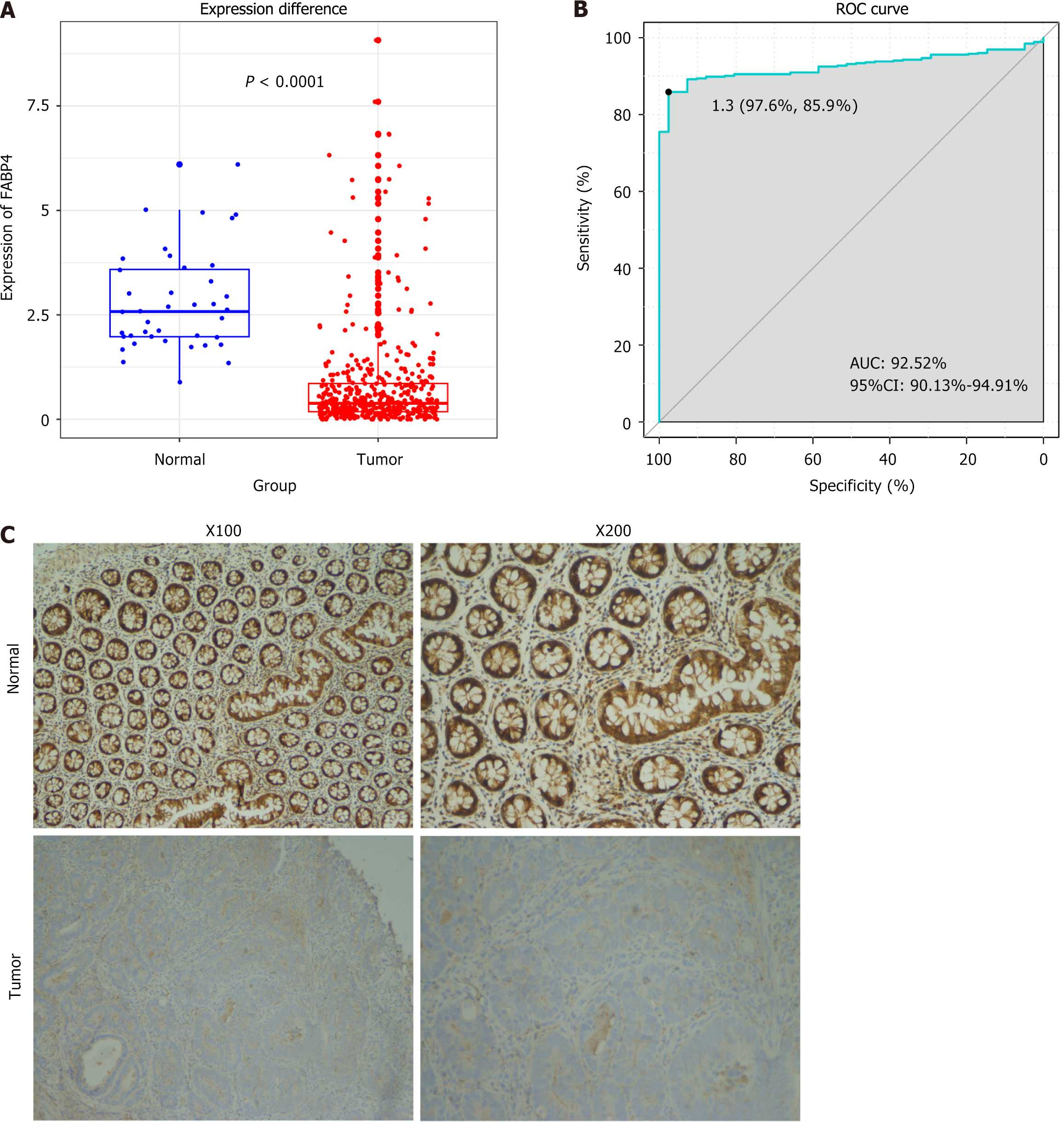
Figure 1 Diagnostic value of fatty acid-binding protein 4 expression in colon adenocarcinoma.
A: Comparison of fatty acid-binding protein 4 (FABP4) mRNA expression levels in colon adenocarcinoma (COAD) tissues and normal tissues; B: Receiver operating characteristic curve for the diagnostic efficacy of FABP4; C: The FABP4 protein levels in COAD tissues and adjacent noncancerous colon tissues were determined via immunohistochemical staining at magnifications of × 100 and × 200. COAD: Colon adenocarcinoma; ROC: Receiver operating characteristic; AUC: Area under the curve.
