Copyright
©The Author(s) 2021.
World J Gastrointest Oncol. Dec 15, 2021; 13(12): 2129-2148
Published online Dec 15, 2021. doi: 10.4251/wjgo.v13.i12.2129
Published online Dec 15, 2021. doi: 10.4251/wjgo.v13.i12.2129
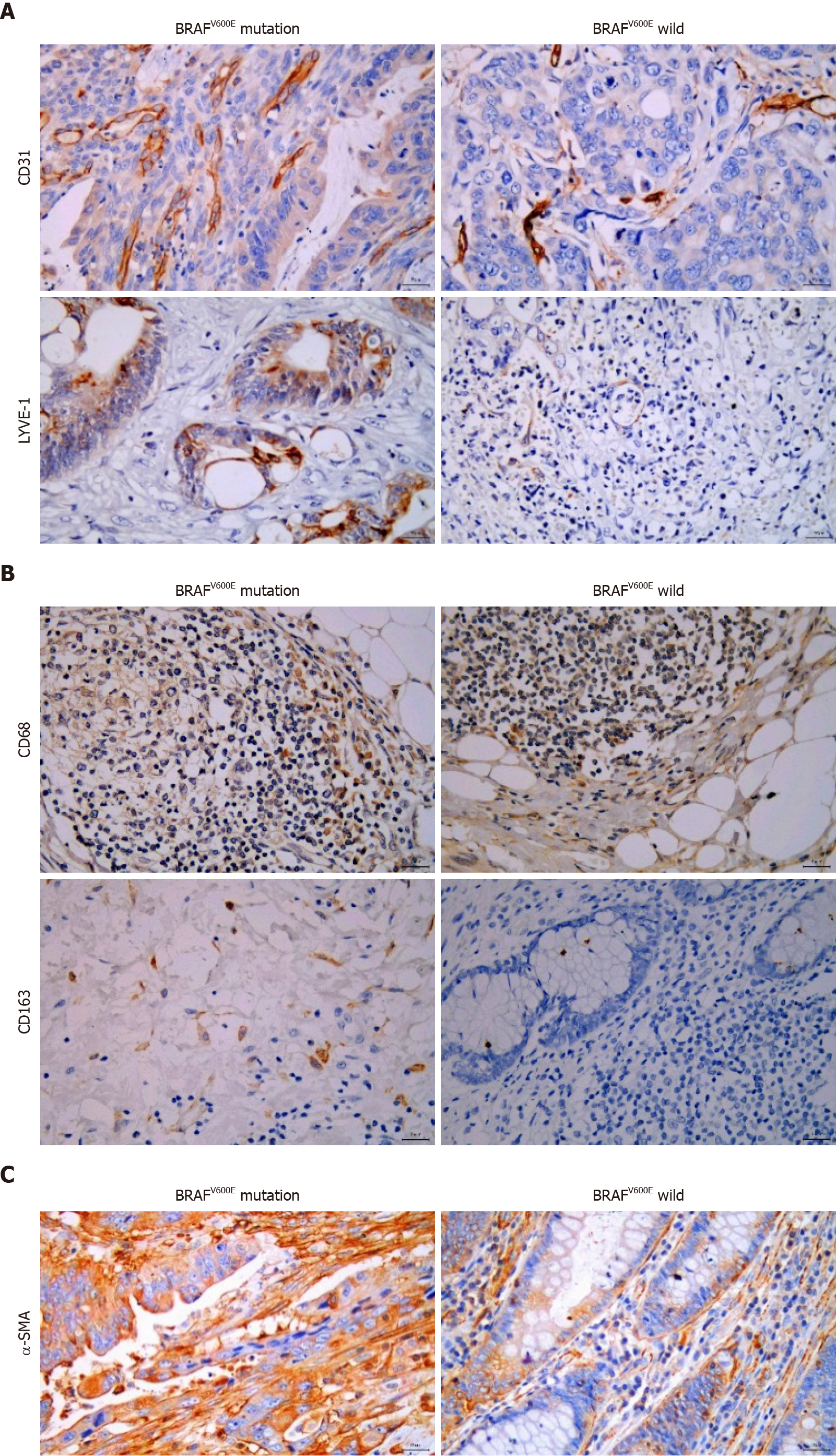
Figure 1 Relationship between BRAFV600E mutation and microvascular density, microlymphatic vessel density, tumor-associated macrophages, and cancer-associated fibroblasts.
