Copyright
©The Author(s) 2024.
World J Gastrointest Oncol. Jun 15, 2024; 16(6): 2592-2609
Published online Jun 15, 2024. doi: 10.4251/wjgo.v16.i6.2592
Published online Jun 15, 2024. doi: 10.4251/wjgo.v16.i6.2592
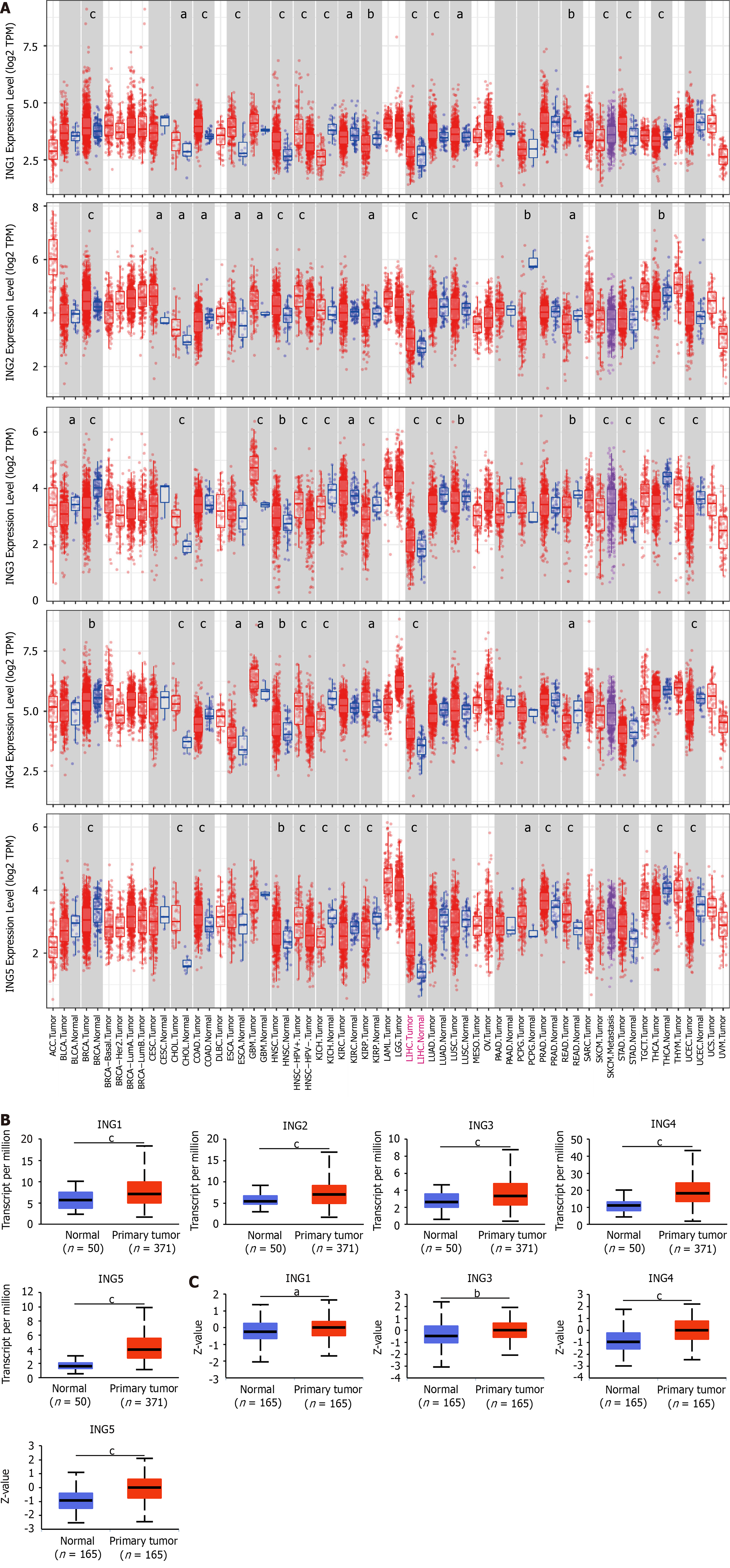
Figure 1 Differential expression of ING family genes in normal and cancer tissues.
