Copyright
©The Author(s) 2021.
World J Gastrointest Oncol. Dec 15, 2021; 13(12): 2101-2113
Published online Dec 15, 2021. doi: 10.4251/wjgo.v13.i12.2101
Published online Dec 15, 2021. doi: 10.4251/wjgo.v13.i12.2101
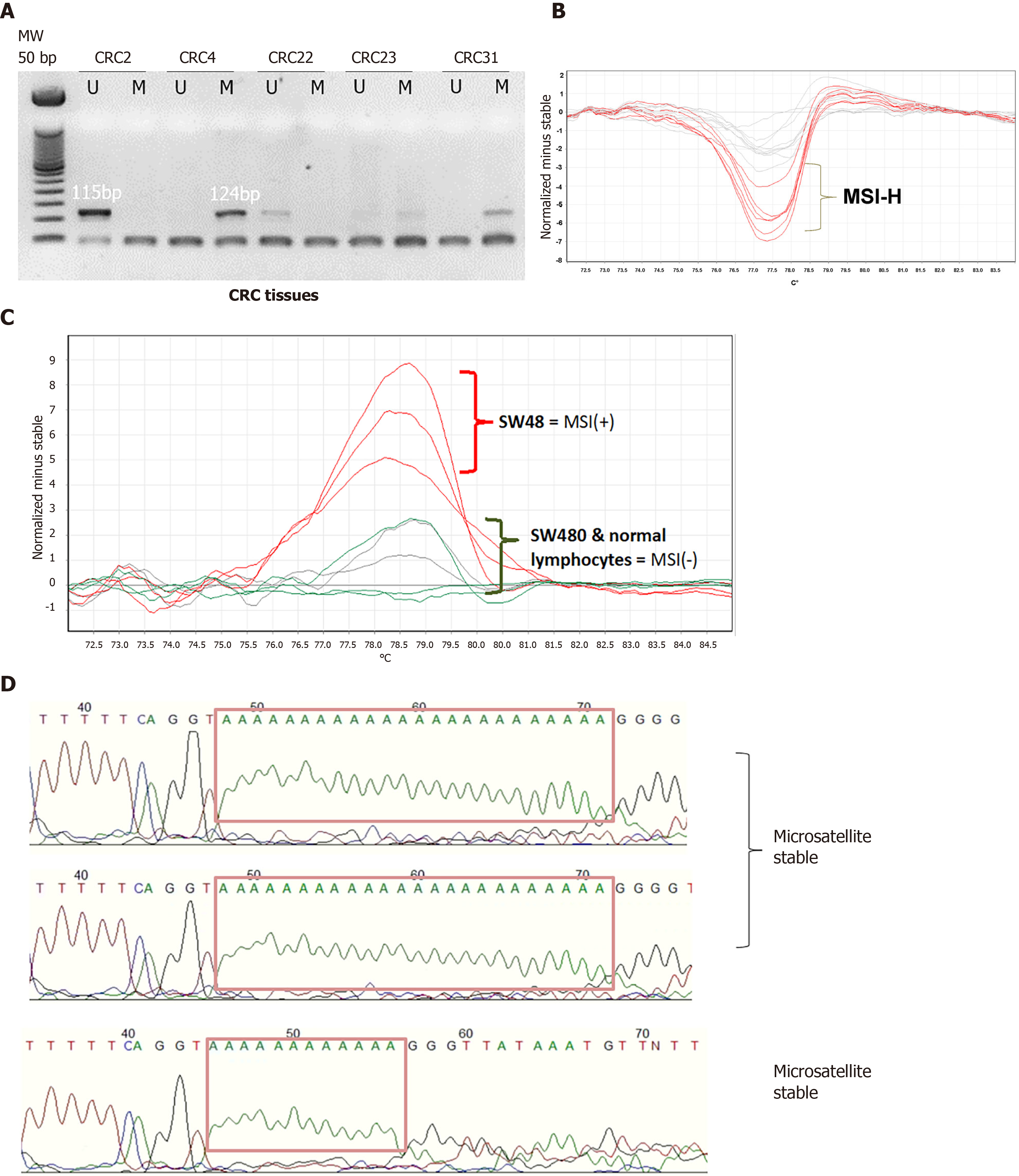
Figure 1 Determination of hMLH1 methylation by methylation-specific polymerase chain reaction and microsatellite instability status by high resolution melting analysis.