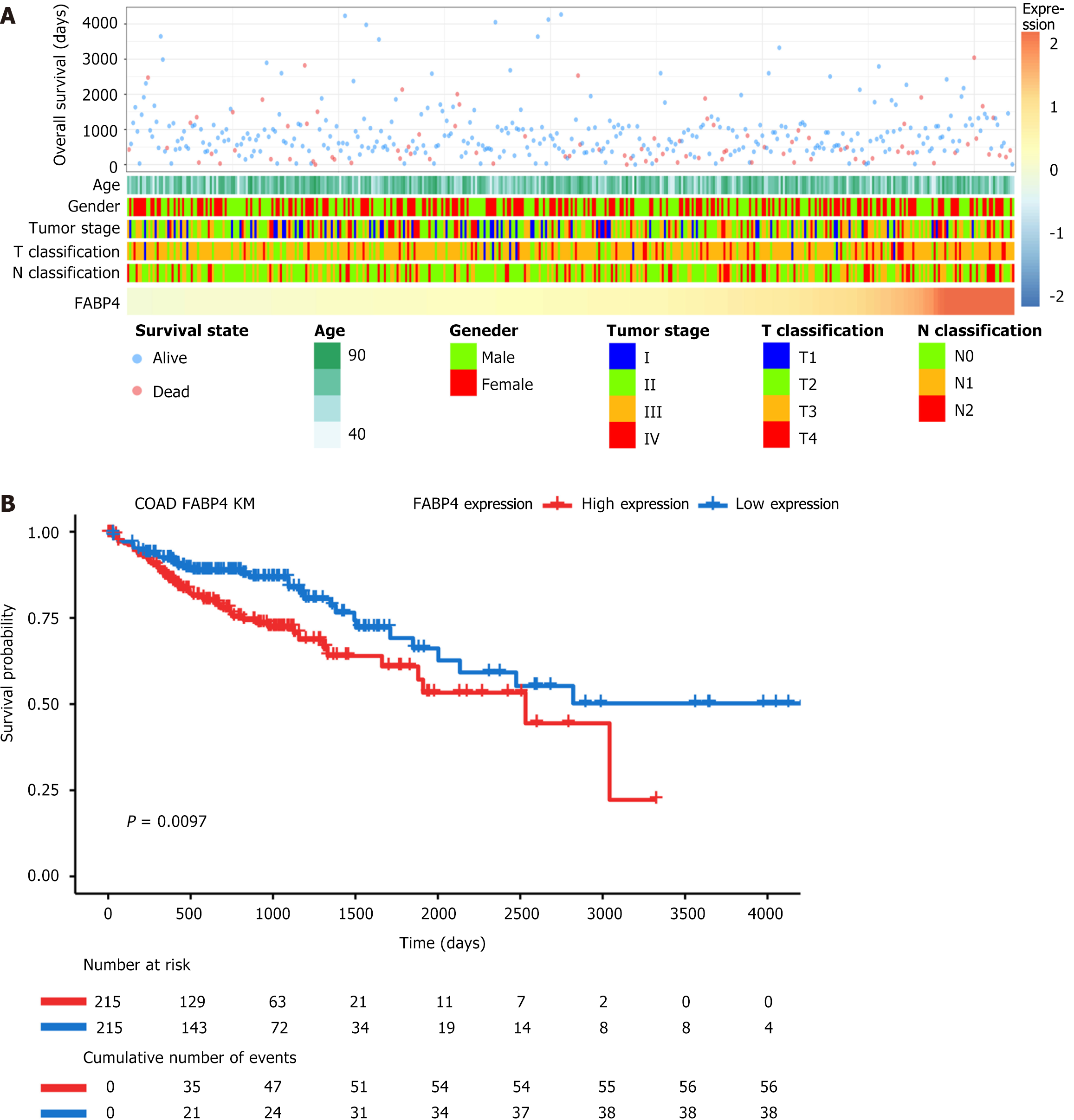
Figure 2 Relationships between fatty acid-binding protein 4 expression and clinical factors in patients with colon adenocarcinoma and Kaplan-Meier survival analysis of patients stratified according to fatty acid-binding protein 4 expression.
A: The relationships between fatty acid-binding protein 4 (FABP4) expression and various clinical factors are ranked in ascending order of FABP4 expression; B: Kaplan-Meier survival curves of the FABP4 high-expression group and low-expression group. The cutoff for each group was the median expression of FABP4. COAD: Colon adenocarcinoma.