A: Expression of ING family genes in different tumor types using Tumor Immune Estimation Resource 2.0 (data from The Cancer Genome Atlas); B: Expression of ING family genes in liver cancer (LIHC) using the University of Alabama at Birmingham Cancer database; C: Protein expression of ING family members in normal and primary LIHC tissues based on the Clinical proteomic tumor analysis consortium dataset. aP < 0.05; bP < 0.01; cP < 0.001.
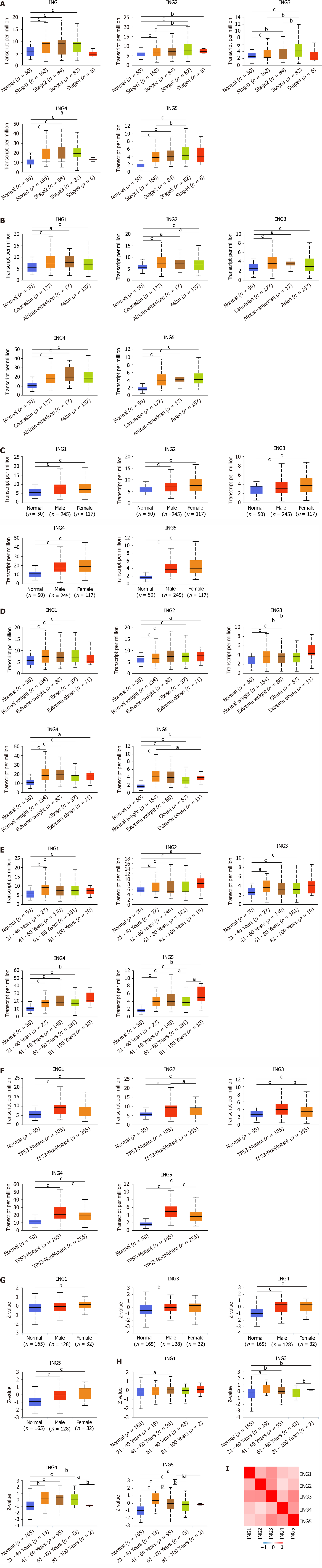
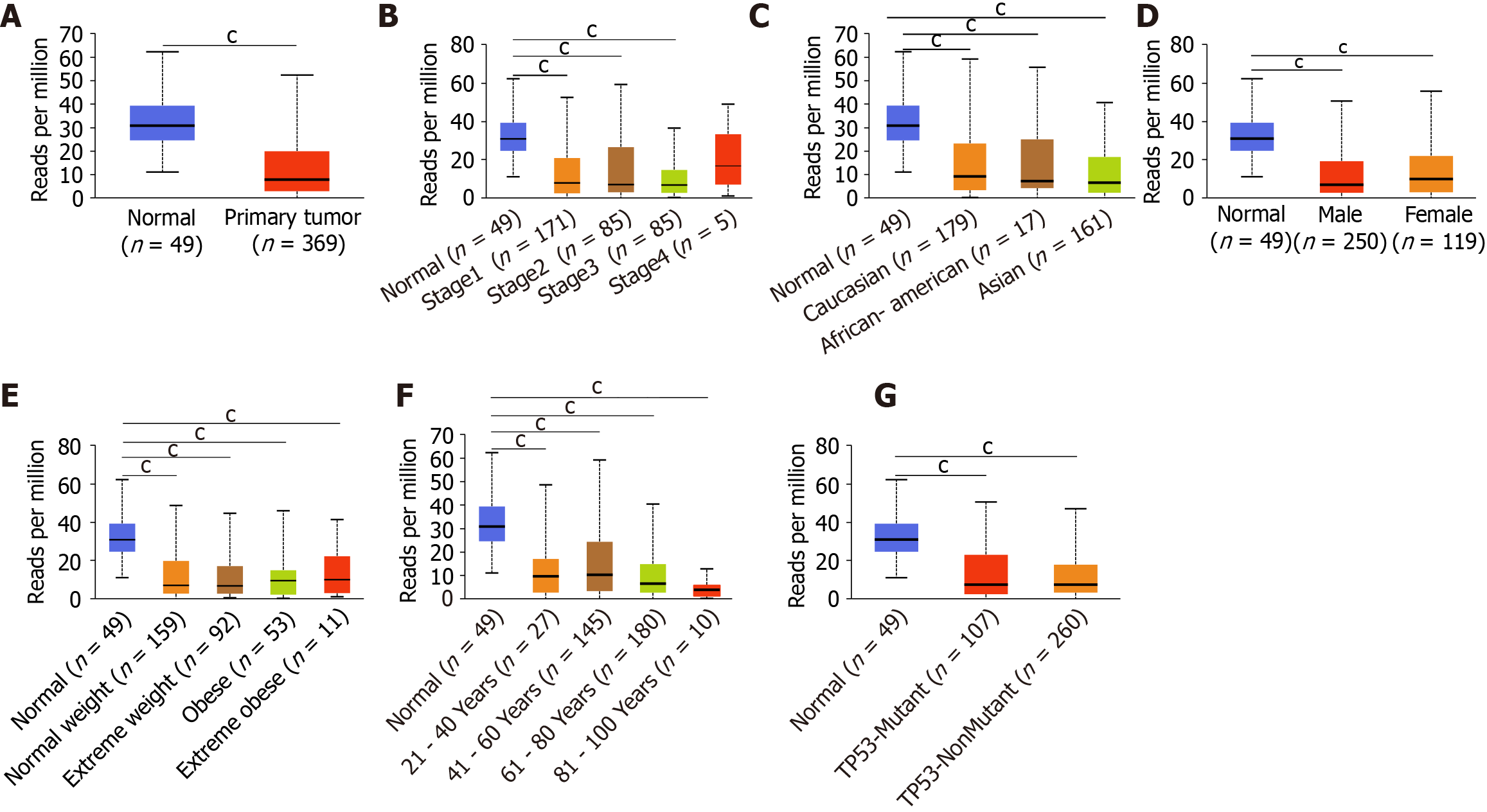
Figure 2 The relationships between the mRNA/protein expression of ING family members and clinicopathological parameters in liver cancer.
