Copyright
©The Author(s) 2024.
World J Gastrointest Oncol. Jul 15, 2024; 16(7): 3032-3054
Published online Jul 15, 2024. doi: 10.4251/wjgo.v16.i7.3032
Published online Jul 15, 2024. doi: 10.4251/wjgo.v16.i7.3032
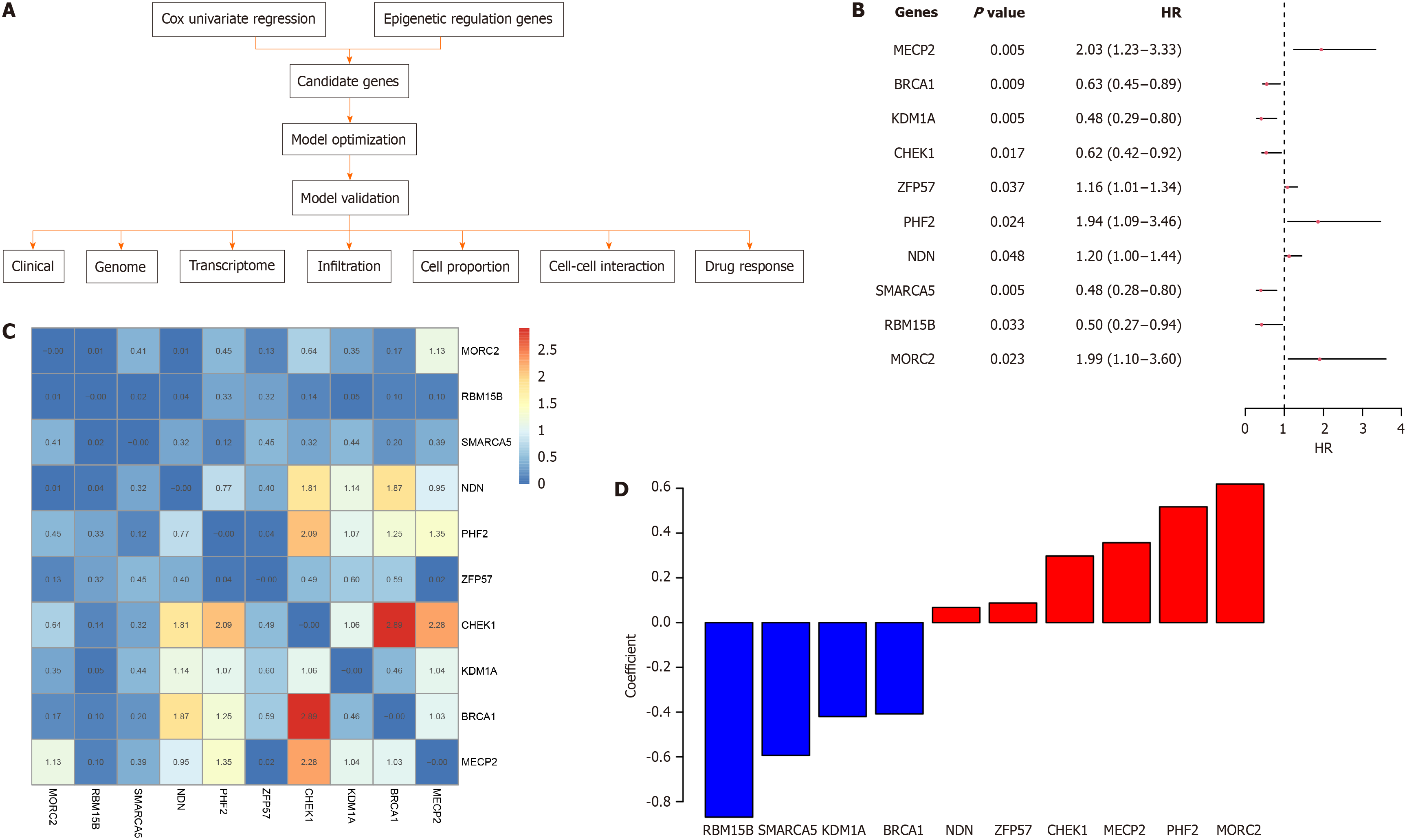
Figure 1 Candidate genes for the signature.
