Copyright
©The Author(s) 2019.
World J Gastrointest Oncol. Feb 15, 2019; 11(2): 91-101
Published online Feb 15, 2019. doi: 10.4251/wjgo.v11.i2.91
Published online Feb 15, 2019. doi: 10.4251/wjgo.v11.i2.91
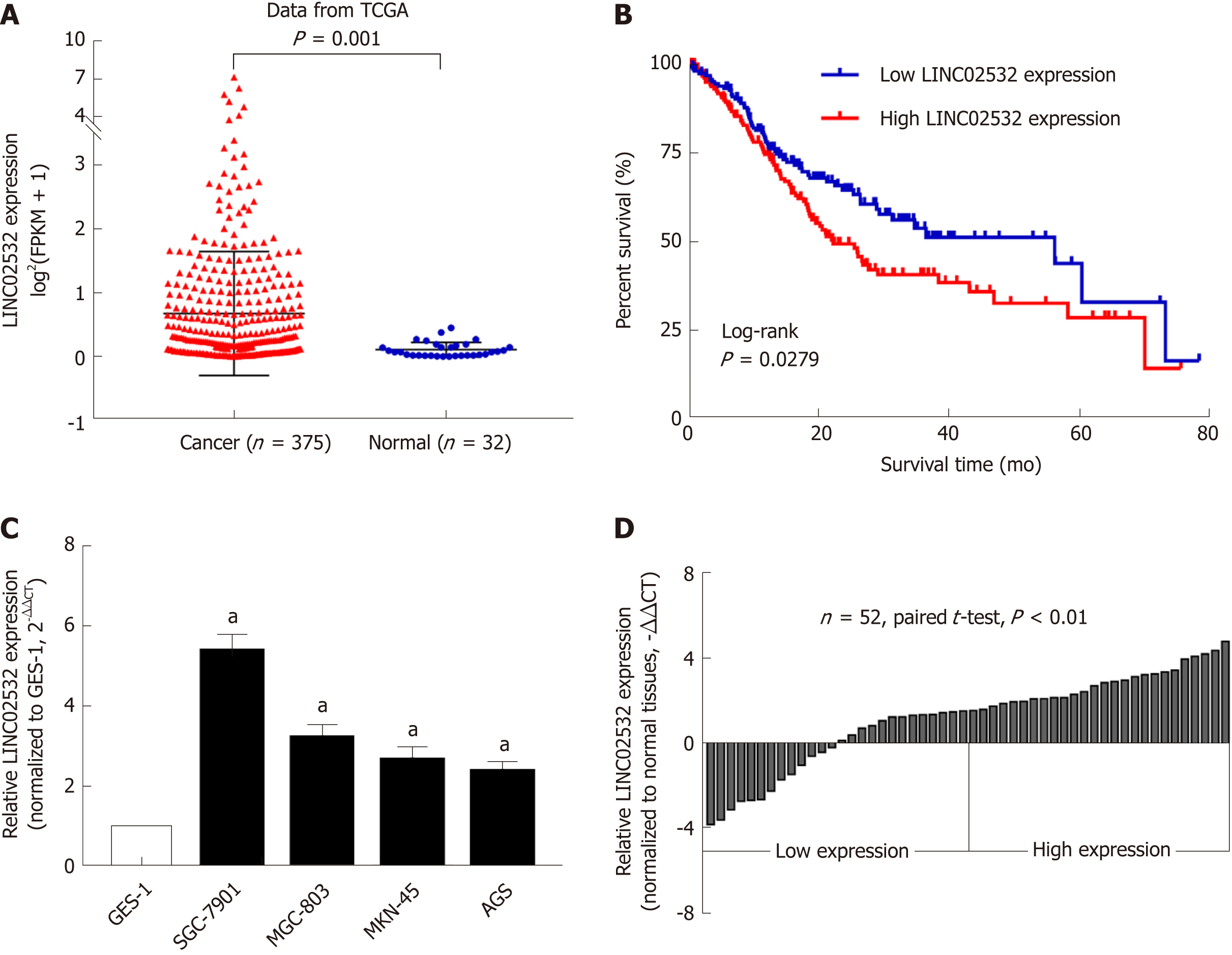
Figure 1 LINC02532 was upregulated in gastric cancer and associated with poor prognosis.