A: Between mRNA expression of ING family members and individual cancer stages; B: Between mRNA expression of ING family members and race; C: Between mRNA expression of ING family members and patient sex; D: Between mRNA expression of ING family members and patient body weight; E: Between mRNA expression of ING family members and patient age; F: Between mRNA expression of ING family members and TP53 mutation status; G: Between protein expression of ING family members and patient sex; H: Between protein expression of ING family members and patient age; I: Correlation of mRNA expression in different ING family members. aP < 0.05; bP < 0.01; cP < 0.001.
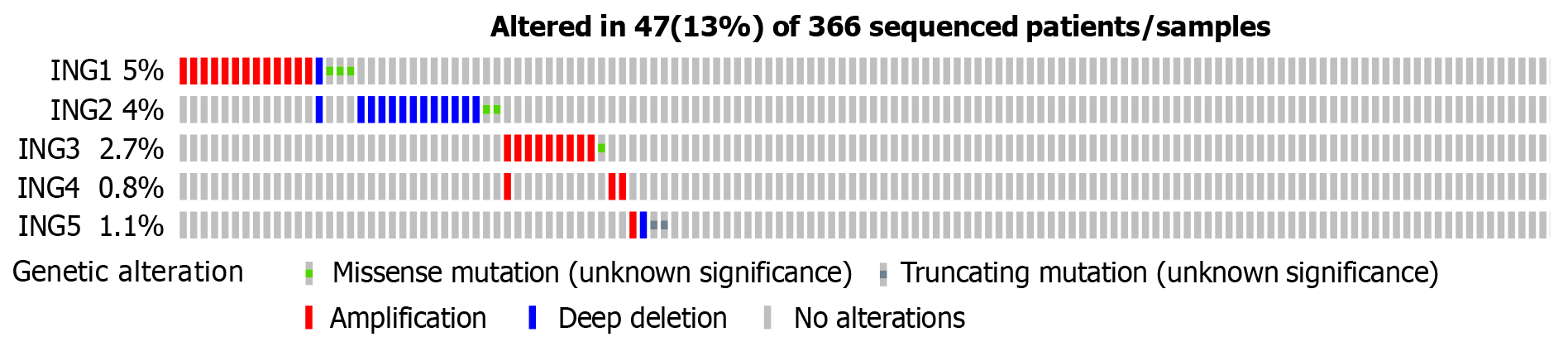
Figure 3 Genetic alterations of ING family genes in patients with liver cancer.