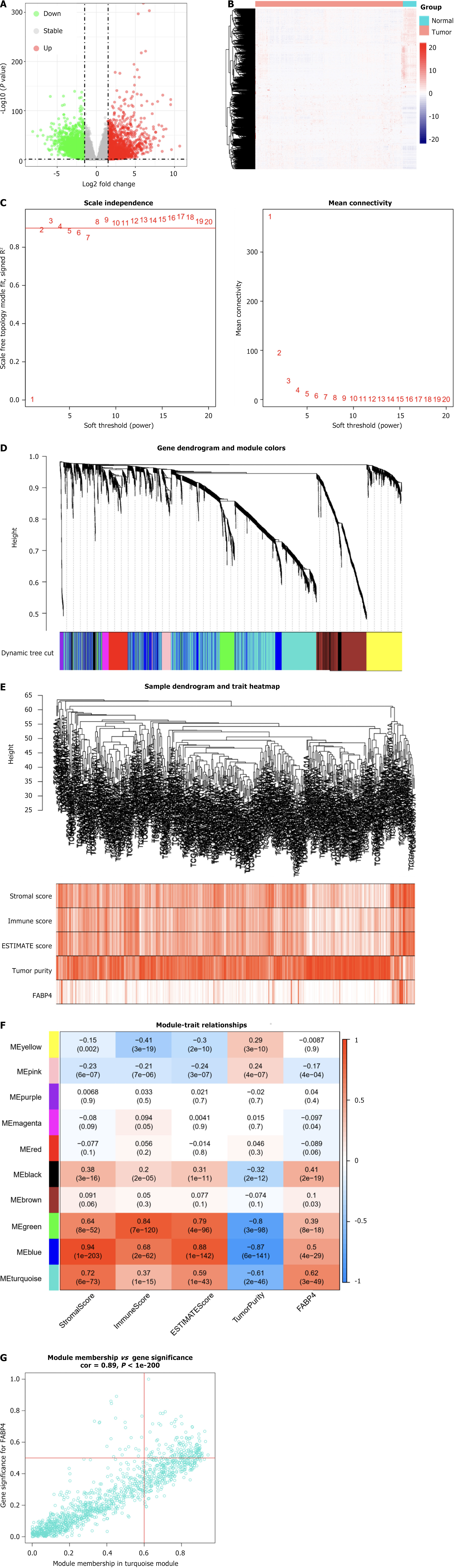
Figure 3 Screen for immune modules and genes related to fatty acid-binding protein 4 in colon adenocarcinoma tissues.
A: Volcano plot of differentially expressed genes (DEGs) between colon adenocarcinoma (COAD) tissues and normal tissues in The Cancer Genome Atlas (TCGA) database; B: Heatmap of DEGs between COAD tissues and normal tissues in TCGA database; C: Calculation of the scale-free fit indices of various soft-thresholding powers (β) and analysis of the mean connectivity of various soft-thresholding powers (β); D: DEGs were clustered based on the dissimilarity measure (1-TOM) and divided into 10 modules; E: Clustering dendrogram of COAD patients; F: A correlation heatmap between module eigengenes and immune parameters (fatty acid-binding protein 4 was used as the main research object) in patients with COAD; G: Scatter plot of the turquoise module eigengenes. COAD: Colon adenocarcinoma; DEGs: Differentially expressed genes; TCGA: The Cancer Genome Atlas.