A: Gel images showing presence of 124 bp product indicating an unmethylated hMLH1 and the 115 bp product for the methylated hMLH1 in clinical samples; B: Normalized difference curves of clinical samples showing microsatellite instability-high profiles in red; C: Difference curve of the high resolution melting analysis; D: Electropherograms showing deletion of 12 adenine repeats in microsatellite instable control SW48 using BAT26 compared to the 24 adenine repeats in stable control SW480 and lymphocytes from a normal individual. MS-PCR: Methylation specific polymerase chain reaction; HRM: High resolution melting; MSI: Microsatellite instability; MSI-H: Microsatellite instability-high; CRC: Colorectal cancer.
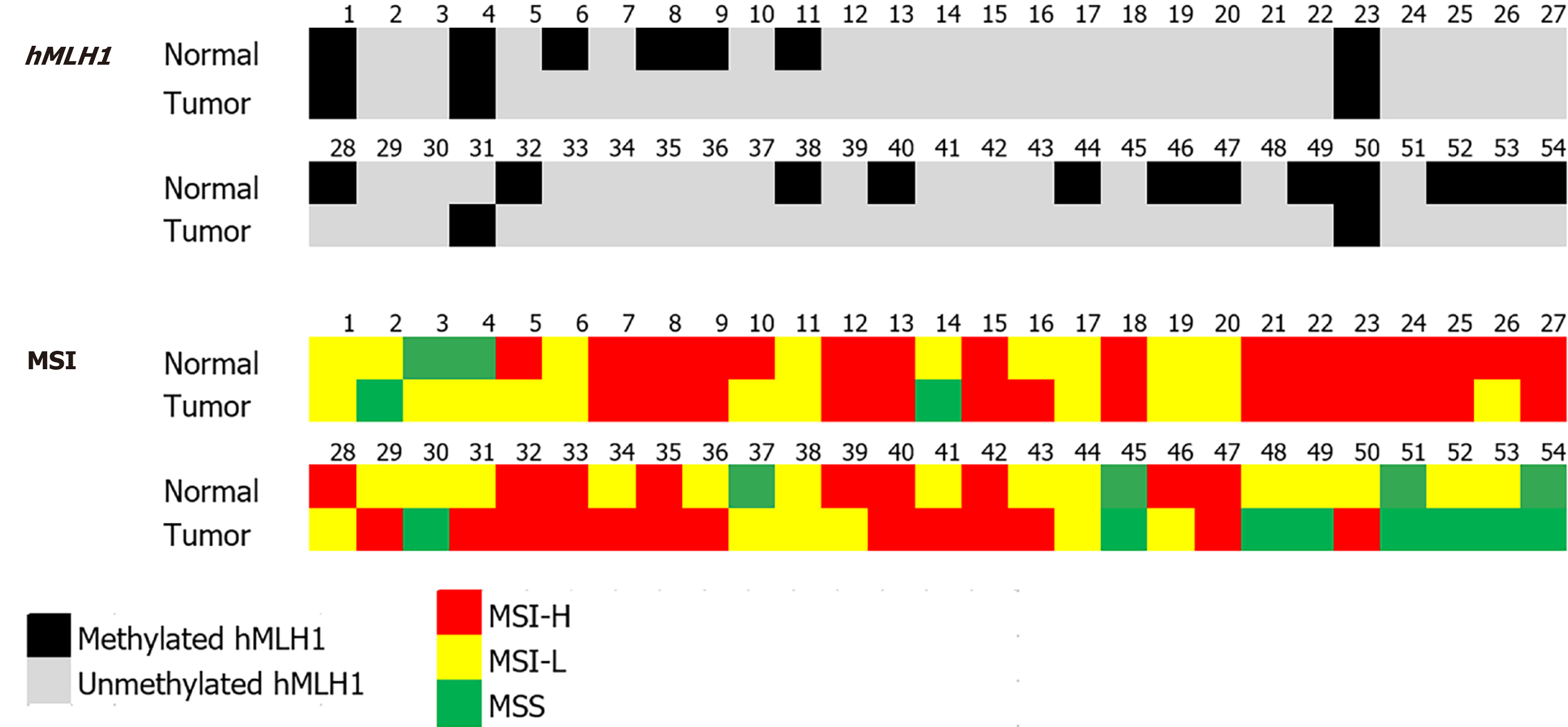
Figure 2 hMLH1 methylation and microsatellite instability analysis results of paired normal and tumor tissues.
hMLH1 methylation analysis was determined using bisulfite converted genomic DNA and methylated and unmethylated primer sets for methylation specific polymerase chain reaction. Microsatellite instability profiling was done by high-resolution melting analysis using the five Bethesda panel of markers. MSI: Microsatellite instability; MSI-H: Microsatellite instability-high; MSI-L: Microsatellite instability-low; MSS: Microsatellite stable.