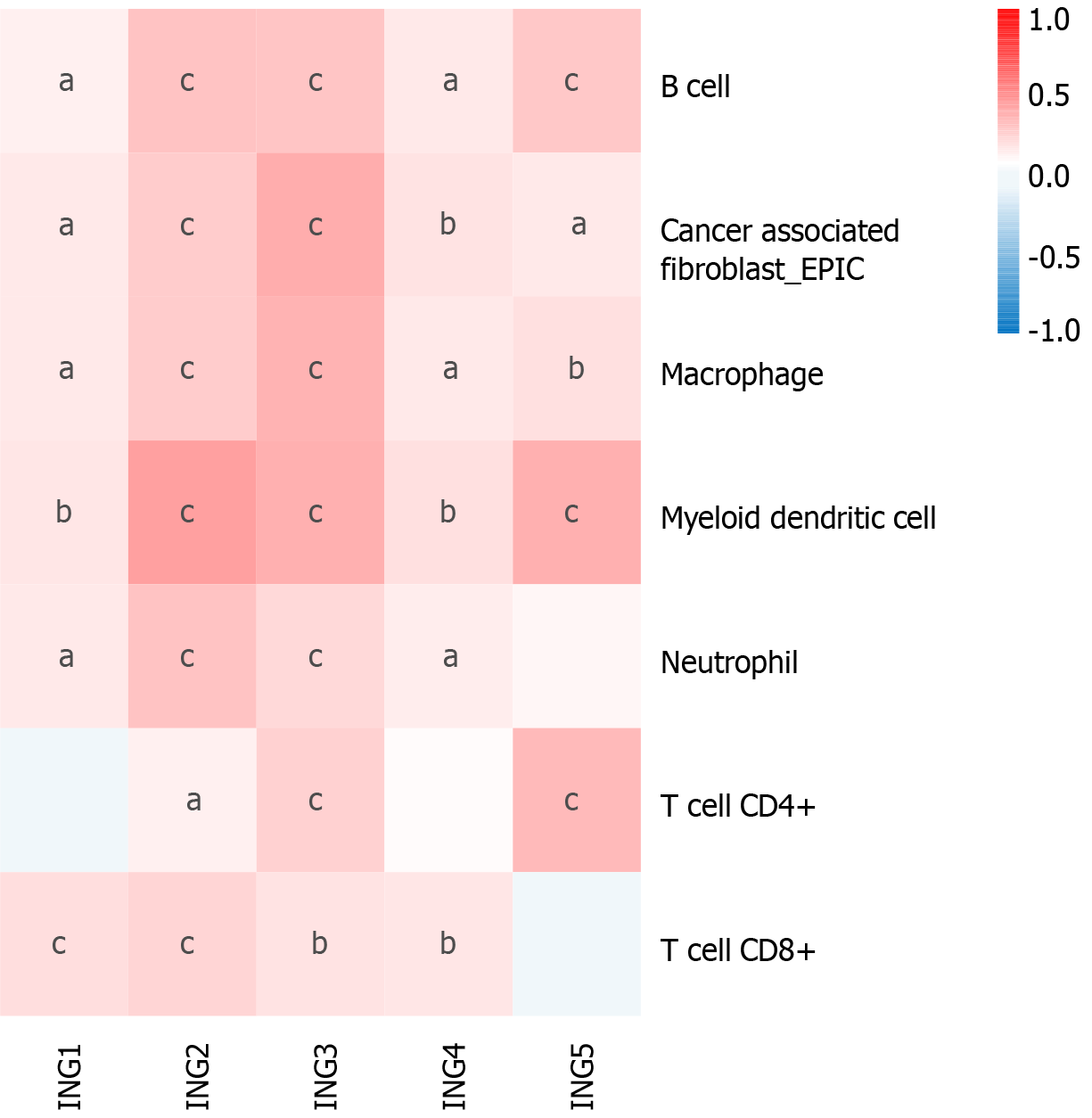
Figure 4 Correlation between expression of ING family genes and B cells, cancer-associated fibroblasts, myeloid dendritic cells, neutrophils, CD4+ T cells, CD8+ T cells, and macrophage infiltration levels in liver cancer.
aP < 0.05; bP < 0.01; cP < 0.001.