A: Expression of CD31 and lymphatic vessel endothelial hyaluronic acid receptor-1 (LYVE-1) in BRAFV600E mutant and BRAF wild type colorectal cancer (CRC) tissues (CD31 labeled microvascular density, and LYVE-1 labeled microlymphatic vessel density; immunohistochemistry, × 400); B: Expression of CD68 and CD163 proteins in BRAFV600E mutant and BRAF wild type CRC tissues (CD68 labeled TAMs, and CD163 labeled M2 subtype macrophages; immunohistochemistry, × 400); C: Expression of α-SMA protein in BRAFV600E mutant and BRAF wild type CRC tissues (α-SMA labeled CAFs; immunohistochemistry, × 400).
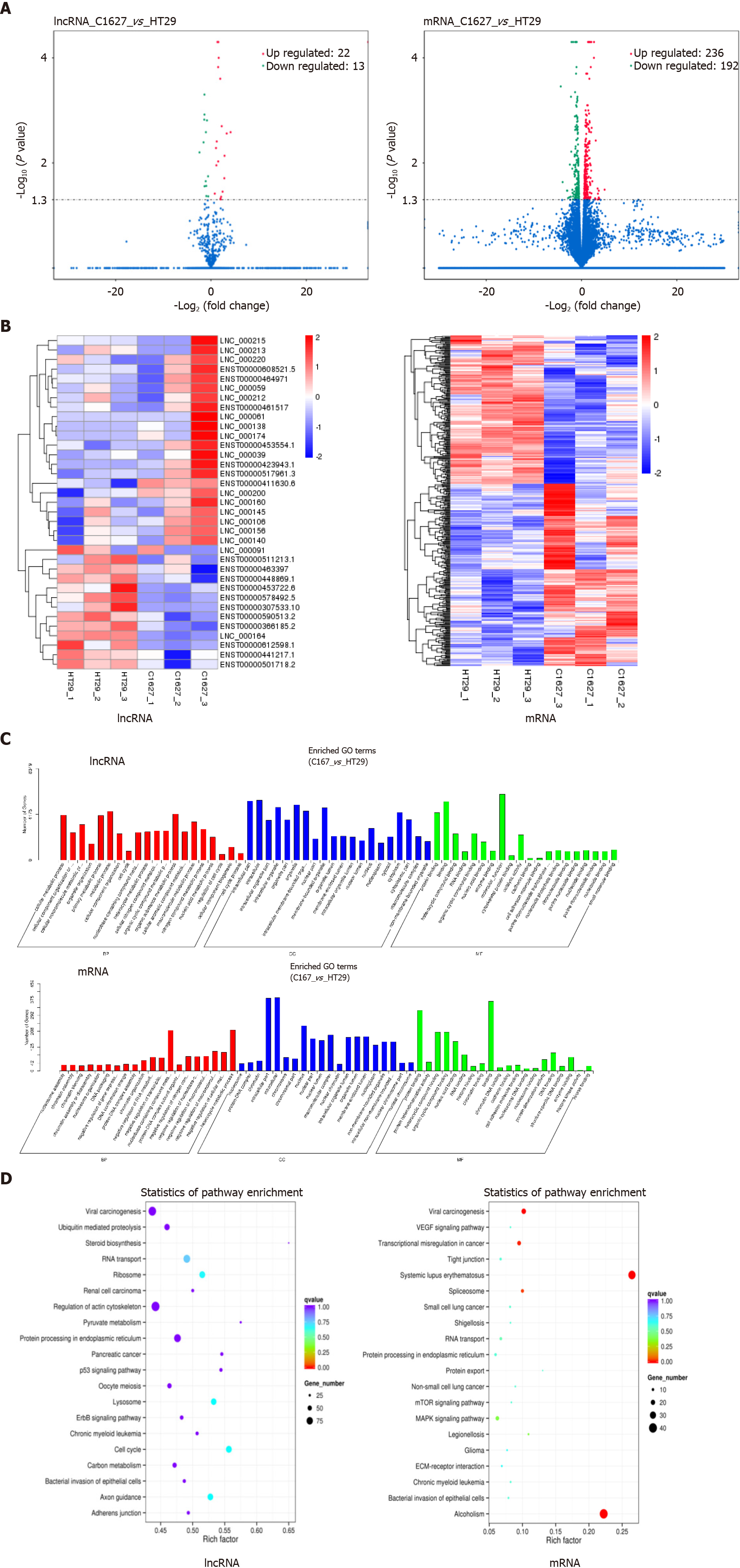
Figure 2 Screening and functional analysis of differentially expressed long noncoding RNAs and mRNAs in exosomes of BRAFV600E mutant colorectal cancer cells and those with BRAF gene silencing.