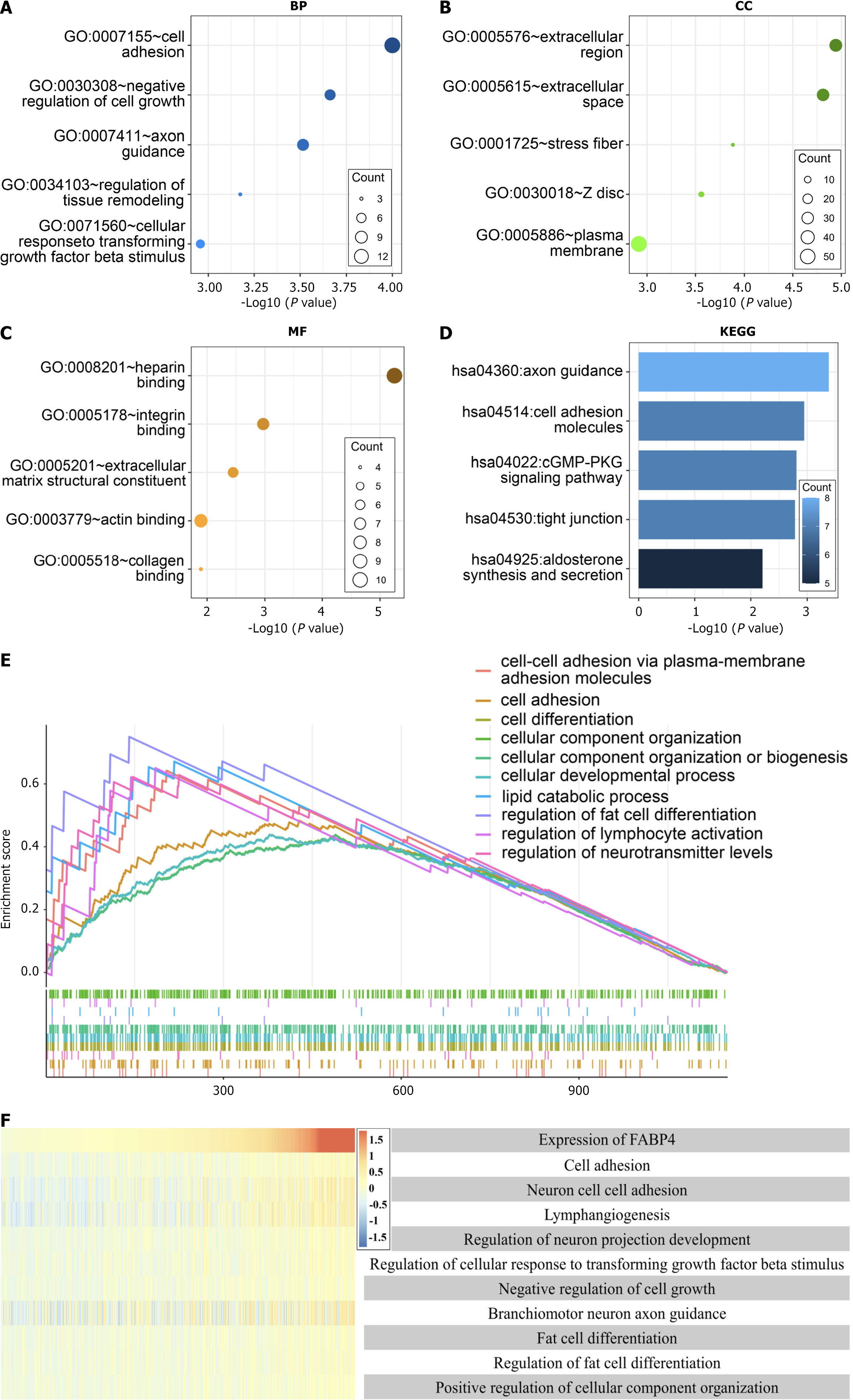
Figure 4 Results of the Gene Ontology and Kyoto Encyclopedia of Genes and Genomes functional enrichment analyses, gene set enrichment analysis and gene set variation analysis.
A-C: Analysis of the biological process, cellular component and molecular function terms of genes strongly associated with fatty acid-binding protein 4 (FABP4); D: Kyoto Encyclopedia of Genes and Genomes analysis of genes strongly associated with FABP4; E: GSEA of genes highly correlated with FABP4; F: Correlation analysis between FABP4 expression and functional enrichment scores. The heatmap shows the expression of FABP4 and the enrichment scores of the functions of the genes in each patient in The Cancer Genome Atlas database. The samples were arranged in ascending order of FABP4 expression. TCGA: The Cancer Genome Atlas; KEGG: Kyoto Encyclopedia of Genes and Genomes; FABP4: Fatty acid-binding protein 4; CC: Cellular component; BP: Biological process; MF: Molecular function.