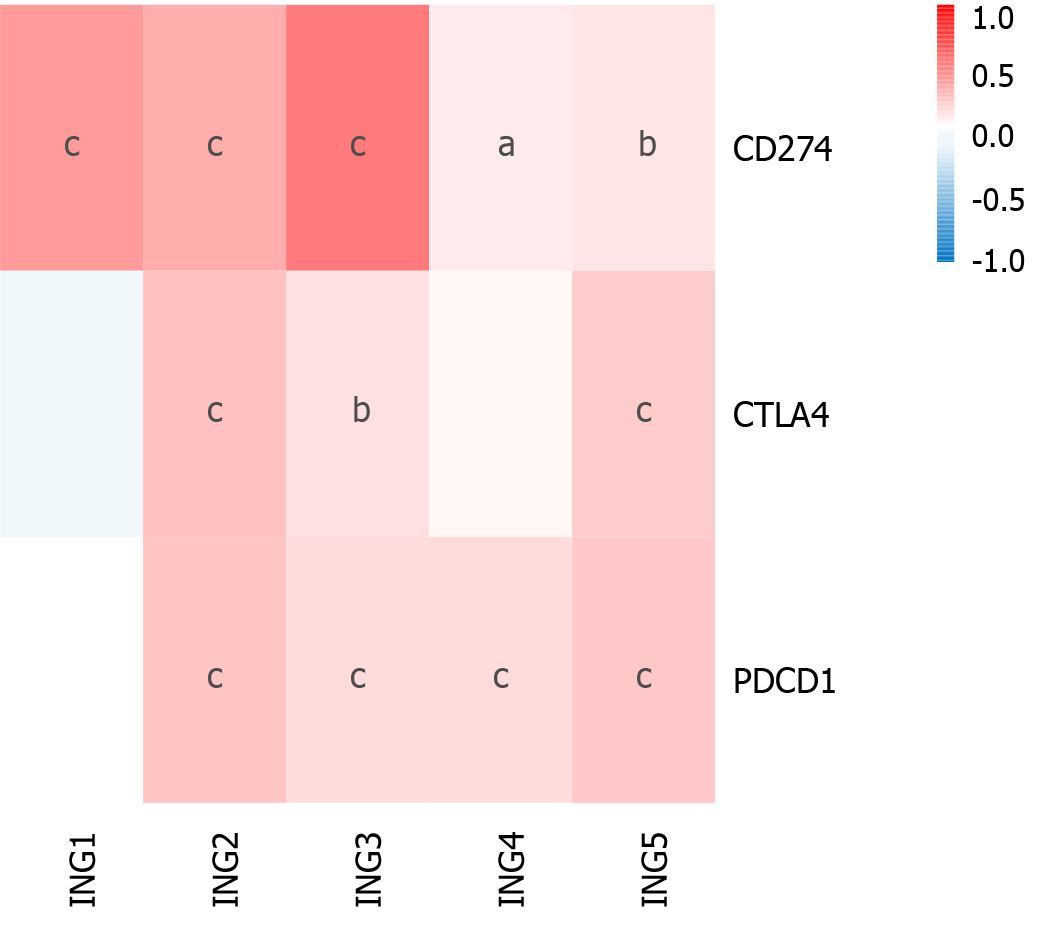
Figure 5 Correlation between expression of ING1 and immune checkpoint genes (PDCD1, CTLA4 and CD274) in liver cancer.
aP < 0.05; bP < 0.01; cP < 0.001.