A: Transcriptomics analysis of differentially expressed long noncoding RNAs (lncRNAs) and mRNAs in the exosomes of HT29 and 1627 cells. Volcano maps are shown (the x axis represents the multiple of difference, and the y axis P values); B: Hierarchical cluster analysis of differentially expressed lncRNAs and mRNAs (red represents upregulated expression, and blue represents downregulated expression); C: GO enrichment analysis of differentially expressed lncRNA target genes and mRNAs; D: KEGG pathway analysis of differentially expressed lncRNA target genes and mRNAs. LncRNA: Long noncoding RNA.
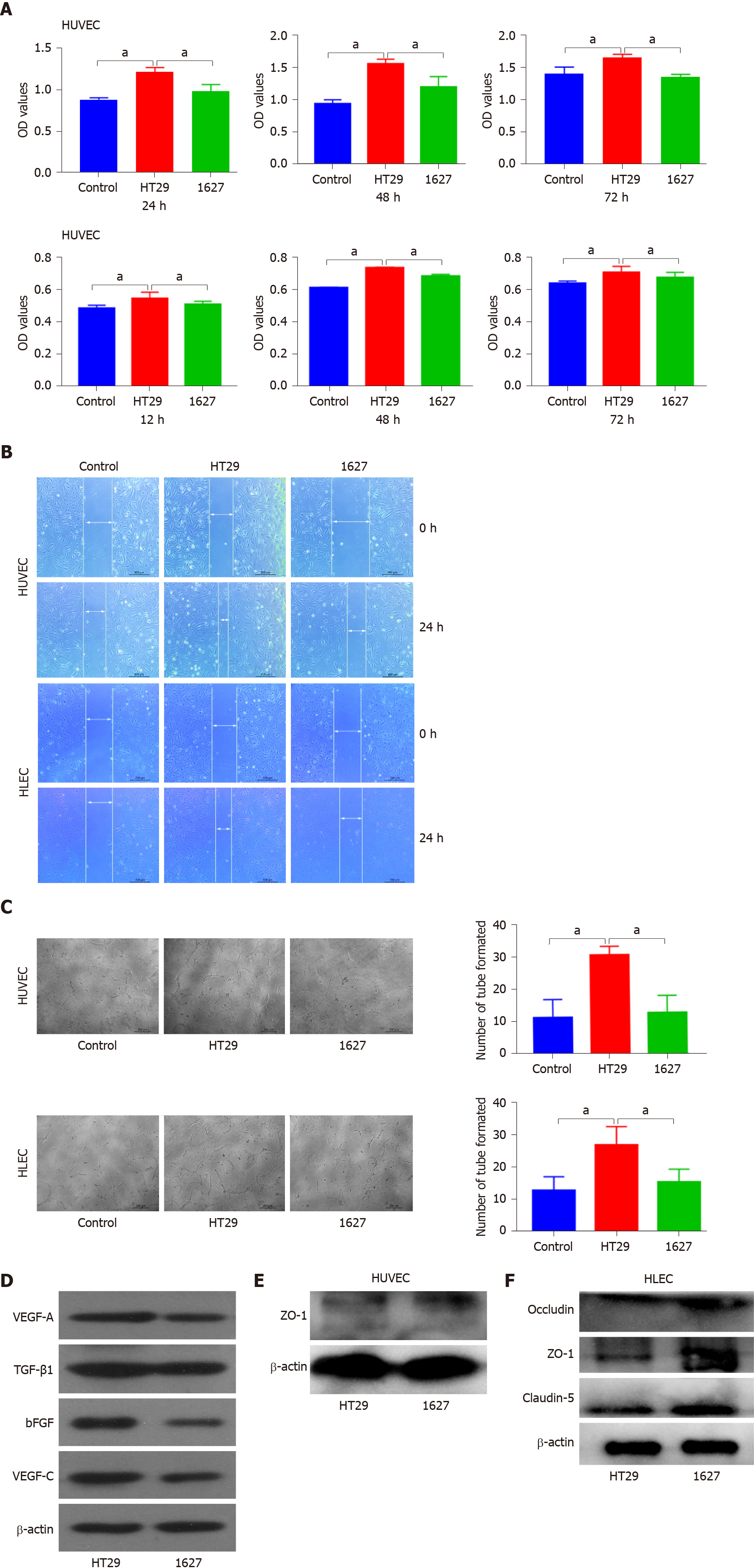
Figure 3 Effect of BRAFV600E mutant colorectal cancer-cell-derived exosomes on the proliferation, migration, tube formation, and protein expression of human umbilical vein endothelial cells and human lymphatic endothelial cells.
A: Detection of changes in proliferation of human umbilical vein endothelial cells (HUVECs) induced by exosomes derived from HT29 and 1627 cells at 24, 48, and 72 h by CCK8 assays. Changes in the proliferation of human lymphatic endothelial cells (HLECs) induced by exosomes derived from HT29 and 1627 cells were detected at 12, 24, and 36 h (the control group was treated with PBS); B: Detection of effects of HT29 and 1627 cell-derived exosomes on the migration of HUVECs and HLECs by scratch test (× 50); C: After 24 h of incubation, the effect of HT29 and 1627 cell-derived exosomes on tube formation ability of HUVECs and HLECs was observed (× 100); D: ELISA was used to detect expression of VEGF-A, TGF-β1, bFGF, and VEGF-C proteins in exosomes derived from HT29 and 1627 cells; E: ELISA was used to detect the effect of exosomes derived from HT29 and 1627 cells on expression of ZO-1 proteins in HUVECs; F: ELISA was used to detect the effect of exosomes derived from HT29 and 1627 cells on expression of ZO-1, occludin, and claudin-5 proteins in HLECs. aP < 0.05. HUVECs: Human umbilical vein endothelial cells; HLECs: Human lymphatic endothelial cells.