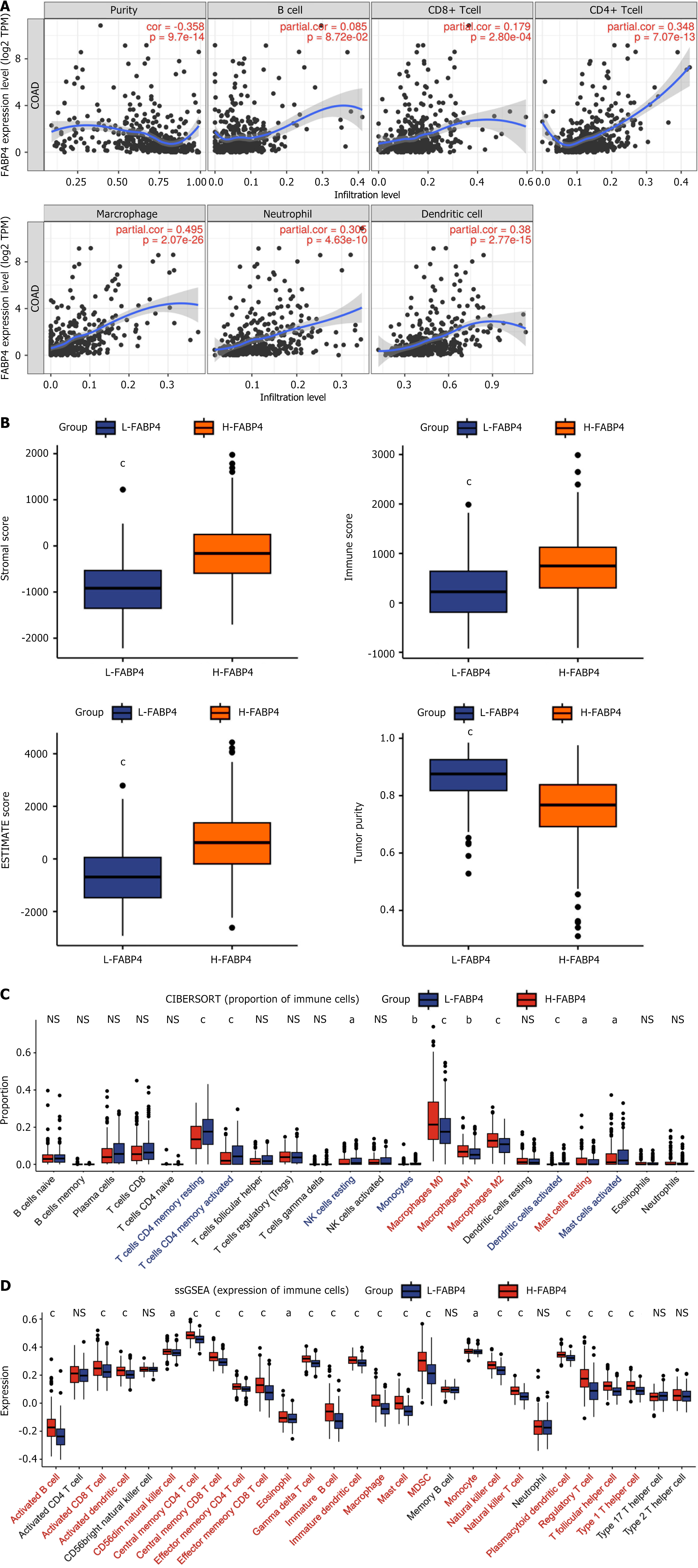
Figure 5 Comparison of immune characteristics associated with fatty acid-binding protein 4 expression.
A: The expression level of fatty acid-binding protein 4 (FABP4) was significantly positively correlated with the infiltration level of immune cells in colon adenocarcinoma; B: Comparisons of the stromal score, immune score, ESTIMATE score, and tumor purity between the high FABP4 expression group and the low FABP4 expression group; C: Proportion of immune cells in the high FABP4 expression group and low FABP4 expression group; D: Comparison of immune cell infiltration between the high FABP4 expression group and the low FABP4 expression group. P values are labeled with asterisks. NS: Not significant; aP < 0.05; bP < 0.01; and cP < 0.001; COAD: Colon adenocarcinoma; ESTIMATE: Estimation of STromal and Immune cells in MAlignant Tumor tissues using Expression data; ssGSEA: Single-sample gene set enrichment analysis.