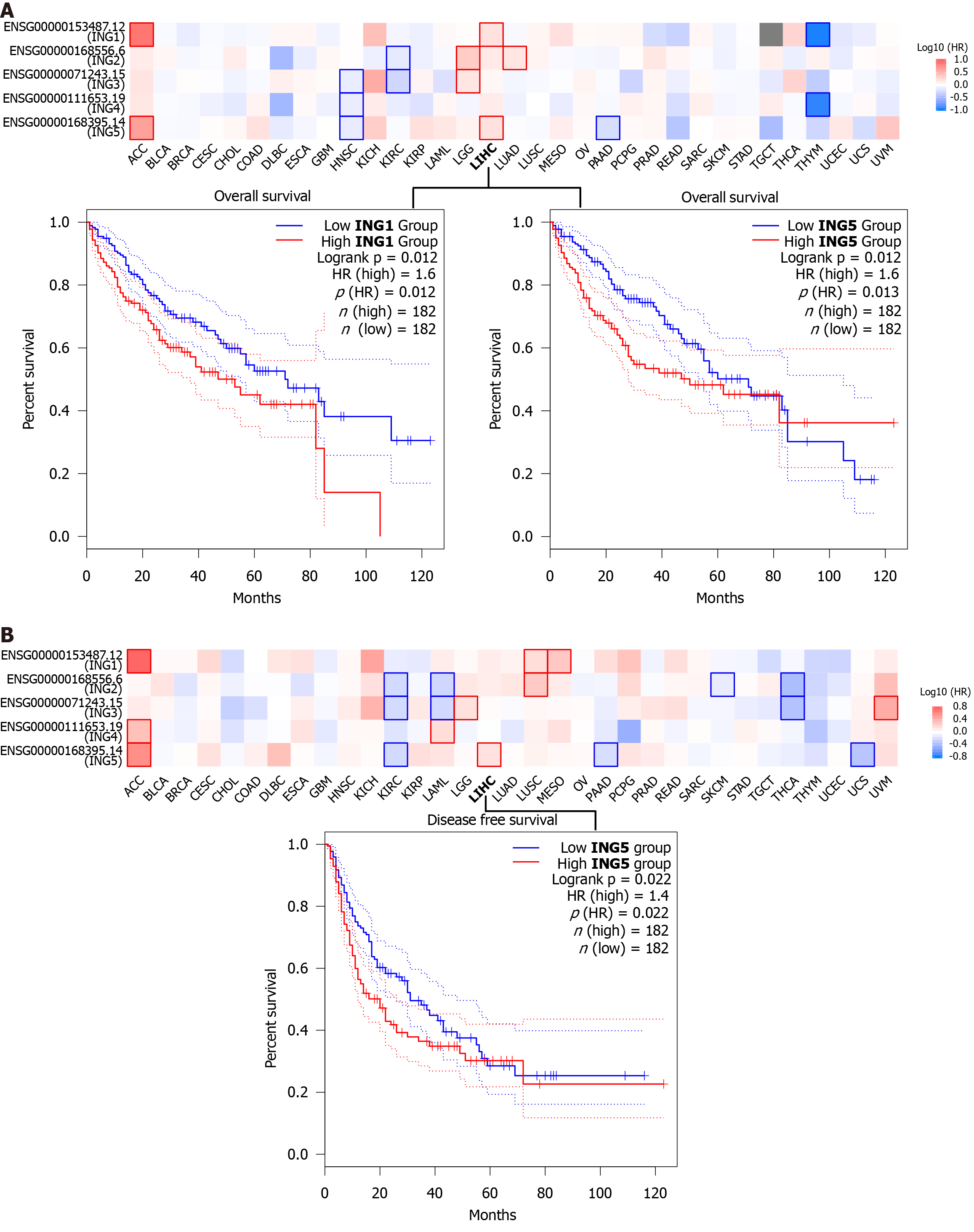
Figure 6 Correlation between expression of ING family genes and prognosis of cancers in The Cancer Genome Atlas using the GEPIA2 tool.
A: Overall survival analysis; B: Disease-free survival analysis. The survival chart and Kaplan-Meier curve with positive results were shown. HR: Hazard ratio.
