Copyright
©The Author(s) 2025.
World J Gastrointest Oncol. Feb 15, 2025; 17(2): 92437
Published online Feb 15, 2025. doi: 10.4251/wjgo.v17.i2.92437
Published online Feb 15, 2025. doi: 10.4251/wjgo.v17.i2.92437
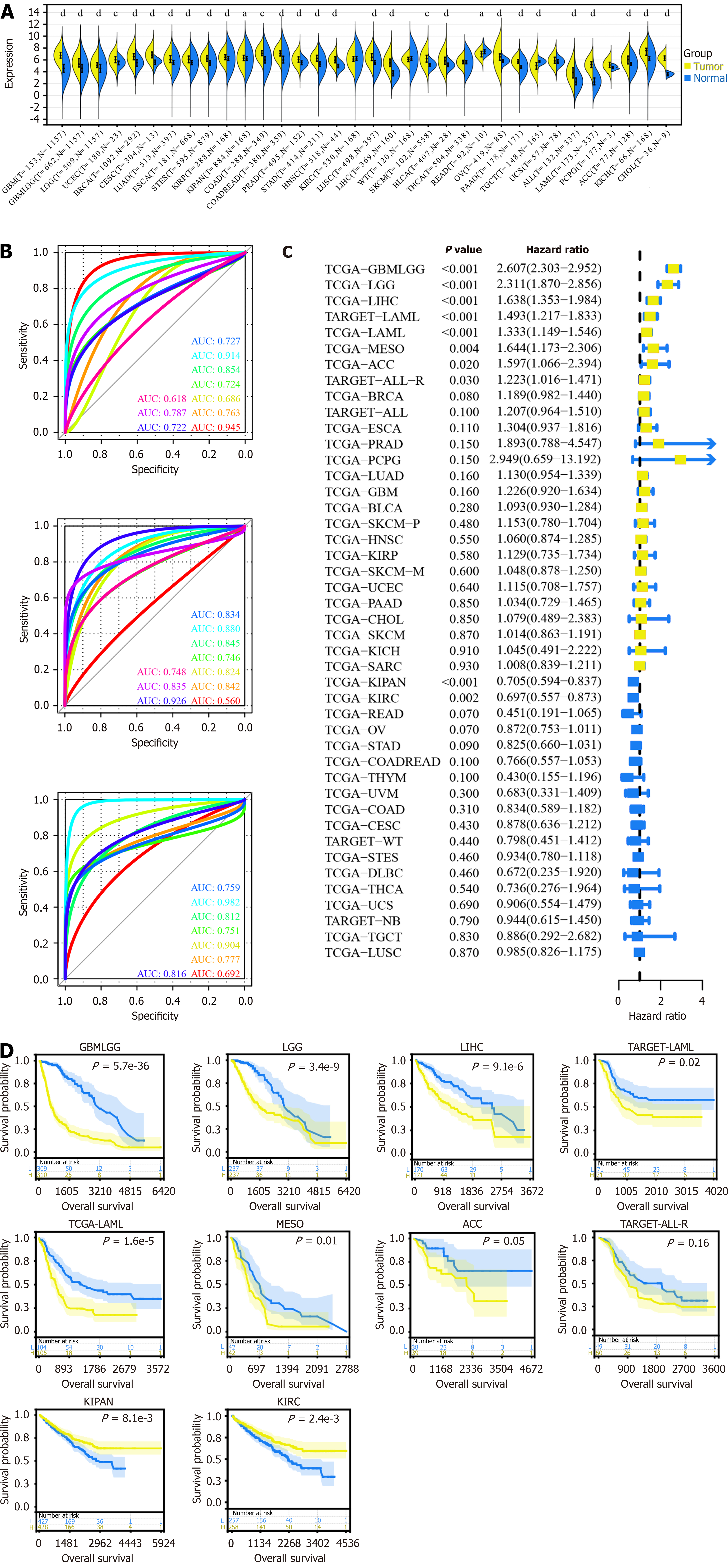
Figure 1 Putative clinicopathological significance of transmembrane protein 106C in pan-cancers.
A: Relative expression values of transmembrane protein 106C (TMEM106C) in pan-cancers; B: Discriminatory ability of TMEM106C in pan-cancers; C: Univariate Cox regression analysis of TMEM106C in pan-cancers; D: Kaplan-Meier survival analysis of TMEM106C in pan-cancers. P values of Wilcoxon test. aP < 0.05. bP < 0.01. cP < 0.001. dP < 0.0001. ACC: Adrenocortical carcinoma; ALL: Acute lymphoblastic leukemia; AUC: Area under the curve; BLCA: Bladder urothelial carcinoma; BRCA: Breast invasive carcinoma; CESC: Cervical squamous cell carcinoma and endocervical adenocarcinoma; CHOL: Cholangiocarcinoma; COAD: Colon adenocarcinoma; COADREAD: Colon adenocarcinoma/rectum adenocarcinoma; DLBC: Lymphoid neoplasm diffuse large B-cell lymphoma; ESCA: Esophageal carcinoma; GBM: Glioblastoma multiforme; GBMLGG: Glioblastoma and low-grade glioma; H: Transmembrane protein 106C-high expression group; HNSC: Head and neck squamous cell carcinoma; KICH: Kidney chromophobe; KIPAN: Pan-kidney cohort (kidney chromophobe + kidney renal clear cell carcinoma + kidney renal papillary cell carcinoma); KIRC: Kidney renal clear cell carcinoma; KIRP: Kidney renal papillary cell carcinoma; L: Transmembrane protein 106C-low expression group; LAML: Acute myeloid leukemia; LGG: Brain lower grade glioma; LIHC: Liver hepatocellular carcinoma; LUAD: Lung adenocarcinoma; LUSC: Lung squamous cell carcinoma; MESO: Mesothelioma; NB: Neuroblastoma; OV: Ovarian serous cystadenocarcinoma; PAAD: Pancreatic adenocarcinoma; PCPG: Pheochromocytoma and paraganglioma; PRAD: Prostate adenocarcinoma; READ: Rectum adenocarcinoma; SARC: Sarcoma; STAD: Stomach adenocarcinoma; SKCM: Skin cutaneous melanoma; STES: Stomach and esophageal carcinoma; TARGET: Therapeutically applicable research to generate effective treatments; TCGA: The cancer genome atlas; TGCT: Testicular germ cell tumors; THCA: Thyroid carcinoma; THYM: Thymoma; UCEC: Uterine corpus endometrial carcinoma; UCS: Uterine carcinosarcoma; UVM: Uveal melanoma; WT: High-risk Wilms tumor.