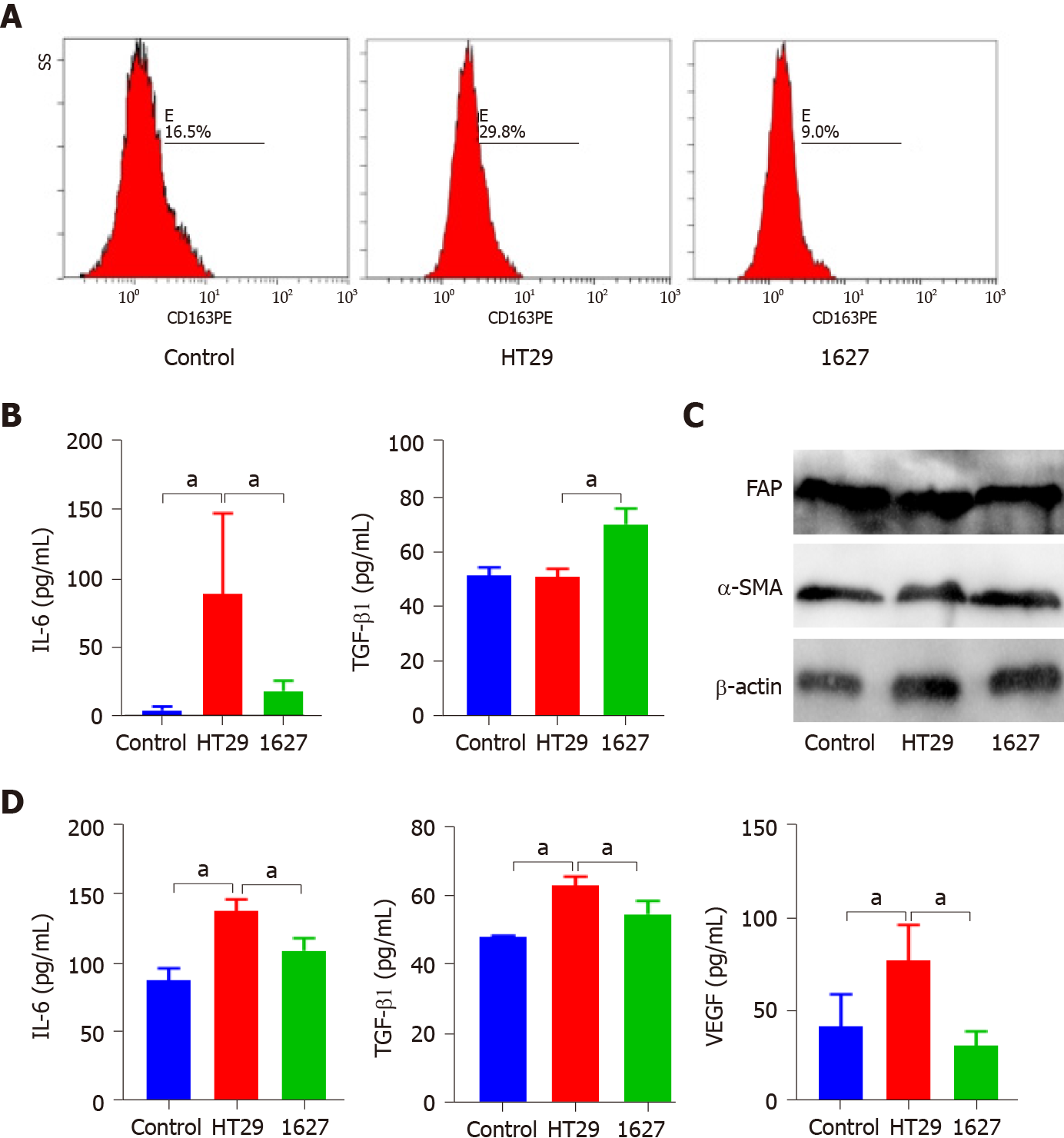
Figure 4 Effect of exosomes derived from BRAFV600E mutant colorectal cancer cells on the phenotype and secretion function of tumor-associated macrophages and cancer-associated fibroblasts.
A: Effect of silencing BRAF gene on expression of CD163 in macrophages induced by exosomes derived from BRAFV600E mutant colorectal cancer cells; B: ELISA was used to detect expression of interleukin (IL)-6 and transforming growth factor (TGF)-β1 in the supernatant of THP-1 cells; C: Western blotting was used to detect expression of FAP and α-SMA in MRC-5 cells; D: ELISA was used to detect expression of IL-6, TGF-β1, and vascular endothelial growth factor in the supernatant of MRC-5 cells. aP < 0.05. VEGF: Vascular endothelial growth factor; IL: Interleukin; TGF: Transforming growth factor.
- Citation: Zhi J, Jia XJ, Yan J, Wang HC, Feng B, Xing HY, Jia YT. BRAFV600E mutant colorectal cancer cells mediate local immunosuppressive microenvironment through exosomal long noncoding RNAs. World J Gastrointest Oncol 2021; 13(12): 2129-2148
- URL: https://www.wjgnet.com/1948-5204/full/v13/i12/2129.htm
- DOI: https://dx.doi.org/10.4251/wjgo.v13.i12.2129