A: Schema of this study; B: Hazard ratio, confidence interval, and P-value of each candidate gene; C: Cox univariate Correlation of the candidate genes, the number and color indicate the correlation P values of gene pairs; D: Coefficient of each gene in the model.
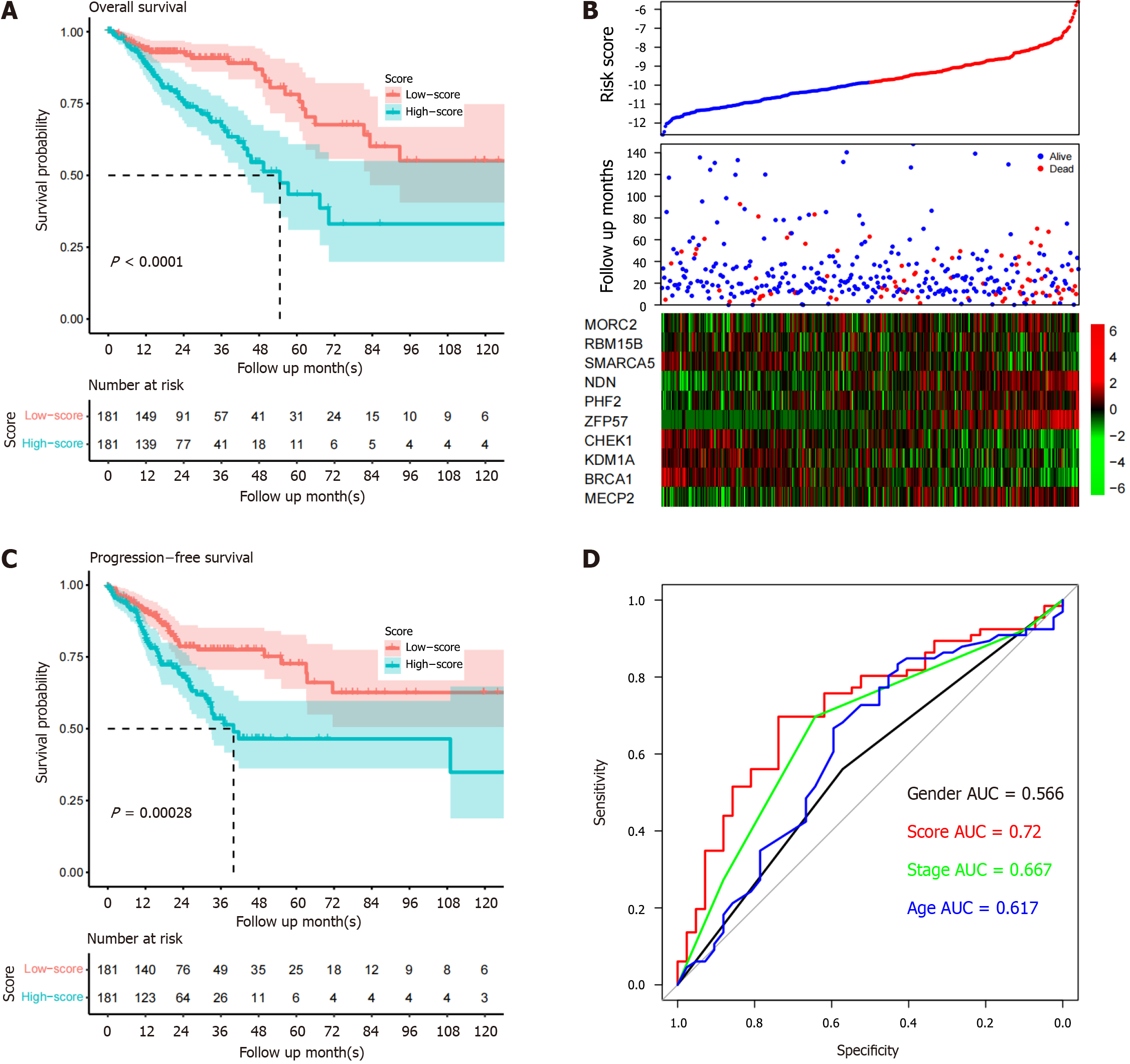
Figure 2 Signature performance in the TCGA dataset.