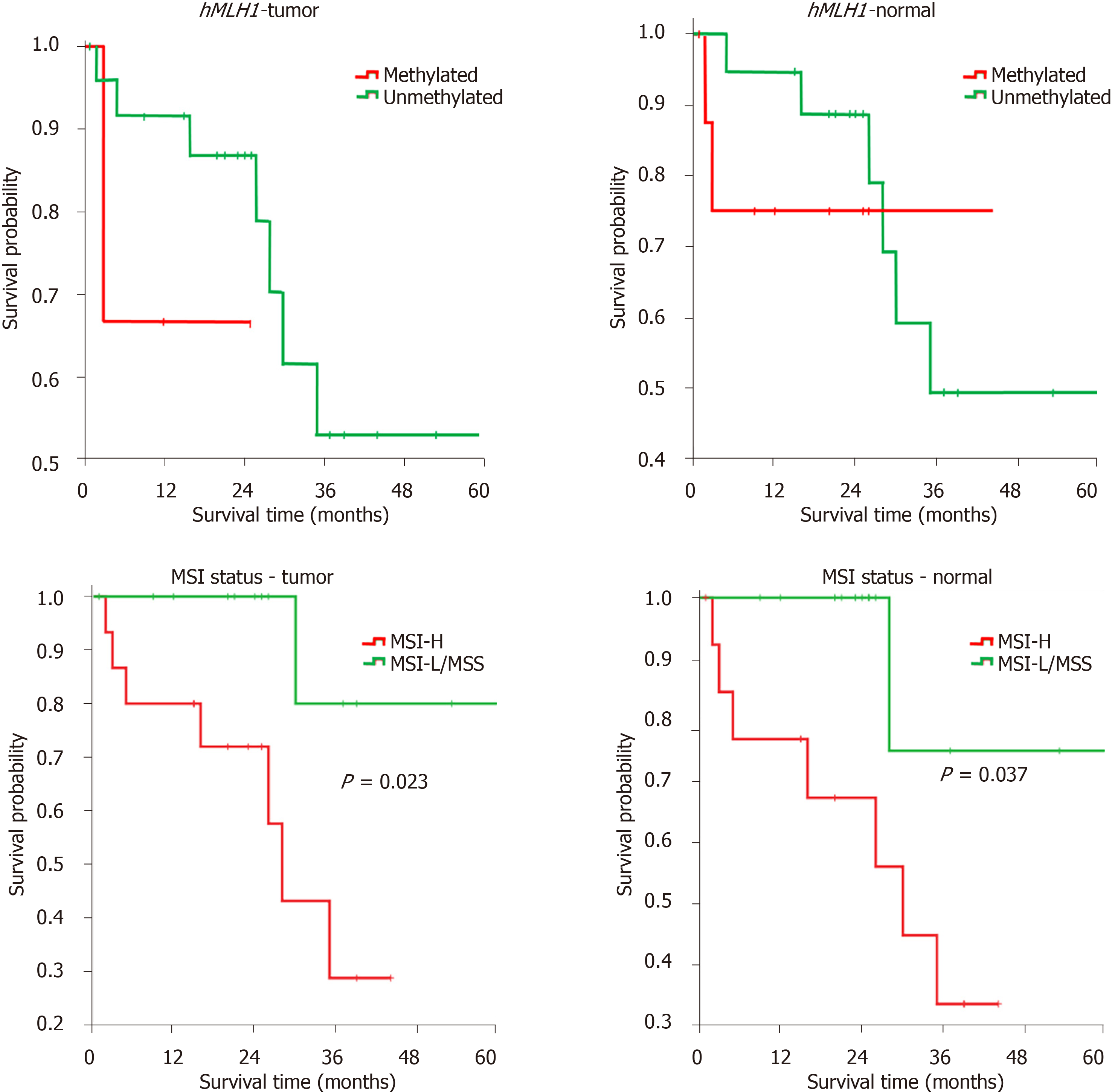
Figure 3 hMLH1 methylation and microsatellite instability analysis for patients’ prognosis.
Kaplan-Meier analysis of colorectal cancer patients’ overall survival in 60-mo of patients’ follow-up after surgery. MSI: Microsatellite instability.
- Citation: Cabral LKD, Mapua CA, Natividad FF, Sukowati CHC, Cortez ER, Enriquez MLD. MutL homolog 1 methylation and microsatellite instability in sporadic colorectal tumors among Filipinos. World J Gastrointest Oncol 2021; 13(12): 2101-2113
- URL: https://www.wjgnet.com/1948-5204/full/v13/i12/2101.htm
- DOI: https://dx.doi.org/10.4251/wjgo.v13.i12.2101