A: Expression levels of LINC02532 in gastric cancer (GC) and normal tissue based on the data from the Cancer Genome Atlas database; B: Kaplan-Meier survival analysis showed that patients with high LINC02532 expression had a poorer prognosis than those with low expression; C: Compared with GES-1, LINC02532 was significantly upregulated in four GC cell lines; D: LINC02532 was upregulated in 52 GC tissues. Expression level data are presented as mean ± SE. aP < 0.05 was considered statistically significant. GC: Gastric cancer; TCGA: The Cancer Genome Atlas; GES-1: Gastric epithelium cell line.
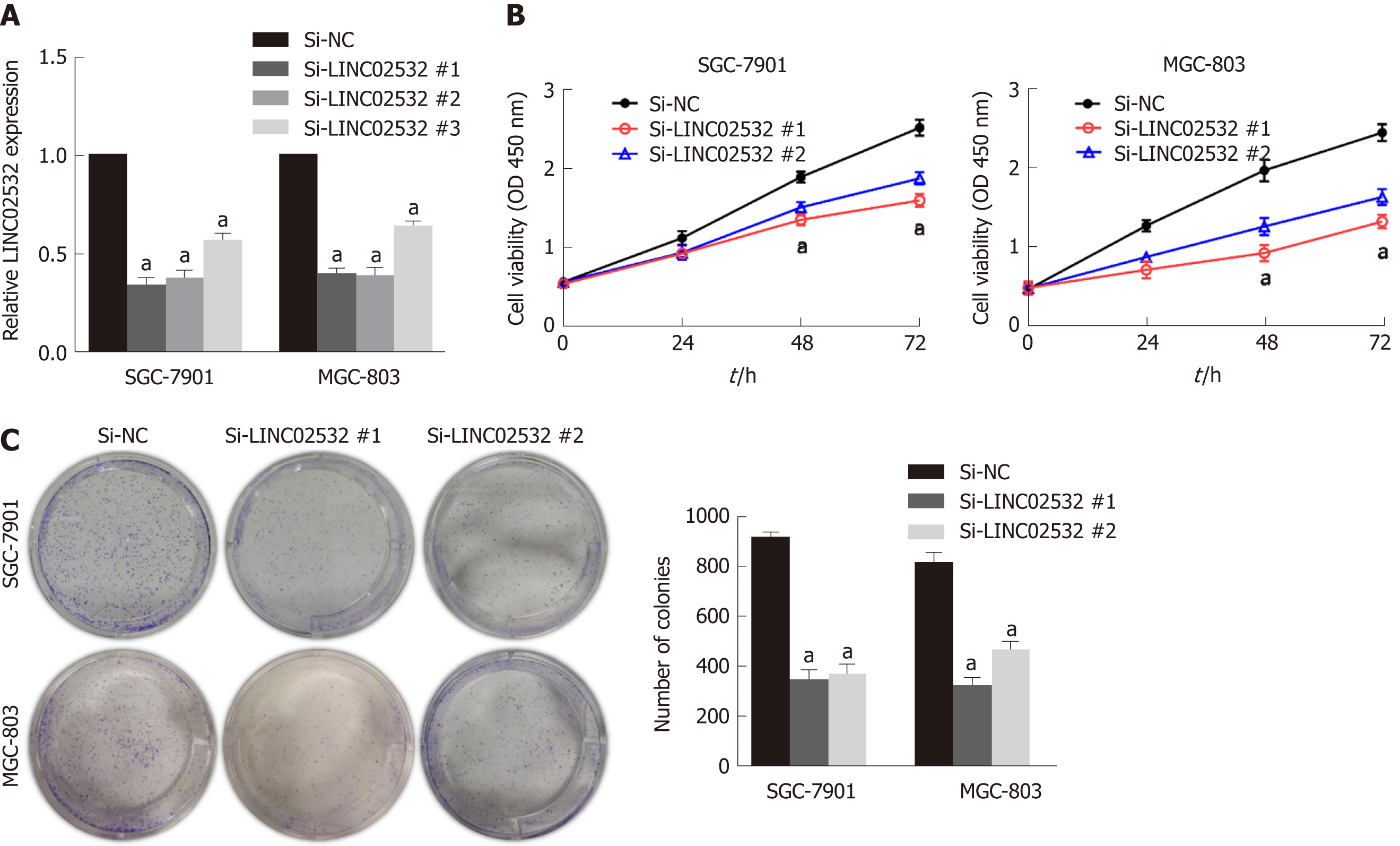
Figure 2 LINC02532 promotes gastric cancer cell proliferation.
A: The relative expression of LINC02532 in SGC-7901 and MGC-803 treated with three siRNAs; B: Cell counting kit-8 assay indicated that LINC02532 knockdown suppressed cell proliferation; C: Colony formation assays showed that LINC02532 knockdown inhibited gastric cancer cell proliferation. Data are presented as mean ± SE. aP < 0.05 was considered statistically significant.