A: Overall survival period of high-risk samples is significantly shorter compared to low-risk samples; B: Heatmap of candidate genes suggests that high-risk samples have high expression of certain genes; C: Progression-free survival of group samples is similar to the overall survival pattern; D: Five-year survival receiver operating characteristic curve of the model and other clinical indicators. AUC: Area under the curve.
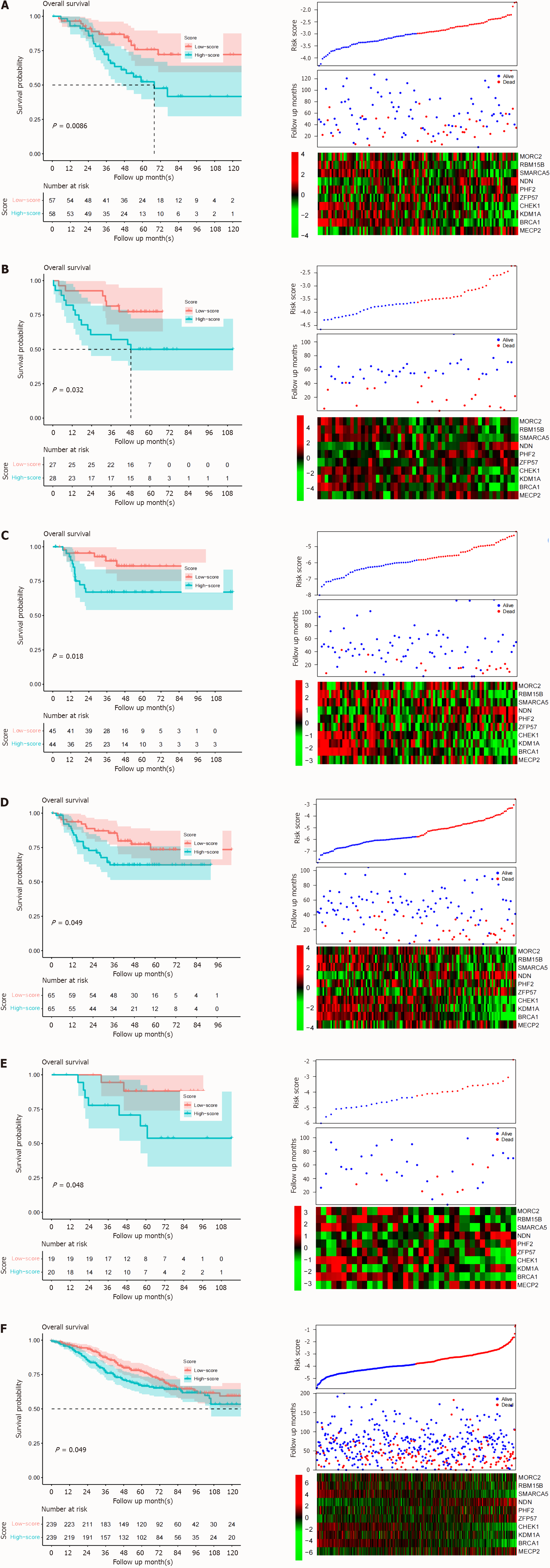
Figure 3 Signature verification.
The performance of the signature was assayed from GEO database. A: GSE17526; B: GSE17537; C: GSE33113; D: GSE37892; E: GSE39048; F: GSE39582. Similar to the training dataset, the upper panel is the survival curve, the middle is the follow-up information and sorted risk scores. The bottom panel is the candidate gene expression heatmap.