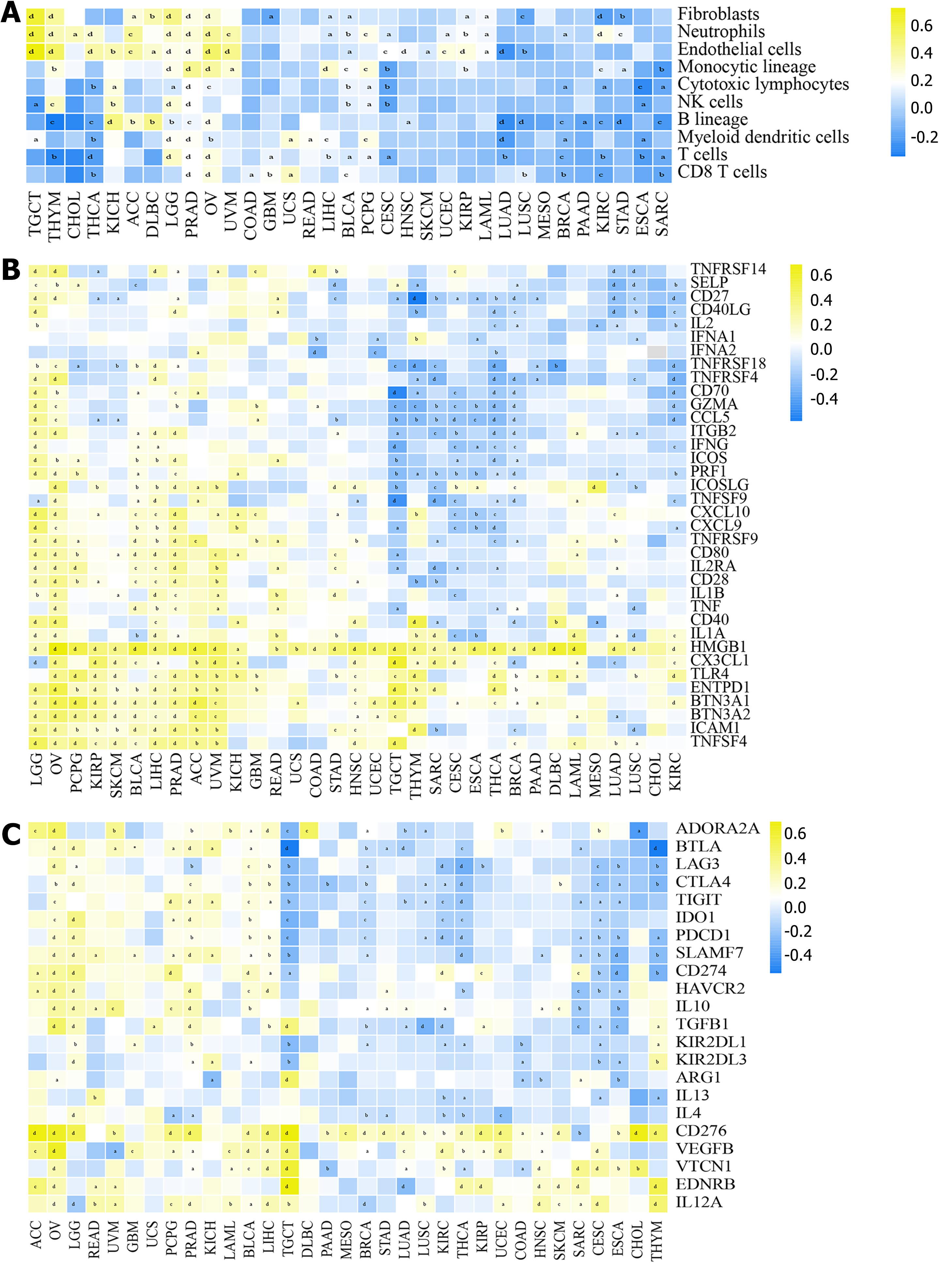
Figure 2 Association between transmembrane protein 106C and tumor microenvironment of pan-cancers.
A: Correlation between transmembrane protein 106C (TMEM106C) and immune-stromal cells in pan-cancers; B: Correlation between TMEM106C and stimulatory immune-related genes in pan-cancers; C: Correlation between TMEM106C and inhibitory immune-related genes in pan-cancers. aP < 0.05. bP < 0.01. cP < 0.001. dP < 0.0001. ACC: Adrenocortical carcinoma; BLCA: Bladder urothelial carcinoma; BRCA: Breast invasive carcinoma; CD: Cluster of differentiation; CESC: Cervical squamous cell carcinoma and endocervical adenocarcinoma; CHOL: Cholangiocarcinoma; COAD: Colon adenocarcinoma; DLBC: Lymphoid neoplasm diffuse large B-cell lymphoma; ESCA: Esophageal carcinoma; GBM: Glioblastoma multiforme; GBMLGG: Glioblastoma and low-grade glioma; HNSC: Head and neck squamous cell carcinoma; IL: Interleukin; KICH: Kidney chromophobe; KIRC: Kidney renal clear cell carcinoma; KIRP: Kidney renal papillary cell carcinoma; LAML: Acute myeloid leukemia; LGG: Brain lower grade glioma; LIHC: Liver hepatocellular carcinoma; LUAD: Lung adenocarcinoma; LUSC: Lung squamous cell carcinoma; MESO: Mesothelioma; NK: Natural killer; OV: Ovarian serous cystadenocarcinoma; PAAD: Pancreatic adenocarcinoma; PCPG: Pheochromocytoma and paraganglioma; PRAD: Prostate adenocarcinoma; READ: Rectum adenocarcinoma; SARC: Sarcoma; STAD: Stomach adenocarcinoma; SKCM: Skin cutaneous melanoma; TGCT: Testicular germ cell tumors; THCA: Thyroid carcinoma; THYM: Thymoma; TNF: Tumor necrosis factor; UCEC: Uterine corpus endometrial carcinoma; UCS: Uterine carcinosarcoma; UVM: Uveal melanoma.