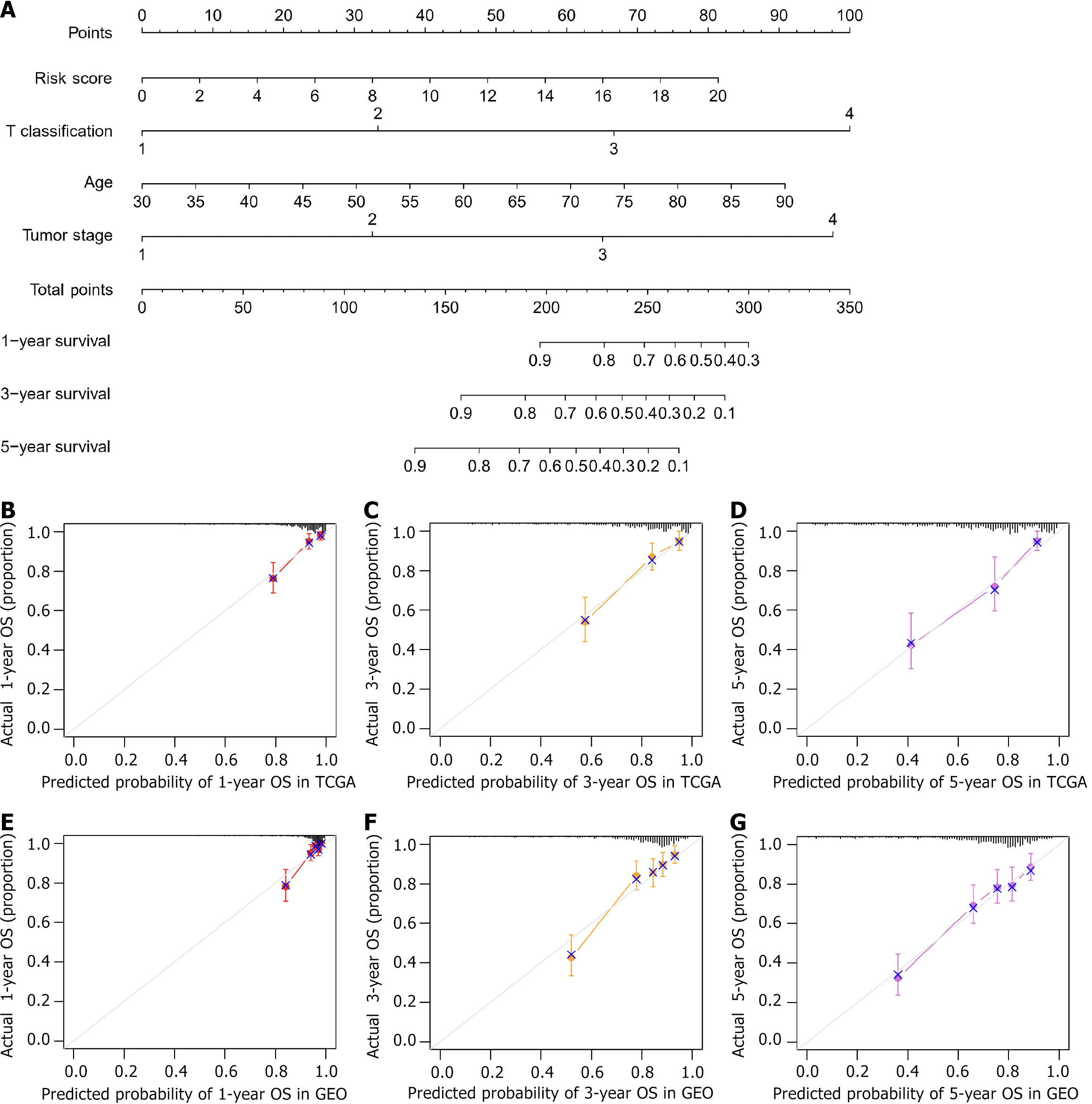
Figure 6 Prognostic value of the nomogram based on the fatty acid-binding protein 4 risk score in colon adenocarcinoma patients.
A: A nomogram was constructed to predict the 1-year, 3-year, and 5-year overall survival (OS) rates by combining the risk score, T classification, age, and tumor stage; B-D: Calibration curves of the nomogram for predicting and observing 1-year, 3-year, and 5-year OS in The Cancer Genome Atlas training cohort. The dashed line at 45° indicates a perfect prediction; E-G: Calibration curves of the nomogram for predicting and observing 1-year, 3-year, and 5-year OS in the Gene Expression Omnibus validation cohort. COAD: Colon adenocarcinoma; TCGA: The Cancer Genome Atlas; OS: Overall survival; GEO: Gene Expression Omnibus.
- Citation: Zhang Y, Zhu WL, Wu M, Gao TY, Hu HX, Xu ZY. Using bioinformatics methods to elucidate fatty acid-binding protein 4 as a potential biomarker for colon adenocarcinoma. World J Gastrointest Oncol 2025; 17(4): 103113
- URL: https://www.wjgnet.com/1948-5204/full/v17/i4/103113.htm
- DOI: https://dx.doi.org/10.4251/wjgo.v17.i4.103113