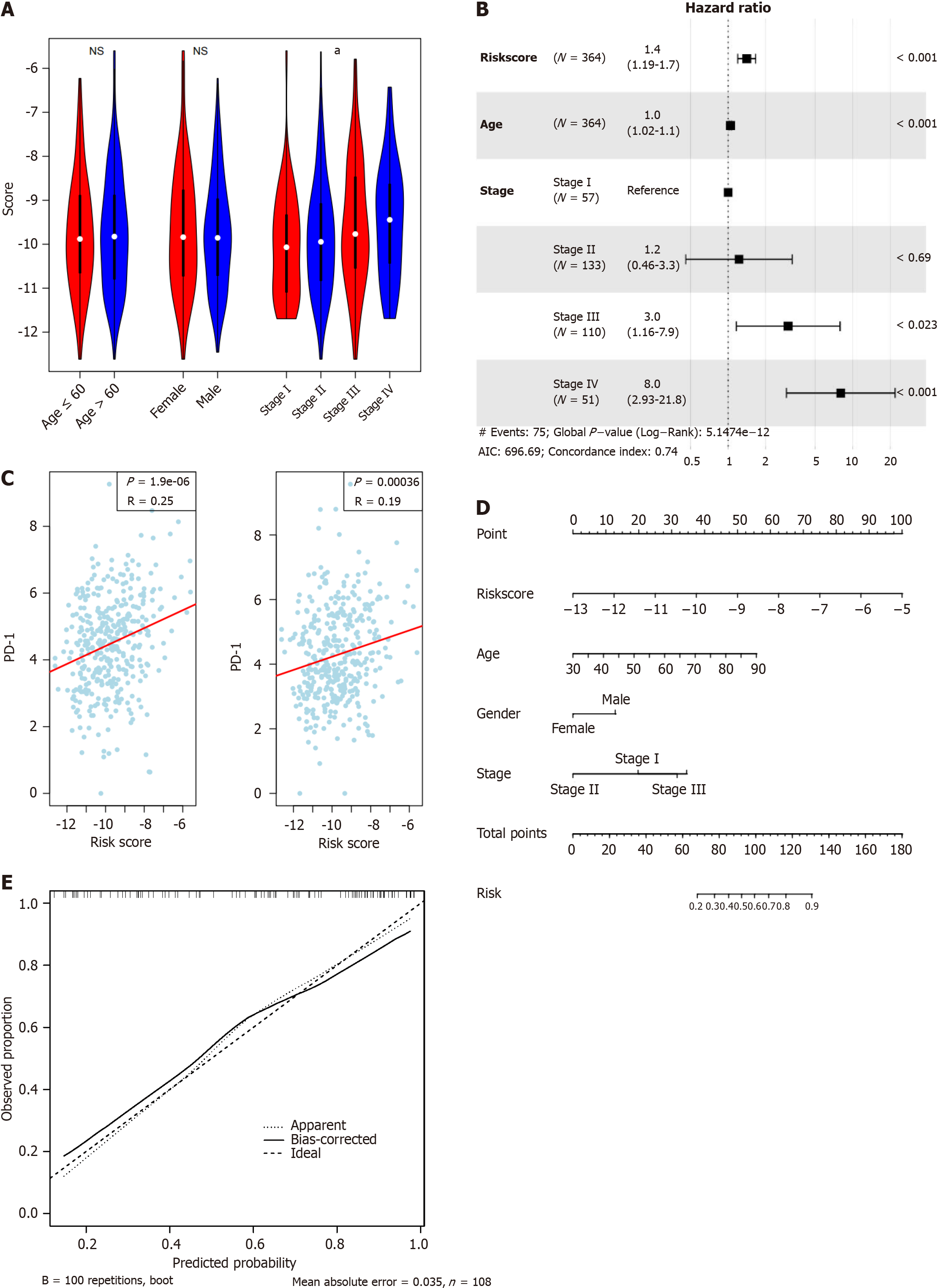
Figure 4 Clinical association of the signature.
A: The signature is statistically correlated with TNM stage, but not age and gender; B: Cox multivariate regression of the signature and clinical indicators suggests that the signature is an independent indicator for survival; C: The signature is positively associated with both PD-1 and PD-L1 gene expression; D: Nomogram for five-year survival; E: Calibrate curve revealed the signature is a valuable marker for predicting survival of colorectal cancer. aP < 0.05; cP < 0.001.