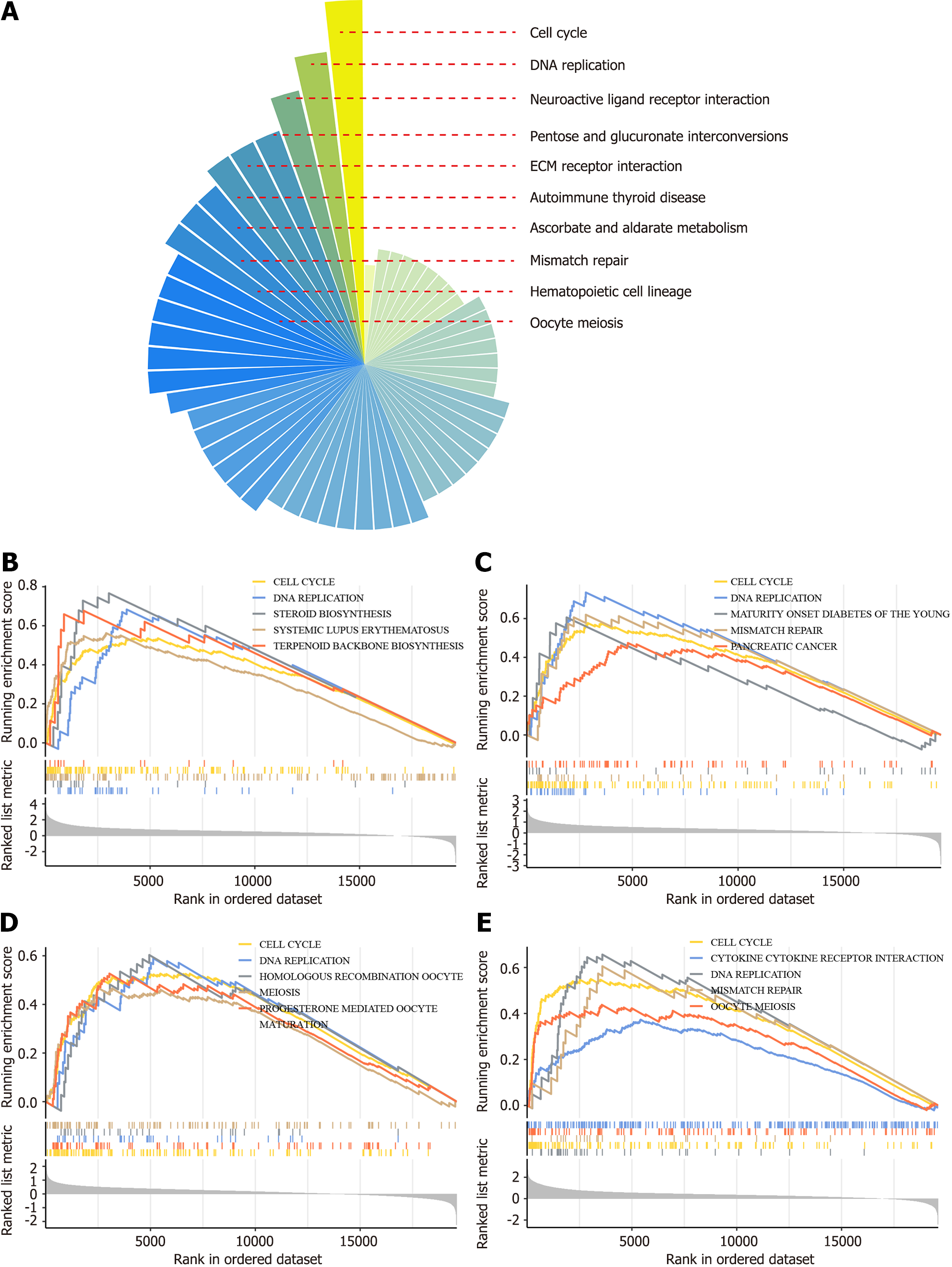
Figure 3 Prospective molecular mechanisms of transmembrane protein 106C underlying pan-cancers.
A: Gene set enrichment analysis was conducted across 33 cancer types to rank the enriched transmembrane protein 106C (TMEM106C) pathways; B-E: Gene set enrichment analysis results for TMEM106C in adrenocortical carcinoma, bladder cancer, breast cancer, and hepatocellular carcinoma, respectively. ECM: Extracellular matrix.
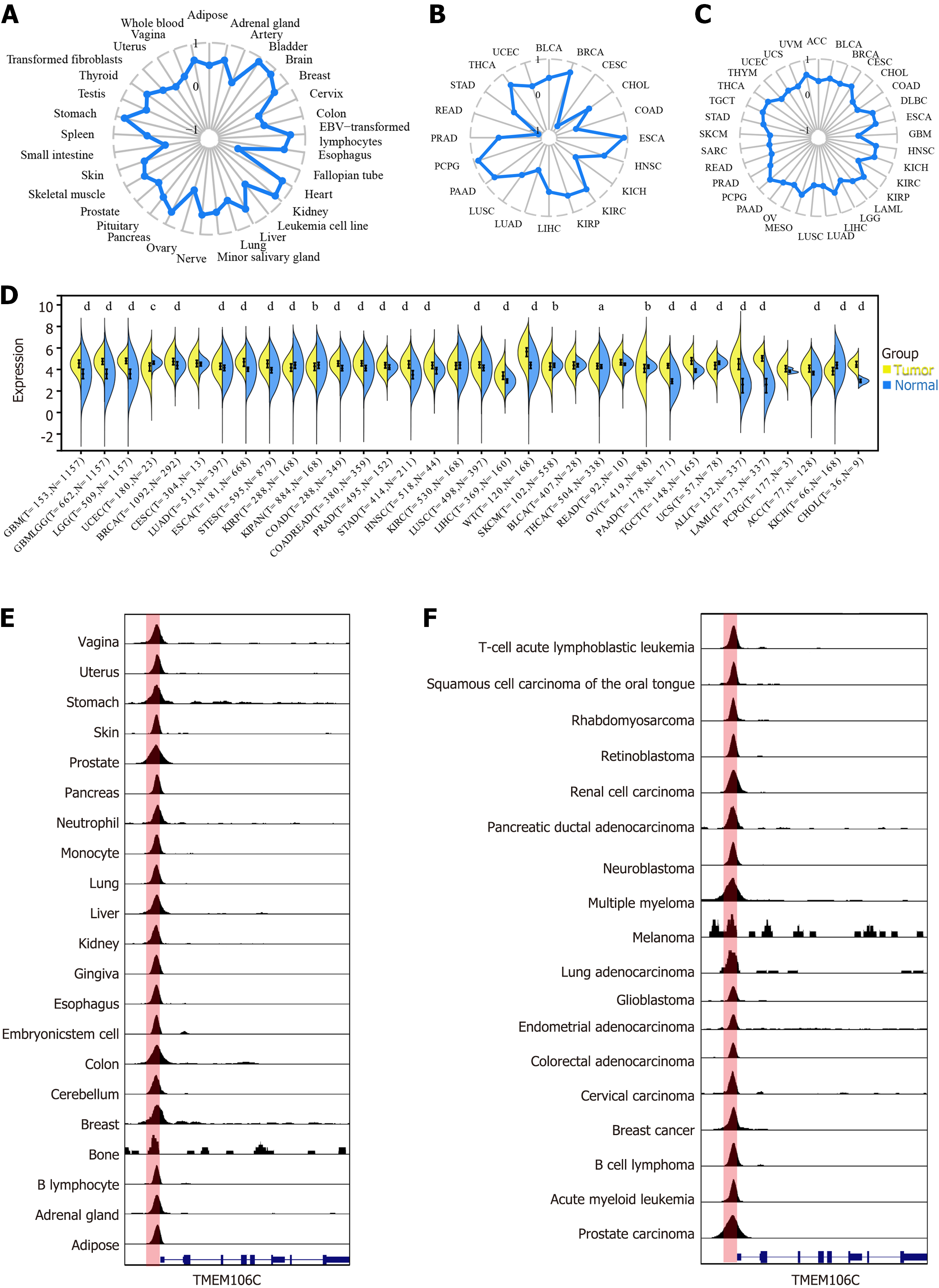
Figure 4 Upstream transcriptional regulation of transmembrane protein 106C in pan-cancers.
With a regulatory potential score of 0.98, CCCTC-binding factor (CTCF) was predicted as the most likely transcriptional factor regulating transmembrane protein 106C (TMEM106C). A-C: Significant positive correlation was observed between CTCF and TMEM106C in diverse normal and cancer tissue; D: CTCF was significantly overexpressed in pan-cancers; E and F: Chromatin immunoprecipitation followed by sequencing analysis revealed significant binding peaks of the CTCF factor in the upstream promoter region of TMEM106C. aP < 0.05. bP < 0.01. cP < 0.001. dP < 0.0001. ACC: Adrenocortical carcinoma; BLCA: Bladder urothelial carcinoma; BRCA: Breast invasive carcinoma; CESC: Cervical squamous cell carcinoma and endocervical adenocarcinoma; CHOL: Cholangiocarcinoma; COAD: Colon adenocarcinoma; COADREAD: Colon adenocarcinoma/rectum adenocarcinoma; DLBC: Lymphoid neoplasm diffuse large B-cell lymphoma; ESCA: Esophageal carcinoma; GBM: Glioblastoma multiforme; GBMLGG: Glioblastoma and low-grade glioma; HNSC: Head and neck squamous cell carcinoma; KICH: Kidney chromophobe; KIPAN: Pan-kidney cohort (kidney chromophobe + kidney renal clear cell carcinoma + kidney renal papillary cell carcinoma); KIRC: Kidney renal clear cell carcinoma; KIRP: Kidney renal papillary cell carcinoma; LAML: Acute myeloid leukemia; LGG: Brain lower grade glioma; LIHC: Liver hepatocellular carcinoma; LUAD: Lung adenocarcinoma; LUSC: Lung squamous cell carcinoma; MESO: Mesothelioma; NB: Neuroblastoma; OV: Ovarian serous cystadenocarcinoma; PAAD: Pancreatic adenocarcinoma; PCPG: Pheochromocytoma and paraganglioma; PRAD: Prostate adenocarcinoma; READ: Rectum adenocarcinoma; SARC: Sarcoma; STAD: Stomach adenocarcinoma; SKCM: Skin cutaneous melanoma; STES: Stomach and esophageal carcinoma; TGCT: Testicular germ cell tumors; THCA: Thyroid carcinoma; THYM: Thymoma; UCEC: Uterine corpus endometrial carcinoma; UCS: Uterine carcinosarcoma; UVM: Uveal melanoma; WT: High-risk Wilms tumor.
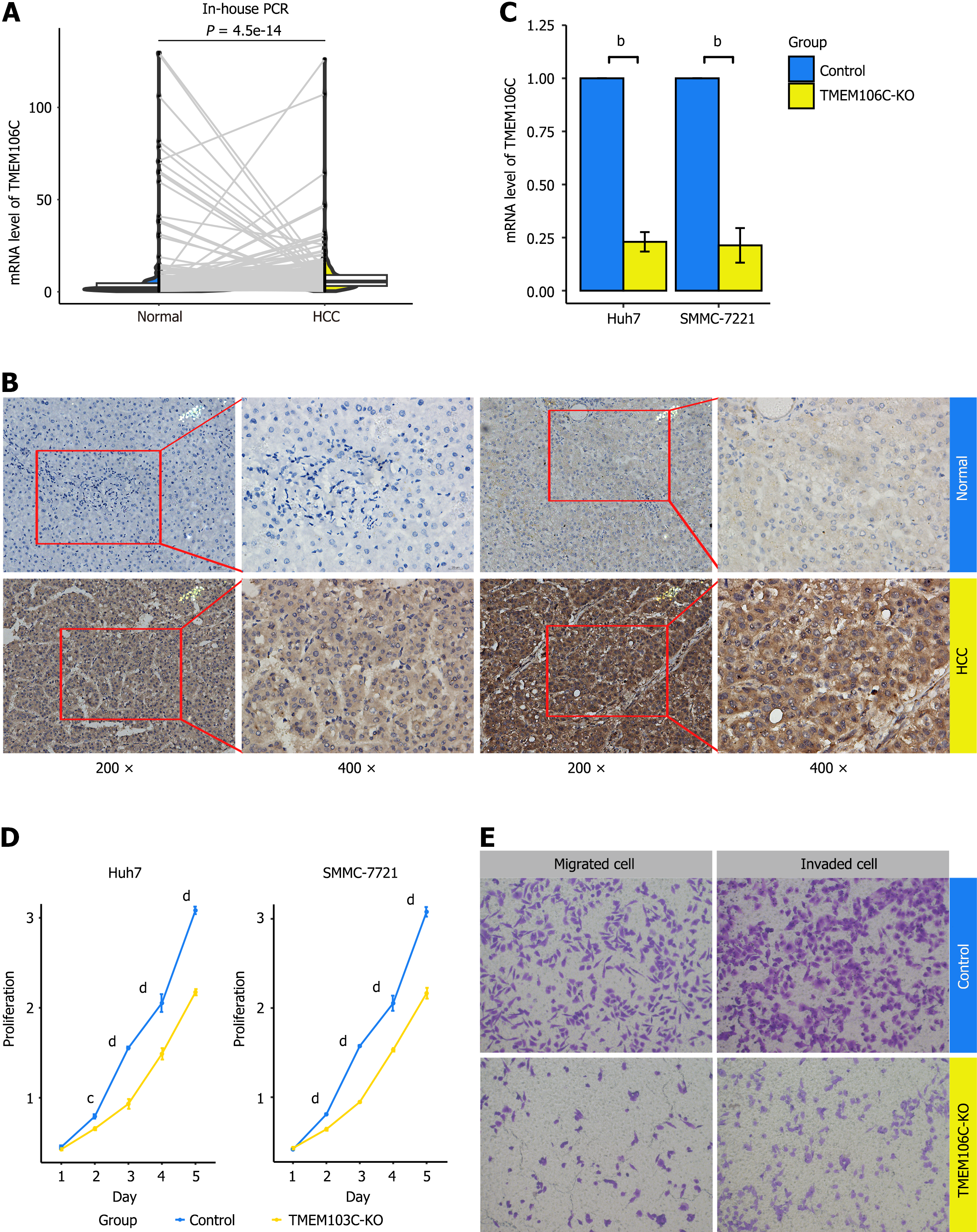
Figure 5 Biological functions of transmembrane protein 106C in hepatocellular carcinoma cells.
A: Transmembrane protein 106C (TMEM106C) mRNA was significantly overexpressed in 248 hepatocellular carcinoma (HCC) tissue compared to 248 paired normal liver tissue samples; B: TMEM106C protein was expressed in the cytoplasm and membranes of HCC tumor cells; C: TMEM106C was successfully knocked out using the clustered regularly interspaced short palindromic repeats gene editing technology; D and E: TMEM106C knockout significantly attenuated the proliferation, migrated and invaded ability of HCC cells. bP < 0.01. cP < 0.001. dP < 0.0001. HCC: Hepatocellular carcinoma; KO: Knockout; PCR: Polymerase chain reaction; TMEM106C: Transmembrane protein 106C.
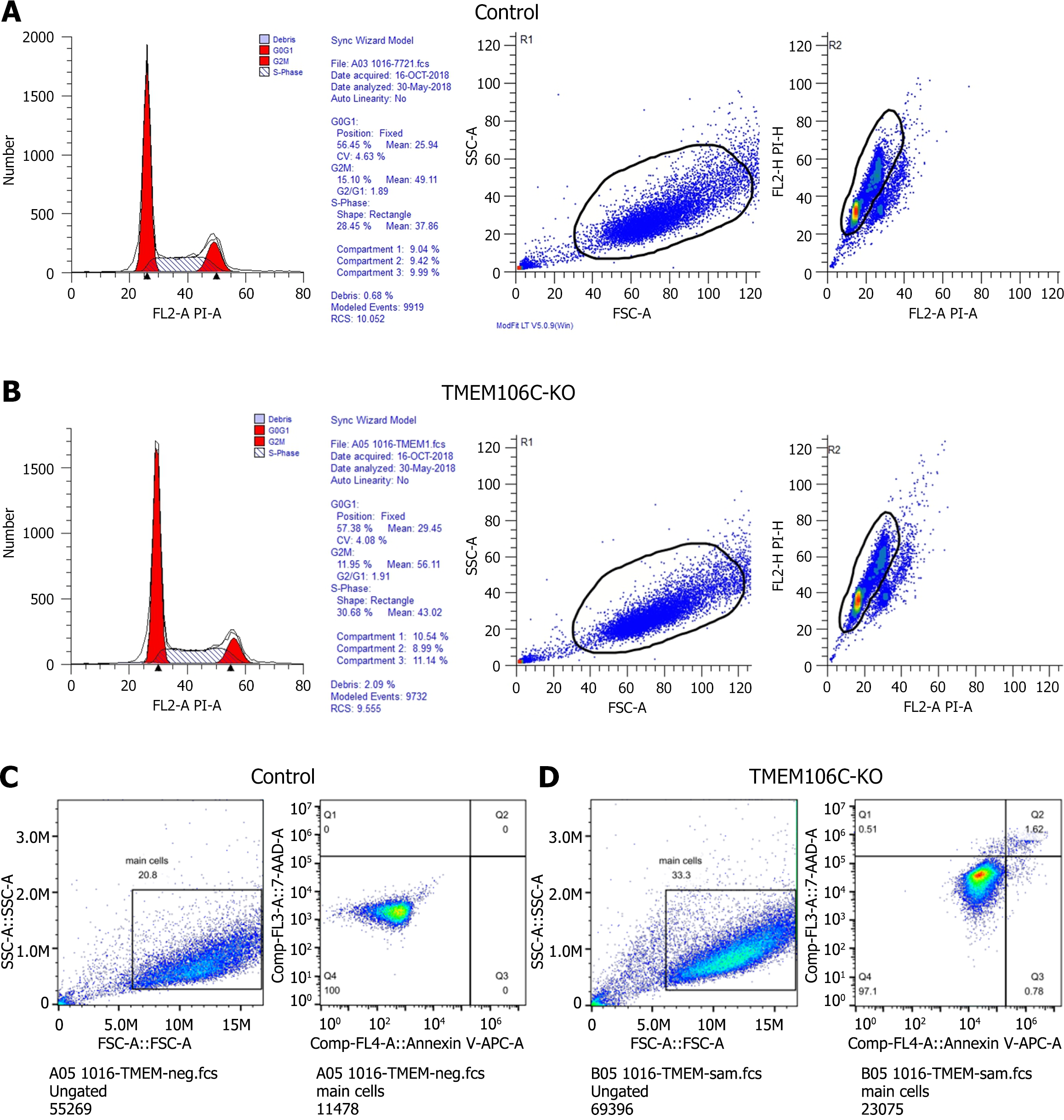
Figure 6 Biological functions of transmembrane protein 106C in hepatocellular carcinoma cells.