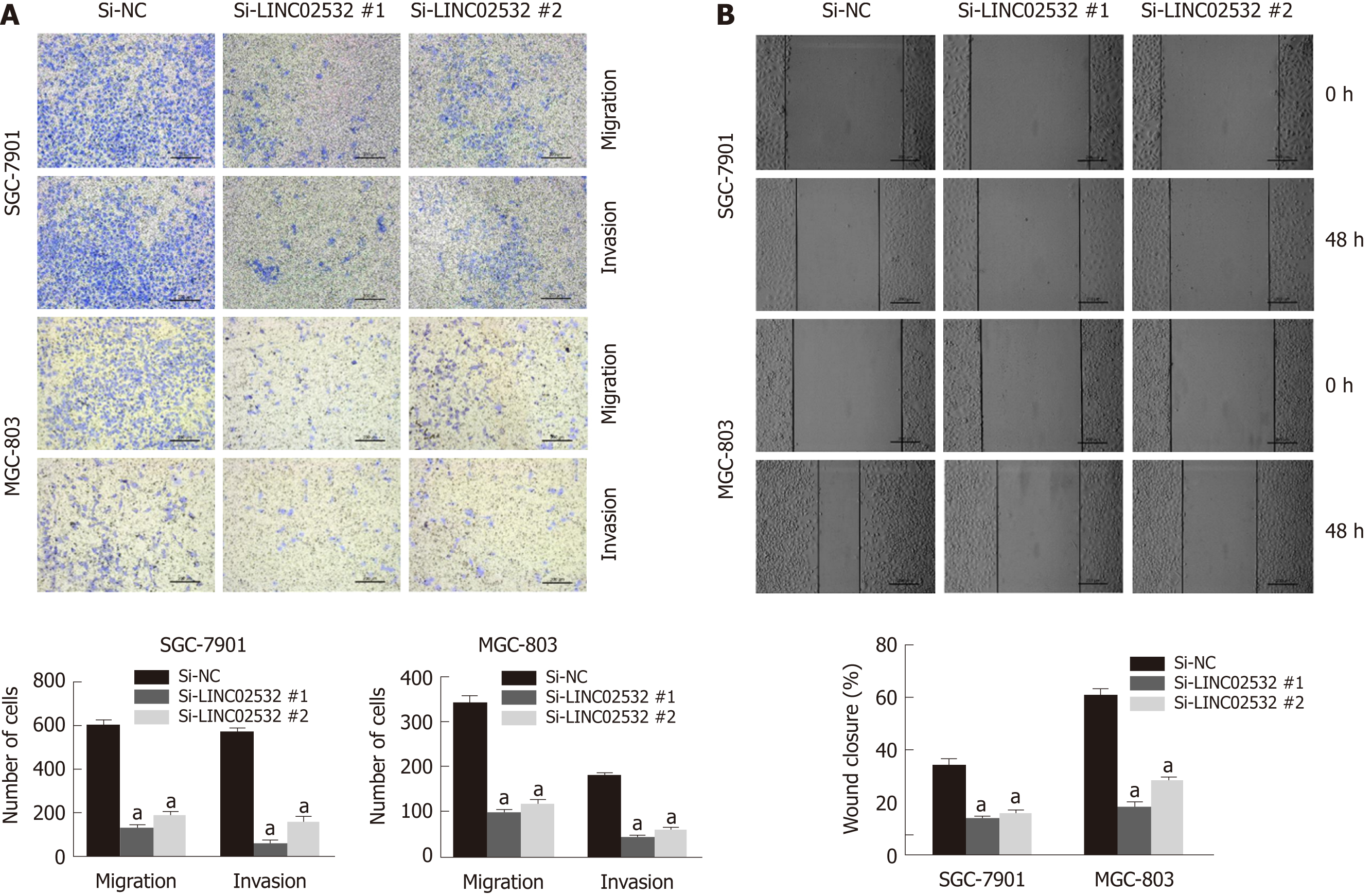
Figure 3 LINC02532 promotes gastric cancer cell migration and invasion.
A: Transwell assays with or without Matrigel were performed to assess the capacity of cell invasion and migration, respectively. The results revealed that LINC02532 knockdown promoted gastric cancer cell migration and invasion; B: The wound healing assay further displayed that LINC02532 knockdown promoted gastric cancer cell migration. aP < 0.05 was considered statistically significant.