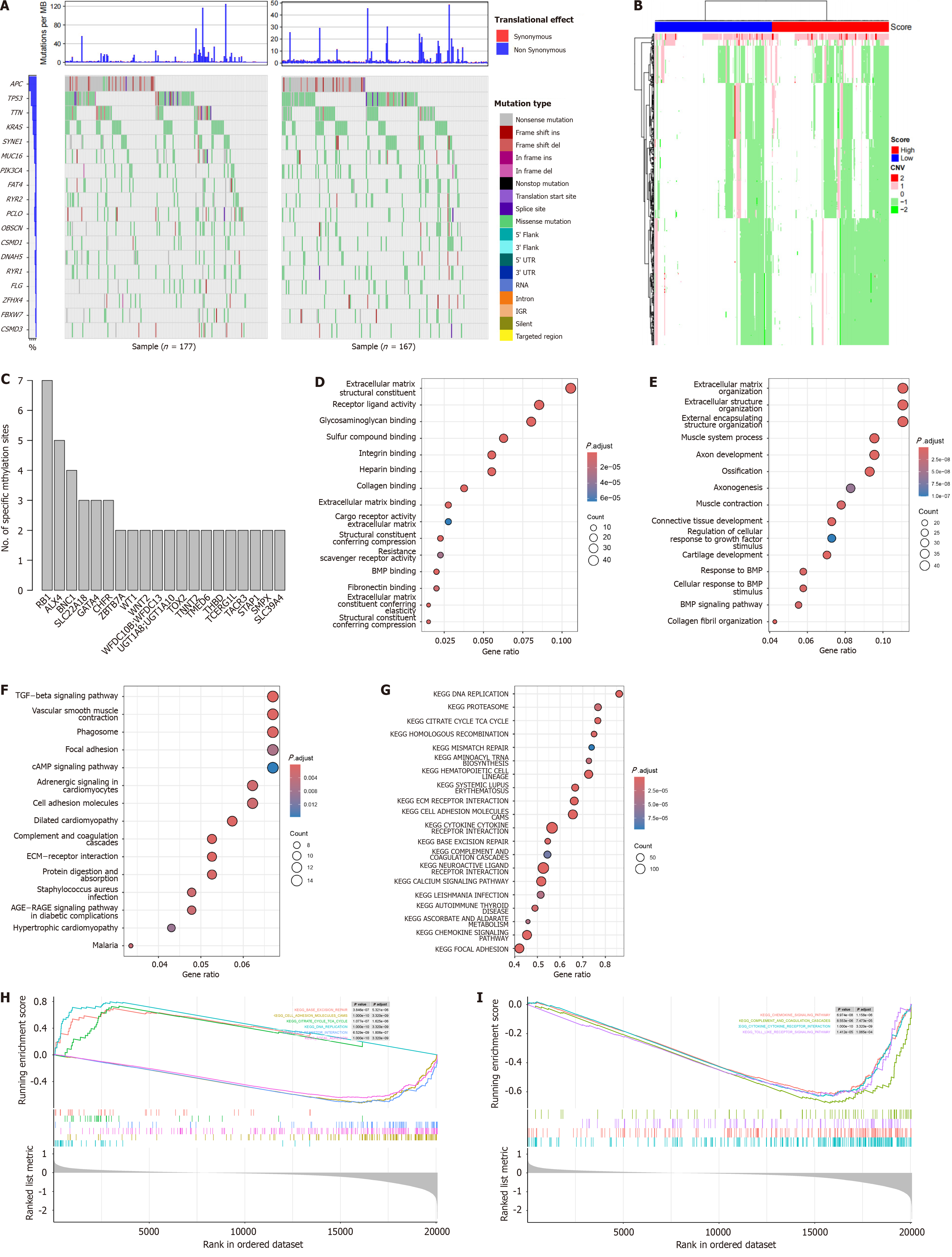
Figure 5 Genome and transcriptome feature of the signature.
A: Mutational landscape of the high and low-risk samples; B: Differential copy number variation was shown; C: Enriched differentially methylated sites across genes were visualized; D: Gene Ontology analyses of differentially expressed genes in molecular function; E: Biological process; F: Kyoto Encyclopedia of Genes and Genomes pathways were shown; G: Gene set enrichment analysis revealed that the enriched pathways; H: Canonical cancer-related pathways; I: Immune-related pathways.