A and B: Transmembrane protein 106C (TMEM106C) knockout arrested the cell cycle process during the DNA synthesis phase; C and D: TMEM106C knockout increased both early and late apoptotic rates of hepatocellular carcinoma cells. FL2-A: The area under the curve of the fluorescence signal detected in the FL2 channel; KO: Knockout; PI-A: The area under the curve of the fluorescence signal detected by the flow cytometer after propidium iodide staining; SSC-A: The area under the curve of the side scatter signal; TMEM106C: Transmembrane protein 106C.
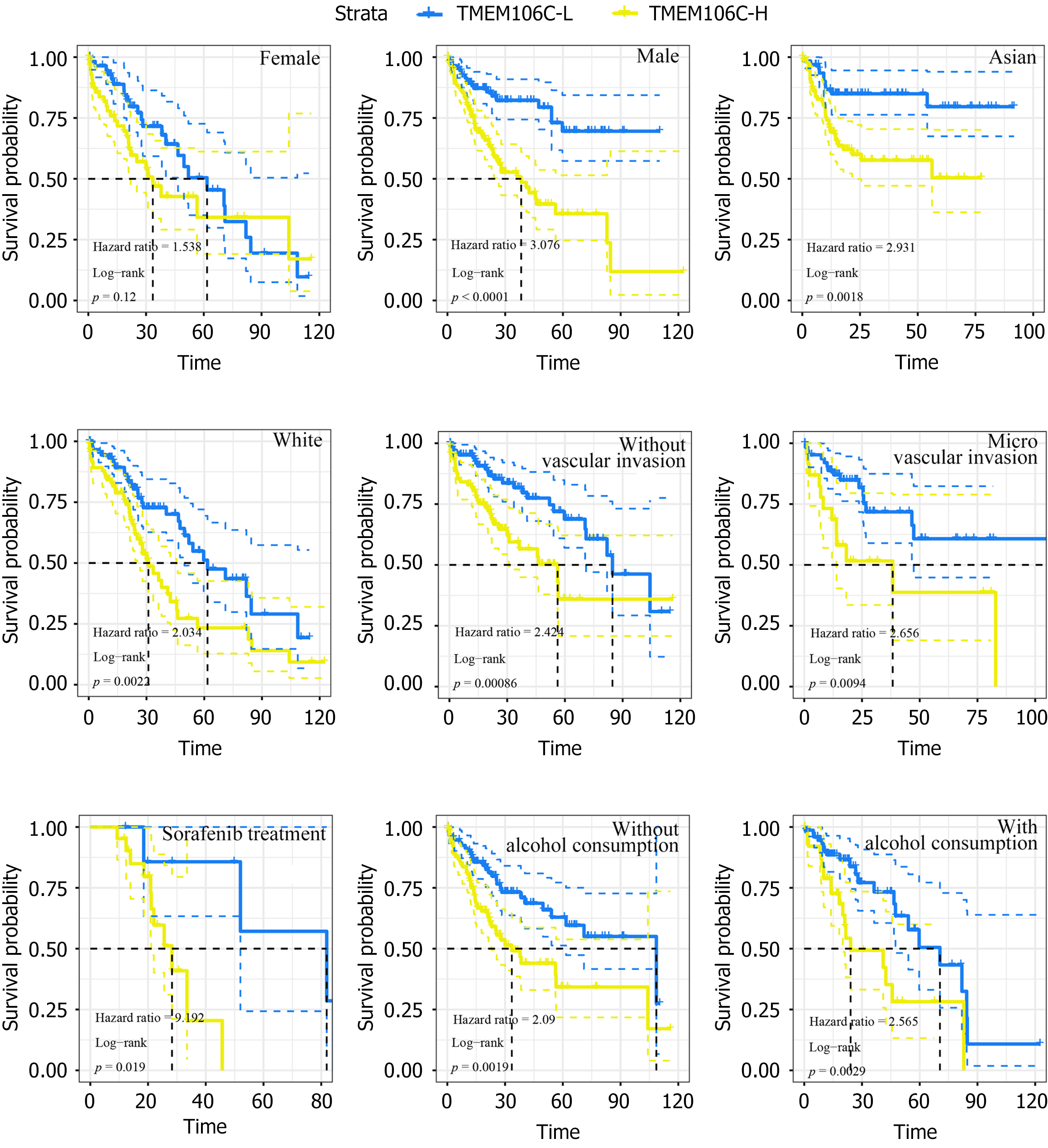
Figure 7 Prognostic value of transmembrane protein 106C in hepatocellular carcinoma patients.
TMEM106C-L: Transmembrane protein 106C-low expression group; TMEM106C-H: Transmembrane protein 106C-high expression group.