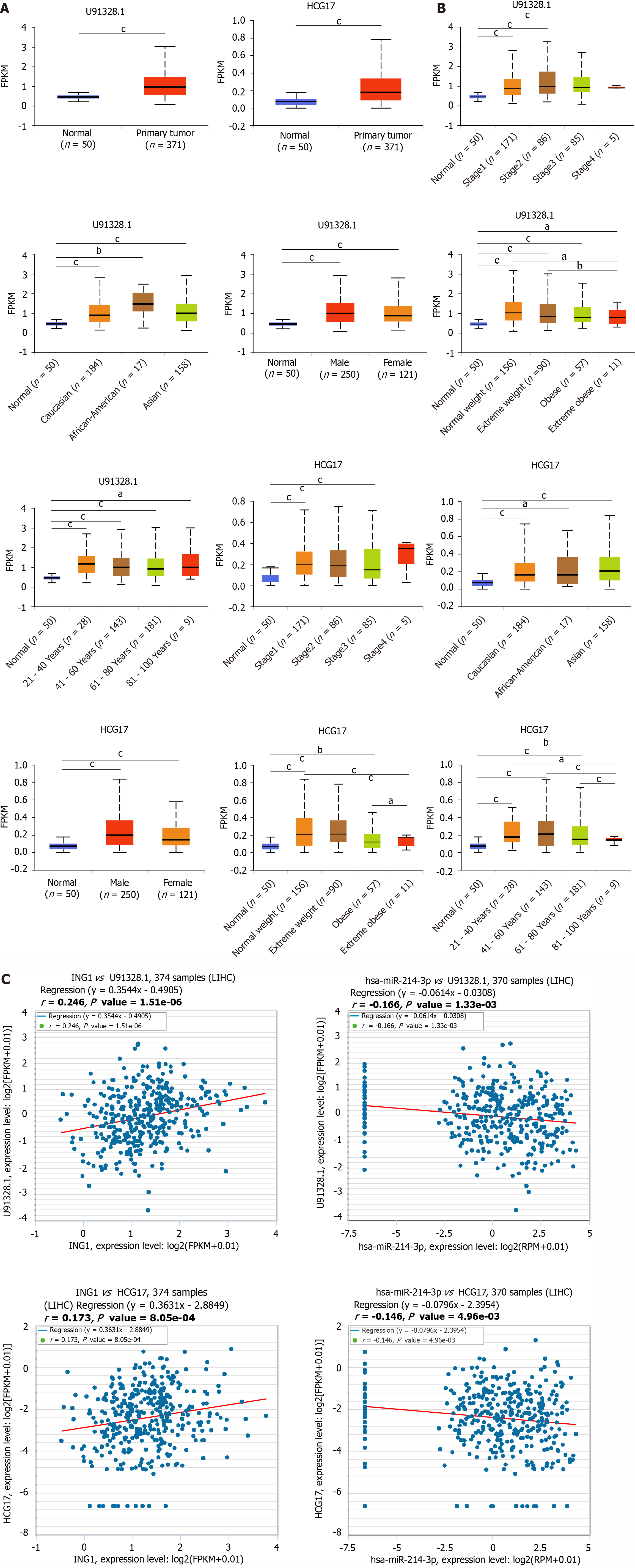
Figure 7 Relationships between hsa-miR-214-3p expression and clinicopathological parameters in liver cancer.
A: Differential expression of hsa-miR-214-3p between normal and cancer tissue; B: Between expression of hsa-miR-214-3p and individual cancer stages; C: Between expression of hsa-miR-214-3p and patient race; D: Between expression of hsa-miR-214-3p and patient gender; E: Between expression of hsa-miR-214-3p and patient weight; F: Between expression of hsa-miR-214-3p and patient age; G: Between expression of hsa-miR-214-3p and TP53 mutation status. cP < 0.001.
Figure 8 Upstream long noncoding RNAs prediction analysis of hsa-miR-214-3p.