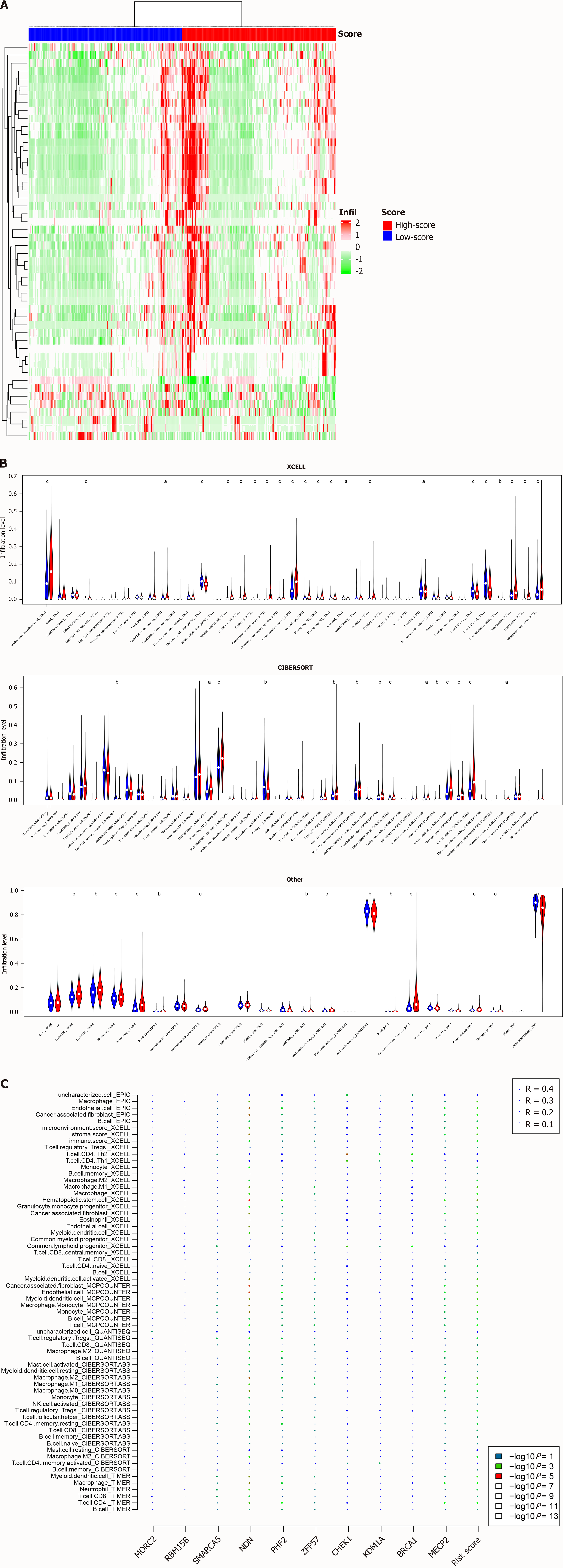
Figure 6 Immune abundance and the signature.
A: Differentially infiltrated cell types in TCGA dataset were identified; B: A high proportion of cell types according to different algorithms were significantly infiltrated between high and low-risk groups; C: correlation analyses revealed that the correlation was contributed by several genes. aP < 0.05, bP < 0.01, cP < 0.001.
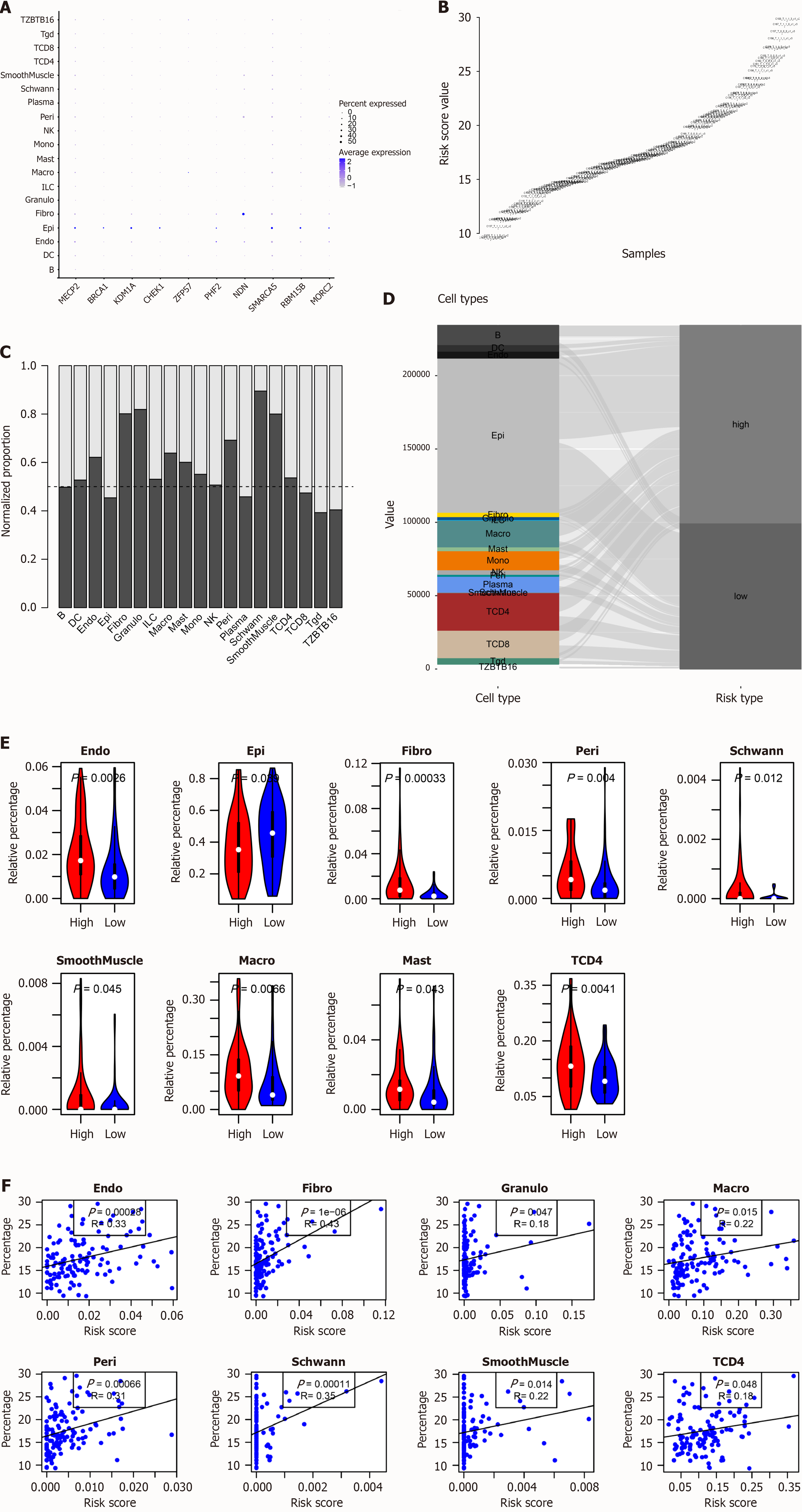
Figure 7 Single cell infiltration of the signature.