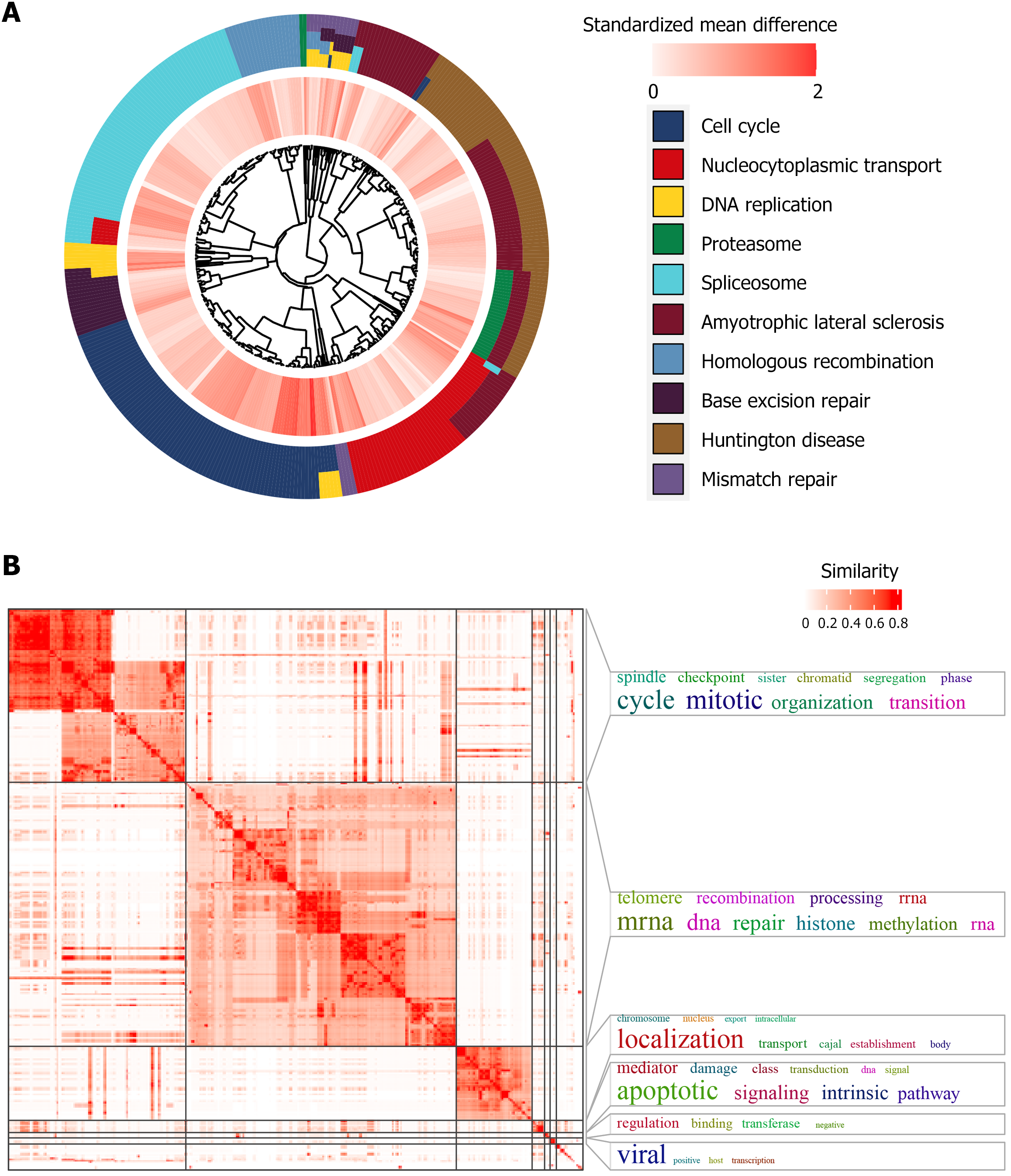
Figure 8 Functional enrichment of transmembrane protein 106C in hepatocellular carcinoma tissue.
A: Kyoto encyclopedia of genes and genomes; B: Gene ontology.
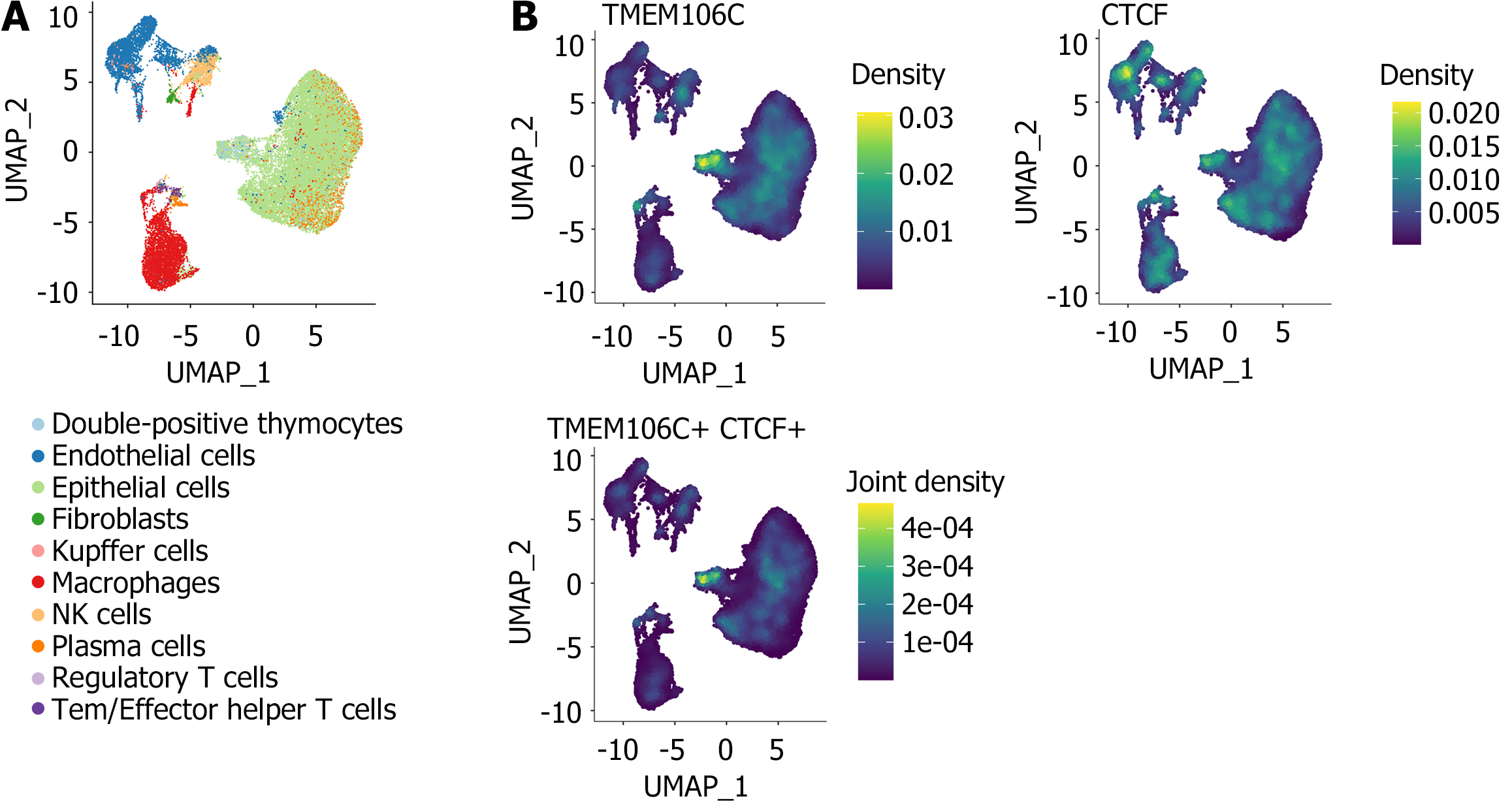
Figure 9 The putative CCCTC-binding factor-transmembrane protein 106C regulatory axis in single cells of hepatocellular carcinoma.