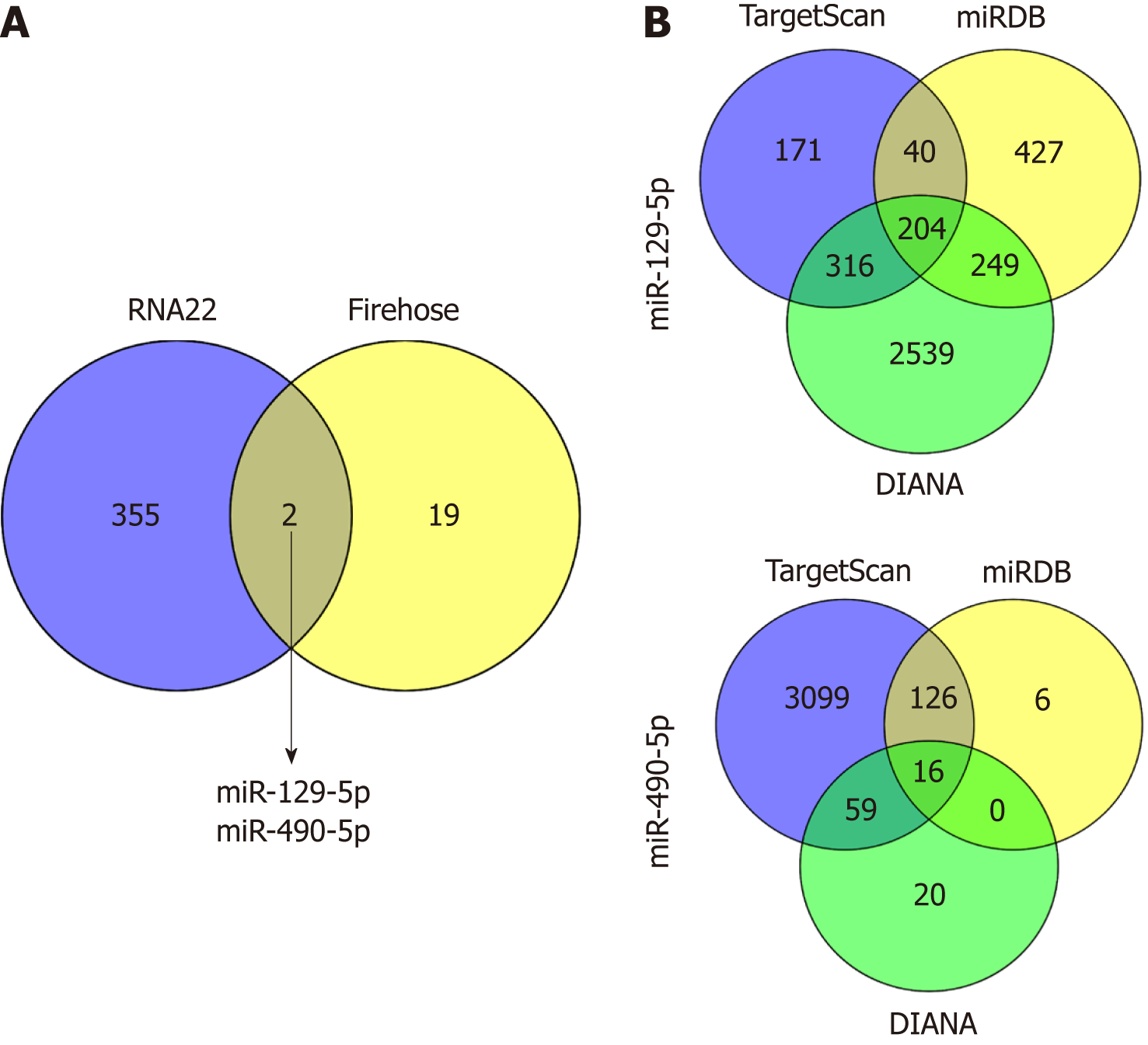
Figure 4 Venn diagrams for obtaining objective miRNAs and target genes.
A: miR-129-5p and miR-490-5p, which are both potential targets of LINC02532 and are downregulated in gastric cancer were identified based on the data from RNA22 and Firehose; B: The target genes of miR-129-5p and miR-490-5p were predicted by three software programs (TargetScan, miRDB, and DIANA).