A: The candidate genes were mainly expressed in ‘basal’ cells; B: After calculating risk scores of each sample, the normalized proportion of high/low-risk groups was visualized; C: A bar plot; D: Sankey diagram; E: The signature is significantly associated with several cell types according to the single cell sequencing data by dividing high and low-risk groups; F: The signature is significantly associated with several cell types according to the single cell sequencing data by Pearson correlation.
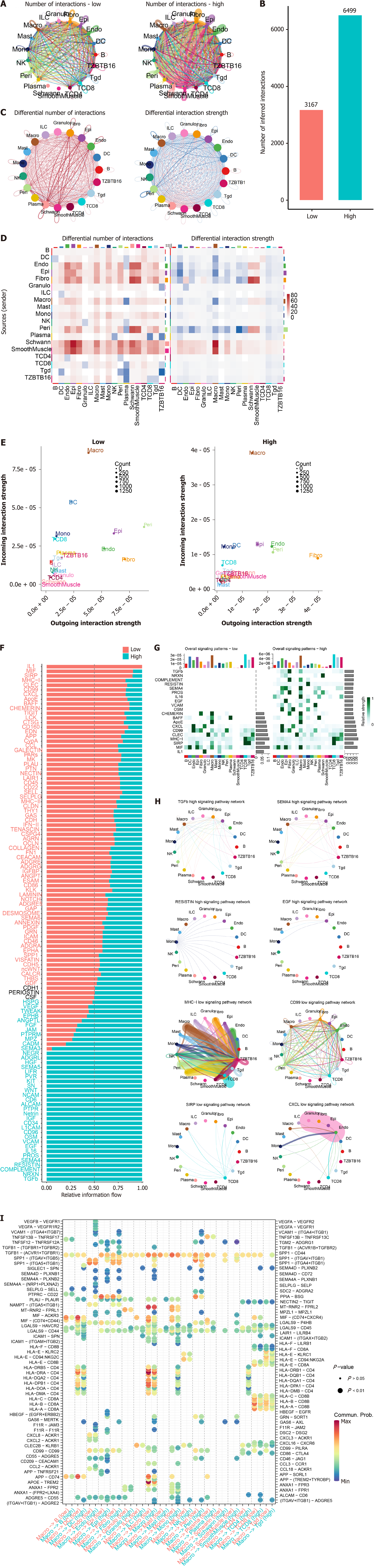
Figure 8 Cell-cell interaction and the signature.