A: Differential expression of two long noncoding RNAs (lncRNAs) in cancer and normal tissues; B: Expression of two lncRNAs in other clinicopathological parameters; C: Correlation analysis between lncRNAs and ING1 or between hsa-miR-214-3p and lncRNAs in liver cancer. aP < 0.05; bP < 0.01; cP < 0.001. LIHC: Liver cancer.
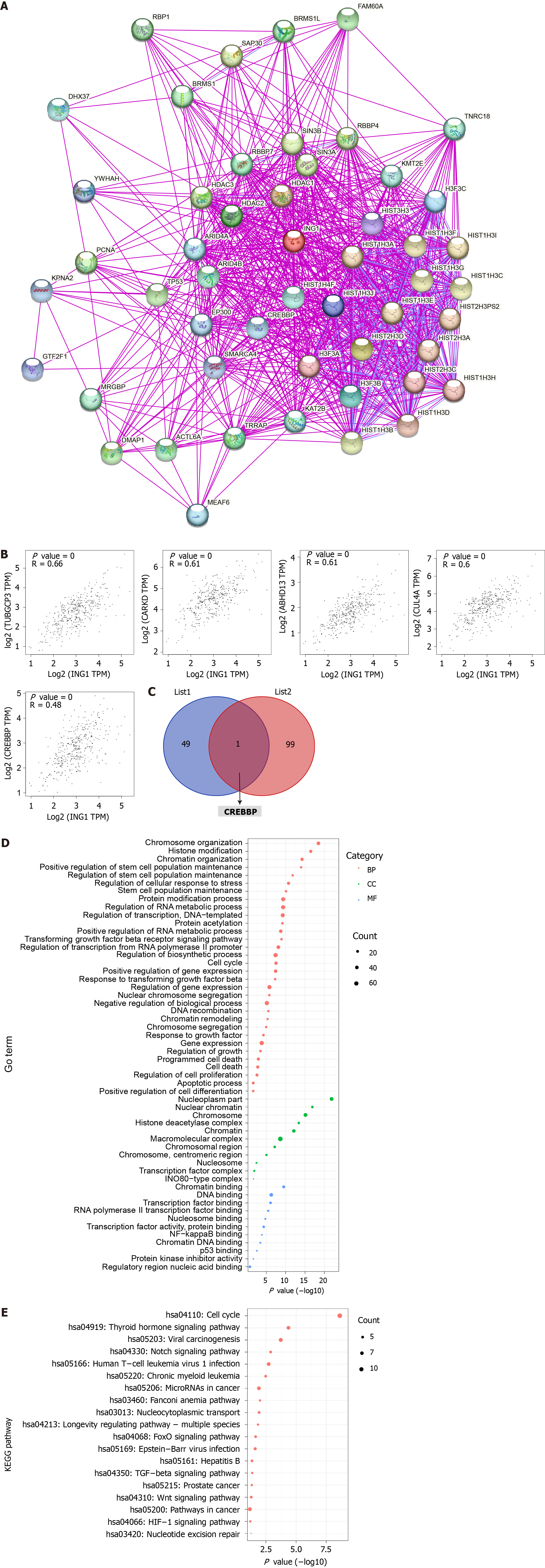
Figure 9 Functional analysis of ING1-related genes.
A: ING1 binding protein using STRING database; B: Expression correlation between ING1 and selected target genes, including TUBGCP3, CARKD, ABHD13, CUL4A and CREBBP; C: Intersection analysis of the binding genes and associated genes of ING1. List 1: ING1 binding proteins; List 2: Top 100 genes associated with ING1 expression; D: Gene Ontology analysis based on the ING1-binding and interaction genes; E: Kyoto Encyclopedia of Genes and Genomes pathway analysis based on the ING1-binding and interaction genes. BP: Biological processes; CC: Cellular components; MF: Molecular functions; GO: Gene Ontology; KEGG: Kyoto Encyclopedia of Genes and Genomes.
- Citation: Liu SC. Comprehensive analysis of clinical and biological value of ING family genes in liver cancer. World J Gastrointest Oncol 2024; 16(6): 2592-2609
- URL: https://www.wjgnet.com/1948-5204/full/v16/i6/2592.htm
- DOI: https://dx.doi.org/10.4251/wjgo.v16.i6.2592