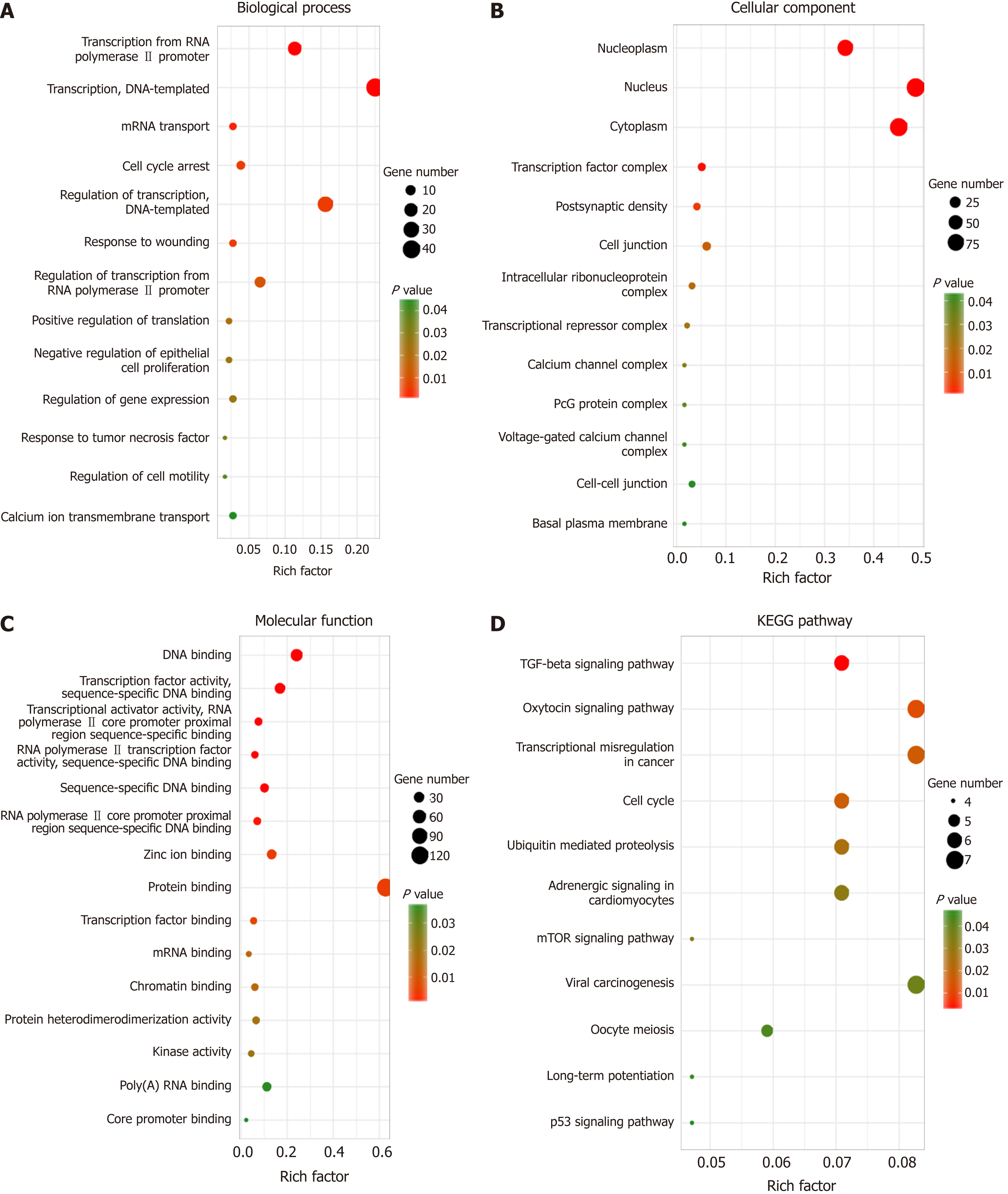
Figure 5 Functional and KEGG pathway enrichment analyses of target genes.
A: The biological process analysis indicated that the genes were enriched during the cell cycle arrest phase, in response to wounding, tumor necrosis factors, and regulation of cell motility; B: Cellular component analysis suggested that genes were present in the nucleus and cytoplasm; C: Molecular function analysis showed that these genes mainly participated in protein binding, transcription factor binding, and kinase activity; D: KEGG pathway analysis revealed that genes were involved in transcriptional misregulation in cancer, cell cycle, and TGF-beta, mTOR, and p53 signaling pathways.
- Citation: Zhang C, Ma MH, Liang Y, Wu KZ, Dai DQ. Novel long non-coding RNA LINC02532 promotes gastric cancer cell proliferation, migration, and invasion in vitro. World J Gastrointest Oncol 2019; 11(2): 91-101
- URL: https://www.wjgnet.com/1948-5204/full/v11/i2/91.htm
- DOI: https://dx.doi.org/10.4251/wjgo.v11.i2.91