A: Annotation of 30896 single cells in hepatocellular carcinoma; B: Significant co-expression trend observed between CCCTC-binding factor and transmembrane protein 106C in malignant epithelial cells. CTCF: CCCTC-binding factor; NK: Natural killer; TMEM106C: Transmembrane protein 106C; UMAP: Uniform manifold approximation and projection.
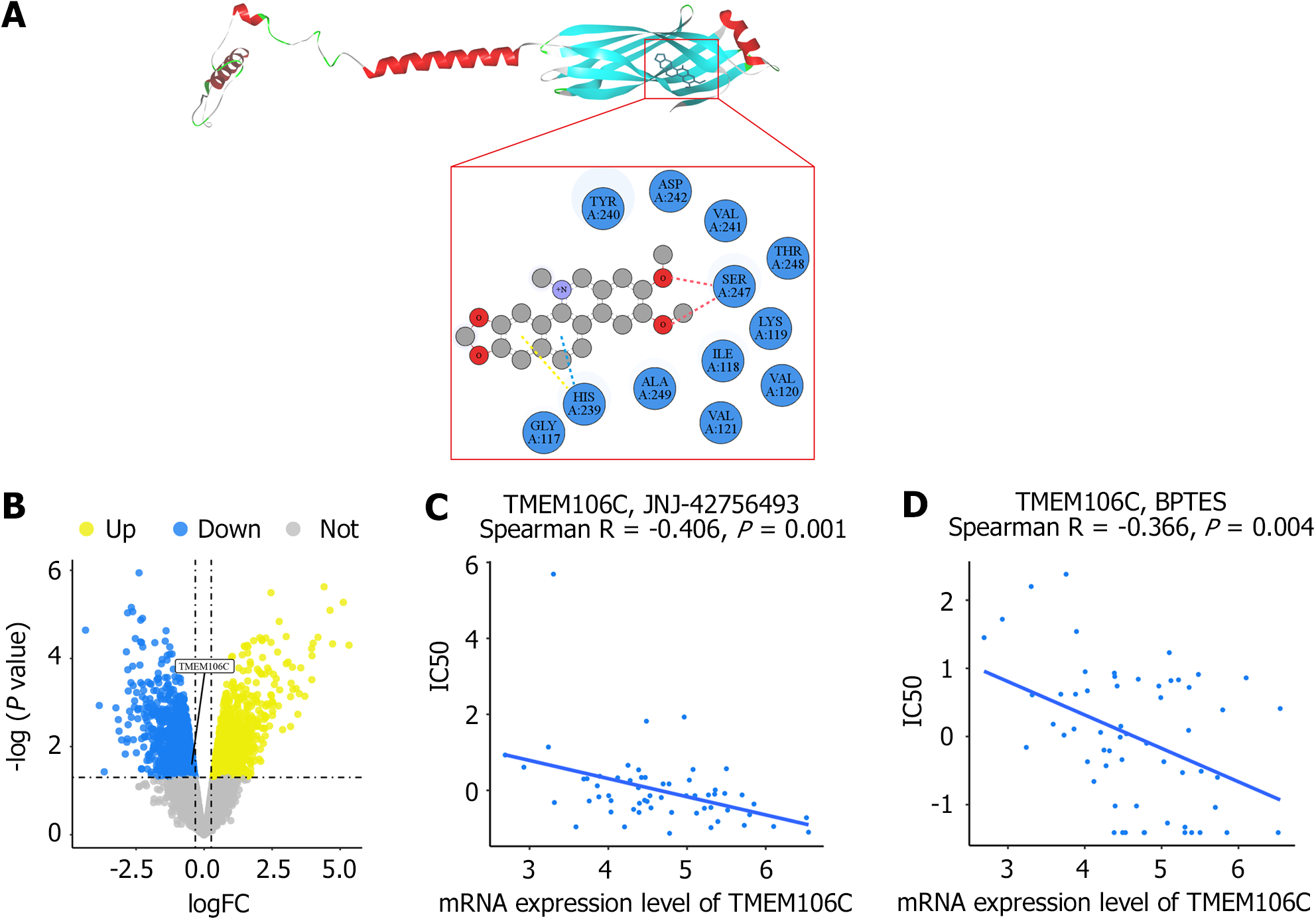
Figure 10 Transmembrane protein 106C as a putative therapeutic target for hepatocellular carcinoma.
A: The predicted transmembrane protein 106C (TMEM106C) protein was docked with the nitidine chloride ligand, revealing conventional hydrogen bonds (indicated by red dotted lines), pi-donor hydrogen bonds (indicated by blue dotted lines), and pi-pi T-shaped molecular interactions (indicated by yellow dotted lines); B: Treatment with nitidine chloride down-regulated TMEM106C mRNA expression in a nude mice hepatocellular carcinoma model; C and D: TMEM106C expression level was found to be significantly correlated with drug sensitivity of anti-hepatocellular carcinoma agents, including a multi-target tyrosine kinase inhibitor, JNJ-42756493 (erdafitinib), and a glutaminase inhibitor, bis-2-(5-phenylacetamido-1,3,4-thiadiazol-2-yl) ethyl sulfide. ALA: Alanine; ASP: Aspartic acid; BPTES: Bis-2-(5-phenylacetamido-1,3,4-thiadiazol-2-yl) ethyl sulfide; GLY: Glycine; HIS: Histidine; IC50: Half-maximal inhibitory concentration; ILE: Isoleucine; LYS: Lysine; SER: Serine; THR: Threonine; TMEM106C: Transmembrane protein 106C; TYR: Tyrosine; VAL: Valine.
- Citation: Li JD, He RQ, Dang YW, Huang ZG, Xiong DD, Zhang L, Du XF, Chen G. Unveiling expression patterns, mechanisms, and therapeutic opportunities of transmembrane protein 106C: From pan-cancers to hepatocellular carcinoma. World J Gastrointest Oncol 2025; 17(2): 92437
- URL: https://www.wjgnet.com/1948-5204/full/v17/i2/92437.htm
- DOI: https://dx.doi.org/10.4251/wjgo.v17.i2.92437