A: Although most interactions exist in both low and high-risk samples; B: The interaction number is significantly different; C and D: Despite that the interaction number is higher in high-risk samples, the interaction strength is lower; E: The macrophage is noticed as the most significantly altered immune cell among the interactions; F: The pathway level interaction information flow was visualized for both groups, overall; G: Top 10 for each group; H: Among these interactions, it is noticed that macrophage is especially active in high-risk samples while interactions involved CD8+ T cell were increased in the low-risk group; I: Specific pathways and cell type interaction landscape also revealed that macrophage is activated in high-risk samples.
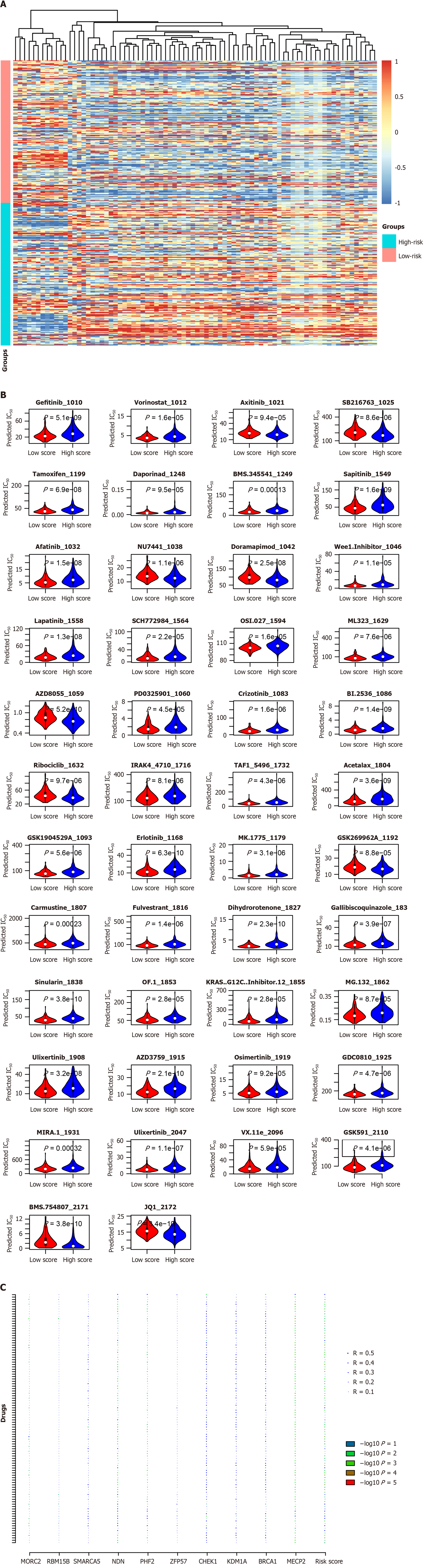
Figure 9 Drug response and the signature.
A: Drugs with different predicted IC50 values were identified and visualized with a heatmap; B: Detailed information was showed with vioplot; C: The drug response prediction was mostly contributed by NDN, PHF2, and MECP2 according to the correlation analyses.
- Citation: Liu HX, Feng J, Jiang JJ, Shen WJ, Zheng Y, Liu G, Gao XY. Integrated single-cell and bulk RNA sequencing revealed an epigenetic signature predicts prognosis and tumor microenvironment colorectal cancer heterogeneity. World J Gastrointest Oncol 2024; 16(7): 3032-3054
- URL: https://www.wjgnet.com/1948-5204/full/v16/i7/3032.htm
- DOI: https://dx.doi.org/10.4251/wjgo.v16.i7.3032